Abstract
Purpose
Epidermal growth factor receptor (EGFR) is considered a potential therapeutic target for anti-EGFR therapy in triple-negative breast cancer (TNBC). However, the frequency of EGFR gene mutation in TNBC is low and varies with ethnicity. This study aimed to investigate the incidence of EGFR gene mutation in TNBC.
Methods
EGFR protein expression was evaluated by immunohistochemistry in tissue microarrays of 493 TNBC cases using four different primary antibodies, which included mutation-specific antibodies. For cases with an immunoreactivity level ≥1+, we performed pyrosequencing analysis for EGFR gene mutation. A case was considered mutation-positive when its mutation frequency minus its limit of detection (LOD) was >10%. Cases with mutation frequency higher than LOD were assessed for EGFR gene mutation status using the Cobas assay and by peptide nucleic acid-mediated polymerase chain reaction (PNA-clamping).
Results
Among 493 TNBCs, 148 (30.0%) exhibited staining ≥1+ for EGFR, including 78 with 1+, 49 with 2+, and 21 with 3+. Positive EGFR expression (≥2+) was significantly associated with lymphovascular invasion (p=0.010), but not with overall survival (p=0.444) or disease-free survival (p=0.388). None of the 493 TNBCs harbored an EGFR gene mutation. Among 148 cases with an EGFR staining result ≥1+, five (3.4%) showed mutation frequencies (4.4%–10.9%) higher than LOD (2.6%–4.3%) in exons 19 (L747_P753>Q) or 21 (L858R and L861Q) as determined by pyrosequencing. However, Cobas and PNA-clamping failed to detect the presence of EGFR gene mutation in these five cases.
Conclusion
No activating mutation of EGFR gene of clinical significance was observed in 148 TNBC cases using three commercially available methods. Thus, EGFR gene mutation appears to be an extremely rare event in patients with TNBC.
Keywords: Breast neoplasms, Epidermal growth factor receptor, Mutation, Triple negative breast neoplasms
INTRODUCTION
Triple-negative breast cancer (TNBC) accounts for approximately 15% to 30% of all breast cancers, and it is more associated with early recurrence and poor prognosis than non-TNBC [1,2]. TNBC is clinically detected by the lack of estrogen receptor (ER) and progesterone receptor (PR) expression, and the absence of amplification or overexpression of human epidermal growth factor receptor 2 (HER2). The absence of these markers entails that hormonal therapies and HER2-targeted therapies are ineffective in TNBC. Therefore, many scientists and oncologists are exploring novel molecular targets that predict response to therapy in TNBC.
The human epidermal growth factor receptor (EGFR) family consists of four EGFRs: EGFR (ErbB2 or HER1), HER2 (ErbB2), HER3 (ErbB3) and HER4 (ErbB4) [3]. These are transmembrane glycoproteins composed of an extracellular ligand binding domain, a transmembrane domain, and an intracellular domain with tyrosine kinase activity. Ligand binding to the EGFR extracellular domain leads to the phosphoryl-ation of multiple tyrosine residues in the intracellular domain. This EGFR activation induces cell proliferation, motility, and survival (protection against apoptosis) by activating downstream signaling pathways such as phosphatidylinositol-3-kinase, c-Jun N-terminal kinase, and mitogen-activated protein kinase pathways [1,3]. Somatic mutations leading to EGFR overexpression and consistent EGFR receptor activation have been reported in several malignancies, including lung cancer [4], colorectal cancer [5], and glioblastoma multiforme [6].
Activated EGFR and HER2 are well-known therapeutic targets in lung and breast cancer, respectively. EGFR-targeted therapies using small molecule tyrosine kinase inhibitors (TKIs) or anti-EGFR monoclonal antibodies (MAbs) can inhibit cell proliferation and induce apoptosis by downregulation of downstream signaling cascades [7,8]. In patients with non-small cell lung cancer (NSCLC), in-frame deletions in exon 19 and the L858R point mutation in exon 21 predict responsiveness to anti-EGFR therapy, and are the most prevalent EGFR gene mutations (accounting for 80%–90% of all EGFR gene mutations) [8]. The EGFR protein is variously overexpressed in breast cancer, especially in TNBC, with a frequency ranging from 13% to 78%, depending on the primary antibodies used [1]. Therefore, EGFR is considered an attractive therapeutic target for EGFR inhibitors in TNBC. Many clinical trials have been conducted with TKIs and MAbs in TNBC, but response rates have been disappointing, and as a result, no anti-EGFR drug is currently approved for TNBC [1,3]. However, the majority of clinical trials on responsiveness to anti-EGFR therapy did not consider the EGFR gene mutation status in TNBC. Amplification or activating mutations of EGFR have rarely been reported in TNBC, and the underlying mechanisms of protein overexpression have not been well established [1]. Nonetheless, patients with activating mutations of EGFR gene should be identified to determine whether anti-EGFR therapy might be effective in TNBC.
In NSCLC, activating mutations of EGFR gene have been observed more frequently in Asian populations than in Western populations [8], and it has been suggested that the incidence of EGFR gene mutation in TNBC also exhibits geographic and ethnic variations. Previous studies have failed to find activating mutations of EGFR gene in TNBC samples from European, Australian, and Japanese patients [9,10,11,12,13,14,15], though American study groups have reported frequencies of 0% and 3.4% for an EGFR exon 21 mutation in TNBC [16,17]. On the other hand, Chinese groups have reported frequencies of 10% and 11.4% for EGFR gene mutations in exons 19 and 21 [18,19], respectively, whereas Korean studies have reported EGFR gene mutations in exons 19 and 21 in 1% to 2% of TNBCs [20,21].
In this study, we aimed to evaluate the incidence of EGFR gene mutation in TNBC patients to determine whether the frequency of EGFR gene mutation is ethnicity dependent and whether EGFR could be used as a biomarker for anti-EGFR therapy in TNBC.
METHODS
Selection of cases and tissue microarray construction
Initially, 567 TNBC cases were selected using reports from the pathology archives of Yeungnam University Hospital, Daegu, Korea. The surgical specimens had been resected between January 1995 and December 2009. Based on a review of hematoxylin and eosin (H&E) stained slides, the most representative formalin-fixed and paraffin-embedded (FFPE) tumor blocks were selected to construct tissue microarrays (TMAs) for each case. Tumor areas identified on H&E slides were marked on the corresponding FFPE tissue blocks. Core punches (Quick-Ray® Manual Tissue Microarrayer; Unitma, Seoul, Korea) were gently pushed into tumor areas, and cores (2 mm) were transferred into the holes of Quick-Ray® recipient blocks (Unitma). Two different cores were obtained per tumor block to address tumor heterogeneity.
Medical records, pathology reports, and H&E slides were reviewed to obtain clinicopathological information, including age at diagnosis, tumor size, lymph node status, lymphovascular invasion, histological grade, Ki-67 labeling index, and the presence of recurrence or metastasis. Information on the cause of death was obtained from the microdata service system provided by Statistics Korea (http://mdis.kostat.go.kr). Overall survival was defined as the time between surgical resection and death or last follow-up and disease-free survival was defined as the time between surgical resection and locoregional recurrence, distant metastasis, death, or last follow-up.
This study was approved by the Institutional Review Board of Yeungnam University Hospital (YUH-13-0475-O75), which waived the requirement for informed consent.
EGFR immunohistochemistry
Immunohistochemistry (IHC) for ER, PR, and HER2 using 4 µm-thick-TMA sections was performed, as described previously, to confirm triple-negative status [22]. For cases equivocal for HER2 staining, silver-enhanced in situ hybridization for HER2 was performed using an INFORM® HER2 DNA probe (Ventana Medical Systems, Tucson, USA).
IHC for EGFR was performed on TMA slides using antibodies from different clones: 2-18C9 (EGFR pharmDx; Dako, Glostrup, Denmark), 3C6 (Ventana Medical Systems), SP111 (anti-EGFR E746-A750 del; Ventana Medical Systems), and SP125 (anti-EGFR L858R; Ventana Medical Systems) (Table 1). Except for EGFR pharmDx, IHC was performed on an automated Benchmark® platform (Ventana Medical Systems) using the UltraView™ universal DAB detection kit (Ventana Medical Systems).
Table 1. The epidermal growth factor receptor antibodies and staining conditions used in this study.
| Clone | Manufacturer | Dilution | Antigen retrieval | Incubation time | Detection kit |
|---|---|---|---|---|---|
| 2-18C9 | Dako, Glostrup, Denmark | Predilution | Proteinase K, 5 min | 30 min | PharmDx Kit |
| 3C6 | Ventana Medical Systems, Tucson, USA | Predilution | Protease I, 8 min | 20 min | UltraView™ DAB |
| SP111 | Ventana Medical Systems, Tucson, USA | Predilution | Standard*, CC1 | 100 min | UltraView™ DAB |
| SP125 | Ventana Medical Systems, Tucson, USA | Predilution | Standard*, CC1 | 100 min | UltraView™ DAB |
CC1=cell conditioning 1 solution.
*Standard antigen retrieval condition is 100℃, 60 minutes in cell conditioning 1 solution, and this procedure was performed in Benchmark® autoimmunostainer.
We regarded the summed tumor area of two consecutive tumor cores as the total tumor area (100%) and interpreted the immunohistochemical results of EGFR as follows: 0, no staining or faint membranous staining in <10% of tumor cells; 1+, weak membranous staining in ≥10% of tumor cells; 2+, moderate membranous staining in ≥10% of tumor cells; or 3+, strong membranous staining in ≥10% of tumor cells [21]. The highest score for the four different antibodies was regarded to be representative. For statistical analyses, we dichotomized the cases as negative (0 or 1+) or positive (2+ or 3+) for EGFR expression.
EGFR gene mutation analysis
Pyrosequencing analysis was used to detect EGFR gene mutations for cases with EGFR scores ≥1+. Results were confirmed by peptide nucleic acid-mediated polymerase chain reaction (PNA-clamping) and by the Cobas 4800 EGFR assay.
Pyrosequencing
Pyrosequencing was performed in a PyroMark Q24 MDx (Qiagen, Germantown, USA) using a therascreen EGFR Pyro Kit (Qiagen, Hilden, Germany). This kit allows quantitative measurement of mutations in codons 719 (exon 18), 768 and 790 (exon 20), and 858–861 (exon 21), along with deletions and complex mutations in exon 19 (Table 2).
Table 2. List of EGFR gene mutations detected by three different methods.
| EGFR | Pyrosequencing | PNA-clamping | Cobas assay |
|---|---|---|---|
| Exon 18 | G719A | G719A | G719A |
| G719S | G719S | G719C | |
| G719C | G719C | G719S | |
| Exon 19 | E746_A750del | E746_A750del | E746_T751 > A |
| E746_A751delinsI | E746_T751 > I | L747_T751del | |
| E746_S752del | E746_T751del | L747_E749del | |
| E746_T751delinsA | E746_T751 > A | E746_T751 > I | |
| E746_S752 > A | E746_S752 > A | E746_T751del | |
| E746_S752delinsV | E746_S752 > V | E746_S752 > A | |
| E746_A750del | E746_A750del | E746_S752 > D | |
| E746_S752delinsD | E746_S752 > D | L747_A750 > P | |
| L747_A750 > P | L747_A750 > P | L747_T751 > Q | |
| L747_T751delinsD | L747_T751 > Q | L747_P753 > Q | |
| L747_E749del | L747_E749del | L747_T751 > S | |
| L747_T751del | L747_T751del | K745_E749del | |
| L747_S752del | L747_S752del | E746_A750 > IP | |
| L747_E749del;A750P | L747_ A750 > P | E746_T751 > V | |
| L747_P753delinsQ | L747_P753 > Q | E746_T751 > IP | |
| L747_T751delinsS | L747_T751 > S | E746_S752 > I | |
| L747_P753delinsS | L747_P753 > S | L747_S752 > Q | |
| L747_T751del | L747_T751del | L747S | |
| L747_T751delinsP | L747_T751 > P | ||
| E746_S752 > I | |||
| E746_P753 > VS | |||
| L747_T751del | |||
| L747_S752 > Q | |||
| Exon 20 | T790M | T790M | T790M |
| S768I | S768I | S768I | |
| A767_V769dupASV | H773_V774insH | Insertions | |
| H773dupH | P772_H773insTTP | ||
| D770_N771insG | P772_H773insGNP | ||
| H773L | |||
| H773_V774insPH | |||
| V774_C775insHV | |||
| Exon 21 | L858R | L858R | L858R |
| L861Q | L861Q |
EGFR=epidermal growth factor receptor; PNA-clamping=peptide nucleic acid-mediated polymerase chain reaction; del=deletion; delins=deletioninsertion; dup; duplication; ins=insertion.
Unstained TMA sections (10 µm) were deparaffinized in xylene, hydrated using a graded alcohol series, and then stained with hematoxylin for 10 seconds. Tumor tissues were manually microdissected from TMA slides, and incubated in 70 µL of InstaGene™ Matrix (Bio-Rad, Hercules, USA) and 0.7 µL of proteinase K at 56℃ for 60 minutes. After boiling at 100℃ for 10 minutes, the mixtures were centrifuged at 4℃ and 12,000×g for 10 minutes. Supernatants were subjected to polymerase chain reaction (PCR). DNA quantities and qualities were measured using a NanoPhotometer™ (Implen, München, Germany). Four PCR tubes containing the PCR master mix, PCR primers for one of the four regions, and template DNA (200–300 ng) (total volume 25 µL) were prepared for each case. PCR was performed in a S-1000 thermal cycler (Bio-Rad) using the following PCR conditions; initial denaturation at 95℃ for 15 minutes, 42 amplification cycles (95℃ for 20 seconds, 53℃ for 30 seconds, 72℃ for 20 seconds), and a final extension at 72℃ for 5 minutes. A wild-type control was included in every PCR setup. PCR products were immobilized on Streptavidin Sepharose® high performance beads, purified, washed, and denatured on the PyroMark Q24 Workstation. Next, five sequencing primers for codons 719, 768, 790, and 858 to 861, and exon 19 del were annealed to the purified single-stranded PCR products. Pyrosequencing was performed on the PyroMark Q24 system according to the manufacturer's instructions. All samples were interpreted with respect to limits of detection (LOD). A case was considered mutation-positive when its mutation frequency minus its LOD was higher than 10%, as is performed for NSCLC. However, in cases with a mutation frequency higher than the LOD, EGFR mutation status was confirmed using the Cobas assay and by PNA-clamping.
Cobas 4800 EGFR assay
The Cobas® EGFR mutation kit (Roche Molecular Systems Inc., Branchburg, USA) can detect mutations in exons 18 (G719A, G719C, and G719S), 19 (deletions and complex mutations), 20 (S768I, T790M, and insertions), and 21 (L858R) of EGFR by a multiplex allele-specific PCR-based assay (Table 2). Briefly, an unstained section (10 µm) from a tumor block was deparaffinized, and the tumor area was manually microdissected. The Cobas DNA sample preparation kit (Roche Molecular Systems Inc.) was used for DNA extraction. PCR was performed using the Cobas z480 analyzer (Roche Molecular Systems Inc.) and a minimum of 150 ng DNA per case. Results were analyzed and reported automatically.
PNA-clamping
PNA-clamping analysis was performed at the manufacturer's laboratory (Panagene Inc., Daejeon, Korea) using the PNA Clamp™ EGFR mutation detection kit (Panagene Inc.), which detects 29 EGFR gene mutations (Table 2). We provided five FFPE tissue sections (10 µm) from the whole tumor block of each case for testing. A case was considered positive for EGFR gene mutation if the delta threshold cycle value-1 (ΔCt-1=standard Ct-sample Ct) was >2.0 or if the ΔCt-2 (sample Ct-non-PNA Ct) was <3.0 when ΔCt-1 was between 0 and 2.0.
Statistical analysis
Statistical analyses were performed using IBM SPSS version 23.0 for Windows (IBM Corp., Armonk, USA). Associations between EGFR expression and clinicopathological characteristics were analyzed using the chi-square test or Fisher exact test. Survival curves were plotted using the Kaplan-Meier method and the significances of differences between survival curves were determined using the log-rank test. All tests were two-sided, and p-values <0.05 were considered statistically significant.
RESULTS
Patient characteristics
Of the 567 cases considered, 493 TNBCs were included in the study. After repeating IHC for ER, PR and HER2, 74 cases were excluded owing to ER or PR positivity (n=41), HER2 positivity (n=27), or no available invasive tumor (n=6) in TMA cores.
The median patient age was 47 years (range, 25–83 years). Of the 493 patients, 248 (50.3%) underwent breast conserving surgery and 245 (49.7%) underwent mastectomy. The majority of the cases were invasive carcinoma of no special type (432, 87.6%), medullary (19, 3.9%), or metaplastic (13, 2.6%). The remaining cases included nine apocrine, four micropapillary, three papillary, three lobular, one adenoid cystic, and nine mixed type carcinomas. The histological grades were as follows: 3 in 451 (91.5%), 2 in 38 (7.7%), and 1 in 4 (0.8%) cases. Regarding tumor size, 216 (43.8%) were pT1, and 256 (51.9%) were pT2. Lymph node metastasis was present in 177 cases (35.9%). Concerning adjuvant treatment, 473 patients (95.9%) received chemotherapy and 277 (56.2%) received radiation therapy. Postoperative follow-up periods ranged from 1 to 238 months (median, 115 months).
EGFR immunohistochemistry
Results of IHC using EGFR pharmDx were as follows: 0 in 377 (76.5%), 1+ in 58 (11.8%), 2+ in 42 (8.5%), and 3+ in 16 (3.2%) cases (Figure 1). In total, 76 cases (15.4%) had an immunoreactivity ≥1+ for the 3C6 EGFR antibody and these included 39 (7.9%) 1+, 19 (3.9%) 2+, and 18 (3.7%) 3+. Among the 493 TNBCs, only four (0.8%) were 1+ for the SP111 (anti-del E746-A750) antibody. For the SP125 (anti-L858R) antibody, 14 cases (2.8%) were 1+ and one (0.2%) was 2+. When the highest scores of the four EGFR antibodies were taken as representative, 148 cases (30%) scored ≥1+, and these included 78 (15.8%) 1+, 49 (9.9%) 2+, and 21 (4.3%) 3+ cases (Table 3).
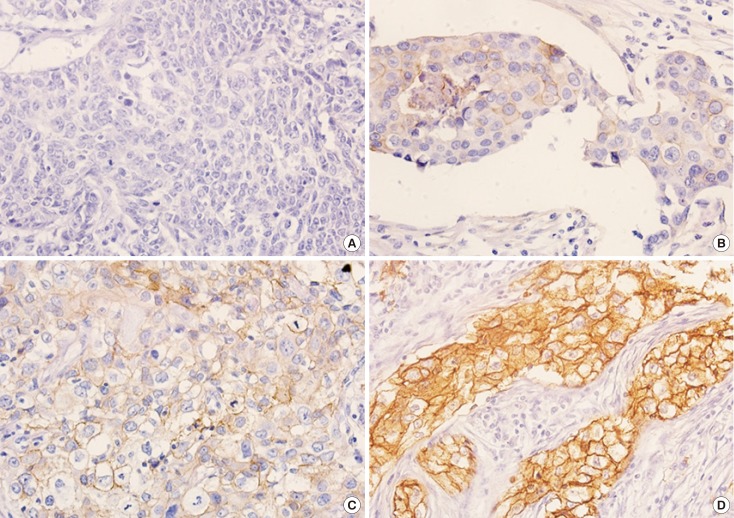
Figure 1. Immunohistochemical results for epidermal growth factor receptor (EGFR) protein as determined using pharmDx (2-18C9). (A) No staining or faint membranous staining in <10% of tumor cells (score 0), (B) faint membranous staining in ≥10% of tumor cells (score 1+), (C) moderate staining in ≥10% of tumor cells (score 2+), (D) strong membranous staining of ≥10% of tumor cells (score 3+) (original magnification ×400, in all figures).
Table 3. Immunohistochemical results of four different EGFR antibodies.
| Score | EGFR IHC (n=493), No. (%) | hIHC score* No. (%) | |||
|---|---|---|---|---|---|
| 2-18C9 | 3C6 | SP111 | SP125 | ||
| 0 | 377 (76.5) | 417 (84.6) | 489 (99.2) | 478 (97.0) | 345 (70.0) |
| 1+ | 58 (11.8) | 39 (7.9) | 4 (0.8) | 14 (2.8) | 78 (15.8) |
| 2+ | 42 (8.5) | 19 (3.9) | 0 | 1 (0.2) | 49 (9.9) |
| 3+ | 16 (3.2) | 18 (3.7) | 0 | 0 | 21 (4.3) |
EGFR=epidermal growth factor receptor; IHC=immunohistochemistry.
*hIHC score was defined as the highest immunoreactivity score among the four EGFR antibodies.
Relations between EGFR expression and clinicopathological parameters
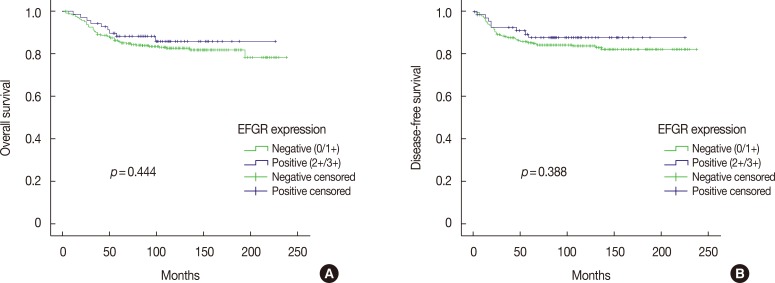
EGFR expression was found to be significantly associated with the presence of lymphovascular invasion (p=0.010). However, no relationship was observed between EGFR expression and other clinicopathological variables such as tumor size, lymph node status, stage, histologic grade, p53 expression, and the Ki-67 proliferation index (Table 4). In addition, no association was detected between positive EGFR expression and overall survival (p=0.444) or disease-free survival (p=0.388) (Figure 2).
Table 4. Associations between EGFR expression and clinicopathological characteristics in 493 patients with triple-negative breast cancer.
| Characteristic | No. (%) | EGFR expression | p-value | |
|---|---|---|---|---|
| Negative (0/1+) No. (%) |
Positive (2+/3+) No. (%) |
|||
| Tumor size (cm) | 0.488 | |||
| ≤2 | 216 (43.8) | 188 (44.4) | 28 (40.0) | |
| >2 | 277 (56.2) | 235 (55.6) | 42 (60.0) | |
| Lymph node metastasis | 0.298 | |||
| Absent | 316 (64.1) | 275 (65.0) | 41 (58.6) | |
| Present | 177 (35.9) | 148 (35.0) | 29 (41.4) | |
| Stage | 0.215 | |||
| I | 164 (33.3) | 144 (34.0) | 20 (28.6) | |
| II | 253 (51.3) | 217 (51.3) | 36 (51.4) | |
| III | 76 (15.4) | 62 (14.7) | 14 (20.0) | |
| Histologic grade | 0.347 | |||
| 1 & 2 | 42 (8.5) | 34 (8.0) | 8 (11.4) | |
| 3 | 451 (91.5) | 389 (92.0) | 62 (88.6) | |
| Lymphovascular invasion | 0.010 | |||
| Absent | 294 (59.6) | 262 (61.9) | 32 (45.7) | |
| Present | 199 (40.4) | 161 (38.1) | 38 (54.3) | |
| p53 expression | 0.496 | |||
| Absent | 251 (50.9) | 218 (51.5) | 33 (47.1) | |
| Present | 242 (49.1) | 205 (48.5) | 37 (52.9) | |
| Ki-67 proliferation index (%) | 0.592 | |||
| ≤20 | 45 (9.2) | 38 (9.0) | 7 (10.0) | |
| >20 | 440 (89.2) | 378 (89.4) | 62 (88.6) | |
| Unknown | 8 (1.6) | 7 (1.7) | 1 (1.4) | |
EGFR=epidermal growth factor receptor.
Figure 2. Kaplan-Meier survival curves for epidermal growth factor receptor (EGFR) protein expression. (A) Overall survival, (B) disease-free survival.
EGFR gene mutation analysis
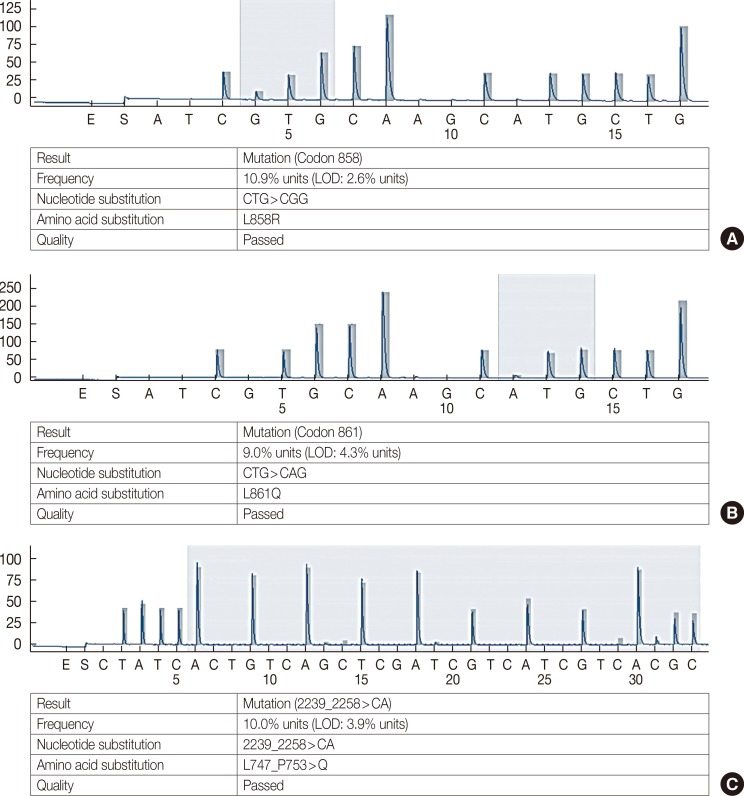
Of the 493 TNBC cases, 148 with a representative IHC score ≥1+ were included in the pyrosequencing based EGFR gene mutation analysis. When the criterion used for NSCLC was applied, no TNBC showed an EGFR gene mutation. Of the 148 cases, five (3.4%) had mutation frequencies higher than their LODs, whereas three had an L861Q mutation in EGFR exon 21 with mutation frequencies of 4.4%, 8.1%, and 9.0% (LOD, 4.3%). Another had an L858R mutation in exon 21 (mutation frequency, 10.9%; LOD, 2.6%), and the remaining one had an exon 19 deletion, L747_P753>Q (mutation frequency, 10.0%; LOD, 3.9%) (Figure 3). The case harboring the L858R mutation had a SP125 IHC staining score of 1+, and the case with the exon 19 deletion was negative for SP111 IHC. Of the three cases with an L861Q mutation, one was 1+ for SP125, and another two were 2+ for EGFR pharmDx (one of these two was also 2+ for 3C6) (Table 5).
Figure 3. Representative pyrosequencing results for EGFR gene mutations. (A) Case with 10.9% mutation frequency for the L858R mutation in EGFR exon 21. (B) Case with 9.0% mutation frequency for the L861Q mutation in EGFR exon 21. (C) Case with 10% mutation frequency for exon 19 deletion, L747_P753>Q. The numbers on the Y axis represent relative fluorescence unit.
EGFR=epidermal growth factor receptor; LOD=limit of detection.
Table 5. Summary of the five triple-negative breast cancer cases studied by pyrosequencing, PNA-clamping and Cobas assay.
| Case No. | EGFR immunohistochemistry | EGFR gene mutation | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Immunostaining score | Pyrosequencing | PNA-clamping | Cobas assay | ||||||
| 2-18C9 | 3C6 | SP111 | SP125 | Site | Frequency (%) | LOD (%) | |||
| 1 | 2+ | 2+ | 0 | 0 | Exon 21, L861Q | 4.4 | 4.3 | Wild | Wild |
| 2 | 0 | 0 | 0 | 1+ | Exon 21, L858R | 10.9 | 2.6 | Wild | Wild |
| 3 | 0 | 0 | 0 | 1+ | Exon 21, L861Q | 9.0 | 4.3 | Wild | Wild |
| 4 | 2+ | 0 | 0 | 0 | Exon 21, L861Q | 8.1 | 4.3 | Wild | Wild |
| 5 | 0 | 0 | 0 | 1+ | Exon 19, L747_P753 > Q | 10.0 | 3.9 | Wild | Wild |
PNA-clamping=peptide nucleic acid-mediated polymerase chain reaction; EGFR=epidermal growth factor receptor; LOD=limit of detection.
The five cases with mutation frequencies higher than their LODs were subjected to Cobas EGFR assays, but no EGFR gene mutation was detected. Because the L861Q mutation in exon 21 was not covered by the Cobas assay, PNA-clamping was also performed for these five cases, but all were reported as wild type.
DISCUSSION
The incidence of EGFR gene mutations in NSCLC has been reported to be 20% to 50% in Asian and 15% to 20% in Western patients [8], whereas in breast cancer (mostly TNBC) the reported incidences range between 0 and 11.4%, depending on the ethnicity and the methods of detection used (Table 6). Although direct DNA sequencing has been widely used, there is no gold standard method for detecting EGFR gene mutations. To overcome the disadvantages of direct sequencing, such as low sensitivity and the requirement for a high percentage of mutant alleles from high quality DNA, highly sensitive detection methods requiring less DNA were recently developed. Pyrosequencing, PNA-clamping, and Cobas EGFR assays are the commercially available EGFR gene mutation detection methods that have been optimized for FFPE samples. Although these methods are highly sensitive, rapid, and can be performed with ease in clinical practice, they only cover mutations in specific sites of the EGFR using available primers; they cannot detect rare or new mutations with uncertain clinical significance.
Table 6. Summary of previous studies for EGFR gene mutation in triple-negative breast cancer.
| Author (year) | Specimen | Population | Method | Result | |
|---|---|---|---|---|---|
| Reis-Filho et al. (2006) [16] | 47 | Metaplastic carcinomas | British | Direct sequencing | No mutations |
| Toyama et al. (2008) [13] | 58 | TNBCs | Japanese | Real-time PCR | No mutations |
| Jacot et al. (2011) [15] | 229 | TNBCs | French | Direct sequencing | No mutations |
| Lv et al. (2011) [18] | 139 | Breast carcinomas | Chinese | Real-time PCR | 1.4% (10% in TNBCs), 19del and L858R |
| Teng et al. (2011) [19] | 70 | TNBCs | Chinese | Direct sequencing | 11.4%, 19del and L858R |
| Martin et al. (2012) [11] | 38 | TNBC with basal-like features | Switzerland | Direct sequencing | No mutations |
| Grob et al. (2012) [10] | 65 | TNBCs | German | Direct sequencing | No mutations |
| Santarpia et al. (2012) [17] | 116 | TNBCs | American | Sequenom technology | 3.4%, Mutation types, not specified |
| Direct sequencing | |||||
| Kim et al. (2013) [20] | 105 | TNBCs | Korean | PNA-clamping | 1.0%, 19del |
| Secq et al. (2014) [12] | 47 | TNBCs | French | Direct sequencing | No mutations |
| Tilch et al. (2014) [14] | 50 | TNBCs | Australian | Direct sequencing | No mutations |
| 57 | Basal-like breast carcinomas | ||||
| Nakajima et al. (2014) [9] | 55 | TNBCs | Japanese | SmartAmp2 method | No mutations |
| Park et al. (2014) [21] | 151 | TNBCs | Korean | Direct sequencing | 2.6%, G719A, V786M, L858R |
REGFR=epidermal growth factor receptor; TNBC=triple-negative breast cancer; PCR=polymerase chain reaction; del=deletion; PNA-clamping=peptide nucleic acid-mediated polymerase chain reaction.
In this study, we performed an EGFR gene mutation analysis in 148 TNBCs expressing the EGFR protein (≥1+) using pyrosequencing and checked the meaningful cases using Cobas and PNA-clamping. Pyrosequencing, Cobas, and PNA-clamping have been shown to have equal or better detection rates for EGFR gene mutations in FFPE samples of NSCLC than those of direct sequencing. Furthermore, mutation-positive patients detected using these methods have been reported to show excellent responses to EGFR TKIs [23,24]. Pyrosequencing can detect and characterize EGFR gene mutations precisely, and quantify the percentages of mutant alleles in clinical samples containing at least 20% tumor cells [25]. We performed manual microdissection for DNA extraction to increase the tumor cell content, and obtained pyrosequencing results from all TMA cores included. Alternative methods such as the Cobas assay and PNA-clamping are known to detect mutation with high sensitivity (~1% of tumor cells). According to the guidelines issued by the manufacturer of the therascreen EGFR Pyro Kit, five cases (3.4%) showed mutation frequencies higher than the LOD by pyrosequencing. One case showed a potential low-level mutation (mutation frequency, 4.4%), and the remaining four cases represented low-level mutations (8.1%–10.9%). However, no mutation was detected by Cobas and PNA-clamping in confirmatory testing. Our laboratory has 4 years of experience with pyrosequencing for the detection of EGFR gene mutations in NSCLC. During this period, we modified the cutoff for positive mutation by conducting inter- and intra-laboratory validation tests using the same and different methods. At present, we consider a case as positive for mutation when its mutation frequency minus its LOD is >10% in NSCLC. Because we did not have any experience of EGFR gene mutation testing by pyrosequencing in breast cancer, we selected cases with a mutation frequency higher than the LOD and confirmed their mutation status using alternative methods. When the positive criterion for NSCLC was applied in the present study, the five TNBCs were found to be negative for EGFR gene mutation. It is possible that these five cases still had low amounts of clinically insignificant mutant EGFR alleles. Nonetheless, it appears that the failure or low response rates in anti-EGFR therapy may be due to the rarity of EGFR gene mutation in TNBC.
In addition to EGFR activating mutations, EGFR amplification and high polysomy have been reported in 0% to 24% and 7% to 64% of TNBCs, respectively [1,21]. Furthermore, the relation between EGFR expression and molecular alterations of EGFR is controversial. In lung and breast cancers, EGFR overexpression was not found to be associated with EGFR gene mutation status [19,26], but was associated with high EGFR copy number (amplification or polysomy) in TNBC [21]. We did not examine the EGFR copy number changes in the present study. The prognostic significance of EGFR expression is also controversial. Park et al. [21] reported that EGFR overexpression (2+ or 3+) was significantly associated with lower stage, but not with other clinicopathologic variables or clinical outcomes in TNBC. In the present study, we observed that while EGFR overexpression was associated with lymphovascular invasion, it was not predictive of patient survival, which concurs with the findings of a previous study [21]. Therefore, we believe that EGFR protein expression alone has limited value in terms of predicting prognosis in TNBC.
Recently, mutation-specific antibodies targeting E746-A750del (exon 19) and L858R (exon 21) (the two most common mutations in EGFR) were developed. Both antibodies show high sensitivity, specificity, and positive predictive value for detecting these specific mutations in NSCLC [27,28]. These antibodies were tested in 300 breast carcinomas, including 220 TNBCs [29]. Of these 300 breast carcinomas, five (2%) showed 1+ and two (1%) showed 2+ immunostaining for the L858R antibody, but all cases were negative for the E746-A750 antibody. Of the 220 TNBCs, only one (0.5%) showed 1+ staining for the L858R antibody. Furthermore, molecular analysis failed to detect any mutation in the two cases with 2+ immunostaining for L858R. In the present study, 0.8% (four cases) of TNBC scored 1+ for E746-A750 antibody, and 2.8% (14 cases) and 0.2% (one case) scored 1+ and 2+ for the L858R antibody, respectively. The three cases that scored 1+ for the L858R antibody represented point mutations in exon 21 (L858R or L861Q) or deletion in exon 19 (L747_P753>Q) with low mutation frequencies as determined by pyrosequencing, but these mutations were not detected by Cobas or PNA-clamping. Furthermore, the remaining 16 cases with 1+ or 2+ scores for mutation-specific antibodies were negative for EGFR gene mutation by pyrosequencing analysis. These results suggest that mutation-specific antibodies are not specific for the diagnosis of EGFR gene mutation, and that TNBCs usually overexpress the wild-type EGFR protein.
It has been suggested that the frequencies of EGFR gene mutations in TNBCs show geographic and ethnic differences [30], similar to that in NSCLC. Although previous studies were limited by small sample numbers, activating EGFR gene mutations were not identified in Japanese, European, or Australian patients with TNBC [9,10,11,12,13,14,15]. In contrast, two Chinese studies reported activating mutation of EGFR gene at a frequency of about 10% [18,19]. Previous Korean studies identified EGFR gene mutations in exons 19 and 21 in 1% to 2% of TNBCs [20,21]. However, we did not find any EGFR gene mutation of clinical significance using three above-mentioned clinically available detection methods. Accordingly, we conclude that EGFR gene mutation is a rare event in TNBC worldwide, and that the relatively high incidence of EGFR gene mutation in the Chinese population needs to be verified by further study using a large cohort of patients and standardized methods.
Footnotes
This study was supported by a Yeungnam University Medical Center grant (2013).
CONFLICT OF INTEREST: The authors declare that they have no competing interests.
References
- 1.Nakai K, Hung MC, Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res. 2016;6:1609–1623. [PMC free article] [PubMed] [Google Scholar]
- 2.Kim MJ, Ro JY, Ahn SH, Kim HH, Kim SB, Gong G. Clinicopathologic significance of the basal-like subtype of breast cancer: a comparison with hormone receptor and HER2/neu-overexpressing phenotypes. Hum Pathol. 2006;37:1217–1226. doi: 10.1016/j.humpath.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scagliotti GV, Selvaggi G, Novello S, Hirsch FR. The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res. 2004;10(12 Pt 2):4227s–4232s. doi: 10.1158/1078-0432.CCR-040007. [DOI] [PubMed] [Google Scholar]
- 5.Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23:1803–1810. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 6.Fuller GN, Bigner SH. Amplified cellular oncogenes in neoplasms of the human central nervous system. Mutat Res. 1992;276:299–306. doi: 10.1016/0165-1110(92)90016-3. [DOI] [PubMed] [Google Scholar]
- 7.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 8.Cheng L, Alexander RE, Maclennan GT, Cummings OW, Montironi R, Lopez-Beltran A, et al. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012;25:347–369. doi: 10.1038/modpathol.2011.215. [DOI] [PubMed] [Google Scholar]
- 9.Nakajima H, Ishikawa Y, Furuya M, Sano T, Ohno Y, Horiguchi J, et al. Protein expression, gene amplification, and mutational analysis of EGFR in triple-negative breast cancer. Breast Cancer. 2014;21:66–74. doi: 10.1007/s12282-012-0354-1. [DOI] [PubMed] [Google Scholar]
- 10.Grob TJ, Heilenkötter U, Geist S, Paluchowski P, Wilke C, Jaenicke F, et al. Rare oncogenic mutations of predictive markers for targeted therapy in triple-negative breast cancer. Breast Cancer Res Treat. 2012;134:561–567. doi: 10.1007/s10549-012-2092-7. [DOI] [PubMed] [Google Scholar]
- 11.Martin V, Botta F, Zanellato E, Molinari F, Crippa S, Mazzucchelli L, et al. Molecular characterization of EGFR and EGFR-downstream pathways in triple negative breast carcinomas with basal like features. Histol Histopathol. 2012;27:785–792. doi: 10.14670/HH-27.785. [DOI] [PubMed] [Google Scholar]
- 12.Secq V, Villeret J, Fina F, Carmassi M, Carcopino X, Garcia S, et al. Triple negative breast carcinoma EGFR amplification is not associated with EGFR, Kras or ALK mutations. Br J Cancer. 2014;110:1045–1052. doi: 10.1038/bjc.2013.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toyama T, Yamashita H, Kondo N, Okuda K, Takahashi S, Sasaki H, et al. Frequently increased epidermal growth factor receptor (EGFR) copy numbers and decreased BRCA1 mRNA expression in Japanese triple-negative breast cancers. BMC Cancer. 2008;8:309. doi: 10.1186/1471-2407-8-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tilch E, Seidens T, Cocciardi S, Reid LE, Byrne D, Simpson PT, et al. Mutations in EGFR, BRAF and RAS are rare in triple-negative and basal-like breast cancers from Caucasian women. Breast Cancer Res Treat. 2014;143:385–392. doi: 10.1007/s10549-013-2798-1. [DOI] [PubMed] [Google Scholar]
- 15.Jacot W, Lopez-Crapez E, Thezenas S, Senal R, Fina F, Bibeau F, et al. Lack of EGFR-activating mutations in European patients with triple-negative breast cancer could emphasise geographic and ethnic variations in breast cancer mutation profiles. Breast Cancer Res. 2011;13:R133. doi: 10.1186/bcr3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reis-Filho JS, Pinheiro C, Lambros MB, Milanezi F, Carvalho S, Savage K, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol. 2006;209:445–453. doi: 10.1002/path.2004. [DOI] [PubMed] [Google Scholar]
- 17.Santarpia L, Qi Y, Stemke-Hale K, Wang B, Young EJ, Booser DJ, et al. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res Treat. 2012;134:333–343. doi: 10.1007/s10549-012-2035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv N, Xie X, Ge Q, Lin S, Wang X, Kong Y, et al. Epidermal growth factor receptor in breast carcinoma: association between gene copy number and mutations. Diagn Pathol. 2011;6:118. doi: 10.1186/1746-1596-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng YH, Tan WJ, Thike AA, Cheok PY, Tse GM, Wong NS, et al. Mutations in the epidermal growth factor receptor (EGFR) gene in triple negative breast cancer: possible implications for targeted therapy. Breast Cancer Res. 2011;13:R35. doi: 10.1186/bcr2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y, Kim J, Lee HD, Jeong J, Lee W, Lee KA. Spectrum of EGFR gene copy number changes and KRAS gene mutation status in Korean triple negative breast cancer patients. PLoS One. 2013;8:e79014. doi: 10.1371/journal.pone.0079014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ, Kim YJ, et al. High EGFR gene copy number predicts poor outcome in triple-negative breast cancer. Mod Pathol. 2014;27:1212–1222. doi: 10.1038/modpathol.2013.251. [DOI] [PubMed] [Google Scholar]
- 22.Choi JE, Kang SH, Lee SJ, Bae YK. Androgen receptor expression predicts decreased survival in early stage triple-negative breast cancer. Ann Surg Oncol. 2015;22:82–89. doi: 10.1245/s10434-014-3984-z. [DOI] [PubMed] [Google Scholar]
- 23.Lee HJ, Xu X, Kim H, Jin Y, Sun P, Kim JE, et al. Comparison of direct sequencing, PNA clamping-real time polymerase chain reaction, and pyrosequencing methods for the detection of EGFR mutations in non-small cell lung carcinoma and the correlation with clinical responses to EGFR tyrosine kinase inhibitor treatment. Korean J Pathol. 2013;47:52–60. doi: 10.4132/KoreanJPathol.2013.47.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon SH, Choi YD, Oh IJ, Kim KS, Choi H, Chang J, et al. Peptide nucleic acid clamping versus direct sequencing for the detection of EGFR gene mutation in patients with non-small cell lung cancer. Cancer Res Treat. 2015;47:661–669. doi: 10.4143/crt.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dufort S, Richard MJ, Lantuejoul S, de Fraipont F. Pyrosequencing, a method approved to detect the two major EGFR mutations for anti EGFR therapy in NSCLC. J Exp Clin Cancer Res. 2011;30:57. doi: 10.1186/1756-9966-30-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li AR, Chitale D, Riely GJ, Pao W, Miller VA, Zakowski MF, et al. EGFR mutations in lung adenocarcinomas: clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression. J Mol Diagn. 2008;10:242–248. doi: 10.2353/jmoldx.2008.070178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brevet M, Arcila M, Ladanyi M. Assessment of EGFR mutation status in lung adenocarcinoma by immunohistochemistry using antibodies specific to the two major forms of mutant EGFR. J Mol Diagn. 2010;12:169–176. doi: 10.2353/jmoldx.2010.090140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim CH, Kim SH, Park SY, Yoo J, Kim SK, Kim HK. Identification of EGFR mutations by immunohistochemistry with EGFR mutation-specific antibodies in biopsy and resection specimens from pulmonary adenocarcinoma. Cancer Res Treat. 2015;47:653–660. doi: 10.4143/crt.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen YH, Brogi E, Hasanovic A, Ladanyi M, Soslow RA, Chitale D, et al. Immunohistochemical staining with EGFR mutation-specific antibodies: high specificity as a diagnostic marker for lung adenocarcinoma. Mod Pathol. 2013;26:1197–1203. doi: 10.1038/modpathol.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamy PJ, Jacot W. Worldwide variations in EGFR somatic mutations: a challenge for personalized medicine. Diagn Pathol. 2012;7:13. doi: 10.1186/1746-1596-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]