Abstract
Immortality is an essential characteristic of cancer cells; a recent transcriptomic study of epithelial cell immortalization has linked epigenetic silencing of the long noncoding RNA Mortal Obligate RNA Transcript (MORT; alias ZNF667-AS1) to this process. This study evaluated the epigenetic and transcriptional state of MORT in two premalignant conditions—ductal carcinomas in situ and colon adenomas. Results show that MORT silencing is an early epigenetic event in human carcinogenesis, likely occurring near the point where premalignant cells gain immortality; this epigenetic silencing is maintained throughout malignant transformation and metastatic growth. Additional associations between MORT loss and clinical and molecular features of breast tumors showed that silencing of MORT occurs predominantly in luminal, receptor-positive breast cancer; is associated with overexpression of CCND1 and mutations of GATA3; and is negatively correlated with TP53 mutations. Taken in toto, MORT silencing occurs early in breast carcinogenesis, probably during cellular immortalization, and precedes the development of invasive luminal breast cancer.
Keywords: Breast neoplasms, Carcinogenesis, DNA methylation, Gene silencing, Long noncoding RNA
Immortality is an essential characteristic of cancer cells; our recent transcriptomic study of in vitro immortalization of human mammary epithelial cells (HMEC) reported on the epigenetic silencing of the long noncoding RNA (lncRNA) Mortal Obligate RNA Transcript (MORT; alias ZNF667-AS1) during the transition of the cells from mortal to immortal [1]. MORT is expressed in all normal, mortal human cell types analyzed to date, but is epigenetically silenced in the majority of human cancers and human cancer cell lines [1]; there is little evidence suggesting genetic mechanisms of inactivation. The experimental finding that MORT is silenced at the mortal-immortal boundary of HMEC, prior to and independent of malignant conversion, suggests that MORT silencing is an early epigenetic event in human carcinogenesis that occurs prior to malignant transformation. Since a large fraction of clinical carcinoma in situ lesions—including ductal carcinoma in situ (DCIS) [2]—show evidence of immortality in the absence of a frank malignancy [3,4,5], it is predicted that the epigenetic silencing of MORT is a common feature of this premalignant state. Breast cancer is a heterogeneous disease, but our previous analysis was not focused on MORT silencing in individual breast cancer subtypes and could not reveal if MORT silencing was an early event in clinical carcinogenesis.
Using molecular and clinical data from publicly accessible databases, we set out to evaluate the epigenetic and transcriptional status of MORT in premalignant tumors. Consistent with previous laboratory studies, our data analysis shows that DNA methylation-mediated silencing of MORT is found in both breast DCIS and colonic adenomas; this moves the timing of MORT epigenetic silencing further back to clinical premalignant conditions. The epigenetic silencing of MORT in premalignancy is maintained throughout progression towards the invasive and metastatic disease; no evidence was found for genetic mechanisms of MORT silencing. The DNA methylation-mediated silencing of MORT is significantly associated with luminal, receptor positive breast cancer; in contrast, basal triple negative breast cancers do not generally display MORT methylation or gene silencing. These early epigenetic events and the cellular mechanisms that they influence may provide new targets for cancer prevention or treatment.
DNA methylation (Illumina HumanMethylation450), gene expression (RNASeqV2), genetic (copy number variation, mutation), and available clinical data for breast tumor samples were downloaded from The Cancer Genome Atlas (TCGA). All data were analyzed with the R programming software [6] using custom scripts. MORT mean β values and mean reads per kilobase per million (RPKM) values were calculated as described previously [1]. Only nonsilent mutations were considered in the gene mutation analysis. The Spearman's rank correlation coefficient (rho) was calculated using the cor.test function. All tests between groups were performed using the wilcox.test function. When appropriate, multiple testing corrections were performed according to Benjamini and Hochberg [7]. PAM50 tumor subtype classification was used as described previously [8]. In addition to TCGA, further Illumina HumanMethylation450 datasets for breast (GSE60185 [9], GSE66313 [10], and GSE58999 [11]) and colon (GSE48684 [12] and GSE77954 [13]) cancer samples were downloaded from Gene Expression Omnibus (GEO). Where necessary, M values were converted to β values using the formula 2M/(1+2M). The reduced representation bisulfite sequencing (RRBS) and RNA-seq DCIS datasets (GSE69993 and GSE69240 [14]) were downloaded from GEO. For RRBS, the mean methylation value for the 19 covered CpGs within the MORT CpG island promoter was used for analysis.
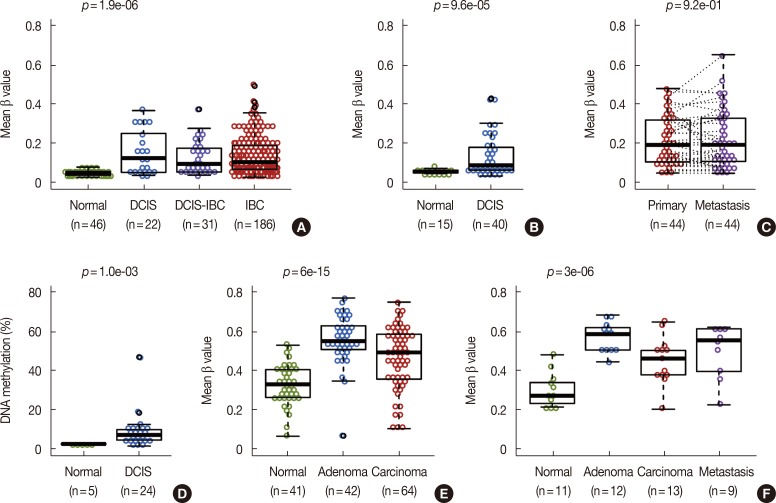
We have previously reported that MORT is epigenetically silenced by DNA methylation in breast cancer and 14 out of 16 other common human tumors investigated. To determine the potential timing of MORT silencing during human breast carcinogenesis, we analyzed the DNA methylation state of the MORT promoter in DCIS samples from two published, independent cohorts [9,10]. The results show a significant increase in DNA methylation of the MORT promoter in DCIS compared with normal samples (Figure 1A, B); MORT methylation did not appear to increase further during progression from DCIS to invasive breast cancer (IBC) (Figure 1A), nor during progression to the metastatic disease, as illustrated by data from a cohort [11] of pairs of primary and metastatic breast tumors (Figure 1C). A third cohort of DCIS, analyzed by RRBS and RNA-seq [14], also showed a significant increase in MORT methylation (Figure 1D) and MORT silencing in a substantial portion of the samples (Supplementary Figure 1, available online). Together, these results show that MORT silencing is an early epigenetic event that occurs during human breast carcinogenesis.
Figure 1. MORT gene silencing is early event in cancer. (A, B) DNA methylation of the MORT promoter in two independent cohorts of breast ductal carcinoma in situ (DCIS) patients. (C) DNA methylation of the MORT promoter in 44 pairs of primary breast tumors and lymph node metastasis. (D) DNA methylation of the MORT promoter as determined by reduced representation bisulfite sequencing in a DCIS cohort. (E, F) DNA methylation of the MORT promoter in two independent cohorts of colon adenomas and carcinomas. All data except (D) were obtained using the Illumina Human Methylation 450 platform. Where more than two groups are presented the p-value is for in situ lesions versus normal samples.
IBC=invasive breast cancer.
We extended our analysis of premalignant lesions and analyzed the DNA methylation state of the MORT promoter in two independent cohorts of colon adenoma samples [12,13]. Although not equivalent to DCIS, colonic adenomas are early premalignant lesions in colorectal cancer and therefore might display MORT silencing if MORT silencing occurs early in carcinogenesis. The DNA methylation level of the MORT promoter in both adenoma cohorts was significantly higher than in normal colon samples; there were no further increases in MORT methylation throughout progression of the adenoma to invasive colon cancer, similar to the results for DCIS and IBC (Figure 1E, F). These results broaden the range of premalignant conditions where MORT is silenced and provide additional evidence that MORT silencing is an early epigenetic event in human carcinogenesis.
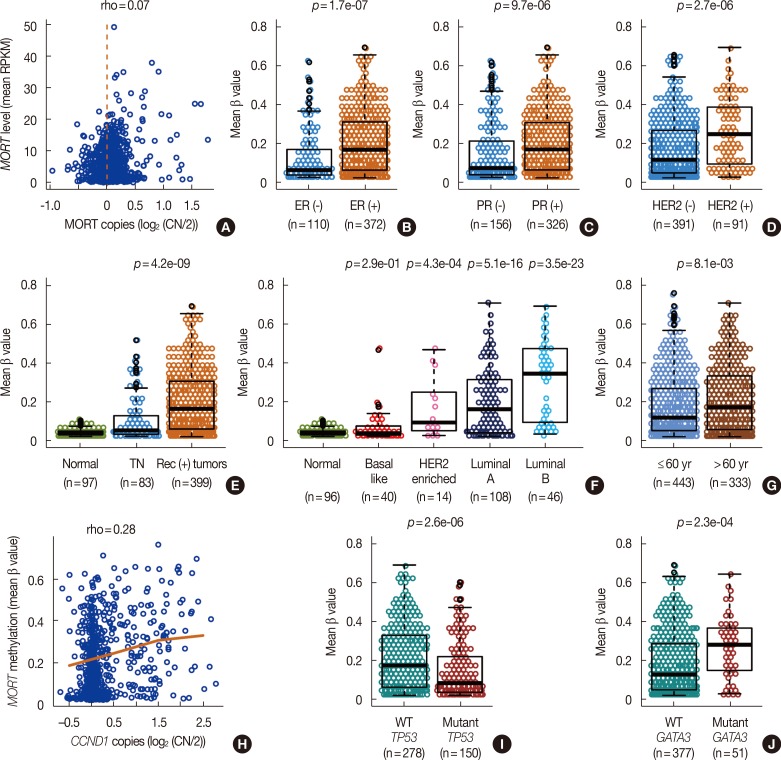
Because breast cancer is a heterogeneous disease, we sought to identify any associations between MORT loss and other clinical and molecular features of breast tumors. Figure 2A presents analysis performed to assess whether gene deletion is an additional mechanism of MORT loss in cancer, in addition to aberrant DNA methylation. The results show that MORT loss rarely occurs due to gene deletion (Figure 2A). MORT loss is therefore predominantly due to epigenetic silencing and increased DNA methylation of the MORT promoter; MORT promoter methylation is a good indicator of MORT silencing that could be used as a surrogate for MORT loss in samples where expression data are not available. When we tested the association between MORT methylation and clinical variables, we found a strong link between MORT methylation and the presence of hormone receptors. Tumors positive for individual receptors have a much higher frequency of MORT promoter methylation (Figure 2B-D) and MORT silencing (Supplementary Figure 2A-C, available online). In contrast, triple negative tumors have a low frequency of MORT methylation (Figure 2E) and near normal MORT RNA levels (Supplementary Figure 2D, available online). These data predict that MORT is predominantly silenced in luminal breast cancers; indeed, when we classified the breast tumors into subtypes according to PAM50 [15], luminal B, luminal A, and human epidermal growth factor receptor 2 (HER2)-enriched tumors had a large proportion of MORT methylation, while basal-like tumors showed no significant difference in MORT methylation or expression compared to normal control samples (Figure 2F, Supplementary Figure 2E, available online). Since some breast cancer subtypes are linked to age, we analyzed the relationship between MORT level and patient age. There is a moderate but notable increase in MORT methylation and decrease in MORT expression in tumors from patients over 60 years old (Figure 2G, Supplementary Figure 2F, available online), which is consistent with evidence of higher incidence of luminal tumors in older women [16]. Together, these results indicate that epigenetic silencing of MORT occurs predominantly in luminal, receptor-positive breast cancers.
Figure 2. DNA methylation mediated MORT gene silencing is linked to luminal, receptor positive breast cancers. (A) MORT expression level plotted versus MORT copy number. DNA methylation of the MORT promoter in receptor negative and receptor positive tumors estrogen receptor (ER) (B), progesterone receptor (PR) (C), and human epidermal growth factor receptor 2 (HER2) (D). (E) DNA methylation of the MORT promoter in triple negative (TN) tumors, tumors positive for at least one receptor and a group of normal samples (p-value is for contrast TN vs. receptor (+) tumors). (F) DNA methylation of the MORT promoter in breast tumors classified into subtypes according to PAM50 (p-values are for contrast of individual tumor subtypes vs. normal). (G) DNA methylation of the MORT promoter in samples from patients >60 years of age compared to those from patients ≤60. (H) Association between amplification of the cyclin D1 locus and DNA methylation of the MORT promoter. (I) DNA methylation of the MORT promoter in samples with wild type TP53 compared to samples with mutated TP53. (J) DNA methylation of the MORT promoter in samples with wild type GATA3 compared to samples with mutated GATA3.
RPKM=reads per kilobase per million; CN=copy number; WT=wild type.
We investigated potential associations between MORT methylation and genetic features in breast tumors. When we correlated MORT methylation with the copy number variation data on the whole genome scale, amplification of a locus containing genes CCND1 and ORAOV1 on chromosome 11 showed the strongest positive correlation (rho=0.28, adj. p=4.95e–11) with MORT promoter methylation (Figure 2H). Of these two amplified genes, only the CCND1 transcript level was significantly negatively correlated with the MORT transcript level (rho=–0.35, adj. p=8.01e–31). CCND1 is a known cancer driver linked to luminal tumors [8] and therefore its overexpression, which inactivates the RB pathway [17], might be also implicated in MORT silencing. Finally, we analyzed the links between MORT methylation and mutations in 23 genes frequently mutated in breast cancer [8]. Mutations in two of these genes were strongly linked to the level of MORT methylation. Mutation of TP53 is negatively correlated to MORT methylation (Figure 2I)—tumors carrying a TP53 mutation usually have normal levels of MORT (Supplementary Figure 2G, available online). GATA3 is frequently mutated in tumors that have reduced MORT expression (Supplementary Figure 2H, available online)—there is a positive correlation between GATA3 mutation and MORT methylation (Figure 2J). This is consistent with evidence showing that TP53 mutations are enriched in basal-like triple negative tumors, while GATA3 mutations are found predominantly in luminal tumors [8]. Overall, these findings suggest that MORT silencing in luminal breast tumors is connected to overexpression of CCND1. The lack of MORT silencing in basal tumors that frequently carry TP53 mutations indicates that MORT might act in a pathway that requires functional TP53, as the cells with nonfunctional TP53 can “afford” to express MORT and become immortal. The dissociation between TP53 mutations and MORT silencing is consistent with the in vitro immortalization model used in the discovery of MORT, where the immortal HMEC retained wild-type TP53 [18].
Overall, our analysis found that, as predicted from the in vitro model of HMEC immortalization, aberrant methylation of the MORT CpG island promoter occurs early in human breast carcinogenesis, likely at or near the point where premalignant cells gain immortality; this epigenetic silencing is maintained throughout malignant transformation and metastatic growth. In breast cancer, MORT gene silencing occurs predominantly in hormone receptor positive luminal tumors, while triple negative basal-like tumors usually show normal levels of MORT expression. Thus, taken in toto, these results are consistent with MORT silencing occurring early in breast carcino-genesis, during cellular immortalization, and before the development of luminal IBC. Delineating the molecular functions of the MORT lncRNA could provide new strategies for therapeutic or chemopreventive interventions.
ACKNOWLEDGMENTS
The results shown here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/).
Footnotes
This work was supported by the Margaret E. and Fenton L. Maynard Endowment for Breast Cancer Research and Center grant ES006694.
CONFLICT OF INTEREST: The authors declare that they have no competing interests.
SUPPLEMENTARY MATERIALS
MORT is silenced in a large fraction of ductal carcinoma in situ (DCIS) samples. The boxplots show MORT RNA level in a group of 10 normal organoids and 25 DCIS samples.
MORT gene silencing is associated with the luminal, receptor positive tumor subtypes. MORT RNA level in tumor groups negative and positive for receptors estrogen receptor (ER) (A), progesterone receptor (PR) (B), and human epidermal growth factor receptor 2 (HER2) (C). (D) MORT RNA level in triple negative (TN) tumors compared to tumors positive in at least one receptor and a group of normal samples (p-value is for contrast between TN vs. receptor (+) tumors). (E) MORT RNA level in breast tumors classified into subtypes according to PAM50 (p-values are for contrast of individual tumor subtypes vs. normal). (F) MORT RNA level in samples from patients older than 60 years compared to those from patients up to 60 years old. (G) MORT RNA level in samples with wild type TP53 compared to samples with mutated TP53. (H) MORT RNA level in samples with wild type GATA3 compared to samples with mutated GATA3.
RPKM=reads per kilobase per million; WT=wild type.
References
- 1.Vrba L, Garbe JC, Stampfer MR, Futscher BW. A lincRNA connected to cell mortality and epigenetically-silenced in most common human cancers. Epigenetics. 2015;10:1074–1083. doi: 10.1080/15592294.2015.1106673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allred DC, Mohsin SK. Biological features of premalignant disease in the human breast. J Mammary Gland Biol Neoplasia. 2000;5:351–364. doi: 10.1023/a:1009573710675. [DOI] [PubMed] [Google Scholar]
- 3.Yashima K, Milchgrub S, Gollahon LS, Maitra A, Saboorian MH, Shay JW, et al. Telomerase enzyme activity and RNA expression during the multistage pathogenesis of breast carcinoma. Clin Cancer Res. 1998;4:229–234. [PubMed] [Google Scholar]
- 4.Shpitz B, Zimlichman S, Zemer R, Bomstein Y, Zehavi T, Liverant S, et al. Telomerase activity in ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 1999;58:65–69. doi: 10.1023/a:1006394209922. [DOI] [PubMed] [Google Scholar]
- 5.Umbricht CB, Sherman ME, Dome J, Carey LA, Marks J, Kim N, et al. Telomerase activity in ductal carcinoma in situ and invasive breast cancer. Oncogene. 1999;18:3407–3414. doi: 10.1038/sj.onc.1202714. [DOI] [PubMed] [Google Scholar]
- 6.R: a language and environment for statistical computing. R Foundation for Statistical Computing. 2017. [Accessed April 27th]. http://www.R-project.org/
- 7.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995;57:289–300. [Google Scholar]
- 8.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischer T, Frigessi A, Johnson KC, Edvardsen H, Touleimat N, Klajic J, et al. Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol. 2014;15:435. doi: 10.1186/s13059-014-0435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson KC, Koestler DC, Fleischer T, Chen P, Jenson EG, Marotti JD, et al. DNA methylation in ductal carcinoma in situ related with future development of invasive breast cancer. Clin Epigenetics. 2015;7:75. doi: 10.1186/s13148-015-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyngold M, Turcan S, Giri D, Kannan K, Walsh LA, Viale A, et al. Remodeling of the methylation landscape in breast cancer metastasis. PLoS One. 2014;9:e103896. doi: 10.1371/journal.pone.0103896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y, Wong CJ, Kaz AM, Dzieciatkowski S, Carter KT, Morris SM, et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology. 2014;147:418–429. doi: 10.1053/j.gastro.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu X, Sandmann T, Frierson H, Jr, Fu L, Fuentes E, Walter K, et al. Integrated genomic analysis of colorectal cancer progression reveals activation of EGFR through demethylation of the EREG promoter. Oncogene. 2016;35:6403–6415. doi: 10.1038/onc.2016.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abba MC, Gong T, Lu Y, Lee J, Zhong Y, Lacunza E, et al. A molecular portrait of high-grade ductal carcinoma in situ. Cancer Res. 2015;75:3980–3990. doi: 10.1158/0008-5472.CAN-15-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins EO, Deal AM, Anders CK, Prat A, Perou CM, Carey LA, et al. Age-specific changes in intrinsic breast cancer subtypes: a focus on older women. Oncologist. 2014;19:1076–1083. doi: 10.1634/theoncologist.2014-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garbe JC, Vrba L, Sputova K, Fuchs L, Novak P, Brothman AR, et al. Immortalization of normal human mammary epithelial cells in two steps by direct targeting of senescence barriers does not require gross genomic alterations. Cell Cycle. 2014;13:3423–3435. doi: 10.4161/15384101.2014.954456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MORT is silenced in a large fraction of ductal carcinoma in situ (DCIS) samples. The boxplots show MORT RNA level in a group of 10 normal organoids and 25 DCIS samples.
MORT gene silencing is associated with the luminal, receptor positive tumor subtypes. MORT RNA level in tumor groups negative and positive for receptors estrogen receptor (ER) (A), progesterone receptor (PR) (B), and human epidermal growth factor receptor 2 (HER2) (C). (D) MORT RNA level in triple negative (TN) tumors compared to tumors positive in at least one receptor and a group of normal samples (p-value is for contrast between TN vs. receptor (+) tumors). (E) MORT RNA level in breast tumors classified into subtypes according to PAM50 (p-values are for contrast of individual tumor subtypes vs. normal). (F) MORT RNA level in samples from patients older than 60 years compared to those from patients up to 60 years old. (G) MORT RNA level in samples with wild type TP53 compared to samples with mutated TP53. (H) MORT RNA level in samples with wild type GATA3 compared to samples with mutated GATA3.
RPKM=reads per kilobase per million; WT=wild type.