Abstract
Generation of tumor-specific T cells is critically important for cancer immunotherapy1,2. A major challenge in achieving a robust T cell response is the spatio-temporal orchestration of antigen cross-presentation in antigen presenting cells (APCs) with innate stimulation. Here we report a minimalist nanovaccine by a simple physical mixture of an antigen with a synthetic polymeric nanoparticle, PC7A NP, which generated a strong cytotoxic T cell response with low systemic cytokine expression. Mechanistically, PC7A NP achieved efficient cytosolic delivery of tumor antigens to APCs in draining lymph nodes leading to increased surface presentation while simultaneously activating type I interferon-stimulated genes. This effect was dependent on STING but not Toll-like receptor or MAVS pathway. Nanovaccine produced potent tumor growth inhibition in melanoma, colon cancer, and human papilloma virus-E6/E7 tumor models. Combination of PC7A nanovaccine with an anti-PD-1 antibody showed great synergy with 100% survival over 60 days in a TC-1 tumor model. Rechallenging of these tumor-free animals with TC-1 cells led to complete inhibition of tumor growth, suggesting generation of long-term antitumor memory. The STING-activating nanovaccine offers a simple, safe and robust strategy in boosting anti-tumor immunity for cancer immunotherapy.
Cancer immunotherapy using nanoparticle vaccines (nanovaccines) is an emerging area with recent advances focusing on the co-delivery of antigens and adjuvants3,4. Spatio-temporal control of antigen transport to the secondary lymphoid organs, cytosolic delivery and cross-presentation in the APCs in coordination with innate stimulation is essential to achieve robust tumor-specific T cell response. Although nanoparticles (<50 nm in diameter) can selectively accumulate inside the lymph nodes4,5, few studies had shown their ability to simultaneously promote antigen presentation and stimulate innate immune response without incorporation of adjuvants (e.g., CpG, Poly(I:C)). Recently, our lab has developed a library of ultra-pH sensitive (UPS) nanoparticles (20–50 nm in diameter) that are finely tunable in a broad range of physiological pH (4-7.4)6. Once taken up by cells, these nanoparticles can buffer the luminal pH of endocytic organelles at specific pH values7. Inspired by “proton sponge” polymers for cytosolic delivery of biologics8 and small nanoparticle size for lymph node targeting, we performed an in vivo screening of UPS nanoparticles to evaluate their abilities in generating cytotoxic T lymphocyte (CTL) response. The UPS library consists of copolymers containing tertiary amines with linear or cyclic side chains (Fig. 1a and Supplementary Fig. 1). Ovalbumin (OVA) was used as a model antigen. OVA loading efficiency was measured to be >75% across different polymer nanoparticles (Supplementary Fig. 2).
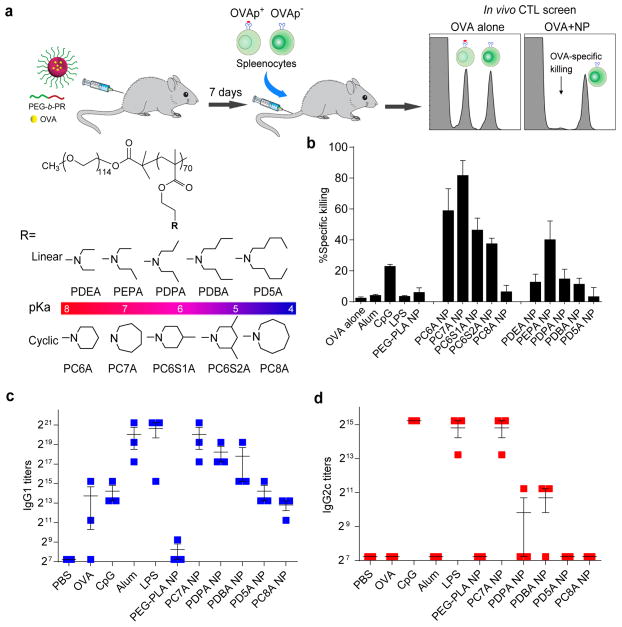
Figure 1. PC7A NP induces robust antigen-specific CTL and Th1 responses.
a, Schematic of CFSE method to screen for polymer structures that generate strong OVA-specific CTL response. OVA was used as a model antigen (10 μg) and loaded in different polymer NPs (30 μg). b, Quantitative comparison of OVA-specific CTL responses in different NP groups (n=3 for each group) identified PC7A NP as the best candidate. OVA-specific productions of IgG1 (c) and IgG2c (d) as induced by different vaccine groups. PC7A NP produced broad CTL, Th1 and Th2 responses comparable to or better than the known adjuvants in each category. In b, c and d, representative data from three independent experiments are presented as means ± s.e.m..
OVA-specific CTL response was quantified by an in vivo CTL assay (Fig. 1a). Results show that PC7A NP allowed the highest OVA-specific splenocyte killings (82%) (Fig. 1b). PC7A NP yielded approximately two-fold stronger CTL response over PC6S1A or PEPA NPs with comparable pKa’s (6.9–7.0). In the linear amine series, PEPA had the highest CTL response. These data suggest that both transition pH (i.e., 6.9 that targets early endosomal pH) and polymer architecture (i.e., cyclic seven-membered ring of PC7A) are important to induce strong CTL response. Conventional PEG-b-poly(D, L-lactic acid) (PEG-PLA) micelles had a weak response (4.2%)9. OVA-PC7A NP induced approximately 20-fold higher CTL response over OVA-Alum (4.3%) or OVA-LPS that stimulates the TLR4 pathway (3.7%) 10, and 3.6-fold higher than OVA-CpG that stimulates the TLR9 pathway (23%)11. OVA-specific antibody responses from the sera of immunized mice showed that mice vaccinated with PC7A NP generated similar titers of OVA-specific IgG1 response comparable to those by Alum or LPS (Fig. 1c). PC7A NP also generated similar titers of OVA-specific IgG2c antibody to those immunized with OVA plus CpG or LPS (Fig. 1d). Altogether, we conclude that PC7A NP was able to induce a robust antigen-specific CTL, Th1 and Th2 responses with comparable or better efficacy than several established adjuvants.
To image nanoparticle transport into draining lymph nodes (dLNs), we labeled PC7A copolymer with indocyanine green and quantified PC7A NP (29 nm in diameter) biodistribution after subcutaneous injection at the tail base. Results show efficient accumulation of PC7A NP in the peripheral lymph nodes at 24 h (Supplementary Figure 3a). Other organs did not show significant accumulation. To investigate the ability of PC7A NP for antigen delivery, we first verified that OVA can be encapsulated in the PC7A NP by strong FRET effect, and the encapsulation was relatively stable in 5% serum over 24 h (Supplementary Fig. 2). We then used Alexa Fluor 647-labelled OVA with and without PC7A NP encapsulation and harvested the dLNs 24 h after subcutaneous injection. Flow cytometry quantified the percentage of OVA-positive cells in CD8α+ and CD8α− dendritic cells (DCs) and macrophages. All three cell populations showed a significantly higher OVA accumulation by PC7A-mediated delivery over OVA alone (Fig. 2a). LN-resident CD8α+ DC cells were known to be important for induction of CTL response12. The amount of OVA-positive CD8α+ DCs increased 29 folds in OVA-PC7A NP group over the OVA only control.
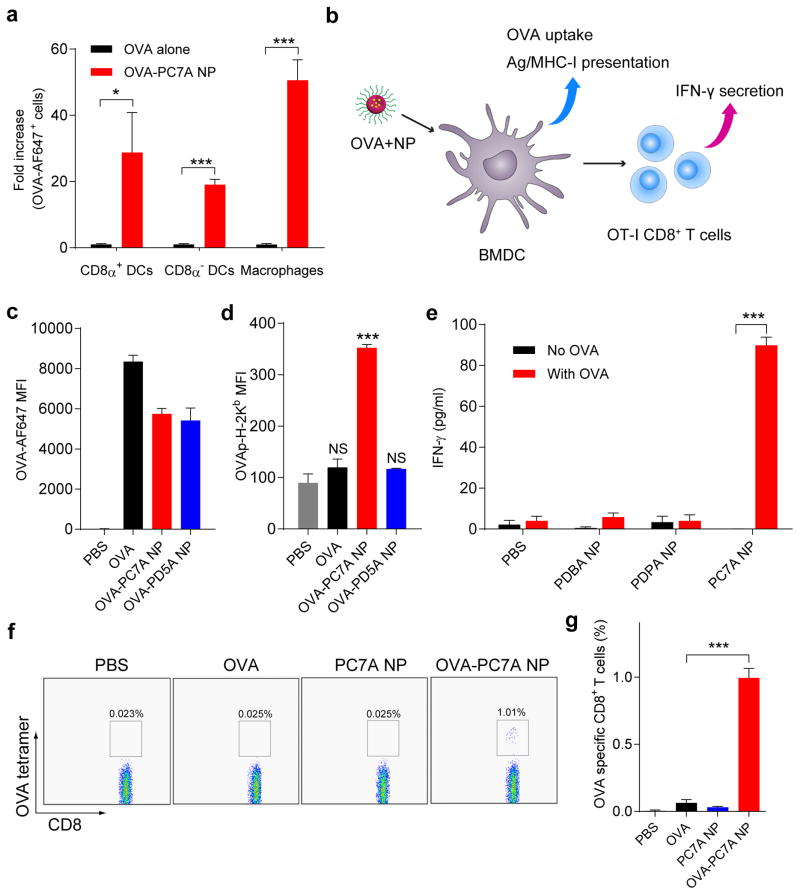
Figure 2. PC7A NP improves antigen delivery and cross-presentation in APCs and stimulates CD8 T cell responses.
a, Quantification of OVA-positive cells in three APC subtypes inside lymph nodes 24 h after subcutaneous injection of AF647-OVA-PC7A NP at the tail base of C57BL/6 mice (n=5). b, Schematic of detection of antigen cross-presentation in BMDCs and CD8+ T cell activation in vitro. c, Quantification of AF647-OVA uptake in BMDCs by flow cytometry after incubation with AF647-OVA alone, AF647-OVA-PC7A NP or AF647-OVA-PD5A NP for 4h. Mean fluorescence intensity (MFI) of AF647-OVA+ cells in BMDCs was determined (n=3). d, Levels of antigen presentation on H-2Kb in BMDCs induced by PC7A or PD5A NP (n=3). e, IFN-γ secretion by OT-I CD8+ T cells after incubating OT-I CD8+ T cells with BMDCs treated with different OVA-NPs (n=3). f, Representative flow dot plots of H-2kb/SIINFEKL tetratmer staining of CD8+ T cells in spleen. g, Percentage of OVA (SIINFEKL) specific CD8+ T cells was measured by flow cytometry (n=4). In a, c–e and g, representative data from three independent experiments are presented as means ± s.e.m.. Statistical significance was calculated by Student’s t-test, ***P<0.001, **P<0.01, *P<0.05. NS, not significant.
We investigated the ability of PC7A NP on cytosolic delivery and cross-presentation of antigens13. Incubation of OVA-PC7A NP with bone marrow derived dendritic cells (BMDCs) showed similar amount of antigen uptake compared to OVA-PD5A NP and less than the OVA only group (Fig. 2c). In contrast, OVA peptide (SIINFEKL)-MHC-I complex had 3-fold increase in antigen cross-presentation by PC7A NP over the two control groups (Fig. 2d). Using an in vitro OT-I CD8+ T cell priming assay, BMDCs treated with OVA-PC7A NP dramatically increased the IFN-γ secretion of CD8+ T cells isolated from OT-I mice over the other control groups (Fig. 2e). This result was further supported by in vivo CD8+ T cell priming assay, where OVA epitope (SIINFEKL) specific CD8+ T cells showed 15-fold higher proliferation in OVA-PC7A NP group over OVA only group (Fig. 2f, g). Endosomal disruption for cytosolic delivery was first indicated by a hemolysis assay in red blood cells (RBCs) at different pH values14. Results showed that PC7A NP had no hemolytic effect at pH 7.4 but induced strong hemolysis (~90%) at pH below 7.0 upon micelle dissociation. PD5A NP did not show any observable RBC hemolysis in the same pH range (Supplementary Figure 4a–b). PC7A NP was able to deliver increased amount of redox-activatable dye-labelled OVA into the cell cytosol compared to the PD5A NP control by confocal microscopy (Supplementary Fig. 4c)15.
Costimulatory signals (e.g., CD80/86) and cytokines are also necessary to induce a strong tumor-specific CTL response16. At 24 h post immunization with OVA-PC7A NP, inguinal LNs increased in size compared to OVA alone (Supplementary Fig. 3b). The total cell number in the inguinal LNs from OVA-PC7A NP-treated mice increased by >2 folds over the controls (Supplementary Fig. 3c). Flow cytometry analysis showed significantly higher expressions of CD86 in different subgroups of APCs from mice treated by OVA-PC7A NP over three other control groups (Fig. 3a and Supplementary Fig. 3d). Type I IFNs have been shown to boost the effectiveness of the CD8+ T cell response 17,18. We examined the expressions of IFN-stimulated genes (ISGs) in the local tissues19 over time after subcutaneous injection of PC7A NP. Poly(I:C) was used as a positive control20. Poly(I:C) was able to elicit higher response in IRF7 and CXCL10 expressions from 2 to 8 h than PC7A NP. At 24 h, PC7A NP produced stronger responses than the Poly(I:C) or PD5A NP group (Fig. 3b–c).
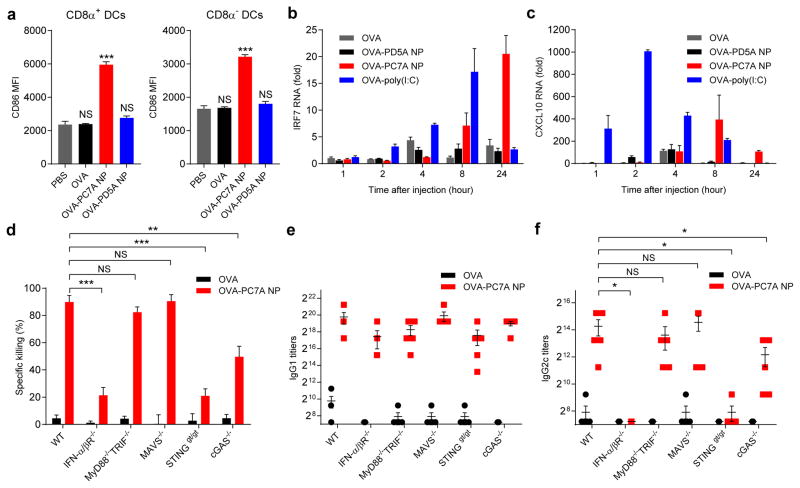
Figure 3. PC7A NP activates APCs in draining lymph nodes and stimulates STING-dependent adaptive immune responses.
a, Expression of co-stimulator CD86 on CD8α+ and CD8α- DCs in inguinal lymph nodes 24 h after injection of nanovaccine (n=5 for each group). Data on macrophages and B cells are shown in Supplementary Figure 3d. b, c, Measurement of expression levels of interferon-stimulated genes (IRF7 and CXCL10) at injection site by qPCR (n=6). d, Quantitative comparison of OVA-specific CTL responses in different knockout mouse groups (n=5 for each group). IgG1 (e) and IgG2c (f) antibody titers in the serum were determined by ELISA (n=5 for each group). Data are presented as means ± s.e.m.. Statistical significance was calculated by Student’s t-test, ***P<0.001, **P<0.01, *P<0.05. NS, not significant.
To validate the effect of type I IFN pathway on CTL response, we measured the OVA-specific CTL and Th1 response in IFN receptor (IFN-α/βR−/−) knockout mice. Data show majority of the CTL/Th1 response was abolished in IFN-α/βR−/− mice compared to wild type control (Fig. 3d–f), consistent with the ISG expression data. Toll-like receptors (TLR), MAVS and STING are known to activate the type I interferon pathways17,21. Immune response in MyD88−/−/TRIF−/−, MAVS−/− or STINGgt/gt mice showed that T cell response was not dependent on TLR or MAVS, whereas STINGgt/gt mice almost recapitulated the phenotypes in IFN-α/βR−/− mice (Fig. 3d–f). Cyclic GMP-AMP synthase (cGAS) can sense cytosolic DNA and produce cyclic GMP-AMP (cGAMP), which subsequently activates STING, leading to induction of type I IFNs22. Additional studies in cGAS−/− mice showed CTL response was partially dependent on cGAS. The roles of STING and cGAS in ISG induction were further confirmed by in vitro cell culture data using bone marrow derived macrophages (BMDMs) and human monocyte THP-1 cells (Supplementary Fig. 5a–b). To evaluate the role of cytosolic DNA in cGAS-dependent STING activation, we transfected DNase I into BMDMs before PC7A treatment23. The PC7A-induced ISG level in wildtype BMDMs decreased to almost the same level as in cGAS−/− BMDMs (Supplementary Fig. 5c). For cGAS-independent STING activation, we performed a STING pulldown assay using biotin-conjugated PC7A NP incubated with THP-1 cells. Results show that only PC7A-biotin was able to retain STING but not PD5A-biotin and PC7A-only (biotin free) controls (Supplementary Fig. 5d). Further pulldown of purified C-terminal domain (CTD, 139-397 AAs) of STING (Supplementary Fig. 5e) suggests direct binding between STING and PC7A. Isothermal calorimetry analysis showed a dissociation constant (Kd) of 1.3 μM (Supplementary Fig. 5f–g). This interaction is weaker compared to cGAMP binding to STING (Kd =9.6 nM). Negligible binding was found between PC7A and bovine serum albumin in the negative control. Despite such evidence of a specific interaction between PC7A and STING, further structural and functional studies are required to determine if PC7A can activate STING through direct binding.
To identify which cell populations are responsible for nanoparticle uptake and STING activation, Cy5-labeled PC7A polymer was employed to quantify NP uptake and phosphorylated IRF3 (pIRF3) was used to detect the activation of STING-type I IFN pathway24. Flow cytometry analysis showed that in LNs 24 h post injection, NP+ cells had significantly elevated pIRF3 expressions over NP− cells (Supplementary Fig. 6a). Furthermore, 87% of NP+ cells expressed a DC cell marker (CD11c+) as corroborated by MHC-II+ expression (Supplementary Fig. 6b). The same analysis was performed on cell suspensions from the injection site (Supplementary Fig. 6c). Data show CD45+ leukocytes internalized significantly higher amount of PC7A NPs than CD45− cells. In the CD45− cells, we did not observe any significant increase in the pIRF3 levels. In the CD45+ cells, significantly elevated pIRF3 levels were found in the NP+ cells over NP− cells. About 95% of CD45+NP+ cells had CD11c+ expressions (lower panel in Supplementary Fig. 6d). These data illustrate APCs especially DCs were the major cell population that took up PC7A NPs and subsequently activated STING-type I IFN pathway.
STING activation has been reported to induce immune regulatory responses such as elevated expressions of indoleamine 2,3-dioxygenase 1 (IDO-1)25. We compared the IDO-1 and CXCL10 expression profiles treated with the whole panel of nanoparticles. Results show that the lack of CTL activity by nanoparticles other than PC7A NP is not a result of elevated IDO-1 expression but rather due to the lack of STING activation (Supplementary Fig. 5i–j). IDO enzyme activity in mice treated with subcutaneous injections of selected polymers (PC7A, PD5A or PEPA NP, 150 μg, 5-fold of vaccine dose) showed no statistical significance (Supplementary Fig. 5h). The elevated IDO expression by PC7A NP may warrant future studies of potential immune checkpoint effect following T cell activation.
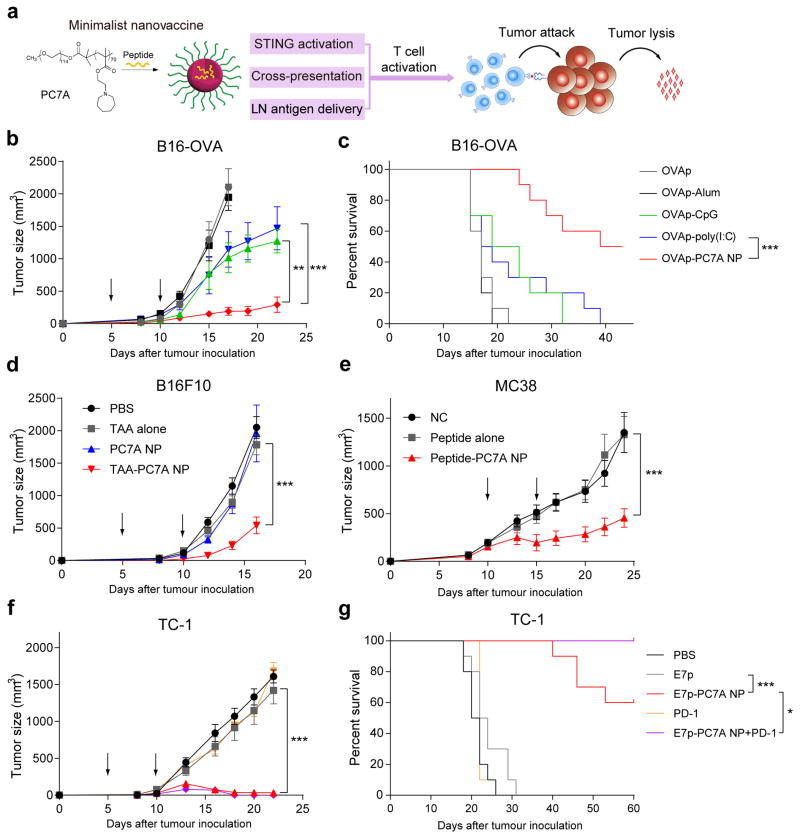
Based on the above characteristics (Fig. 4a), we investigated the antitumor efficacy of PC7A nanovaccine in several tumor models. In the B16-OVA melanoma model, a physical mixture of an antigenic peptide (OVA257-280, 0.5 μg) with PC7A NP (30 μg) was formulated. Different nanovaccine groups were subcutaneously injected 5 days after tumor inoculation, followed by a booster shot 5 days later (Fig. 4b). In the PBS control group, all the animals died in 20 days. OVA257-280 alone, PC7A NP alone or OVA257-280-PD5A NP groups did not offer any significant tumor growth inhibition or survival benefit over the PBS control (Supplementary Fig. 7a–b). OVA257-280-CpG and OVA257-280-Poly(I:C) groups conferred a minor degree of immune protection (Fig. 4b–c). In contrast, OVA257-280-PC7A NP achieved the maximum therapeutic efficacy, where 50% of animals survived over 40 days. In B16-F10 melanoma, we used a cocktail of either tumor associated antigens (Gp10021-41, Trp1214-237, Trp2173-196) or neoantigens (Obsl1T1764M, Kif18bK739N, Def8R255G)26 in PC7A NP (0.5 μg for each peptide, 30 μg polymer). PC7A vaccination significantly slowed the growth of B16F10 tumors over antigen only, PC7A only and non-treated controls (Fig. 4d and Supplementary Fig. 7c). In the colon cancer MC38 model, we selected three tumor neoantigens (Reps1P45A, AdpgkR304M, Dpagt1V213L)27. Data also show significantly improved tumor growth inhibition (Fig. 4e). Finally, we employed human papilloma virus (HPV) E6/7 TC-1 tumors28,29. Using an E7-derived peptide E743-62, 50% of mice are tumor free 60 days after treatment with E743-62-PC7A NP (Fig. 4f–g and Supplementary Fig. 8e). Combination of PC7A nanovaccine with an anti-PD-1 antibody showed synergy in both B16-OVA melanoma and TC-1 tumor models (Fig. 4g and Supplementary Fig. 8). In the TC-1 model, 100% of the animals survived for over 60 days and 90% animals were tumor free (Supplementary Fig. 8e). Both B16OVA and TC-1 tumor models showed mild PD-L1 expressions on tumor cells while certain subtype of myeloid cells had high PD-L1 expressions over the isotype control (Supplementary Fig. 8d, f). These data support the nanovaccine synergy with anti-PD1 therapy. Meanwhile, anti-PD-1 therapy alone did not lead to significantly improved antitumor effect in either model similar to other reports30,31. Tumor-free mice were rechallenged with 1×106 TC-1 tumor cells 82 days after tumor inoculation. Data show that previously treated, tumor-free mice were resistant to the newly inoculated tumor cells whereas such tumors grew robustly in naïve mice and surgically cured mice (Supplementary Figure 7e). These results suggest a long-term antitumor response induced by the nanovaccine, which likely activate memory T cells. Analyses of systemic cytokines/chemokines of mice treated with PC7A NP (150 μg, 5-fold of vaccine dose) showed much less systemic cytokine levels compared to Poly(I:C) control (Supplementary Figure 9). Histology analysis of major organs (e.g., liver, spleen, kidney, heart) did not show any observable toxicity in mice treated with repeated injections of PC7A nanovaccine (150 μg, 5-fold of vaccine dose, Supplementary Figure 10). These data demonstrate the safe and efficacious antitumor immunity of PC7A nanovaccine at a small antigen dose (0.5 μg) and its notable synergy with a checkpoint inhibitor.
Figure 4. PC7A nanovaccine inhibits tumor growth and prolongs survival in tumor bearing mice.
a, Schematic of the minimalist design of PC7A nanovaccine. b, c C57BL/6 mice (n=10 per group) inoculated with 1.5×105 B16-OVA tumor cells were treated with OVA peptide, PC7A nanovaccine, CpG, poly(I:C) and Alum plus peptide (0.5 μg). Tumor growth (b) and Kaplan–Meier survival curves (c) of tumor-bearing mice were shown. (d) Tumor growth inhibition study of B16F10 melanoma. C57BL/6 mice (n=10 per group) inoculated with 1.5×105 B16F10 tumor cells were treated with a cocktail of tumor associated antigens (Gp10021-41, Trp1214-237, Trp2173-196) in PC7A NP at specific time point indicated by the arrows. (e) Tumor growth inhibition study of MC38 colon cancer in C57BL/6 mice. Mice (n=10 per group) inoculated with 1.0×106 MC38 tumor cells were treated with a cocktail of neoantigens (Reps1P45A, AdpgkR304M, Dpagt1V213L) in PC7A NP, and nanovaccine was administered on day 10 and 15 in established tumors (100–200 mm3). In the HPV tumor model, tumor growth inhibition (f) and survival data (g) in C57BL/6 mice (n=10 per group) were analyzed after tumor inoculation with 1.5×105 TC-1 tumor cells. In b and d–f, data are presented as means ± s.e.m.. Statistical significance was calculated by Student’s t-test, ***P<0.001, **P<0.01, *P<0.05. Statistical significance for survival analysis in c and g was calculated by the log-rank test, ***P<0.001, **P<0.01, *P<0.05.
In summary, we discovered a synthetic nanoparticle that not only enhances antigen delivery and cross-presentation but also stimulates the STING pathway to boost antitumor immunity for cancer immunotherapy. The simplicity, robust T cell activation and synergy with checkpoint inhibition make the PC7A nanovaccine an attractive candidate for clinical development. This nanovaccine platform can be rapidly adopted to incorporate many existing tumor-associated antigens as well as a growing number of tumor neoantigens32,33. The unique characteristics of PC7A NP also allow it to package microbial antigens as vaccines for the prevention and treatment of infectious diseases.
Online Methods
Materials
Monomers 2-(diethylamino)ethyl methacrylate (DEA-MA) and 2-aminoethyl methacrylate (AMA-MA) were purchased from Polyscience Company. Ovalbumin and OVA257-263, CpG ODN were purchased from Invivogen. Imject Alum was purchased from Thermo Scientific, LPS and Poly(I:C) were purchased from Sigma-Aldrich. OVA257-280 (SIINFEKLTEWTSSNV MEERKIKV), E743-62 (GQAEPDRAHYNIVTFCCKCD), E749-57 (RAHYNIVTF), Gp10021-41(VGALEGSRNQDWLGVPRQLVT), Trp1214-237(SHEGPAFLTWHRYHLLQLERDMQE), Trp2173-196(QPQIANCSVYDFFVWLHYYSVRDT), Obsl1T1764M (EGVELCPGNKYEMRRHG TTHSLVIHD), Kif18bK739N(PSKPSFQEFVDWENVSPELNSTDQPFL), Def8R255G(SHCHWNDLAVIPAGVVHNWDFEPRKVS), Reps1P45A (GRVLELFRAAQLANDVVLQIMELCGAT R), AdpgkR304M(GIPVHLELASMTNMELMSSIVHQQVFPT), Dpagt1V213L(EAGQSLVISASII VF NLLELEGDYR) were synthesized by Biomatik. PEG-PLA was purchased from Advanced Polymer Materials Inc. (Montreal, QC, Canada). Other solvents and reagents were purchased from Sigma-Aldrich or Fisher Scientific Inc.
Syntheses of methacrylate monomers
Monomers including 2-(ethylpropylamino) ethyl methacrylate (EPA-MA), 2-(dipropylamino) ethyl methacrylate (DPA-MA), 2-(dibutylamino) ethyl methacrylate (DBA-MA) and 2-(dipentylamino) ethyl methacrylate (D5A-MA), 2-(pentamethyleneimino)ethyl methacrylate (C6A-MA), 2-(hexamethyleneimino)ethyl methacrylate (C7A-MA) were synthesized following previous publications34,35. New monomers including 2-(heptamethyleneimino)ethyl methacrylate (C8A-MA), 2-(4-methylpiperidineleneimino)ethyl methacrylate (C6S1A-MA), 2-(3,5-dimethylpiperidineleneimino)ethyl methacrylate (C6S2A-MA) were synthesized following a previously published procedure35. Below are the chemical characterizations of the new monomers:
2-(Heptamethyleneimino)ethyl methacrylate (C8A-MA)
1H NMR (TMS, CDCl3, ppm): 6.10 (br, 1H, CHH=C(CH3)-), 5.54 (br, 1H, CHH=C(CH3)-), 4.19 (t, J = 6.1 Hz, 2H, -OCH2CH2N-), 2.77 (t, J = 6.1 Hz, 2H, -OCH2CH2N-), 2.60 (td, 4H, -N(CH2CH2CH2)2CH2), 1.94 (m, 3H, CH2=C(CH3)-), 1.61 (tdd, 2H, -N(CH2CH2CH2)2CH2), 1.54 (td, 8H, -N(CH2CH2CH2)2CH2).
2-(4-Methylpiperidineleneimino)ethyl methacrylate (C6S1A-MA)
1H NMR (TMS, CDCl3, ppm): 6.07 (br, 1H, CHH=C(CH3)-), 5.53 (br, 1H, CHH=C(CH3)-), 4.26 (m, 2H, -OCH2CH2N-), 2.88 (m, 2H, -OCH2CH2N-), 2.65 (m, 2H, -N(CHHCH2)2CHCH3), 2.04 (tt, 2H, -N(CHHCH2)2CHCH3), 1.92 (m, 3H, CH2=C(CH3)-), 1.59 (m, 2H, -N(CH2CHH)2CHCH3), 1.31 (m, 1H, -CHCH3), 1.21 (m, 2H, -N(CH2CHH)2CHCH3), 0.89 (d, 3H, -CHCH3).
2-(3,5-Dimethylpiperidineleneimino)ethyl methacrylate (C6S2A-MA)
1H NMR (TMS, CDCl3, ppm): 6.09 (br, 1H, CHH=C(CH3)-), 5.55 (br, 1H, CHH=C(CH3)-), 4.28 (t, 2H, -OCH2CH2N-), 2.85 (ddt, 2H, -OCH2CH2N-), 2.66 (t, 2H, -N(CHHCHCH3)2CH2), 1.94 (m, 3H, CH2=C(CH3)-), 1.68 (m, 3H, -N(CH2CHCH3)2CHH), 1.57 (t, 2H, -N(CH2CHCH3)2CH2), 0.93 (d, 1H, -N(CH2CHCH3)2CHH), 0.84 (d, 3H, -N(CH2CHCH3)2CH2).
Syntheses of PEG-b-PR block copolymers
PEG-b-PR copolymers were synthesized by atom transfer radical polymerization (ATRP) following procedures previously reported36. PEG-b-PDPA is used as an example to illustrate the procedure. First, DPA-MA (7 mmol), PMDETA (0.1 mmol), and MeO-PEG114-Br (0.1 mmol) were dissolved in 2-propanol (2 mL) and DMF (2 mL). After three cycles of freeze-pump-thaw, CuBr (0.1 mmol) was added. The polymerization was carried out at 40 °C for 10 hours. The reaction mixture was passed through a neutral Al2O3 column, the residue was dialyzed in distilled water and lyophilized. The obtained polymers were characterized by 1H NMR and gel permeation chromatography (GPC).
Syntheses of PEG-b-(PR-r-Dye) block copolymers
AMA-MA was introduced in the PEG-b-PR copolymers for the conjugation of dyes followed the similar procedure34,35. After synthesis, PEG-b-(PR-r-AMA) (10 mg) and Dye-NHS (1.5 equivalences) was both dissolved in DMF. After overnight reaction, the copolymers were purified by preparative GPC.
Preparation of nanoparticles
Micelles were prepared following a solvent evaporation method35. In the example of PEG-b-PC7A, copolymer was dissolved in methanol and then added into distilled water under sonication. Methanol was removed using ultrafiltration tube (MW = 100 KD). The nanoparticles were characterized by dynamic light scattering (DLS, Malvern MicroV model, He-Ne laser, λ= 632 nm) for hydrodynamic diameter (Dh).
OVA loading and stability studies
OVA loading efficiency inside nanoparticles was measured by an ultrafiltration method. Briefly, nanoparticles (300 μg/mL) were mixed with AF647-labelled OVA (100 μg/mL) for 30 min. Free OVA was removed by ultrafiltration tube with a molecular weight cutoff of 100 kD. The concentration of free OVA was measured on a Hitachi fluorometer (F-7500 model) with excitation wavelength at 640 nm. The loading efficiency was calculated using the equation below:
To evaluate the loading stability, OVA-loaded PC7A nanoparticles were incubated in PBS buffer (pH 7.4) containing 5% fetal bovine serum over different times. Free OVA was separated and determined as described above.
The FRET experiment was further designed to investigate the polymer and OVA interactions. Typically, Cy3.5-conjugated PC7A (100 μg/mL) was incubated with AF647-labelled OVA (20ug/mL) in PBS buffer (pH = 7.4). After 30 min incubation, the fluorescence emission spectra were obtained on a Hitachi fluorometer (F-7500 model). The samples were excited at 590 nm, and the emission spectra were collected from 600 to 750 nm.
Animals and cells
All animal procedures were performed with ethical compliance and approval by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center. Female C57BL/6 mice (6–8 weeks) were obtained from the UT southwestern breeding core. INF-α/βR−/−mice were kindly provided by Dr. David Farrar (UT Southwestern). STINGgt/gt mice, MyD88−/−mice, TRIF−/−mice C57BL/6- Tg (TcraTcrb)1100Mjb/J (CD45.2, H-2b ) (OT-I) mice, C57BL/6- CD45.1 mice were purchased from the Jackson laboratory. MyD88−/−/TRIF−/−mice were crossed in our lab. cGas−/− mice were generated as previously described37. All these strains were maintained on C57BL/6J background. For each experiment, mice were randomly allocated by blinded investigators to each group. STINGgt/gt and cGAS−/− BMDMs were derived from corresponding knockout mice, and then cultured in M-CSF containing medium for 6-7days. THP-1 cells were purchased from ATCC and we established THP-1 cell lines stably expressing shRNA targeting hSTING and hcGAS as described before38. B16-OVA cells were kindly provided by Dr. Patrick Hwu at MD Anderson Cancer Center, TC-1 cells were kindly provided by Dr. T. C. Wu at John Hopkins University, MC38 cells were kindly provide by Dr. Yangxin Fu(UT Southwestern). All cell lines were routinely tested using mycoplasma contamination kit (R&D). Cells were cultured in complete medium (DMEM, 10% fetal bovine serum, 100 U/mL penicillin G sodium and 100 μg/mL streptomycin (Pen/Strep), MEM non-essential amino acids (all from Invitrogen), and 20 μM β-mercaptoethanol(β-ME)) at 37°C in 5% CO2 and the normal level of O2.
In vivo cytotoxicity killing assay
Groups of C57BL/6 mice were injected (OVA 10 μg plus nanoparticles 30 μg or other adjuvants with the same dose) subcutaneously at the tail base of C57BL/6 mice. Imject Alum (4 mg per mouse, 50μl:50μl mixture with the antigen solution) was used by volume ratio as recommended by manufacture. One week later, naïve C57BL/6 mice were sacrificed and spleenocytes were collected. Half of the spleenocytes were pulsed with OVA257-263 or E749-57 peptides for 2 h in complete media at 37 °C. The unpulsed and peptide-pulsed cells were labeled with 0.5 or 0.05 μM Carboxyfluorescein succinimidyl ester (CFSE), respectively, in serum free media for 15 mins. Equal numbers (1×107) of CFSElow (OVA pulsed cells) and CFSEhigh (unpulsed cells) were mixed together and injected intravenously into the immunized mice. After 16 h, the blood from treated mice was collected and subjected to flow cytometry analysis. The number of CFSEhigh and CFSElow was determined and used to calculate the percentage of OVA peptide-pulsed target cell killing. Specific killing was defined as percentage of specific lysis = [1−non-transferred control ratio/experimental ratio] x100.
ELISA assay
For antibody detection, groups of C57BL/6 mice were immunized with different vaccines on day 0 and 14. On day 21, 50 μL of blood was drawn from tail vein and the antigen-specific IgG1 and IgG2c in serum were measured by ELISA. For ELISA assay, flat-bottomed 96-well plates (Nunc, Rochester, NY) were precoated with OVA protein at a concentration of 0.5 μg protein/well in 50 mM carbonate buffer (pH 9.6) at 4 °C overnight, which were then blocked with 5% Glycine. Antisera obtained from immunized animals were serially diluted from 102 to 106 in PBS-0.05% Tween (PBS-T), pH 7.4, and were added to the wells and incubated at 37 °C for 1 h. Goat anti-mouse IgG1 and IgG2c (HRP) (Abcam, Cambridge, MA) were used at a dilution of 1:10,000 in PBS-T–1% BSA for labeling. After adding the HRP substrates, optical densities were determined at a wavelength of 450 nm in an ELISA plate reader (Bio-Rad, Hercules, CA).
Lymph node imaging assay
To investigate whether NP can accumulate in the draining lymph nodes, we labeled the PC7A copolymer with indocyanine green (ICG, λex/λem = 800/820 nm). ICG-encoded PC7A NP (30 μg per mice) was injected subcutaneously at the tail base of C57BL/6 mice. NP distribution was imaged using a clinical camera (SPY Elite®). Animals were sacrificed at 24h after injection of NP, major organs and inguinal and axillary LNs were excised and imaged.
In vivo cell uptake assay
For antigen delivery assay, subcutaneous injections at tail base of C57BL/6 mice were performed with PBS alone, OVA-AF647, or nanoparticle plus OVA-AF647 treatments. At 24 h post injection, mice were sacrificed and inguinal lymph nodes were removed, teased with 26 gauge needles and then passed through a 70-μm cell strainer (BD) to recover a cell suspension. The lymph node cell suspension was stained with PI and anti-CD11c-FITC, anti-CD11b-pacific blue, anti-B220-APC-Cy7, anti-CD8a-PE-Cy7. Four major APCs populations (CD8α+DC cells (CD11c+CD11b−B220−CD8α+), CD8α−DC cells (CD11c+ CD8α−), macrophage cells (CD11b+ CD11c− B220−), B cells (B220+ CD11c−)) were analyzed for the OVA-AF647 positive cells. APC maturation was measured by staining with anti-CD86-PE.
For nanoparticle uptake and STING activation assay, subcutaneous injections at tail base of C57BL/6 mice were carried out with PBS alone, or PC7A-Cy5 (30 μg) treatments. At 24 h post injection, mice were sacrificed, inguinal lymph nodes and subcutaneous tissue were removed, and digested in collagenase IV (Sigma-Aldrich) solution for 25 mins at 37°C. Tissue was then passed through a 70-μm cell strainer (BD) to recover a cell suspension. All the cell suspension were stained with PI and anti-CD11c-FITC, anti-MHCII-BV605, anti-CD45.2-Apc-Cy7. For intracellular pIRF3 staining, cells were permeabilized by the Fixation/Permeabilization kit (BD Cat#554714). After blocking with mouse serum, cells were stained with pIRF3 antibody (Cell Signaling, Cat # 4947) and subsequently were stained with anti-rabbit IgG-PE secondary antibody (Biolegend). Flow cytometry (LSRII, BD) was performed on stained cell suspensions and analyzed with FlowJo software (Tree Star Inc. Ashland, OR).
In vitro cell uptake and cross presentation assay
Bone marrow-derived dendritic cells were generated by culturing bone marrow cells flushed from femurs of C57BL/6J mice in DC media: DMEM supplemented with 10% FBS, pen/strep, sodium pyruvate and 20 ng/mL GM-CSF. Media was half replaced every 2 days; non-adherent and loosely adherent immature dendritic cells (DCs) were collected on day 6 and phenotyped by determining expression of CD11c (routinely 60–80% CD11c+). OVA-AF647 (2ug/mL) or mixture of OVA-AF647 with different nanoparticles (50 μg/mL) was incubated with murine bone marrow-derived dendritic cells (BMDCs) at 37°C for 4 hours and quantified using the mean fluorescence intensity (MFI) of cells by FACS. For cross presentation assay, BMDCs were incubated with OVA alone or mixture of OVA with different nanoparticles at 37°C for 18–20 hours, then the OVA257-267 presented on the MHC-I on cell surface were detected by monoclonal antibody 25mAb-D1.16, an antibody specifically recognize OVA peptide SIINFEKL bound to H-2Kb.
In vitro CD8+ T cell priming assay
To evaluate antigen presentation by OVA-NP-pulsed BMDCs, IFN-γ secretion by primed OT-I T cells was used to quantify CD8+ T-cell activation. Briefly, BMDCs were incubated with 3 μg/mL OVA alone or mixture of OVA with different nanoparticles (50 μg/mL) at 37°C for 18 h. CD8+ T lymphocytes from OT-I mice were selected by magnetic separation (MACS system; Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s indications. The purity of CD8+ T lymphocytes was >95%. CD8+ T cells were plated at 2 × 105 cells/well in 96-well plates (Costar; Corning, Inc., Corning, NY) in RPMI media containing 10% FCS and 2 × 105 unpulsed, or antigen-pulsed BMDCs that were added for 24 h. Cell culture supernatants were collected and analyzed for cytokine content using mouse TH1/TH2 9-Plex Ultra-sensitive Kit (Meso Scale Discovery). Samples were run in triplicate.
In vivo CD8+ T cell priming assay
Spleens were harvested from B6 CD45.2+ OT-I mice, CD8+ T cells from cell suspensions were isolated by magnetic bead separation on a MACS column. 5 × 104 OT-1 CD8+ T cells were transferred into B6 CD45.1+ mice via intravenous (i.v.) injection and allowed to acclimate for 1 day prior to immunization. 1 day later, groups of CD45.1+ mice were immunized with PBS alone, OVA(10 μg), or nanoparticle (30 μg) plus OVA subcutaneously at the tail base. One week later, spleens were harvested and dispersed into single-cell suspensions, stained with anti-CD8-PE-cy7, APC-conjugated H-2Kb/OVA (SIINFEKL) tetramer (NIH) for flow cytometry analysis.
Hemolysis assay
The capacity of polymers to promote pH-dependent disruption of lipid bilayer membranes was assessed via a red blood cell hemolysis assay as previously described14. Polymers were incubated for 1 h at 37 °C in the presence of mouse erythrocytes at 20 μg/mL in 100 mM sodium phosphate buffer (supplemented with 150 mM NaCl) in the pH range of the endosomal processing pathway (7.4, 7.2, 7.0, 6.8, 6.6, and 6.4). The extent of cell lysis (i.e. hemolytic activity) was determined spectrophotometrically by measuring the amount of hemoglobin released (A541 nm). Hemolytic activity was normalized to a 100% lysis control (1% Triton X-100 treated red blood cells). Samples were run in triplicate.
Flow cytometry
Antibodies were purchased from Biolegend. The following primary antibodies were used: anti-CD16/CD32 (Biolegend, Cat#: 101301, clone: 93), anti-CD8-PE-cy7 (Biolegend, Cat#: 100721, clone: 53-6.7), anti-CD11c-FITC (Biolegend, Cat#: 117305, clone: N418) and anti-CD11b-Pacif blue (Biolegend, Cat#: 101223, clone: M1/70), anti-B220-APC-cy7 (Biolegend, Cat#: 103223, clone: RA3-6B2), anti-CD86-PE (Biolegend, Cat#: 105007, clone: GL-1), anti- H-2Kb bound to SIINFEKL-APC (Biolegend, Cat#:141605, clone:25-D1.16), anti-CD45.2-APC (Biolegend, Cat#:109814, clone:104), anti-CD45.2-APCcy7 (Biolegend, Cat#:109823, clone:104), anti-PD-L1-PE (Biolegend, Cat#:124307, clone:10F.9G2), isotype control-PE (Biolegend, Cat#:400607, clone:RTK4530), anti-F4/80-PE/cy7 (Biolegend, Cat#:123113, clone:BM8), anti-Gr-1-FITC (Biolegend, Cat#:108419, clone:RB608C5), anti-MHCII-BV605 (Biolegend, Cat#:107639, clone:M5/114.15.2), anti-rabbit IgG-PE (Biolegend, Cat#:406421, clone:poly4064). Flow data were acquired on a BD™ LSR II flow cytometer and analyzed using Flowjo software.
RT-PCR
Subcutaneous tissues were taken at indicated time points after injection with OVA-PC7A NP (OVA 10 μg, PC7A 150 μg) or the same dose of different adjuvants. To obtain BMDM, about 1 × 107 bone marrow cells were cultured in DMEM containing 10% FBS, antibiotics and conditioned media from L929 cell culture. After 6 to 7 days, mature macrophages were harvested and cultured on 12-well plates for experiments5. Total RNAs were extracted by TRIzol (Invitrogen) from cells or tissues according to the manufacturer’s instructions. q-RT-PCR were performed as previously described37,38. Samples were run in triplicate. The following primers were used for q-RT-PCR.
mIRF7: ATGCACAGATCTTCAAGGCCTGGGC; GTGCTGTGGAGTGCACAGCGGAAGT;
mCXCL10: GCCGTCATTTTCTGCCTCA; CGTCCTTGCGAGAGGGATC;
mIDO-1: CGGACTGAGAGGACACAGGTTAC; ACACATACGCCATGGTGATGTAC;
mRPL19: AAATCGCCAATGCCAACTC; TCTTCCCTATGCCCATATGC;
hCXCL10: TGGCATTCAAGGAGTACCTC; TTGTAGCAATGATCTCAACACG;
hIDO-1: GCCAGCTTCGAGAAAGAGTTG; ATCCCAGAACTAGACGTGCAA;
hGAPDH: ATGACATCAAGAAGGTGGTG; CATACCAGGAAATGAGCTTG.
DNase I transfection
BMDMs were transfected with 5 μg of DNase I (Roche) by transfection reagent DOTAP (Roche) according to manufacturer’s instructions. After incubating the cells with DOTAP-DNase I or DOTAP alone for 1hr, cells were washed to remove excess of transfection reagent and enzyme and were then incubated with PC7A at 400 μg/ml for 9 hr. CXCL10 were measured by RT-PCR.
STING pulldown assay
To investigate the STING interaction with PC7A copolymer, we labeled the PC7A copolymer with biotin (2–3 biotins per polymer chain). For THP-1 cell pulldown assay, PC7A-biotin (200 μg/ml) was incubated with THP-1 cells for 8 hrs, and then cells were collected and lysed in RIPA buffer (Sigma R0278). Lysates were precipitated with streptavidin-modified dynabeads (BD 557812). Samples were analyzed using SDS-PAGE and Western blots by rabbit anti-STING antibody (Cell Signaling, Cat # 13647). For STING protein pull down assay, human STING CTD (139–379) expression and purification had been described before39, PC7A-biotin(50 μg/mL) was incubated with STING CTD(1 μg/ml) for 3hrs in PBS (pH=6.8), PEPA-biotin in PBS (pH=6.8) and PD5A-biotin in PBS (pH=4.4) with the same concentration were used as control groups, and then precipitated with streptavidin-modified dynabeads. Samples were analyzed using SDS-PAGE and Western blots.
Isothermal Titration Calorimetry (ITC)
ITC was employed to measure the binding affinities between STING CTD and PC7A polymers or cGAMP using a VP-ITC microcalorimeter (GE Healthcare), ITC of PC7A-bovine serum albumin (BSA) was used as a negative control. The titrations were performed at 20 °C in the buffer containing 25 mM HEPES (pH 6.8), 150 mM NaCl. Thirty-two injections were performed with 4 min spacing time. The titration traces were integrated by NITPIC, and then the curves were fitted by SEDFIT.39 The figures were prepared using GUSSI (http://biophysics.swmed.edu/MBR/software.html).
IDO enzyme activity assay in tissues
Bacterial pDNA (pEGFPN1, Clontech) was prepared using an endotoxin-free Kit (Qiagen). Mice were intravenously injected with 30 μg pDNA mixed with in vivo-jetPEI (Polyplus -transfection, N:P =8) or were injected s.c. with NPs (150 μg, 5-fold of vaccine dose). IDO activity was measured as described in previous reports40,41. Enzyme activity was expressed as the product content per hour per gram of tissue protein.
Immunization and tumor therapy experiments
Six to eight week old mice (n=10 for each group) were injected subcutaneously with B16-OVA or B16F10 melanoma cells(1.5 × 105), or TC-1 cells(1.5 × 105), MC38 cells(5 × 105) into the right flank of mice. Animals were immunized with subcutaneous injection at the tail base of antigen-polymer NP (0.5 μg per antigen peptide, PC7A NP 30 μg) or the same dose of different adjuvants as described in the main text. Or at day 3, 6, 9 and 12, some groups were i.p. injected with 200 μg checkpoint inhibitors (anti-mPD-1, BioXcell, BE0146) for comparison or synergy evaluation. The tumor growth was subsequently measured twice a week using a digital caliper and calculated as 0.5×length × width2 by blinded investigators. Mice were killed when tumor surface area reached 1500mm3, the end point of tumor detection is 2 fold of the longest survival time (LST) of control group, so around 40 days for melanoma tumor model, and around 60 days for TC-1 and MC38 tumor models.
For PD-L1 expression analyses, tumor tissues were digested by 1 mg/mL collagenase IV (Sigma-Aldrich) and 0.2 mg/mL DNase I (Sigma-Aldrich) for 45 minutes at 37°C. Cells were then stained with antibodies against PD-L1, CD11b, Gr-1, F4/80, CD11c, and CD45 (Biolegend).
Statistical analysis
Based on pilot immunization and tumor treatment studies, we used group sizes of 3–6 animals/group for immunogenicity measurements and 10 animals/group for tumor therapy experiments. Statistical analysis was performed using Microsoft Excel and Prism 5.0 (GraphPad). Data are expressed as means ± s.e.m. Data were analyzed by Student’s t test. Variance similarity test (f-test) was performed before t-test. All t-tests were one-tailed and unpaired, and were considered statistically significant if p< 0.05, (*, p<0.05; **, p<0.01; ***, p<0.001 unless otherwise indicated). The survival rates of the two groups were analyzed using a log-rank test and were considered statistically significant if p< 0.05.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01AI093967 to Z.J.C., R01EB013149 and R01CA129011 to J.G.) and Cancer Prevention and Research Institute of Texas (RP120718-P3 and RP150498 to Z.J.C). Z.J.C. is an Investigator of Howard Hughes Medical Institute. Animal imaging work is supported by the UT Southwestern Small Animal Imaging Resource Grant (U24 CA126608) and Simmons Cancer Center Support Grant (P30 CA142543). We thank Z. Zeng for cell culture, Q. Wei for polymer synthesis, T. Zhao for animal imaging. We thank N. Yan for helpful discussions on STING activation, T.C. Wu for kindly providing the TC-1 tumor cells, P. Hwu for the B16-OVA cancer cells, Y. Peng for vaccine safety analysis, S. Tso for ITC experiment, and the molecular pathology core of UT Southwestern for tissue toxicity analysis.
Footnotes
AUTHOR CONTRIBUTIONS
M.L. and Z.W. designed and performed majority of the experiments, analyzed the data, and wrote the first draft of the paper; H.W. made the initial observation of the immune stimulatory activity of PC7A and performed the experiments on CD8 T cell activation by PC7A in vitro and in vivo; H.C. analyzed innate cytokine expression in local tissues and cell lines; Z.L. assisted with the CTL and Th1 experiments; Y.L and D.M assisted with the PC7A and STING interaction experiments; G.H. and C.W. assisted animal efficacy evaluation; X.C. assisted transgenic mice studies; Y.F. assisted the PD-L1 expression in B16 and TC-1 tumors; M.R.P., J.L. and A.F. contributed to experimental designs; Z.J.C. and J.G. supervised all the experiments and revised the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
Data Availability Statement
The datasets generated and/or analyzed during the study are available from the corresponding authors upon request.
References
- 1.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuai R, Ochyl LJ, Bahjat KS, Schwendeman A, Moon JJ. Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat Mater. 2016 doi: 10.1038/nmat4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H, et al. Structure-based programming of lymph-node targeting in molecular vaccines. Nature. 2014;507:519–522. doi: 10.1038/nature12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy ST, et al. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat Biotechnol. 2007;25:1159–1164. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 6.Ma X, et al. Ultra-pH-sensitive nanoprobe library with broad pH tunability and fluorescence emissions. J Am Chem Soc. 2014;136:11085–11092. doi: 10.1021/ja5053158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, et al. A nanobuffer reporter library for fine-scale imaging and perturbation of endocytic organelles. Nat Commun. 2015;6:8524. doi: 10.1038/ncomms9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maldonado RA, et al. Polymeric synthetic nanoparticles for the induction of antigen-specific immunological tolerance. Proc Natl Acad Sci U S A. 2015;112:E156–165. doi: 10.1073/pnas.1408686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 11.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 12.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heath WR, et al. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol Rev. 2004;199:9–26. doi: 10.1111/j.0105-2896.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 14.Wilson JT, et al. pH-Responsive nanoparticle vaccines for dual-delivery of antigens and immunostimulatory oligonucleotides. ACS Nano. 2013;7:3912–3925. doi: 10.1021/nn305466z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, et al. A Redox-Activatable Fluorescent Sensor for the High-Throughput Quantification of Cytosolic Delivery of Macromolecules. Angew Chem Int Ed Engl. 2017;56:1319–1323. doi: 10.1002/anie.201610302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liechtenstein T, Dufait I, Lanna A, Breckpot K, Escors D. Modulating Co-Stimulation during Antigen Presentation to Enhance Cancer Immunotherapy. Immunol Endocr Metab Agents Med Chem. 2012;12:224–235. doi: 10.2174/187152212802001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15:405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 18.Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF. Type I interferon response and innate immune sensing of cancer. Trends Immunol. 2013;34:67–73. doi: 10.1016/j.it.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 21.Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13:543–551. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- 22.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll EC, et al. The Vaccine Adjuvant Chitosan Promotes Cellular Immunity via DNA Sensor cGAS-STING-Dependent Induction of Type I Interferons. Immunity. 2016;44:597–608. doi: 10.1016/j.immuni.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo SR, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemos H, Huang L, McGaha TL, Mellor AL. Cytosolic DNA sensing via the stimulator of interferon genes adaptor: Yin and Yang of immune responses to DNA. Eur J Immunol. 2014;44:2847–2853. doi: 10.1002/eji.201344407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreiter S, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 28.Sun YY, et al. Local HPV Recombinant Vaccinia Boost Following Priming with an HPV DNA Vaccine Enhances Local HPV-Specific CD8+ T Cell Mediated Tumor Control in the Genital Tract. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-15-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Zhou H, Wang W, Fu YX, Zhu M. A novel dendritic cell targeting HPV16 E7 synthetic vaccine in combination with PD-L1 blockade elicits therapeutic antitumor immunity in mice. Oncoimmunology. 2016;5:e1147641. doi: 10.1080/2162402X.2016.1147641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice AE, et al. An HPV-E6/E7 immunotherapy plus PD-1 checkpoint inhibition results in tumor regression and reduction in PD-L1 expression. Cancer Gene Ther. 2015;22:454–462. doi: 10.1038/cgt.2015.40. [DOI] [PubMed] [Google Scholar]
- 31.Holmgaard RB, Zamarin D, Munn DH, Wolchok JD, Allison JP. Indoleamine 2,3-dioxygenase is a critical resistance mechanism in antitumor T cell immunotherapy targeting CTLA-4. J Exp Med. 2013;210:1389–1402. doi: 10.1084/jem.20130066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 33.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou K, et al. Tunable, ultrasensitive pH-responsive nanoparticles targeting specific endocytic organelles in living cells. Angew Chem Int Ed Engl. 2011;50:6109–6114. doi: 10.1002/anie.201100884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou K, et al. Multicolored pH-tunable and activatable fluorescence nanoplatform responsive to physiologic pH stimuli. J Am Chem Soc. 2012;134:7803–7811. doi: 10.1021/ja300176w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsarevsky NV, Matyjaszewski K. “Green” atom transfer radical polymerization: from process design to preparation of well-defined environmentally friendly polymeric materials. Chem Rev. 2007;107:2270–2299. doi: 10.1021/cr050947p. [DOI] [PubMed] [Google Scholar]
- 37.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins AC, et al. Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host Microbe. 2015;17:820–828. doi: 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang L, et al. Engineering DNA nanoparticles as immunomodulatory reagents that activate regulatory T cells. J Immunol. 2012;188:4913–4920. doi: 10.4049/jimmunol.1103668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The Absence of IDOHoshi M et al. The absence of IDO upregulates type I IFN production, resulting in suppression of viral replication in the retrovirus-infected mouse. J Immunol. 2010;185:3305–3312. doi: 10.4049/jimmunol.0901150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.