Abstract
Biological differences exist between strains of laboratory mice, and it is becoming increasingly evident that there are differences between substrains. In the C57BL/6 mouse, the primary substrains are called 6J and 6N. Previous studies have demonstrated that 6J and 6N mice differ in response to many experimental models of human disease. The aim of our study was to determine if differences exist between 6J and 6N mice in terms of their response to acute carbon tetrachloride (CCl4) exposure. Mice were given CCl4 once and were euthanized 12 to 96 h later. Relative to 6J mice, we found that 6N mice had increased liver injury but more rapid repair. This was because of the increased speed with which necrotic hepatocytes were removed in 6N mice and was directly related to increased recruitment of macrophages to the liver. In parallel, enhanced liver regeneration was observed in 6N relative to 6J mice. Hepatic stellate cell activation occurred earlier in 6N mice, but there was no difference in matrix metabolism between substrains. Taken together, these data demonstrate specific and significant differences in how the C57BL/6 substrains respond to acute CCl4, which has important implications for all mouse studies utilizing this model.
Key words: Carbon tetrachloride, Hepatic injury, Liver regeneration, Hepatic inflammation
INTRODUCTION
Different strains of laboratory mice exhibit genotypic and phenotypic differences, which can greatly impact experimental outcomes1,2. Recently, mice of the same strain but different substrain have been evaluated for possible differences in several experimental models. Substrains are defined as branches of an inbred strain that are either known or suspected to be genetically different from the original inbred strain3. The commonly used C57BL/6 strain has two distinct substrains that were established after a breeding pair moved from The Jackson Laboratory (Jackson) to the National Institutes of Health (NIH) in the 1950s3,4. These substrains are called C57BL/6J (6J, Jackson) and C57BL/6N (6N, NIH). After separation, the 6J mice developed a spontaneous mutation in the nicotinamide nucleotide transhydrogenase (Nnt) gene5,6. This mutation results in a deletion of exons 7–11 of the Nnt gene and leads to a nonfunctional protein5. The NNT enzyme is partially responsible for antioxidant defense within the mitochondria. It is located on the inner mitochondrial membrane and converts NADP+ to NADPH using the proton gradient and NADH6,7. The NADPH produced acts as a cofactor for glutathione reductase to convert oxidized glutathione (GSSG) to reduced glutathione (GSH)8,9.
Recent studies report that 6J and 6N mice differ in alcohol preference10,11 and in response to diet-induced obesity12,13. Although NNT activity and oxidative stress have been linked to some of the differences seen in these models, microarray studies identified several other genetic differences between substrains, making it difficult to say that the nonfunctional NNT is solely responsible for all the observed differences11,14.
Substrain differences are also important to consider when using genetically modified mice. Specifically, two separate labs reported conflicting results on the role JNK2 plays in acetaminophen (APAP)-induced liver injury. Nakagawa et al.15 reported that Jnk2 −/− mice are protected from APAP-induced liver injury, while Bourdi et al.16 reported that Jnk2 −/− mice have exacerbated liver injury relative to wild-type mice. It was determined that each lab used a different B6 substrain as a control and that these two substrains (6N and 6J) differentially responded to APAP exposure17. This raises the question of a potential differential response between these substrains in other commonly used mouse models of liver injury.
Given the findings in APAP-induced liver injury, we hypothesized that a differential response would occur between 6N and 6J mice after acute CCl4 exposure, another commonly used model of liver injury. CCl4 causes centrilobular necrosis shortly after exposure18. This liver injury and its subsequent repair are analogous to the well-established model of wound healing that occurs in the skin; the stages include inflammation, regeneration, and matrix remodeling19,20. In the liver, inflammatory chemokines and cytokines are synthesized primarily by liver-resident macrophages, Kupffer cells, which, in turn, recruit circulating neutrophils and monocytes to the liver that continue to synthesize inflammatory mediators. These infiltrating inflammatory cells can exacerbate injury as well as remove dead and dying hepatocytes and, therefore, also actively participate in wound healing21–25. Following injury, the liver regenerates to restore normal mass and function26,27. Similar to skin wound healing, matrix remodeling occurs at the later stages of liver repair. This involves matrix synthesis, primarily by activated hepatic stellate cells, as well as matrix metabolism, primarily by macrophages28,29. Here we explored whether differences exist in hepatic injury and wound healing, including the inflammation, regeneration, and matrix remodeling stages, following acute CCl4 exposure between 6J and 6N mice.
MATERIALS AND METHODS
Materials
Olive oil and carbon tetrachloride were purchased from Sigma-Aldrich (St. Louis, MO, USA), and Buprenex (buprenorphine HCl) was manufactured by Reckitt Benckiser Healthcare UK, Ltd. (Hull, UK) and distributed by Reckitt Benckiser Pharmaceuticals, Inc. (Richmond, VA, USA). The anesthetic used was a mixture containing ketamine (Akom, Inc., Decator, IL, USA), xylazine (KetaVed; Vedco, Inc., St. Joseph, MO, USA), and acepromazine (Vedco, Inc.).
Primary antibodies used include cytochrome P450 2E1 (CYP2E1; ab28146; Abcam, Cambridge, MA, USA), proliferating cell nuclear antigen (PCNA; clone PC10; EMD-Millipore, Billerica, MA, USA), cyclin D1 (CCND1; clone 92g2; Cell Signaling Technology, Danvers, MA, USA), α-smooth muscle actin (αSMA; clone 1A4; Abcam), Ki-67 (ab66155; Abcam), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; clone 14C10; Cell Signaling Technology), F4/80 (MCA497; Bio-Rad, Hercules, CA, USA), NQO1 (N5288; Sigma-Aldrich), goat anti-mouse IgG–HRP (sc-2005; Santa Cruz Biotechnology, Santa Cruz, CA, USA), goat anti-rabbit IgG–HRP (ab97080; Abcam), biotinylated goat anti-rat (Vector), and donkey anti-rabbit IgG–Alexa Fluor 488 conjugate (a-21206; Life Technologies, Waltham, MA, USA).
Animal Care and Use
Animal protocols were approved by the University of Kansas Medical Center’s (KUMC) Institutional Animal Care and Use Committee (IACUC). Male C57BL/6J (6J) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and C57BL/6N (6N) mice were purchased from Charles River (Wilmington, MA, USA). Mice were exposed to CCl4 within 1 week of arriving at KUMC via intraperitoneal injection. CCl4 was diluted 1:3 in olive oil and delivered at a concentration of 0.4 mg/g body weight. A subcutaneous injection of Buprenex preceded CCl4 exposure as an anesthetic per KUMC IACUC recommendation. Control mice received the anesthetic and an olive oil injection. Mice were euthanized 12, 24, 48, 72, or 96 h after exposure (n = 5–6 mice per group). Liver and plasma were collected from each mouse as described previously30.
Genotyping
All mice were genotyped for Nnt status using DNA isolated from flash-frozen liver, using the Qiagen DNeasy Blood and Tissue Kit (Valencia, CA, USA) and polymerase chain reaction (PCR; Invitrogen PCR reagents) as previously described31. In brief, the reaction mixture contained final concentration of 1× PCR buffer, 2.5 mM MgCl2, 0.67 mM dNTPs, 1.25 U Platinum Taq Polymerase, 1.0 μM Nnt-Com (5′-GTAGGGCCAACTGTTTCTGCATGA-3′), 0.67 μM Nnt-Mut (5′-GTGGAATTCCGCTGAGAGAACTCTT-3′), 0.33 μM Nnt wild type (5′-GGGCATAGGAAGCAAATACCAAGTTG-3′), 2 μl of isolated DNA, and water to bring volume to 25 μl. An MJ Research PTC-200 thermocycler (Bio-Rad) programmed to initial melt for 5 min at 95°C; 35 cycles of 45 s at 95°C, 30 s at 58°C, 45 s at 72°C; final extension for 5 min at 72°C was used to amplify PCR products. The products were loaded and separated on a 1% agarose gel containing ethidium bromide and imaged using Gel Doc EQ (Bio-Rad). All mice were found to have expected Nnt genotype; mice purchased from The Jackson Laboratory were Nnt mutant, and mice purchased from Charles River were Nnt wild type.
Liver Injury and Triglyceride Assessment
Plasma alanine aminotransferase (ALT) activity was determined using commercially available reagents (Sekisui Diagnostics, Exton, PA, USA) and calculated using the extinction coefficient method. Total hepatic triglyceride levels were determined using GPO reagent (Pointe Scientific, Canton, MI, USA) 24 h after the liver was digested with 3 M KOH in 65% ethanol for 1 h at 70°C. A curve using known concentrations of GPO triglyceride standard was generated and used to calculate unknown triglyceride content in each liver sample.
Histological Analysis
Liver tissue was fixed in 10% formalin for 18–24 h and then placed in 70% ethanol at 4°C until it was processed using an automated tissue processor (ASP3005; Leica, Buffalo Grove, IL, USA) and embedded in paraffin. Five-micrometer-thick tissue sections were stained with hematoxylin and eosin (H&E) using a Leica Autostainer XL and coverslipper (CV5030; Leica). A board-certified pathologist examined H&E-stained liver sections. After examining the entire liver section, the area of necrosis was determined as a percentage of total area. Necrosis was defined as regions of tissue that were hypereosinophilic and lacked nuclei. The pathologist had no knowledge of what experimental group each slide came from. For infiltrating cell count, five pericentral necrotic areas per sample were measured; the area of the central vein was subtracted to calculate net necrotic area. The infiltrating, nonparenchymal cell nuclei were then counted in each necrotic area. An EVOS FL Auto Cell Imaging System (Thermo Fisher) was used to complete this analysis.
RNA Isolation, cDNA Synthesis, and Real-Time Polymerase Chain Reaction (PCR)
Liver pieces were placed in RNA later solution (Ambion, Grand Island, NY, USA) immediately following dissection to stabilize RNA. Tissue was homogenized using a bead homogenizer (FastPrep 24; MP Biomedicals, Solon, OH, USA) in RLT buffer (RNeasy Mini Kit; Qiagen) containing β-mercaptoethanol (βME). For RNA isolation from hepatocytes, 350 μl of RLT + βME per 3 × 106 cells was added to pelleted cells. The hepatocytes were further disrupted by passing this lysate through an 18-gauge needle four times, followed by four times through a 23-gauge needle. The remainder of the procedure was the same for whole liver and isolated primary hepatocytes. RNA was isolated using an RNeasy Mini Kit, and a RETROscript Kit (Life Technologies/Ambion) was used to reverse transcribe RNA to cDNA. Real-time PCR was used to evaluate hepatic transcript levels using a Bio-Rad CFX384 machine and the 2−ΔΔCt method. Results were normalized to Rn18s (housekeeping gene) and expressed as fold change over each substrain’s baseline (oil treated) value. Primer sequences are found in Table 1. All primer sequences, unless otherwise stated, came from the Primer Bank (https://pga.mgh.harvard.edu/primerbank/)32–34. Each gene was analyzed on a single 384-well plate that included cDNA from every mouse in this study, therefore eliminating plate-to-plate variation.
Table 1.
Primer Sequences Used for Real-Time PCR
| Gene Name | Protein Name | Sequence Source | Forward Primer | Reverse Primer |
|---|---|---|---|---|
| Acta2 | αSMA | Primer Bank ID: 31982518b1 | GTCCCAGACATCAGGGAGTAA | TCTATCGGATACTTCAGCGTCA |
| Ccl2 | CCL2/MCP1 | Pascual et al. (2011) | AGGTCCCTGTCATGCTTCTG | TCTGGACCCATTCCTTCTTG |
| Ccnd1 | CCND1/CyclinD1 | Pritchard et al. (2011) | CAGAAGTGCGAAGAGGAGGTC | TCATCTTAGAGGCCACGAACAT |
| Col1a1 | Col1a1 | Stefanovic and Stefanovic (2012) | ATGTTCAGCTTTGTGGACCTC | CAGAAAGCACAGCACTCGC |
| Cxcr2 | CXCR2 | Primer Bank ID: 6753456a1 | ATGCCCTCTATTCTGCCAGAT | GTGCTCCGGTTGTATAAGATGAC |
| Cxcl2 | CXCL2/MIP2 | Tang et al. (2013) | GCGCCCAGACAGAAGTCATAG | AGCCTTGCCTTTGTTCAGTATC |
| Cyp2e1 | CYP2E1 | Primer Bank ID: 57634519b1 | CATCACCGTTGCCTTGCTTG | CAGATGGATACGAGGAGGAGG |
| Emr1 | F4/80 | Primer Bank ID: 183583543b1 | CTGCACCTGTAAACGAGGCTT | TTGAAAGTTGGTTTGTCCATTGC |
| Gclc | GCLC | Primer Bank ID: 33468897A1 | GGGGTGACGAGGTGGAGTA | GTTGGGGTTTGTCCTCTCCC |
| Nqo1 | NQO1 | Primer Bank ID: 161621259b1 | AGGATGGGAGGTACTCGAATC | TGCTAGAGATGACTCGGAAGG |
| Pcna | PCNA | Primer Bank ID: 7242171a1 | TTTGAGGCACGCCTGATCC | GGAGACGTGAGACGAGTCCAT |
| Tnfα | TNF-α | Pritchard et al. (2010) | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
Immunoblotting
Liver pieces were flash frozen in liquid nitrogen immediately following euthanasia, and lysates were prepared as previously described35. Protein concentration was determined using a BCA assay, and 40 μg of total protein was resolved on a 10% SDS-PAGE gel. Proteins were transferred to PVDF membranes using semidry transfer, blocked for 1 h in 5% BSA, and incubated overnight at 4°C in primary antibody with immunoreactivity to the protein of interest. Blots were incubated with HRP-conjugated secondary antibodies, and enhanced chemiluminescent substrate (GE Healthcare, Piscataway, NJ, USA) was used to generate luminescence, which was captured on radiographic film. Band density was quantified using ImageJ (NIH, Bethesda, MD, USA) and normalized to the housekeeping protein, GAPDH, before calculating the fold difference of hepatic protein content between substrains.
CYP2E1 Activity Assay
CYP2E1 activity was determined as previously described, with the following adjustments30. Briefly, microsomes were isolated from whole liver. The reaction mixture consisted of 100 μg of protein, 4 μl of p-nitrophenol, 10 μl of phosphate buffer (4 ml of 1 M K2HPO4 + 1 ml of 1 M KH2PO4, pH 7.4), water to bring volume to 100 μl, and 10 μl of 11 mM NADPH. The hydroxylation of p-nitrophenol to p-nitrocatechol was used to calculate CYP2E1 activity using the extinction coefficient method. Activity is expressed as nm/min/mg total protein.
Hepatocyte Isolation and Culture
Ten- to 12-week-old C57BL/6N and C57BL/6J mice were anesthetized with a mixture of ketamine (200 mg/kg) and xylazine (10 mg/kg). Once mice were anesthetized, the abdomen was shaved using animal clippers. Mice were then affixed to a surgical platform using laboratory tape, and the abdomen was cleaned using 70% ethanol. The peritoneal cavity was then exposed, and the intestines were moved to one side using a sterile cotton swab. Appropriate liver lobes were then moved to expose the inferior vena cava and the portal vein. Next, using a pair of sterile curved forceps, a sterile suture was directed under the vena cava and tied loosely around the vein. The vena cava was then cannulated with a 22-gauge IV catheter, the needle was removed, and ligature was tightened around the catheter. Perfusion tubing was then attached to the catheter, and perfusion was started (8.2 ml/min, 100 ml total) using perfusion buffer (1× HBSS Ca2+, Mg2+ free, 100 U/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES). Immediately after starting the perfusion, the portal vein was cut to allow outflow of perfusate, and then the diaphragm was cut, and the superior vena cava was clamped using small hemostats. Perfusion continued using 100 ml of a second buffer [1× HBSS with Ca2+/Mg2+, 100 U/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES, 0.025 mg/ml Liberase (Roche)]. Once complete, perfusion was halted, and the liver was dissected out of the mouse and placed into a sterile beaker containing 20–30 ml of ice-cold disruption buffer (1× HBSS Ca2+, Mg2+ free, 100 U/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES, 1 × 10−7 M insulin). The liver was then cut into large pieces using sterile surgical scissors, and then sterile forceps were used to gently agitate the dissected liver pieces in the ice-cold disruption buffer to liberate hepatic cells from the digested hepatic extracellular matrix. The cell suspension was then passed through three different sterile filters (100-, 70-, and 30-μm mesh sizes) into successive 50-ml centrifuge tubes. The volume of the cell suspension was then brought to 50 ml using ice-cold disruption buffer and centrifuged at 50 × g for 5 min at 4°C. The supernatant was removed by aspiration, and cells were washed two more times in the same buffer. After the third wash, hepatocytes were resuspended in complete Williams E medium (Williams E, 100 U/ml penicillin, 100 U/ml streptomycin, 100 nM insulin, 2 mM GlutaMAX, and 5% FBS). Viable cells were counted using a hemacytometer after staining with trypan blue and plated on collagen-coated plates at 0.5 × 106 cells per well in a 24-well plate. Remaining hepatocytes were snap frozen in liquid nitrogen for analysis of Cyp2e1 expression.
Carbon Tetrachloride Exposure, In Vitro, and Sample Collection
Two to 3 h after plating, the medium was aspirated and the cells were washed with 1 ml of prewarmed D-PBS; this was repeated twice. After the second wash, 1 ml of medium (Williams E, 100 U/ml penicillin, 100 U/ml streptomycin, 100 nM insulin, 2 mM GlutaMAX, 5% FBS, 1% DMSO) with or without CCl4 (0, 1, 5, or 10 mM) was added to each well. One concentration of CCl4 was used per plate, and each plate was sealed using Parafilm to limit interplate exposure to volatilized CCl4 and placed back into a humidified 5% CO2 incubator. Media addition to each plate was staggered by 15 min to allow for the collection of images, culture supernatants, and cell lysates 24 h after CCl4 exposure. A single representative image was taken from each of two replicate wells per treatment using a Zeiss Axio Observer A.1 inverted microscope (Peabody, MA, USA) and Olympus DP71 camera operated by cellSens imaging software (Olympus, Center Valley, PA, USA). The entire culture medium (1 ml) was removed, placed into a 1.5-ml microfuge tube, and snap frozen in liquid nitrogen. Hepatocytes remaining in the well were lysed in 200 μl of ice-cold cell lysis buffer (25 mM HEPES, 5 mM EDTA, 0.1% CHAPS, 1 μg/ml pepstatin, 0.5 μg/ml leupeptin, 2 μg/ml aprotinin, 1× complete EDTA-free Protease Inhibitor Cocktail). The wells were scraped using a pipette tip to ensure release of hepatocytes from the bottom of each well. Then the lysate was transferred to a 1.5-ml microfuge tube and allowed to sit at room temperature for 5 min prior to snap freezing in liquid nitrogen. Both cell culture media and cell lysates were stored at −80°C until use. Hepatocytes from one 6N and one 6J mouse were used in each experiment, and the experiment was repeated three times on 3 separate days with two technical replicates completed each day.
Lactate Dehydrogenase (LDH) Assay
All samples were thawed only once, and cell death was determined as described previously36. In brief, cell lysates were sonicated 2 × 3 s and centrifuged for 20 min at 20,000 × g at 4°C. A reaction buffer containing 9.64 mM KH2PO4, 50.42 mM K2HPO4, 0.91 mM pyruvate, and 2.17 mM NADH-Na2 was used for this assay. For LDH release in media, 100 μl of media was combined with 700 μl of reaction mixture, and the kinetics of the reaction was measured at 340 nm and the difference in the absorbance determined over time. For LDH contained in cells, 30 μl of lysate plus 730 μl of reaction mixture were used. The death ratio was determined by the amount of LDH release in the medium divided by the total LDH in the media and cell lysate. The data are presented as the fold change over no CCl4 treatment, and experimental replicates are graphed.
Hepatic Leukocyte Esterase [Chloracetate Esterase (CAE)] Localization and Quantification
CAE was detected in liver sections from all mice using a naphthol-AS-D CAE kit according to the manufacturer’s instructions (Sigma-Aldrich). Briefly, formalin-fixed, paraffin-embedded liver tissues were cut into 5-μm-thick sections, heated to 60°C for 20 min, deparaffinized in three changes of SafeClear (5 min each), and then rehydrated in a series of graded ethanols. After staining, sections were dehydrated and mounted for analysis. Quantification of CAE+ cells was performed by an individual blinded to substrain and time point. Because of the small number of CAE+ cells per 200× image, all CAE+ cells in one entire liver section per mouse were counted.
F4/80 Immunofluorescence
Formalin-fixed paraffin sections were cut to 5 μm. Paraffin was removed by SafeClear (Fisher), and tissues were rehydrated by putting slides through a series of ethanols (100% 2 × 3 min, 95% 1 × 3 min, 85% 1 × 3 min, 70% 1 × 3 min, 50% 1 × 3 min) and then into reverse osmosis water. Slides were immersed in 1× Reveal Decloacker (BioCare Medical, Concord, CA, USA) and heated in a microwave for 2 min at 50% power followed by 7 min at 10% power then allowed to cool for 1 h at room temperature. Slides were washed in TBS-T, then endogenous peroxidase was blocked using 3% H2O2 for 15 min, followed by another wash in TBS. Endogenous avidin and biotin were blocked using an avidin/biotin blocking kit. Slides were rinsed in TBS after both avidin- and biotin-blocking steps, and 10% goat serum and 0.4% Triton X-100 in TBS were used to block nonspecific binding in each section. F4/80 primary antibody was diluted in a blocking solution at a concentration of 1:100 and applied to sections overnight at 4°C. The following day, primary antibody was removed, and slides were washed in TBS-T and incubated in biotinylated anti-rat secondary antibody diluted 1:100 in a blocking solution for 2 h at room temperature. Following a wash in TBS-T, slides were incubated in an avidin/biotin complex to amplify signal, washed again in TBS-T, and a second amplification step was completed using a tyramide signal amplification (TSA) kit (PerkinElmer, Waltham, MA, USA) diluted 1:400. Nuclei were stained with 4′,6-diamidino-2-phenylidole (DAPI) following a wash in TBS-T, and sections were covered with a glass coverslip and then sealed with clear nail polish. Three to five nonoverlapping 200× images were taken of each section using an Olympus BX51 microscope with an Olympus BH2RFLT3 burner and Olympus DP71 camera operated by the DP Controller software (Olympus, Waltham, MA, USA). ImageJ was used to quantify the area and intensity of positive staining above a threshold that remained constant for all images.
Ki-67 Immunofluorescence
Liver pieces were embedded in optimal cutting temperature (OCT) matrix (Sakura TissueTek, Torrance, CA, USA), then sections were cut at 6 μm and allowed to warm to room temperature for 2 min. Sections were then fixed using 10% formalin for 10 min. Triton X-100 (0.1%) in phosphate-buffered saline (PBS) was applied for 15 min, and then slides were washed in PBS 2 × 5 min. Ten percent donkey serum was used to block sections for 1 h at room temperature followed by Ki-67 primary antibody diluted 1:500 in 1% donkey serum overnight at 4°C. The following day, slides were washed in PBS 2 × 5 min, incubated with Alexa Flour 488-conjugated donkey anti-rabbit secondary antibody diluted 1:500 in PBS for 1 h. Following 3 × 2 min PBS washes, DAPI was applied as a counterstain in aqueous mounting media. Three nonoverlapping images were taken of each section using an Olympus BX51 microscope with an Olympus BH2RFLT3 burner and Olympus DP71 camera at 200× using the DP Controller software (Olympus). Ki-67+ cells were manually counted using GFP fluorescent images and ImageJ.
αSMA Immunohistochemistry
Formalin-fixed, paraffin-embedded liver tissues were cut into 5-μm sections, deparaffinized using Citrisolv (Fisher Scientific, Waltham, MA, USA), and rehydrated in a series of graded ethanols. Antigens were exposed using 10% Reveal Decloaker (BioCare Medical). Slides were washed in Tris-buffered saline (TBS) 2 × 15 min, and endogenous peroxidase activity was blocked using 3% H2O2 for 15 min. Following a wash in TBS, endogenous avidin and biotin (Vector Laboratories, Burlingame, CA, USA) were blocked. Mouse-on-mouse (MOM) (Vector Laboratories) IgG-blocking diluent was applied to sections for 1 h. Next, slides were washed in TBS, and MOM-blocking diluent was applied for 5 min. αSMA antibody was diluted 1:100 and applied to sections for 30 min. Slides were washed in TBS, and then biotin-conjugated anti-mouse IgG reagent (MOM kit) was applied to slides for 10 min. Following a TBS wash, the signal was amplified using avidin/biotin–HRP complex (Vectastain ABC; Vector Laboratories). 3,3′-Diaminobenzidine was used to detect positive staining, and hematoxylin was used as a counterstain. Slides were immersed in a bluing reagent, dehydrated in a series of graded ethanol and Citrisolv, and sealed using glass coverslips and Cytoseal mounting medium (Thermo Fisher Scientific). Five nonoverlapping 200× images were taken of each section using an Olympus BX51 microscope and Olympus DP71 camera operated by the DP Controller software (Olympus). ImageJ was used to quantify the area of positive staining above a threshold that remained constant for all images.
In Situ Zymography
Matrix remodeling was evaluated using in situ zymography as described previously30. In brief, 7-μm frozen sections were incubated overnight in developing buffer containing dye-quenched gelatin (Oregon Green 488; Life Technologies/Molecular Probes). The following day, the developing buffer was removed, and DAPI mounting medium was used as a counterstain. Three to five nonoverlapping images per sections were taken at 200× magnification using an Olympus BX51 microscope with an Olympus BH2RFLT3 burner and Olympus DP71 camera using the DP Controller software. ImageJ was used to quantify the area and intensity of positive signal over a threshold that remained constant for every image.
Data Analysis and Statistics
All experiments were completed within 4 weeks. The two-factor factorial design was used in order to conduct the study. Over the course of the 4 weeks, mice of each substrain (6J and 6N) at different CCl4 exposure time points, and control mice, were randomly arranged in a factorial design, that is, each replicate of the experiment contains mouse–treatment combinations. Two-way analysis of variance method was used to analyze the data. Fold change mRNA results were log transformed in order to satisfy normality assumption before analyzing the data. Post hoc pairwise comparisons with multiple testing adjustments were completed using Tukey’s method. The comparisons having an adjusted value of p < 0.05 were considered statistically significant. For the cases having only two groups, Student’s t-test was used to determine significance. A Pearson’s correlation was done to determine the relationship between necrotic area and number of infiltrating cells. Analyses were carried out using SAS software version 9.4 (SAS Institute, Cary, NC, USA) or GraphPad Prism version 6 (La Jolla, CA, USA). Results are presented as mean ± standard error mean in the figures.
RESULTS
Liver Injury and Steatosis After CCl4 Exposure
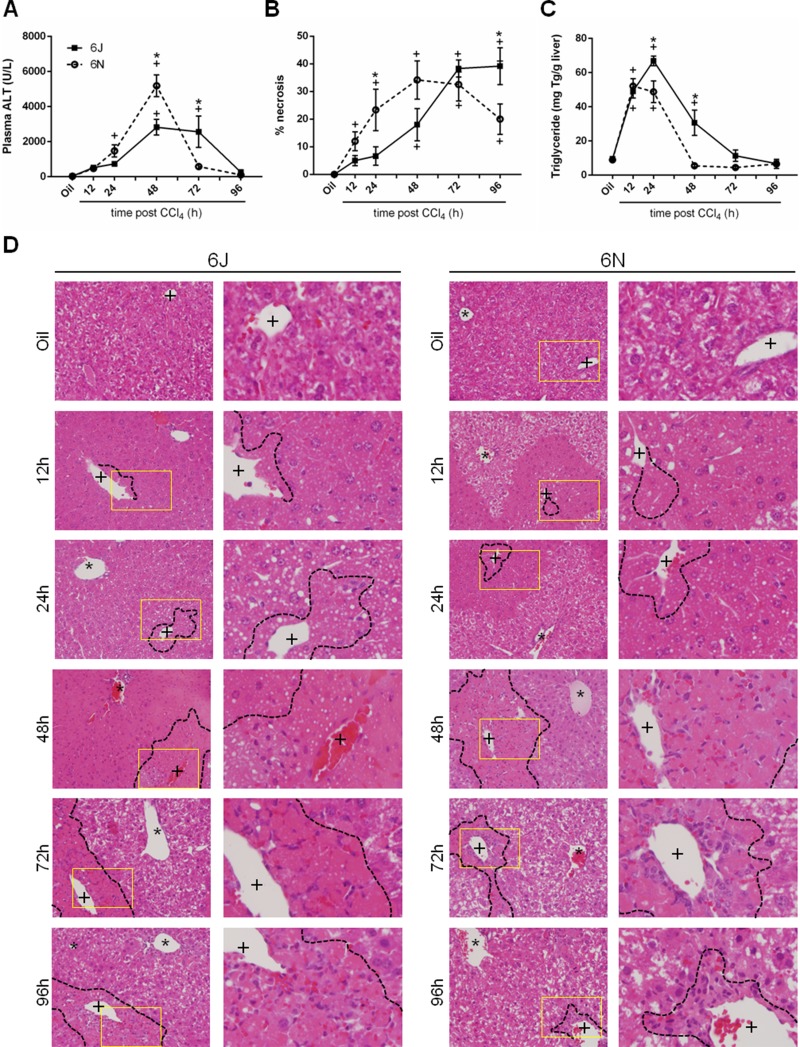
To determine if 6N and 6J mice have a differential response to acute CCl4 exposure, liver injury was evaluated by measuring plasma ALT activity. This enzyme is released into circulation by dead and dying hepatocytes. Both strains had increased plasma ALT activity following CCl4 exposure, but 6N mice had greater plasma ALT activity compared to 6J mice 48 h after CCl4 exposure (Fig. 1A). By 72 h after CCl4 exposure, ALT activity in 6N mice was not different than baseline, while 6J remained elevated (Fig. 1A). By 96 h after CCl4 exposure, plasma ALT returned to baseline in 6J mice (Fig. 1A). Because CCl4 causes centrilobular necrosis, we also evaluated liver injury by histopathology. Necrosis increased in both substrains following CCl4; however, the area of necrosis increased in 6N mice 12 h following CCl4 exposure, while the area of necrosis did not increase above baseline in the 6J mice until 48 h after CCl4 exposure (Fig. 1B and D). At 96 h after CCl4 exposure, 6N mice had less necrosis compared to 6J mice (Fig. 1B and D). Together, these data demonstrate that 6N mice had increased injury, but also a faster recovery from that injury, after acute CCl4 exposure.
Figure 1.
Hepatic injury following CCl4 exposure. Mice were exposed to CCl4 and euthanized 12, 24, 48, 72, or 96 h later. (A) Plasma alanine aminotransferase (ALT) was determined by enzymatic assay. (B) Quantification of necrosis evaluated by a board-certified pathologist blinded to experimental group. (C) Total hepatic triglyceride content was determined by a biochemical assay. Data are presented as mean ± standard error of mean. (D) Representative histology of hematoxylin and eosin (H&E)-stained slides showing necrosis around the central veins (+) of the liver; asterisks (*) indicate portal veins. A black dashed line outlines necrotic areas, defined as hypereosinophilic and lacking hepatocyte nuclei. The yellow-boxed area is enlarged in the right next to each image to better depict steatosis at each time point. Throughout the article, the black squares/bars/solid lines represent data from 6J mice, and white circles/bars/dashed lines represent data from 6N mice. n = 5–6. *p ≤ 0.05 when comparing substrains at a single time point; +p ≤ 0.05 when comparing the indicated CCl4 time point to the oil (control) of the same substrain.
Hepatic triglyceride accumulation, measured biochemically and evident in H&E-stained liver sections, also increased in both strains following CCl4 exposure (Fig. 1C and D). Contrary to plasma ALT levels and area of necrosis, 6J mice had more hepatic triglyceride than 6N mice 24 h post-CCl4 exposure. While the triglyceride levels in 6N mice returned to baseline by 48 h after CCl4 exposure, levels in 6J mice did not return to baseline until 72 h after CCl4 exposure (Fig. 1C and D).
Baseline CYP2E1 Protein and Activity
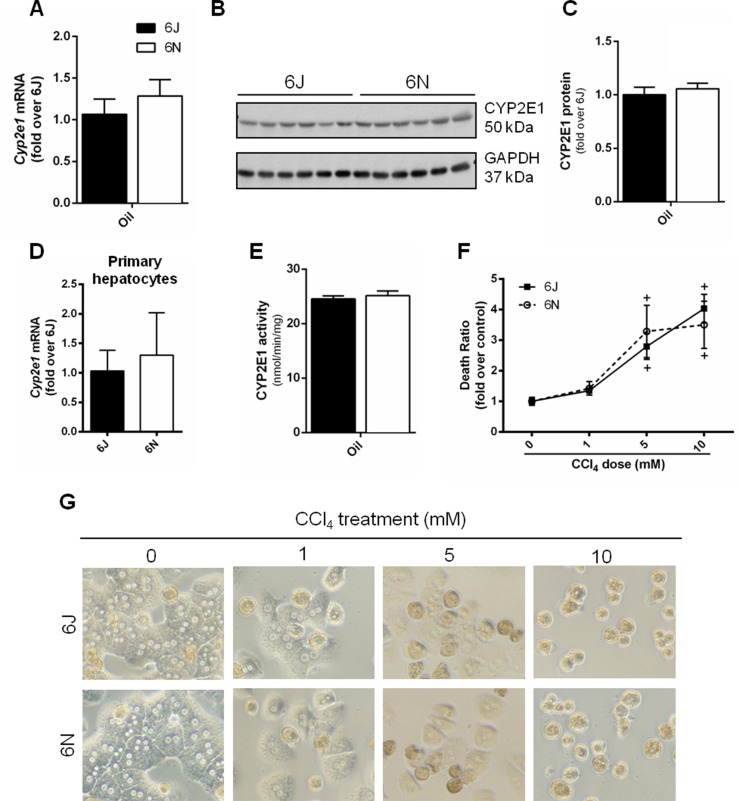
In order for CCl4 to induce liver injury, it must undergo bioactivation by CYP2E118. Therefore, to eliminate the possibility that differential CYP2E1 activity accounted for variation in the liver injury between substrains, we measured baseline hepatic CYP2E1. There was no difference in hepatic CYP2E1 mRNA or protein levels between 6N and 6J mice (Fig. 2A–C) or in Cyp2e1 mRNA from isolated primary hepatocytes (Fig. 2D). Similarly, there was no difference in the CYP2E1 activity between the two substrains as assessed by the hydroxylation of p-nitrophenol to p-nitrocatechol using microsomes isolated from whole liver (Fig. 2E). These findings suggest that the observed differences in liver injury were not due to substrain-specific differences in CCl4 bioactivation.
Figure 2.
CYP2E1 protein content and activity, in vitro CCl4 exposure. Control mice given an olive oil injection and euthanized 72 h later were evaluated for (A) Cyp2e1 mRNA and (B, C) CYP2E1 protein from whole liver. (D) Cyp2e1 expression was determined in isolated hepatocytes from mice that did not receive olive oil injection. (E) CYP2E1 activity in microsomes isolated from whole liver of mice that received olive oil injection was determined by the hydroxylation of p-nitrophenol to p-nitrocatechol. (F, G) Primary hepatocytes were isolated from 6N and 6J mice and exposed to 0, 1, 5, and 10 mM CCl4 for 24 h. (F) Death ratio was calculated from the amount of lactate dehydrogenase (LDH) released in the media versus the total amount of LDH in the cells and media. (G) Cell morphology was evaluated in micrographs taken at the end of the 24-h incubation.
In Vitro Hepatocyte Sensitivity to CCl4
To determine if the increased liver injury observed in 6N mice was due to an increase in hepatocyte sensitivity, primary hepatocytes from 6N and 6J mice were exposed to CCl4 for 24 h at three different CCl4 concentrations. The higher concentrations (5 and 10 mM) induced hepatocyte cell death detectable by LDH release 24 h after CCl4 exposure (Fig. 2F); morphological changes were obvious at all CCl4 concentrations (Fig. 2G). However, there was no difference in the cell death between the two substrains (Fig. 2F). This suggests that increased hepatocyte sensitivity to CCl4 is not responsible for the increased injury observed in the 6N mice compared to the 6J mice.
Antioxidant Defense Following CCl4
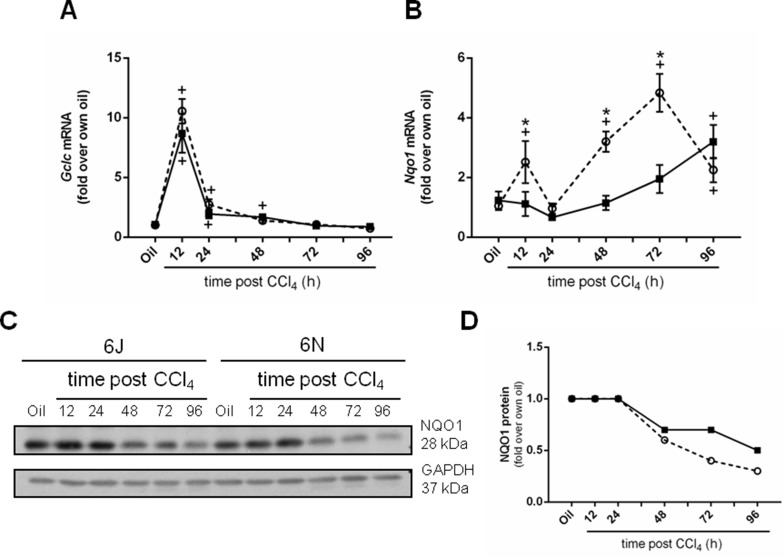
6J mice have a mutation in the Nnt gene, which can attenuate antioxidant defense6,37,38. However, reduced NNT activity does not make 6J mice more sensitive to CCl4-mediated liver injury as one might expect. In fact, the reverse was true: 6N mice are more sensitive to CCl4-induced liver injury than 6J mice (Fig. 1A). Therefore, we evaluated two additional parameters associated with antioxidant defense to determine if they were increased in 6J mice to compensate for lack of functional NNT and perhaps responsible for the observed protection from CCl4-induced liver injury. Gclc, glutamate-cysteine ligase catalytic domain, is the rate-limiting step in glutathione synthesis and is important for antioxidant defense after CCl4-induced liver injury39,40. Gclc mRNA increases rapidly in liver in both substrains of mice early following CCl4 exposure and is not different between substrains at any time point (Fig. 3A). Nqo1, NAD(P)H dehydrogenase, is another vital part of the liver’s antioxidant defense against many hepatotoxins41. While hepatic Nqo1 mRNA is increased in 6N mice compared to the 6J mice 12, 48, and 72h after CCl4 (Fig. 3B), hepatic NQO1 protein levels in 6N mice do not parallel these changes. In fact, NQO1 protein decreases over time in both substrains (Fig. 3C and D). Although a trend to a decrease in NQO1 protein is observed in 6N mice 72 and 96 h after CCl4 exposure, NQO1 protein is not different between substrains when plasma ALT levels are different between substrains (compare Fig. 1A and Fig. 3C and D). Together, these data suggest that antioxidant defense is equivalent between 6J and 6N mice following CCl4 exposure and does not account for observed differences in CCl4-induced liver injury between substrains.
Figure 3.
Selected hepatic antioxidant molecules. Mice were exposed to CCl4 and euthanized 12, 24, 48, 72, or 96 h later, and the hepatic transcript accumulation of (A) Gclc and (B) Nqo1 was determined. Individual mouse liver samples were pooled to evaluate NQO1 protein concentration throughout the time course. (C) Immunoblot and (D) densitometry of the immunoblot shown in (C). n = 5–6 per group. *p ≤ 0.05 when comparing substrains at a single time point; +p ≤ 0.05 when comparing the indicated CCl4 time point to the oil (control) of the same substrain.
Inflammation Following CCl4 Exposure: Hepatic Neutrophil Accumulation
Because hepatocyte sensitivity to CCl4 and antioxidant defenses were not different between substrains, we hypothesized that another mechanism must be responsible for increased liver injury observed in 6N mice. Neutrophils are recruited rapidly after CCl4-mediated injury and can exacerbate hepatocyte cell death24,25; therefore, we measured several neutrophil-related markers in 6J and 6N mice. First, we evaluated Cxcl2 (MIP2) and Cxcr2, a major neutrophil chemotactic protein and its receptor, respectively, at the mRNA level. Cxcl2 transcripts increased in 6J and 6N mice 12 h after CCl4 and are not different between substrains (Fig. 4A). However, 24 and 48 h after CCl4 exposure, 6N mice have increased Cxcl2 expression (Fig. 4A). Cxcr2 is increased 12 h after CCl4 and remains elevated in both strains until 48 h after CCl4 exposure (Fig. 4B). Thereafter, Cxcr2 transcripts decrease in 6N mice but remain elevated in 6J (Fig. 4B).
Figure 4.
Hepatic neutrophil accumulation after CCl4 exposure. Mice were exposed to CCl4 and euthanized 12, 24, 48, 72, or 96 h later. (A) Cxcl2 and (B) Cxcr2 transcripts were quantified in whole-liver cDNA samples using real-time polymerase chain reaction (PCR). (C) Quantification of chloracetate esterase (CAE) staining used to quantify the number of neutrophils in the liver at baseline (oil) and 12 and 24 h after CCl4. (D) Representative images of CAE+ cells with neutrophil morphology (pink staining). Black dotted line is outlining CAE+ cells and red solid line is are of image enlarged in far right panel. The + indicates central veins in the images. n = 5–6 per group. *p ≤ 0.05 when comparing substrains at a single time point; +p ≤ 0.05 when comparing the indicated CCl4 time point to the oil (control) of the same substrain.
To further evaluate neutrophils, we localized CAE, a leukocyte esterase, in livers from 6J and 6N mice. CAE was not found in control livers from either substrain (Fig. 4C and D). However, CAE+ cells were apparent in livers from both substrains 12 and 24 h after CCl4 exposure (Fig. 4C and D). Although there was a trend to an increase in CAE+ cells in livers from 6N mice when compared to 6J mice 24 h after CCl4, this difference was not significant (p = 0.0682) (Fig. 4C and D). No CAE+ cells were found in livers from mice 48, 72, or 96 h after CCl4 exposure (data not shown). Taken together, these data suggest that neutrophil-mediated hepatocyte injury may partially contribute to increased liver injury observed in 6N mice. It is interesting to note that CAE staining data do not parallel hepatic Cxcr2 transcript levels (Fig. 4B). This suggests that the sustained rise in Cxcr2 mRNA observed, in particular, in 6J mice is due to Cxcr2 expression in other hepatic cells, such as hepatocytes, as observed following APAP overdose and ischemia–reperfusion-induced liver injury42,43.
Inflammation After CCl4 Exposure: Hepatic Macrophage Accumulation
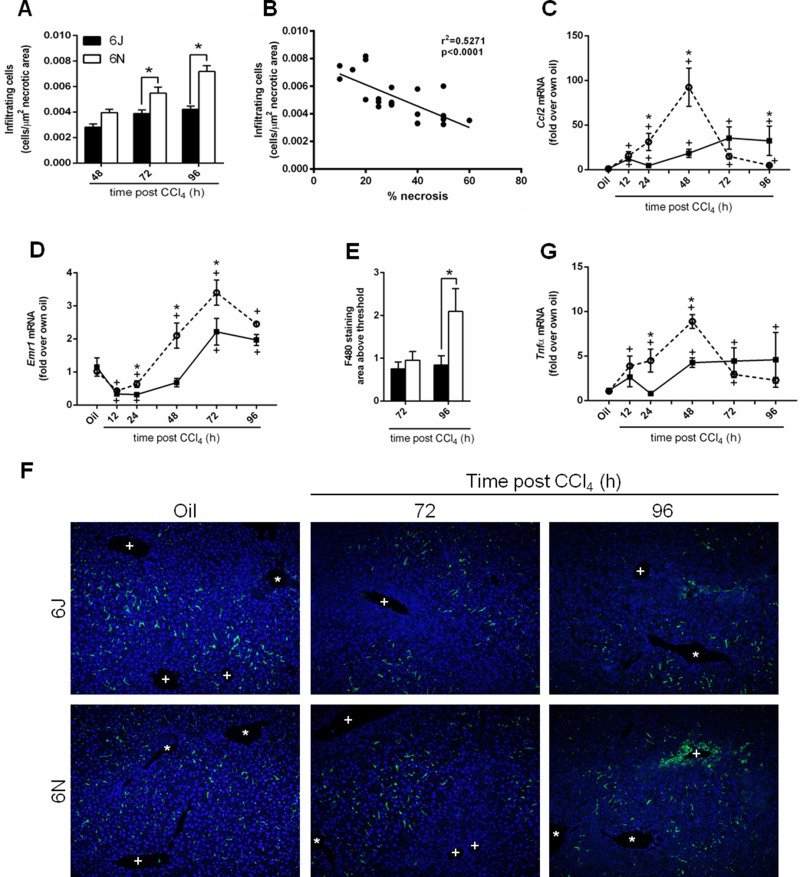
Macrophages are also recruited to the liver after injury and can play roles in sustaining injury as well as promoting repair22,44. Because we observed a more rapid removal of necrotic tissue in livers from 6N mice (Fig. 1B and D), we hypothesized that cell recruitment was increased in this substrain and contributed to more rapid repair, compared to 6J mice. To determine whether this relationship existed, we first counted the number of nonparenchymal cells within the pericentral (necrotic) areas in livers from 6J and 6N mice. There were very few nonparenchymal cells within the necrotic area 12 and 24 h after CCl4 (Fig. 1D, not quantified); however, by 48 h, nonparenchymal cells increased in necrotic areas from both substrains (Figs. 1D and 5A), and at 72 and 96 h after CCl4 exposure, 6N mice had more necrosis-infiltrating cells compared to 6J mice (Figs. 1D and 5A). While infiltration of cells into necrotic areas 72 and 96 h after CCl4 does not support a role for inflammation-mediated exacerbation of liver injury in 6N mice, it does support a role for these cells in removal of necrotic hepatocytes. In fact, the number of infiltrating cells was negatively correlated with the area of necrosis (Fig. 5B).
Figure 5.
Infiltrating cells and macrophages in liver after CCl4 exposure. (A) The number of nonparenchymal cells infiltrating the necrotic area was counted in livers from mice euthanized 48, 72, and 96 h after CCl4 exposure. (B) The number of cells was correlated to the percent necrosis determined from H&E-stained sections. Indices of macrophage accumulation were evaluated by determining hepatic transcript accumulation of (C) Ccl2 and (D) Emr1 by real-time PCR. (E) Quantification of F4/80+ macrophages determined by immunofluorescence 72 and 96 h after CCl4 exposure. (F) Representative F4/80 immunofluorescence images. In each image, central veins are marked with a +, and portal veins are marked with a *. (G) Quantification of Tnfα transcripts by real-time PCR. n = 5–6 per group. *p ≤ 0.05 when comparing substrains at a single time point; +p ≤ 0.05 when comparing the indicated CCl4 time point to the oil (control) of the same substrain.
Next, we wanted to verify that macrophages were associated with this repair response. After CCl4 exposure, resident hepatic macrophages and other liver cells produce chemokines that attract peripheral monocytes into the liver, leading to an increase in hepatic macrophage number43. Ccl2 is the gene that encodes monocyte chemoattractant protein 1, a chemokine that aids in the recruitment of inflammatory cells, including macrophages. Hepatic Ccl2 transcripts increased above baseline 12 h after CCl4 exposure in both 6N and 6J mice (Fig. 5C). However, 6N mice had increased Ccl2 transcripts 24 and 48 h after CCl4 exposure relative to 6J mice at the same time point (Fig. 5C). Conversely, 96 h after CCl4 exposure, Ccl2 transcripts were higher in 6J mice relative to 6N mice (Fig. 5C). To assess hepatic macrophage content, we examined expression of Emr1, the F4/80 gene, a common mouse macrophage marker. Emr1 transcripts initially decreased in both substrains following CCl4 exposure (Fig. 5D). By 48 h after CCl4 exposure, Emr1 transcripts increased above baseline in 6N mice (Fig. 5D). However, it was not until 72 h after CCl4 exposure that Emr1 transcripts increased above baseline in 6J mice; Emr1 transcripts were still greater in 6N mice at this time point (Fig. 5D). Consistent with earlier and increased Emr1 transcript levels, 6N mice exhibited increased F4/80+ staining compared to 6J mice 96 h after CCl4 exposure (Fig. 5E and F).
Tumor necrosis factor-α (Tnfα) is a cytokine produced mainly by macrophages43. TNF-α can induce hepatocyte death45,46 or drive liver regeneration47,48, depending on cellular context. Therefore, we evaluated Tnfα transcripts in liver after CCl4 exposure. Tnfα increased above baseline in 6N mice earlier (12 h) and was greater than in 6J mice (24 and 48 h) (Fig. 5G). While Tnfα transcripts returned to baseline by 96 h in 6N mice, these transcripts remained elevated in 6J mice (Fig. 5G).
Together, all of these data suggest that 6N mice exhibited a greater capacity to recruit cells, including macrophages, into the liver, where they participated in more rapid removal of necrotic tissue, hastening liver repair after CCl4 exposure. However, increased cell recruitment observed in 6N mice may have also promoted increased liver injury, perhaps through increased TNF-α-induced hepatotoxicity.
Liver Regeneration Following CCl4 Exposure
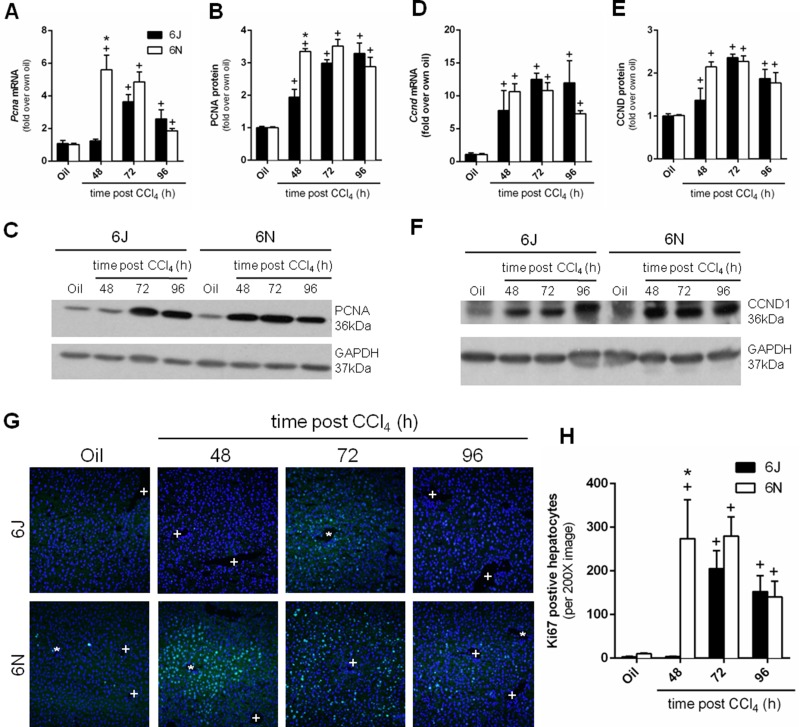
Following the massive necrosis caused by CCl4 and subsequent inflammation, the liver regenerates to restore normal mass and function26,27. This occurs at the later time points, 48, 72, and 96 h, after CCl4 exposure. We evaluated regeneration by analyzing hepatic gene expression and protein levels of proliferating cell nuclear antigen (Pcna), a DNA protein clamp essential for replication, and cyclin D1 (Ccnd1), a protein required for cell cycle progression. Hepatic Pcna transcript levels increased in both genotypes following CCl4 exposure. The increase in the 6N mice occurred earlier, at 48 h after CCl4 exposure, compared to 6J mice, which increased 72 h after CCl4 exposure (Fig. 6A). PCNA protein increased in both substrains 48 h after CCl4 exposure; however, in parallel to mRNA data, 6N had higher PCNA levels compared to 6J mice (Fig. 6B and C). The PCNA protein remained elevated in both substrains 96 h after CCl4 exposure. Ccnd1 expression increased following CCl4 exposure in both substrains 48 h after CCl4 exposure; there was no difference in Ccnd1 transcript levels between 6N and 6J mice (Fig. 6D). CCND1 protein levels also increased in both substrains following CCl4 exposure; however, there was no difference between substrains at any time point (Fig. 6E and F). Ki-67, which is present throughout the active phases of the cell cycle, increased in both substrains following CCl4 exposure, however with different kinetics (Fig. 6G and H). Specifically, Ki-67+ cells were present 48 h after CCl4 exposure in 6N mice but were not present until 72 h after CCl4 exposure in 6J mice (Fig. 6G and H). Together, these data suggest that 6N mice initiated regeneration earlier than 6J mice.
Figure 6.
Hepatic regeneration following CCl4 exposure. Mice were exposed to CCl4 and euthanized 48, 72, or 96 h later. Pcna was evaluated at both the (A) mRNA and (B, C) protein level by real-time PCR or Western blotting, respectively. Ccnd1 was evaluated at both the (D) mRNA and (E, F) protein level. (G) Ki-67 immunofluorescence was performed to identify proliferating hepatocytes. 4′,6-Diamidino-2-phenylidole (DAPI) was used to visualize nuclei. In each image, central veins are marked with a +, and portal veins are marked with a *. (H) Quantification of Ki-67+ cells. n = 5–6 per group. *p ≤ 0.05 when comparing substrains at a single time point; +p ≤ 0.05 when comparing the indicated CCl4 time point to the oil (control) of the same substrain.
Matrix Remodeling Following CCl4 Exposure
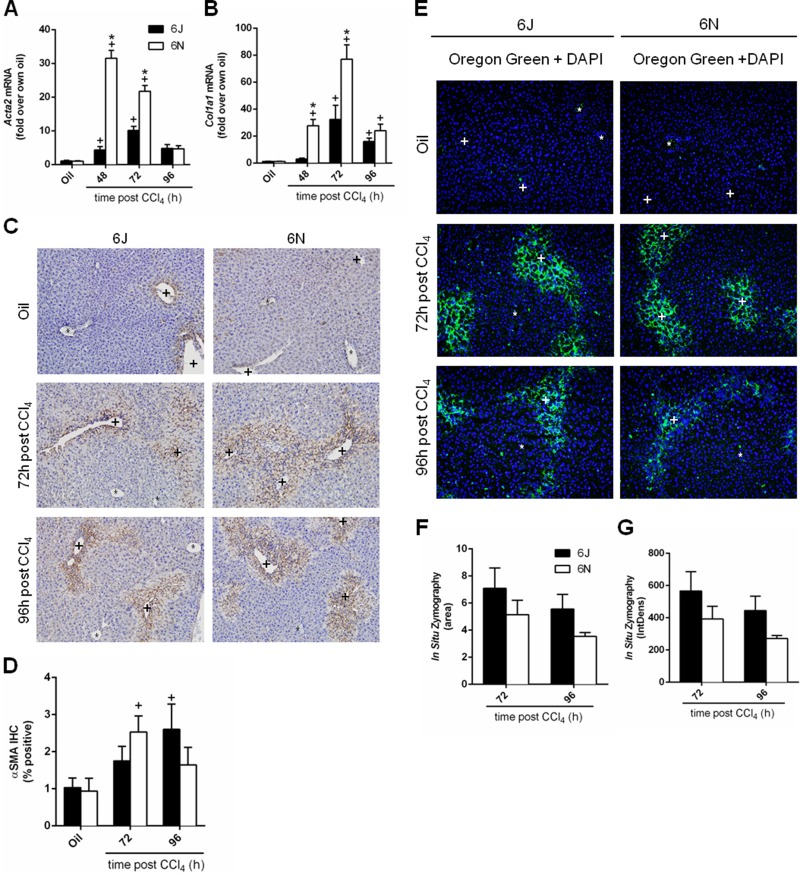
The final stage of wound healing is matrix remodeling (matrix synthesis and degradation) and, similar to regeneration, occurs at 48, 72, and 96 h after CCl4 exposure20,49. Activated hepatic stellate cells (HSCs) contribute to matrix synthesis29. HSCs transdifferentiate into myofibroblasts and begin expressing αSMA (Acta2) and synthesizing type 1 collagen (Col1a1)29. Hepatic Acta2 transcript levels increased following CCl4 exposure in both 6J and 6N mice; Acta2 transcript levels were higher in 6N mice 48 h after CCl4 exposure when compared to 6J mice (Fig. 7A). Expression of Col1a1 also increased in both substrains following CCl4 exposure and was higher in 6N mice at 48 and 72 h after CCl4 exposure compared to 6J mice (Fig. 7B). Immunohistochemistry revealed that αSMA increased in both substrains following CCl4 exposure (Fig. 7C and D). However, αSMA increased over baseline in 6N mice at 72 h after CCl4 exposure, while αSMA protein did not increase above baseline in 6J mice until 96 h after CCl4 exposure (Fig. 7C and D). At this point, αSMA levels were not different from baseline in 6N mice (48 h is not included because of high nonspecific staining in the necrotic tissue) (Fig. 7C and D). Matrix synthesis is opposed by matrix metabolism, which can be evaluated using in situ zymography50,51. Very little matrix metabolism occurred at baseline (Fig. 7E, top, not quantified) or early time points (not shown), but matrix metabolism did occur 72 and 96 h after CCl4 exposure. However, matrix metabolism was not different (area or intensity) between 6J and 6N mice at either time point (Fig. 7F and G). These data together suggest that matrix remodeling was only partially different (synthesis but not metabolism) between substrains.
Figure 7.
Matrix remodeling following CCl4 exposure. Mice were exposed to CCl4 and euthanized 48, 72, or 96 h later. Hepatic transcript accumulation of (A) Acta2 and (B) Col1a1 was evaluated by real-time PCR. (C) Immunohistochemistry for αSMA, the Acta2 gene product, was performed and (D) quantified as the percent area of positive staining in each image. (E) In situ zymography was used to detect matrix metabolism in frozen liver sections at baseline and at 72 and 96 h after CCl4 exposure. The green fluorescence indicates areas of matrix metabolism. DAPI was used to visualize nuclei. (F) The area and (G) intensity above a set threshold were quantified after in situ zymography. Baseline matrix metabolism was too low to be quantified. Central veins are marked with a +, and portal veins are marked with a *. n = 5–6 per group. *p ≤ 0.05 when comparing substrains at a single time point; +p ≤ 0.05 when comparing the indicated CCl4 time point to the oil (control) of the same substrain.
DISCUSSION
The goal of our study was to determine whether C57BL/6 substrains differed in their response to a single exposure to CCl4. We evaluated indices central to liver injury and wound healing, including inflammation, regeneration, and matrix remodeling. Similar to APAP-induced liver injury17, we report here that 6N mice have increased injury compared to 6J mice; this increased injury was not due to differences in CYP2E1 bioactivation or hepatocyte sensitivity to CCl4 between substrains, but might be due to increased numbers of neutrophils and/or earlier and greater TNF-α production observed in 6N mice.
Paradoxically, the increase in TNF-α in 6N mice may have also contributed to the more rapid onset of liver regeneration in 6N mice, relative to 6J mice, after CCl4 exposure. This apparent dichotomy may be due, in part, to the location where hepatocytes are exposed to TNF-α. For example, the stressed hepatocytes found immediately around the necrotic pericentral area may be more sensitive to TNF-α-mediated cell death, while the hepatocytes found in the periportal area may be more sensitive to TNF-α-mediated promotion of liver regeneration. Our own data support this notion, as the area of necrosis was greater in 6N mice compared to 6J mice 48 h after CCl4 (Fig. 1B and D), while at the same time point, hepatocytes in the periportal areas of 6N expressed Ki-67 while 6J mice did not (Fig. 6G and H); these phenomena occurred when Tnfα expression was greatest in 6N liver (Fig. 5G). Subsequently, increased infiltration of nonparenchymal cells, including macrophages, 72 and 96 h after CCl4, was correlated to more rapid clearing of the necrotic tissue in the 6N mice compared to the 6J mice. While matrix synthesis began earlier in 6N mice, matrix metabolism was similar between the 6J mice and the 6N mice. Therefore, despite increased liver injury in 6N mice, liver repair was more rapid when compared to repair in the less-injured 6J mice. This is likely due to the more rapid inflammatory, regenerative, and HSC responses observed in 6N mice.
Because CCl4 causes mitochondrial damage, mutations in the Nnt gene would be expected to exacerbate injury due to the decreased antioxidant capacity. Despite this, and similar to APAP-induced liver injury, we report here that liver injury is increased in 6N mice. Cellular lipid membranes are disrupted after CCl4 bioactivation. This bioactivation also causes mitochondrial damage and oxidative stress reflected in a decrease in mitochondrial GSH/GSSG ratio39,40. In addition, CCl4-induced liver damage can be attenuated using antioxidants to raise the mitochondrial GSH/GSSG ratio. This, in turn, decreases cell damage and lowers plasma ALT levels39,40. Nnt mutant mice have a deceased basal GSH/GSSG ratio. Thus, it should follow that the Nnt mutant mice would have increased injury6. However, this does not occur after APAP17 or CCl4-induced liver injury (described in this study), which suggests that factors other than the mutated Nnt may play a more prominent role in substrain-specific responses to hepatic injury triggered by APAP and CCl4. We also report that there is no difference in hepatocyte sensitivity to cell death caused by CCl4, in vitro, which further supports the idea that other factors, besides the mutated Nnt gene, are responsible for increased sensitivity of 6N mice to CCl4-induced liver injury.
Studies demonstrating that 6J mice are more sensitive to oxidative stress contradict our findings that 6N mice exhibited increased injury following acute CCl4 exposure. In a number of model systems, mutated Nnt results in several redox alterations, including higher rates of H2O2 release and the spontaneous oxidation of NADPH, along with a reduced GSH/GSSG ratio6,37,38. Mutant Nnt is a genetic modifier in two separate, transgenic mice. Bcl2l2 encodes for BCL-W, which protects cells from apoptosis during cellular and oxidative stress52. When Bcl2l2 −/− mice are on an Nnt mutant (6J) background, they display increased embryonic lethality compared to Bcl2l2 −/− on an Nnt wild-type (6N) background31,52. Similarly, mice deficient in mitochondrial superoxide dismutase (Sod2 −/−) exhibit enhanced tissue damage and increased lethality shortly after birth when they are on an Nnt mutant background compared to Sod2 −/− mice on an Nnt wild-type background5. In contrast to what is observed after APAP overdose17,53 or after CCl4 in the present study, these studies show that a mutant Nnt leads to increased sensitivity to oxidative stress.
Additional studies comparing 6J and 6N mice in models of diet-induced obesity show that the Nnt mutation is linked to glucose intolerance and reduced insulin secretion13,54. Although both substrains are sensitive to diet-induced obesity, after high-fat diet (60% of calories from fat) feeding, 6J mice gain more weight and display increased glucose intolerance compared to 6N mice12. Rescuing the mutated Nnt back to wild type using bacterial artificial chromosomes improves glucose tolerance54. These data suggest that the mutated Nnt gene itself is at least partially responsible for the difference in glucose intolerance between the 6J and 6N mice54. However, after feeding these substrains a diet that contains 45% calories from fat, 6N mice display greater hepatic inflammation despite having a functional NNT enzyme54, which is similar to what we observe after CCl4 exposure. Collectively, and similar to APAP overdose17 and to CCl4 exposure, a mutant Nnt can help protect mice from liver inflammation despite leading to impaired glucose tolerance. When thinking about these published studies and the data presented here, one must begin to consider what other factors, besides a mutated Nnt, are contributing to substrain-specific differences in various animal models of human disease, particularly when inflammation, liver damage, and repair are concerned.
Additional investigations document single nucleotide polymorphisms (SNPs) between the two substrains, which may account for differences between 6J and 6N mice, dependent or independent of Nnt status. Indeed, 11 different SNPs exist between 6J and 6N mice3,4. Most of the SNPs map to noncoding regions of the genome, but five SNPs are linked to specific genes including Naaladl2 (N-acetylated α-linked acidic dipeptidase-like 2), Aplp2 (amyloid precursor-like protein 2), Lims2 (LIM and senescent cell antigen-like-containing domain protein 1), Fgf14 (fibroblast growth factor 14), and Snap29 (synaptosomal-associated protein 29)3,4. Although it is beyond the scope of this study, it is possible that one or more of these SNPs alone or together with the well-established Nnt mutation may contribute to the differences seen between the 6J and 6N mice following acute CCl4 exposure.
Another explanation for the, at first, unexpected results found here may be explained by “hormesis.” Hormesis can be defined as low doses of a stressor being beneficial to survival, while high doses of the same stressor are detrimental. It often refers to a chemical insult but can also refer to adaptations to stress55,56. If hormesis is considered in this scenario, 6J mice, which have had a nonfunctional NNT enzyme, have likely adapted to the lower antioxidant capacity in other ways. Therefore, they can withstand higher oxidative stress, relative to nonadapted mice, resulting in reduced cell death and subsequently limited inflammatory response. It is not until an injurious insult has overcome this new homeostasis that robust injury and inflammation occur. This concept has been applied to several phenomena in toxicology57,58. To determine if 6J mice compensated for reduced NNT activity by increasing other antioxidant defenses according to the principals of hormesis, we evaluated Gclc and Nqo1 mRNA and NQO1 protein; we found little difference in these antioxidant molecules between the substrains. While beyond the scope of this study, further studies should evaluate additional antioxidant defense pathways in 6J and 6N mice to determine if differences in the observed antioxidant response contribute to differences in liver injury and subsequent inflammation noted in these substrains.
Our thorough characterization of differences between substrains after acute CCl4 exposure is a critical first step in understanding differences between 6J and 6N mice. Our study shows, for the first time, that despite the fact that CCl4-induced liver injury is worse in 6N mice, liver repair was more rapid when compared to repair in the less-injured 6J mice. This is likely due to the more robust inflammatory, regenerative, and HSC responses observed in 6N mice, but other mechanisms may also be involved. In addition to exploration of whether hormesis is involved in protecting 6J mice from CCl4-induced liver injury relative to 6N mice suggested above, it might be beneficial to determine if a zone-specific role for TNF-α exists in driving hepatocyte death versus hepatocyte proliferation 48 h after CCl4 exposure. Additionally, further evaluation of the role neutrophils play in exacerbation of liver injury in 6N mice is also warranted. These would be excellent next steps to understanding substrain differences in this commonly used model of acute hepatotoxin exposure.
Our study highlights the importance of identifying and choosing the correct control substrain when using genetically modified mice in studies that utilize acute, and likely chronic, CCl4. Similar to the contradictory reports on Jnk2 −/− mice in APAP-induced liver injury, comparing a genetically modified mouse to the wrong C57BL/6 substrain after CCl4 exposure may lead to data misinterpretations. We urge researchers to report not only what strain but also what substrain of mouse is used in experiments. This consideration is vitally important in this new age of experimental rigor, transparency, and reproducibility required by the NIH59,60.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH), National Center for Research Resources (P20 RR021940), the National Institute of General Medical Sciences (P20 GM103549), the National Institute of Environmental Health Sciences “Training Program in Environmental Toxicology” (T32 ES007079), the National Institutes of Alcohol Abuse and Alcoholism (R00 AA17918), an NIH Clinical and Translational Science Award grant (UL1 TR000001, formerly UL1RR033179) awarded to the University of Kansas Medical Center (KUMC), and an internal Lied Basic Science Grant Program of the KUMC Research Institute. In addition, funds from the Center for Reproductive Health after Disease from the National Centers for Translational Research in Reproduction and Infertility (P50 HD076188) and the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK098414) were used to complete the work in this study. A special thanks to the KUMC’s Laboratory Animal Resource husbandry staff, veterinary technicians, veterinarians, Office of Animals Welfare, and Institutional Animal Care and Use Committee for the care of our animals. Thanks to the KUMC Pharmacology, Toxicology, and Therapeutics COBRE Cell Isolation Core for the preparation of primary hepatocytes from 6J and 6N mice. Thanks also to members of the KUMC Pharmacology, Toxicology, and Therapeutics Department, in particular Dr. Ben Woolbright (for helpful discussions and LDH assay training), Dr. Partha Kasturi (for the hormesis idea), and Dr. Yuxia Zhang (for the use of her Zeiss Axio Observer A.1 inverted microscope, Olympus DP71 camera, and cellSens imaging software).
REFERENCES
- 1. Reynolds J. Strain differences and the genetic basis of experimental autoimmune anti-glomerular basement membrane glomerulonephritis. Int J Exp Pathol. 2011;92(3):211–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walkin L, Herrick SE, Summers A, Brenchley PE, Hoff CM, Korstanje R, Margetts PJ. The role of mouse strain differences in the susceptibility to fibrosis: A systematic review. Fibrogenesis Tissue Repair 2013;6(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mekada K, Abe K, Murakami A, Nakamura S, Nakata H, Moriwaki K, Obata Y, Yoshiki A. Genetic differences among C57BL/6 substrains. Exp Anim. 2009;58(2):141–9. [DOI] [PubMed] [Google Scholar]
- 4. Zurita E, Chagoyen M, Cantero M, Alonso R, Gonzalez-Neira A, Lopez-Jimenez A, Lopez-Moreno JA, Landel CP, Benitez J, Pazos F and others. Genetic polymorphisms among C57BL/6 mouse inbred strains. Transgenic Res. 2011;20(3):481–9. [DOI] [PubMed] [Google Scholar]
- 5. Huang TT, Naeemuddin M, Elchuri S, Yamaguchi M, Kozy HM, Carlson EJ, Epstein CJ. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum Mol Genet. 2006;15(7):1187–94. [DOI] [PubMed] [Google Scholar]
- 6. Ronchi JA, Figueira TR, Ravagnani FG, Oliveira HC, Vercesi AE, Castilho RF. A spontaneous mutation in the nicotinamide nucleotide transhydrogenase gene of C57BL/6J mice results in mitochondrial redox abnormalities. Free Radic Biol Med. 2013;63:446–56. [DOI] [PubMed] [Google Scholar]
- 7. Hoek JB, Rydstrom J. Physiological roles of nicotinamide nucleotide transhydrogenase. Biochem J. 1988;254(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vogel R, Wiesinger H, Hamprecht B, Dringen R. The regeneration of reduced glutathione in rat forebrain mitochondria identifies metabolic pathways providing the NADPH required. Neurosci Lett. 1999;275(2):97–100. [DOI] [PubMed] [Google Scholar]
- 9. Dalton TP, Chen Y, Schneider SN, Nebert DW, Shertzer HG. Genetically altered mice to evaluate glutathione homeostasis in health and disease. Free Radic Biol Med. 2004;37(10):1511–26. [DOI] [PubMed] [Google Scholar]
- 10. Ramachandra V, Phuc S, Franco AC, Gonzales RA. Ethanol preference is inversely correlated with ethanol-induced dopamine release in 2 substrains of C57BL/6 mice. Alcohol Clin Exp Res. 2007;31(10):1669–76. [DOI] [PubMed] [Google Scholar]
- 11. Mulligan MK, Ponomarev I, Boehm SL 2nd, Owen JA, Levin PS, Berman AE, Blednov YA, Crabbe JC, Williams RW, Miles MF, Bergeson MS. Alcohol trait and transcriptional genomic analysis of C57BL/6 substrains. Genes Brain Behav. 2008;7(6):677–89. [DOI] [PubMed] [Google Scholar]
- 12. Nicholson A, Reifsnyder PC, Malcolm RD, Lucas CA, MacGregor GR, Zhang W, Leiter EH. Diet-induced obesity in two C57BL/6 substrains with intact or mutant nicotinamide nucleotide transhydrogenase (Nnt) gene. Obesity (Silver Spring) 2010;18(10):1902–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Freeman H, Shimomura K, Cox RD, Ashcroft FM. Nicotinamide nucleotide transhydrogenase: A link between insulin secretion, glucose metabolism and oxidative stress. Biochem Soc Trans. 2006;34(Pt 5):806–10. [DOI] [PubMed] [Google Scholar]
- 14. Heiker JT, Kunath A, Kosacka J, Flehmig G, Knigge A, Kern M, Stumvoll M, Kovacs P, Bluher M, Kloting N. Identification of genetic loci associated with different responses to high-fat diet-induced obesity in C57BL/6N and C57BL/6J substrains. Physiol Genomics 2014;46(11):377–84. [DOI] [PubMed] [Google Scholar]
- 15. Nakagawa H, Maeda S, Hikiba Y, Ohmae T, Shibata W, Yanai A, Sakamoto K, Ogura K, Noguchi T, Karin M and others. Deletion of apoptosis signal-regulating kinase 1 attenuates acetaminophen-induced liver injury by inhibiting c-Jun N-terminal kinase activation. Gastroenterology 2008;135(4):1311–21. [DOI] [PubMed] [Google Scholar]
- 16. Bourdi M, Korrapati MC, Chakraborty M, Yee SB, Pohl LR. Protective role of c-Jun N-terminal kinase 2 in acetaminophen-induced liver injury. Biochem Biophys Res Commun. 2008;374(1):6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bourdi M, Davies JS, Pohl LR. Mispairing C57BL/6 substrains of genetically engineered mice and wild-type controls can lead to confounding results as it did in studies of JNK2 in acetaminophen and concanavalin A liver injury. Chem Res Toxicol. 2011;24(6):794–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weber LW, Boll M, Stampfl A. Hepatotoxicity and mechanism of action of haloalkanes: Carbon tetrachloride as a toxicological model. Crit Rev Toxicol. 2003;33(2):105–36. [DOI] [PubMed] [Google Scholar]
- 19. Pellicoro A, Ramachandran P, Iredale JP, Fallowfield JA. Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat Rev Immunol. 2014;14(3):181–94. [DOI] [PubMed] [Google Scholar]
- 20. Balaji S, Watson CL, Ranjan R, King A, Bollyky PL, Keswani SG. Chemokine involvement in fetal and adult wound healing. Adv Wound Care (New Rochelle) 2015;4(11):660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiso K, Ueno S, Fukuda M, Ichi I, Kobayashi K, Sakai T, Fukui K, Kojo S. The role of Kupffer cells in carbon tetrachloride intoxication in mice. Biol Pharm Bull. 2012;35(6):980–3. [DOI] [PubMed] [Google Scholar]
- 22. Zigmond E, Samia-Grinberg S, Pasmanik-Chor M, Brazowski E, Shibolet O, Halpern Z, Varol C. Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. J Immunol. 2014;193(1):344–53. [DOI] [PubMed] [Google Scholar]
- 23. Mochizuki A, Pace A, Rockwell CE, Roth KJ, Chow A, O’Brien KM, Albee R, Kelly K, Towery K, Luyendyk JP, Copple BL. Hepatic stellate cells orchestrate clearance of necrotic cells in a hypoxia-inducible factor-1alpha-dependent manner by modulating macrophage phenotype in mice. J Immunol. 2014;192(8):3847–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moles A, Murphy L, Wilson CL, Chakraborty JB, Fox C, Park EJ, Mann J, Oakley F, Howarth R, Brain J, Masson S, Karin M, Seki E, Mann DA. A TLR2/S100A9/CXCL-2 signaling network is necessary for neutrophil recruitment in acute and chronic liver injury in the mouse. J Hepatol. 2014;60(4):782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramaiah SK, Jaeschke H. Role of neutrophils in the pathogenesis of acute inflammatory liver injury. Toxicol Pathol. 2007;35(6):757–66. [DOI] [PubMed] [Google Scholar]
- 26. Cienfuegos JA, Rotellar F, Baixauli J, Martinez-Regueira F, Pardo F, Hernandez-Lizoain JL. Liver regeneration—The best kept secret. A model of tissue injury response. Rev Esp Enferm Dig. 2014;106(3):171–94. [PubMed] [Google Scholar]
- 27. Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Transl Res. 2014;163(4):352–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo J, Friedman SL. Hepatic fibrogenesis. Semin Liver Dis. 2007;27(4):413–26. [DOI] [PubMed] [Google Scholar]
- 29. Friedman SL. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88(1):125–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCracken JM, Jiang L, Deshpande KT, O’Neil MF, Pritchard MT. Differential effects of hyaluronan synthase 3 deficiency after acute vs chronic liver injury in mice. Fibrogenesis Tissue Repair 2016;9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Navarro SJ, Trinh T, Lucas CA, Ross AJ, Waymire KG, Macgregor GR. The C57BL/6J mouse strain background modifies the effect of a mutation in Bcl2l2. G3 (Bethesda) 2012;2(1):99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31(24):e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics 2008;9:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spandidos A, Wang X, Wang H, Seed B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38(Database issue):D792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pritchard MT, McMullen MR, Stavitsky AB, Cohen JI, Lin F, Medof ME, Nagy LE. Differential contributions of C3, C5, and decay-accelerating factor to ethanol-induced fatty liver in mice. Gastroenterology 2007;132(3):1117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bajt ML, Knight TR, Lemasters JJ, Jaeschke H. Acetaminophen-induced oxidant stress and cell injury in cultured mouse hepatocytes: Protection by N-acetyl cysteine. Toxicol Sci. 2004;80(2):343–9. [DOI] [PubMed] [Google Scholar]
- 37. Arkblad EL, Tuck S, Pestov NB, Dmitriev RI, Kostina MB, Stenvall J, Tranberg M, Rydstrom J. A Caenorhabditis elegans mutant lacking functional nicotinamide nucleotide transhydrogenase displays increased sensitivity to oxidative stress. Free Radic Biol Med. 2005;38(11):1518–25. [DOI] [PubMed] [Google Scholar]
- 38. Sheeran FL, Rydstrom J, Shakhparonov MI, Pestov NB, Pepe S. Diminished NADPH transhydrogenase activity and mitochondrial redox regulation in human failing myocardium. Biochim Biophys Acta 2010;1797(6–7):1138–48. [DOI] [PubMed] [Google Scholar]
- 39. Wong HS, Chen JH, Leong PK, Leung HY, Chan WM, Ko KM. beta-sitosterol protects against carbon tetrachloride hepatotoxicity but not gentamicin nephrotoxicity in rats via the induction of mitochondrial glutathione redox cycling. Molecules 2014;19(11):17649–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ip SP, Ko KM. The crucial antioxidant action of schisandrin B in protecting against carbon tetrachloride hepatotoxicity in mice: A comparative study with butylated hydroxytoluene. Biochem Pharmacol. 1996;52(11):1687–93. [DOI] [PubMed] [Google Scholar]
- 41. Hwang JH, Kim YH, Noh JR, Gang GT, Kim KS, Chung HK, Tadi S, Yim YH, Shong M, Lee CH. The protective role of NAD(P)H:quinone oxidoreductase 1 on acetaminophen-induced liver injury is associated with prevention of adenosine triphosphate depletion and improvement of mitochondrial dysfunction. Arch Toxicol. 2015;89(11):2159–66. [DOI] [PubMed] [Google Scholar]
- 42. Hogaboam CM, Bone-Larson CL, Steinhauser ML, Lukacs NW, Colletti LM, Simpson KJ, Strieter RM, Kunkel SL. Novel CXCR2-dependent liver regenerative qualities of ELR-containing CXC chemokines. FASEB J. 1999;13(12):1565–74. [DOI] [PubMed] [Google Scholar]
- 43. Dixon LJ, Barnes M, Tang H, Pritchard MT, Nagy LE. Kupffer cells in the liver. Compr Physiol. 2013;3(2):785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. You Q, Holt M, Yin H, Li G, Hu CJ, Ju C. Role of hepatic resident and infiltrating macrophages in liver repair after acute injury. Biochem Pharmacol. 2013;86(6):836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seki E, Brenner DA, Karin M. A liver full of JNK: Signaling in regulation of cell function and disease pathogenesis, and clinical approaches. Gastroenterology 2012;143(2):307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Malhi H, Guicciardi ME, Gores GJ. Hepatocyte death: A clear and present danger. Physiol Rev. 2010;90(3):1165–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Webber EM, Bruix J, Pierce RH, Fausto N. Tumor necrosis factor primes hepatocytes for DNA replication in the rat. Hepatology 1998;28(5):1226–34. [DOI] [PubMed] [Google Scholar]
- 48. Nishiyama K, Nakashima H, Ikarashi M, Kinoshita M, Nakashima M, Aosasa S, Seki S, Yamamoto J. Mouse CD11b+ Kupffer cells recruited from bone marrow accelerate liver regeneration after partial hepatectomy. PLoS One 2015;10(9):e0136774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deshpande KT, Liu S, McCracken JM, Jiang L, Gaw TE, Kaydo LN, Richard ZC, O’Neil MF, Pritchard MT. Moderate (2%, v/v) ethanol feeding alters hepatic wound healing after acute carbon tetrachloride exposure in mice. Biomolecules 2016;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lindsey M, Wedin K, Brown MD, Keller C, Evans AJ, Smolen J, Burns AR, Rossen RD, Michael L, Entman M. Matrix-dependent mechanism of neutrophil-mediated release and activation of matrix metalloproteinase 9 in myocardial ischemia/reperfusion. Circulation 2001;103(17):2181–7. [DOI] [PubMed] [Google Scholar]
- 51. Yan SJ, Blomme EA. In situ zymography: A molecular pathology technique to localize endogenous protease activity in tissue sections. Vet Pathol. 2003;40(3):227–36. [DOI] [PubMed] [Google Scholar]
- 52. Ross AJ, Waymire KG, Moss JE, Parlow AF, Skinner MK, Russell LD, MacGregor GR. Testicular degeneration in Bclw-deficient mice. Nat Genet. 1998;18(3):251–6. [DOI] [PubMed] [Google Scholar]
- 53. Duan L, Davis JS, Woolbright BL, Du K, Cahkraborty M, Weemhoff J, Jaeschke H, Bourdi M. Differential susceptibility to acetaminophen-induced liver injury in sub-strains of C57BL/6 mice: 6N versus 6J. Food Chem Toxicol. 2016;98(Pt B):107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Freeman HC, Hugill A, Dear NT, Ashcroft FM, Cox RD. Deletion of nicotinamide nucleotide transhydrogenase: A new quantitive trait locus accounting for glucose intolerance in C57BL/6J mice. Diabetes 2006;55(7):2153–6. [DOI] [PubMed] [Google Scholar]
- 55. Milisav I, Poljsak B, Suput D. Adaptive response, evidence of cross-resistance and its potential clinical use. Int J Mol Sci. 2012;13(9):10771–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bhakta-Guha D, Efferth T. Hormesis: Decoding two sides of the same coin. Pharmaceuticals (Basel) 2015;8(4):865–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Counts JL, Goodman JI. Principles underlying dose selection for, and extrapolation from, the carcinogen bioassay: Dose influences mechanism. Regul Toxicol Pharmacol. 1995;21(3):418–21. [DOI] [PubMed] [Google Scholar]
- 58. Williams GM, Iatropoulos MJ. Alteration of liver cell function and proliferation: Differentiation between adaptation and toxicity. Toxicol Pathol. 2002;30(1):41–53. [DOI] [PubMed] [Google Scholar]
- 59. Omary MB, Cohen DE, El-Omar EM, Jalan R, Low MJ, Nathanson MH, Peek RM Jr., Turner JR. Not all mice are the same: Standardization of animal research data presentation. Gastroenterology 2016;150(7):1503–4. [DOI] [PubMed] [Google Scholar]
- 60. Collins F, Tabak L. Policy: NIH plans to enhance reproducibility. Nature 2014;505:612–3. [DOI] [PMC free article] [PubMed] [Google Scholar]