Abstract
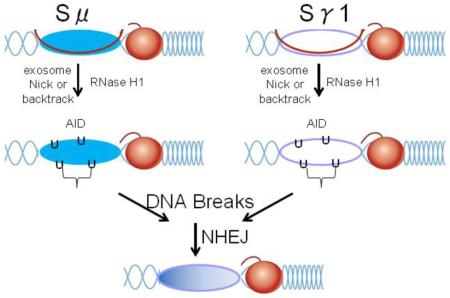
R-loops, three strand structures consisting of mRNA hybridized to the complementary DNA and a single stranded DNA loop, are formed in switch regions on the heavy chain immunoglobulin locus. To determine if R-loops have a direct effect on any of the steps involved in isotype switching, we generated a transgenic mouse that over expressed RNase H1, an enzyme that cleaves the RNA of RNA/DNA hybrids in B cells. R-loops in the switch μ region were depleted by 70% in ex vivo activated splenic B cells. Frequencies of isotype switching to IgG1, IgG2b, IgG2c, and IgG3 were the same as C57BL/6 control cells. However, somatic hypermutation was increased specifically on the transcribed strand from μ-γ joins, indicating that R-loops limit AID access to the transcribed DNA strand. Our data suggest that, in the normal G+C rich context of mammalian class switch recombination regions, R-loops are obligatory intermediates. Processing of the R-loops is needed to remove RNA allowing AID to promote somatic hypermutation on both DNA strands to generate double-strand DNA breaks for efficient class switch recombination. One of the two cellular RNases H may assist in this process.
Graphical abstract
Introduction
Antibodies comprise heavy and light chains and are extraordinarily diverse in primary amino acid sequences permitting recognition of a very large variety of antigens. In the mouse, the building blocks of the heavy chain reside on the centromere distal region of chromosome 12. The entire region is almost 3 Mb in length and contains both variable (V) and constant (C) regions. During B cell development, rearrangement of the heavy chain occurs with one of many V segments joined to one of several diverse (D) sequences and a joining (J) sequence. Most of the myriad of antibody differences are established during VDJ recombination. The recombined VDJ exon can be transcribed to express immunoglobulin with the C region of the heavy chain class M (Cμ) to produce IgM. Cμ is one of eight similar C regions, all of which reside in a 200 Kb region downstream of the Cμ exons. Changing from Cμ to one of the other C regions produce different immunoglobulin isotypes altering antibody function to more appropriately eliminate pathogens encountered in different parts of the animal.
Changing isotype occurs by a process called Class Switch Recombination (CSR), the ultimate step of which involves breaking and rejoining the DNA thereby removing the region between Cμ and a downstream C region and connecting the transcription unit of VDJ to the new C exons. Transcription of the DNA at the two sites of DNA breaks is necessary with different cytokines stimulating transcription at each target switch region. Transcripts from these sites form R-loops, in which the nascent RNA is hybridized to its complementary DNA and the opposite DNA strand is in single form [1]. These structures allow access by Activation Induced (cytosine) Deaminase (AID), which deaminates cytosine to uracil within single stranded DNA of VDJ and switch (S), regions, to produce nucleotide substitutions called somatic hypermutation (SHM) and DNA strand breaks [2-4]. In particular, the S regions in mice are approximately 3-9 kb long and precede the C genes on the heavy chain locus. They are composed of repetitive sequences containing clusters of 3-4 G:C base pairs interspersed by WGCW (W = A/T) motifs in which AID deaminates the cytosine. Mutations and double strand breaks are common in S regions, and donor and acceptor breaks are then rejoined by non-homologous end joining mechanisms. For example, a double strand break in the switch region (Sμ) in front of Cμs can be joined to a break in the switch region (Sγ) in front of Cγ>, resulting in a IgM to IgG switched isotype [5].
SHM occurs on both transcribed and non-transcribed strands of DNA, indicating that both are in single stranded form at some time. When R-loops were first described in the switch regions of B cells, it was suggested that RNase H could play an important role, perhaps altering the frequency of isotype switching [6]. In mouse and other metazoans, there are two types of RNase H (H1 and H2), both of which are required for viability [7]. For RNA/DNA hybrids, including R-loops, either enzyme could eliminate the RNA portion of the hybrid. RNase H1 acts as a monomer while RNase H2 is a heterotrimeric enzyme making the former more amenable for over expression. However, the RNA exosome has been implicated in removing the R-loop RNA, in the mouse CH12F3 B lymphoma cells and in mouse B cells [8]. Moreover, the absence of RNA exosome function in B cells results in accumulation of RNA/DNA hybrids that act as long non-coding RNA enhancers [9].
Almost uniformly, studies examining R-loops in other DNA sequences invoke the R-loops formed during isotype switching as a known example of in vivo R-loops [10]. Support that putative R-loops are true R-loops often relies on over expressing RNase H1 to partially reverse a phenotype or eliminate detection of R-loops by the RNA/DNA specific S9.6 antibody [11, 12]. Here we report that very high levels of the nuclear form of mouse RNase H1 decreased R-loops in activated B cells to 30% in Sμ and by 50% in β-actin but there was no effect on the frequency of isotype switching. However, SHM mutagenesis of the transcribed DNA strand is increased suggesting that RNA is removed from the R-loop.
Results
Transgenic mouse expressing high levels of the nuclear isoform of RNase H1 in B cells
Construction of Transgenic (Tg) Mouse Expressing RNase H1
Translation of the Rnaseh1 mRNA initiates at each of three AUG start codons [13]. Initiation of translation at the first AUG codes for a seven amino acid upstream open reading frame that limits translation from the second AUG, which produces a protein targeted to the mitochondria. The nuclear form of RNase H1 comes from translation of the third AUG. Translational regulation of Rnaseh1 serves to maintain low levels of RNase H1. When the transcript is produced at high levels, the mitochondrial enzyme causes a growth arrest [13]. Therefore, we over expressed only the nuclear form of RNase H1.
Plasmid P14 TCR β chain cDNA construct B [14] was modified by replacing the T cell receptor (TCR) cDNA with the cDNA of mouse Rnaseh1 starting at the AUG coding for M27. Expression of the nuclear form of RNase H1 was under control of the histocompatibility H-2Kb promoter and Eμ enhancer (Fig. 1a). DNA containing the genes shown in Fig. 1a was injected into zygotes at the pronuclear stage [15] at the NCI Transgenic Mouse Model Laboratory. The injected embryos were surgically transferred into the oviduct of 0.5 days post coitus pseudopregnant female and allowed to go to term. The resulting progeny were tail-clipped at weaning age and screened by Southern blot analysis for the presence of the transgene. Mice obtained from the NCI Transgenic Mouse Model Laboratory were examined for germline transmission resulting in selection of several transgenic strains.
Fig. 1. Transgenic mouse.
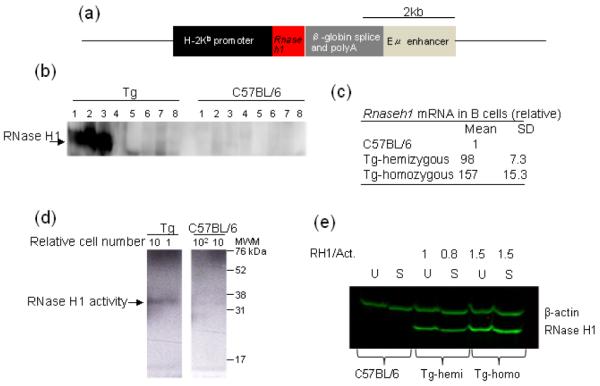
Depiction of DNA construct for expressing RNase H1 in B cells (a). Western blot analysis of RNase H1 protein expression in various tissues in hemizygous Tg and C57BL/6 mice. Lanes marked 1 splenic B cells, 2 thymus, 3 bone marrow, 4 brain, 5, heart, 6 kidney, 7 liver, and 8 lung (b). Quantitative PCR of mRNA in C57BL/6, hemizygous Tg and homozygous Tg mice (c). Autoradiograph (reverse image) of RNase H1 activity of resting B cells from hemizygous Tg and C57 BL/6 mice from B cells. Relative number of cells used is indicated above each lane. MWM – molecular weight makers (d). Western blot analysis of RNase H1 from uninduced resting (U) and 48 h stimulated (S) B cells. RH1/Act.is the ratio of signals for RNase H1 and β-actin in each lane. (e).
One mouse strain exhibits very high levels of RNase H1 expression
We examined B cells from spleens of several strains of mice which exhibited germline transmission for Rnaseh1 mRNA content. The mouse strain used in the experiments presented here exhibited by far the highest over expression providing us with the greatest opportunity to observe any effects of RNase H1 in B cell development. Expression of RNase H1 protein could be detected at high levels in bone marrow, splenic B- and T- cells, but not in other tissues, reflecting the Eμ enhancer regulation (Fig. 1b). The hemizygous (the homologous chromatid does not contain the insert – therefore the Tg is hemizygous not heterozygous) Tg mouse expressed more than 98 times the C57BL/6 level of Rnaseh1 mRNA (Fig. 1c). We found a further increase (almost to 160 fold) when the transgene was homozygous (Fig. 1c). We observed a concordant enzymatic activity increase with mRNA levels as seen in an in situ gel assay in which radio-labelled substrate was present in the gel during electrophoresis of total cell protein. RNase H1 activity results in loss of the substrate from the gel following incubation of the gel in a buffer which allows removal of SDS, refolding of the enzyme and hydrolysis of the embedded RNA of the RNA/DNA hybrid. In Fig. 1d, a negative image is shown for clarity [16]. Although the assay is semi-quantitative, it is clear that the homozygous Tg RNase H1 activity is at least 50 -100 times that of C57BL/6; the latter showing no signal at 100 times the amount of protein applied to the gel (Fig. 1d). The Eμ enhancer is active after in vitro stimulation of B cells, but it was important to confirm that RNase H1 continues to be present at high levels after stimulation. RNase H1 abundance in resting B cells is very high, and following stimulation of B cells with lipopolysaccharide (LPS) and interleukin 4 (IL4) for two days hemizygous and homozygous Tg mice, showed that RNase H1 protein was abundant in both Tg strains (Fig. 1e). B cells from C57BL/6 mice have very low amounts of RNase H1 that are not detectable in the figure. The signal from RNase H1 in hemizygous Tg mice is consistent with the ~100 fold increase in mRNA (Fig. 1c) as is the 1.5 fold higher amount of RNase H1 for the homozygous Tg samples.
Frequencies of isotype switching are unchanged
Despite the high levels of RNase H1 both prior to and after stimulation in vitro, we found that splenic B- and T- cells are present in Tg mice at levels similar to those of C57BL/6 mice (data not shown). To assess frequencies of CSR, we stimulated splenic B cells ex vivo with various combinations of LPS and interleukins to induce switching to various isotypes. Surprisingly, we detected no differences in CSR frequencies to IgG1 in hemizygous (not shown) or homozygous Tg mice. Likewise, switching to IgG2b, IgG2c, and IgG3 were the same in homozygousTg and C57BL/6 mice (Fig. 2a). We therefore conclude that all of the data reported in Fig. 2a are consistent with the unaltered frequencies of CSR in Tg B cells. Previously, we demonstrated a lack of effect on CSR frequencies for B cells in which E. coli RNase HI was expressed at a high level in CH123F3-2 cell line [17]. However, means to demonstrate the bacterial RNase H was active on any substrate in the CH12 cells were unavailable at that time.
Fig. 2. Analysis of steps in isotype switching.
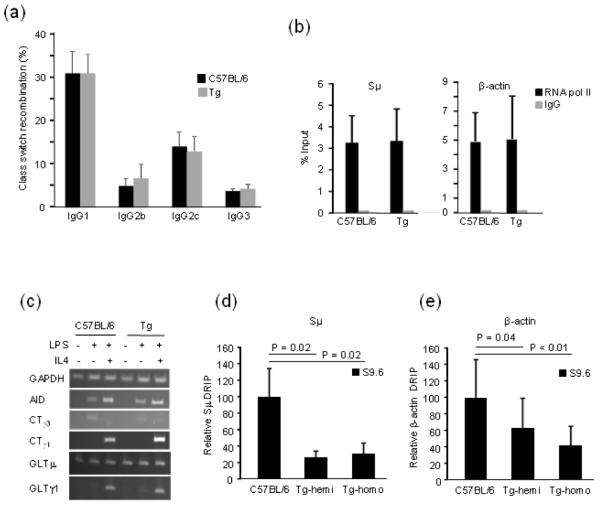
Frequencies of CSR to IgG1, IgG2b, Ig2c and IgG3 for B cells from homozygous Tg and C57BL/6 mice. Error bars signify the SD of values from three independent experiments with one mouse per genotype per experiment (a). Transcriptional profile in C57BL/6 and homozygous Tg mice. ChIP analysis of RNA pol II (black bar) or IgG control (grey bar). Error bars represent the SD of three independent experiments with two mice per genotype (b). Semi-quantitative PCR analyses of AID, CTs and GLTs with GAPDH control. Cells were treated or not with LPS and IL4 as indicated by + or – (c). DNA-RNA hybrid abundance in hemizygous and homozygous Tg mice. DRIP analysis represented as relative percent to C57BL/6 in (d (e) -actin. Black bars represent DRIP with S9.6 antibody. IgG control results are discussed in text. The values are so low they would not be visible in the histograms. Error bars represent the standard deviation of four independent experiments with 2 to 3 mice per experiment for a total of 6 C57BL/6, 4 hemizygous, and 5 homozygous mice. P value was calculated using 2-tailed student T-test with unequal variance.
RNA pol II accumulation, AID, S region transcription and circle transcripts are unchanged
Here, we systematically examined in some detail the various steps involving CSR in an attempt to discover any differences in the Tg mice. Transcription and R-loop formation at the sites of switching are the first action in the process. It has been proposed that the R-loop structure slows down progression of RNA polymerase (pol) II, and attracts co-factors such as Spt5 and RNA exosome to bring in AID to the site [8, 18, 19]. After stimulation of splenic B cells ex vivo with LPS and IL4, we assessed the amount of RNA pol II accumulation at the Sμ and β-actin loci using nuclear run-on (data not shown) and chromatin immunoprecipitation (Fig. 2b). We found no difference between C57BL/6 and Tg mice, indicating overall transcription is not perturbed by the presence of excess RNase H1. Germline transcripts (GLTs) at Sμ and Sγ1 are necessary for switching to IgG1 when B cells are stimulated by LPS and IL4. Consistent with normal CSR levels, similar amounts of germline transcripts of Sμ and Sγ1 were present in B cells in C57BL/6 and Tg mice that had no added cytokine, LPS alone or LPS plus IL4 (Fig. 2c). AID mRNA expression increased similarly in C57BL/6 and Tg B cells when stimulated with LPS plus IL4 to a greater extent than cells stimulated with LPS alone (Fig. 2c), the latter of which stimulates isotype switching to IgG3. Recombination leading to isotype switching produces a second product in the form of a circular DNA molecule from the excised DNA when joining the Sμ and downstream C regions. The promoter upstream of Sμ remains active and produces a new circle transcript (CT) that contains sequences of the S region to which the isotype changes, e.g., IgG1. This transcript is an excellent additional indication of recombination. We detected this type of transcript in both strains of mice when B cells were stimulated by LPS to switch to IgG3 and LPS plus IL4 switching to IgG1 (Fig. 2c). We conclude that the data reported in Fig. 2c are consistent with the unaltered frequencies of CSR for Tg B cells presented Fig. 2a.
R-loops are decreased in the Sμ region
To address whether over expressed levels of RNase H1 were functionally active, we analyzed RNA/DNA hybrid levels during CSR. To examine R-loops in the Sμ region, we measured their relative amounts in C57BL/6 and Tg mice after stimulation of splenic B cells ex vivo with LPS and IL4. R-loops were quantified after immunoprecipitation by DRIP assay using an antibody specific for RNA/DNA hybrids (S9.6) [20]. qPCR was then performed to measure amounts in Sμ, β-actin (promoter region containing R-loops), and β-globin (a gene not expressed in B cells). R-loops were detected in C57BL/6 cells in both Sμ and β-actin regions but not in β-globin (Fig. 2d and e). Using a control IgG antibody, we observed only undetectable levels of R-loops for all three DNA regions (<0.1% of the signal detected for Sμ and β-actin regions; with β-globin not detected with either antibody). To further confirm that the signal was indeed due to R-loops, samples were treated with recombinant RNase H in vitro, followed by precipitation with S9.6 antibody. These samples had a significantly reduced signal compared to untreated samples (data not shown).The amount of R-loops recovered from Sμ was about 50 times higher than from β-actin. In both hemizygous and homozygous Tg mice, R-loops were reduced by 70% at the Sμ locus, and by 50% at the β-actin locus. The residual fraction of R-loops detected in the Sμ region by the S9.6 antibody (Fig. 2d and e) is approximately the same in both hemi- and homo-gzygous Tg mice. Perhaps there is still insufficient RNase H1 to remove all of the hybrid in vivo or that is protected from hydrolysis.
SHM is increased by targeting the transcribed DNA strand
If R-loops are necessary for SHM, RNase H1 removal of RNA from the hybrid could have an effect in frequency or pattern of mutations. We hypothesized that SHM would decrease if after RNA processing by RNase H1, reverting the R-loop to dsDNA. To test this hypothesis, we sequenced the upstream Sμ locus for AID-induced mutations (Fig. 3a). However, rather than a decrease we saw a significant 1.4 fold increase in mutation frequency in homozygous Tg B cells compared to C57BL/6 B cells (Fig. 3b). Interestingly, most of SHM increase was observed on the transcribed strand (Fig. 3c). In particular, the frequency of G to A and G to T increased significantly. Because the data are recorded from the nontranscribed strand by convention, this means that the complementary C on the transcribed strand is undergoing more mutation. We interpret these findings to show that high levels of RNase H1 can remove the RNA annealed to the transcribed strand providing more time and/or opportunity for AID to modify the strand.
Fig. 3. Analysis of Slμ mutations during CSR.
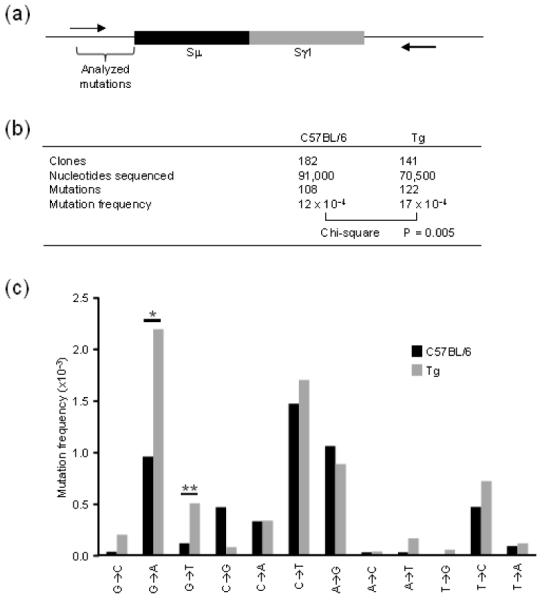
Arrows indicate the location of junction primers within the 5′-flanking region of Sμ and 3′-flanking region of Sγ1. Bracket highlights region of mutational analysis in 500 bp of Sμ sequence (a). Frequency of mutations in C57BL/6 and homozygous Tg mice. Mutations were pooled from 4 independent experiments with 1-2 mice per genotype per experiment. Chi-square analysis was performed comparing mutated to nonmutated nucleotides from each genotype (b). Mutation frequencies for individual mutation type. Frequencies and significance were calculated using SHMTool software. *P = 0.001; **P = 0.035 (c).
Discussion
R-loops have gained prominence as exhibiting both positive and negative effects in many different processes. Involvement of R-loop structures in isotype switching in B cells has been the paradigm for many such studies [10]. RNA/DNA can be followed in purified DNA using the S9.6 anti RNA/DNA antibody [21, 22], and in vivo by over expression of RNase H1 in cells [11]. In many cases, these RNA/DNA hybrids lead to specific types of DNA instability such as recombination [23], collisions between replication and transcription machineries [12, 24], and affecting the utility of antisense therapeutic drugs [25]. Expression of high levels in RNase H1 in E.coli [26], in S. cerevisiae [23, 27], mammalian cells [11, 28], and other organisms has been used to confirm the presence of R-loops in vivo. Decreasing the abundance of R-loop by over expressing RNase H1 indicates (i) there are R-loops present in vivo and (ii) that RNase H1, and likely RNase H2, in normal amounts have no or only limited access to these R-loops.
In this study, we decreased the level of R-loops in mice by over-expressing RNase H1 in B cells to test the dogma that R-loop formation is critical for CSR. The process of CSR may occur in a complex which prevents RNase H1 accessing the RNA/DNA hybrid and would therefore have no effect on CSR. We also considered that increased RNase H1 expression might produce more isotype switching by allowing greater exposure of single strand DNA to AID to initiate breaks. This would suggest that some of the transcription-generated R-loops are never used for CSR. Alternatively, we might have expected a decrease in CSR frequency if the processing of the RNA strand of the R-loop resulted in reformation of duplex DNA eliminating single-stranded DNA substrate for AID. However, we found that there was a significant loss of R-loops, with an increase in SHM, and yet the frequency of isotype switching was unchanged.
It is important to keep in mind that a B cell either switches or it does not and the frequency of isotype switching reflects the number of cells that have undergone CSR. For example, if transcription/R-loop formation occurs in one cell but does not in a second the frequency would be 100% or zero%, respectively. It is likely that high levels of RNase H1 do not affect RNA pol II abundance or the formation of R-loops. However, excess RNase H1 might remove the RNA and collapse the RNA/DNA hybrids. Thus, we believe that the simplest interpretation of our data is that additional removal of the R-loop RNA by RNase H1 on top of the processing by the RNA exosome does not increase the number of dsDNA breaks in the cell. However, the observed loss of RNA/DNA hybrid provides further evidence indicating the presence of R-loops during normal CSR in mouse B cells. After removal of the RNA, the DNA strands remain separated due to the strong secondary structure in the nontranscribed strand, likely forming G-quartets [29]. It then follows that the increase in SHM in the Sμ region is easily explained by the continued single stranded character of both the displaced and transcribed DNA strands. The transcribed strand contains a plethora of complementary C’s for AID deamination, which after RNA removal, become available. Thus, AID produces a significant excess of C deaminations that are reflected in the complementary G to A and G to T increase that we documented (Fig. 3c). There was also a small, but insignificant increase in G to C in the Tg cells.
What is the contribution of RNase H1 vs. RNA exosome in removing RNA from R-loops? Our study suggests the RNA exosome is limited in removal of RNA and that patches of residual hybrid limit SHM but not CSR. High levels of RNase H1 in the Tg B cells may remove these hybrid patches allowing AID greater or earlier access to the nontranscribed strand as evidence for the increase in SHM. In the current model for exposing the transcribed DNA strand for AID deamination, removal of the RNA annealed to the transcribed DNA requires the RNA exosome, which gains access to the RNA through interactions of AID, Spt5 and RNA pol II [9, 30]. The RNA exosome is a very abundant complex necessary for degradation of many types of RNAs [31]. How the exosome gains access to the 3′ RNA end required for its activity is unclear. Possibly the small amount of RNase H1 in B cells could provide such ends. The low levels of RNase H1 may also serve to increase stabilities of R-loops present in many other sites in the genome. Our data demonstrate that R-loops exist in vivo and RNase H1 increases access of AID to the template strand. This increases SHM indicating that, in the normal G+C rich context of mammalian class switch recombination, R-loops are obligatory intermediates for CSR.
Materials and Methods
Construction of the Tg mouse
The cDNA of mouse Rnaseh1 gene was inserted in the plasmid P14 TCR β chain cDNA construct B [14] replacing the TCR cDNA. The DNA including the H-2Kb promoter, Rnaseh1, μ-globin splice and polyA and Eμ enhancer (see Fig. 1a) was injected into zygotes at the pronuclear stage as described in results.
Mice
Mice of both genders were used at 2-6 months of age. All animal procedures were reviewed and approved by the Animal Care and Use Committees of the National Institute of Child Health and Human Development, and the National Institute on Aging.
Ex vivo stimulation
Resting splenic B cells were collected by negative selection with anti-CD43 and anti-CD11b magnetic beads (Miltenyi Biotec) and cultured in RPMI media (Invitrogen) containing 10% (v/v) fetal bovine serum (Sigma-Aldrich), 100 U/ml penicillin-streptomycin (Invitrogen), 2 mM glutamine (Invitrogen), and 50 μM β-mercaptoethanol (Sigma-Aldrich). Cells were plated at 0.5 × 106 cells/ml in 24-well plates and stimulated with 5 μg/ml LPS (E. coli serotype 0111:B4; Sigma-Aldrich) to induce IgG3; LPS plus 5 ng/ml recombinant IL4 (Biolegend) for IgG1; LPS plus 2 ng/ml TGF-β (R&D Systems) for IgG2b; and LPS plus 25 ng/ml IFN-γ (R&D Systems) for IgG2c. Flow cytometry analysis of switched populations was conducted after 4 days using cells stained with FITC- or PerCP-labeled anti-B220 (clone RA3-6B2, eBioscience), and either allophycocyanin-conjugated anti-IgG1 (clone M1-14D12, eBioscience), recombinant PE-conjugated IgG2b, IgG2c, or IgG3 antibodies (Southern Biotech).
Westerns and RNase H1 activity
Resting splenic B cells and B cells stimulated by LPS and IL4 as described above were collected and lysed in a buffer containing 50 mM Tris pH 7.5, 10% glycerol, I mM 2-mercaptoethanol, 2% SDS by boiling for five minutes. Equivalent amount of proteins were loaded as determined by using an anti β-actin antibody (abcam). Detection of RNase H1 with an anti RNase H1 antibody (abcam) was performed using the Odissey Clx Imaging System and quantification of the relative amount of proteins was done using the Image Studio Lite software. Proteins of B cells from two different mice for each of C57BL/6, hemizygous and homozygous Tg mice were analyzed. For detection of RNase H1 in different animal tissues, tissues were homogenized to obtain individual cells. Equal numbers of cells of each tissue were boiled in SDS-sample buffer and loaded in gel. Detection was performed using Pierce ECL Plus Western Blotting system.
In-gel RNase H activity assay was performed using the method described previously [16].
PCR analysis
Analysis of transcripts by RT-PCR was conducted following the method described in Kinoshita et al.[32] with modifications. Resting B cells were isolated from mouse spleens by B cells isolation kit (BD bioscience) and stimulated by culturing in complete RPMI medium supplemented with LPS or LPS/IL4. Total RNA was extracted from the stimulated and unstimulated B cells by using Ambion’s RNAqueous-4PCR Total RNA Isolation Kit according to the manufacturer’s instruction. cDNA synthesis and RT-PCR was done using the Avian HS RTPCR Kit from Sigma according to the instruction manual. Reverse transcription was performed by using 2 μg of total RNA in 20 μl reaction volume. RT-PCR was performed by using 4 μl of the reverse transcription reaction mixture in 50 μl reaction volume. PCR was initiated from denaturing step of 94°C for 5 min and followed by 35 cycles of amplification step (94°C for 30s, 58°C for 30s, 68°C for 30s). PCR products were separated by agarose gel electrophoresis. Primer sets for AID, CTγ1 and CTγ3 transcripts are from Kinoshita et al. [32] and for GAPDH, GLTμ and GLTγ1 transcripts are from Reina San Martin et al.[33].
Quantitative RT-PCR was performed using LightCycler 480 SYBR green 1 master mix (Roche) and LightCycler 480 real-time PCR instrument (Roche). The primers used for Rnaseh1 mRNA quantification have been previously described [13].
DNA/RNA immunoprecipitation (DRIP)
DRIP analysis was performed using a modified protocol [20]. B cells were stimulated for 2 days in LPS and IL4. Cells were collected and genomic DNA was isolated by standard proteinase K digestion and phenol chloroform extraction. 100 μg of genomic DNA was treated with S1 nuclease (1U/μl) for 1 hr in 250 μl containing 1x S1 nuclease buffer and 0.3 M NaCl. DNA was precipitated to remove buffer and resuspended in 120 μl TE buffer. Samples were subject to 2 rounds of 7½ minute sonication using a biodisruptor on full power to sheer DNA. 5 μg of genomic DNA was incubated with antibody pre-bound to protein-G dynabeads and incubated for 2 hrs at 4°C. Antibodies against RNA/DNA hybrid, S9.6 was purified from ATCC HB8730. Cells were cultured in 400 ml Iscove’s MDM, 1 ml 500X hypoxanthine and thymidine, 100 ml horse serum and 5 ml Pen/strep. Supernatant was purified over protein G Sepharose column. For RNase H controls, samples were treated prior to pulldown with 12.5 units of RNase H (NEB). S9.6 IgG or control total rabbit IgG (Sigma) were used at 2.5 μg per reaction. Samples were washed sequentially in 1 ml of: 1) FA buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 2% triton-X-100, 0.2% sodium deoxycholate, and 0.2% SDS), 2) FAN buffer (1x FA with 650 mM NaCl), 3) LiCl buffer (10mM Tris, 0.25 M LiCl, 0.5% Igepal (NP-40), 0.5% sodium deoxycholate, and 1mM EDTA), 4) two times in TE buffer for 5 min. at 4°C. Bound fraction was eluted in 300 μl of elution buffer (100 mM sodium bicarbonate and 1% SDS) with incubate at 65°C for 2 hrs while mixing. Eluted material was phenol chloroform extracted and ethanol precipitated. Precipitated material was resuspended in 50 μl nuclease-free H2O. DRIP efficiency was analyzed by real time PCR using Power SYBR green matermix (Invitrogen) using the same primers as ChIP experiments. Percent input was calculated against 1:100 dilution of starting material using traditional ΔCt methods.
Chromatin immunoprecipitation
μChIP experiments were performed as previously described [34]. Briefly, 0.5 million activated B cells were isolated, washed twice in PBS, and crosslinked with 1% formaldehyde for 10 minutes at room temperature. Cells were washed, resuspended in 120 μl lysis buffer, and sonicated to an average DNA size of 500 bp. Chromatin was incubated with antibody pre-bound to protein-G dynabeads (Invitrogen) and incubated for two hours at 4°C. Antibodies against RNA pol II (Millipore) or control total rabbit IgG (Sigma) were used at 2.5 μg per reaction. Beads were washed extensively and bound fraction was isolated by heating in elution buffer. ChIP efficiency was analyzed by realtime PCR using Power SYBR green matermix (Invitrogen) using primers in Sμ; Sg-fwd 5′-CTGCCTACACTGGACTGTTCTGAGC-3′ and Sg-rev 5′-CAGCTCACCCCATCTCACCCCATC-3′, β-actin; Bac-fwd 5′-CACCCGCGAGCACAGCTTC-3′ and Bac-rev 5′-GGATCACTCAGAACGGACACCATG-3′, β-globin; Bglo-fwd 5′-GCCTTGCCTGTTCCTGCTC-3′ and Bglo-rev 5′- CAGACCATAAACTGTATTTTTCTTATT-3′.
Hypermutation analysis
Activated B cells were isolated after 4 day stimulation with LPS and IL4. Purified DNA was subjected to Sμ-Sγ1 junctional amplification as previously described [35]. Mutations were analyzed in sequences which contained no recombination event within the first 500 bp of Sμsequence. Individual mutation frequencies for the full mutation spectra were calculated using SHMTool software [36].
Highlights.
Importance of R-loops in isotype switching in B cells
New transgenic mouse overexpressing RNase H1
Decrease in R-loops does not alter frequency of switching
Somatichypermutation increased on transcribed DNA strand
Acknowledgments
This work was supported the Intramural Research Programs of the National Institutes of Health (NIH), National Institute on Aging (P.J.G.), and National Institute of Child Health and Human Development (R.J.C.). We thank Drs. Ed Max and Aruvathani Arudchandran for help in the early stages of this project, to Dr. Doug Koshland for providing the procedure for DRIP-seq prior to its publication and the lab of Dr. Herbert C. Morse III for many helpful discussion and information.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–51. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- [2].Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- [3].Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin Isotype Switching Is Inhibited and Somatic Hypermutation Perturbed in UNG-Deficient Mice. Curr Biol. 2002;12:1748–55. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- [4].Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, et al. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol. 2011;12:70–6. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Stavnezer J, Schrader CE. IgH Chain Class Switch Recombination: Mechanism and Regulation. J Immunol. 2014;193:5370–8. doi: 10.4049/jimmunol.1401849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Daniels GA, Lieber MR. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23:5006–11. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Basu U, Meng F-L, Keim C, Grinstein V, Pefanis E, Eccleston J, et al. The RNA Exosome Targets the AID Cytidine Deaminase to Both Strands of Transcribed Duplex DNA Substrates. Cell. 2011;144:353–63. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–89. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–97. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- [11].Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–9. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hamperl S, Cimprich KA. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst) 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Suzuki Y, Holmes JB, Cerritelli SM, Sakhuja K, Minczuk M, Holt IJ, et al. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol Cell Biol. 2010;30:5123–34. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pircher H, Mak TW, Lang R, Ballhausen W, Rüedi E, Hengartner H, et al. T cell tolerance to Mlsa encoded antigens in T cell receptor V beta 8.1 chain transgenic mice. EMBO J. 1989;8:719–27. doi: 10.1002/j.1460-2075.1989.tb03431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Overbeek PA. Transgenic Animal Technology. Third Edition Elsevier; London: 2014. 3 - Factors Affecting Transgenic Animal Production A2 - Pinkert, Carl A; pp. 71–107. [Google Scholar]
- [16].Crouch RJ, Arudchandran A, Cerritelli SM. RNase H1 of Saccharomyces cerevisiae: methods and nomenclature. Methods Enzymol. 2001;341:395–413. doi: 10.1016/s0076-6879(01)41166-9. [DOI] [PubMed] [Google Scholar]
- [17].Lee C-G, Kinoshita K, Arudchandran A, Cerritelli SM, Crouch RJ, Honjo T. Quantitative Regulation of Class Switch Recombination by Switch Region Transcription. J Exp Med. 2001;194:365–74. doi: 10.1084/jem.194.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, et al. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–44. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, et al. Activation Induced Cytidine Deaminase Targets DNA at Sites of RNA Polymerase II Stalling by Interaction with Spt5. Cell. 2010;143:122–33. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016;30:1327–38. doi: 10.1101/gad.280834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, et al. Characterization of monoclonal antibody to DNA · RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89:123–30. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- [22].Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res. 2006;34:e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huertas P, Aguilera A. Cotranscriptionally Formed DNA:RNA Hybrids Mediate Transcription Elongation Impairment and Transcription-Associated Recombination. Mol Cell. 2003;12:711–21. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- [24].Poveda AM, Le Clech M, Pasero P. Transcription and replication: breaking the rules of the road causes genomic instability. Transcription. 2010;1:99–102. doi: 10.4161/trns.1.2.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wu H, Lima WF, Zhang H, Fan A, Sun H, Crooke ST. Determination of the Role of the Human RNase H1 in the Pharmacology of DNA-like Antisense Drugs. J Biol Chem. 2004;279:17181–9. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- [26].Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Nat Acad Sci USA. 1995;92:3526–30. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wahba L, Amon Jeremy D, Koshland D, Vuica-Ross M. RNase H and Multiple RNA Biogenesis Factors Cooperate to Prevent RNA:DNA Hybrids from Generating Genome Instability. Mol Cell. 2011;44:978–88. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chédin F, et al. Cotranscriptional R-loops are the main cause of estrogen-induced DNA damage. eLife. 2016;5:e17548. doi: 10.7554/eLife.17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29:3124–33. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Casellas R, Basu U, Yewdell WT, Chaudhuri J, Robbiani DF, Di Noia JM. Mutations, kataegis, and translocations in B lymphocytes: towards a mechanistic understanding of AID promiscuous activity. Nat Rev Immunol. 2016;16:164–76. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol. 2016;17:227–39. doi: 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- [32].Kinoshita K, Harigai M, Fagarasan S, Muramatsu M, Honjo T. A hallmark of active class switch recombination: Transcripts directed by I promoters on looped-out circular DNAs. Pro Nat Acad Sci USA. 2001;98:12620–3. doi: 10.1073/pnas.221454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX Is Required for Recombination Between Immunoglobulin Switch Regions but Not for Intra-Switch Region Recombination or Somatic Hypermutation. J Exp Med. 2003;197:1767–78. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Maul RW, Cao Z, Venkataraman L, Giorgetti CA, Press JL, Denizot Y, et al. Spt5 accumulation at variable genes distinguishes somatic hypermutation in germinal center B cells from ex vivo-activated cells. J Exp Med. 2014;211:2297–306. doi: 10.1084/jem.20131512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wu X, Stavnezer J. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J Exp Med. 2007;204:1677–89. doi: 10.1084/jem.20070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Maccarthy T, Roa S, Scharff MD, Bergman A. SHMTool: a webserver for comparative analysis of somatic hypermutation datasets. DNA Repair (Amst) 2009;8:137–41. doi: 10.1016/j.dnarep.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]