Abstract
The eukaryotic RNA exosome is a well-conserved protein complex with ribonuclease activity implicated in RNA metabolism. Various families of non-coding RNAs (ncRNAs) have been identified as substrates of the complex, underscoring its role as a ncRNA processing/degradation unit. However, the role of RNA exosome and its RNA processing activity on DNA mutagenesis/alteration events have not been investigated until recently. B lymphocytes use two DNA alteration mechanisms, class switch recombination (CSR) and somatic hypermutation (SHM), to re-engineer their antibody gene expressing loci until a tailored antibody gene for a specific antigen is satisfactorily generated. CSR and SHM require the essential activity of the DNA activation induced cytidine deaminase (AID). Causing collateral damage to the B cell genome during CSR and SHM, AID induces unwanted (and sometimes oncogenic) mutations at numerous non-immunoglobulin gene sequences. Recent studies have revealed that AID’s DNA mutator activity is regulated by the RNA exosome complex, thus providing an example of a mechanism that relates DNA mutagenesis with RNA processing. Here, we review the emergent functions of RNA exosome during CSR, SHM and other chromosomal alterations in B cells, and discuss implications relevant to mechanisms that maintain B cell genomic integrity.
Keywords: RNA exosome, AID, class switch recombination, somatic hypermutation, translocation, R-loops, transcription, genome integrity
Graphical abstract
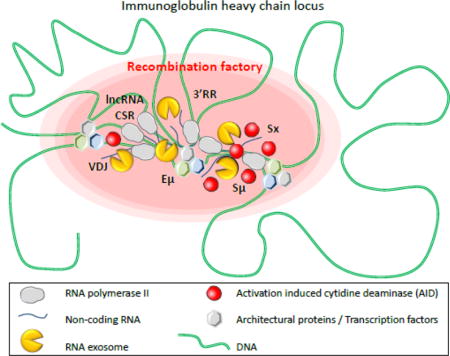
Introduction
Genomic integrity is crucial for cell homeostasis and several mechanisms converge to detect and repair genomes to avoid deleterious effects1. Mutagenic agents can be environmental (UV, chemical, viral, etc.) or endogenous (ROS, replication error, etc.); importantly, transcription itself can induce genome instability by opening chromatin and exposing single strand DNA2. The immune system negates a potentially infinite number of pathogens via mechanisms requiring tailor-made receptors and antibody molecules that recognize pathogen-derived antigens (Ag) with high specificity. However, the number of genes is limited and vastly disproportionate to the number of encoded receptors. Evolution has selected elegant recombination mechanisms to generate Ag receptor diversity from a limited set of genes. In mammals, the first mechanism to create Ag receptor diversity is common to both types of T and B lymphocytes, occurs in the thymus ‘T’ and bone marrow ‘B’ respectively, and is called V(D)J recombination. During V(D)J recombination, one variable (V), eventually one diversity (D), and one junction (J) genes are randomly assembled by recombination activating genes (RAG) recombinases to shape the Ag binding site and a first Ag receptor repertoire3. These recombination steps are tightly regulated by numerous criteria including loci nuclear localization4, transcription factor- and architectural protein-induced chromosomal looping which allows promoter/enhancer interactions and synapsis between recombination sequences5, chromatin remodeling6, and DNA demethylation7. Globally, these changes induce accessible and transcribed chromatin during VDJ recombination in B lymphocytes8–13. As B cell development continues, B cells traverse to secondary lymphoid organs formed in the spleen, tonsil, or gut and undergo a second round of immunoglobulin (Ig) gene diversification steps that depends on the expression of the activation induced cytidine deaminase (AID) protein14–16. This enzyme initiates mutations and double strand breaks (DSB) for somatic hypermutation (SHM) and class switch recombination (CSR) processes respectively, vastly increasing the diversity of B cell Ag receptors (BCR) and antibodies. Again, several layers of regulation ensure spatial and temporal specificity of these recombination steps, now implying the RNA exosome as a master regulator.
Somatic hypermutation, class switch recombination, and B cell fate
Following V(D)J recombination, B cells express a membrane IgM molecule, associated with Igα and Igβ signaling subunits to form the BCR. Transcription initiates from the V promoters to polyadenylation sites of constant region exon(s); thereafter, RNA splicing joins VDJ to IgM (and IgD in immature B cells) exons or VJ to an IgL constant exon for IgH and IgL mature transcripts, respectively. Ig chains are then translated and associate as double heterodimer proteins, with the VDJ and VJ associated to form the variable region, creating an Ag binding site of predefined specificity, while the IgH constant region allows recruitment of specific immune system actors (Figure 1, top). Antigen binding to the BCR, in coordination with co-stimulatory receptors, induces signaling, B cell proliferation, and activation that eventually lead to AID expression. AID introduces mutations at VDJ and VJ genes leading to SHM, and a germinal center-based process leads to selection of somatically mutated B cell clones that have the highest affinity for the Ag, allowing affinity maturation (Figure 1, middle).
Figure 1. Genomic organization of the IgH locus and immunoglobulin structure.
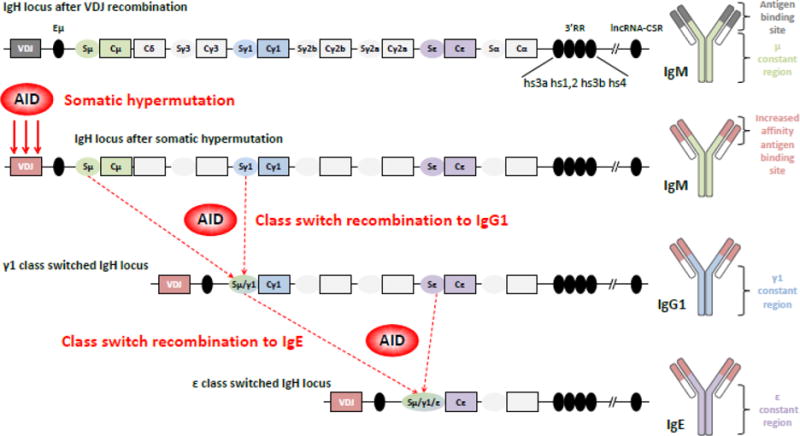
Top. The mouse IgH locus is represented after VDJ recombination (not to scale). The recombined VDJ gene and constant (C) genes are represented as outlined boxes. Horizontally-oriented ovals portray switch (S) regions (preceded by promoters and I exons, not shown) preceding each constant gene (excepted Cδ). Black ovals portray regulatory elements: intronic enhancer μ (Eμ), 3′ regulatory region (3′RR) and the newly identified lncRNA-CSR region, located approximately 2.6 mega-bases downstream of the 3′RR. The resulting immunoglobulin (Ig) protein is IgM, shown on the right.
Middle. After B cell activation, AID is expressed and induces its mutagenic activity on the VDJ gene, allowing production of IgM antibodies with increased affinity for the antigen. Then AID is targeted to S regions to initiate double strand breaks, thereby permitting recombination between two S regions, here Sμ and Sγ1. This process generates high affinity IgG1 antibodies and a diversified B cell repertoire.
Bottom. Cells can re-express AID after re-exposure to antigen and undergo a second class switch recombination event, from γ1 to ε here, to produce high affinity IgE antibodies (direct class switching from μ to ε is also possible). All these AID-mediated events shape the Ig repertoire to improve antigen affinity and to adapt the Ig class for an optimal immune response.
These cells can also undergo CSR after AID activity on switch donor (Sμ) and switch acceptor regions (Sx), leading to DNA double strand breaks. For example, a Th2 orientated immune response induces CSR from IgM to IgG1 to produce high affinity switched antibodies (Figure 1, bottom). These IgG1 class switched cells have the ability to undergo a second round of CSR to IgE, after re-exposure to Ag, to generate high affinity IgE antibodies (Figure 1, bottom), although cells can also directly switch from IgM to IgE to generate low affinity IgE antibodies17, these pathways resulting from a probabilistic transcription of Iγ and Iε non-coding transcripts18. Thus, from one V(D)J recombined clone, AID can generate many unswitched and switched sub-clones with increased Ag affinity, generating Ig diversity and optimal immune responses. Sub-clones differentiate into memory B cells and short-lived or long-lived plasma cells to ensure cellular and humoral memory and to secrete specific antibodies. IgM+ memory B cells persist for long periods of time and are able to re-initiate the germinal center reaction to permit new rounds of SHM and CSR19. Class-switched BCRs govern B cell fate by providing specific signals from their specific intra-cellular tails, globally promoting plasma cell differentiation for IgA20, IgG121,22 and IgE23,24 BCR. The IgE BCR itself dramatically impairs cellular mobility and induces apoptosis25,26 in order to avoid IgE memory which is potentially hazardous, as IgE-class antibodies are implicated in allergic reactions and anaphylactic shock. A new study also has demonstrated that AID, by introducing mutations in V(D)J genes, creates poly-autoreactive cells which are selected against over time, decreasing the longevity of such memory B cells22. Finally, a new AID-dependent recombination event, joining Sμ and switch-like regions located in the 3′RR at the end of the IgH locus, deletes all IgH constant genes and induces B cell apoptosis27, again regulating the B cell compartment. So AID and B cell fate are intrinsically linked (for review28) and precise mechanisms governing AID activity, subsequent B cell outcome, and humoral immune responses are important fields of investigation.
The developmentally-regulated introduction of DNA mutations and breaks into the genome is an unusual process and is potentially catastrophic if not carefully controlled. Thus, AID expression, localization, activity, and targeting need to be carefully regulated by the B cell both intrinsically and within the germinal center environment29–31.
Interestingly immunoglobulin genes themselves have specific features restricting AID activity specifically to sites of DNA recombination. Some of these features, that are relevant to ncRNA transcription and RNA exosome function, are discussed below.
DNA elements in the immunoglobulin loci that express RNA exosome substrate non-coding RNAs
Ig genes include the Ig heavy (IgH) and Ig light chain (IgL) loci located on different chromosomes. IgL genes undergo VJ recombination during early B cell development and SHM in activated B cells while IgH genes undergo VDJ recombination, SHM, and also CSR, allowing the rearrangement of the Ig constant region and replacing Cμ with Cγ, Cα or Cε. The V(D)J genes span approximately 3 million base pairs in their unrecombined (unrearranged, germline) configuration and create a functional V(D)J cassette after V(D)J recombination. Global IgH organization is very complex with a total of 195 VH, 10 DH, 4 JH gene segments32, 8 constant (C) genes preceding by switch (S) sequences (excepted for Cδ), with the entire locus suffused with promoters, enhancers, insulators and recombination sequences (Figure 1 shows a simplified scheme of the mouse IgH locus after VDJ recombination). Main enhancers include the Eμ intronic enhancer, downstream JH genes, and the 3′ regulatory region (3′RR) at the end of the constant genes. Eμ was the first mammalian enhancer described33,34 and mainly acts during the pro-B cell developmental stage by modulating V(D)J recombination35. The 3′RR is considered a B cell specific “super-enhancer” (SE) and is a cluster of multiple DNase hypersensitive site (hs) enhancers: hs3a; hs1,2; hs3b and hs4 (for review36), which can be grouped into two distinct modules (the proximal element including hs3a, hs1,2, hs3b and a palindromic structure, and a distal element including hs4) acting in a relay-race to ensure fine-tuned BCR expression in naïve B cells and Ag dependent locus remodeling in mature stages37. These later events include SHM and CSR which are both severely decreased in the absence of the complete 3′RR super-enhancer38,39, whereas individual or combinatorial deletion of hypersensitivity sites leads to variable but less severe phenotypes40–43. A newly identified regulatory region, lncRNA-CSR, interacting with the 3′RR also has also been discovered approximately 2.6 mega-base pairs downstream (Figure 1)44.
Along with the progression of CSR and SHM, different mandatory non-coding transcripts are induced at S and enhancer regions allowing both AID targeting and concomitant AID substrate generation. A mechanistic role of RNA exosome function in both AID targeting and AID substrate generation is emerging, and these mechanisms depend upon the 3′-end RNA processing/degradation of some transcripts that are expressed at the immunoglobulin loci. We will review specifically these newly identified functions of RNA exosome in B cells, restricting physiological AID activity to Ig genes.
AID recruitment to the genome: legitimate on-targeting versus hazardous off-targeting
It is believed that during interphase chromosomes are confined in specific chromosomal territories of the nucleus45 (Figure 2, top). B cells differentiate into heterogeneous subtypes (pro-B, pre-B, follicular, germinal center, plasmablast, plasma cell, etc.) and can remodel their genomes, making nuclear organization and particularly Ig locus positioning critical. Thus, during B cell development and differentiation, Ig loci are localized in different nuclear territories, undergo contraction and looping, thereby allowing long-range interactions and specific configurations during precisely coordinated time-windows. During CSR both IgH chromosomes are activated, undergo germline transcription46, and are positioned in euchromatin domains47. This phenomenon results in similar AID targeting on both V(D)J genes during hypermutation48,49 and inter-allelic CSR in rabbits50, mice51, and humans52. Inter-chromosomal recombination are also observed between IgH and IgL loci (physiologically targeted by AID) and non-Ig genes (such as c-myc, a frequent off-target of AID and translocated in Burkitt lymphoma),53 via poorly understood mechanisms. Although some studies have found a correlation between nuclear proximity and inter-chromosomal translocations54,55, other complex parameters also have been shown to play major roles. Translocation hotspots have been mapped using different strategies: transcription56 and/or transcription at start site modifications57 and/or level of DNA damage58 at breakpoints as other parameters influence frequency of long distance chromosomal translocations. New findings have further refined these maps of recurrent translocation regions and implicated role of B cell super-enhancers and regulatory clusters59, antisense transcription at bidirectionally transcribed loci leading to ssDNA structures44,60, RNA polymerase collisions during convergent transcription61, or alternative Z-DNA structures62. So rather than only nuclear proximity, other parameters also contribute to AID “on-targeting” and legitimate repair versus “off-targeting” and translocations. Given that lymphocyte populations are highly heterogeneous in term of developmental stage, lifetime, localization, cell-cycle, Ag receptor specificity and Ag exposure (revealed by single-cell analysis of lymphocyte diversity63), it is conceivable that variation will occur in genomic organization and particularly in epigenetic chromatin states between subpopulations. So translocations occurring at different chromosomal positions could reflect this in vivo heterogeneity with specific subsets prone to precise off-targeting.
Figure 2. RNA exosome-dependent resolution of R-loops during class switch recombination.
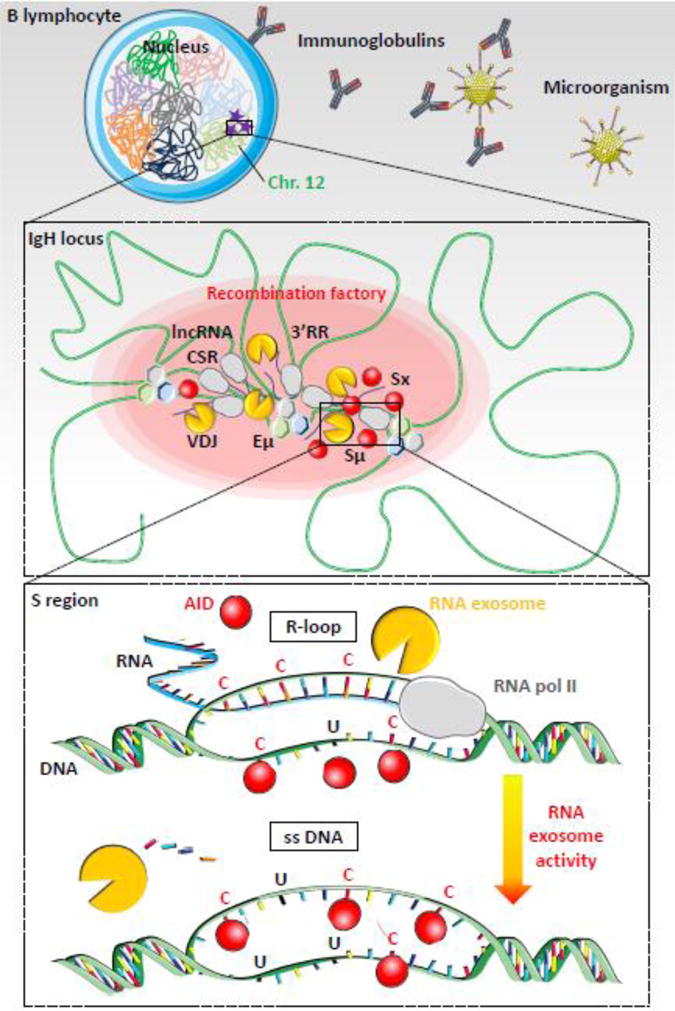
Top. B lymphocyte is shown with nucleus and chromosomes folded in specific chromosomal territories. Mouse IgH loci are located on chromosome 12 (green). AID mutagenic activity is represented as purple stars. Microorganism (for example: an adenovirus) invasion is represented with immunoglobulin production to neutralize pathogen (figure not to scale).
Middle. Chromatin (green) is folded in several loops, induced by transcription factors and architectural proteins, like CTCF, cohesin, mediator or other (hexagons), which bind DNA sequences from distant elements, creating topologically associated domains. Transcription is induced at switch regions and enhancers by RNA polymerase II (grey) producing non-coding RNA (blue lines). RNA exosome (yellow) regulates enhancer activity by degrading eRNA and removes RNA from DNA/RNA hybrids at switch donor (Sμ) and acceptor (Sx) regions. Regulatory elements Eμ, 3′ regulatory region (3′RR), lncRNA-CSR, Iμ and Ix create a hub connecting both switch regions in close conformation and establishing a synapsis for CSR. AID (red) targets switch regions leading to double strand breaks, DNA repair and CSR. These events occur into “recombination factory” (red oval).
Bottom. Switch regions are transcribed by RNA polymerase II (grey). Stalled RNA polII generates RNA/DNA hybrids (R-loops, blue-green hybrid DNA-RNA strand) that prevent DNA accessibility to AID on the transcribed strand. AID indeed targets cytosines (C) on the accessible single strand DNA (ssDNA) of the non-transcribed strand but cannot access the cytosines involved in the R-loops. However, RNA exosome degrades the RNA hybridized to the DNA, generating a new ssDNA and allowing AID accessibility to the second DNA strand. The cytosines are then deaminated to uraciles (U) on both strands and processed by DNA repair mechanisms, leading to double strand breaks in S regions. DNA repair then joins the two S regions to create a new recombined IgH locus utilizing a new constant gene.
Despite all the low off-targeting frequency in the B cell genome, AID targeting still is most robust and specific at the Ig loci genes. Thus, AID activity and Ig chromosomes are intrinsically coupled via one, or potentially many, mechanisms that are not fully understood.
Long-range interactions and DNA element synapsis for optimal CSR
Besides their linear sequences, genes are folded in more complex and specific 3D chromatin structures to ensure fine-tuning of gene expression64. A closer view of chromosomes reveals chromosomal folding into “topologically associated domains” in which long-range interactions between regulatory elements create DNA loops, associating promoters and enhancers and also creating synapsis for VDJ recombination65 and CSR66 (Figure 2, middle). These loops are created by interactions between DNA sequences and transcription factors and/or architectural proteins. For example CTCF-binding elements and CTCF proteins create loops that confine RAG activity during VDJ recombination. Deletion of such elements disrupts RAG on- and off-targeting in B cells67. An intergenic control region, located between V and D genes and containing two CTCF-binding elements, mediates long distance IgH chromosomal loops and ensures balance in Ag receptor repertoire68. Other architectural proteins likewise contribute to IgH locus contraction and long-range interactions at different stages of B cell development and recombination. For example, during CSR, cohesin complex proteins and mediator complex proteins are dynamically recruited to the IgH locus to ensure long-range interactions and to complete CSR, and their absence disregulates CSR69,70. Globally, several proteins are implicated in control of IgH long-range interactions, including Pax5, YY1, EZH2, Ikaros, condensin and the above-mentioned CTCF, cohesin and mediator complexes, at different levels of structural organization71. These factors contribute to IgH locus contraction and interactions of regulatory regions, recombination substrates, and proteins during VDJ recombination and CSR, establishing some “recombination factories” prone to RAG or AID mediated recombination (Figure 2, middle). AID is the mandatory factor for CSR, it initiates the generation of DSB and, due to the consequence of generating DNA lesions, facilitates the recruitment of DNA damage repair factors. Additionally, some studies have suggested that AID, via its C-terminal, plays a direct role in the recruitment of DNA damage repair factors such as DNA-PKcs72. Regulatory elements are also important to control and complete CSR. Insertion of a cassette including S regions at the IgL locus (an AID physiological target), strongly induces AID targeting (mutations and internal deletions to these S regions) but very few long distance CSR-like events are observed between them, probably because of the absence of the 3′RR on the IgL containing chromosome73, suggesting additional levels of regulation to create an efficient synapsis for CSR.
So globally it appears important to create a bona fide “synapsis” for CSR, which implies that specific DNA sequences functionally interact with proteins, regulatory regions, AID and its cofactors, including now the RNA exosome.
The RNA exosome complex: current understanding of structure and function
RNA exosome is an essential protein complex that maintains the homeostasis of various RNA species in the cell. Since the identification of eukaryotic RNA exosome as an RNA processing complex in 1997, it has been demonstrated to be involved in multiple events of RNA metabolism. RNA exosome participates in RNA processing and surveillance mechanisms such as the maturation of rRNAs, snRNAs, snoRNAs, mRNAs and ncRNAs and degradation of aberrant RNAs in the nucleus (Figure 3)74–81. RNA exosome also participates in translational quality control and regulated turn-over in processes such as no go decay (NGD), nonsense-mediated mRNA decay (NMD), nonstop mRNA decay (NSD), deadenylated RNA, histone mRNA and ARE-containing mRNA in the cytoplasm82–90. Although these various RNA processing events have been reported to involve RNA exosome, primarily from literature generated using yeast as a model system, it has been difficult to unravel which effects are caused by RNA exosome deletion and which from the indirect effects of RNA complex deletion. In addition, which of these functions are also seen in mammalian cells that have a disproportionately large non-coding genome, unlike yeast, remains to be systematically investigated.
Figure 3. RNA exosome structure and functions.
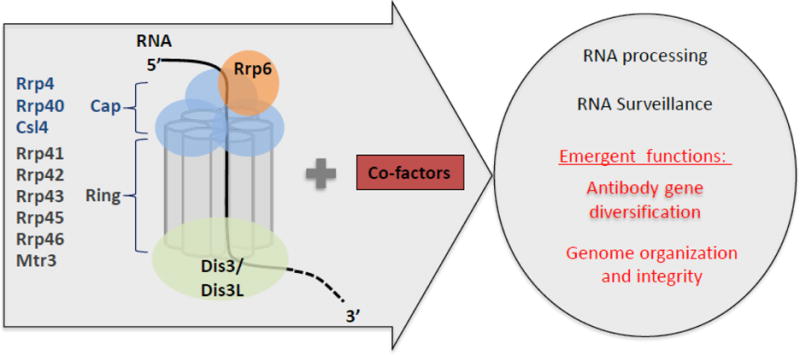
The eukaryotic RNA exosome and cofactor complex are represented in the arrow. The RNA exosome core consists of a ring (Rrp41, Rrp42, Rrp43, Rrp45, Rrp46 and Mtr3) and a cap (Rrp4, Rrp40 and Csl4) structure and two catalytic subunits, Rrp6 and Dis3. General RNA exosome functions (black text) and newly identified functions (red text) are shown in the circle.
Eukaryotic RNA exosome is composed of highly conserved core subunits and catalytic subunits (Figure 3). Core subunits composing the ring structure include Rrp41, Rrp45 (also known as PM/Scl75), Rrp42, Rrp43 (also known as OIP2), Mtr3 and Rrp46 and the cap structure is composed of Rrp4, Rrp40 and Csl491. Catalytic subunits Rrp44 (also known as Dis3) and Rrp6 are bound to core subunits92. Rrp6 bound on the cap structure of the RNA exosome core possesses 3′ to 5′ exonuclease activity and Rrp44 bound “under” the ring structure of the core has both 3′ to 5′ exonuclease and endonuclease activities (Figure 3)93–96. Although eukaryotic RNA exosome core subunits are highly conserved, recent studies have shown substantial differences in catalytic subunits and cofactors between the yeast and human RNA exosome complexes. For example, three human Dis3 isoforms (hDis3, hDis3L1 and hDis3L2) were identified unlike the sole Rrp44/Dis3 moiety in yeast, and hDis3 and hDis3L1 associate with core subunits exclusively97,98. Furthermore, the subunits tend to localize to different compartment, the nucleus for hDis3 and the cytoplasm for hDis3L1 and hDis3L2 while in yeast the sole isoform can position itself in both compartments97,98. Human and yeast Dis3 have both endo- and exo-ribonuclease activities unlike human Dis3L198. hRrp6/PM/Scl100 degrades structured RNA substrates more effectively than yRrp699 and while both human and yeast Rrp6 have the same exonuclease activity, only hRrp6 is localized in nucleus and cytoplasm unlike yeast Rrp6100. Recently, cofactors of human RNA exosome have been identified as being important for targeting various RNA substrates. The nuclear exosome targeting (NEXT) complex containing hMtr4, ZCCHC8 and RBM7 was identified as a human nuclear exosome cofactor, responsible for degrading promoter upstream transcripts (PROMPTs)75. The proposed TRAMP-like complex consisting of hMtr4, ZCCHC7 (Air2p) and PAPD5/hTRF4-2 (Trf4p) in human cells was found to be associated with hRrp6101–103. Rrp47 (C1D), MPP6 and hMtr4 are also known as cofactors which recruit hRrp6 to RNA substrates101,104. Accordingly, it seems that human RNA exosome has functions which have diverged from those observed in yeast RNA exosome, possibly to accommodate the new requirements that evolved with the increased size and complexity of the human genome.
The role of RNA exosome in affecting DNA regulation, either via a transcription-coupled mechanism or via a post-transcriptional mechanism that depends on non-coding RNA processing/trimming with eventual effect on DNA mutagenesis is only beginning to be understood. New mammalian models have now been developed to investigate these questions and to extend the list of RNA exosome functions.
New mouse models to reveal unexpected RNA exosome functions
Mouse models have been generated to evaluate the contribution of different RNA exosome subunits deletion in B lymphocytes and ES cells. These mice express the Exosc10 or Exosc3 COIN (conditional inversion) alleles which are functional before cre-mediated inversion and inactivation44,60. Then a reporter fluorescent protein is expressed after this inversion process, allowing monitoring of the inactivation efficiency as well as sorting of the KO cells. “Exotome” (transcriptome of RNA exosome deficient cells) analyses from these mice reveal accumulation of new RNA exosome substrates including lncRNAs, antisense RNAs and eRNAs. [These data are available in a easily analyzable format on the exotome browser: https://rabadan.c2b2.columbia.edu/cgi-bin/hgGateway?hgsid=258127] Functional studies reveal the importance of RNA exosome in the prevention of genomic instability at eRNAs expressing regions. In the absence of RNA exosome these enhancer sequences accumulate R-loops (detected by DRIP assay) and DNA DSB (evidenced by γH2AX ChIP experiments) and failed to accumulate chromatin silencing markers (H3K9me2 and HP1γ ChIP experiments). Importantly some of these x-eRNAs expressing divergently transcribed enhancers are translocation hot-spots with the IgH locus in B cells (Birc3 and Ncoa3 enhancers). Molecular analyses reveal super-enhancer characteristics of some of these expressing enhancers44.
The lncRNA-CSR has been identified as a regulatory element associated noncoding RNA, using this strategy. The lncRNA-CSR region accumulates transcripts in the absence of Exosc3 or Exosc10 in B cells. This region interacts with hs4 located in the IgH 3′RR SE, as demonstrated directly by 3C experiments. CRISPR/Cas9 mediated deletion of lncRNA-CSR in CH12F3 B cell lines impairs this interaction, decreases germline transcripts at S acceptor region Sα, and CSR to IgA. These results demonstrate for the first time that a very far (~2.6 mega-base pairs) regulatory region, outside of the IgH locus, is able to regulate activity of a super-enhancer and functionally affect CSR44. This region is important to create and/or stabilize chromosomal loops, implicating 3′RR and probably Eμ, Sμ and Sα in CSR synapsis and optimal IgA CSR. Impact of lncRNA-CSR deletion on CSR to other classes has to be investigated in different models (because CH12F3 cells can only switch to IgA). Potentially, other regions may contribute to control of 3′RR activity, making it possible that one region is specific purely for one Ig class or alternatively that one region is specific for all isotypes. The lncRNA-CSR region could also contribute to control of SHM, VDJ recombination, and B cell development and differentiation by modulating 3′RR activity, Eμ activity, or other regulatory regions. Surprisingly, all of the hypersensitivity sites on the 3′RR and the lncRNA-CSR locus accumulate DNA double strand breaks in B cells27,44, that are detectable by “Translocation Capture” technique based assays 44. Thus, these RNA exosome substrate expressing enhancers/super-enhancers are not only vital cogs in antibody gene diversification control mechanisms but also susceptible sites for genomic instability due to programmed DNA recombination induced collateral damage. Interestingly, a long non-coding RNA (ncRNA) has been identified as an important partner to establish and maintain long-range interactions at the HOXA locus105. So, mechanisms of interaction, such as RNA/protein, RNA/RNA or RNA/protein/RNA could be involved in chromosomal looping. RNA exosome could be involved in the control of such non-coding transcripts and could participate in long-range interactions globally across the genome or specifically at the IgH locus during CSR.
RNA exosome cooperates with AID to promote DNA deamination during SHM and CSR
Another prerequisite for CSR is non-coding transcription of S targeted regions, which could be induced after chromosomal looping or participate in loop creation and/or maintenance. This transcription is induced by specific transcription factors that bind I promoters and activate germline transcription106. The resulting germline transcripts spans the I exon (upstream of the S region) as well as the S region, linking transcription, AID targeting, and CSR106. Epigenetic changes are also induced by the 3′RR, in particular at S acceptor regions107 and could participate in folding and/or recruitment of AID and cofactors. It has been proposed that intronic switch ncRNA, generated during this non-coding transcription and be processed into a G-quadruplex RNA structure, can be bound by AID and targets it to S regions in a sequence-specific manner108. The transcription of highly repetitive S regions creates DNA:RNA hybrids, called R-loops, and G-quartets, stabilizing ssDNA of the non-transcribed strand (Figure 2, bottom, upper panel). S regions (1 to 12kb) are prone to form R-loops, due to their highly repetitive G-rich composition, sense and anti-sense transcription109, and frequent RNA polII pausing (brought about by the G-rich composition). Moreover, the suitability of S regions for CSR is determined by their ability to form R-loops and the number of AID target sites110. RNA polII pausing is induced by Spt5111 and contributes to the creation and maintenance of these structures. Importantly, the paused polymerases generate ssDNA, an efficient target for AID. RNA polII abundance also correlates with SHM in germinal center cells112. At this time AID can act on the non-transcribed strand but cannot access the transcribed strand, although access to the transcribed strand is necessary to introduce DSB at S regions.
To better understand this paradoxical situation, AID co-immunoprecipitation experiments were undertaken which identified RNA exosome as an AID partner during CSR113. In fact, as we will see, RNA exosome degrades these hybridized ncRNAs, giving AID access to the DNA and permitting DNA deamination of both DNA strands60,113 (Figure 2, bottom, lower panel). DNA repair then introduces DSB in these S regions and finally CSR results114. This new RNA exosome function allows degradation of the RNA bound to the transcribed strand DNA during CSR through RNA exosome’s 3′-5′ exoribonucleolytic activity. During CSR, RNA exosome is recruited to S regions by an AID-dependent and transcription-dependent mechanism. In vitro deamination assays demonstrate that RNA exosome cooperates with AID to deaminate dsDNA cytidines on SHM substrates (RGYW-rich artificial substrates), R-loops, and core Sμ substrates. Importantly, strand specific assays show that RNA exosome is required to deaminate the template strand and in the absence of RNA exosome (Rrp40 subunit) CSR is decreased in vitro113. Exosc3 mouse model confirms and extends these data in vivo and ex vivo60. These Exosc3 deficient B cells also are defective in CSR ex vivo and SHM in vivo, over-express non-coding germline transcripts, and accumulate RNA:DNA hybrids at S regions, decreasing AID accessibility to the hybridized DNA strand and subsequent CSR60.
Recently, it has been demonstrated that antisense transcription can promote R-loops115. Genome-wide analysis further reveals that R-loops are enriched at sites expressing antisense RNA116 and are sensitive to RNA exosome. Globally, R-loops are more widespread and have more functions than previously predicted (for review117). Importantly, the study of Exosc3 deficient transcriptomes has revealed new RNA exosome substrates, including transcription start site-associated antisense transcripts (xTSS-RNAs) and antisense RNAs inside the bodies of genes (asRNAs) that can be longer than 500 bps and are transcribed bidirectionally/divergently from cognate coding gene transcripts. Strikingly these xTSS-RNAs and asRNAs are associated with AID off-target translocation hot-spots (and other sites of DNA double strand breaks) across the B cell genome. ChIP experiments demonstrate AID recruitment to some of these divergently transcribed regions. In the absence of RNA exosome these promoters and gene bodies accumulate R-loops, directly detected by DRIP assays or indirectly by H3S10ph ChIP or RPA-seq data (RPA binds to DNA breaks118), and deletion of these regions decreases AID off-targeting60. So RNA exosome has an important function in removing non-coding transcripts initiating from bidirectionally transcribed regions, thereby avoiding R-loop accumulation, and accordingly limiting AID off-targeting, and genomic instability in these regions.
AID-independent mechanisms of R-loop/RNA exosome associated DNA damage in B cells
DNA/RNA hybrids induced genome structural alterations, like R-loops or G-quartets, expose single-strand DNA to enzymes, such as AID in B cells, but also to other deaminases such as the APOBEC family119 or various mutagenic agents120, inducing genome mutagenesis. R-loops can also create genome instability by mechanisms associated with transcription-associated recombination (for review121) and those that are eventually mediated by transcription-coupled nucleotide excision repair122. Finally R-loop structures can induce RNA pol II stalling and collision with replisome, in head-on or co-directional orientation, perturbing fork progression123 and inducing DNA damage and genomic instability124. Different factors contribute to remove R-loops, avoid their accumulation and these deleterious effects, including topoisomerases, helicases, RNase H family of enzymes (for reviews117,125) and the relationship of these pathways with RNA exosome function will be an important topic of investigation in the future. It is likely that many mutations, DNA double strand breaks and translocations seen in the B cell genome are not associated with AID’s DNA mutagenic activity; and conversely, RNA exosome mediated processing of ncRNAs in the B cell genome may be equally important for preventing DNA insults by various other mechanisms that are not directly associated with AID in B cells44.
Conclusion
RNA exosome functions were first established in the context of yeast and Drosophila studies, revealing important roles in RNA metabolism and processing. Investigating RNA exosome’s contributions in mammalian B cells during CSR reveals new and unexpected functions as an AID mandatory co-factor for optimal physiological DNA recombination. Conditional KO of different RNA exosome subunits in ES cells and B lymphocytes demonstrates its crucial role in maintenance of genome stability, chromatin modification, and control of enhancer activity. Further investigations should extend the list of RNA exosome co-factors and functions in both global and specific biological pathways.
Acknowledgments
We would like to acknowledge European Commission (LTFCOFUND2013, GA-2013-609409) support from Marie Curie Actions for BL’s fellowship (EMBO fellowship ALTF 906-2015). We acknowledge Servier Medical Art (http://www.servier.fr/smart/banque-dimages-powerpoint) for illustration database. During the preparation of this article, research in the Basu laboratory was funded by grants from NIAID (4RO1A1099195-01A1), NIH (1DP2OD008651-01), the Leukemia and Lymphoma Society of America and the Pershing Square Sohn Cancer Research Alliance.
Abbreviations used
- AID
activation induced cytidine deaminase
- CSR
class switch recombination
- SHM
somatic hypermutation
- Ag
antigen
- Ig
immunoglobulin
- BCR
B cell Ag receptor
- hs
DNase hypersensitive site
- ncRNA
non-coding RNA
- S
switch
- SE
super-enhancer
- DSB
double strand break
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network. Nat Rev Genet. 2013;14:62–75. doi: 10.1038/nrg3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaillard H, Aguilera A. Transcription as a Threat to Genome Integrity. Annu Rev Biochem. 2016;85:291–317. doi: 10.1146/annurev-biochem-060815-014908. [DOI] [PubMed] [Google Scholar]
- 3.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 4.Chaumeil J, Skok JA. A new take on v(d)j recombination: transcription driven nuclear and chromatin reorganization in rag-mediated cleavage. Front Immunol. 2013;4:423. doi: 10.3389/fimmu.2013.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feeney AJ. Epigenetic regulation of antigen receptor gene rearrangement. Curr Opin Immunol. 2011;23:171–177. doi: 10.1016/j.coi.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin-Klein R, Bergman Y. Epigenetic regulation of monoallelic rearrangement (allelic exclusion) of antigen receptor genes. Front Immunol. 2014;5:625. doi: 10.3389/fimmu.2014.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reth MG, Alt FW. Novel immunoglobulin heavy chains are produced from DJH gene segment rearrangements in lymphoid cells. Nature. 1984;312:418–423. doi: 10.1038/312418a0. [DOI] [PubMed] [Google Scholar]
- 9.Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 10.Corcoran AE, Riddell A, Krooshoop D, Venkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- 11.Bolland DJ, et al. Antisense intergenic transcription in V(D)J recombination. Nat Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 12.Bolland DJ, et al. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol Cell Biol. 2007;27:5523–5533. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi NM, et al. Deep sequencing of the murine IgH repertoire reveals complex regulation of nonrandom V gene rearrangement frequencies. J Immunol. 2013;191:2393–2402. doi: 10.4049/jimmunol.1301279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muramatsu M, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 15.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 16.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 17.Xiong H, Dolpady J, Wabl M, Curotto de Lafaille MA, Lafaille JJ. Sequential class switching is required for the generation of high affinity IgE antibodies. J Exp Med. 2012;209:353–364. doi: 10.1084/jem.20111941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu YL, Stubbington MJT, Daly M, Teichmann SA, Rada C. Intrinsic transcriptional heterogeneity in B cells controls early class switching to IgE. J Exp Med. 2016 doi: 10.1084/jem.20161056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dogan I, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 20.Duchez S, et al. Premature replacement of mu with alpha immunoglobulin chains impairs lymphopoiesis and mucosal homing but promotes plasma cell maturation. Proc Natl Acad Sci USA. 2010;107:3064–3069. doi: 10.1073/pnas.0912393107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin SW, Goodnow CC. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nat Immunol. 2002;3:182–188. doi: 10.1038/ni752. [DOI] [PubMed] [Google Scholar]
- 22.Gitlin AD, et al. Independent Roles of Switching and Hypermutation in the Development and Persistence of B Lymphocyte Memory. Immunity. 2016;44:769–781. doi: 10.1016/j.immuni.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erazo A, et al. Unique maturation program of the IgE response in vivo. Immunity. 2007;26:191–203. doi: 10.1016/j.immuni.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Sullivan BM, Allen CDC. Fluorescent in vivo detection reveals that IgE(+) B cells are restrained by an intrinsic cell fate predisposition. Immunity. 2012;36:857–872. doi: 10.1016/j.immuni.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 25.He JS, et al. The distinctive germinal center phase of IgE+ B lymphocytes limits their contribution to the classical memory response. J Exp Med. 2013;210:2755–2771. doi: 10.1084/jem.20131539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laffleur B, et al. Self-Restrained B Cells Arise following Membrane IgE Expression. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 27.Péron S, et al. AID-driven deletion causes immunoglobulin heavy chain locus suicide recombination in B cells. Science. 2012;336:931–934. doi: 10.1126/science.1218692. [DOI] [PubMed] [Google Scholar]
- 28.Laffleur B, et al. AID-induced remodeling of immunoglobulin genes and B cell fate. Oncotarget. 2014;5:1118–1131. doi: 10.18632/oncotarget.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patenaude AM, Di Noia JM. The mechanisms regulating the subcellular localization of AID. Nucleus. 2010;1:325–331. doi: 10.4161/nucl.1.4.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basu U, Chaudhuri J, Phan RT, Datta A, Alt FW. Regulation of activation induced deaminase via phosphorylation. Adv Exp Med Biol. 2007;596:129–137. doi: 10.1007/0-387-46530-8_11. [DOI] [PubMed] [Google Scholar]
- 31.Keim C, Kazadi D, Rothschild G, Basu U. Regulation of AID, the B-cell genome mutator. Genes Dev. 2013;27:1–17. doi: 10.1101/gad.200014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- 33.Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 34.Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 35.Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci USA. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinaud E, et al. The IgH locus 3′ regulatory region: pulling the strings from behind. Adv Immunol. 2011;110:27–70. doi: 10.1016/B978-0-12-387663-8.00002-8. [DOI] [PubMed] [Google Scholar]
- 37.Garot A, et al. Sequential activation and distinct functions for distal and proximal modules within the IgH 3′ regulatory region. Proc Natl Acad Sci USA. 2016;113:1618–1623. doi: 10.1073/pnas.1514090113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent-Fabert C, et al. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010;116:1895–1898. doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- 39.Rouaud P, et al. The IgH 3′ regulatory region controls somatic hypermutation in germinal center B cells. J Exp Med. 2013;210:1501–1507. doi: 10.1084/jem.20130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manis JP, et al. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J Exp Med. 1998;188:1421–1431. doi: 10.1084/jem.188.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bébin AG, et al. In vivo redundant function of the 3′ IgH regulatory element HS3b in the mouse. J Immunol. 2010;184:3710–3717. doi: 10.4049/jimmunol.0901978. [DOI] [PubMed] [Google Scholar]
- 42.Vincent-Fabert C, et al. Ig synthesis and class switching do not require the presence of the hs4 enhancer in the 3′ IgH regulatory region. J Immunol. 2009;182:6926–6932. doi: 10.4049/jimmunol.0900214. [DOI] [PubMed] [Google Scholar]
- 43.Pinaud E, et al. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15:187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- 44.Pefanis E, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 46.Delpy L, Le Bert M, Cogné M, Khamlichi AA. Germ-line transcription occurs on both the functional and the non-functional alleles of immunoglobulin constant heavy chain genes. Eur J Immunol. 2003;33:2108–2113. doi: 10.1002/eji.200323969. [DOI] [PubMed] [Google Scholar]
- 47.Skok JA, et al. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- 48.Delpy L, Sirac C, Le Morvan C, Cogné M. Transcription-dependent somatic hypermutation occurs at similar levels on functional and nonfunctional rearranged IgH alleles. J Immunol. 2004;173:1842–1848. doi: 10.4049/jimmunol.173.3.1842. [DOI] [PubMed] [Google Scholar]
- 49.Yeap LS, et al. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell. 2015;163:1124–1137. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kingzette M, Spieker-Polet H, Yam PC, Zhai SK, Knight KL. Trans-chromosomal recombination within the Ig heavy chain switch region in B lymphocytes. Proc Natl Acad Sci USA. 1998;95:11840–11845. doi: 10.1073/pnas.95.20.11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynaud S, et al. Interallelic class switch recombination contributes significantly to class switching in mouse B cells. J Immunol. 2005;174:6176–6183. doi: 10.4049/jimmunol.174.10.6176. [DOI] [PubMed] [Google Scholar]
- 52.Laffleur B, et al. Immunoglobulin genes undergo legitimate repair in human B cells not only after cis- but also frequent trans-class switch recombination. Genes Immun. 2014;15:341–346. doi: 10.1038/gene.2014.25. [DOI] [PubMed] [Google Scholar]
- 53.Wang JH, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rocha PP, et al. Close proximity to Igh is a contributing factor to AID-mediated translocations. Mol Cell. 2012;47:873–885. doi: 10.1016/j.molcel.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sklyar I, et al. Distinct Patterns of Colocalization of the CCND1 and CMYC Genes With Their Potential Translocation Partner IGH at Successive Stages of B-Cell Differentiation. J Cell Biochem. 2016;117:1506–1510. doi: 10.1002/jcb.25516. [DOI] [PubMed] [Google Scholar]
- 56.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qian J, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pefanis E, et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meng FL, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khair L, Baker RE, Linehan EK, Schrader CE, Stavnezer J. Nbs1 ChIP-Seq Identifies Off-Target DNA Double-Strand Breaks Induced by AID in Activated Splenic B Cells. PLoS Genet. 2015;11:e1005438. doi: 10.1371/journal.pgen.1005438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 64.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montefiori L, et al. Extremely Long-Range Chromatin Loops Link Topological Domains to Facilitate a Diverse Antibody Repertoire. Cell Rep. 2016;14:896–906. doi: 10.1016/j.celrep.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wuerffel R, et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu J, et al. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 2015;163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas-Claudepierre AS, et al. The cohesin complex regulates immunoglobulin class switch recombination. J Exp Med. 2013;210:2495–2502. doi: 10.1084/jem.20130166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas-Claudepierre AS, et al. Mediator facilitates transcriptional activation and dynamic long-range contacts at the IgH locus during class switch recombination. J Exp Med. 2016;213:303–312. doi: 10.1084/jem.20141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerasimova T, et al. A structural hierarchy mediated by multiple nuclear factors establishes IgH locus conformation. Genes Dev. 2015;29:1683–1695. doi: 10.1101/gad.263871.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zahn A, et al. Activation induced deaminase C-terminal domain links DNA breaks to end protection and repair during class switch recombination. PNAS. 2014;111:E988–E997. doi: 10.1073/pnas.1320486111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bonaud A, et al. Efficient AID targeting of switch regions is not sufficient for optimal class switch recombination. Nat Commun. 2015;6:7613. doi: 10.1038/ncomms8613. [DOI] [PubMed] [Google Scholar]
- 74.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8 S rRNA in Saccharomyces cerevisiae. The EMBO Journal. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lubas M, et al. Interaction Profiling Identifies the Human Nuclear Exosome Targeting Complex. Molecular Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 76.Chlebowski A, Lubas M, Jensen TH, Dziembowski A. RNA decay machines: The exosome. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms. 2013;1829:552–560. doi: 10.1016/j.bbagrm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 77.Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. Transcriptome-wide Analysis of Exosome Targets. Molecular Cell. 2012;48:422–433. doi: 10.1016/j.molcel.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmid M, et al. Rrp6p Controls mRNA Poly(A) Tail Length and Its Decoration with Poly(A) Binding Proteins. Molecular Cell. 2012;47:267–280. doi: 10.1016/j.molcel.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rougemaille M, et al. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Dijk EL, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 81.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 82.Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Orban TI. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Araki Y, et al. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nature Structural & Molecular Biology. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nicholson P, Mühlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem Soc Trans. 2010;38:1615–1620. doi: 10.1042/BST0381615. [DOI] [PubMed] [Google Scholar]
- 87.van Hoof A, Staples RR, Baker RE, Parker R. Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol Cell Biol. 2000;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vasudevan S, Peltz SW, Wilusz CJ. Non-stop decay? a new mRNA surveillance pathway. BioEssays. 2002;24:785–788. doi: 10.1002/bies.10153. [DOI] [PubMed] [Google Scholar]
- 89.Tsuboi T, et al. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 90.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Q, Greimann JC, Lima CD. Reconstitution, Activities, and Structure of the Eukaryotic RNA Exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 92.Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nature Structural & Molecular Biology. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 93.Chlebowski A, Tomecki R, Gas López ME, Séraphin B, Dziembowski A. Catalytic properties of the eukaryotic exosome. Adv Exp Med Biol. 2011;702:63–78. doi: 10.1007/978-1-4419-7841-7_6. [DOI] [PubMed] [Google Scholar]
- 94.Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A. The eukaryotic RNA exosome: Same scaffold but variable catalytic subunits. RNA Biology. 2011;8:61–66. doi: 10.4161/rna.8.1.14237. [DOI] [PubMed] [Google Scholar]
- 95.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→ 5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 96.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends in Biochemical Sciences. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 97.Staals RH, et al. Dis3-like 1: a novel exoribonuclease associated with the human exosome. The EMBO journal. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tomecki R, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Januszyk K, Liu Q, Lima CD. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA. 2011;17:1566–1577. doi: 10.1261/rna.2763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Dijk EL, Schilders G, Pruijn GJM. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schilders G, van Dijk E, Pruijn GJM. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Research. 2007;35:2564–2572. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fasken MB, et al. Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex. J Biol Chem. 2011;286:37429–37445. doi: 10.1074/jbc.M111.271494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lubas M, et al. Interaction Profiling Identifies the Human Nuclear Exosome Targeting Complex. Molecular Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 104.Schilders G. MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Research. 2005;33:6795–6804. doi: 10.1093/nar/gki982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nwigwe IJ, Kim YJ, Wacker DA, Kim TH. Boundary Associated Long Noncoding RNA Mediates Long-Range Chromosomal Interactions. PLoS ONE. 2015;10:e0136104. doi: 10.1371/journal.pone.0136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 107.Saintamand A, et al. Elucidation of IgH 3′ region regulatory role during class switch recombination via germline deletion. Nat Commun. 2015;6:7084. doi: 10.1038/ncomms8084. [DOI] [PubMed] [Google Scholar]
- 108.Zheng S, et al. Non-coding RNA Generated following Lariat Debranching Mediates Targeting of AID to DNA. Cell. 2015;161:762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Perlot T, Li G, Alt FW. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions. Proc Natl Acad Sci USA. 2008;105:3843–3848. doi: 10.1073/pnas.0712291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang ZZ, et al. The strength of an Ig switch region is determined by its ability to drive R loop formation and its number of WGCW sites. Cell Rep. 2014;8:557–569. doi: 10.1016/j.celrep.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maul RW, et al. Spt5 accumulation at variable genes distinguishes somatic hypermutation in germinal center B cells from ex vivo–activated cells. J Exp Med. 2014;211:2297–2306. doi: 10.1084/jem.20131512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Basu U, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Daniel JA, Nussenzweig A. The AID-induced DNA damage response in chromatin. Mol Cell. 2013;50:309–321. doi: 10.1016/j.molcel.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boque-Sastre R, et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci USA. 2015;112:5785–5790. doi: 10.1073/pnas.1421197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chan YA, et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10:e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 118.Yamane A, et al. RPA accumulation during class switch recombination represents 5′-3′ DNA end resection during the S-G2/M phase of the cell cycle. Cell Rep. 2013;3:138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Knisbacher BA, Gerber D, Levanon EY. DNA Editing by APOBECs: A Genomic Preserver and Transformer. Trends Genet. 2016;32:16–28. doi: 10.1016/j.tig.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 120.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sollier J, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gan W, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stirling PC, et al. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hamperl S, Cimprich KA. Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell. 2016;167:1455–1467. doi: 10.1016/j.cell.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]


