Abstract
Enterococcus mundtii strains isolated from the larval feces of the Mediterranean flour moth Ephestia kuehniella show antimicrobial activity against a broad spectrum of Gram-positive and Gram-negative bacteria. The in vitro probiotic characterization of one isolate revealed a high auto-aggregation score, a hydrophilic cell surface, tolerance for low pH, no hemolytic activity, and susceptibility to all tested antibiotics. We used the red flour beetle Tribolium castaneum, an established model organism, for the in vivo characterization of one probiotic E. mundtii isolate from E. kuehniella larvae. Tribolium castaneum larvae were fed orally with the probiotic isolate or the corresponding supernatant and then infected with either the entomopathogen Bacillus thuringiensis or Pseudomonas entomophila. Larvae exposed to the isolate or the supernatant showed increased survival following infection with B. thuringiensis but not P. entomophila. Heat treatment or treatment with proteinase K reduced the probiotic effect of the supernatant. However, the increased resistance attracts a fitness penalty manifested as a shorter lifespan and reduced fertility. T. castaneum has, pending on further research, the potential as an alternative model for the pre-screening of probiotics.
Keywords: Tribolium castaneum, probiotics, Enterococcus mundtii, in vivo model, antimicrobial, Bacillus thuringiensis
Introduction
All animals are associated with a diverse microbial community that promotes their health (McFall-Ngai et al., 2013; Sommer and Bäckhed, 2013). Any disruption to the population of gut microbiota caused by antibiotics or immune system deficiency therefore reduces the fitness of the host (Lozupone et al., 2012; Modi et al., 2014) whereas the administration of probiotic bacteria can increase fitness (Buffie and Pamer, 2013). Probiotics are defined as “live microorganisms, that when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014).
Probiotics have many applications, including the optimization of growth and survival in animal species used for aquaculture and agriculture (Chaucheyras-Durand and Durand, 2009; Pandiyan et al., 2013), and the prevention or treatment of gastrointestinal tract infections in humans (Deshpande et al., 2010; Kotzampassi and Giamarellos-Bourboulis, 2012). Probiotics have several mechanisms of action, including the production of antimicrobial compounds, the inhibition of virulence genes, the enhancing of epithelial barrier functions or the stimulation of the host immune system (Oelschlaeger, 2010). The best-studied microorganisms with probiotic activity are the bifidobacteria, lactobacilli, enterococci and yeasts (Varankovich et al., 2015). The genus Enterococcus (order lactobacillales) is a controversial group that contains both probiotic strains (Hosseini et al., 2009; Strompfová and Lauková, 2009; Al Atya et al., 2015) and pathogenic strains (Higashide et al., 2005; Martin et al., 2005; Gaspar et al., 2009). Enterococci produce organic acids, hydrogen peroxide and up to four different classes of enterocins (Franz et al., 2007), supporting the call for a legislative framework for probiotics (Papadimitriou et al., 2015).
The adult gut microbiota of mammals is essential for the development of the immune system in the offspring (Round and Mazmanian, 2009; Gomez de Aguero et al., 2016). Animals have evolved different ways to transfer beneficial microbes to their offspring, e.g., female mammals can transfer their own beneficial microbes through milk during lactation (Jost et al., 2014). Birds can transfer their microbes via the eggshell or regurgitation (Godoy-Vitorino et al., 2010; Ruiz-De-Castañeda et al., 2011; Kohl, 2012), and insects can transfer beneficial microbes by trophallaxis or coprophagy (Koch and Schmid-Hempel, 2011; Engel and Moran, 2013; Brune and Dietrich, 2015). Insect feces are not only relevant for microbial transmission but they also fulfill a protective function (Rosengaus et al., 2013; Diehl et al., 2015). This is particularly relevant in the case of storage pests, which defecate and live in the same environment.
In vitro assays for the screening of probiotic microorganisms typically test for antimicrobial activity, microbial colonization, and safety. In vivo assays are also recommended, because probiotics can negatively affect certain host species, as observed in honeybees (Ptaszyńska et al., 2015) and in humans (Besselink et al., 2008). The regulatory framework governing probiotics varies in different countries, making it difficult to find standardized methods (Baldi and Arora, 2015). Caenorhabditis elegans, Drosophila melanogaster, and Galleria mellonella are currently used for the preclinical screening of probiotics, because they are suitable for large-scale screening therefore reduce costs compared to mammalian models (Papadimitriou et al., 2015; Vilela et al., 2015). Invertebrates are also more suitable for ethical reasons and with respect to the 3Rs strategy (Russell and Burch, 1959). Tribolium castaneum is a well-established insect model organism, which is easy and inexpensive to rear in the laboratory. T. castaneum is currently used as a model for infection, for transgenerational effects and for the screening of drugs (Zou et al., 2007; Roth et al., 2010; Milutinović et al., 2014; Bingsohn et al., 2016) but not thus far for the screening of probiotics. This model insect is well-suited for screening based on functional genomics because it benefits from a sequenced genome (Tribolium Genome Sequencing Consortium et al., 2008), well-established RNA interference (RNAi) techniques (Bucher et al., 2002; Knorr et al., 2013), a recently established CRISPER/Cas system for gene knockout (Gilles et al., 2015), and the availability of transcriptome datasets generated under various conditions (Park et al., 2008; Altincicek et al., 2013; Dippel et al., 2014). It is also possible to generate axenic strains, which are useful for the analysis of probiotic microbes in isolation (Futo et al., 2016).
Here, we report the in vitro probiotic characterization of Enterococcus mundtii isolates, sourced from the feces of the Mediterranean flour moth Ephestia kuehniella, a common storage pest. We also investigated the in vivo protective role of one E. mundtii isolate and the corresponding supernatant by oral administration to T. castaneum before challenging the beetles with different entomopathogenic bacteria. We conclude that T. castaneum is suitable as a high-throughput screening platform for the in vivo testing of potential probiotics.
Materials and Methods
Insect Rearing
Ephestia kuehniella larvae (provided by the Julius Kühn-Institut, Berlin, Germany) were kept in glass jars at room temperature in darkness, and were fed on wheat grains (Alnatura, Bickenbach, Germany). T. castaneum Cro1 beetles collected in 2010 (Milutinović et al., 2013) were maintained on a heat-sterilized standard diet of wheat flour (type 550, Alnatura, Bickenbach, Germany) and 5% brewer’s yeast at 32°C and 70% humidity in darkness.
Bacterial Isolation and Identification
Bacteria were isolated from the feces of E. kuehniella larvae. Feces were plated with a spatula tip onto casein soya agar and incubated for 48 h at 30°C. Randomly chosen colonies were re-streaked on de Man, Rogosa and Sharpe (MRS) agar to obtain pure isolates. Isolated strains were used to prepare glycerol stocks at -20°C. All isolates were identified as strains of E. mundtii based on 16S rDNA analysis. Bacterial DNA was amplified using primers p8FPL 5′-AGT TTG ATC CTG GCT CAG-3′ and p806R 5′-GGA CTA CCA GGG TAT CTA AT-3′ (Relman et al., 1992) yielding a product of ∼800 bp. The following PCR program was used: 94°C/7 min; 35 cycles at 94°C/60 s, 55°C/60 s, 72°C/60 s; and a final extension step at 72°C/10 min. PCR products were separated by 1% agarose gel electrophoresis and stained with SYBR Safe (Thermo Fisher Scientific, Waltham, MA, United States). Prior to sequencing (Macrogen Europe, Amsterdam, Netherlands), the PCR products were purified using a DNA purification kit (Macherey-Nagel, Düren, Germany). Sequences were deposited at GenBank (Supplementary Table S1). A bacterial phylogenetic tree was created using the neighbor-joining method (Saitou and Nei, 1987) by aligning the 16S rDNA sequences against known sequences in the NCBI database using MAFFT v7 (Katoh and Standley, 2013).
Antimicrobial Characterization
The E. mundtii isolates were first screened using the agar spot on lawn technique (Schillinger and Lücke, 1989) with the following modifications for antimicrobial activity. Overnight cultures of the isolates grown in MRS medium at 30°C were spotted (7 μl) onto 0.7% MRS agar plates and incubated at 30°C for 24 h under aerobic conditions. The indicator bacteria (Table 1) comprised an overnight culture mixed with 1% lysogeny broth (LB) agar at a final concentration of 1 × 105 cells ml-1. Previously spotted isolates were overlaid with 10 ml of the indicator bacteria. After complete solidification of the upper layer, the plates were incubated for an additional 16 h at 30°C under aerobic conditions. Inhibition was scored positive if the radius of the zone of inhibition around the colonies of the isolates was 1 mm or larger. We carried out three replicates per isolate.
Table 1.
Agar spot on lawn test with E. mundtii isolates against indicator bacteria.
| Isolated E. mundtii strains | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Indicator strains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| Bacillus thuringiensis (DSM 2046) | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Escherichia coli (strain D 31) | + | + | + | + | + | ++ | + | + | + | + | + | + | + | + | + |
| Klebsiella terrigena (DSM 2687) | + | ++ | ++ | ++ | ++ | + | + | + | + | + | + | + | ++ | + | ++ |
| Listeria grayi (DSM 20601) | + | + | + | - | + | + | - | - | + | + | - | - | - | + | + |
| Listeria innocua (DSM 20649) | + | + | + | - | - | + | - | - | - | + | - | - | + | + | + |
| Listeria seeligeri (DSM 20751) | + | - | - | + | + | - | - | - | + | - | - | + | + | + | |
| Micrococcus luteus (DSM 20030) | + | ++ | ++ | ++ | ++ | + | ++ | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ |
| Pseudomonas aeruginosa (DSM 50071) | + | + | - | + | - | - | + | - | - | ++ | - | + | - | + | + |
| Pseudomonas entomophila (DSM 28517) | + | ++ | ++ | + | + | + | + | + | + | + | + | + | ++ | + | ++ |
| Salmonella subterranean (DSM 1620) | + | - | + | + | + | + | - | - | - | + | - | - | + | + | + |
| Staphylococcus aureus (DSM 2569) | + | + | - | - | - | - | - | - | - | - | - | - | - | + | + |
Positive inhibition was scored if the radius of the zone of inhibition around the colonies of the isolates was 3–5.99 mm (++) or 1– 2.99 mm (+) and negative inhibition (-) was scored if the zone was 0–0.99 mm. Scores are based on three replicates. The DSM number refers to strains obtained from the DSMZ strain collection, Braunschweig, Germany.
Inhibitory substances from the isolates were further characterized by the agar well-diffusion assay (AWDA) (Tagg and McGiven, 1971) with the following modifications. E. mundtii isolates were grown under aerobic conditions for 24 h at 30°C in MRS medium. The cell free supernatant (CFS) was obtained by centrifugation (3200 × g, 4°C, 15 min), adjusted to pH 6.5 and passed through a 0.22-μm filter (Carl Roth, Karlsruhe, Germany). Afterward the CFS was concentrated 100-fold, mixed with ethyl acetate (1:1), and shaken vigorously for 1 h. The mixture was stabilized for a further 1 h and then the aqueous phase was removed. The solvent was concentrated under reduced pressure in a rotational evaporator (Büchi Labortechnik AG, Switzerland). To reduce the effect of proteinaceous compounds, 2 ml concentrated CFS was mixed with 80 μl 25 mg ml-1 proteinase K (Sigma-Aldrich, Taufkirchen, Germany) for 1 h at 37°C. To reduce the effect of H2O2, 2 ml concentrated CFS was treated with 40 μl 2 mg ml-1 catalase (Sigma) for 1 h at 37°C. Finally, the heat stability of CFS was tested by heating for 10 min at 98°C. For the AWDA, 40 μl of CFS treated in one of the three ways described above was transferred to 5-mm wells in 1% LB agar plates containing 105 indicator bacteria ml-1. The plates were incubated at 30°C for 16 h. The zones of inhibition were measured in mm using a digital caliper, with three replicates per isolate. Sterile concentrated MRS medium was used as a negative control.
Probiotic Characterization
Auto-aggregation correlates based on bacterial adhesion to host cells were determined as described (Del Re et al., 2000; Al Kassaa et al., 2014). E. mundtii isolates were grown in MRS medium for 16 h at 30°C. Cells were harvested by centrifugation (3200 g, 4°C, 15 min) and washed twice with sterile phosphate buffered saline (PBS; pH 7), re-suspended in PBS and adjusted to 108 cells ml-1. The cell suspension (4 ml) was mixed by vortexing and incubated at room temperature for 24 h. After 5 and 24 h, 100 μl of the upper phase was removed and the absorbance at 600 nm was measured. The percentage of auto-aggregation was calculated as follows:
where OD1 is the optical density at 5 or 24 h and OD0 is the optical density at time point zero. This experiment was performed in triplicate.
To detect bacterial adhesion to solvents, a hydrophobicity assay was carried out as previously described (Rosenberg et al., 1980; Al Kassaa et al., 2014). The bacterial suspension was prepared as described for the auto-aggregation assay, and 3 ml of the suspension was mixed with 1 ml xylene (Carl Roth) by vortexing for 2 min. The aqueous phase was removed after incubating the mixture for 10 min at room temperature, and after incubation for a further 2 h at room temperature the absorbance was measured at 600 nm. The percentage of hydrophobicity was calculated as follows:
where OD1 is the optical density after 2 h and OD0 is the optical density before adding the xylene. This experiment was performed in triplicate.
Isolate tolerance to low pH was determined as described (Hosseini et al., 2009) and reveals how well the bacteria can survive under harsh gut conditions. An overnight culture was washed twice with PBS and then resuspended in PBS acidified to pH 2, 3, and 4, respectively. After incubating for 3 h, the bacteria were plated on LB agar in order to count the number of colony-forming units (CFUs).
Safety Characterization
Hemolytic activity was determined by spotting 7-μl overnight cultures of E. mundtii onto Columbia sheep blood agar plates as a required safety assay (bioMérieux, Marcy-l’Étoile, France). The plates were incubated for 48 h at 30°C. The presence of clear zones around the colonies indicated hemolytic activity (Abriouel et al., 2008). Staphylococcus aureus (DSM 2569) was used as a positive control. The experiment was performed in triplicates.
The sensitivity of the isolates to antibiotics was determined by performing a broth microdilution assay in 96-well plates (Wiegand et al., 2008). The following antibiotic concentration ranges (μg ml-1) was used: ampicillin (0.25–128), rifampicin (0.25–128), erythromycin (0.25–128), kanamycin (2–256), ciprofloxacin (0.25–128); fosfomycin (0.25–128), streptomycin (2–256), tetracycline (0.25–128), and vancomycin (0.25–128). Isolates were grown in LB medium at 30°C overnight and the bacterial suspension was adjusted to 106 cells ml-1. A 50-μl aliquot of the suspension was then mixed with 50 μl of the antibiotic solution in a microtiter plate well and incubated for 16 h at 30°C. The minimal inhibitory concentration (MIC) was defined as the lowest concentration that inhibits visible growth.
In Vivo Characterization Following Entomopathogenic Challenge
For the survival assay, 15 glass jars containing approximately 100 adult T. castaneum (∼1 month old) were transferred to 100 g flour for oviposition. On the third day after oviposition, the eggs were removed by sieving through a 250-μm mesh (Retsch, Haan, Germany). The eggs collected from all jars were mixed, and ∼400 eggs were transferred to the different probiotic diets (see below) each on 150-mm Petri dishes. After 8 days on these six diets, 96 larvae of similar size were transferred individually to the three challenged diets, one per well of a microtiter plate. A total of 1728 larvae were used in this experiment. The plates were sealed with transparent adhesive foil and punctured with small holes for air supply. Survival was monitored daily for 7 days. The experiment was carried out under standard rearing conditions (32°C and 70% humidity).
Diet Preparation
For the preparation of the different probiotic diets, 0.15 g ml-1 of flour (type 405, Alnatura, Bickenbach, Germany) supplemented with 5% brewer’s yeast was mixed with: (1) sterile MRS medium as a negative control; (2) 5 × 109 cells ml-1 of E. mundtii 1 isolate, grown in MRS medium at 30°C overnight, washed with PBS twice, centrifuged (3200 × g, 4°C, 15 min) and re-suspended in MRS medium; (3) crude CFS from the same isolate; (4) CFS heated at 98°C for 20 min; (5) CFS treated with 4 ml proteinase K (25 mg ml-1) for 1 h at 37°C; or (6) CFS adjusted to pH 7. The diet was poured into Petri dishes either (150-mm diameter = 100 ml or 90 mm diameter = 20 ml), dried at 37°C and shredded.
The immune challenged diet containing Bacillus thuringiensis (DSM 2046) was prepared as previously described (Milutinović et al., 2013). We added 0.15 g flour with 5% brewer’s yeast to 1 ml of the bacterial suspension, and adjusted the cell density to 5 × 109 cells ml-1. The diet containing Pseudomonas entomophila (DSM 28517) was prepared by growing P. entomophila overnight at 30°C in LB, centrifuging the suspension (3200 × g, 4°C, 15 min) and washing the pellet twice in PBS before re-suspending in PBS. The cell density was adjusted as described for B. thuringiensis. PBS mixed with flour and yeast was used as negative control. We transferred 40 μl of the suspension to each well of a 96-well plate and dried the plates at 37°C overnight.
In Vivo Characterization of Fitness Parameters
For the longevity experiment, 10 glass jars containing ∼100 adult beetles (∼1 month old) were transferred to 100 g flour for oviposition. After 24 h, the eggs collected from all jars were mixed and ∼100 eggs were transferred to the MRS, CFS or E. mundtii probiotic diets prepared as described above on 90-mm Petri dishes. Five replicates per diet were incubated under standard conditions. Longevity was measured using a thermotolerance assay at 42°C (Grünwald et al., 2013). Twenty-five age-controlled beetles of mixed sex were transferred after 5 days on the standard diet to the same probiotic diet used to rear the larvae. Survival was recorded daily, and dead beetles were removed from the Petri dishes.
The effect of the probiotic diets (MRS, CFS and E. mundtii) on the fitness of T. castaneum was determined by measuring the reproductive success of the beetles. Age-controlled virgin beetles were obtained as described for the longevity experiment with the additional sexing of the pupae. Five days after eclosion, virgin beetles (five of each sex, each diet with five replicates) were allowed to mate for 24 h on 10 g of flour in Falcon tubes covered with breathable tissue. After 24 h, the eggs were counted (fertility) and transferred to a standard diet followed by incubation for 8 days. Hatched larvae were counted to determine fecundity (Bingsohn et al., 2016).
Statistics
Statistical analysis was carried out using R (v3.3.2, R Development Core Team, 2008). The survival data were evaluated using Kaplan Meir statistics in the ‘survival’ package (v 2.40.1 Therneau and Grambsch, 2001) and multiple pairwise comparisons among groups were carried out using log-rank tests. p-values were adjusted using the ‘holm’ correction method. The fertility and fecundity data were analyzed by one-way analysis of variance (ANOVA) and the ‘holm’ correction method.
Results
Identification of Isolates
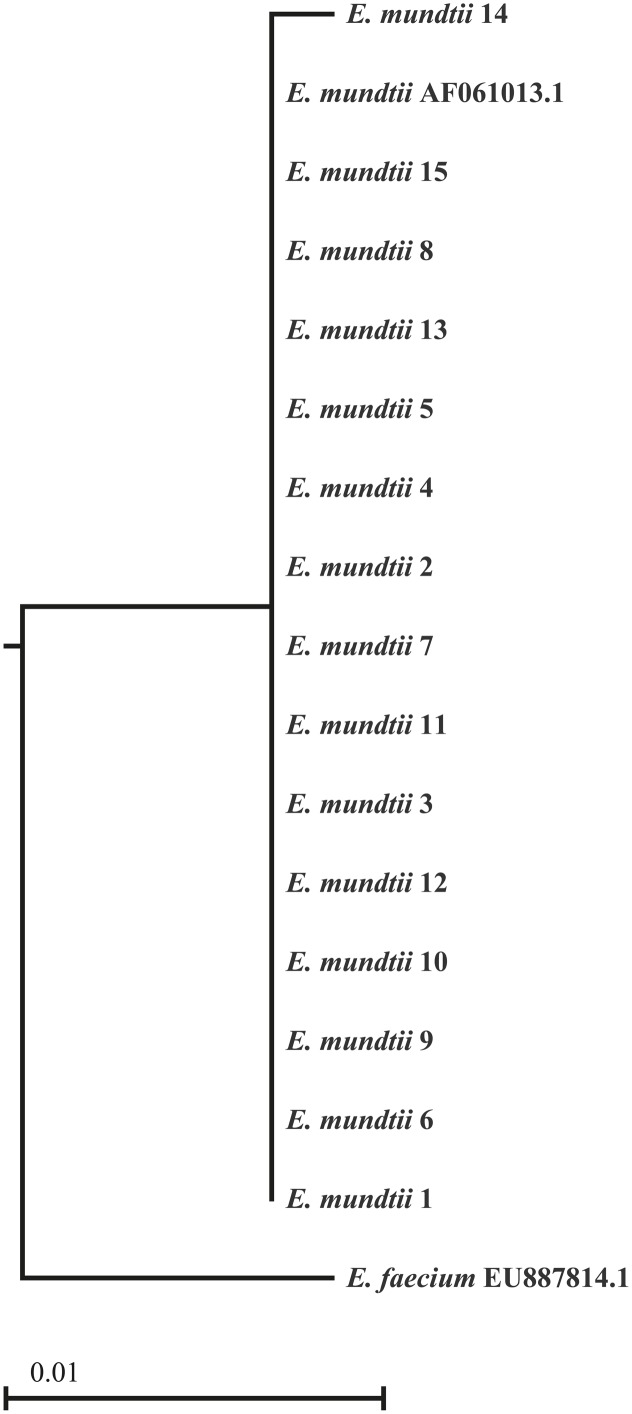
BLAST analysis of partial 16S rDNA sequences from 15 isolates showed 100% identity to E. mundtii. The phylogenetic relationships shown in Figure 1 confirm that the isolates are strains of E. mundtii.
FIGURE 1.
Phylogenetic tree of the 16S rDNA sequences derived from 15 Enterococcus mundtii isolates. The sequences from the isolated strains were aligned with two strains identified by their GenBank accession numbers. The scale is based on the bootstrap values from the neighbor-joining method.
Antimicrobial Characterization
An initial screen using the agar spot on lawn method revealed that all 15 isolates showed antimicrobial activity against a broad spectrum of indicator bacteria (Table 1). Some isolates also showed activity against entomopathogenic B. thuringiensis and P. entomophila, as well as human pathogenic bacteria such as P. aeruginosa and S. aureus. For the further characterization of the antimicrobial compounds, we focused on the CFS of E. mundtii isolate 1 using the AWDA (Table 2) because this isolate showed the best antimicrobial profile in the agar spot on lawn assay. The crude CFS did not show antimicrobial activity against the indicator bacteria, so we decided to concentrate the CFS by 100-fold. The concentrated CFS was able to inhibit the indicator bacteria to different degrees (Table 2). Treatment of the CFS with proteinase K or catalase reduced its antimicrobial activity against some of the indicator bacteria (Table 2).
Table 2.
Agar well-diffusion assay with CFS from E. mundtii isolate 1 with different treatments against indicator bacteria.
| Indicator strains | Crude | 100 × fold | Proteinase K | Catalase | Heated |
|---|---|---|---|---|---|
| Bacillus thuringiensis (DSM 2046) | 0 | 4.75 ± 0.32 | 4.23 ± 0.09 | 4.29 ± 0.12 | 4.58 ± 0.58 |
| Escherichia coli (strain D 31) | 0 | 4.75 ± 0.43 | 4.81 ± 0.69 | 4.33 ± 0.27 | 4.60 ± 0.57 |
| Klebsiella terrigena (DSM 2687) | 0 | 6.3 ± 0.17 | 6.12 ± 0.08 | 4.73 ± 1.00 | 6.14 ± 0.26 |
| Listeria grayi (DSM 20601) | 0 | 2.25 ± 0.53 | 1.02 ± 0.15 | 2.43 ± 0.29 | 2.55 ± 0.06 |
| Listeria innocua (DSM 20649) | 0 | 3.27 ± 0.37 | 0.56 ± 0.96 | 2.82 ± 0.13 | 2.97 ± 0.34 |
| Listeria seeligeri (DSM 20751) | 0 | 2.03 ± 1.19 | 0.97 ± 0.86 | 2.71 ± 0.37 | 2.08 ± 1.05 |
| Micrococcus luteus (DSM 20030) | 0 | 3.92 ± 0.41 | 3.68 ± 0.69 | 3.83 ± 0.46 | 3.69 ± 0.26 |
| Pseudomonas aeruginosa (DSM 50071) | 0 | 4.9 ± 0.22 | 4.49 ± 0.57 | 4.07 ± 0.62 | 4.88 ± 0.39 |
| Pseudomonas entomophila (DSM 28517) | 0 | 3.83 ± 0.42 | 3.68 ± 0.36 | 3.62 ± 0.31 | 3.52 ± 0.38 |
| Salmonella subterranean (DSM 1620) | 0 | 6.72 ± 0.42 | 6.39 ± 0.31 | 6.33 ± 0.49 | 6.48 ± 0.08 |
| Staphylococcus aureus (DSM 2569) | 0 | 0 | 0 | 0 | 0 |
The radius of the zone of inhibition is measured in mm. Results are mean values of three replicates with standard deviations.
Probiotic Characterization
The cell surface properties of E. mundtii 1 isolate are shown in Table 3. The isolate showed a high rate of auto-aggregation after 24 h, indicating a significant level of bacterial adhesion to host cells. However, the low level of adhesion to solvents indicated that the surface was hydrophilic. The viability of E. mundtii 1 was reduced at pH 2 and slightly reduced at pH 3 (Table 4).
Table 3.
Auto-aggregation rate and hydrophobicity of E. mundtii 1.
| E. mundtii 1 | |
|---|---|
| Auto-aggregation (%) 5 h | 12.69 ± 1.28 |
| Auto-aggregation (%) 24 h | 89.66 ± 2.83 |
| Hydrophobicity (%) | 8.30 ± 9.4 |
Table 4.
Effect of low pH on E. mundtii 1.
| E. mundtii 1 | |
|---|---|
| CFU start | 8.50 ± 0.07 |
| CFU pH 2 after 3 h | 4.11 ± 0.04 |
| CFU pH 3 after 3 h | 6.49 ± 0.05 |
| CFU pH 4 after 3 h | 8.31 ± 0.037 |
Results are in log CFU ml-1 with SD.
Safety Characterization
The microdilution broth assay was used to determine MIC values. E. mundtii 1 was susceptible to all the antibiotics listed in Table 5. There was no evidence of hemolytic activity on sheep blood agar plated with any of the 15 E. mundtii isolates.
Table 5.
Antimicrobial susceptibility of E. mundtii 1.
| Antimicrobial agent | MIC (μg ml-1) |
|---|---|
| Ampicillin | <0.25 |
| Ciprofloxacin | <0.25 |
| Erythromycin | <0.25 |
| Fosfomycin | 4 |
| Kanamycin | 32 |
| Rifampicin | <0.25 |
| Streptomycin | 64 |
| Tetracycline | <0.25 |
| Vancomycin | <0.25 |
In Vivo Characterization
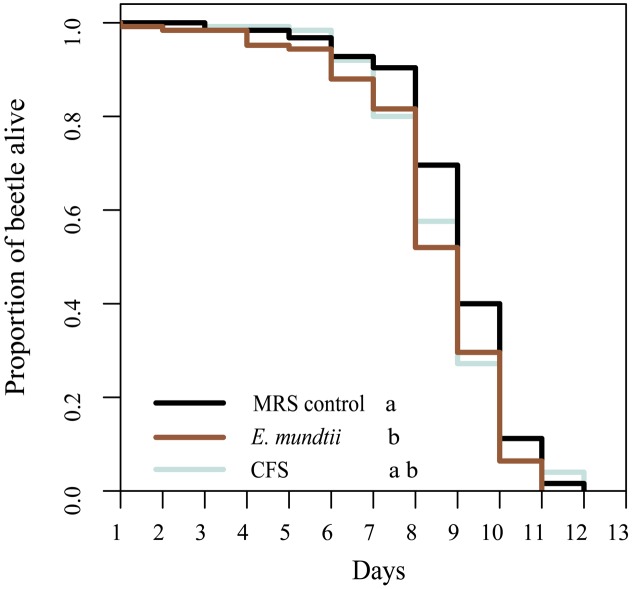
The potential probiotic effect of the E. mundtii 1 isolate (and the corresponding supernatant) was tested by measuring the survival of T. castaneum when challenged with a pathogen in the presence or absence of the isolate. There were no significant differences in survival when we compared larvae fed on the control diet with or without an earlier exposure to the probiotic diet (χ2 = 5.1, df = 5, p = 0.399) (Figure 2A) indicating that isolate had no adverse effects on the larvae. In contrast, there was a significant increase in survival rates when we compared larvae fed on the diet containing B. thuringiensis with or without an earlier exposure to the probiotic diets (χ2 = 24.8, df = 5, p < 0.0001) (Figure 2B). The above probiotic effect of the CFS was reduced by treatment with proteinase K or by heating to 98°C. In contrast to the diets spiked with B. thuringiensis, there was no significant difference in survival rates when we compared larvae fed on the diet containing P. entomophila with or without an earlier exposure to the probiotic diet (χ2 = 4, df = 4, p = 0.403) (Figure 2C). The precise p-values from multiple pairwise comparisons of the Kaplan Meir curves with ‘holm’ correction are presented in Supplementary Tables S2–S4.
FIGURE 2.
The effect of probiotic diets on Tribolium castaneum survival. Survival rates of larvae raised on different probiotic diets based on flour mixed with (i) MRS (control diet), (ii) E. mundtii, (iii) CFS pH 7, (iv) CFS treated with proteinase K, (v) CFS heated to 98°C, or (vi) crude CFS. Eight days after exposure to the probiotic diets, larvae (n = 96 per treatment) were challenged with (A) the control diet, (B) a diet spiked with Bacillus thuringiensis diet, or (C) a diet spiked with Pseudomonas entomophila. Statistically significant differences in the treatments are indicated by differing lowercase letters (p < 0.05). Precise p-values for the multiple comparisons are presented in Supplementary Tables S1–S3.
The potential impact of the E. mundtii isolate on the longevity of T. castaneum was determined using a thermotolerance assay. We observed a significantly shorter lifespan when beetles were reared on diets containing E. mundtii (χ2 = 6.4, df = 2, p = 0.0398) (Figure 3). However, there were no significant differences in longevity among beetles fed on the CFS diet, the MRS control diet or the E. mundtii diet (Supplementary Table S5). We also investigated the influence of the probiotic diet on the fitness (fertility and fecundity) of T. castaneum and found that the number of eggs differed significantly in a diet-dependent manner (ANOVA df = 2; F = 17,163; p < 0.001) (Supplementary Figure S1). Multiple pairwise comparisons among the diet groups showed that beetles maintained on the probiotic E. mundtii diet laid significantly fewer eggs than beetles on the CFS and control diets (p < 0.001). However, there was no significant difference in fecundity among the diet groups (ANOVA df = 2; F = 0.0327; p = 0.968).
FIGURE 3.
Longevity of T. castaneum. Larvae were raised on different probiotic diets, and age controlled adult beetles were maintained on the same diets. Survival was determined by performing a thermotolerance assay (five replicates with 25 beetles of mixed sex). Statistically significant differences between treatments are indicated by differing lowercase letters (p < 0.05). Precise p-values for the multiple comparisons are presented in Supplementary Table S4.
Discussion
We have shown that probiotic bacteria can be isolated from the feces of storage food insects. Based on the analysis of 16S rDNA sequences we isolated several new strains of E. mundtii, and characterized their probiotic profiles as well as their broad antimicrobial activity against Gram-positive and Gram-negative bacteria. We also show the potential for T. castaneum as a model system for screening of such probiotic bacteria in vivo.
The screen for antimicrobial activity revealed a broad spectrum of activity in all 15 isolates we tested. However, we were unable to detect antimicrobial activity in the crude CFS, in contrast to previous studies of E. mundtii, which reported a broad spectrum of antimicrobial activity also in the crude CFS (Zendo et al., 2005; Schelegueda et al., 2015). We were able to detect a similar antimicrobial profile in the agar spot on lawn assay and CFS only when the latter was concentrated by 100-fold. E. mundtii belongs to the order lactobacillales, whose members are known to produce a variety of heat-stable bacterocins that are sensitive to proteinase K (Franz et al., 2007). However, proteinase K and catalase treatments reduced the antimicrobial activity of only a few of our isolates, and heat treatment had no impact in vitro. By adjusting the pH of the extract, we ruled out the possibility that antimicrobial activity was based on organic acids. The major antimicrobial compounds in the isolates are therefore heat stable and resistant to proteinase K, as previously reported (Silva et al., 1987; Ouzari et al., 2008). Further testing is required to identify and characterize the antimicrobial compounds in detail. In many screens for probiotic bacteria, only the crude CFS is tested for antimicrobial activity, which may lead to false negative results given that many genes remain silent under standard laboratory cultivation conditions and the corresponding compounds may not be synthesized (Nett et al., 2009; Seyedsayamdost, 2014; Rutledge and Challis, 2015).
To further characterize the probiotic profile of one of our isolates we investigated the surface properties of E. mundtii isolate 1. This isolate showed a high level of auto-aggregation after 24 h and a low level of solvent adhesion. Auto-aggregation is related to the ability of bacterial cells to adhere to epithelial cells and form colonies (Kos et al., 2003), which is a prerequisite for probiotic bacteria because this is how they colonize the gut. The low solvent adhesion indicates the presence of a hydrophilic cell surface based on polysaccharides (Collado et al., 2007, 2008). E. mundtii 1 isolate was able to survive in a low-pH environment, making the isolate resistant to the harsh conditions in the digestive system (Conway et al., 1987; Jena et al., 2013). It is also important to evaluate the safety of probiotic bacteria, especially in the genus Enterococcus, because the closely related species E. faecalis possesses hemolytic activity (De Vuyst et al., 2003). We could not detect hemolytic activity in any of our isolates. There are also concerns that Enterococci may be able to transfer antibiotic resistance genes (Palmer et al., 2010), but E. mundtii isolate 1 was susceptible to all the antibiotics we tested.
In vitro assays allow the initial preselection of potentially probiotic strains, but in vivo testing is also necessary to check for systemic interactions (Papadimitriou et al., 2015). We introduced the model organism T. castaneum as a novel in vivo probiotic screening platform. T. castaneum is an established model organism that can be used for infection assays and has the capacity for immune priming (Roth et al., 2010; Knorr et al., 2015; Futo et al., 2016). The feeding of T. castaneum larvae with either E. mundtii 1 or the corresponding CFS protected them against the entomopathogen B. thuringiensis but not against P. entomophila. A similar result was reported for C. elegans, i.e., feeding with Gram-positive probiotics resulted in protection only against Gram-positive pathogens and not against Gram-negative pathogens (Kim and Mylonakis, 2012). The crude CFS showed no in vitro antimicrobial activity but nevertheless resulted in a protective function in vivo, indicating that the probiotic properties are not based on antimicrobial activity. Both E. mundtii and B. thuringiensis are Gram-positive bacteria, which should activate the same host immune responses upon contact, in contrast to Gram-negative bacteria as P. entomophila (Lemaitre and Hoffmann, 2007; Yokoi et al., 2012). Although the in vivo protective effect of the CFS was reduced by heating to 98°C or treatment with proteinase K, the loss of activity was not significant. This may indicate that the protective function of the CFS is based on bacterocins, which are thought to act as signaling peptides for communication with other bacteria or the host immune system (Cotter et al., 2013). The diverse class of bacterocins includes proteinase K-sensitive as well as heat-labile compounds (Yang et al., 2014). Alternatively, Gram-positive cell wall compounds such as peptidoglycans and lipoteichoic acid are thought to activate the immune system (Leulier et al., 2003; Rao and Yu, 2010). Probiotic bacteria are widely used in aquaculture as food supplements and for the white leg shrimp Litopenaeus vannamei, where the supernatant of a probiotic bacterial culture also protects the hosts from pathogens and induces specific immune system genes (Sha et al., 2016). Further research is required to investigate the protective function of the supernatant in more detail, and to determine whether the protective function involves the modulation of the immune system or alters the dynamics of the larval gut microbiota.
Although, our isolate increased host resistance toward an entomopathogen there was a trade-off in terms of beetle fitness. Certain probiotic bacteria have been shown to increase the lifespan of C. elegans (Grompone et al., 2012; Park et al., 2015) but our isolate had the opposite effect on T. castaneum. Furthermore the fertility of the beetle was also reduced following treatment with the E. mundtii isolate. One potential explanation for this phenomenon is the recently observed translocation of bacteria from the T. castaneum gut to the eggs, where they elicit an innate immune response that triggers a fitness penalty (Knorr et al., 2015). The potential for such adverse effects highlights the importance of in vivo assays for the characterization of probiotic bacteria.
Our results confirm that T. castaneum is suitable as an alternative model invertebrate for pre-screening in probiotic research, for human application pending on further confirmation in the mouse model. The short generation time and longevity of T. castaneum makes it particularly suitable for long-term studies of probiotics, including potential transgenerational effects and the influence of probiotics on fitness parameters. Here, T. castaneum also becomes of interest as test organism with respect to edible insects being closely related to Tenebrio molitor, the most promising candidate in mass rearing of insects for feed and food currently (Grau et al., 2017). Our E. mundtii isolate showed beneficial probiotic properties in vitro and partly also in vivo, suggesting that the feces of insect food pests could be a good source for probiotic bacteria in the future.
Author Contributions
Planned and conceived the experiments: TG and GJ. Performed the experiments: TG. Analyzed the experiments: TG and GJ. Drafted the manuscript and contributed to the data interpretation: TG, AV, and GJ. All authors read, critically revised and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Richard M. Twyman for editing the manuscript and Tilottama Biswas and Sara DeLeon for their valuable help.
Footnotes
Funding. The project was founded within the LOEWE Center for Insect Biotechnology and Bioresources (ZIB), granted by the German state of Hessen’s excellence initiative. GJ was additionally funded by a grant of the Volkswagenstiftung.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01261/full#supplementary-material
References
- Abriouel H., Omar N. B., Molinos A. C., López R. L., Grande M. J., Martínez-Viedma P., et al. (2008). Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 123 38–49. 10.1016/j.ijfoodmicro.2007.11.0671016/j.ijfoodmicro.2007.11.067 [DOI] [PubMed] [Google Scholar]
- Al Atya A. K., Drider-Hadiouche K., Ravallec R., Silvain A., Vachee A., Drider D. (2015). Probiotic potential of Enterococcus faecalis strains isolated from meconium. Front. Microbiol. 6:227 10.3389/fmicb.2015.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Kassaa I., Hamze M., Hober D., Chihib N.-E., Drider D. (2014). Identification of vaginal lactobacilli with potential probiotic properties isolated from women in north lebanon. Microb. Ecol. 67 722–734. 10.1007/s00248-014-0384-7 [DOI] [PubMed] [Google Scholar]
- Altincicek B., Elashry A., Guz N., Grundler F. M. W., Vilcinskas A., Dehne H.-W. (2013). Next Generation sequencing based transcriptome analysis of septic-injury responsive genes in the beetle Tribolium castaneum. PLoS ONE 8:e52004 10.1371/journal.pone.0052004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi A., Arora M. (2015). Regulatory categories of probiotics across the globe: a review representing existing and recommended categorization. Indian J. Med. Microbiol. 33:2 10.4103/0255-0857.150868 [DOI] [PubMed] [Google Scholar]
- Besselink M. G., van Santvoort H. C., Buskens E., Boermeester M. A., van Goor H., Timmerman H. M., et al. (2008). Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet 371 651–659. 10.1016/S0140-6736(08)60207-X [DOI] [PubMed] [Google Scholar]
- Bingsohn L., Knorr E., Vilcinskas A. (2016). The model beetle Tribolium castaneum can be used as an early warning system for transgenerational epigenetic side effects caused by pharmaceuticals. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 18 57–64. 10.1016/j.cbpc.2016.03.0021016/j.cbpc.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Brune A., Dietrich C. (2015). The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu. Rev. Microbiol. 69 145–166. 10.1146/annurev-micro-092412-155715 [DOI] [PubMed] [Google Scholar]
- Bucher G., Scholten J., Klingler M. (2002). Parental RNAi in Tribolium (Coleoptera). Curr. Biol. CB 12 R85–R86. 10.1016/S0960-9822(02)00666-8 [DOI] [PubMed] [Google Scholar]
- Buffie C. G., Pamer E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13 790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaucheyras-Durand F., Durand H. (2009). Probiotics in animal nutrition and health. Benef. Microbes 1 3–9. 10.3920/BM2008.1002 [DOI] [PubMed] [Google Scholar]
- Collado M. C., Meriluoto J., Salminen S. (2007). Measurement of aggregation properties between probiotics and pathogens: in vitro evaluation of different methods. J. Microbiol. Methods 71 71–74. 10.1016/j.mimet.2007.07.0051016/j.mimet.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Collado M. C., Meriluoto J., Salminen S. (2008). Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 226 1065–1073. 10.1007/s00217-007-0632-x [DOI] [Google Scholar]
- Conway P. L., Gorbach S. L., Goldin B. R. (1987). Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J. Dairy Sci. 70 1–12. 10.3168/jds.S0022-0302(87)79974-3 [DOI] [PubMed] [Google Scholar]
- Cotter P. D., Ross R. P., Hill C. (2013). Bacteriocins - a viable alternative to antibiotics? Nat. Rev. Microbiol. 11 95–105. 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- De Vuyst L., Foulquié Moreno M. R., Revets H. (2003). Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int. J. Food Microbiol. 84 299–318. 10.1016/S0168-1605(02)00425-7 [DOI] [PubMed] [Google Scholar]
- Del Re B., Sgorbati B., Miglioli M., Palenzona D. (2000). Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 31 438–442. 10.1046/j.1365-2672.2000.00845.x [DOI] [PubMed] [Google Scholar]
- Deshpande G., Rao S., Patole S., Bulsara M. (2010). Updated meta-analysis of probiotics for preventing necrotizing enterocolitis in preterm neonates. Pediatrics 125 921–930. 10.1542/peds.2009-1301 [DOI] [PubMed] [Google Scholar]
- Diehl J. M., Körner M., Pietsch M., Meunier J. (2015). Feces production as a form of social immunity in an insect with facultative maternal care. BMC Evol. Biol. 15:40 10.1186/s12862-015-0330-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippel S., Oberhofer G., Kahnt J., Gerischer L., Opitz L., Schachtner J., et al. (2014). Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genomics 15:1141 10.1186/1471-2164-15-1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P., Moran N. A. (2013). Functional and evolutionary insights into the simple yet specific gut microbiota of the honey bee from metagenomic analysis. Gut Microbes 4 60–65. 10.4161/gmic.22517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C. M. A. P., Van Belkum M. J., Holzapfel W. H., Abriouel H., Gálvez A. (2007). Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol. Rev. 31 293–310. 10.1111/j.1574-6976.2007.00064.x [DOI] [PubMed] [Google Scholar]
- Futo M., Armitage S. A. O., Kurtz J. (2016). Microbiota plays a role in oral immune priming in Tribolium castaneum. Front. Microbiol. 6:1383 10.3389/fmicb.2015.01383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar F., Teixeira N., Rigottier-Gois L., Marujo P., Nielsen-LeRoux C., Crespo M. T. B., et al. (2009). Virulence of Enterococcus faecalis dairy strains in an insect model: the role of fsrB and gelE. Microbiology 155 3564–3571. 10.1099/mic.0.030775-0 [DOI] [PubMed] [Google Scholar]
- Gilles A. F., Schinko J. B., Averof M. (2015). Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Development 142 2832–2839. 10.1242/dev.125054 [DOI] [PubMed] [Google Scholar]
- Godoy-Vitorino F., Goldfarb K. C., Brodie E. L., Garcia-Amado M. A., Michelangeli F., Domínguez-Bello M. G. (2010). Developmental microbial ecology of the crop of the folivorous hoatzin. ISME J. 4 611–620. 10.1038/ismej.2009.147 [DOI] [PubMed] [Google Scholar]
- Gomez de Aguero M., Ganal-Vonarburg S. C., Fuhrer T., Rupp S., Uchimura Y., Li H., et al. (2016). The maternal microbiota drives early postnatal innate immune development. Science 351 1296–1302. 10.1126/science.aad2571 [DOI] [PubMed] [Google Scholar]
- Grau T., Vilcinskas A., Joop G. (2017). Sustainable farming of the mealworm Tenebrio molitor for the production of food and feed. Z. Für Naturforschung C 10.1515/znc-2017-0033 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Grompone G., Martorell P., Llopis S., González N., Genovés S., Mulet A. P., et al. (2012). Anti-inflammatory Lactobacillus rhamnosus CNCM I-3690 strain protects against oxidative stress and increases lifespan in Caenorhabditis elegans. PLoS ONE 7:e52493 10.1371/journal.pone.0052493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald S., Stellzig J., Adam I. V., Weber K., Binger S., Boll M., et al. (2013). Longevity in the red flour beetle Tribolium castaneum. Genes Nutr. 8 439–448. 10.1007/s12263-012-0330-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashide T., Takahashi M., Kobayashi A., Ohkubo S., Sakurai M., Shirao Y., et al. (2005). Endophthalmitis caused by Enterococcus mundtii. J. Clin. Microbiol. 43 1475–1476. 10.1128/JCM.43.3.1475-1476.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Guarner F., Reid G., Gibson G. R., Merenstein D. J., Pot B., et al. (2014). Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11 506–514. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- Hosseini S. V., Arlindo S., Böhme K., Fernández-No C., Calo-Mata P., Barros-Velázquez J. (2009). Molecular and probiotic characterization of bacteriocin-producing Enterococcus faecium strains isolated from nonfermented animal foods. J. Appl. Microbiol 107 1392–1403. 10.1111/j.1365-2672.2009.04327.x [DOI] [PubMed] [Google Scholar]
- Jena P. K., Trivedi D., Thakore K., Chaudhary H., Giri S. S., Seshadri S. (2013). Isolation and characterization of probiotic properties of Lactobacilli isolated from rat fecal microbiota: probiotic lactobacilli from rat gutflora. Microbiol. Immunol. 57 407–416. 10.1111/1348-0421.120541111/1348-0421.12054 [DOI] [PubMed] [Google Scholar]
- Jost T., Lacroix C., Braegger C. P., Rochat F., Chassard C. (2014). Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 16 2891–2904. 10.1111/1462-2920.12238 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Mylonakis E. (2012). Caenorhabditis elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain NCFM enhances gram-positive immune responses. Infect. Immun. 80 2500–2508. 10.1128/IAI.06350-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr E., Bingsohn L., Kanost M. R., Vilcinskas A. (2013). “Tribolium castaneum as a model for high-throughput RNAi screening,” in Yellow Biotechnology II Advances in Biochemical Engineering/Biotechnology, ed. Vilcinskas A. (Berlin: Springer; ), 163–178. 10.1007/10_2013_208 [DOI] [PubMed] [Google Scholar]
- Knorr E., Schmidtberg H., Arslan D., Bingsohn L., Vilcinskas A. (2015). Translocation of bacteria from the gut to the eggs triggers maternal transgenerational immune priming in Tribolium castaneum. Biol. Lett. 11:20150885 10.1098/rsbl.2015.0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H., Schmid-Hempel P. (2011). Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. U.S.A. 108 19288–19292. 10.1073/pnas.1110474108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. D. (2012). Diversity and function of the avian gut microbiota. J. Comp. Physiol. B 182 591–602. 10.1007/s00360-012-0645-z [DOI] [PubMed] [Google Scholar]
- Kos B., Šušković J., Vuković S., Šimpraga M., Frece J., Matošić S. (2003). Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 94 981–987. 10.1046/j.1365-2672.2003.01915.x1046/j.1365-2672.2003.01915.x [DOI] [PubMed] [Google Scholar]
- Kotzampassi K., Giamarellos-Bourboulis E. J. (2012). Probiotics for infectious diseases: more drugs, less dietary supplementation. Int. J. Antimicrob. Agents 40 288–296. 10.1016/j.ijantimicag.2012.06.006 [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25 697–743. 10.1146/annurev.immunol.25.022106.141615 [DOI] [PubMed] [Google Scholar]
- Leulier F., Parquet C., Pili-Floury S., Ryu J.-H., Caroff M., Lee W.-J., et al. (2003). The drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4 478–484. 10.1038/ni922 [DOI] [PubMed] [Google Scholar]
- Lozupone C. A., Stombaugh J. I., Gordon J. I., Jansson J. K., Knight R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489 220–230. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B., Garriga M., Hugas M., Aymerich T. (2005). Genetic diversity and safety aspects of enterococci from slightly fermented sausages. J. Appl. Microbiol. 98 1177–1190. 10.1111/j.1365-2672.2005.02555.x [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M., Hadfield M. G., Bosch T. C., Carey H. V., Domazet-Lošo T., Douglas A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S. A. 110 3229–3236. 10.1073/pnas.1218525110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinović B., Fritzlar S., Kurtz J. (2014). Increased survival in the red flour beetle after oral priming with bacteria-conditioned media. J. Innate Immun. 6 306–314. 10.1159/000355211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinović B., Stolpe C., Peuβ R., Armitage S. A. O., Kurtz J. (2013). the red flour beetle as a model for bacterial oral infections. PLoS ONE 8:e64638 10.1371/journal.pone.0064638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S. R., Collins J. J., Relman D. A. (2014). Antibiotics and the gut microbiota. J. Clin. Invest. 124 4212–4218. 10.1172/JCI72333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett M., Ikeda H., Moore B. S. (2009). Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 26 1362–1384. 10.1039/b817069j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaeger T. A. (2010). Mechanisms of probiotic actions – A review. Int. J. Med. Microbiol. 300 57–62. 10.1016/j.ijmm.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Ouzari H., Najjari A., Amairi H., Gtari M., Hassen A., Boudabous A. (2008). Comparative analysis of Lactococcus lactis bacteriocins and preliminary characterisation of a new proteinase K resistant lactococcin member. Ann. Microbiol. 58 83–88. 10.1007/BF03179449 [DOI] [Google Scholar]
- Palmer K. L., Kos V. N., Gilmore M. S. (2010). Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 13 632–639. 10.1016/j.mib.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandiyan P., Balaraman D., Thirunavukkarasu R., George E. G. J., Subaramaniyan K., Manikkam S., et al. (2013). Probiotics in aquaculture. Drug Invent. Today 5 55–59. 10.1016/j.dit.2013.03.003 [DOI] [Google Scholar]
- Papadimitriou K., Zoumpopoulou G., Foligné B., Alexandraki V., Kazou M., Pot B., et al. (2015). Discovering probiotic microorganisms: in vitro, in vivo, genetic and omics approaches. Front. Microbiol. 6:58 10.3389/fmicb.2015.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. R., Oh S., Son S. J., Park D.-J., Oh S., Kim S. H., et al. (2015). Bacillus licheniformis isolated from traditional korean food resources enhances the longevity of Caenorhabditis elegans through serotonin signaling. J. Agric. Food Chem. 63 10227–10233. 10.1021/acs.jafc.5b03730 [DOI] [PubMed] [Google Scholar]
- Park Y., Aikins J., Wang L. J., Beeman R. W., Oppert B., Lord J. C., et al. (2008). Analysis of transcriptome data in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 38 380–386. 10.1016/j.ibmb.2007.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptaszyńska A. A., Borsuk G., Zdybicka-Barabas A., Cytryñska M., Małek W. (2015). Are commercial probiotics and prebiotics effective in the treatment and prevention of honeybee nosemosis C? Parasitol. Res. 115 397–406. 10.1007/s00436-015-4761-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rao X.-J., Yu X.-Q. (2010). Lipoteichoic acid and lipopolysaccharide can activate antimicrobial peptide expression in the tobacco hornworm Manduca sexta. Dev. Comp. Immunol. 34 1119–1128. 10.1016/j.dci.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman D. A., Schmidt T. M., MacDermott R. P., Falkow S. (1992). Identification of the uncultured bacillus of whipple’s disease. N. Engl. J. Med. 327 293–301. 10.1056/NEJM199207303270501 [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Gutnick D., Rosenberg E. (1980). Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9 29–33. 10.1111/j.1574-6968.1980.tb05599.x1111/j.1574-6968.1980.tb05599.x [DOI] [Google Scholar]
- Rosengaus R. B., Mead K., Du Comb W. S., Benson R. W., Godoy V. G. (2013). Nest sanitation through defecation: antifungal properties of wood cockroach feces. Naturwissenschaften 100 1051–1059. 10.1007/s00114-013-1110-x [DOI] [PubMed] [Google Scholar]
- Roth O., Joop G., Eggert H., Hilbert J., Daniel J., Schmid-Hempel P., et al. (2010). Paternally derived immune priming for offspring in the red flour beetle, Tribolium castaneum. J. Anim. Ecol. 79 403–413. 10.1111/j.1365-2656.2009.01617.x [DOI] [PubMed] [Google Scholar]
- Round J. L., Mazmanian S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9 313–323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-De-Castañeda R., Vela A. I., Lobato E., Briones V., Moreno J. (2011). bacterial loads on eggshells of the pied flycatcher: environmental and maternal factors. Condor 113 200–208. 10.1525/cond.2011.100035 [DOI] [Google Scholar]
- Russell W. M. S., Burch R. L. (1959). The Principles of Humane Experimental Technique. London: Methuen. [Google Scholar]
- Rutledge P. J., Challis G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13 509–523. 10.1038/nrmicro3496 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schelegueda L. I., Vallejo M., Gliemmo M. F., Marguet E. R., Campos C. A. (2015). Synergistic antimicrobial action and potential application for fish preservation of a bacteriocin produced by Enterococcus mundtii isolated from Odontesthes platensis. LWT - Food Sci. Technol. 64 794–801. 10.1016/j.lwt.2015.06.017 [DOI] [Google Scholar]
- Schillinger U., Lücke F. K. (1989). Antibacterial activity of Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 55 1901–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost M. R. (2014). High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. U.S.A. 111 7266–7271. 10.1073/pnas.1400019111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Y., Wang L., Liu M., Jiang K., Xin F., Wang B. (2016). Effects of lactic acid bacteria and the corresponding supernatant on the survival, growth performance, immune response and disease resistance of Litopenaeus vannamei. Aquaculture 452 28–36. 10.1016/j.aquaculture.2015.10.014 [DOI] [Google Scholar]
- Silva M., Jacobus N. V., Deneke C., Gorbach S. L. (1987). Antimicrobial substance from a human Lactobacillus strain. Antimicrob. Agents Chemother. 31 1231–1233. 10.1128/AAC.31.8.1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Bäckhed F. (2013). The gut microbiota — masters of host development and physiology. Nat. Rev. Microbiol. 11 227–238. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- Strompfová V., Lauková A. (2009). Enterococci from piglets—probiotic properties and responsiveness to natural antibacterial substances. Folia Microbiol. (Praha.) 54 538–544. 10.1007/s12223-009-0078-6 [DOI] [PubMed] [Google Scholar]
- Tagg J., McGiven A. R. (1971). Assay system for bacteriocins. Appl. Microbiol. 21:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T. M., Grambsch P. M. (2001). Modeling Survival Data: Extending the Cox Model, 2nd Edn New York, NY: Springer. [Google Scholar]
- Tribolium Genome Sequencing Consortium Richards S., Gibbs R. A., Weinstock G. M., Brown S. J., Denell R., et al. (2008). The genome of the model beetle and pest Tribolium castaneum. Nature 452 949–955. 10.1038/nature067841038/nature06784 [DOI] [PubMed] [Google Scholar]
- Varankovich N. V., Nickerson M. T., Korber D. R. (2015). Probiotic-based strategies for therapeutic and prophylactic use against multiple gastrointestinal diseases. Front. Microbiol. 6:685 10.3389/fmicb.2015.00685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela S. F., Barbosa J. O., Rossoni R. D., Santos J. D., Prata M. C., Anbinder A. L., et al. (2015). Lactobacillus acidophilus ATCC 4356 inhibits biofilm formation by C. albicans and attenuates the experimental candidiasis in Galleria mellonella. Virulence 6 29–39. 10.4161/21505594.2014.981486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand I., Hilpert K., Hancock R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3 163–175. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- Yang S.-C., Lin C.-H., Sung C. T., Fang J.-Y. (2014). Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front. Microbiol. 5 683 10.3389/fmicb.2014.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi K., Koyama H., Minakuchi C., Tanaka T., Miura K. (2012). Antimicrobial peptide gene induction, involvement of Toll and IMD pathways and defense against bacteria in the red flour beetle, Tribolium castaneum. Results Immunol. 2 72–82. 10.1016/j.rinim.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zendo T., Eungruttanagorn N., Fujioka S., Tashiro Y., Nomura K., Sera Y., et al. (2005). Identification and production of a bacteriocin from Enterococcus mundtii QU 2 isolated from soybean. J. Appl. Microbiol. 99 1181–1190. 10.1111/j.1365-2672.2005.02704.x [DOI] [PubMed] [Google Scholar]
- Zou Z., Evans J. D., Lu Z., Zhao P., Williams M., Sumathipala N., et al. (2007). Comparative genomic analysis of the Tribolium immune system. Genome Biol. 8:R177 10.1186/gb-2007-8-8-r177 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.