Abstract
Beneficial aspects of endophytic microorganisms have motivated researchers to explore plant endophytic world. The present study was aimed to isolate and characterize the seed-borne endophytic bacteria from diverse maize genotypes. Eighty maize seed endophytic bacteria (MSEB), isolated from 30 maize genotypes, were characterized using polyphasic approach. The dendrograms and phylogenetic tree generated on the basis of ARDRA analysis and metabolic profiling of endophytic bacteria revealed genotypic and biochemical diversity among MSEB. The 16S rDNA sequence analysis revealed Bacillus as the most dominant encountered genus affiliated with Phylum Firmicutes. Few isolates belonged to genus Staphylococcus, whereas one isolate was identified as Corynebacterium sp. under Phylum Actinobacteria. Majority of the MSEB isolates exhibited antagonism against phytopathogenic fungi, production of ammonia, and secretion of lytic enzymes; some isolates also exhibited indole acetic acid production, the traits of which can be helpful in endophytic establishment and advantageous to the host plant. Besides, many MSEB exhibited tolerance to salinity (10%), osmotic stress (40% PEG6000), and temperature (60 °C), indicating their possible application under stress conditions. Endophytic nature of the selected MSEB isolates was confirmed by tracking their presence in shoots, leaves, and roots of the host seedlings with the help of biochemical marker (rifampicin resistance). Thus, the MSEB identified in the present study can be explored as potential bioinputs for improving plant growth and productivity under stressed conditions, besides helping in understanding the plant–endophyte interactions.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0860-0) contains supplementary material, which is available to authorized users.
Keywords: Seed endophytes, Maize, NTSYSpc, Abiotic stress
Introduction
The term “endophyte” (endon Greek for within; phyton for plant) refers to diverse microbes, most frequently fungi and bacteria, that survive and colonize internal tissues of host plant causing no apparent or immediate disease symptoms (Bacon and White 2000). A large number of culturable and unculturable endophytic bacterial species have been reported from different plant tissues, such as seeds, roots, stems, leaves, pollen, etc. during their life-cycles (Azevedo et al. 2000). Endophytes offer a wide range of benefits to plants such as growth promotion (Sturz et al. 2000), induction of plant defense mechanisms (Senthilkumar et al. 2007), production of anti-herbivory compounds (Sullivan et al. 2007), nutrition acquisition (Jha and Kumar 2007), and tolerance to biotic and abiotic stresses (Bacon et al. 2015). Besides, they produce a plethora of secondary metabolites of potential application in medicine, agriculture, and industry (Strobel et al. 2004).
Endophytic microorganisms may be transmitted either vertically (direct transfer from parent to progeny through seed) or horizontally (plant to plant) by entering the plant tissue through root zone or through aerial portions such as flowers, stems, cotyledons, etc. (James et al. 2002). It is now understood that plants and the seeds produced by them have co-evolved with diverse microorganisms (Nelson 2004). Numerous studies have provided evidence that seeds harbor a diverse endophytic microbial community (Truyens et al. 2015). Seed-associated microorganisms have been reported to play a role in the preservation and germination of the seed (Chee-Sanford et al. 2006).
Maize (Zea mays), also known as queen of Cereals, is the major source of carbohydrates, cultivated globally on nearly 150 M ha area and contributing approximately 36% (782 mt) to the global grain production (Parihar et al. 2011). In India, maize is the third most important food crop after rice and wheat. However, maize production is affected by numerous biotic and abiotic factors. Use of microbial based strategies can be ecofriendly and economic alternative to chemical inputs in combating the effect of biotic and abiotic stresses on agricultural productivity. Therefore, the present investigation was carried out to elicit the diversity of maize seed endophytic bacteria (MSEB) using molecular and biochemical approaches and to study their functional traits helpful for plant growth promotion.
Materials and methods
Isolation of seed borne endophytes
Thirty different maize genotypes from discrete sources viz., Directorate of Maize Research, New Delhi, India, regional station of National Bureau of Plant Genetic Resources, Hyderabad, and regional centre of CIMMYT, Hyderabad, were used to isolate maize seed endophytic bacteria (MSEB) (Table 1, supplementary file). A protocol was standardized for isolation of MSEB. Briefly, five seeds per genotype were treated with 0.1% of HgCl2 for 2 min under shaking conditions. After decanting mercuric chloride solution, the seeds were immersed in 95% ethanol for 4 min followed by repeated washings (ten times for complete removal of traces of mercuric chloride and ethanol) with sterile distilled water. To verify the success of the sterilization process, an aliquot of the final rinse was pour plated on two different media, i.e., TSA (Trypticase soy agar for fast growers) and R2A (Reasoner's 2A agar for slow growers). The plates were incubated at 28 ± 1 °C for 7 days to check the surface sterilization efficacy. Surface sterilized seeds per genotype were soaked in 5 ml sterile distilled water for 12 h. An aliquot of this water was again pour plated on TSA and R2A and two surface sterilized seeds per genotype were placed on the bacterial growth media to check the surface sterilization efficacy. The remaining three seeds were triturated gently with sterile motor and pestle. The macerate was pour plated on TSA and R2A (Johnston-Monje and Raizada 2011). The plates were incubated at 28 ± 1 °C and morphologically different colonies were picked and purified by re-streaking on respective media. Pure cultures of putative MSEB were maintained on nutrient agar (NA) slants at 4 °C and as glycerol stocks (60%) at −40 °C for further use.
Cultural and morphological characters
The morphological characterization was done by recording observations on color, size and other characteristics (form, margin, elevation and pigmentation) of colonies on agar medium, by following Bergey’s Manual of Determinative Bacteriology (Holt et al. 1994).
Biochemical characterization
Biochemical characterization of the isolates was done on the basis of their ability to utilize 35 different carbon substrates by using a Hi-Carbohydrate kit (HIMEDIA), as per manufacturer’s instructions. The carbon source utilization profile of the isolates was transformed into binary matrices, analyzed by using NTSYSpc and dendrogram was generated.
Molecular characterization
The total Genomic DNA was extracted from overnight raised cultures of MSEB isolates in nutrient broth at 28 ± 1 °C according to the protocol described by (Chen and Kuo1993). The isolated DNA was eluted in sterile Milli-Q water, visualized on 0.8% agarose gel by gel electrophoresis and stored at 4 °C for further use. Genomic DNA was subjected to polymerase chain reaction (PCR) for amplification of 16S rDNA in a 50-µl final volume containing 3 µl (~10.0 ng) total DNA, 0.2 µM each of universal forward (5′AGAGTTTGATCCTGGCTCAG 3′) and reverse (5′AAGGAGGTGATCCAGCCGCA3′) primer pair, 20 µL PCR master mix (Qiagen). A negative control (PCR mixture without DNA) was included in all PCR amplifications under standard conditions (initial denaturation 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 1 min 30 s and final extension at 72 °C for 7 min) using a Veriti 96-well PCR System model 9902 (Applied Biosystems, Singapore) to amplify a ~1500 bp fragment. The aliquots (5 µl) of PCR product were electrophoresed and visualized on 1.2% (w/v) agarose gel (Sambrook et al. 1989).
Amplified ribosomal DNA restriction analysis (ARDRA)
The 16S rDNA amplicon was subjected to ARDRA by following the standard protocol (Laguerre et al. 1994). Briefly, the amplified product was digested individually with 1U of restriction enzymes EcoRI, MspI and HaeIII according to manufacturer’s (GeNeiTM, India) instructions. The digested product was electrophoresed on 2.5% (w/v) agarose gel, stained with ethidium bromide and visualized using a gel documentation system. A 100-bp ladder was used as a DNA marker. Restriction pattern of amplified 16S rDNA with each of the three restriction enzymes was recorded and restriction profiles generated. The restriction profiles obtained with each of the three restriction enzymes were assembled to obtain a single combined banding pattern for each isolate. The restriction profiles (individual and combined) obtained were transformed into binary matrices and analyzed by using NTSYSpc “Numerical Taxonomy System for Personal Computer” version 2.02e. (Rohlf 1988), generating the dendrograms and the principal component analysis, with Jaccard (J) coefficient and cluster analysis was performed by the un-weighted pair group method arithmetic average (UPGMA).
Identification of MSEB based on 16S rDNA sequence
Representative isolates from different sub-clusters in the dendrogram, generated based on ARDRA (three restriction enzymes) were selected for sequencing of 16S rDNA for molecular identification. The PCR product was purified and sequenced by (Genes & Life Healthcare Pvt. Ltd., Hyderabad, India) Applied Biosystems platform. The sequences obtained were evaluated in NCBI-BLASTN against the existing GenBank database to know the identity of the isolates. ClustalW program (www.ebi.ac.uk) was used to align these sequences. Based on the 16S rDNA sequences, the evolutionary history of 35 isolates was inferred using the neighbor-joining method (Saitou and Nei 1987). The analysis involved 53 16S rDNA sequences including reference sequences. All positions containing gaps and missing data were eliminated. There were a total of 1342 positions in the final dataset. Evolutionary analysis was conducted in MEGA6 Tamura et al. (2013). The partial 16S rDNA sequences were submitted to NCBI, GenBank and accession numbers were obtained.
In vitro Screening of MSEB for abiotic stress tolerance
In order to screen the isolates for drought stress tolerance, nutrient broth amended with polyethylene glycol (PEG 6000) (10, 20, 30 and 40%) was inoculated with 1% of overnight raised culture broth. Growth of the isolates at various stress levels was estimated by measuring the optical density (OD) at 600 nm after incubation at 28 °C for 48 h, under shaking conditions (120 rpm). For studying thermo-tolerance, the overnight raised cultures were spotted on nutrient agar plates and observed for positive growth after incubation at different temperatures (30, 35, 40, 45, 50, 55 and 60 °C) for 2–4 days. The isolates were spotted on nutrient agar plates containing sodium chloride (2, 4, 6, 8 and 10%) and incubated at 28 °C for 2–4 days, to study the salinity tolerance level. All the experiments were performed in triplicate.
Screening of MSEB for plant growth promoting traits
All the 80 endophytic bacterial isolates were screened in vitro for exhibition of plant growth promoting traits like production of ammonia, siderophore, indole acetic acid (IAA), and nutrient (phosphorous, zinc and potassium) solubilization. For testing ammonia production, briefly, 50 µL of overnight raised cultures were inoculated in 10 ml of peptone water (peptone 10 g/L, NaCl 5 g/L, Agar 15 g/L, pH 7) and incubated at 28 °C for 72 h, followed by the addition of 1 ml Nessler’s reagent. Development of yellow to brown color indicated the production of ammonia (Dye 1962). For siderophore production, overnight raised cultures were spot inoculated onto nutrient agar plates amended with chrome azurol S (CAS). The plates were incubated at 28 °C and observed for the appearance of yellow to orange halo around the bacterial growth (Schwyn and Neilands 1987). For IAA production, the test organisms were spot inoculated on LB agar amended with tryptophan and incubated at 28 °C for 48 h. Post incubation, 2 to 3 drops of orthophosphoric acid were added around the bacterial colonies followed by placing a 82-mm-diameter disc of whatman grade 1 filter paper overlaid with 2 ml of Salkowski reagent (2% 0.5 M HCl in 35% perchloric acid) (Brick et al. 1991). The plates were incubated at room temperature for 30 min. Isolates producing IAA were identified by the appearance of a characteristic red halo on the filter paper in the vicinity of colony.
For testing phosphate solubilization, 4 µL of overnight raised bacterial culture was spotted on National Botanical Research Institute’s phosphate growth medium (NBRIP) containing insoluble tricalcium phosphate (0.2%) and incubated at 28 °C for 3–4 days (Pikovskaya 1948). The plates were observed for the appearance of clear zone of solubilization around the bacterial colonies. Potassium solubilisation was tested by spotting 4 µL of overnight raised culture on Aleksandrov agar plates having potassium aluminium silicate as a source of insoluble inorganic potassium (0.2%) and incubated at 28 °C for 3–7 days (Hu et al. 2006). The plates were observed for the appearance of clear zone of solubilization around the bacterial colonies. For zinc solubilization, 4 µL of overnight raised culture was spotted on basal media amended with 0.2% of insoluble Zinc (ZnO, ZnCO3) and incubated at 28 °C for 3–7 days. The appearance of clear zones around colonies indicated the ability of microbes to solubilize zinc (Saravanan et al. 2004).
Antagonism against plant pathogenic fungi
MSEB were screened in vitro for antagonistic activity against three broad host range soil-borne fungal pathogens viz. Rhizoctonia solani (root rot in maize), Sclerotia rolfsii (collar rot in maize) and Macrophomina phaseolina (charcoal rot in maize) by dual culture assay method. A 5-mm mycelial agar disc of fungal pathogen was placed on centre of MDA (malt dextrose agar) plate using a sterile borer. Aliquots (4 µL) of overnight grown bacterial cultures were spotted 2 cm away from the centre. Plates were incubated for 7 days at 25 °C and growth inhibition of fungal pathogen by MSEB was observed.
Production of lytic enzymes by MSEB
The MSEB isolates were screened for the production of lytic enzymes (amylase, esterase, lipase, protease, cellulase and pectinase) by using suitable substrates. For amylase activity the overnight raised bacterial cultures were spot inoculated on starch agar plates and incubated at 28 °C for 48 h followed by flooding with lugal’s iodine. Clear zone around the growth of colony indicated amylase production (Sahu et al. 2005). Esterase activity was determined by spot inoculating the overnight raised bacterial cultures on media composed of (g/L) peptone, 10; NaCl, 5; CaCl2, 0.1; Tween 80, 1% (v/v), agar–agar, 15; pH, 7.4. The plates were incubated at 28 °C for 5 days and observed for the presence of white precipitate around the grown colony. For lipase activity, Tween 80 was replaced with Tween 20 in the above mentioned method (Plou et al. 1998). For testing proteolytic activity, media containing nutrient broth 8 g/L, glucose 1 g/L, agar 15 g/L (pH 8) was autoclaved and mixed with 15 ml of separately autoclaved skimmed milk and poured in Petri plates. The overnight raised bacterial cultures were spot inoculated on these plates followed by incubation at 28 °C for 7 days. The plates were observed for the presence of a clear zone around the colony (Smibert and krieg 1994). Cellulase activity was determined according to method described by Emmyrafedziawati and Stella (2015). The test organism was spot inoculated on media plate containing carboxy methyl cellulose (CMC, high viscosity) and incubated at 28 °C for 7 days. After incubation, plates were flooded with Gram’s iodine and observed for development of clearing zone around the colony after 30–60 min of incubation. Pectinase activity of organisms were determined by spot inoculation of test cultures on media containing pectin as sole carbon source and incubated at 28 °C for 3 days. Pectin utilization was detected by flooding the culture plates with freshly prepared potassium iodide solution (Hankin et al. 1971).
Tracking of tagged (rifampicin resistant) MSEB in the host plant
Rifampicin-resistant mutants of selected wild-type MSEB Bacillus species were generated by spontaneous mutagenesis by spread plating overnight raised bacterial cultures on nutrient agar plates amended with 100 µg mL−1 of rifampicin followed by incubation at 28 ± 2 °C. The colonies appearing on rifampicin-amended medium were subsequently transferred 20 times on the same medium to check the stability of rif mutants (Glandorf et al. 1992). The mutants exhibiting morphological and biochemical (PGP traits) similarity were selected for plant bioassay. For plant bioassay, surface sterilized maize seeds (Bioseed 9681) were imbibed (1 h) in culture broth (108 cells per ml) of rif mutant MSEB strains and placed in test tubes (150 × 30 mm) containing soft agar (0.8%) under sterile conditions. The tubes were sealed with cotton plugs. After 5–6 days when shoot length reached test tube neck, the cotton plugs were removed and wide-mouthed tubes were inverted on tubes carrying seedlings and fixed with the help of tape to maintain the sterile conditions, in order to avoid aerial contamination of plant shoots. After 15 days of germination, the seedlings were removed aseptically and surface sterilized followed by macerated root, stem and leaf parts separately in sterile water. The macerate was placed in sterile perti plates followed by pouring of growth media containing rifampicin (100 µg/ml). After incubation, the plates were observed for growth of similar morphotypes as inoculated. The experiment was repeated under unsterile soil conditions in glass bottles covered with polybags.
Results
Isolation of endophytic bacteria from maize seed
Successful seed surface sterilization was confirmed by absence of microbial growth on the plates containing final wash, water from overnight soaking of seeds and surface sterilized seeds, after 7 days of incubation. Morphologically different colonies appearing on two growth media (TSA and R2A agar) from the seed macerate of each maize genotype were picked, purified and maintained. A total of 80 putative endophytic bacteria were isolated from 30 different maize genotypes (Table 1, supplementary file).
Molecular characterization and ARDRA
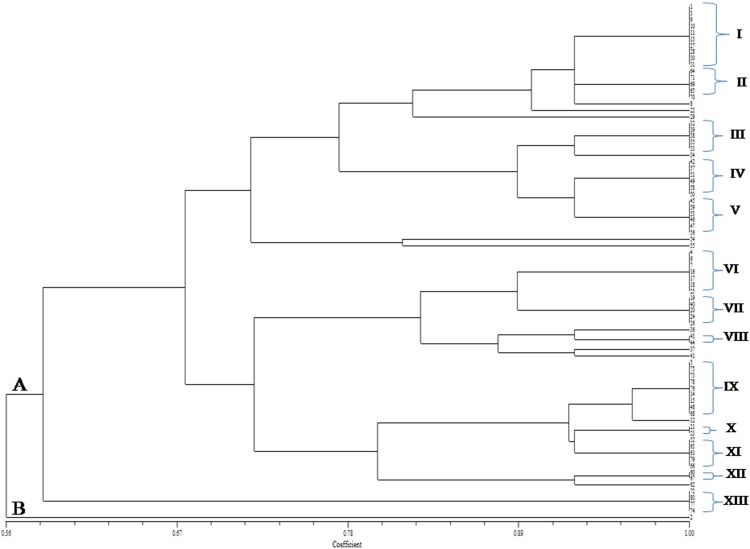
The PCR amplification of 16SrDNA sequence resulted in amplicon of ~1.5 kb for different isolates. The dendrogram generated using NTSYSpc version 2.02e software based on ARDRA sub-clustering pattern revealed molecular diversity among 80 MSEB. Dendrogram based on EcoRI restriction analysis revealed three major clusters (I, II and III) consisting of 33, 31 and 16 isolates, respectively, at a minimum similarity level of 33% (Fig. 5, supplementary file), Dendrogram based on MspI restriction analysis formed two major clusters (A and B) at a similarity level of 46% which were further sub clustered into I, II, III and IV consisting of 32, 24, 17, 4 isolates, respectively (Fig. 6, supplementary file), and the dendrogram based on Hae III restriction analysis formed two major clusters (A and B) at a similarity level of 59% which were further sub clustered into I, II, III, IV, V, VI and VII consisting of 21, 18, 2, 7, 21, 5 and 3 isolates, respectively (Fig. 7, supplementary file). For the better understanding and accuracy of diversity pattern, combined dendrogram was generated considering all three restriction enzymes. Two major clusters (A and B) at a similarity level of 56% were generated and were further sub clustered into 12 different clusters (I, II, III, IV, V,VI, VII, VIII, IX, X, XI, XII and XIII) consisting of 10, 5, 5, 6, 6, 7, 5, 2, 9, 2, 5, 2 and 4 isolates, respectively (Fig. 1).
Fig. 1.
Consensus dendrogram generated by NTSYSpc on the basis of combined 16S rDNA restriction profile (EcoRI, MspI and HaeIII enzymes) of MSEB
Biochemical profile of the MSEB
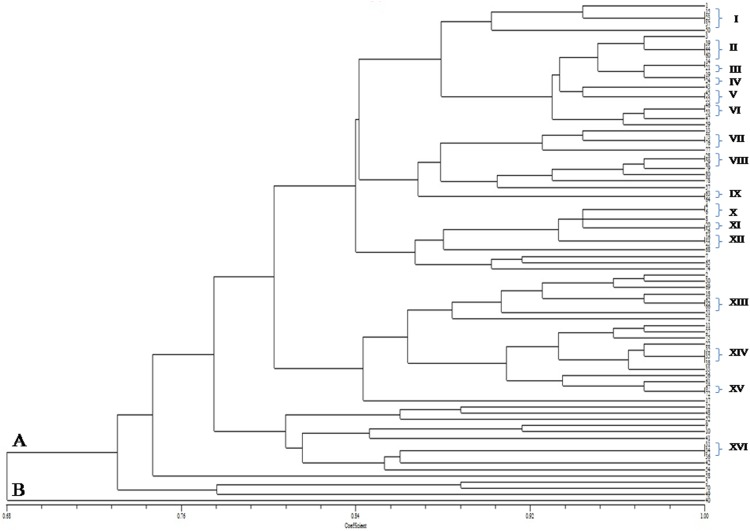
The dendrogram generated based on biochemical profile (utilization of 35 carbohydrate sources) placed 80 MSEB into two major clusters (A and B) at a similarity level of 68% which were further sub clustered into I, II, III, IV, V, VI, VII, VIII, IX, X, XI, XII, XIII, XIV, XV and XVI consisting of 5, 3, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 2, and 3 isolates, respectively (Fig. 2), thus indicating high variability among isolates with respects to metabolic efficiency.
Fig. 2.
Dendrogram generated on the basis of carbon source utilization pattern of MSEB by using NTSYSpc
Identification of MSEB based on 16S rDNA sequencing
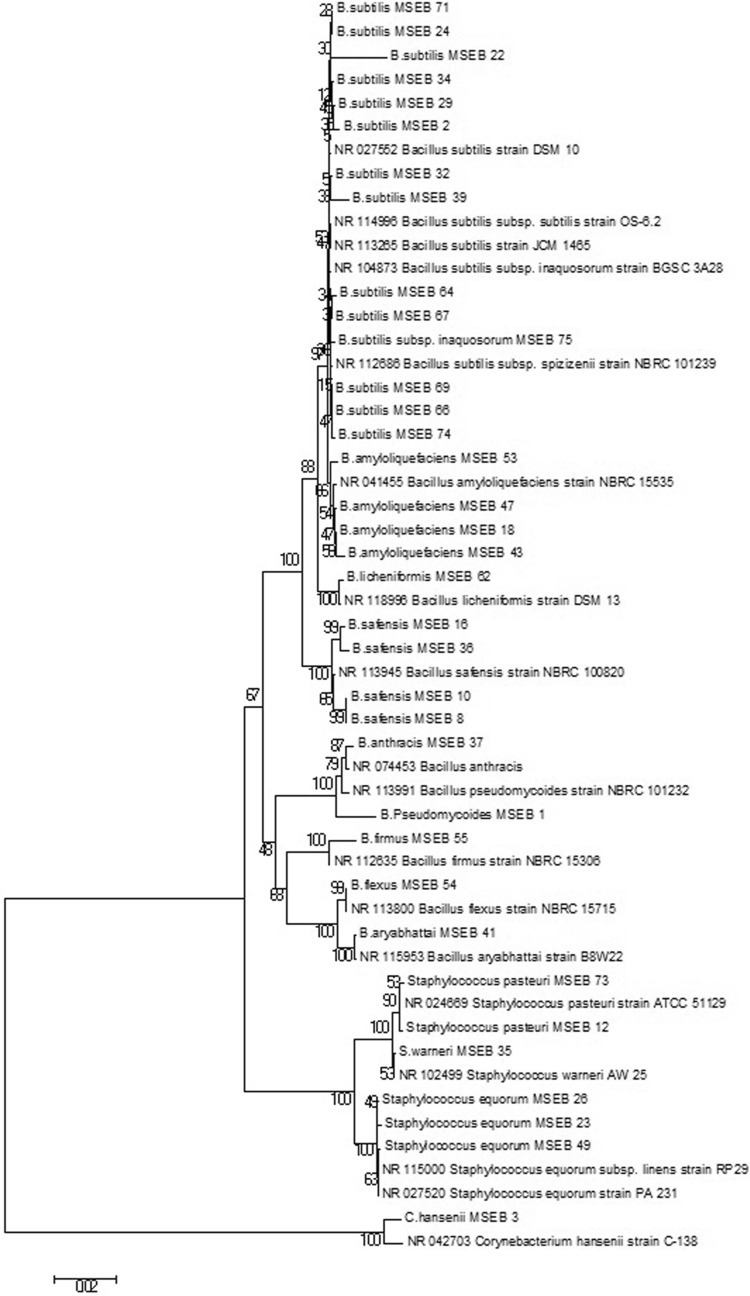
Overall, 35 isolates, identified based on 16S rDNA sequencing, showed 99–100% similarity with closest phylogenetic neighbours in GenBank database of NCBI. Of the 35 isolates, the most dominant encountered group of isolates were affiliated to genera Bacillus followed by Staphylococcus under phylum Firmicutes, whereas one isolate was affiliated to genus Corynebacterium under phylum Actinobacteria. The distribution was genetically diverse on several species and sub species level of Bacillus and Staphylococcus. Thirty-five selected MSEB isolates were identified as B. subtilis (7 isolates), B. subtilis subsp. spizizenii (3 isolates), B. subtilis subsp. inaquosorum (4 isolates), B. safensis (4 isolates), B. amyloliquefaciens (4 isolates), B. flexus (1 isolate), B. firmus (1 isolate), B. licheniformis (1 isolate), B. pseudomycoides (1 isolate), B. aryabhattai (1 isolate), B. anthracis (1 isolate), Staphylococcus warneri (1 isolate), S. equorum subsp. linens (2 isolates), S. equorum (1 isolate), S. pasteuri (2 isolate) and Corynebacterium hansenii (1 isolate). The sequence data were deposited to NCBI GenBank under the accession numbers KP261051 to KP261083, and KP063587 (Table 1). Phylogenetic tree generated based on 16S rDNA sequences, of 35 isolates, by neighbour-joining analysis revealed the relationship among different isolates. Distinct clusters of genus Bacillus could be delineated which in most cases corresponded to established species. The first largest group included nine strains of Bacillus spp. The second group included four strains of Staphylococcus spp., while the third group was more distant which included Corynebacterium sp. strain (Fig. 3).
Table 1.
Identification of MSEB, functional traits and tolerance to abiotic stress conditions
| Isolate number | ARDRA cluster no. | Similarity in NCBI data base | Query Cover (%) | Identity (%) | NCBI accession number | Functional trait exhibited in vitro | Tolerance to abiotic stress conditions |
|---|---|---|---|---|---|---|---|
| MSEB 1 | 1 | B. pseudomycoides NBRC 101232 | 100 | 98 | KP063587 | Ant, Amm, IAA, Pro, Li, Est, Amy, Cell, Pec | PEG (30%), Temp (45 °C), NaCl (10%) |
| MSEB 10 | 1 | B. safensis NBRC 100820 | 100 | 99 | KP261051 | Ant, Amm, Amy | PEG (30%), Temp (45 °C), NaCl (10%) |
| MSEB 3 | 1 | C. hansenii C-138 | 100 | 99 | KP063588 | Ant, Amm, Est, pec, | Temp (45 °C), NaCl (10%) |
| MSEB 8 | 2 | B. safensis NBRC 100820 | 100 | 99 | KP261052 | Ant, Amm, Est, Li, Pro, Pec | PEG (30%), Temp (45 °C), NaCl (10%) |
| MSEB 69 | 2 | B. subtilis subsp. spizizenii NBRC 101239 | 99 | 99 | KP261053 | Amm, IAA, Amy, Lip, Pro, Cell, Pec | PEG (30%), Temp (60 °C), NaCl (10%) |
| MSEB 64 | 2 | B. subtilis subsp. spizizenii NBRC 101239 | 99 | 99 | KP261054 | Amm, IAA, Amy, Est, Lip, Pro, Pec | PEG (40%), Temp (60 °C), NaCl (10%) |
| MSEB 71 | 2 | B. subtilis subsp. inaquosorum BGSC3A28 | 100 | 99 | KP261055 | Amm, Amy, Lip, Pro, Cell, Pec | PEG (30%), Temp (60 °C), NaCl (8%) |
| MSEB 22 | 2 | B. subtilis subsp. subtilis OS-6.2 | 100 | 98 | KP261056 | Ant, Amm, Amy, Lip, Pro, Cell, Pec | PEG (30%), Temp (45 °C), NaCl (6%) |
| MSEB 29 | 2 | B. subtilis subsp. subtilis OS-6.2 | 100 | 99 | KP261057 | Ant, Amm, Amy, Est, Lip, Pro, Cell | PEG (40%), Temp (45 °C), NaCl (8%) |
| MSEB 32 | 3 | B. subtilis subsp. subtilis OS-6.2 | 100 | 99 | KP261058 | Ant, Amm, IAA, Amy, Est, Lip, Pro, Pec | PEG (40%), Temp (45 °C), NaCl (8%) |
| MSEB 39 | 3 | B. subtilis JCM 1465 | 100 | 99 | KP261059 | Ant, Amm, Amy, Est, Lip, Pro, Cell, Pec | PEG (40%), Temp (45 °C), NaCl (8%) |
| MSEB 35 | 3 | S. warneri AW 25 | 100 | 99 | KP261060 | Ant, Amm, Est, Pro | PEG (40%), Temp (45 °C), NaCl (6%) |
| MSEB 34 | 3 | B. subtilis subsp. subtilis OS-6.2 | 100 | 99 | KP261061 | Ant, Amm | PEG (40%), Temp (45 °C), NaCl (6%) |
| MSEB 49 | 4 | S. equorum subsp. linens RP29 | 100 | 99 | KP261062 | Amm, Pec | Temp (45 °C), NaCl (10%) |
| MSEB 47 | 5 | B. amyloliquefaciens NBRC 15535 | 99 | 99 | KP261063 | Ant, Amm, Amy, Lip, Pro, Cell, Pec | PEG (40%), Temp (50 °C), NaCl (10%) |
| MSEB 53 | 5 | B. amyloliquefaciens NBRC 15535 | 100 | 99 | KP261064 | Ant, Amm, Amy, Lip, Pro, Cell, Pec | PEG (40%), Temp (45 °C), NaCl (10%) |
| MSEB 54 | 5 | B. flexus NBRC 15715 | 99 | 99 | KP261065 | Ant, Amm, Est, Lip, Cell, Pec | PEG (40%), Temp (45 °C), NaCl (10%) |
| MSEB 55 | 5 | B. firmus NBRC 15306 | 99 | 99 | KP261066 | Ant, Amm, Amy, Pro, Cell, Pec | PEG (10%), Temp (45 °C), NaCl (10%) |
| MSEB 16 | 6 | B. safensis NBRC 100820 | 100 | 99 | KP261067 | Ant, Amm, Est, Lip, Pro | PEG (20%), Temp (45 °C), NaCl (10%) |
| MSEB 18 | 6 | B. amyloliquefaciens NBRC 15535 | 99 | 99 | KP261068 | Ant, Amm, Amy, Pro, Cell, Pec | PEG (10%), Temp (40 °C), NaCl (10%) |
| MSEB 24 | 7 | B. subtilis DSM 10 | 100 | 99 | KP261069 | Ant, Amm, Amy, Lip, Cell, Pec | PEG (30%), Temp (40 °C), NaCl (10%) |
| MSEB 26 | 7 | S. equorum subsp. linens RP29 | 100 | 99 | KP261070 | Amm, Amy | PEG (30%), Temp (50 °C), NaCl (10%) |
| MSEB 36 | 8 | Bacillus safensis NBRC 100820 | 98 | 99 | KP261071 | Ant, Amm, Amy | PEG (20%), Temp (45 °C), NaCl (10%) |
| MSEB 43 | 8 | B. amyloliquefaciens NBRC 15535 | 99 | 99 | KP261072 | Ant, Amm, Amy, Est, Pro, Cell, Pec | PEG (40%), Temp (55 °C), NaCl (10%) |
| MSEB 41 | 8 | B. aryabhattai B8W22 | 100 | 99 | KP261073 | Ant, Amm, Amy, Est, Cell | PEG (40%), Temp (45 °C), NaCl (10%) |
| MSEB 73 | 9 | S. pasteuri ATCC 51129 | 100 | 99 | KP261074 | Amm, Est, Lip, Pro, Pec | PEG (40%), Temp (60 °C), NaCl (8%) |
| MSEB 75 | 9 | B. subtilis subsp. inaquosorum BGSC 3A28 | 100 | 99 | KP261075 | Amm, IAA, Amy, Est, Lip, Pro, Cell, Pec | PEG (40%), Temp (60 °C), NaCl (6%) |
| MSEB 12 | 9 | S. pasteuri ATCC 51129 | 99 | 99 | KP261076 | Amm, Amy, Pro, Cell, Pec | Temp (45 °C), NaCl (6%) |
| MSEB 23 | 10 | S. equorum PA 231 | 100 | 99 | KP261077 | Amm, Amy, Cell, Pec | PEG (10%), Temp (45 °C), NaCl (8%) |
| MSEB 66 | 11 | B. subtilis subsp. inaquosorum BGSC 3A28 | 100 | 99 | KP261078 | Ant, Amm, IAA, Amy, Est, Lip, Pro, Cell, Pec | PEG (40%), Temp (60 °C), NaCl (10%) |
| MSEB 37 | 8 | B. anthracis | 100 | 99 | KP261079 | Amm, Pro | PEG (30%), Temp (45 °C), NaCl (6%) |
| MSEB 67 | 12 | B. subtilis subsp. inaquosorum BGSC 3A28 | 100 | 99 | KP261080 | Amm, Pro | PEG (10%), Temp (60 °C), NaCl (10%) |
| MSEB 62 | 12 | B. licheniformis DSM 13 | 100 | 99 | KP261081 | Ant, Amm, Est, Lip, Pro, Cell, Pec | PEG (40%), Temp (60 °C), NaCl (10%) |
| MSEB 74 | 13 | B. subtilis subsp. spizizenii NBRC 101239 | 99 | 99 | KP261082 | Amm, IAA, Amy, Est, Lip, Pro, Cell, Pec | PEG (30%), Temp (60 °C), NaCl (10%) |
| MSEB 2 | 13 | B. subtilis DSM 10 | 100 | 98 | KP261083 | Ant, Amm, Amy, Est, Pro, Cell, Pec | PEG (20%), Temp (45 °C), NaCl (10%) |
Ant antagonism, Amm ammonia, IAA indole acetic acid, Pro protease, Li lipase, Est esterase, Amy amylase, Cell cellulase, Pec pectinase, PEG PEG6000, Temp temperature
Fig. 3.
Phylogenetic tree of MSEB based on 16S rDNA gene sequence analysis MEGA6
Screening of maize seed endophytes for abiotic stress tolerance
In vitro screening revealed that of the 80 MSEB 73, 73, 72, 55 and 42 isolates could tolerate salinity levels of 2, 4, 6, 8 and 10%, respectively. Further, 69, 43, 41 and 22 isolates could grow in the nutrient broth amended with 10, 20, 30 and 40% of PEG6000, respectively. Screening at different temperatures revealed that 80, 74, 73, 63, 30, 28, 23 isolates could grow at temperature 30, 35, 40, 45, 50, 55, 60 °C, respectively.
Plant growth-promoting traits and antagonism against fungal phytopathogens
Variation was observed among MSEB isolates with respect to plant growth promoting traits. Ammonia production was observed in all the 80 MSEB isolates, whereas, IAA production was observed in 21% of isolates. None of the isolates could exhibit siderophore production and solubilization of tricalcium phosphate, potassium aluminum silicate, zinc oxide and zinc carbonate. Of the 80, 68 MSEB isolates exhibited in vitro antagonism against one or two or three fungal pathogens. The number of isolates antagonistic towards Rhizoctonia solani, Sclerotia rolfsii and Macrophomina phaseolina were 16, 19 and 33, respectively. Of 80 isolates, 17, 15 and 7 could antagonize one, two or all three of test fungal pathogens, respectively, whereas 12 isolates did not exhibit antagonism against any of the test pathogens.
Lytic enzyme production by MSEB
Number of MSEB isolates positive for amylase, esterase, lipase, protease, cellulose and pectinase was 55, 41, 46, 55, 47 and 53, respectively. Of 80, ten isolates could produce all the six lytic enzymes, whereas two could produce none of the lytic enzymes. Majority of the isolates (51) could produce four or more lytic enzymes.
Endophytic establishment of MSEB in maize seedlings
Selected isolates (MSEB 8, MSEB 17, MSEB 72, MSEB 78) were subjected to spontaneous mutagenesis to generate rifampicin-resistant mutants. The rif mutants when used as seed inoculants could be reisolated from root, stem and leaves of host plant after 15 days of germination under sterile as well as unsterile conditions (Table 2; Fig. 4, supplementary file). The recovery of inoculated rif mutants from different plant parts confirmed the endophytic nature of the strains.
Table 2.
Reisolation of MSEB (rifampicin resistant) from root, stem and leaves of host plant under sterile as well as unsterile conditions
| Rif mutant strains | Cfu/plant (sterile conditions) | Cfu/plant (unsterile conditions) | ||||
|---|---|---|---|---|---|---|
| Root | Shoot | Leaves | Root | Shoot | Leaves | |
| MSEB 72 | TNTC | 25 | 59 | 68 | 21 | 3 |
| MSEB 78 | TNTC | 29 | 4 | 26 | 4 | 13 |
| MSEB 8 | TNTC | 86 | 25 | 48 | 31 | 25 |
| MSEB 17 | 41 | 33 | 20 | 67 | 46 | 21 |
TNTC too numerous to count
Discussion
In the present study, a culture-dependent approach was followed to assess the seed endophytic bacterial diversity of diverse maize seed genotypes using molecular and biochemical approaches. A total of 80 MSEB were isolated from surface sterilized seeds of 30 maize genotypes using two growth media. The 16S rDNA of the isolates was amplified and subjected to restriction by three enzymes. Dendrograms generated on the basis of ARDRA profiles (80 isolates) revealed HaeIII as most efficient restriction enzyme (7 sub clusters) in differentiating the isolates as compared to EcoRI and MspI. Further, dendrogram generated on the basis of combined ARDRA profile (three enzymes) differentiated the isolates into 13 sub clusters, indicating that combined ARDRA can be more discriminatory while studying the microbial diversity (Heyndrickx et al. 1996; Viti and Giovannett 2005). The dendrogram generated on the basis of biochemical profile (carbon source utilization pattern) of isolates revealed even more diversity (16 sub clusters), indicating high phenotypic diversity among MSEB isolates. The results indicate the importance of polyphasic approach in studying the bacterial diversity (Guedes et al. 2008).
Clustering pattern in the dendrogram generated on the basis of ARDRA was in line with that of phylogenetic tree generated based on 16S rDNA sequences. For example, isolates MSEB 64 and MSEB 69 were in same sub cluster II in dendrogram (ARDRA) and also clustered together in phylogenetic tree, while in few cases, phenotypic clusters and position of isolates in phenogram contradicted with their corresponding position in phylogram. For instance, isolate MSEB 2 (B. subtilis) in phenogram (Fig. 1) was found to be much distantly related with all other isolates and showing only 56% similarity, whereas in phylogram (Fig. 3) clustered with other B. subtilis strain, indicating some heterogeneity in its genetic makeup. The observations were supported by in silico ARDRA analysis that exhibited similar results. Similar pattern of results was also reported by earlier workers Bhatt and Singh (2016), who attempted to compare the phenotypic and genotypic diversity of the bacterial isolates from the saline desert by dendrogram and phylogenetic tree, respectively. While the dendrogram supported the clustering and diversity patterns displayed by the phylogenetic tree, in certain cases, distinct dendrogram patterns were evident. Certain bacteria belonging to the same species on the basis of 16S rDNA sequences displayed distinct phenotypic characteristics.
Among the 80 isolates, 35 representative isolates from ARDRA sub clusters were identified on the basis of 16S rDNA sequencing as species of three genera viz. Bacillus, Staphylococcus and Cornybacterium. These bacterial genera have been previously described as microorganisms associated with seeds of other plant species such as eucalyptus, grape vine, cactus and pumpkin (Truyens et al. 2015; Khalaf and Raizada 2016). Bacillus, Staphylococcus and Cornybacterium are Gram-positive, aerobic, endospore-forming bacteria that inhabit different ecological niches such as plants, rhizosphere, soil and water (Borriss 2011). The endospore-forming property is thought to protect the seed inhabitants from changes within the seed during storage, desiccation, seed maturation and germination (Truyens et al. 2015; Khalaf and Raizada 2016). A conspicuous feature of our study is that the genus Bacillus appeared to comprise the culturable core seed microbiota across different maize genotypes tested, indicating their more general association with maize. Bacteria belonging to genus Bacillus have previously been isolated from modern maize and its ancestor teosinte (Figueiredo et al. 2009; Johnston-Monje and Raizada 2011). Bacillus is known to be a seed endophyte with vertical transmission from one plant generation to next (White et al. 2014). Bacteria belonging to genus Bacillus have been reported to facilitate plant growth under adverse conditions besides preventing disease expansion through synthesis of novel compounds and antifungal metabolites (Bent and Chanway 1998) and form important commercial biofertilizers and biopesticides used for crop production (Borriss 2011). Similarly, plant beneficial potential of bacteria belonging to genera Staphylococcus and Corynebacterium has been reported (Akram et al. 2016). Presence of opportunistic human pathogens such as seed endophytes has been reported previously (Berg et al. 2005). However, further research can explore the link, if any, between human pathogens and plants. Isolation of Pantoea sp., Frigoribacterium sp., Microbacterium sp., Bacillus sp., Paenibacillus sp., Stenotrophomonas sp., Enterobacter sp., Sphingomonas sp. and Burkholderia sp. from kernels of different maize cultivars has been reported (Johnston-Monje and Raizada 2011; Truyens et al. 2015). Johnston-Monje and Raizada (2011) observed that seed endophyte community composition varied in relation to plant host phylogeny and there was a core microbiota of endophytes that was conserved in maize seeds across boundaries of evolution, ethnography and ecology. To the best of our knowledge this is the first report on isolation of C. hansenii, B. fermus, B. flexus, S. pasteuri and S. equorum from the surface sterilized maize seed.
The endophytic bacteria may benefit the host through different plant growth-promoting mechanisms. In the present study, 17 isolates (21%) were IAA producers. Bacterial IAA induces better root growth and/or formation of lateral roots/root hairs which can help plant in water and nutrient acquisition. All isolates exhibited production of ammonia. These results are in accordance with previous report by Prasad and Sunayana Dagar (2014). Marques et al. (2009) suggested that bacteria can produce ammonia and provide nitrogen to the host plant although over production of ammonia can serve as a triggering factor for the virulence of opportunistic plant pathogens (Passari et al. 2015). Moreover, Majority of the MSEB isolates exhibited antagonism against tested fungal phytopathogens. Members of Genus Bacillus are well known to produce antifungal metabolites. This trait can be advantageous for the plant particularly at the time of seed germination stage which is more prone to pathogen attack. And probably this can be one of the selection criteria by the host plant while selecting endophytic population. Besides, the MSEB isolates exhibited tolerance to abiotic stresses (salinity, high temperature and drought) indicating their potential application under stress conditions. Abiotic stress tolerance by endophytic bacteria has been previously demonstrated (Sanjay et al. 2014). Endophyte infection conferred population stability in tall fescue during drought stress through improved tiller and whole plant survival (West et al. 1993). Similarly, endophytes have been shown to induce mechanisms of drought avoidance (morphological adaptations), drought tolerance (physiological and biochemical adaptations) and drought recovery in infected grasses (Malinowski and Belesky 2000). Thus the trait of abiotic stress tolerance in endophytes is very relevant in climate change scenario with projections of increased frequencies and intensities of abiotic stresses in near future posing problem to agricultural productivity.
Production of lytic enzymes might help bacteria to gain entry into the plant tissues and establish as endophyte. Besides, lytic enzymes might play important role in controlling plant pathogens. Seed endophytes frequently appear to own amylase activity to make use of starch and recommence growth after long-term survival inside the seeds (Mano et al. 2006). Extracellular enzymes help microbes to degrade plant cell walls to allow entry of root-colonizing endophytes (Hardoim et al. 2008; Truyens et al. 2015). Besides, lytic enzymes also help to construct polysaccharide- and peptide-rich biofilms that help to establish the microbial community and permit attachment to host cells (e.g., rhizosphere surface, inside plants) (Khalaf and Raizada 2016). Moreover, some proteases secreted by certain strains of Bacillus are toxic to nematodes through cuticle degradation activity (Khalaf and Raizada 2016).
The colonization studies of selected MSEB using spontaneous rifampicin-resistant mutants established that the isolates inoculated on seed surface could gain entry into the host plant and aggressively colonized the roots and arial parts of maize seedlings under sterile and unsterile conditions. However, vertical transmission of the MSEB from seed to seed needs to be confirmed through further experiments.
This study has revealed the culturable endophytic bacterial diversity associated with the seeds of 30 different genotypes of the economically important maize. Bacillus was observed to be the most profusely isolated bacterial genus having capability to bestow growth promotion to their hosts. The seeds may be source for these microbes, to assist their host plants to acquire and stimulate growth. Aforesaid research may guide the development of unique biofertilizers, coated onto seeds as stable spores. Future challenge and goal will be to manage endophytic microbial communities to favor plant colonization by beneficial bacteria. This would be amenable when a better knowledge on endophytic ecology and their molecular interactions is attained. The contribution of this research has economic and environmental impact.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure 4. Establishment of MSEB in maize seedlings under sterile and unsterile conditions (TIFF 3871 kb)
Figure 5. Dendrogram of MSEB generated on the basis of 16S rDNA restriction profile (EcoRI) by using NTSYSpc (TIFF 842 kb)
Figure 6. Dendrogram of MSEB generated on the basis of 16S rDNA restriction profile (MspI) by using NTSYSpc (TIFF 842 kb)
Figure 7. Dendrogram of MSEB generated on the basis of 16S rDNA restriction profile (Hae III) by using NTSYSpc (TIFF 894 kb)
Acknowledgements
Authors of this manuscript are thankful to ICAR-AMAAS (Application of Microorganisms in Agriculture and Allied Sectors) for the financial support. Authors are also thankful to the National Coordinator, ICAR-NICRA (National Initiative on Climate Resilient Agriculture) for providing the maize seeds.
Compliance with ethical standards
Conflict of interest
The authors have declared no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0860-0) contains supplementary material, which is available to authorized users.
References
- Akram, Shahid M, Tariq M, Azeem M, Javed MT, Saleem S, Riaz S. Deciphering Staphylococcus sciuri SAT-17 mediated anti-oxidative defense mechanisms and growth modulations in salt stressed maize (Zea mays L.) Front Microbiol. 2016;7(867):1–14. doi: 10.3389/fmicb.2016.00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo JL, Maccheroni W, Jr, Pereira JO, Araújo WL. Endophytic microorganisms: a review on insect control and recent advances on tropical plants. Electron J Biotechnol. 2000;3:e1–e4. doi: 10.2225/vol3-issue1-fulltext-4. [DOI] [Google Scholar]
- Bacon CW, White JF. Microb Endophytes. New York: Marcel Deker Inc; 2000. [Google Scholar]
- Bacon CW, Palencia ER, Hinton DM (2015) Abiotic and biotic plant stress-tolerant and beneficial secondary metabolites produced by endophytic Bacillus species. In: Arora NK (ed) Plant microbes symbiosis: applied facets. Springer, New Delhi, pp 163–177. doi: 10.1007/978-81-322-2068-8_8
- Bent E, Chanway CP. The growth-promoting effects of a bacterial endophyte on lodgepole pine are partially inhibited by the presence of other rhizobacteria. Can J Microbiol. 1998;44:980–988. doi: 10.1139/w98-097. [DOI] [Google Scholar]
- Berg G, Krechel A, Ditz M, Sikora R, Ulrich A, Hallmann J. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol Ecol. 2005;51:215–229. doi: 10.1016/j.femsec.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Bhatt HB and Singh SP (2016) Phylogenetic and phenogram based diversity of haloalkaliphilic bacteria from the saline desert. In: Bhukya B, Tangutur AD (ed) Microbial biotechnology-technological challenges and developmental trends. Apple Academic Press pp 373–386 doi:10.1201/b19978-24
- Borriss R (2011) Use of plant-associated Bacillus strains as biofertilizers and biocontrol agents in agriculture. In: Maheshwari DK (ed) Bacteria in agrobiology: Plant growth responses. Springer Berlin Heidelberg. doi:10.1007/978-3-642-20332-9_3
- Brick JM, Bostock RM, Silversone SE. Rapid in situ assay for indole acetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol. 1991;57:535–538. doi: 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee-Sanford JC, Williams MM, Davis AS, Sims GK. Do microorganisms influence seed-bank dynamics? Weed Sci. 2006;54:575–587. doi: 10.1614/WS-05-055R.1. [DOI] [Google Scholar]
- Chen WP, Kuo TT. A simple and rapid method for the preparation of Gram-negative bacterial genomic DNA. Nucl Ac Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye DW. The inadequacy of the usual determinative tests for identification of Xanthomonas spp. NZT Sci. 1962;5:393–416. [Google Scholar]
- Emmyrafedziawati AKR, Stella M. Hydrolysis of carboxymethyl cellulose (CMC) by Bacillus isolated from compost. J Trop Agric and Fd Sc. 2015;43(2):129–135. [Google Scholar]
- Figueiredo JEF, Gomes EA, Guimarães CT, Lana UGP, Teixeira MA, Lima GVC, Bressan W. Molecular analysis of endophytic bacteria from the genus Bacillus isolated from tropical maize (Zea mays L.) Braz J Microbiol. 2009;40:522–534. doi: 10.1590/S1517-83822009000300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glandorf DCM, Brand I, Bakker PAHM, Schippers B. Stability of rifampicin resistance as a marker for root colonization studies of Pseudomonas putida in the field. Plant Soil. 1992;147:135–142. doi: 10.1007/BF00009379. [DOI] [Google Scholar]
- Guedes HV, dos Santos ST, Perin L, Teixeira KR, Reis VM, Baldani JI. Polyphasic characterization of Gluconacetobacter diazotrophicus isolates obtained from different sugarcane varieties. Braz J Microbiol. 2008;39(4):718–723. doi: 10.1590/S1517-83822008000400023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin L, Zucker M, Sands DC. Improved solid medium for the detection and enumeration of pectolytic bacteria. Appl Microbiol. 1971;22:205–209. doi: 10.1128/am.22.2.205-209.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim PR, von Overbeek LS, von Elsas JD. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Heyndrickx M, Vauterin L, Vandamme P, Kersters K, De Vos P. Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J Microbiol Methods. 1996;26:247–259. doi: 10.1016/0167-7012(96)00916-5. [DOI] [Google Scholar]
- Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST. Bergy’s manual of determinative bacteriology. Baltimore: Williams & Wikins Press; 1994. [Google Scholar]
- Hu XF, Chen J, Guo JF. Two phosphate and potassium solubilizing bacteria isolated from Tiannu mountain, Zhejiang, China. World J Microbiol Biotechnol. 2006;22:983–990. doi: 10.1007/s11274-006-9144-2. [DOI] [Google Scholar]
- James EK, Gyaneshwar P, Mathan N, Barraquio WL, Reddy PM, Iannetta PP, Olivares FL, Ladha JK. Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol Plant Microbe Interact. 2002;15:894–906. doi: 10.1094/MPMI.2002.15.9.894. [DOI] [PubMed] [Google Scholar]
- Jha PN, Kumar A. Endophytic colonization of Typha australis by a plant growth promoting bacterium Klebsiella oxytoca GR 3. J Appl Microbiol. 2007;103:1311–1320. doi: 10.1111/j.1365-2672.2007.03383.x. [DOI] [PubMed] [Google Scholar]
- Johnston-Monje D, Raizada MN. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS One. 2011;6(6):1–22. doi: 10.1371/journal.pone.0020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf EM, Raizada MN. Taxonomic and functional diversity of cultured seed associated microbes of the cucurbit family. BMC Microbiol. 2016;16:1–16. doi: 10.1186/s12866-015-0617-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguerre G, Allard MR, Revoy F, Amarger N. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR amplified 16S rRNA genes. Appl Environ Microbiol. 1994;60:56–63. doi: 10.1128/aem.60.1.56-63.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowski DP, Belesky DP. Adaptations of endophyte infected cool-season grasses to environmental stresses: mechanisms of drought and mineral stress tolerance. Crop Sci. 2000;40:923–940. doi: 10.2135/cropsci2000.404923x. [DOI] [Google Scholar]
- Mano H, Tanaka F, Watanabe A, Kaga H, Okunishi S, Morisaki H. Culturable surface and endophytic bacterial flora of the maturing seeds of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ. 2006;21:86–100. doi: 10.1264/jsme2.21.86. [DOI] [Google Scholar]
- Marques APGC, Pires C, Moreira H, Rangel AOSS, Castro PML. Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biol Biochem. 2009;42:1229–1235. doi: 10.1016/j.soilbio.2010.04.014. [DOI] [Google Scholar]
- Nelson EB. Microbial dynamics and interactions in the spermosphere. Ann Rev Phytopathol. 2004;42:271–309. doi: 10.1146/annurev.phyto.42.121603.131041. [DOI] [PubMed] [Google Scholar]
- Parihar CM, Jat SL, Singh AK, Kumar RS, Hooda KS, Chikkappa GK, Singh DK (2011) Maize production technologies in India. DMR Technical bulletin, Directorate of maize research, Pusa Campus, New Delhi, p 30
- Passari AK, Mishra VK, Gupta VK, Yadav MK, Saikia R, Singh BP. In vitro and in vivo plant growth promoting activities and DNA fingerprinting of antagonistic endophytic actinomycetes associated with medicinal plants. PLoS One. 2015;10(9):1–18. doi: 10.1371/journal.pone.0139468. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pikovskaya RI. Mobilization of phosphorus and soil in connection with the vital activity of some microbial species. Mikrobiologia. 1948;17:362–370. [Google Scholar]
- Plou FJ, Ferrer M, Nuero OM, Calvo MV, Alcalde M, Reyes F, Ballesteros A. analysis of tween 80 as an esterase/lipase substrate for lipolytic activity assay. Biotechnol Tech. 1998;12:183–186. doi: 10.1023/A:1008809105270. [DOI] [Google Scholar]
- Prasad MP, Darar S. Identification and characterization of endophytic bacteria from fruits like avacado and black grapes. Int J Curr Microbiol App Sci. 2014;3(8):937–947. [Google Scholar]
- Rohlf FJ. NTSYSpc Numerical taxonomy and multivariate analysis system. Exeter: Exeter; 1988. [Google Scholar]
- Sahu MK, Sivakumar K, Kannan L. Degradation of organic matters by the extracellular enzymes of actinomycetes isolated from the sediments and mollusks of the vellar estuary. J Aquat Biol. 2005;20:142–144. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. 2. NewYork: Cold Spring Harbor laboratory; 1989. [Google Scholar]
- Sanjay A, Purvi NP, Meghna JV, Rao GG. Isolation and characterization of endophytic bacteria colonizing halophyte and other salt tolerant plant species from coastal Gujarat. Afr J Microbiol Res. 2014;8(17):1779–1788. doi: 10.5897/AJMR2013.5557. [DOI] [Google Scholar]
- Saravanan VS, Subramoniam SR, Raj SA. Assessing in vitro solubilization potential of different zinc solubilizing bacterial (ZSB) isolates. Brazilian J Microbiol. 2004;35(1–2):121–125. doi: 10.1590/S1517-83822004000100020. [DOI] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophore. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Senthilkumar M, Govindasamy V, Annapurna K. Role of antibiosis in suppression of charcoal rot disease by soybean endophyte Paenibacillus sp. HKA 15. Curr Microbiol. 2007;55:25–29. doi: 10.1007/s00284-006-0500-0. [DOI] [PubMed] [Google Scholar]
- Smibert RM, Krieg NR. Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR, editors. Methods for general and molecular bacteriology. Washington: ASM; 1994. pp. 607–654. [Google Scholar]
- Strobel G, Daisy B, Castillo U, Harper J. Natural products from endophytic microorganisms. J Nat Prod. 2004;67:257–268. doi: 10.1021/np030397v. [DOI] [PubMed] [Google Scholar]
- Sturz AV, Christie BR, Nowak J. Bacterial endophytes: potential role in developing sustainable system of crop production. Crit Rev Plant Sci. 2000;19:1–30. doi: 10.1016/S0735-2689(01)80001-0. [DOI] [Google Scholar]
- Sullivan TJ, Rodstrom J, Vandop J, Librizzi J, Graham C, Schardl CL, Bultman TL. Symbiont mediated change in Lolium arundinaceum inducible defenses: evidence from changes in gene expression and leaf composition. New Phytol. 2007;176:673–679. doi: 10.1111/j.1469-8137.2007.02201.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truyens S, Weyens N, Cuypers A, Vangronsveld J. Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep. 2015;7(1):40–50. doi: 10.1111/1758-2229.12181. [DOI] [Google Scholar]
- Viti C, Giovannett L. Characterization of cultivable heterotrophic bacterial communities in Cr-polluted and unpolluted soils using Biolog and ARDRA approaches. Appl Soil Ecol. 2005;28:101–112. doi: 10.1016/j.apsoil.2004.07.008. [DOI] [Google Scholar]
- West CP, Izekor E, Turner KE, Elmi AA. Endophyte effects on growth and persistence of tall fescue along a water supply gradient. Agron J. 1993;85:264–270. doi: 10.2134/agronj1993.00021962008500020019x. [DOI] [Google Scholar]
- White JF, Torres MS, Sullivan RF, Jabbour RE, Chen Q, Tadych M, Irizarry I, Bergen MS, Havkin-Frenkel D, Belanger FC. Occurrence of Bacillus amyloliquefaciens as a systemic endophyte of vanilla orchids. Microsc Res Techniq. 2014;77(11):874–885. doi: 10.1002/jemt.22410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 4. Establishment of MSEB in maize seedlings under sterile and unsterile conditions (TIFF 3871 kb)
Figure 5. Dendrogram of MSEB generated on the basis of 16S rDNA restriction profile (EcoRI) by using NTSYSpc (TIFF 842 kb)
Figure 6. Dendrogram of MSEB generated on the basis of 16S rDNA restriction profile (MspI) by using NTSYSpc (TIFF 842 kb)
Figure 7. Dendrogram of MSEB generated on the basis of 16S rDNA restriction profile (Hae III) by using NTSYSpc (TIFF 894 kb)