Abstract
O-GlcNAcylation of carbohydrate-responsive element-binding protein (ChREBP) is believed as an important modulator of ChREBP activities, however little direct evidence of O-GlcNAcylation on ChREBP and no exact O-GlcNAcylation sites have been reported so far. Here, we validate O-GlcNAcylation on ChREBP in cell-free coupled transcription/translation system and in cells by chemoenzymatic and metabolic labeling, respectively. Moreover, for the first time, we identify O-GlcNAcylation on Ser614 in the C-terminus of ChREBP by mass spectrometry and validate two important sites, Thr517 and Ser839 for O-GlcNAcylation and their function via molecular and chemical biological method. Under high glucose conditions, Ser514 phosphorylation enhances ChREBP O-GlcNAcylation, maintaining the transcriptional activity of ChREBP; Ser839 O-GlcNAcylation is essential for Mlx-heterodimerization and DNA-binding activity enhancement, consequently inducing transcriptional activity. Ser839 O-GlcNAcylation is also crucial for ChREBP nuclear export partially by strengthening interactions with CRM1 and 14-3-3. This work is a detailed study of ChREBP O-GlcNAcylation and highlights the biological consequences of the site-specific O-GlcNAcylation dynamics of ChREBP.
Metabolic syndrome is a collection of abnormalities including obesity, type 2 diabetes mellitus, dyslipidemia, fatty liver, and proinflammatory state (1). Understanding the control of metabolism at the molecular level is of importance.
Glucose is not only an energy source but also a signaling molecule participating in physiological and pathological regulation via glucose signaling pathway (2, 3). Transcription factor carbohydrate-responsive element-binding protein (ChREBP)1 has recently emerged as a major mediator of glucose action and a central transcriptional regulator of glycolysis and de novo fatty acid synthesis in liver (3, 4) and adipose tissue (5) as well as a broader and crucial player in various processes ranging from glucolipotoxicity to apoptosis and/or proliferation in specific cell types (6–9), indicating an important role in developing metabolic syndrome. ChREBP is a basic helix-loop-helix leucine zipper transcription factor that contains several potential functional domains including a nuclear localization signal (NLS), a proline-rich stretch (PRO), a bHLH/ZIP, and a ZIP-like domain (Fig. 1). ChREBP and Max-like protein X (Mlx) form heterodimers (10, 11) and bind to the carbohydrate response element (ChoRE), which is composed of two E-boxes separated by five base pairs (CABGTG-nnCnG-nGnSTG) (12–14). As the glucose level rises, ChREBP undergoes a series of transactivation, allowing ChREBP to translocate into the nucleus and ChREBP/Mlx heterodimer to bind on the ChoRE of its target genes, which are particularly associated with glycolysis (l-type puruvate kinase, l-PK) (4, 15) and lipogenesis (acetyl-CoA carboxylase, ACC; fatty acid synthase, FAS; and stearyl-CoA desaturase 1, SCD1) (11, 13, 15). Because liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in obese ob/ob mice (16, 17), ChREBP could be a potential therapeutic target. Thus, an accurate knowledge of the regulatory mechanisms of ChREBP activity is crucial for the development of pharmacological approaches for the treatment of metabolic syndrome.
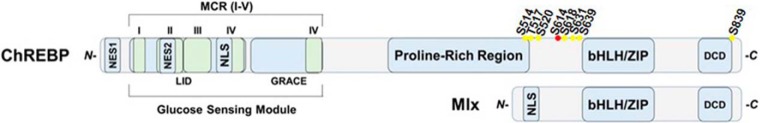
Fig. 1.
Domain structure of ChREBP and Mlx. The C-termini of ChREBP and Mlx are similar and contain a dimerization and cytoplasmic localization domain (DCD) and a basic helix-loop-helix leucine zipper region (bHLH/ZIP). The N terminus of ChREBP contains a glucose-sensing module that is functionally divided into a low-glucose inhibitory domain (LID) and a glucose-response activation conserved element (GRACE), latter of which also harbors the transactivation domain (TAD). The N-terminal region is also called Mondo conserved region (MCR). MCR is further divided into 5 subdomains MCR I-V (marked in aqua). NES: Nuclear export sequences; NLS: Nuclear localization signal. The new ChREBP O-GlcNAcylation site identified by MS in this article is marked with red dot and all sites studied in this work are also marked with yellow dots.
Today, it is well-accepted that specific post-translational modifications on ChREBP such as phosphorylation (18–20) and acetylation (21) contribute to the regulation of glucose and lipid metabolism, the molecular mechanisms enhancing its transcriptional activity in metabolic syndromes, such as obesity and type 2 diabetes, remain largely unknown. O-GlcNAcylation is a dynamic and reversible modification dependent on the activity of O-GlcNAc transferase (OGT) which transfers the monosaccharide N-acetylglucosamine (GlcNAc) to serine/threonine residues on target proteins (22), whereas O-GlcNAcase (OGA) which removes GlcNAc from target proteins (23). Emerging data indicate that O-GlcNAcylation may play an important role in the glucose response, protein stability and transactivation of ChREBP, as well as glucose utilization and lipid synthesis (24–27) in the liver, pancreas, and kidney. The regulation of ChREBP turnover is an area that has not yet been extensively investigated, and the identification of the exact serine and/or threonine residues modified by O-GlcNAcylation within the ChREBP protein will help to unravel the underlying mechanism(s).
Herein we present the direct evidence of O-GlcNAcylation on ChREBP and investigate the biological consequences of site-specific O-GlcNAcylation dynamics of ChREBP.
EXPERIMENTAL PROCEDURES
Metabolic Labeling of ChREBP Protein
Hepa1–6 cells were seeded at a density of 3 × 106 cells/100-mm culture dish. To cells at 90% confluency, Hepa1–6 cells were transiently cotransfected with expression constructs of pDEST26-HA-OGT and pCMVTNT-His-ChREBP-WT using Lipofectamine™ 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Transfection media was replaced with DMEM low-glucose media (with 10% FBS, 100 U/ml penicillin/streptomycin) containing 200 μm Ac36AzGlcNAc (1,000× stock in DMSO) or DMSO vehicle, and cells were incubated for an additional 24 h. Cells were treated with 10 μm Thiamet G 12 h before harvested for further analysis. All treatment did not affect cell viability, as determined by trypan blue exclusion.
In Vitro Cell-free Validation of ChREBP O-GlcNAcylation
In vitro coupled cell-free transcription and translation was performed on expression constructs of His6-tag mouse ChREBP and human OGT using TNT® T7 Quick Coupled Transcription/Translation Systems (Promega, Madison, WI) according to the manufacturer's instructions. In brief, 1 μg of expression construct of mouse ChREBP and 1 μg of that of human OGT were added into two individual transcription/translation reactions, with Halt™ Protease and Phosphatase Inhibitor Mixture (EDTA-free), at 30 °C for 90 min. To the end, two reactions were combined, followed by UDP-GlcNAc (final conc. 100 μm) and PUGNAc (final conc. 10 μm) being added to the mixture. The combined reaction was incubated at 37 °C for 4 h, followed by being treated with 10 units calf intestine phosphatase (New England Biolabs, Ipswich, MA) for an additional 2 h at 37 °C. Proteins were precipitated as described in the supplemental Information. Then, O-GlcNAcylated proteins were chemoenzymatically labeled with GalNAz and biotinylated using copper free click chemistry before Western blot analysis.
Chemoenzymatic Labeling of O-GlcNAcylated Proteins
Chemoenzymatic Labeling was carried out as described previously (28) using Click-iT™ O-GlcNAc Enzymatic Labeling System (Invitrogen) and control experiments were carried out in parallel in the absence of the labeling enzyme GalT Y289L or UDP-GalNAz. Biotinylation was performed using copper-free click chemistry and biotinylated proteins were pull down or analyzed by Western blot as described in the supplemental Information.
Site-Mapping of ChREBP O-GlcNAcylation: Experimental Design and Statistical Rationale
O-GlcNAcylated ChREBP was expressed and purified as described in the Supplemental Information. Thirty micrograms of each obtained protein sample were incubated with freshly made dithiothreitol (DTT, final conc. 100 mm) at 70 °C for 1 h to reduce cysteines, followed by alkylation with freshly prepared iodoacetamide (IA, final conc. 200 mm) for 1 h at room temperature in the dark. The buffer was subsequently exchanged to 25 mm NH4HCO3 using Amicon Ultra Centrifugal Filters (3 kDa molecular weight cutoff, Millipore). Proteolysis was performed using trypsin (Thermo Scientific) at 37 °C overnight with a 25:1 (w/w) protein/trypsin ratio. Samples were boiled and centrifuged, followed by 20 μl of each sample packed into the sample vials.
For each sample, the digest was analyzed using an Orbitrap Fusion Tribrid (Thermo Scientific) mass spectrometer coupled to a Proxeon Easy-nLC 1000 nanoLC system (Thermo Scientific). The peptides (∼1 μg) were desalted on a 100 μm × 3 cm C18 trap column which packed with 5 μm, 120 Å ReproSil-Pur C18-AQ beads (Dr. Maisch, Germany) for 4 min and then separated on a 75 μm × 25 cm nanoLC analytical column packed with 3 μm, 120 Å ReproSil-Pur C18-AQ resin. Mobile phase A was 0.1% formic acid in 2% aqueous acetonitrile, and mobile phase B was 0.1% formic acid in 98% aqueous acetonitrile. A step gradient of 2–7% B, 0–10 min; 7 to 22% B, 10–92 min; 22 to 35% B, 92–105 min; 35 to 90% B, 105–110 min and 90% B, 110–120 min was used. The flow rate was 300 nL/min. The mass spectrometer was operated in the positive ion mode with the following parameter settings: spray voltage, 2.4 kV; capillary temperature, 275 °C; and S-lens level, 60%. The mass acquisition range was set at 350–1550, and the resolution of the orbitrap analyzer for MS analysis was set at 120,000 (m/z = 200) with a 4 × 105 automatic gain control (AGC) target. Top speed mode with a 3 s-cycle was selected for MS2 analysis using orbitrap as the mass analyzer, the resolution was set at 30,000 (m/z = 200) with a 5 × 104 AGC target, and the maximum injection time was set at 150 ms. The parent ions were fragmented by higher-energy collisional dissociation (HCD) with normalized collision energy (NCE) set at 40%. We did three experiments for digest of proteins expressed under the HG (25 nm glucose, H), MG (15 nm glucose, M), or LG (5 nm glucose, L) condition respectively, to get a better identification. There were no extra controls and technical or biological replicates in this LC-MS/MS analysis.
Proteome discoverer (version 1.4) was used for the data processing. The raw data was searched against a Mus musculus UniProt protein database (16,785 proteins, accessed on 2016-05-07) using Sequest HT search engine. Trypsin was selected as the proteolytic enzyme. Parameters for peptide identification were set as follows: precursor mass tolerance, 10 ppm; fragment mass tolerance, 0.02 Da; maximum missed cleavage sites, 2; static modifications, carbamidomethyl modification (+57.021464 Da) of Cys residues; dynamic modifications, oxidation modification (+15.994915 Da) of Met residues, HexNAc modification (+203.079373 Da) of Ser and Thr residues. The search results were validated at 0.1% false discovery rate (FDR) with Percolator. The sites of modification on reported peptides were manually confirmed by comparing experimental sequence ions to theoretical fragment ions generated by Protein Prospector v 5.17.0. The data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (29) with the data set identifier PXD004286.
Statistical Analysis
Protein levels were measured by analyzing the bands density using Quantity One then normalized to the density of β-Actin or α-Tubulin. All experiments were repeated more than three times as biological and technical replicates, and average relative fold changes were calculated. p values were calculated from Student's paired t test when comparing within groups and from Student's unpaired t test when comparing between groups. Two-way analysis of variance (ANOVA) and Bonferroni comparison post-test were performed in the indicated figures when more than two groups were compared.
Others
Detailed methods for isolation and culture of mouse primary hepatocytes, molecular biology, cell culture and transfections, transcriptional reporter gene assays, cell extracts and protein preparation, modulation of cellular O-GlcNAc levels, chemical synthesis and labeling using copper-free click chemistry, protein precipitation using chloroform (CHCl3)/methanol (MeOH) precipitation method, biotin pull-down assay, immunoprecipitation, Western blot, and expression and purification of O-GlcNAcylated ChREBP were described in the supplemental Information.
RESULTS
O-GlcNAcylation of ChREBP Is Dynamically Induced in Hepatocytes
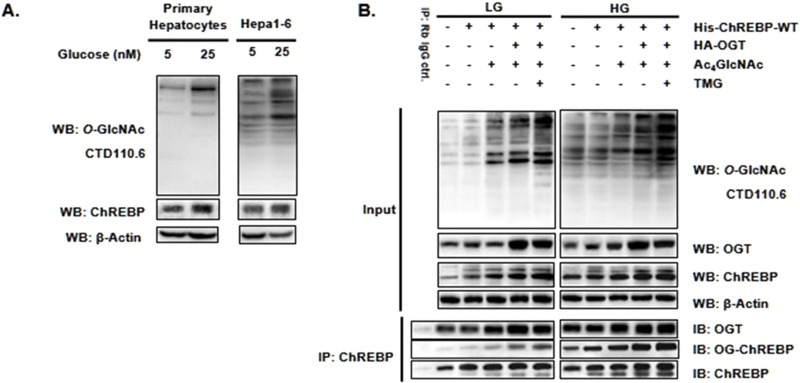
Previous reports have indicated that ChREBP, which acts as a glucose sensor, is regulated by O-GlcNAcylation (24–27), but there is no confirmed evidence for the existence and site-specific function of its O-GlcNAcylation. We sought to begin with confirming the O-GlcNAcylation of ChREBP in hepatocytes. UDP-GlcNAc is biosynthesized from several key metabolites, including glucose via the de novo hexosamine biosynthesis pathway (HBP) and 1,3,4,6-tetra-O-acetyl-2-acetamido-2-deoxy-d-glucopyranose (Ac4GlcNAc) via the GlcNAc salvage HBP (supplemental Scheme S1). Consequently, O-GlcNAcylation can be rapidly induced and is dynamically sensitive to changes in cellular UDP-GlcNAc concentrations (30). We first assessed the increased O-GlcNAcylated protein pools and ChREBP O-GlcNAcylation upon glucose and Ac4GlcNAc treatment in hepatocytes. Blotting with O-GlcNAc antibody, CTD110.6, and ChREBP antibody demonstrated that the global abundance of O-GlcNAcylation and ChREBP levels were slightly increased upon the addition of glucose in both mouse primary hepatocytes and mouse hepatocarcinoma cells, Hepa1–6 (Fig. 2A). We then confirmed the interaction between ChREBP and OGT, as well as the dynamic O-GlcNAcylation on ChREBP in Hepa1–6 cells. The ChREBP-overexpressed Hepa1–6 cells with or without OGT cotransfection were incubated in indicated glucose and Ac4GlcNAc concentrations with or without pharmacological inhibition of OGA using a specific inhibitor Thiamet-G (TMG) (31). The OGT was copurified in ChREBP immunoprecipitates. The results showed an increase of the global O-GlcNAcylation levels upon the addition of glucose and Ac4GlcNAc, whereas OGT overexpression and TMG treatment further enhanced the O-GlcNAc modification (Fig. 2B). Moreover, the O-GlcNAcylated ChREBP was detected using the anti-O-GlcNAc antibody (CTD110.6) on the immunoprecipitated ChREBP. An interaction between ChREBP and OGT was observed and the O-GlcNAcylation on ChREBP was enhanced by OGT overexpression and TMG treatment (Fig. 2B).
Fig. 2.
O-GlcNAc modification of ChREBP in hepatocytes. A, Primary hepatocytes (left) and Hepa1–6 cells (right) were exposed to indicated glucose concentration. The increase of the O-GlcNAcylated protein pools and ChREBP levels were assessed by Western blot (WB). B, ChREBP with (lane 3, 4, 7, 8) or without (lane 1, 2, 5, 6) OGT cotransfected Hepa1–6 cells were incubated in 5 mm (LG, left) or 25 mm (HG, right) glucose condition with or without 200 μm Ac4GlcNAc and treated with 10 μm Thiamet if needed. Coimmunoprecipitation (Co-IP) was performed using anti-ChREBP antibody. The O-GlcNAc-modified protein pools, the levels of ChREBP and OGT were determined by WB, respectively. All data are representative of 3 independent experiments.
In vitro Cell-free and Cell-based Validation of ChREBP O-GlcNAcylation
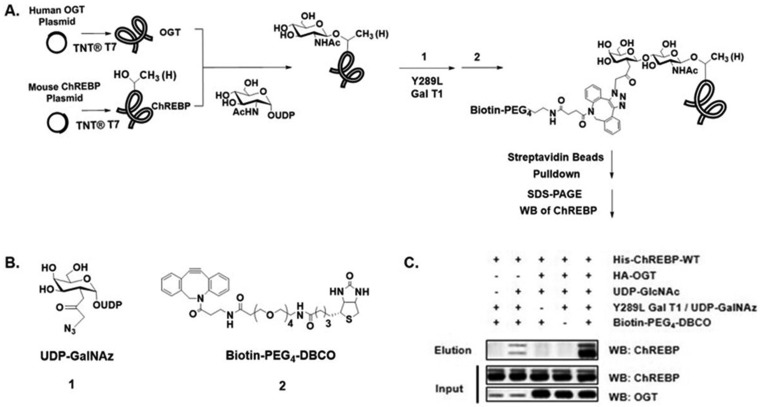
All the previous studies on O-GlcNAcylation of ChREBP used O-GlcNAc antibody CTD110.6 to prove the existence of O-GlcNAc motifs on immunoprecipitated ChREBP from cells or tissues. Considering that the CTD110.6 antibody has been shown to bind to other O- and N-linked glycans (32), further validation of ChREBP O-GlcNAcylation is needed. To further confirm the O-GlcNAcylation modified by OGT on ChREBP, we combined our newly modified cell-free coupled transcription/translation system (TNT® T7) with a well-established chemoenzymatic labeling approach (28) to express ChREBP and OGT, modify proteins with O-GlcNAc, and label O-GlcNAcylated proteins with biotin (Fig. 3A). Immunoblotting of the biotin-labeled proteins with anti-ChREBP antibody showed that only the addition of UDP-GlcNAc caused O-GlcNAc modification of ChREBP, and the OGT administration strongly enhanced this modification (Fig. 3C). The data demonstrated that O-GlcNAcylation of ChREBP was a result of the interaction between ChREBP and OGT.
Fig. 3.
In vitro cell-free validation of ChREBP O-GlcNAcylation. A, Schematic diagram of the method. B, Structures of tool molecules. C, Detection of O-GlcNAcylated ChREBP. Reaction mixture prior to pull down (Input) and the captured proteins (Elution) were immunoblotted with antibodies toward ChREBP and OGT. UDP-GlcNAc addition and OGT administration strongly enhanced the modification (lane 2, 5). Control experiments in absence of Y289L GalT1 (lane 3) or biotin derivate (lane 4) demonstrated selective labeling. All data are representative of 3 independent experiments.
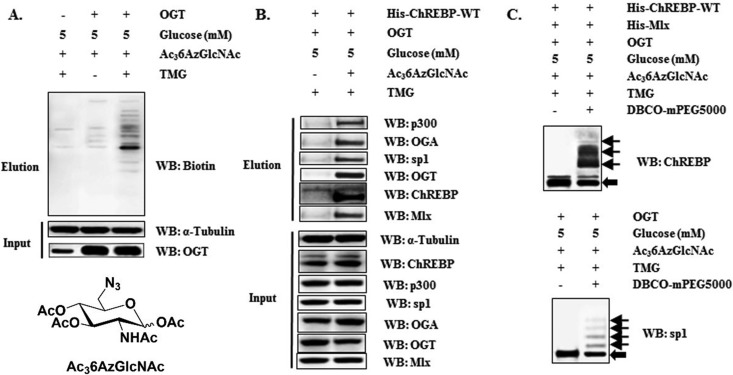
We next examined the dynamic O-GlcNAcylation of ChREBP in cells. This was done via cell-based metabolic labeling of ChREBP with 1,3,4-tris-O-acetyl-6-azido-6-deoxy-N-acetyl-glucosamine (Ac36AzGlcNAc), a synthetic specific metabolic chemical reporter for O-GlcNAcylation (33). Transfected Hepa1–6 cells were cultured in conditioned media with 5 mm glucose and 200 μm Ac36AzGlcNAc, treated with TMG if needed, and the cell lysates were subjected to copper-free click reaction for biotinylation (supplemental Scheme S2). Western blot of biotin-labeled proteins using streptavidin-HRP against biotin revealed labeling of a wide-range of proteins in cells, and OGT overexpression and TMG treatment could largely enhance the efficiency of 6-azido-6-deoxy-N-acetyl-glucosamine (6AzGlcNAc) metabolic incorporation (Fig. 4A). To determine if 6AzGlcNAc could report on O-GlcNAcylation of a certain protein, the eluates were immunoblotted with antibodies against ChREBP and its interactors. As shown in Fig. 4B, both ChREBP and its interactors including p300, OGT, OGA, and Mlx, were captured, indicating that O-GlcNAcylation might occur on these proteins. Specificity protein 1 (SP1)—a known O-GlcNAc modified protein (34)—was detected as a reference and Western blot of input before biotinylation and enrichment exhibited equal protein levels in all samples (Fig. 4B). To validate these findings, we also immunoprecipitated ChREBP and these interactors from the modified and biotinylated lysates respectively using corresponding antibodies, and then immunoblotted biotinylation. Strong biotinylation signals indirectly reflected the direct O-GlcNAcylation on these proteins (supplemental Fig. S1A). ChREBP and these interactors were immunoprecipitated from the natural unlabeled cell lysates as well, and O-GlcNAcylation signals were also detected using anti-O-GlcNAc antibody CTD110.6 (supplemental Fig. S1B). Based on all these data, we claimed that O-GlcNAcylation directly occurred on ChREBP, as well as its interactors, p300, OGT, OGA, and Mlx.
Fig. 4.
Cell-based validation of ChREBP O-GlcNAcylation. A, OGT overexpressed Hepa1–6 cells were exposed to indicated conditions with 200 μm Ac36AzGlcNAc for 24 h. Lysates were assessed for OGT, α-Tubulin, and Eluates for the biotin-modified protein pools by WB. B, Hepa1–6 cells were cotransfected with OGT and ChREBP and exposed to indicated conditions with 200 μm Ac36AzGlcNAc. Eluates from the biotin pull-down assay were assessed for ChREBP, p300, OGA, OGT, Mlx and SP1by WB. All these proteins and α-Tubulin from cell lysates were immunoblotted as references. C, Metabolic labeled cell lysates were reacted with PEG mass-tag and assessed for ChREBP (up) and SP1 (down) respectively by WB. All data are representative of 3 independent experiments.
Simultaneously, to quantify the stoichiometry of ChREBP O-GlcNAcylation, we selectively labeled O-6AzGlcNAc-modified proteins with a 5-kDa polyethylene glycol (PEG) mass tag to shift their molecular mass through copper-free click chemistry. This method we used was modified based on the previous report (35). Immunoblotting with ChREBP antibody enabled the direct visualization of both the non-glycosylated and glycosylated ChREBP. Omission of the PEG mass tag prevented the mass shift and confirmed the specificity of the reaction (Fig. 4C). SP1 was also detected as a reference. All these data confirmed that ChREBP can be directly O-GlcNAcylated by OGT both in vitro and in cells.
O-GlcNAcylation Sites on ChREBP Are Identified by Tandem Mass Spectrometry
To map the sites of O-GlcNAcylation on ChREBP, high-resolution tandem mass spectrometric (MS/MS) analysis was performed. To facilitate mass spectrometry, His-tagged ChREBP was expressed in HEK293T cells in high O-GlcNAcylation conditions and purified using Ni-NTA agarose. After trypsin digestion, the generated peptides were subjected to nanoLC-MS/MS using higher-energy collisional dissociation (HCD) with normalized collision energy (NCE) set at 40%. Database search of the LC-MS/MS data led to the identification of several peptides derived from ChREBP with a sequence coverage of 56% (depicted in supplemental Fig. S8A, shaded gray). We observed four peptides containing a single O-GlcNAcylation site and two peptides containing multiple O-GlcNAcylation sites. Notably, almost all O-GlcNAcylated peptides of ChREBP were detected from digest of proteins expressed under the HG condition and no modification was identified when under the LG condition. The MS/MS for ions of all the O-GlcNAcylated peptides were provided in the supplemental information (supplemental Figs. S2∼S7). All of the O-GlcNAcylated peptides observed exhibited the expected neutral loss of the GlcNAc monosaccharide from precursor peptide and fragment ions upon HCD, as well as generation of three or more diagnostic HexNAc fragment ions at m/z 126, 138, 144, 168, 186, or 204 (36, 37). Interestingly, multiple O-GlcNAcylation sites were found close in ChREBP protein primary sequence, and the peptide 513ASPPTLASATASPTATATAR532 was identified with different O-GlcNAcylation patterns at the same time. Then, we validated these O-GlcNAcylation sites by molecular biological and chemical biological methods.
To confirm our newly found O-GlcNAcylation sites, site-mutation analysis was performed in combination with chemoenzymatic labeling and biotin-pull down assay. Each individual O-GlcNAcylated residue identified in the MS/MS analysis was mutated to an alanine (A) or valine (V) residue to block O-GlcNAcylation. A noteworthy reduction of ChREBP O-GlcNAcylation level was observed corresponding to T517V and S839A mutants. O-GlcNAcylation deficiency at S514, S520, S614, S618, S631, and S639 of ChREBP also caused a certain reduction of protein O-GlcNAcylation (supplemental Fig. S9). All the results gave us much prospect of O-GlcNAcylation sites confirmation by further analysis.
Site-Specific O-GlcNAcylation Deficiency on ChREBP Affects Its Transcriptional Activity
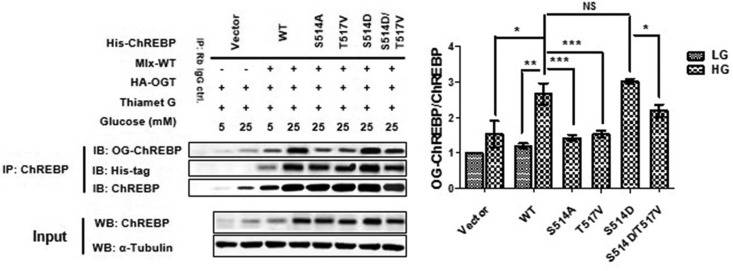
Because all information of the ChREBP O-GlcNAcylation sites in this study was new, we next focused on the biological consequences of the site-specific O-GlcNAcylation dynamics. Previous studies have reported phosphorylations on S514 (38) and S614 (20), which directly competed with O-GlcNAcylation for the target sites, therefore two more mutants with O-GlcNAcylated residue mutated to an aspartate (D) to potentially mimic phosphorylation were constructed. All the ChREBP mutants were then tested for their relative transcriptional activity under HG conditions. Each ChREBP construct was expressed in Hepa1–6 cells as described in supplementary method to ensure that it gave a product with the appropriate size and was expressed at comparable levels. Of all the single-site mutants constructed, transcriptional activities of majority of ChREBP mutants were comparable with that of WT ChREBP (supplemental Fig. S10). Only two mutants, S514D and S614A, showed a more robust activity compared with WT ChREBP. However, the S514A and S614D mutants behaved similarly to WT ChREBP, indicating that O-GlcNAcylation and phosphorylation of these sites alone were not decisive for glucose activation. Two other single mutants, T517V and S839A, displayed a significant decrease in its transcriptional activity (like vector transfection) relative to the WT ChREBP, suggesting that O-GlcNAcylation status at these sites might modulate ChREBP activity more essentially. Given the modest effect of other mutations on ChREBP transcriptional activity, they were not pursued further in this work. Overall, we concluded that the O-GlcNAcylation status of T517 and S839 was critical for the glucose activation of ChREBP under HG conditions, and probably O-GlcNAcylation on S514 and S614 was important for competition with phosphorylation and optimal activation, but not indispensable.
Ser514 Phosphorylation Enhances ChREBP O-GlcNAcylation
In our MS/MS data, two O-GlcNAcylation patterns were found for the peptide 513ASPPTLASATASPTATATAR532 at the same time (supplemental Figs. S2 and S6), indicating a regulation mechanism for O-GlcNAcylation site selection. A previous study has provided information on phosphorylation of S514 by the mammalian target of rapamycin complex 1 (mTORC1), which is up-regulated in response to rapamycin (38), we want to investigate the interference between Ser514 phosphorylation and Thr517 O-GlcNAcylation. Immunoprecipitated ChREBP or its mutants from transfected HEK293T cells were immunoblotted for the O-GlcNAcylation level. Under the HG conditions, the S514D mutant maintained, but the T517V one significantly decreased, ChREBP O-GlcNAcylation levels when compared with that of the wild type ChREBP (Fig. 5). It was suggested that phosphorylation at S514 of ChREBP could enhance the O-GlcNAcylation of the full-length protein, and T517 was an important site for ChREBP O-GlcNAcylation levels. To know whether S514 phosphorylation directly enhanced O-GlcNAcylation at T517, a two-site mutant S514D/T517V was employed to remedy a lack of T517 O-GlcNAcylation antibody for the detection of ChREBP O-GlcNAcylation level variations. The S514D/T517V mutant had lower O-GlcNAcylation level compared with both that of the S514D one and that of WT ChREBP, but still higher than that of the T517V one (Fig. 5). It was indicated that S514 phosphorylation could enhance ChREBP O-GlcNAcylation at many sites other than T517, and T517 of ChREBP was an important site for ChREBP O-GlcNAcylation.
Fig. 5.
Interference between Ser514 phosphorylation and Thr517 O-GlcNAcylation. Either wild type (WT) ChREBP or one of the site-mutated ChREBP mutants overexpressed HEK293T cells were cotransfected with OGT and Mlx under the indicated conditions. Immunoprecipitation was performed using anti-ChREBP antibody. The levels of ChREBP and O-GlcNAcylated ChREBP were determined by WB. α-Tubulin and ChREBP from cell lysates were immunoblotted as references. Each site-specific-mutated ChREBP mutants gave a product with the appropriate size and was expressed at comparable levels relative to WT ChREBP under HG conditions. Left lane shows a representative WB result of 7 independent experiments. Right lane shows WB result in statistical result. Error bars denote the mean ± S.E., n = 7. * p < 0.05; ** p < 0.01; *** p < 0.001; NS: no significance.
O-GlcNAcylation at C-Terminus of ChREBP Is Important for Its Function
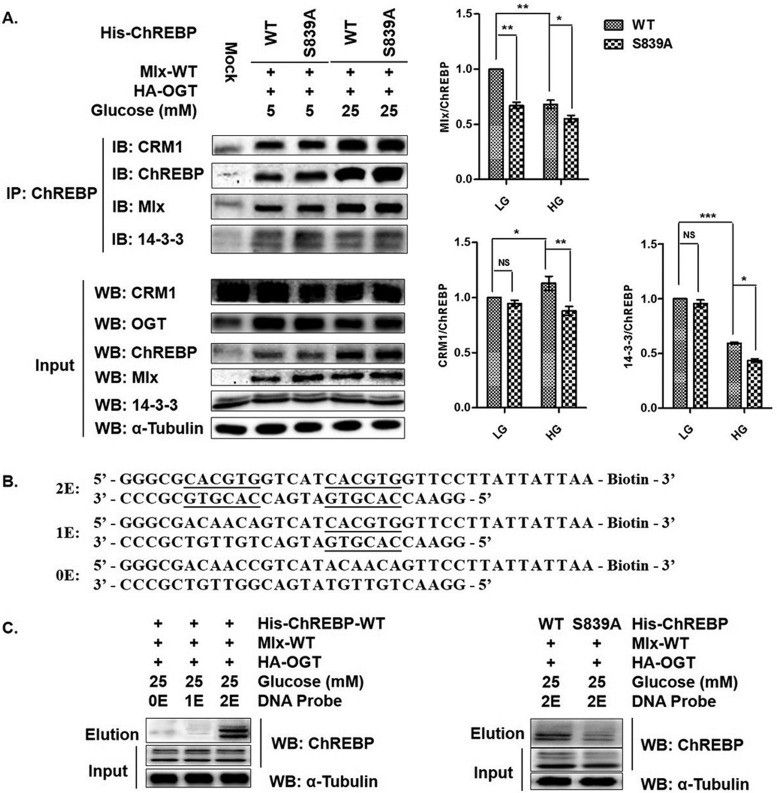
The result that transcriptional activity of S839A ChREBP significantly decreased versus WT ChREBP shown in supplemental Fig. S10, suggested a latent mechanism for O-GlcNAcylation-dependent regulation of ChREBP activity to a certain extent. Because the C-terminal region of ChREBP has been implicated in DNA-binding and Mlx-heterodimerization, we first analyzed the effect of deficient O-GlcNAcylation at S839 of ChREBP on interaction between ChREBP and Mlx. HEK293T cells were cotransfected with expression constructs of ChREBP (WT or S839A), OGT and Mlx, and incubated in the LG or HG conditions with TMG treatment for 12 h before harvest. Coimmunoprecipitation was performed in cell lysates using ChREBP antibody and the eluates were assessed for ChREBP and Mlx using corresponding antibodies. Fig. 6A showed less copurified Mlx with S839A mutant versus the wild type, indicating a weaken interaction between ChREBP and Mlx under HG condition without O-GlcNAcylation at S839 of ChREBP.
Fig. 6.
O-GlcNAcylation at Ser839 of ChREBP is important for its function. A, Either WT ChREBP or S839A ChREBP mutant was coexpressed in HEK293T with OGT and Mlx, and incubated under the indicated conditions for 12 h with TMG treatment. Coimmunoprecipitation was performed using anti-ChREBP antibody and the eluates were assessed for ChREBP and its interactors, Mlx, CRM1, and 14-3-3 by WB. Cell lysates were also immunoblotted as references. Left lane shows a representative WB result of 6 independent experiments. Right lane shows WB result in statistical result. Error bars denote the mean ± S.E., n = 6. * p < 0.05; ** p < 0.01; *** p < 0.001; NS: no significance. B, Designed DNA-binding probes for ChREBP, whose E-boxes of the glucose-responsive binding element—ChoRE—were underlined. C, Cells were transfected like A and incubated under the HG condition. Cell lysates were pulled down using biotinylated DNA probes and streptavidin-beads. Eluates and cell lysates were assessed for ChREBP by WB. α-Tubulin from cell lysates was immunoblotted as a reference. Left lane shows only 2E DNA probe could pull down ChREBP, right lane shows DNA-binding differences between WT ChREBP and S839A mutant. All data are representative of 3 independent experiments.
We next designed a biotinylated DNA probe (Fig. 6B) to pull down ChREBP from HG-incubated, ChREBP (WT or S839A), OGT and Mlx cooverexpressed HEK293T cell lysates. As shown in Fig. 6C, only the 2E DNA probe containing two tandem E-boxes could pull down wild type ChREBP, whereas E-box substitutions (1E and 0E) were totally deficient in binding to ChREBP (Fig. 6C, left). When using the 2E DNA probe to pull down WT ChREBP and S839A ChREBP, there was a significant reduction of eluted S839A ChREBP compared with WT ChREBP (Fig. 6C, right). These results demonstrated that deficient O-GlcNAcylation at S839 of ChREBP impaired its DNA binding activity, maybe because of attenuate Mlx-heterodimerization.
Although S839 lies in the C-terminal region of ChREBP, our coimmunoprecipitation experiment in Fig. 6A showed that O-GlcNAcylation deficiency at this site did reduce the interactions of ChREBP with CRM1 and 14-3-3, which were reported to bind to the N-terminal region of ChREBP and assist translocation of ChREBP from nucleus to cytoplasm (39–41). To see whether S839A mutation alters ChREBP nucleocytoplasmic shuttling, immunofluorescence studies were performed to track the ChREBP proteins in Hepa1–6 cells transfected with His-tagged WT ChREBP or the S839A mutant (supplemental Fig. S11). The confocal microscopy images showed a marked increase in nuclear accumulation of S839A ChREBP under the HG condition compared with that of WT ChREBP, potentially owing to the weakened assistant from CRM1 and 14-3-3. These observations indicated that O-GlcNAcylation at S839 of ChREBP was important for its function performing.
DISCUSSION
ChREBP has emerged as a major player in the development of metabolic syndrome, but the molecular mechanisms regulating glucose-dependent activation of ChREBP remain largely unknown. Because of the lack of specific detection and identification methods for O-GlcNAcylation sites on ChREBP, the question how O-GlcNAcylation regulates ChREBP activity has troubled researchers for years. This is the first work to more specifically modulate and validate O-GlcNAcylation on ChREBP using chemical biology methods besides the antibody-based method.
Ac4GlcNAc treatment provides a complementary method for modulating ChREBP O-GlcNAcylation in cells via the GlcNAc salvage HBP, which might have the less disturbance on the cell metabolism compared with glucose. Using our modified in vitro transcription/translation method, we detected strong O-GlcNAcylation signal on ChREBP and its interactors. Although the other groups reported using in vitro transcription/translation method to get O-GlcNAcylated proteins before (42, 43), our method is more rapid and potent for studying protein-protein interactions and protein modifications without the additional administration of purified OGT protein. Application of the specific metabolic reporter for O-GlcNAcylation-Ac36AzGlcNAc (33) in Hepa1–6 cells enables the O-GlcNAc-selective labeling of ChREBP as well as the subsequent detection and enrichment of O-GlcNAcylated ChREBP, which provides a more specific and efficient way compared with the well-accepted antibody-based method.
Moreover, mass spectrometry provides the most straightforward and detailed insight into the O-GlcNAcylation on proteins despite of the difficulty to get enough modified target proteins and the lability of O-GlcNAcylation during sample preparation and detection. Fortunately, we detected 6 O-GlcNAcylated peptides of ChREBP from the coverage of peptides for all ChREBP MS/MS (shaded yellow in supplemental Fig. S8A). Additional regions of ChREBP that contain serine and threonine residues are not represented in our analysis (unshaded in supplemental Fig. S8A), and any modifications present on these peptides would have been missed. Our work is the study basis for the mechanism by which O-GlcNAcylation regulates ChREBP signaling. For example, previous studies have shown that S514 and S614 of ChREBP could be phosphorylated by mTORC1 (38) and glycogen synthase kinase 3 (GSK3) (20) respectively, O-GlcNAcylations at these two sites could directly compete with phosphorylation and consequently alter ChREBP activity. To offset our failure to get site information using ETD, we combined site-mutation analysis with chemoenzymatic labeling and biotin-pull down assay to validate our newly found O-GlcNAcylation sites. Because ChREBP O-GlcNAcylation was studied using mass spectrometry combined with molecular and chemical biological methods for the first time, we focused on the biological consequences of the site-specific O-GlcNAcylation dynamics. The location of each O-GlcNAcylation site with respect to functional domains of ChREBP was shown in Fig. 1. All sites were found cluster in the C-terminal region of ChREBP outside the bHLH/ZIP domain and DCD, which mediates DNA-binding and Mlx-heterodimerization. The C-terminal region of ChREBP was a region remarkably conserved between ChREBP from a wide variety of species and between ChREBP and its paralogue, MondoA (44, 45) (supplemental Fig. S8B). O-GlcNAcylation sites in this region may be important for ChREBP transcriptional activity. Therefore, we employed a l-PK gene reporter assay to investigate effects of site-specific O-GlcNAcylation dynamics on ChREBP transcriptional activity. According to the reference, retreatment of cells with the HG condition caused a rapid increase in l-PK mRNA, with a maximal level reached after 24 h, and the abundance of l-PK mRNA returned rapidly to low levels after the HG condition removed (3). Our analysis was conducted at 24 h after treatment with HG, at which time transcription was strongly induced. Our results highlight the regulatory role of site-specific O-GlcNAcylation at Thr517 and Ser839 of ChREBP in cells. Clearly, further analysis will be necessary to distinguish between these possibilities. Expression rates stimulated by glucose of several genes belonging to the glycolytic/lipogenic pathways are various in different cells, so that there may be disputable activity tendency using different gene reporters or even the same reporter at different time point. A consensus is clear that further analysis will be necessary to distinguish the mechanisms by which ChREBP differ from one gene to another when stimulated by glucose.
Although the previous work has shown the enhancement of ChREBP transcriptional activity for target genes by O-GlcNAcylation (25), the regulatory molecular mechanism is unknown till now. Our current data shows that the deficient O-GlcNAcylation at Ser839 nearly abolish ChREBP transcriptional activity (supplemental Fig. S10). The attenuated interaction between ChREBP and its partner, Mlx (Fig. 6A), as well as the disrupted DNA binding activity (Fig. 6B), are all direct explanations for the decreased target gene expression. Like many transcription factors, ChREBP is modulated by various post-translational modifications (PTMs). Regarding the C-terminal region of ChREBP, several PKA modification sites, such as S626/T666, negatively regulate ChREBP nuclear import and DNA-binding activity, respectively (19). In addition to phosphorylation regulation, HAT activity-containing transcriptional coactivator p300 associates with ChREBP and acetylates Lys672 in the bHLHZ domain and promotes ChREBP DNA binding (21). In current study, we found Ser839 O-GlcNAcylation as an additional site-specific regulatory PTM, which is necessary for ChREBP function by enhancing Mlx-heterodimerization and DNA binding activity, consequently potentiating the induction of target genes.
Moreover, we found O-GlcNAcylation deficiency at S839 of ChREBP also affects its nuclear export relative to WT ChREBP under the HG condition (supplemental Fig. S11). It should be noted that the putative nuclear export sequence (NES) or nuclear localization signal (NLS) lies in the N terminus of ChREBP, known as a glucose-sensing module (GSM), and Ser839 is far away from these regions. The basis of its increased nuclear accumulation is investigated via coimmunoprecipitation experiment, and weakened interactions of ChREBP with CRM1 and 14-3-3 account for this increased nuclear retention to some extent (Fig. 6A). The C-terminus of ChREBP, MondoA and Mlx are similar and contain a dimerization and cytoplasmic localization domain (DCD). Because conserved domains at the C-terminus of MondoA and Mlx control their subcellular localization (46), conserved domains of ChREBP and Mlx may also contribute to their localization (supplemental Fig. S12). The relative rates of nuclear import and export of a given protein contribute to its subcellular localization at steady state. Our data demonstrate that O-GlcNAcylation at S839 of ChREBP is necessary for its nuclear export, but the molecular mechanism for the enhancement of its interactions with CRM1 and 14-3-3 is still confusing.
In this study, we also found phosphorylation at Ser514 can enhance O-GlcNAcylation of ChREBP and T517 is an important site for ChREBP O-GlcNAcylation. Because phosphorylation at Ser514 by mTORC1 can be up-regulated by Rapamycin (38), a well-known inducer of cell autophagy though mTOR pathway, our finding proposes the new question for us: what's the role of ChREBP and its O-GlcNAcylation in autophagy? Further studies will be in need to address this question.
This work sheds light on the regulatory molecular mechanism of ChREBP O-GlcNAcylation, elucidating the importance of site-specific O-GlcNAcylation for maintaining protein structure, aiding for DNA-binding, responding to HG-dependent transcriptional activation, and thus potentially triggering its target protein signaling, which may render cells susceptible to hyperglycemia. These findings could help us to understand the role of glucose metabolism in metabolic syndrome and suggest potential targets to prevent the development of glucotoxicity and lipotoxicity.
DATA AVAILABILITY
The data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (29) with the data set identifier PXD004286.
Supplementary Material
Acknowledgments
We thank G.-J. Boons for gifting us the expression construct of OGT plasmid.
Footnotes
Author contributions: A.Y. and X.Y. designed research; A.Y. and D.L. performed research; A.Y., L.C., and X.Y. analyzed data; A.Y. and X.Y. wrote the paper.
* This work was financially supported by the grants (2013CB910700, 2012CB822100) from the Ministry of Science and Technology of China, and the National Natural Science Foundation of China (21232002).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ChREBP
- carbohydrate-responsive element-binding protein
- Mlx
- Max-like protein X
- NLS
- nuclear localization signal
- bHLH/ZIP
- basic-helix-loop-helix-leucine zipper
- ChoRE
- carbohydrate response element
- L-PK
- L-type puruvate kinase
- ACC
- acetyl-CoA carboxylase
- FAS
- fatty acid synthase
- SCD1
- stearyl-CoA desaturase 1
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase
- HBP
- hexosamine biosynthesis pathway.
REFERENCES
- 1. Eckel R. H., Grundy S. M., and Zimmet P. Z. (2005) The metabolic syndrome. Lancet 365, 1415–1428 [DOI] [PubMed] [Google Scholar]
- 2. Foufelle F., Gouhot B., Pegorier J. P., Perdereau D., Girard J., and Ferre P. (1992) Glucose stimulation of lipogenic enzyme gene expression in cultured white adipose tissue. A role for glucose 6-phosphate. J. Biol. Chem. 267, 20543–20546 [PubMed] [Google Scholar]
- 3. Girard J., Ferre P., and Foufelle F. (1997) Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annu. Rev. Nutr. 17, 325–352 [DOI] [PubMed] [Google Scholar]
- 4. Yamashita H., Takenoshita M., Sakurai M., Bruick R. K., Henzel W. J., Shillinglaw W., Arnot D., and Uyeda K. (2001) A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. U.S.A. 98, 9116–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Herman M. A., Peroni O. D., Villoria J., Schon M. R., Abumrad N. A., Bluher M., Klein S., and Kahn B. B. (2012) A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 484, 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tong X., Zhao F., Mancuso A., Gruber J. J., and Thompson C. B. (2009) The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 106, 21660–21665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Metukuri M. R., Zhang P., Basantani M. K., Chin C., Stamateris R. E., Alonso L. C., Takane K. K., Gramignoli R., Strom S. C., O'Doherty R. M., Stewart A. F., Vasavada R. C., Garcia-Ocaña A., and Scott D. K. (2012) ChREBP mediates glucose-stimulated pancreatic β-cell proliferation. Diabetes 61, 2004–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soggia A., Flosseau K., Ravassard P., Szinnai G., Scharfmann R., and Guillemain G. (2012) Activation of the transcription factor carbohydrate-responsive element-binding protein by glucose leads to increased pancreatic β-cell differentiation in rats. Diabetologia 55, 2713–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guillemain G., Filhoulaud G., Da Silva-Xavier G., Rutter G. A., and Scharfmann R. (2007) Glucose is necessary for embryonic pancreatic endocrine cell differentiation. J. Biol. Chem. 282, 15228–15237 [DOI] [PubMed] [Google Scholar]
- 10. Stoeckman A. K., Ma L., and Towle H. C. (2004) Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. J. Biol. Chem. 279, 15662–15669 [DOI] [PubMed] [Google Scholar]
- 11. Ishii S., Iizuka K., Miller B. C., and Uyeda K. (2004) Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc. Natl. Acad. Sci. U.S.A. 101, 15597–15602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shih H. M., Liu Z., and Towle H. C. (1995) Two CACGTG motifs with proper spacing dictate the carbohydrate regulation of hepatic gene transcription. J. Biol. Chem. 270, 21991–21997 [DOI] [PubMed] [Google Scholar]
- 13. Ma L., Robinson L. N., and Towle H. C. (2006) ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J. Biol. Chem. 281, 28721–28730 [DOI] [PubMed] [Google Scholar]
- 14. Jeong Y. S., Kim D., Lee Y. S., Kim H. J., Han J. Y., Im S. S., Chong H. K., Kwon J.-K., Cho Y. H., Kim W. K., Osborne T. F., Horton J. D., Jun H. S., Ahn Y. H., Ahn S. M., and Cha J. Y. (2011) Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PLoS ONE 6, e22544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iizuka K., Bruick R. K., Liang G., Horton J. D., and Uyeda K. (2004) Deficiency of carbohydrate response element-binding protein (ChREBP. reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. U.S.A. 101, 7281–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dentin R., Benhamed F., Hainault I., Fauveau V., Foufelle F., Dyck J. R., Girard J., and Postic C. (2006) Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes 55, 2159–2170 [DOI] [PubMed] [Google Scholar]
- 17. Iizuka K., Miller B., and Uyeda K. (2006) Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am. J. Physiol. Endocrinol. Metab. 291, E358–E364 [DOI] [PubMed] [Google Scholar]
- 18. Tsatsos N. G., and Towle H. C. (2006) Glucose activation of ChREBP in hepatocytes occurs via a two-step mechanism. Biochem. Biophys. Res. Commun. 340, 449–456 [DOI] [PubMed] [Google Scholar]
- 19. Kawaguchi T., Takenoshita M., Kabashima T., and Uyeda K. (2001) Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc. Natl. Acad. Sci. U.S.A. 98, 13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsatsos N. G., Davies M. N., O'Callaghan B. L., and Towle H. C. (2008) Identification and function of phosphorylation in the glucose-regulated transcription factor ChREBP. Biochem. J. 411, 261–270 [DOI] [PubMed] [Google Scholar]
- 21. Bricambert J., Miranda J., Benhamed F., Girard J., Postic C., and Dentin R. (2010) Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Invest. 120, 4316–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haltiwanger R. S., Blomberg M. A., and Hart G. W. (1992) Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase. J. Biol. Chem. 267, 9005–9013 [PubMed] [Google Scholar]
- 23. Dong D. L., and Hart G. W. (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-β-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 [PubMed] [Google Scholar]
- 24. Sakiyama H., Fujiwara N., Noguchi T., Eguchi H., Yoshihara D., Uyeda K., and Suzuki K. (2010) The role of O-linked GlcNAc modification on the glucose response of ChREBP. Biochem. Biophys. Res. Commun. 402, 784–789 [DOI] [PubMed] [Google Scholar]
- 25. Guinez C., Filhoulaud G., Rayah-Benhamed F., Marmier S., Dubuquoy C., Dentin R., Moldes M., Burnol A. F., Yang X., Lefebvre T., Girard J., and Postic C. (2011) O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 60, 1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ido-Kitamura Y., Sasaki T., Kobayashi M., Kim H. J., Lee Y. S., Kikuchi O., Yokota-Hashimoto H., Iizuka K., Accili D., and Kitamura T. (2012) Hepatic FoxO1 integrates glucose utilization and lipid synthesis through regulation of Chrebp O-glycosylation. PLoS ONE 7, e47231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park M. J., Kim D. I., Lim S. K., Choi J. H., Han H. J., Yoon K. C., and Park S. H. (2014) High glucose-induced O-GlcNAcylated carbohydrate response element-binding protein (ChREBP) mediates mesangial cell lipogenesis and fibrosis: the possible role in the development of diabetic nephropathy. J. Biol. Chem. 289, 13519–13530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Clark P. M., Dweck J. F., Mason D. E., Hart C. R., Buck S. B., Peters E. C., Agnew B. J., and Hsieh-Wilson L. C. (2008) Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J. Am. Chem. Soc. 130, 11576–11577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen D. L., Gloster T. M., Yuzwa S. A., and Vocadlo D. J. (2012) Insights into O-linked N-acetylglucosamine ([0–9]O-GlcNAc) processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates. J. Biol. Chem. 287, 15395–15408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., and Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490 [DOI] [PubMed] [Google Scholar]
- 32. Isono T. (2011) O-GlcNAc-specific antibody CTD110.6 cross-reacts with N-GlcNAc2-modified proteins induced under glucose deprivation. PLoS ONE 6, e18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chuh K. N., Zaro B. W., Piller F., Piller V., and Pratt M. R. (2014) Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification. J. Am. Chem. Soc. 136, 12283–12295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jackson S. P., and Tjian R. (1988) O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell 55, 125–133 [DOI] [PubMed] [Google Scholar]
- 35. Rexach J. E., Rogers C. J., Yu S. H., Tao J., Sun Y. E., and Hsieh-Wilson L. C. (2010) Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol. 6, 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao P., Viner R., Teo C. F., Boons G. J., Horn D., and Wells L. (2011) Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J. Proteome Res. 10, 4088–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Myers S. A., Daou S., Affar el B, Burlingame A. (2013) Electron transfer dissociation (ETD): the mass spectrometric breakthrough essential for O-GlcNAc protein site assignments-a study of the O-GlcNAcylated protein host cell factor C1. Proteomics 13, 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Demirkan G., Yu K., Boylan J. M., Salomon A. R., and Gruppuso P. A. (2011) Phosphoproteomic profiling of in vivo signaling in liver by the mammalian target of rapamycin complex 1 (mTORC1). PLoS ONE 6, e21729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukasawa M., Ge Q., Wynn R. M., Ishii S., and Uyeda K. (2010) Coordinate regulation/localization of the carbohydrate responsive binding protein (ChREBP) by two nuclear export signal sites: discovery of a new leucine-rich nuclear export signal site. Biochem. Biophys. Res. Commun. 391, 1166–1169 [DOI] [PubMed] [Google Scholar]
- 40. Merla G., Howald C., Antonarakis S. E., and Reymond A. (2004) The subcellular localization of the ChoRE-binding protein, encoded by the Williams-Beuren syndrome critical region gene 14, is regulated by 14-3-3. Hum. Mol. Genet. 13, 1505–1514 [DOI] [PubMed] [Google Scholar]
- 41. Sakiyama H., Wynn R. M., Lee W. R., Fukasawa M., Mizuguchi H., Gardner K. H., Repa J. J., and Uyeda K. (2008) Regulation of nuclear import/export of carbohydrate response element-binding protein (ChREBP): interaction of an α-helix of ChREBP with the 14-3-3 proteins and regulation by phosphorylation. J. Biol. Chem. 283, 24899–24908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Starr C. M., and Hanover J. A. (1990) Glycosylation of nuclear pore protein p62. Reticulocyte lysate catalyzes O-linked N-acetylglucosamine addition in vitro. J. Biol. Chem. 265, 6868–6873 [PubMed] [Google Scholar]
- 43. Ortiz-Meoz R. F., Merbl Y., Kirschner M. W., and Walker S. (2014) Microarray discovery of new OGT substrates: the medulloblastoma oncogene OTX2 is O-GlcNAcylated. J. Am. Chem. Soc. 136, 4845–4848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Billin A. N., Eilers A. L., Coulter K. L., Logan J. S., and Ayer D. E. (2000) MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol. Cell Biol. 20, 8845–8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cairo S., Merla G., Urbinati F., Ballabio A., and Reymond A. (2001) WBSCR14, a gene mapping to the Williams–Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum. Mol. Genet. 10, 617–627 [DOI] [PubMed] [Google Scholar]
- 46. Eilers A. L., Sundwall E., Lin M., Sullivan A. A., and Ayer D. E. (2002) A novel heterodimerization domain, CRM1, and 14-3-3 control subcellular localization of the MondoA-Mlx heterocomplex. Mol. Cell Biol. 22, 8514–8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (29) with the data set identifier PXD004286.