Abstract
Crohn's Disease (CD) is a relapsing inflammation of the gastrointestinal tract that affects a young working age population and is increasing in developing countries. Half of all sufferers will experience stricturing or fistulizing intestinal complications that require extensive surgical interventions and neither genes nor clinical risk factors can predict this debilitating natural history. We applied discovery and verification phase studies as part of an NCI-FDA modeled biomarker pipeline to identify differences in the low-mass (<25kDa) blood-serum proteome between CD behavioral phenotypes. A significant enrichment of epithelial component proteins was identified in CD patients with intestinal complications using quantitative proteomic profiling with label-free Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS). DAVID 6.7 (NIH) was used for functional annotation analysis of detected proteins and immunoblotting and multiple reaction monitoring (MRM) to verify a priori findings in a secondary independent cohort of complicated CD (CCD), uncomplicated inflammatory CD (ICD), Th1/17 pathway inflammation controls (rheumatoid arthritis), inflammatory bowel disease controls (ulcerative colitis), and healthy controls. Seventy-six high-confidence serum proteins were modulated in CCD versus ICD by LC-MS/MS (p < 0.05, FDR q<0.01), annotating to pathways of epithelial barrier homeostasis (p < 0.01). In verification phase, a putative serology panel developed from discovery proteomics data consisting of desmoglein-1, desmoplakin, and fatty acid-binding protein 5 (FABP5) distinguished CCD from all other groups (p = 0.041) and discriminated complication in CD (70% sensitivity and 72.5% specificity at score ≥1.907, AUC = 0.777, p = 0.007). An MRM assay secondarily confirmed increased FABP5 levels in CCD (p < 0.001). In a longitudinal subanalysis-cohort, FABP5 levels were stable over a two-month period with no behavioral changes (p = 0.099). These studies along the biomarker development pipeline provide substantial proof-of-principle that a blood test can be developed specific to transmural intestinal injury. Data are available via the PRIDE proteomics data repository under identifier PXD001821 and PeptideAtlas with identifier PASS00661.
Crohn's disease (CD)1 is a progressive Inflammatory Bowel Disease (IBD) in which more than half of all patients will experience a stricturing (SCD) or fistulizing (FCD) complication within 10 years from diagnosis (1, 2). The cause of progression to complicated (SCD and FCD) disease (CCD) is unknown and can only be diagnosed through colonoscopy or cross-sectional radiological imaging (2, 3). By that time, irreversible and cumulative damage has occurred and the ensuing surgeries, prolonged hospitalizations, and disability make up a significant component of the overall disease burden of CD (4, 5). A young age at diagnosis, positive anti-Saccharomyces cerevisiae antibody (ASCA) serology, ileal disease, and perianal disease are risk factors for CCD, however their predictive accuracies remain unclear (2, 3, 6). Genotyping also only accounts for 13.6% of the variance in CD, which makes further prediction for CCD challenging (3, 7). Robust time-sensitive predictors of disease course are needed to be able to evaluate the efficacy of early escalation or “Top-down” therapies, which may stand the best chance for changing the natural history of CD (2).
Proteins are the mechanistic components that directly lead to phenotypic manifestations (8, 9). Blood serum contains up to 10,000 proteins and has unique access to the full-thickness of intestinal tissues through the microvasculature, which distinguishes it from current modalities of gastrointestinal tract monitoring that are retrospective and macroscopic in nature (2, 3). This makes the blood serum a distinct source of in vivo, quantitative and real-time disease behavior markers in this context. Eighty percent of the serum protein concentration is comprised of albumin and immunoglobulins, with unique small molecular weight proteins existing at low concentrations (<1 ng/ml), including C-reactive protein (CRP), interleukins and peptide hormones (10, 11). This dynamic range makes the serum “proteome” both technically challenging to explore and in need of special statistical power proficiencies (8, 9, 12). In this study, we examine the low-mass (<25kDa) serum protein fraction between CD behavioral phenotypes in discovery and qualification phase proteomic studies of the National Cancer Institute-Food and Drug Administration (NCI-FDA) developed clinical biomarker pipeline (13). The low-mass serum fraction of CD behavioral phenotypes is enriched using in-solution electrophoresis for untargeted “shotgun” analysis by label-free quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS), and immunoblotting and multiple reaction monitoring (MRM) MS is used to confirm LC-MS/MS findings in qualification phase (12).
An enrichment of epithelial barrier component proteins is reported in the low-mass serum fraction of CCD, and a putative 3-protein serology panel that distinguishes CCD from inflammatory CD (ICD), Th1/17 inflammation pathway controls (rheumatoid arthritis (RA)), IBD controls (ulcerative colitis (UC)), and healthy controls (C) is established. In keeping with the NCI-FDA biomarker workflow, a NCI and Clinical Proteomic Tumor Analysis Consortium (CPTAC)-guideline Tier-2 MRM assay was developed for biomarker candidate fatty acid-binding protein 5 (FABP5) (13, 14). MRM assay was used to confirm serum FABP5 differences between CCD, ICD, RA, UC, and controls, and to investigate intraindividual FABP5 variation in a longitudinal subanalysis cohort.
EXPERIMENTAL PROCEDURES
Study Population and Sample Collection
Serum samples were obtained from subjects recruited from Concord Repatriation General Hospital and Bankstown-Lidcombe Hospital, Sydney Australia. CD, UC, and RA subjects were recruited from IBD and rheumatology ambulatory clinics respectively, and controls from patients without gastrointestinal diseases who underwent endoscopy (e.g. for screening or exclusion of gastrointestinal bleeding) with normal findings. To test biomarker specificity, RA patients were selected as positive inflammatory controls as the disease shares certain Th1/17 response pathways with CD (15). IBD diagnoses were confirmed by histological and endoscopic criteria and RA by rheumatoid arthritis classification criteria of at least 6 months duration. All CD subjects had their behavioral phenotype confirmed by a gastroenterologist with radiologic and/or endoscopic evidence within 30 days from blood sampling as part of their routine care. CCD was defined as presence of active intestinal complications (untreated/balloon dilated strictures and nonhealed abscess/fistulas). ICD subjects with concomitant perianal disease were excluded as a potential confounder. Disease-specific activity indices for CD, UC, and RA (Crohn's Disease Activity Index (CDAI), Partial Mayo, and 28-Joint Disease Activity Score (DAS28), respectively) with paired biochemical markers of inflammation (CRP and Erythrocyte Sedimentation Rate (ESR)) were collected. Twenty milliliters of peripheral blood were drawn from consenting subjects in two 20 ml SST tubes. Serum was separated from blood cells by centrifugation at 1400 rpm immediately after collection and frozen at −80 °C. All participants gave written informed consent.
Ethics
This study was approved by the Sydney Local Health District Human Research Ethics Committee (Approval code: CH62/6/2011–154).
Extraction of Low-Mass Serum Fraction
In-solution electrophoresis was used to partition the 1–25 kDa protein component from serum samples as previously described (16, 17). Briefly, Pooled serum samples were created by combining 30 μl aliquots of individual patient samples based on CD behavioral phenotype, and protease inhibitor (Roche, Basel, Switzerland) was added to pooled serum samples according to manufacturer recommendations and treated serums were then diluted with 150 μl of 180 mm Tris/20 mm EACA/4 M urea buffer, pH 10.2; in a 1:1 ratio. Protease inhibited serum samples were loaded onto a ProteomeSep electrophoresis instrument (NuSep, Sydney, Australia). Serum samples were separated into 1–25 kDa, 26–45 kDa, 46–65 kDa, and 66–125 kDa partitions and the 1–25 kDa (low-mass) fraction was collected for LC-MS/MS.
Label-Free LC-MS/MS
Low-mass fraction serum samples were prepared with C18 stage tips (Thermo Scientific, IL, USA) according to manufacturer recommendations except for an 80% Acetonitrile, 0.1% formic acid elution buffer. Dried peptides were re-solubilized in 50 μl of 50 mm NH4HCO3, pH 8.0. Trypsin was then added to the samples at an enzyme to protein ratio of ∼1:100 and incubated overnight at 37 °C. Following digestion, 4 μl of formic acid was added to stop the reaction and samples were dried. Samples were then resuspended in 10 μl 2% CH3COOH, 0.1% formic acid prior to LC-MS/MS.
One microliter (10%) of each sample was injected onto the nano-LC for analysis. An LTQ-FT Ultra mass spectrometer (Thermo Electron, Bremen, Germany) was used to analyze the serum samples. The sample contents were separated by nano-LC using an Ultimate 3000 HPLC and autosampler (Dionex, Amsterdam, Netherlands). Samples were loaded onto a micro C18 precolumn (500 μm × 2 mm, Bruker, CA) with Buffer B (98% H20, 2% CH3CN, 0.1% TFA) at 10 μl/min. After a 4-min wash the precolumn was switched (Valco 10 port valve, Dionex) into line with a fritless nano column (75 μm i.d × 10 cm) containing reverse-phase C18 media (5 μm, 200Å Magic, Bruker). Peptides were eluted using a linear gradient of Buffer A (98% H20, 2% CH3COOH, 0.01% HFBA) to Buffer B (98% CH3CN, 2% H20, 0.01% HFBA) at 250 nL/min over 60 min. The column tip was positioned ∼0.5 cm from the heated capillary (T = 200 °C) of the LTQ-FT and 1800V was applied to a low volume tee (Upchurch Scientific, WA). The instrument was operated in Data-Dependent Acquisition (DDA) mode with positive ions generated by electrospray. A survey scan over a 350–1750 mass-to-charge ratio (m/z) range was acquired in the FT ICR cell. Collision-induced dissociation was used by the linear ion trap in which up to 8 of the most abundant ions (>2000 counts) with charge states of ≥ (M+2H)+2 were successively isolated and fragmented. Mass-to-charge ratios selected for MS/MS were dynamically excluded for 60 s. All samples were analyzed in technical triplicates.
LC-MS/MS Spectra Analysis
LTQ-FT *.RAW data files were imported into Progenesis LC-MS v4.0 (Progenesis QI) (Waters, MA). Mass spectra from each run were aligned to a reference sample by matching consistent m/z ions by retention time in a pair-wise fashion. Quantitation of ions was achieved by gain factor calculation (log-scale abundance ratio scaling factor) against the reference sample. Within-group CV% of ions was first evaluated and ions that were significantly changing within the group by one-way Analysis of Variance (ANOVA) (p < 0.05) were discarded from subsequent between-group analyses. Statistical comparisons between groups were performed by one-way ANOVA with multiple testing correction by FDR q value significance set at < 0.01(18). Mass spectra of significantly modulating ions were then target-decoy searched against the Swiss-prot protein knowledgebase (Release 2011_04, Apr 2011, containing >526,900 sequence entries) using the Mascot v2.3 search engine (Matrix Science, MA) with 5ppm (peptide) and 0.5Da (ms/ms) error tolerance settings. Mascot search parameters were set to “All-species,” with no enzyme and variable modifications to: cysteine (acrylamide); methionine (oxidation); serine, threonine, and tyrosine (phosphorylation). Peptides with anion scores >20 were considered for identification. Peptide-sequence matches and protein inferences were then mapped back to quantitative ion data and summary protein-level statistics were calculated (fold change, one-way ANOVA p value, q value and power).
Functional Annotation Analysis
Functional annotation analysis was performed with DAVID v6.7 (The Database for Annotation, Visualization, and Integrated Discovery, NIH). A homo sapien gene list was used as the background comparator for the submitted data set and the functional annotation chart and functional annotation clustering tools were examined to determine enriched biological features in the LC-MS/MS data. For the functional annotation chart an EASE score (modified Fisher's exact test) cutoff of <0.01 after Benjamini-Hochberg (BH) correction was set as the significance threshold for enrichment of each identified Gene Ontology (GO) term (19). For functional annotation clustering, the identified biological module groups were evaluated by their enrichment score (determined by -log transformation of the mean of all individual p values for terms within the cluster), and the significance of the module's enrichment was determined by a BH-corrected EASE score of <0.01 for their top GO term (by EASE score) within the group (19). Enrichment values of individual GO terms are reported as fold change, EASE scores and Benjamini-Hochberg corrected p values, and biological module clusters as enrichment scores (19). Three proteins were then selected as preliminary candidates for qualification phase immunoblot and MRM assay development.
MRM
A NCI-CPTAC Tier 2 MRM assay was developed for FABP5 as an essential part of the NCI-FDA biomarker pipeline workflow (14). Briefly, LC-MS/MS sequenced FABP5 peptides were searched against the National Center for Biotechnology Information (NCBI) Protein BLAST database for uniqueness, and a sequence-specific precursor/fragment ion profile (transition list) was developed for a proteotypic peptide in Skyline SRM environment v1.4 (MacCoss Lab, UW). The MRM method was then validated by extensive iterative experimentation. Tier-2 level quantification was performed using a Reverse-Polynomial Dilution (RPD) calibration (20). Serum samples were analyzed on a 4000QTrap (ABSCIEX, MA) mass spectrometer coupled to a Dionex Ultimate 3000 liquid chromatograph (Thermo Fisher Scientific). Pertinent assay information has been reported in compliance with NCI-CPTAC and the National Heart, Lung and Blood Institute's (NHLBI) Proteomics Centers' recommendations (supplementary Experimental Procedures) (14).
Immunoblot Assay
Dot blots for desmoglein-1 (DSG1), desmoplakin (DSK), and fatty acid-binding protein 5 (FABP5) proteins were performed in serum samples following standard procedures (21). Briefly, 5 μl of serum was dotted onto PVDF membranes, incubated in primary and secondary antibodies and developed. Quantitation was performed using ImageJ 1.48v (NIH).
Experimental Design and Statistical Rationale
This study was conducted in accordance with the biomarker development pipeline outlined by Rifai et al. (13). The 2013 NCI-FDA workshop statistical model used to determine verification phase sample size (based on a biomarker candidate signal difference of greater than 5.0 standard deviations between groups and >80% power in verification phase) (12). Values are expressed as mean ± S.E. All variables were evaluated for normality of distribution using the Shapiro-Wilk test of normality and the appropriate statistical test consequently applied: One-way ANOVA was used to compare biomarker values between categorical independent variables with Gaussian distributions and Kruskal Wallis H test for those with non-Gaussian distributions. A Pearson's product-moment correlation and Spearman's rank-order correlation were used for equivalent continuous variables. Independent variables that met an association probability of p < 0.1 were then entered into a multiple regression analysis using the enter method to determine their predictive ability for biomarker values with all other clinical factors held constant. Discriminant function analysis was used to determine discriminability of DSG1, DSK, and FABP5 for CCD and the subsequent discriminant function coefficients were then used to weight each protein value contribution to a combined biomarker panel score—hereafter referred to as the Serum Epithelial Components (SEC) score. Receiver operating characteristic (ROC) was used to determine classification ability of biomarker values and SEC score for CCD. After applying a Shapiro-Wilk test of normality on the difference between paired samples, a paired t test was used to evaluate intra-individual variations in biomarker concentrations in the longitudinal cohort. SPSS 20 (IBM) was used for statistical analyses. GraphPad Prism 6 (GraphPad Software, CA) and TIBCO Spotfire (Tibco, MA) were used for graphical presentation of results.
Further details on experimental procedures, statistical analyses and SEC score calculation are provided in supplementary Materials. The LC-MS/MS data in this manuscript has been deposited at the PRIDE proteomics data repository (The European Bioinformatics Institute, Cambridge, UK) with the data set identifier PXD001821 at (http://www.ebi.ac.uk/pride/archive/). The Multiple Reaction Monitoring (MRM) Proteomics data set has been deposited at PeptideAtlas (The Institute for Systems Biology, WA) with the identifier PASS00661 (http://www.peptideatlas.org/PASS/PASS00661).
RESULTS
One hundred and thirteen subjects consented to this study. Samples from 12 control subjects were excluded because of significant findings at endoscopy (i.e. ulcers, atypia), and 7 CD subjects were excluded from qualification phase analyses because of the presence of skin extraintestinal manifestations (i.e. psoriasis, erythema nodosum) as possible confounding factors. Independent age and sex-matched cohorts were used for each set of analyses. A total of 94 subjects were included in this study (Table I).
Table I. Subject Characteristics by Study. C = Controls, ICD = Inflammatory Crohn's, SCD = Stricturing Crohn's, FCD = Fistulizing Crohn's, CCD = Complicated (SCD and FCD) Crohn's, UC = Ulcerative colitis, RA = Rheumatoid arthritis. Dichotomous variables (medications, gender, resection) were compared between groups by chi-square test and continuous variables (age, age at disease onset, duration of disease, CRP, ESR, disease activity score) by kruskal wallis. Continuous variables are reported as median ± interquartile range. Controls were excluded from comparisons based on disease activity, medications, and resections. C, UC, and RA were excluded from comparisons based on age at disease onset, disease duration, and ileal disease.
| Cases | Male | Female | Age (years) | Age at disease onset (years) | Duration of disease (years) | CRP (mg/L) | ESR (mm/hr) | CDAI/Partial MAYO/DAS28 | Biologics (N) | Steroids (N) | Immuno-modulators (N) | Mesalazines (N) | Resection (N) | Ileal disease (N) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery Phase | (n = 34) | (n = 17) | (n = 17) | ||||||||||||
| ICD | 16 | 9 | 7 | 31 ± 13 | 28 ± 21 | 6 ± 4 | 13 ± 21 | 26 ± 51.5 | 114.5 ± 130.7 | 5 | 8 | 8 | 14 | 7 | 5 |
| SCD | 9 | 2 | 7 | 49 ± 23 | 32 ± 14 | 7 ± 2 | 6 ± 8 | 19 ± 17.5 | 41 ± 98.5 | 2 | 4 | 4 | 2 | 6 | 6 |
| FCD | 9 | 6 | 3 | 27 ± 4 | 24 ± 4 | 3 ± 2 | 9 ± 5 | 28 ± 7 | 130 ± 41 | 6 | 3 | 6 | 5 | 4 | 8 |
| p value between groups | 0.133 | 0.890 | 0.203 | 0.195 | 0.698 | 0.799 | 0.094 | 0.111 | 0.673 | 0.73 | 0.005 | 0.506 | 0.015 | ||
| Qualification Phase | (n = 50) | (n = 23) | (n = 27) | ||||||||||||
| C | 10 | 5 | 5 | 39 ± 36 | NA | NA | 0.45 ± 0.05 | 6 ± 1 | NA | 0 | 0 | 0 | 0 | 0 | NA |
| UC | 10 | 5 | 5 | 31 ± 8 | NA | NA | 5.1 ± 14.3 | 21.5 ± 29.5 | 6.5 ± 6.8 | 0 | 7 | 7 | 6 | 0 | NA |
| ICD | 10 | 5 | 5 | 34 ± 14 | 31 ± 19 | 7 ± 2 | 14.6 ± 15.1 | 18 ± 14 | 124.6 ± 47.3 | 2 | 3 | 4 | 3 | 2 | 4 |
| CCD | 10 | 5 | 5 | 35 ± 17 | 30 ± 9 | 11 ± 13 | 4.6 ± 4.3 | 13 ± 3 | 159.2 ± 223 | 7 | 2 | 6 | 2 | 7 | 6 |
| RA | 10 | 3 | 7 | 50 ± 18 | NA | NA | 2.5 ± 4.3 | 13.5 ± 10.2 | 2.0 ± 1.4 | 3 | 5 | 7 | 0 | 0 | NA |
| p value between groups | 0.863 | 0.036a | 0.531 | 0.629 | 0.265 | 0.763 | 0.095d | 0.006b | 0.110 | 0.475 | 0.060 | 0.002c | 0.371 | ||
| Longitudinal CD subanalysis cohorte | (n = 5) | (n = 3) | (n = 2) | ||||||||||||
| 1st sample | 5 | 3 | 2 | 39 ± 19 | 30 ± 9 | 16 ± 13 | 4.2 ± 4.6 | 9 ± 5.2 | 142 ± 109 | 4 | 2 | 2 | 4 | 4 | 4 |
| 2ndsample | 5 | 3 | 2 | 39 ± 19 | 30 ± 9 | 16 ± 13 | 1.4 ± 1.1 | 10 ± 7 | 216 ± 108 | 5 | 2 | 3 | 4 | 4 | 4 |
aUC subjects were significantly younger than RA subjects (p < 0.01). There were no other differences in age between groups.
bFrequency of biologics was significantly higher in CCD (p < 0.01).
cFrequency of resections was significantly higher in CCD compared to UC and ICD (p < 0.05) (RA and C were excluded from comparison). Only UC, ICD and CCD were compared for frequency of mesalazine therapy.
dComparison between groups was made by categorical sorting of individual disease scores into remission, mild, moderate, and severe disease categories based on respective scoring reference ranges.
eThe longitudinal subanalysis cohort consisted of CCD subjects sampled twice over 54 ± 14 days with continual active complication(s) (confirmed by radiologic/endoscopic evidence on the day of each blood draw as part of routine care).
Inflammatory and Complicated Crohn's Disease Have Distinct Low-Mass Serum Proteomes
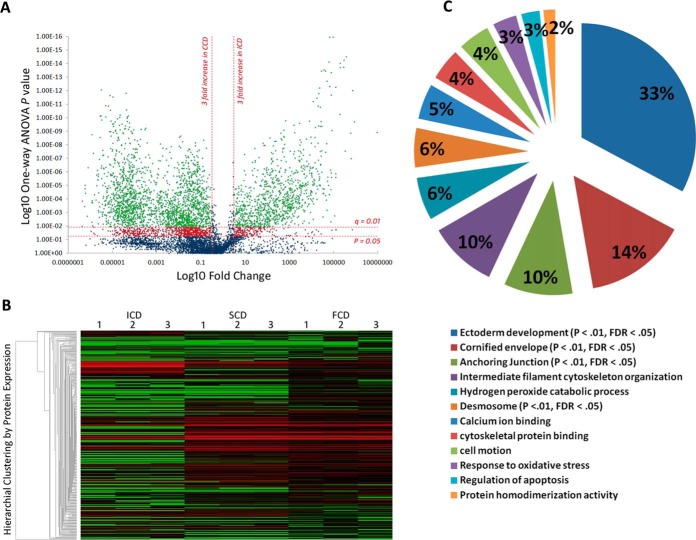
7099 common ion features with MS/MS spectra were detected across all samples (Fig. 1A). A total 16,287 ms/ms spectra was available for the 7099 features and the median within-group % coefficient of variation (%CV) for these features was 16.5%. The 7099 features sequenced to 2164 unique peptides (FDR = 5.3%) which mapped to 348 proteins (Fig. 1B). Three thousand nine hundred and sixty ions (859 peptides) were significantly changing between behavioral phenotypes by MS1 ion intensity quantitation (p < 0.05) (Fig. 1A). After controlling for multiple testing (FDR q<0.01) and statistical power (>80% - based on sample size, signal delta (Δ) between groups, %CV within group), the portion of significantly changing ions was 3133 (716 peptides) (Fig. 1A). This mapped to 172 identified proteins that were significantly modulated between ICD and CCD (p < 0.05, q<0.01, power >80%) (Fig. 1B; supplemental Fig. S2). Protein modulation between SCD and FCD was less pronounced (n = 118, p < 0.05, q<0.01, power>80%).
Fig. 1.
Discovery phase Proteomics identifies distinct low-mass serological protein profiles in CD behavioral phenotypes. A, Volcano plot summarizing low-mass molecular ion differences between ICD and CCD. Data points in the lower center area of the plot have a fold change close to 1 and a p value approaching 1, indicating no significant change. Points in the upper left and right quadrants indicate significant up- and downregulation. Points in green are ions significantly changing after FDR correction (q value) (n = 3960 ions sequenced to 859 peptides) and points in red are significantly changing but discarded to prevent type 1 error. B, Heatmap visualization of low-mass serological protein profiles of CD phenotypes. Each column represents a technical replicate of a pooled phenotypic sample, and each row is a protein (n = 348). The protein list is arranged by hierarchal clustering to form groups of like measurements. Red indicates up-regulation within the data set, black represents a median quantitative value and green indicates downregulation. Significantly enriched clusters of proteins in SCD and FCD (n = 161, p < 0.05, FDR q = 0.01) can be seen in the center of the graphic quantitatively visualized by red fill. C, Modules of biological function clusters annotated to the 172 modulating protein data set between CCD and ICD represented as percentages of total enrichment. The modules are identified by the top GO term annotated to that cluster. Significantly enriched biological functions (when compared with the background homo sapien genome) are indicated by an EASE score p value <0.05 and an FDR p value <0.01 after Benjamini-Hochberg correction.
Low-mass Serum Proteome of Complicated Crohn's Disease Enriched with Epithelial Component Proteins
Functional annotation clustering discriminated the 172 protein data set into 12 clusters of enriched biological module groups (Fig. 1C). Four groups were significantly enriched using a Benjamini-Hochberg -corrected EASE score cutoff of p < 0.01 (Fig. 1C). Within the 4 significant clusters the top 5 GO terms (by fold enrichment) associated with up-regulated proteins in CCD were: desmosome, cornified envelope, ectoderm development, apical junction complex, and keratinocyte differentiation (BH-corrected EASE score p < 0.001) (Table II). The 11 downregulated proteins in CCD in the 172-protein data set were not found to have significant functional annotations (supplemental Table S2). Ninety-six single peptide-identified up-regulated proteins in CCD that did not annotate to enriched biological module groups were discarded from further analysis and a final high-confidence data set of 76 modulating proteins with functional annotation information was finalized (supplemental Table S2). Annotated spectra for pertinent single-peptide identified proteins are provided in the supplemental Material accompanying this manuscript (supplemental Fig. S3–S16).
Table II. Enriched Gene Ontology (GO) terms in Complicated Crohn's disease (CCD). 32 unique proteins of the 172 significantly changing protein dataset between Inflammatory Crohn's and CCD (Supplementary Table I) were annotated across the top 10 GO terms by functional cluster analysis (EASE score p < 0.05, Benjamini-Hochberg correction p < 0.01). Please note that proteins may be annotated to more than one GO term.
| GO term | Protein count | Fold enrichment | EASE score | Benjamini-Hochberg corrected p value |
|---|---|---|---|---|
| Desmosome | 6 | 120.0 | 7.60E-10 | 1.50E-08 |
| Cornified envelope | 4 | 75.0 | 1.70E-05 | 1.50E-04 |
| Ectoderm development | 15 | 29.0 | 1.50E-17 | 7.40E-15 |
| Apical junction complex | 7 | 29.0 | 9.50E-08 | 1.20E-06 |
| Keratinocyte differentiation | 5 | 29.0 | 2.10E-05 | 2.70E-03 |
| Intermediate filament | 11 | 25.0 | 6.60E-12 | 3.30E-10 |
| Cell to cell junction | 8 | 17.0 | 2.20E-07 | 2.20E-06 |
| Anchoring junction | 7 | 17.0 | 2.50E-06 | 2.30E-05 |
| Epithelium development | 7 | 12.0 | 1.90E-05 | 3.10E-03 |
| Cytoskeleton | 21 | 6.3 | 4.30E-13 | 4.30E-11 |
Desmosomal and Anti-Microbial Proteins Selected for Qualification Phase Proteomics
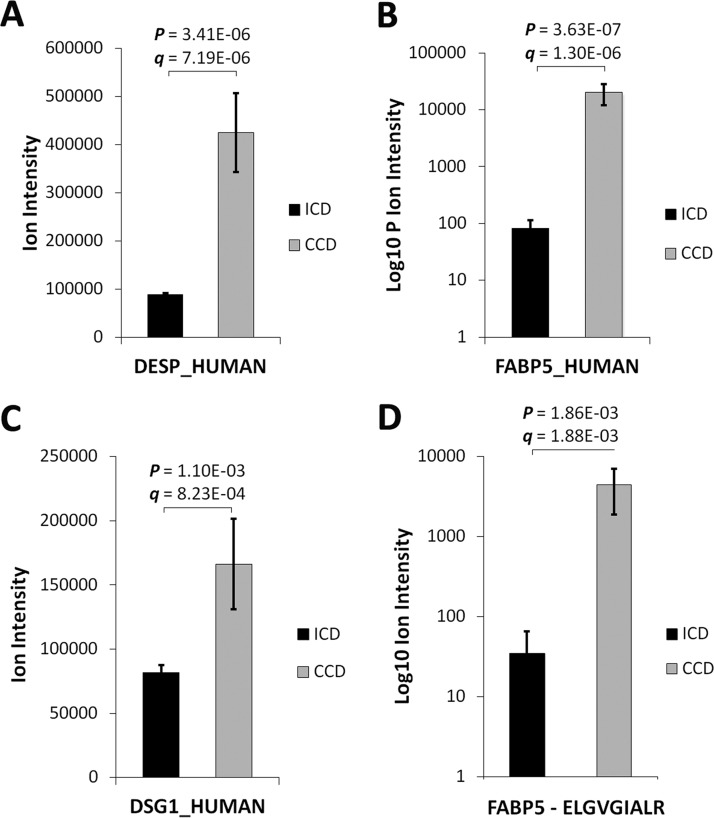
DSG1, DSK and FABP5 proteins were selected for immunoblot qualification based on significant differential regulation (p < 0.01) by label-free LC-MS/MS (Fig. 2A–2C) and inclusion in the significantly enriched clusters and GO terms by functional annotation analysis (BH-corrected EASE score p < 0.01) (Fig. 1C; Table II). Applying the NCI-FDA workshop statistical model, markers expressed in >80% of samples in a 16:18 case/control (ICD versus CCD) discovery phase study with a signal Δ ≥5.0 standard deviations would yield >80% power in verification phase with a minimum assay n = 20 (12). The mean signal Δ for the 3-protein panel in this study was 5.1. FABP5 was further chosen for tier 2 MRM assay development based on significant differential regulation at the peptide level (p < 0.01) by a high-confidence sequenced proteotypic peptide (Fig. 2D), its high discriminant function coefficient for complication (see “Results,” Serum Epithelial Proteins Discriminative of CCD), unique immunological and cell trauma properties (Fig. 7C) (22–24), and established stability in a healthy population-based cohort (25).
Fig. 2.
Serum levels of DSG1, DSK and FABP5 are increased in CCD by label-free LC-MS/MS. Relative A, DSK B, FABP5, and C, DSG1 levels in CCD and ICD. D, A FABP5 proteotypic peptide (ELGVGIALR) was sequenced by LC-MS/MS and contributed to FABP5 quantitative comparisons between ICD and CCD (FABP5 identified and quantified by 3 nonconflicting peptides).
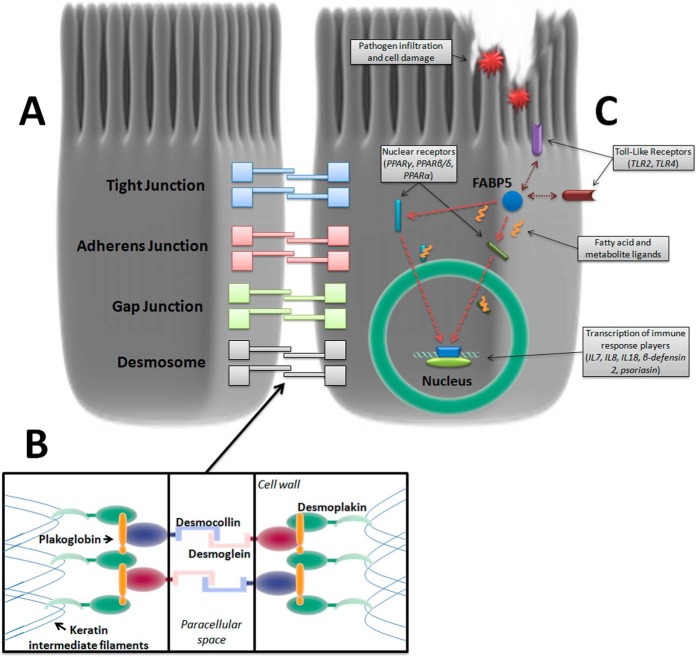
Fig. 7.
Cell-adhesion complexes and biomarker candidates in the intestinal epithelium. A, A range of junctional complexes bind intestinal epithelial cells together to form the epithelial sheet. Desmosomes are the strong mechanical linkage complexes that hold the basal ends of cells together with extracellular and cytoplasmic components: B, Desmoglein and Desmocollin fasten adjacent cells together in the paracellular space whereas Desmoplakin anchors the intercellular binding structure to each cell's intermediate filament lattice structure. C, Fatty Acid-Binding Protein 5 (FABP5) is a cytoplasmic protein that has significant crosstalk with Toll-Like Receptor (TLR) pathogen recognition activation and chaperones a wide array of ligands to specific nuclear receptors during cell trauma, regulating cytokine and antibacterial peptide production. Listed in the graphic are some of the interacting and target receptors of FABP5 and subsequently produced immune response factors. Double arrows indicate TLRs mutually activated with FABP5 and single arrows indicate target nuclear receptors that FABP5 trafficks their payload to for transcription of immune response protein/peptides in the cell nucleus.
Serum Epithelial Proteins Discriminative of CCD
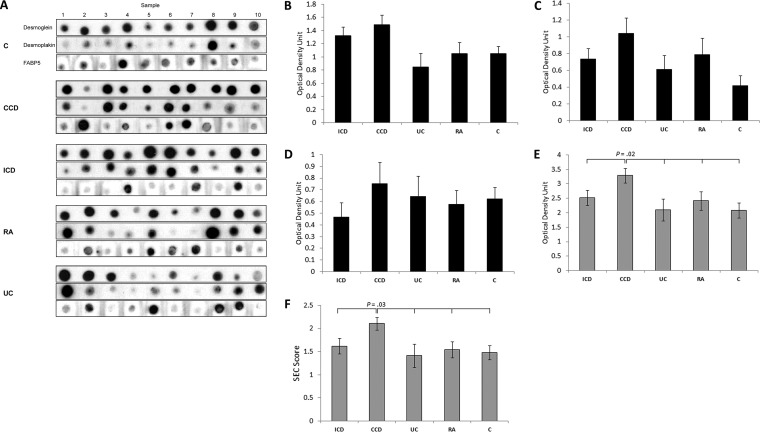
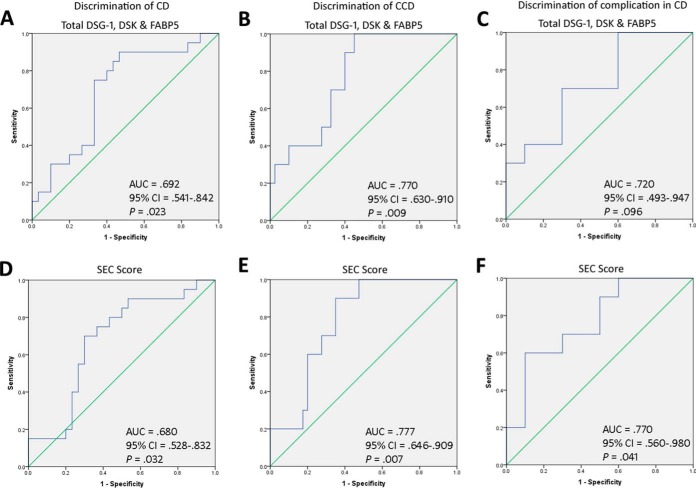
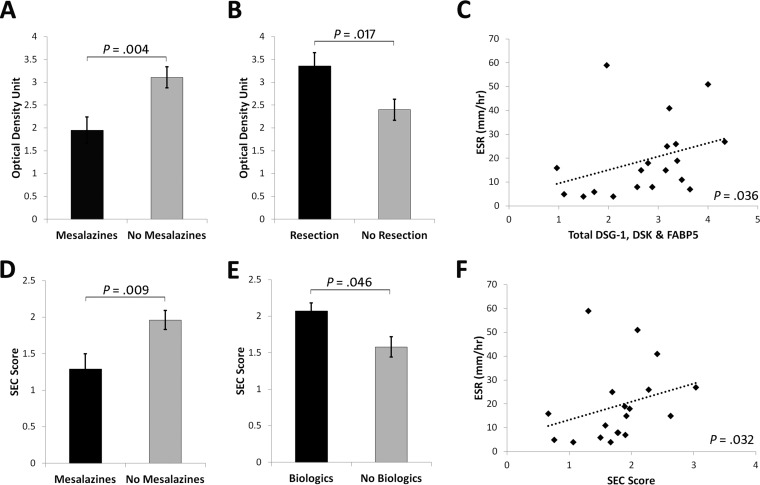
Total DSG1, DSK, and FABP5 was increased in CCD (3.29 ± 0.25 optical density unit) compared with ICD (2.52 ± 0.26), UC (2.10 ± 0.38), RA (2.42 ± 0.32), and Cs (2.09 ± 0.25) (p = 0.022) (Fig. 3E). By ROC curve, total DSG1, DSK and FABP5 could significantly discriminate CD and CCD from all other conditions, and had 90% sensitivity and 60% specificity at a cutoff value of ≥2.584 for CCD discrimination (Fig. 4A–4B). Classification ability of total DSG1, DSK, and FABP5 for complicated phenotypes within CD did not reach statistical significance (Fig. 4C). Total DSG1, DSK, and FABP5 also positively correlated with ESR (p = 0.036), and was lower in those on mesalazine therapy (p = 0.004) and in those without previous resection(s) (p = 0.017) (Fig. 5A–5C). Total DSG1, DSK, and FABP5 was not associated with any other clinical factors: CRP (p = 0.630), CDAI (p = 0.582), biologics (p = 0.120), corticosteroids (p = 0.132), or immunomodulators (azathioprine/6-mercaptopurine/methotrexate) (p = 0.918). Neither age (p = 0.915), gender (p = 0.145), age at diagnosis (p = 0.667), duration of disease (p = 0.931), nor ileal disease location (p = 0.684) were associated with total DSG1, DSK and FABP5. ESR, mesalazine therapy, and previous resection(s) were hence entered into a multiple regression model to determine independent predictiveness for biomarker values. The model was not a significant predictor (p = 0.173) of total DSG1, DSK, and FABP5 values. ESR and previous resection(s) approached significance as independent predictors (p = 0.069 and p = 0.052, respectively), whereas mesalazines did not (p = 0.938).
Fig. 3.
Verification of epithelial-component biomarker candidates in Complicated Crohn's disease in a second cohort. A, Serum dot blots ordered by group. B, Normalized relative levels of DSG1, C, DSK, and D, FABP5 between comparative groups. E, Total DSG1, DSK and FABP5 levels and the weighted F, SEC score between groups. One-way ANOVA p values are stated where statistically significant (p < 0.05).
Fig. 4.
Classification ability of serological epithelial component proteins for Complicated Crohn's disease. Total DSG1, DSK, and FABP5 levels were able to classify: A, CD against RA, UC, and control groups, B, CCD against ICD, RA, UC, and controls, and C, CCD against ICD. The putative Serological Epithelial Component (SEC) score, developed using discriminant function coefficients of the biomarker candidates, improved classification of CD and CCD in all cases: D, CD against RA, UC, and control groups, E, CCD against ICD, RA, UC, and controls, and F, CCD against ICD.
Fig. 5.
Significant associations between biomarker candidates and clinical characteristics. A, Total DSG1, DSK, and FABP5 levels were lower in subjects on mesalazines (3.11 ± 0.23 versus 1.95 ± 0.29), and in subjects without B, previous resection(s) (3.36 ± 0.29 versus 2.40 ± 0.23). C, Total DSG1, DSK, and FABP5 levels were also moderately correlated with ESR (r = 0.484). D, The SEC score remained lower in subjects on mesalazines (1.96 ± 0.13 versus 1.29 ± 0.21) and was E, higher in subjects on biological therapies (2.07 ± 0.11 versus 1.58 ± 0.14). F, SEC score also maintained a moderate correlation with ESR (r = 0.492).
DSG1, DSK, and FABP5 was a significant CCD classification model by discriminant function analysis (p = 0.036). Standardized discriminant function coefficients of 0.726 (DSG1), 0.284 (DSK) and 0.971 (FABP5) were hence used to construct the SEC score (See supplemental Materials - Experimental Procedures). SEC score was significantly higher in CCD (p = 0.032) (Fig. 3F) and could significantly discriminate CD (p = 0.023) and CCD (p = 0.007) against all other groups (Fig. 4D–4E). SEC score also improved classification of complicated phenotypes within CD (p = 0.041) (Fig. 4F) with 70% sensitivity and 70% specificity at a cutoff value of 1.907. The positive and negative predictive value at this cutoff was 71.8% and 70.7%, respectively. The specificity could be improved to 90% at an SEC cutoff of 1.999, with 60% sensitivity and 86% positive and 69% negative predictive values. SEC score remained correlated with ESR (p = 0.032) and lower in subjects on mesalazines (p = 0.009) (Fig. 5D and 5F). SEC score was also significantly higher in those on biologics (p = 0.046) (Fig. 5E). SEC score was not associated with any other relevant clinical factors: resection(s) (p = 0.163), ileal disease (p = 0.415), age (p = 0.920), gender (p = 0.563), steroidal therapy (p = 0.340), immunomodulators (p = 0.426), CRP (p = 0.497), CDAI (p = 0.997), age at diagnosis (p = 0.373), or duration of disease (p = 0.923). ESR, mesalazine therapy, and biologics was not a significant predictive model for SEC score by multiple regression (p = 0.761), and neither ESR (p = 0.761), mesalazine therapy (p = 0.761), nor biologics (p = 0.761) were independent predictors of SEC score. Individual DSG1, DSK, and FABP5 values were all increased in CCD compared with ICD, UC, RA, and Cs, but the differences did not reach significance alone (p = 0.051, p = 0.103, p = 0.708, respectively) (Fig. 3B–3D).
Serum FABP5 Levels Increased in CCD and Stable in a Longitudinal Cohort
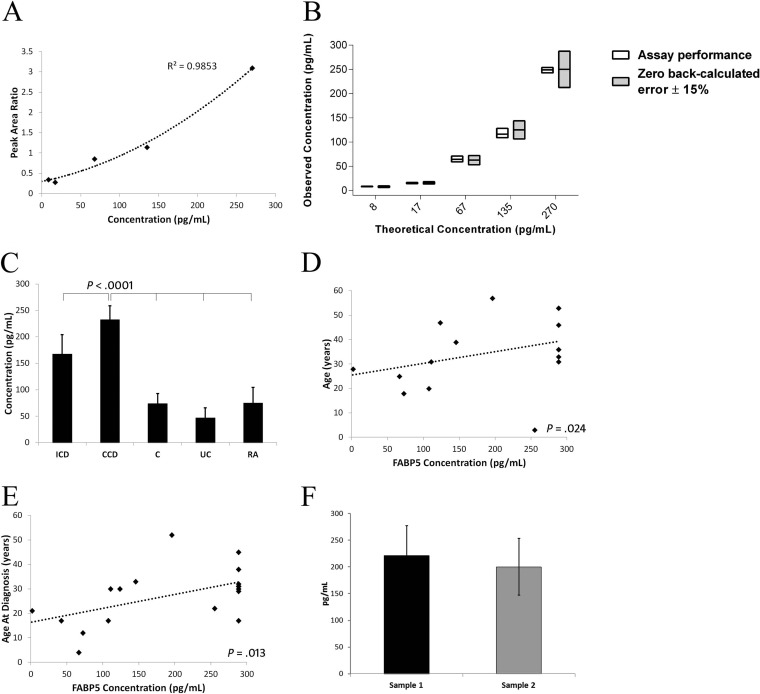
An MRM assay for FABP5 was developed that met NCI-CPTAC Tier-2 specifications (supplementary Experimental Procedures; supplemental Table S1; Fig. 6A–6B) (14). Increases in serum FABP5 levels in CCD identified by label-free LC-MS/MS and immunoblotting were further verified by MRM assay; with FABP5 levels higher in CCD (232.37 ± 26.15pg/ml) compared with ICD (167.41 ± 36.85), UC (46.95 ± 19.22), RA (74.79 ± 29.46), and Cs (74.04 ± 18.93) (p < 0.001) (Fig. 6C). FABP5 levels were moderately correlated with age (p = 0.024) and age at diagnosis (p = 0.013; Fig. 6D–6E) but not with duration of disease (p = 0.728) nor inflammation and disease activity markers: CRP (p = 0.062), ESR (p = 0.368), or CDAI (p = 0.179). FABP5 levels alone could not discriminate CCD from ICD by ROC curve (p = 0.290). There were also no differences in FABP5 based on previous resections (p = 0.529), or current biologic (p = 0.297), steroid (p = 0.074), immunomodulator (p = 0.884) or mesalazine therapy (p = 0.520). FABP5 levels were additionally unrelated to gender (p = 0.706) or ileal disease (p = 0.647). FABP5 levels were not significantly different in five CCD subjects who gave serial blood samples (54 ± 14 days between sampling) with continual active complication(s) (p = 0.829) (Fig. 6F).
Fig. 6.
FABP5 levels in CCD verified by NCI-CPTAC Tier-2 level MRM assay. A, Calibration curve for FABP5 MRM assay. A coefficient of determination of r2 = 0.99 was achieved using a polynomial regression equation (x = −18.475y2 + 149.37y - 34.974) for a 1–300 pg/ml range of quantification. B, Quantitative performance of the FABP5 assay against a low-error criterion of zero back-calculated error ± 15%. The back-calculated error margin of the assay was 1.7% and precision %CV was 12.4. The interday full analytical replicate %CV was 10.7. C, Serum FABP5 levels were increased in CCD compared with all other groups. D, FABP5 levels were moderately correlated with age (rs = 0.521), and E, age at diagnosis (rs = 0.561). F, FABP5 levels were stable in active CCD subjects sampled at two time points between 54 ± 14 days (221.45 ± 55.33 versus 200.13 ± 52.84, p = 0.829).
DISCUSSION
In this study an increase of circulating epithelial component proteins was identified in CCD using discovery and qualification phase proteomics studies as part of an NCI-FDA modeled clinical biomarker pipeline (12, 26). This finding supports the dysregulation of epithelial barrier function in CD (27–29), and presents substantial evidence that a blood test can be developed representative of transmural intestinal injury. Stricturing and fistulizing intestinal complications begin with deep tissue damage following chronically sustained inflammation, thus a measure of transmural tissue integrity may be able to provide early detection of impending complication development.
Increased epithelial component proteins detected in discovery phase included major desmosomal components (DSG1, DSK, desmocollins, plakoglobin), desmosome-associated intermediate filaments (keratins, filaggrins), and immunoregulatory epithelial proteins (FABP5, Peroxiredoxin-1, Serpin B3). These proteins may be liberated in transmural intestinal injury either as a primary pathogenic mechanism or secondary consequence, and become detectable in the blood. Three epithelial components in particular, DSG1, DSK, and FABP5, were confirmed to be increased in a second independent qualification cohort of CCD against ICD and UC (as intestinal inflammation controls), RA (Th1/17 systemic inflammatory control), and healthy controls. An MRM assay was then developed for FABP5 as part of the NCI-FDA biomarker pipeline that further confirmed increased levels in CCD and viability on longitudinal follow up.
DSG1 and DSK are desmosomal proteins essential to the mechanical linkage of intestinal epithelial cells.(28) DSG1 is a cadherin that binds the intercellular space between neighboring epithelial cells along with its counterpart desmocollin-1 (28). DSK anchors the DSG/desmocollin-plakoglobin/plakophilin intercellular complex to the intracellular intermediate filament lattice structure (Fig. 7A–7B) (30). Lastly, FABP5 is a cytoplasmic protein expressed in epithelial cells throughout the gastrointestinal tract, and is part of the FABP superfamily of intracellular fatty-acid chaperones proposed to be markers of pathogen infiltration and tissue injury by cell leak (22, 31). FABP5 regulates the expression of several cytokines and antibacterial peptides by delivering fatty acid ligands to target nuclear receptors, and interacts with Toll-Like Receptor (TLR) activation to form part of the innate immune response (Fig. 7C) (23–25). FABP family members FABP1 and 2 have also been shown to predict poorer clinical outcomes in necrotizing enterocolitis (22). Taken together, DSG1, DSK and FABP5 may therefore be involved in the progressive intestinal injury associated with the development of CD complications via their effects on intestinal cell adhesion, intercellular intermediate filament structure binding and innate immunity, respectively. The panel of DSG1, DSK, and FABP5, and the composite SEC score were able to discriminate CCD from ICD and all other groups. UC and ICD typically do not involve gross deformation of tissue beyond the muscularis mucosae and intestinal epithelial component proteins may therefore not be released into systemic circulation (4). The SEC score was also independent of acute inflammation as measured by CRP, which may further indicate that it is a direct measure of barrier function and tissue injury rather than an acute-phase inflammatory response (32). Construction of the SEC score was guided by standardized discriminant function coefficient calculations as part of a discriminant function analysis, which determines the strength and direction of each individual biomarker in contributing to the maximum discriminability of the overall model for complication in CD. This allows an objective assessment of the efficacy of each candidate to the biomarker panel, with the potential to remove candidates that negatively affect discriminability. DSG1, DSK, and FABP5 all exhibited positive coefficients for CCD discrimination (0.726, 0.284, and 0.971, respectively) and were hence included in the current working SEC score. Inclusion of a greater number of novel basal epithelial component proteins will likely increase the performance of the SEC score and the positive results obtained in qualification phase thus far justify the development of MRM and antibody-based quantification methods for more novel biomarker candidates as part of the continual application of the biomarker development pipeline Fig. 7. Increase of FABP5 in CCD was confirmed by MRM assay, and the longitudinal stability of FABP5 levels in the presence of active complications was also demonstrated. The high sensitivity and tight margin-of-error of a validated Tier 2 MRM assay allows quantification of very low concentration biomarkers and is a crucial aspect of the research assay optimization phase in the NCI-FDA biomarker pipeline (12, 13, 20). The Tier 2 assay developed in this study attained a quantification range of 1–300pg/ml, achieving a significant practical capability that must be met in order to detect subclinical transformations that may begin with small (and previously undetectable) concentration changes (20, 33). Intermediate filament proteins are released in large quantities into circulation from epithelial cells undergoing cell death (34, 35), and early detection of serologically increased intestinal epithelial components may have the potential to objectively measure transmural damage that precedes clinical manifestation of complication. This may be a time-sensitive and individual-specific indicator, in contrast to clinical risk factors.
Stricturing and fistulizing intestinal complications begin with deep tissue damage following chronically sustained inflammation. It stands to logic that a measure of transmural tissue integrity may be able to prognose impending complication development. Despite this seemingly straightforward concept, prior to this work there had been relatively few investigations into ways in which the full-thickness of the intestinal tissue could be monitored to predict the development of stricturing and fistulizing complications. The reason for this is because of the difficulty in evaluating subclinical tissue changes in vivo. Examinations under anesthesia and radiological/ultrasound imaging can only diagnose intestinal complications after the fact. Urine lactulose/mannitol ratio can evaluate small bowel permeability, but not the integrity of the large intestine and thus limits its sensitivity for complication. In fact, the bloodstream may be one of the only mediums capable of prognosis because of its circulation in the microvasculature, making it accessible to all layers and segments of the intestinal tract. Although a multitude of independent studies have established both the dysregulation of intestinal epithelium components in CD and the ability to assay bloodstream products translocated from the intestinal lumen, the notion of measuring intestinal tissue changes in a blood test remains open to certain skepticism because its collection is removed from the site of pathology (27–29, 36–38). The increase of circulating epithelial components in CCD identified in this study against ICD, UC, RA, and nondisease controls (to the exclusion of other epithelial conditions), substantially suggest that extensive injury to intestinal tissue is responsible for this enrichment. However future studies correlating serological epithelial component levels with matched immunohistochemical evidence from CCD tissues could further confirm this concept.
Sample size errors may be present in this study, although verification of LC-MS/MS results in a second independent cohort of sex and age-matched inflammatory and differential IBD controls using antibody-based techniques confirmed the significant findings. The explicit exclusion of patients with concurrent skin diseases and other relevant comorbidities (i.e. COPD involving airway epithelium damage) strongly suggest that the increase of epithelial barrier components observed in CCD are likely to be of intestinal origin. However, control groups including disorders involving aberrant epithelial cell death (i.e. eczema) may be needed in future studies to evaluate how the presence of such extraintestinal conditions may impact the clinical utility of the biomarker panel. As part of qualification phase analyses, biomarker values were also evaluated against relevant clinical markers, IBD history and medication use. Total DSG1, DSK, and FABP5 levels and SEC score were higher in subjects not on mezalazine therapy (Fig. 5A and 5D). This may be because of the higher number of UC subjects on mesalazines (n = 6, though not significantly different from other groups) where tissue injury is more superficial and intestinal epithelial components may not cross into systemic circulation, or cross over at the same rates as in CD. Another possibility could be attributed to the ability of mesalazines to inhibit epithelial proliferation by reducing signaling and progenitor cell activation of epithelial b-catenin (the adherens junction's cadherin linkage protein) (Fig. 7A) (39), which it could also do for desmosomal cadherins. This is less likely however, as mesalazines effectively induce mucosal healing in UC but are not known to do so in CD, which supports the notion that mesalazines can restore apical paracellular linkage complexes (adherens junctions) and thus attenuate superficial inflammation, but not the basal complexes (desmosomes) that become disarrayed in deep tissue inflammation (40, 41). This would further indicate the specificity of the biomarker panel for CD, as it is primarily composed of deep tissue desmosomal component biomarkers. ESR, biologics use, and past surgical resections were also variably associated with total DSG1, DSK, and FABP5 levels, and the SEC score. These clinical factors, together with mesalazine therapy, lost significant associations with total DSG1, DSK, and FABP5 levels and the SEC score when entered into a multivariate model. This is most likely because of the sample size in qualification phase of this study which was not designed to evaluate these novel biomarker associations. Independent sample sizes must be calculated to evaluate these novel associations in ongoing verification phase analyses of the biomarker pipeline, and the final verification phase cohort must be large enough to authenticate each of these associations. This is an important consideration moving forward as it may affect how the biomarker panel should be interpreted in a disease population on variable therapeutic regimens and with varying clinical histories.
The current results substantiate the genomic and functional evidence that have established the dysregulation of the intestinal epithelium and epithelial junctional complexes in CD (27–29). Continuation of studies along the biomarker pipeline is required to determine whether there is a detectable serological increase of epithelial components in the initial phases of transmural injury, which could prognose manifestation of intestinal complications. Regardless of whether epithelial defects are a primary or secondary effect of a complicated natural history of CD, circulating epithelial components could be clinically useful indicators of disease progression. The crux lies in the detectability of epithelial changes before clinical manifestation of intestinal complications. The sensitivity and large quantification range of MRM (capable of pico- to femto- liters of quantification) meets this technical requirement. Indeed, correlation of serum FABP5 levels with age and age at diagnosis may suggest that epithelium perturbations are an early and perhaps predisposing event in complicated CD. Specific analysis of the FABP5 transcription pathway in CD could elucidate this potential mechanism of stricturing and fistulizing complications.
In conclusion, discovery and qualification phase proteomic studies have identified epithelial barrier component proteins that discriminate CCD from ICD, RA, UC, and controls. The preliminary biomarker panel presented here demonstrates the proof-of-principle for a serological test to assess transmural disease behavior in CD. This could provide an individualized indication for timely aggressive immunosuppressive therapy.
DATA AVAILABILITY
Further details on experimental procedures, statistical analyses and SEC score calculation are provided in supplementary Materials. The LC-MS/MS data in this manuscript has been deposited at the PRIDE proteomics data repository (The European Bioinformatics Institute, Cambridge, UK) with the data set identifier PXD001821 at (http://www.ebi.ac.uk/pride/archive/). The Multiple Reaction Monitoring (MRM) Proteomics data set has been deposited at PeptideAtlas (The Institute for Systems Biology, WA) with the identifier PASS00661 (http://www.peptideatlas.org/PASS/PASS00661).
Supplementary Material
Acknowledgments
We thank Dr. Bain Shenstone and his team for assistance in recruiting rheumatoid arthritis subjects for this study. We would like to thank the South West Sydney Local Health District from whom Valerie Wasinger and Rupert Leong received funds for this project, the Australian Government for Rupert Leong's fellowship through the National Health and Medical Research Council, and UNSW Australia, South West Sydney Clinical School for scholarship funding for Yunki Yau.
Footnotes
Author contributions: Y.Y.Y., R.W.L., and V.C.W. designed research; Y.Y.Y., A.P., D.R., and V.C.W. performed research; V.C.W. contributed new reagents or analytic tools; Y.Y.Y. analyzed data; Y.Y.Y. wrote the paper; R.W.L. critical review and editing of manuscript; V.C.W. critical review and editing of the manuscript.
* This work was supported by funds from the South West Sydney Local Health District (VW and RL) and UNSW Australia, South West Sydney Clinical School (YY).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CD
- Crohn's disease
- DAS28
- 28-Joint Disease Activity Score
- CPTAC
- Clinical Proteomic Tumor Analysis Consortium
- CCD
- complicated Crohn's disease
- CRP
- C-reactive protein
- CDAI
- Crohn's Disease Activity Index
- DSG1
- desmoglein-1
- DSK
- desmoplakin
- ESR
- erythrocyte sedimentation rate
- FABP5
- fatty acid binding protein 5
- FDA
- Food and Drug Administration
- FDR
- false discovery rate
- FCD
- fistulizing Crohn's disease
- GO
- Gene Ontology
- IBD
- inflammatory bowel disease
- ICD
- inflammatory Crohn's disease
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MRM
- multiple reaction monitoring
- NCI
- National Cancer Institute
- RPD
- reverse-polynomial dilution
- RA
- rheumatoid arthritis
- SCD
- stricturing Crohn's disease
- UC
- ulcerative colitis.
REFERENCES
- 1. Satsangi J., Silverberg M. S., Vermeire S., and Colombel J. F. (2006) The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 55, 749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thia K. T., Sandborn W. J., Harmsen W. S., Zinsmeister A. R., and Loftus E. V. Jr. (2010) Risk factors associated with progression to intestinal complications of Crohn's disease in a population-based cohort. Gastroenterology 139, 1147–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ryan J. D., Silverberg M. S., Xu W., Graff L. A., Targownik L. E., Walker JR, Carr R., Clara I., Miller N., Rogala L., and Bernstein C. N. (2013) Predicting complicated Crohn's disease and surgery: phenotypes, genetics, serology and psychological characteristics of a population-based cohort. Alimentary Pharmacol. Therapeutics 38, 274–283 [DOI] [PubMed] [Google Scholar]
- 4. Nielsen O. H., Rogler G., Hahnloser D., and Thomsen O. O. (2009) Diagnosis and management of fistulizing Crohn's disease. Nat. Clin. Pract. Gastroenterol. Hepatol. 6, 92–106 [DOI] [PubMed] [Google Scholar]
- 5. Miheller P., Kiss L. S., Juhasz M., Mandel M., and Lakatos P. L. (2013) Recommendations for identifying Crohn's disease patients with poor prognosis. Expert Rev. Clin. Immunol. 9, 65–76 [DOI] [PubMed] [Google Scholar]
- 6. Antunes O., Filippi J., Hebuterne X., and Peyrin-Biroulet L. (2014) Treatment algorithms in Crohn's - up, down or something else? Clin. Gastroenterol. 28, 473–483 [DOI] [PubMed] [Google Scholar]
- 7. Ananthakrishnan A. N., Huang H., Nguyen D. D., Sauk J., Yajnik V., and Xavier R. J. (2014) Differential effect of genetic burden on disease phenotypes in Crohn's disease and ulcerative colitis: analysis of a North American cohort. Am. J. Gastroenterol. 109, 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ventham N. T., Kennedy N. A., Nimmo E. R., and Satsangi J. (2013) Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology. 145, 293–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lazaridis K. N., and Juran B. D. (2005) American Gastroenterological Association future trends committee report: the application of genomic and proteomic technologies to digestive disease diagnosis and treatment and their likely impact on gastroenterology clinical practice. Gastroenterology 129, 1720–1752 [DOI] [PubMed] [Google Scholar]
- 10. Adkins J. N., Varnum S. M., Auberry K. J., Moore R. J., Angell N. H., Smith R. D., et al. (2002) Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics 1, 947–55 [DOI] [PubMed] [Google Scholar]
- 11. Tang H. Y., Beer L. A., and Speicher D. W. (2011) In-depth analysis of a plasma or serum proteome using a 4D protein profiling method. Methods Mol. Biol. 728, 47–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skates S. J., Gillette M. A., LaBaer J., Carr S. A., Anderson L., Liebler D. C., et al. (2013) Statistical design for biospecimen cohort size in proteomics-based biomarker discovery and verification studies. Journal of proteome research. 12, 5383–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rifai N., Gillette M.A., and Carr S. A. (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotech. 24, 971–83 [DOI] [PubMed] [Google Scholar]
- 14. Carr S. A., Abbatiello S. E., Ackermann B. L., Borchers C., Domon B., Deutsch E. W., et al. (2014) Targeted peptide measurements in Biology and Medicine: Best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics 13, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho J. H. (2008) The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 8, 458–466 [DOI] [PubMed] [Google Scholar]
- 16. Ly L., and Wasinger V. C. (2008) Peptide enrichment and protein fractionation using selective electrophoresis. Proteomics 8, 4197–4208 [DOI] [PubMed] [Google Scholar]
- 17. Wasinger V. C., Yau Y., Duo X., Zeng M., Campbell B., Shin S., et al. (2016) Low mass blood peptides discriminative of inflammatory bowel disease (IBD) severity: A quantitative proteomic perspective. Mol. Cell. Proteomics 15, 256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bickel D. R. (2013) Simple estimators of false discovery rates given as few as one or two p-values without strong parametric assumptions. Statist. Appl. Gen. Mol. Biol. 12, 529–543 [DOI] [PubMed] [Google Scholar]
- 19. Huang, da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 20. Yau Y. Y., Duo X., Leong R. W. L., and Wasinger V. C. (2015) Reverse-polynomial dilution calibration methodology extends lower limit of quantification and reduces relative residual error in targeted peptide measurements in blood plasma. Mol. Cell. Proteomics 14, 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abcam®. Dot Blot Protocol May 5th 2014. Available from: http://www.abcam.com/ps/pdf/protocols/Dot%20blot%20protocol.pdf. [Google Scholar]
- 22. Ng E. W., Poon T. C., Lam H. S., Cheung H. M., Ma T. P., Chan K. Y., et al. (2013) Gut-associated biomarkers L-FABP, I-FABP, and TFF3 and LIT score for diagnosis of surgical necrotizing enterocolitis in preterm infants. Ann. Surgery 258, 1111–1118 [DOI] [PubMed] [Google Scholar]
- 23. Gally F., Chu H. W., and Bowler R. P. (2013) Cigarette smoke decreases airway epithelial FABP5 expression and promotes Pseudomonas aeruginosa infection. PloS One 8, e51784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adachi Y., Hiramatsu S., Tokuda N., Sharifi K., Ebrahimi M., Islam A., et al. (2012) Fatty acid-binding protein 4 (FABP4) and FABP5 modulate cytokine production in the mouse thymic epithelial cells. Histochem. Cell Biol. 138, 397–406 [DOI] [PubMed] [Google Scholar]
- 25. Ishimura S., Furuhashi M., Watanabe Y., Hoshina K., Fuseya T., Mita T., et al. (2013) Circulating levels of fatty acid-binding protein family and metabolic phenotype in the general population. PloS One 8, e81318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodriguez H., Tezak Z., Mesri M., Carr S. A., Liebler D. C., Fisher S. J., et al. (2010) Analytical validation of protein-based multiplex assays: a workshop report by the NCI-FDA interagency oncology task force on molecular diagnostics. Clin. Chem. 56, 237–243 [DOI] [PubMed] [Google Scholar]
- 27. VanDussen K. L., Liu T. C., Li D., Towfic F., Modiano N., Winter R., et al. (2014) Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn's disease. Gastroenterology 146, 200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petit C. S., Barreau F., Besnier L., Gandille P., Riveau B., Chateau D., et al. . Requirement of cellular prion protein for intestinal barrier function and mislocalization in patients with inflammatory bowel disease. Gastroenterology 143, 122–132 [DOI] [PubMed] [Google Scholar]
- 29. Tong M., Li X., Wegener, Parfrey L., Roth B., Ippoliti A., Wei B., et al. (2013) A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PloS One 8, e80702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fuchs E., and Raghavan S. (2002) Getting under the skin of epidermal morphogenesis. Nature reviews Genetics 3, 199–209 [DOI] [PubMed] [Google Scholar]
- 31. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., et al. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 32. Peyrin-Biroulet L., Reinisch W., Colombel J. F., Mantzaris G. J., Kornbluth A., Diamond R., et al. (2014) Clinical disease activity, C-reactive protein normalisation and mucosal healing in Crohn's disease in the SONIC trial. Gut 63, 88–95 [DOI] [PubMed] [Google Scholar]
- 33. Henderson P., van Limbergen J. E., Schwarze J., and Wilson D. C. (2011) Function of the intestinal epithelium and its dysregulation in inflammatory bowel disease. Inflammatory Bowel Dis. 17, 382–395 [DOI] [PubMed] [Google Scholar]
- 34. Kramer G., Erdal H., Mertens H. J., Nap M., Mauermann J., Steiner G., et al. (2004) Differentiation between cell death modes using measurements of different soluble forms of extracellular cytokeratin 18. Cancer Res. 64, 1751–1756 [DOI] [PubMed] [Google Scholar]
- 35. Luft T., Conzelmann M., Benner A., Rieger M., Hess M., Strohhaecker U., et al. . Serum cytokeratin-18 fragments as quantitative markers of epithelial apoptosis in liver and intestinal graft-versus-host disease. Blood 110, 4535–4542 [DOI] [PubMed] [Google Scholar]
- 36. Dubinsky M. C., Lin Y. C., Dutridge D., Picornell Y., Landers C. J., Farrior S., et al. (2006) Serum immune responses predict rapid disease progression among children with Crohn's disease: immune responses predict disease progression. Am. J. Gastroenterol. 101, 360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carvalho B. M., and Abdalla Saad M. J. (2013) Influence of gut microbiota on subclinical inflammation and insulin resistance. Mediators of Inflammation 13, 986734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Presley L. L., Ye J., Li X., Leblanc J., Zhang Z., Ruegger P. M., et al. (2012) Host-microbe relationships in inflammatory bowel disease detected by bacterial and metaproteomic analysis of the mucosal-luminal interface. Inflammatory Bowel Dis. 18, 409–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown J. B., Lee G., Managlia E., Grimm G. R., Dirisina R., Goretsky T., Cheresh P., Blatner N. R., Khazaie K., Yang G. Y., Li L., and Barrett T. A. (2010) Mesalamine Inhibits Epithelial β-Catenin Activation in Chronic Ulcerative Colitis. Gastroenterology 138, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lim W. C., Wang Y., MacDonald J. K., and Hanauer S. (2016) Aminosalicylates for induction of remission or response in Crohn's disease. Cochrane Database Syst. Rev. 3, CD008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Papi C., Fasci-Spurio F., Rogai F., Settesoldi A., Margagnoni G., and Annese V. (2013) Mucosal healing in inflammatory bowel disease: treatment efficacy and predictive factors. Digestive Liver Dis. 45, 978–985 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further details on experimental procedures, statistical analyses and SEC score calculation are provided in supplementary Materials. The LC-MS/MS data in this manuscript has been deposited at the PRIDE proteomics data repository (The European Bioinformatics Institute, Cambridge, UK) with the data set identifier PXD001821 at (http://www.ebi.ac.uk/pride/archive/). The Multiple Reaction Monitoring (MRM) Proteomics data set has been deposited at PeptideAtlas (The Institute for Systems Biology, WA) with the identifier PASS00661 (http://www.peptideatlas.org/PASS/PASS00661).