Abstract
Nε-Acetylation of lysine residues represents a frequently occurring post-translational modification widespread in bacteria that plays vital roles in regulating bacterial physiology and metabolism. However, the role of lysine acetylation in cyanobacteria remains unclear, presenting a hurdle to in-depth functional study of this post-translational modification. Here, we report the lysine acetylome of Synechococcus sp. PCC 7002 (hereafter Synechococcus) using peptide prefractionation, immunoaffinity enrichment, and coupling with high-precision liquid chromatography-tandem mass spectrometry analysis. Proteomic analysis of Synechococcus identified 1653 acetylation sites on 802 acetylproteins involved in a broad range of biological processes. Interestingly, the lysine acetylated proteins were enriched for proteins involved in photosynthesis, for example. Functional studies of the photosystem II manganese-stabilizing protein were performed by site-directed mutagenesis and mutants mimicking either constitutively acetylated (K99Q, K190Q, and K219Q) or nonacetylated states (K99R, K190R, and K219R) were constructed. Mutation of the K190 acetylation site resulted in a distinguishable phenotype. Compared with the K190R mutant, the K190Q mutant exhibited a decreased oxygen evolution rate and an enhanced cyclic electron transport rate in vivo. Our findings provide new insight into the molecular mechanisms of lysine acetylation that involved in the negative regulation of oxygen evolution in Synechococcus and creates opportunities for in-depth elucidation of the physiological role of protein acetylation in photosynthesis in cyanobacteria.
Nε-lysine acetylation is a reversible and highly dynamic post-translational modification (PTM)1 in both eukaryotes and prokaryotes. Being different from the irreversible Nα-acetylation, it is dynamically catalyzed by protein acetyltransferases and deacetylases (1). Acetyltransferases transfer an acetyl moiety from acetyl-coenzyme A to the ε-amino group of lysine residue in proteins, and the process can be reversed by deacetylases. Recently, the exploration of a large data set of acetylation sites has been challenging because of the substoichiometric nature of this PTM (2). By using high-efficiency immunoaffinity enrichment strategies and state-of-the-art liquid chromatography-tandem mass spectrometry (LC-MS/MS), comprehensive analysis of lysine acetylation has been extensively studied in both eukaryotes (3–9) and prokaryotes, including Escherichia coli (10–12), Salmonella enterica (13), Geobacillus kaustophilus (14), Thermus thermophilus (15), Bacillus subtilis (16), Erwinia amylovora (17), Streptomyces roseosporus (18), Mycobacterium tuberculosis H37Ra (19), M. tuberculosis Rv (20), Vibrio parahemeolyticus (21), Synechocystis sp. PCC 6803 (hereafter Synechocystis) (22), Bacillus amyloliquefaciens (23), and Spiroplasma eriocheiris (24). These analyses have shown that lysine acetylation is an evolutionarily, highly conserved PTM and is of great importance in the regulation of diverse cellular processes, such as carbon metabolism and photosynthesis, in Synechocystis (22). Even now, lysine-acetylated proteome data for cyanobacteria are scarce.
Cyanobacteria constitute a large and morphologically diverse group among prokaryotes and can produce oxygen (25). Based on the endosymbiosis theory, plastids in eukaryotic algae and higher plants were derived from the ancestral cyanobacteria (26). Recently, cyanobacteria have been employed to produce renewable biofuels in an economically effective and environmentally sustainable manner (27). Synechococcus with high glycogen production capacity may become a suitable candidate for the development of biofuels (28–29). In addition, it is one of the most studied model cyanobacterium with respect to photosynthesis under high-intensity light and high salinity as well as other metabolic processes (28, 30–31). The complete genome sequence of Synechococcus has been determined (see http://www.ncbi.nlm.nih.gov/), and it can be easily genetically transformed with a versatile system (32), making it a model organism to study biotechnological applications and photosynthesis (28). Our previous study has shown that lysine acetylation plays a very important role in carbon metabolism and photosynthesis in Synechocystis (22). Nevertheless, the molecular mechanism of lysine acetylation in the regulation of photosynthesis remains unclear in cyanobacteria.
To gain insights into the biological significance of lysine acetylation in the photosynthetic prokaryote Synechococcus, we analyzed its acetylproteome using immunoaffinity enrichment strategies integrated with high-accuracy LC-MS/MS and characterized the acetylation events in this model system. In this study, we identified 1653 unique lysine acetylation sites from 802 proteins. Interestingly, many acetylated proteins associated with photosynthesis were identified, followed by validation of enhancement of acetylation status in the whole cell lysate under high-intensity light exposure using Western blotting analysis. Functional study revealed that acetylation at K190 of the photosystem II (PSII) manganese-stabilizing protein (PsbO) negatively regulated oxygen evolution. These results provide clues to oxygen evolution process regulated by PsbO acetylation in cyanobacteria.
EXPERIMENTAL PROCEDURES
Cell Culture and Protein Extraction
The Synechococcus strain from Pasteur Culture Collection was cultured in A+ medium, gassing with 1% CO2 (v/v) in filtered air. The strain was grown at 38 °C under continuous illumination (33) (normal light: 250 μmol photons/m2/s; high-intensity light: 2000 μmol/m2/s). Cells were harvested in the exponential phase (OD730 ≈ 0.8) and then exposed to various stresses for 12 h, including A5 (-A5), calcium (-Ca), iron (-Fe), phosphate (-P), and nitrogen (-N) deficiencies. For high-intensity light treatment, cells in the exponential phase were exposed to 2000 μmol photons/m2/s for 30 min. For high salinity treatment and photoheterotrophic growth conditions (+Gly), cells in the exponential phase were resuspended in A+ medium containing 2.5 m NaCl for 30 min or enriched in A+ medium containing 10 mm glycerol, respectively (28). Following this, nicotinamide (10 mm) was added to the cultures for an additional 30 min to inhibit the endogenous protein deacetylase activities. The cells were harvested by centrifugation (6000 × g at 4 °C for 5 min), washed with A+ medium, and resuspended in lysis buffer containing 20 mm Tris-Cl (pH 7.5), 150 mm NaCl, 10 mm nicotinamide, 1% Triton X-100, and 1× protease inhibitor mixture (Thermo Fisher Scientific, Waltham, MA). The suspension was sonicated (JY92-IIN Ultrasonic Homogenizer; Ningbo Scientz Biotechnology Co., Ltd., Ningbo, Zhejiang, China) at an output of 135 W for 30 min (5 s on followed by 5 s off) on ice-water. Cellular debris was removed by centrifugation (3000 × g at 4 °C for 10 min), and the supernatant was stored at −80 °C. The protein concentration was determined by Bradford assay (Bio-Rad, Hercules, CA).
In-solution Trypsin Digestion, High-performance Liquid Chromatography Fractionation, and Enrichment of Acetylated Lysine Peptides
Total proteins (2 mg) were reduced, alkylated, and subjected to trypsin digestion, and the peptides were desalted and fractionated. Protein A/G agarose-conjugated antiacetyllysine antibody (PTM Biolabs, Inc., Chicago, IL) was used to enrich the acetylated peptides, as previously described (22).
LC-MS/MS Analysis
The enriched peptides were analyzed by a Q-Exactive mass spectrometer (Thermo Fisher Scientific) with an easy nLC-1000 system (Thermo Fisher Scientific) and at a linear gradient from 6 to 90% solvent B (98% acetonitrile/0.1% formic acid, v/v) over 40 min. Mass spectrometer was operated in a data-dependent mode with an automatic switch between MS and MS/MS acquisition. Full MS spectra from 350 to 1800 mass-to-charge ratio (m/z) were acquired at a resolution of 70,000 at m/z = 200 in profile mode. Following every survey scan, up to 20 most intense precursor ions were picked for MS/MS fragmentation by higher energy C-trap dissociation with 28% normalized collision energy. Lock mass at m/z 445.12003 was enabled for full MS scan. The dynamic exclusion duration was set to 15 s with a repeat count of 1 exclusion window. The electrospray voltage applied was 2 kV. Automatic gain control was used to prevent overfilling of the ion trap, and 5 × 104 ions were accumulated for generation of MS/MS spectra.
Data Analysis
The mass spectra were searched against the Synechococcus database (/cyanobase/GCA_000019485.1/genes.faa/release date 2008/03/14; 3186 entries in the database) combined with the standard MaxQuant contaminants database using MaxQuant with integrated Andromeda v1.5.3.30 search engine (34). Enzyme specificity was set to full cleavage by trypsin, with two maximum missed cleavage sites permitted. Minimum peptide length was set to 6. Mass tolerances of precursor and fragment ion were set to 6 ppm and 0.05 Da, respectively. Carbamidomethylation of cysteine was set as a fixed modification, whereas oxidation of methionine, deamidation of asparagine and glutamine, and N-terminal acetylation and acetylation of lysine were set as variable modifications. The estimated FDR thresholds for proteins, peptides, and modification sites were specified at maximum 1%. The lysine-acetylated peptides identified were manually inspected using a modified method to exclude isobaric trimethylation (35). Moreover, all acetylation sites assigned to the peptide C-terminal were removed. All the raw data were deposited in a publicly accessible database PeptideAtlas (www.peptideatlas.org) (36).
Bioinformatics Analysis
The identified acetylproteins were grouped into biological process and molecular function classes based on the Gene Ontology (GO) terms using Blast2GO (37–40). The protein subcellular localization was analyzed by PSORTb v3.0 program (41). GO enrichment was performed by BiNGO v2.44 (42–43). The aligning peptide sequences with 12 residues surrounding the acetylated lysine residue were extracted, excluding all peptides where the acetylated lysine was located less than seven amino acids from the C-terminal end of the peptide, and these lysine-acetylated motifs were analyzed by motif-x (44). A heat map was generated to further analyze lysine acetylation sites (45–47). Secondary structures were predicted using NetSurfP (48–49). Conservation of acetylproteins across species was determined using BLASTP (50). KEGG pathway was analyzed using KEGG mapper (51–52). The three-dimensional structure of PsbO was modeled using CPHmodels-3.0 (53). The interaction network analysis was performed using STRING (54) and further analyzed using MCODE (55).
Production of Polyclonal Antibodies against Photosystem I Subunit VII (PsaC) and PsbO, Immunoprecipitation, and Western Blotting Analysis
Anti-PsaC and anti-PsbO polyclonal antibodies were generated by PEPTBIO Inc. (Wuhan, Hubei, China), against the synthetic peptides CKAGQIASSPRTED and GAVFLDPKARGTASG, respectively. Immunoprecipitation using anti-PsbO antibody and Western blotting analysis using anti-PsbO, anti-PsbC, and antiacetyllysine antibodies were performed as previously described (22).
Amino Acid Sequences Alignment of PsbO
Amino acid sequences of PsbO protein from Synechococcus sp. PCC 7002 (GI:501262885), Synechocystis sp. PCC 6803 (GI:16331068), Nostoc sp. PCC 7120 (GI:499307220), Synechococcus elongatus PCC 7942 (GI:499562747), Gloeobacter violaceus PCC 7421 (GI:499456216), Chlamydomonas reinhardtii (GI:159473144), Arabidopsis thaliana (GI:15240013), Solanum tuberosum (GI:809113), Triticum aestivum (GI:21844), Spinacia oleracea (GI:21283), and Pisum sativum (GI:344004) were aligned by CLC Main Workbench (Qiagen Co., Aarhus, Denmark).
Construction of PsbO Point Mutants by Site-directed Mutagenesis
The kanamycin resistance cassette was inserted intoSYNPCC7002_A0269 gene to create a knockout mutant (ΔpsbO) and into SYNPCC7002_A1479 gene to generate a neutral platform as the alternative wild type (WT*). The trans-complemented psbO mutant and site-directed mutants were assembled with the cpcBA promoter, derived from Synechocystis (PcpcBA), in the front of SYNPCC7002_A0269 gene and the chloramphenicol resistance cassette at the termini. The site-directed mutagenesis was carried out with QuickChangeII mutagenesis kit (Agilent Technologies, Santa Clara, CA), according to manufacturer instructions. Transformation of the Synechococcus strain was performed as previously reported (32). Cells were screened on A+ agar plates with chloramphenicol (25 μg/ml) or kanamycin (50 μg/ml). Transformants were then verified by PCR and direct DNA sequencing. Primers used in this study are listed in supplemental Table S10.
Oxygen Evolution Rate Analysis
Photosynthetic oxygen evolution rates were measured by a Clark-type oxygen electrode (Hansatech Instruments Ltd., Norfolk, UK). Cells of all strains were collected in the exponential phase and resuspended in fresh A+ media, then diluted to a final OD730 = 1 for oxygen evolution measurements. For temperature treatments, cells were incubated at 28, 33, 38, and 43 °C for 10 min. Cell suspensions were added to the electrode chamber with continuous stirring for each experiment. The various electrode chamber temperatures were maintained by a circulating water bath. Oxygen evolution rate was measured after 2 min in the dark with stimulating irradiance of 500 μmol photons/m2/s and 10 mm NaHCO3.
Chlorophyll Fluorescence Analysis
Chlorophyll fluorescence measurements were made using a pulse-amplitude fluorimeter DUAL-PAM-100 (Heinz Walz GmbH, Effeltrich, Germany). All the mutants were collected in their exponential phase and diluted to OD730 = 1 for chlorophyll fluorescence analysis. Before measurements, cells were dark-adapted for 15 min at room temperature. The intensity of the actinic light was maintained at 50 μmol photons/m2/s. The light intensity dependence of electronic conversion efficiency of photosystem II was detected with the stepwise increase in photosynthetically active radiation (0–1000 μmol photons/m2/s) through the fluorescence measure mode of a dual channel. For recording the transient post-illumination increase, the saturating pulse and actinic light lasting for 2 min were terminated and the slow kinetics had been allowed to run for around 10 min.
Experimental Design and Statistical Rationale
The wild-type strain of Synechococcus was prepared from the same lysates as those used for the study of acetylation proteome by Yang et al. (47). Specifically, acetylated peptides from whole cell lysates were fractionated and enriched using immunoaffinity enrichment strategies. Eight samples were analyzed by high-accuracy nanoflow LC-MS/MS technology (Fig. 1B). FDR thresholds for proteins, peptides, and modification sites were set to maximum 1%. Data source name of the proteome of Synechococcus sp. PCC 7002 was GCA_000019485.1 (release date 2008/03/14) with 3186 entries in the database searched. GO analysis was performed using Blast2GO (41) and GO enrichment (42) using Hypergeometric test. Benjamini-Hochberg FDR correction was carried out at a significance level of 0.05. Lysine-acetylated motifs were analyzed by motif-x (44) at a significance level of 0.000001. Secondary structures were predicted using NetSurfP (48), with p values calculated by Wilcoxon test.
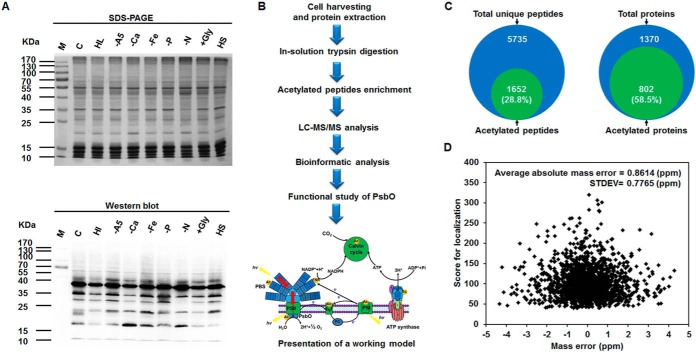
Fig. 1.
Synopsis of the acetylome analysis in Synechococcus sp. PCC 7002. A, Western blotting analysis of lysine acetylation profiles with antiacetyllysine antibodies in response to different stress conditions. Equal amounts of protein lysates (20 μg) were extracted from the cells under standard culture conditions (C), high-intensity light stress for 30 min (HL), A5 deficiency (-A5), calcium deficiency (-Ca), iron deficiency (-Fe), phosphate deficiency (-P), nitrogen deficiency (-N), glycerol addition (+Gly), and high salinity stress for 30 min (HS). Coomassie blue staining of the gel acts as a loading control. B, Workflow for the acetylome analysis in Synechococcus. Proteins form exponentially growing cells were extracted and peptides cleaved by trypsin were prefractionated using in-solution digestion. Lysine-acetylated peptides were enriched using acetyl-lysine antibody and subsequently analyzed by nano-LC-MS/MS. Bioinformatic analysis and functional analysis of photosystem II manganese-stabilizing protein (PsbO) were performed. C, Pie chart illustrates the percentage of lysine-acetylated peptides and proteins compared with all identified peptides and proteins, respectively. D, Mass error distribution of the lysine-acetylated peptides.
In the functional study, three replicates were prepared and analyzed for each of the ten biological samples: wild type (WT), WT* (SYNPCC7002_A1479 as a neutral platform), ΔpsbO, trans-complemented psbO mutant, and the site-directed mutants under the control of PcpcBA, including samples with a nonacetylated arginine to construct the mutants K99R, K190R, and K219R or a mimic-acetylated glutamine to construct K99Q, K190Q, and K219Q. The following analyses were performed on the samples: immunoprecipitation and Western blotting analysis of PsbO acetylation level; construction of ΔpsbO, WT*, trans-complemented psbO mutant, and site-directed mutants by site-directed mutagenesis; followed by growth rate, oxygen evolution rate, and chlorophyll fluorescence analyses. Mean and standard deviation of the three independent experiments are presented and the statistical tests used to analyze the data are indicated in the respective figures and/or manuscript sections.
RESULTS
Establishment of the Synechococcus Acetylome
To characterize the extent of lysine acetylation in Synechococcus, Western blotting analysis of whole cell lysates using antiacetyllysine antibody was conducted. It showed multiple lysine-acetylated protein bands under different culture conditions (Fig. 1A), suggesting the wide distribution of diverse acetylated proteins and a regulated process of lysine acetylation in this organism. To further comprehensively identify the large-scale data set of lysine-acetylated sites on Synechococcus proteins, the total proteins from different growth conditions before the total protein extracts were isolated, and the acetylated peptides were enriched using antiacetyllysine antibody. Then, the enriched modified peptides and sites were analyzed by a nanoflow LC-MS/MS (Fig. 1B). The suspicious acetylated peptides with unclear MS/MS spectra were manually removed (supplemental Table S1). In this study, we detected 802 proteins acetylated at 1653 unique sites. It is noteworthy that the identified acetylated proteins comprising 25.2% (802/3186) of Synechococcus proteome exceed the maximum previous findings of acetylome in bacteria (18, 20–22). Synechococcus proteomic data also showed that 28.8% acetylated peptides and 58.5% acetylated proteins were enriched, compared with the identified total unique peptides and proteins, respectively (Fig. 1C). In addition, we estimated the mass accuracy of the modified peptide and the average absolute mass error was 0.8614 ppm (Fig. 1D). We also found that the average acetylation degree was 1.97. Of all these acetylated proteins, ∼55.1% had a single acetylated site, 21.1% carried two acetylated sites, 10% contained three sites, and 6.6% carried four sites. Notably, 58 proteins revealed five or more acetylated sites. For instance, phycobilisome core-membrane linker phycobiliprotein with 17 acetylation sites was the most intensively acetylated protein in this research. This data set provides a comprehensive view of lysine acetylation in Synechococcus.
Functional Classification of Acetylproteins
To better understand the role of identified acetylproteins in Synechococcus, we classified these proteins according to their subcellular localization, biological process, and molecular function (supplemental Fig. S1 and supplemental Table S2). Among the 802 acetylated proteins, 569 (70.9%) were functionally annotated to various biological processes. Many acetylated proteins were involved in cellular amino acid metabolism, translation, generation of precursor metabolites and energy, carbohydrate metabolism, photosynthesis, and other processes. With respect to the annotated molecular function, the acetylated proteins were assigned to several subcategories as ion binding, 252; oxidoreductase activity, 102; and lyase activity, 53. Based on subcellular localization, 491 (61.2%) acetylproteins were in the cytoplasm, 100 were assigned to cytoplasmic membrane, 10 to periplasm, 6 to outer membrane, and 3 to extracellular space. However, the localization of 22.9% acetylproteins was not predicted. We also performed a statistical test by applying a multiple testing correction, and the results have been presented in supplemental Table S3. We conducted GO enrichment analyses to examine which functional categories were targeted for lysine acetylation (supplemental Fig. S2 and supplemental Table S4). GO enrichment analysis of biological process category demonstrated that acetylproteins involved in cellular metabolism (p value = 1.12E-02), metabolism (p value = 1.12E-02), cellular process (p value = 1.12E-02), and primary metabolism (p value = 3.52E-02) were significantly enriched. These findings indicated the essential regulatory roles of lysine acetylated proteins in cellular metabolism in Synechococcus.
Analysis of Lysine Acetylation Sites
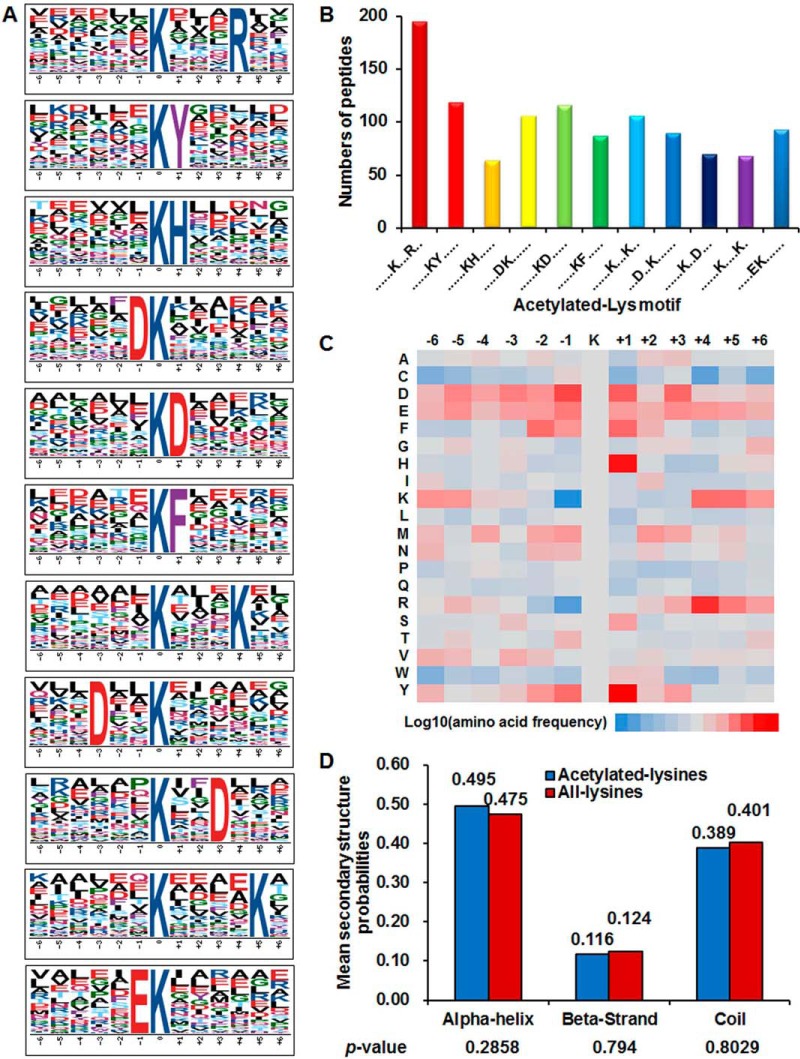
To evaluate the nature of acetylated lysine in Synechococcus, we compared the occurrences of 12 amino acid residues surrounding the acetylation site with those of all lysine that occur in the Synechococcus proteome. Previous studies have found preferences for amino acid residues around the acetylated lysine in prokaryotes (11, 18–22, 24). Herein, we defined 11 enriched acetylation site motifs for 1103 unique sites, accounting for 66.8% of the sites identified, namely KacXXXR, KacY, KacH, DKac, KacD, KacF, KacXXXK, DXXKac, KacXXD, KacXXXXK, and EKac (Kac, acetylated lysine; X, random amino acid residue; Fig. 2A; supplemental Table S5), and they exhibit distinct abundances (Fig. 2B). Consistent with previous findings (18, 20–22, 24), for three of these motifs, a positively charged lysine or arginine enrichment was observed on the +4 or +5 position toward the C terminus. In addition, histidine (H), phenylalanine, or tyrosine (Y) on the +1 position toward the C terminus appeared frequently, as previously described in bacteria (11, 18, 21, 23–24). The motifs KacH, KacY, and KacF were also conserved in strawberry (56), implying some conserved motifs surrounding acetylated sites are shared by plants and bacteria. Interestingly, two negatively charged residues, including glutamate (E) on −1 position toward N terminus and aspartate (D) on −1/+1 or −4/+4 position toward N or C terminus, were observed. These results suggested that negatively or positively charged or aromatic amino acid residues might be functionally important for acetylation. To our knowledge, five of these motifs, including E on −1 position and D on −1/+1 or −4/+4 position, were the first significant motifs identified in bacteria. Considering that acetylated peptides with these three motifs account for 28.5% of all identified peptides, we speculate that proteins with such motifs might have some special functions in this model system. These results were also determined by an inspection of the intensity map analysis (Fig. 2C). We observed that the residues H and Y are preferred at +1 position of acetylpeptides. The different enrichment of amino acid residues surrounding acetylation sites implies unique substrate preferences in Synechococcus and reflects the conservation of several target sequences of bacterial lysine acetyltransferase.
Fig. 2.
Analysis of lysine acetylation sites. A, Probability logos of significantly enriched acetylation site motifs after aligning peptide sequences, including 12 residues surrounding the acetylated lysine residue, using motif-x. B, The number of identified peptides containing acetylated lysine in each motif. C, The relative abundance of amino acid residues flanking the acetylation sites represented by an intensity map. The intensity map shows the relative abundance for ±6 amino acids from the Synechococcus lysine-acetylated site. The colors in the intensity map represent the log10 of the ratio of frequencies within acetyl-13-mers versus nonacetyl-13-mers (red shows enrichment, blue shows depletion). D, Distribution of secondary structures containing lysine acetylation sites. The different secondary structures (α-helix, β-strand and -coil) of acetylated lysine identified in this study were compared with the secondary structures of all lysine identified through proteomics.
To investigate the local secondary structures of acetylproteins surrounding acetylation sites, we performed a structural analysis of the lysine-acetylated proteins. As demonstrated, ∼49.5% of the modified sites were in regions of α-helices, 192 sites were in β-sheets, and 643 were in the unstructured regions (Fig. 2D). A similar distribution pattern of secondary structures was found in acetylated lysine in the Synechocystis proteome (22), which differs from most other bacterial acetylomes (18, 20, 23). Distinct from other bacteria, we deduced that acetylation prefers the α-helix structures in cyanobacteria.
Comparative Analysis with Other Bacterial Acetylome
Lysine acetylation is an evolutionarily conserved PTM in both prokaryotes and eukaryotes (1). To determine the conservation of the identified acetylated proteins, we performed a systematic analysis of the orthologs of acetylproteins by comparing the Synechococcus acetylproteome to several bacterial acetylproteomes (11–12, 15–17, 57). Orthologs of 512 (63.8%) acetylated proteins could be detected in the other bacterial acetylproteomes. Among the 802 acetylated proteins, 397 shared common sequences with Synechocystis sp. PCC 6803, whereas 219, 114, 108, 91, 37, 154, 139, and 97 proteins had orthologs in the acetylproteome of E. coli, T. thermophilus, S. enterica, E. amylovora, Leptospira interrogans, B. subtilis, Mycoplasma pneumonia, and G. kaustophilus, respectively (supplemental Table S6). Generally, the Synechococcus acetylome exhibited greater similarity to Gram-negative bacteria (486 proteins) than to Gram-positive bacteria (256 proteins). The comparative analysis of acetylproteomes revealed that many of the acetylated proteins might serve a conserved role in cellular metabolism and translation, which is consistent with these microorganisms with characterized acetylproteomes. These results point to an evolutionarily conserved and potentially vital role of acetylation in cellular function.
Protein-protein Interaction Network of the Identified Acetylproteins
To further understand the cellular processes regulated by acetylation, the STRING database was used to analyze the interaction network of these identified acetylproteins. The network showed that 748 acetylated proteins were network nodes, with 7474 physical and functional interactions (supplemental Table S7). Twenty-eight highly interconnected clusters of acetylated proteins were retrieved (supplemental Table S8). The top two clusters consisted of numerous acetylproteins involved in protein synthesis and photosynthesis (supplemental Fig. S3), with 1138 and 358 direct physical or indirect functional interactions, respectively, indicating the important regulatory roles of acetylation modification in the two processes. Interestingly, they have also been assigned to ribosome, aminoacyl-tRNA biosynthesis, and photosynthesis pathways via KEGG pathway analysis (supplemental Table S9).
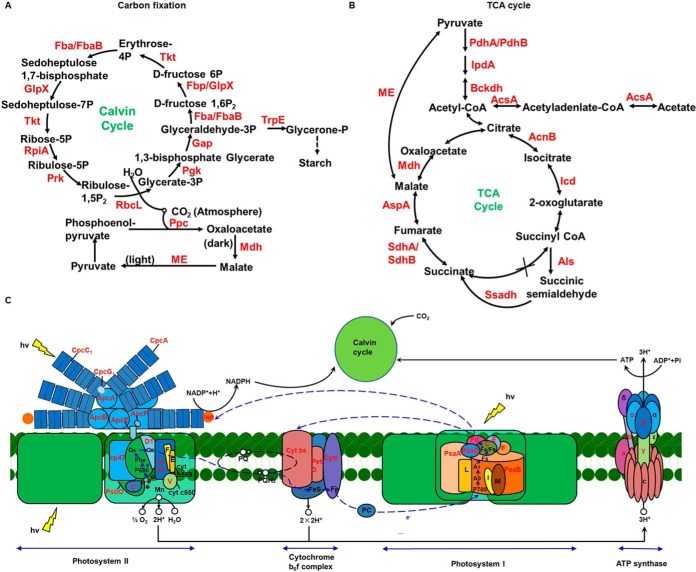
Lysine acetylation has been shown to play a major role in metabolism regulation in prokaryotes (18–22). In the Synechococcus acetylproteomes, many of the identified acetylated proteins were involved in metabolic processes consistent with previous findings. As shown in supplemental Table S9, 232 acetylated proteins were found to be involved in various metabolic pathways. Nearly all enzymes involved with each step of central metabolism were identified to be acetylated, indicating the role of reversible lysine acetylation in the regulation and control of cellular enzymatic activity at PTM level in this cyanobacterium. Moreover, 43 acetylated proteins were involved in carbon metabolism, 9 were photosynthesis-antenna proteins, 27 involved in photosynthesis, 14 involved with carbon fixation photosynthetic organisms, 17 in glycolysis/gluconeogenesis, and 10 in citrate cycle (Fig. 3). Consistent with the Synechococcus proteogenomic study (58), our data also revealed the potential role of protein acetylation in photosynthesis (supplemental Fig. S3).
Fig. 3.
Schematic illustration of lysine acetylation events involved in carbon fixation (A), tricarboxylic acid cycle (B), and photosynthetic pathway (C). The lysine-acetylated proteins are labeled in red.
Amino Acid Sequences Alignment of PsbO
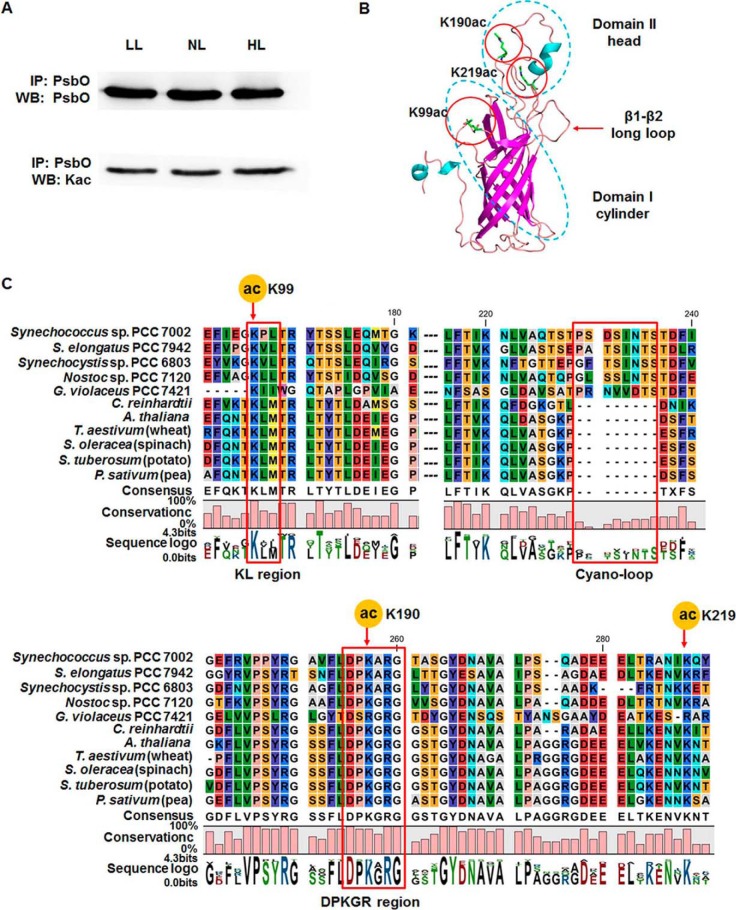
The manganese (Mn)-stabilizing protein PsbO in the PSII complex in all oxygenic photosynthetic organisms plays central roles in maintaining the integrity and activity of the Mn cluster and is essential for optimal oxygen evolution (59). To validate the acetylation events in PsbO, we carried out Western blotting analysis of the whole cell lysate under different conditions using pan antiacetylation antibody and anti-PsbO antibody. Our findings confirmed PsbO acetylation through immunoprecipitation; nevertheless, PsbO acetylation levels remained unchanged under different light intensities (Fig. 4A). To better understand the potential role of acetylation in PsbO conformation, the three-dimensional structure of PsbO was modeled, and the location of the three identified acetylation sites have been marked accordingly in Fig. 4B. The conservation of amino acid sequences of PsbO among species from cyanobacteria to higher plants was analyzed. Fig. 4C shows multiple alignments of partial sequences of PsbO from five cyanobacteria and six higher plants. This comparison indicates that cyanobacteria have one distinct region differing from higher plants, presented as the cyano-loop labeled in a rectangle. Although multiple alignments showed that PsbO was poorly conserved, researchers have identified five highly conserved regions within this protein among oxygenic organisms (60). Two acetylated sites of PsbO, including K99 and K190, were in KL and DPKGR conserved regions, respectively. Considering the evolutionary conservation of acetylation in both prokaryotes and eukaryotes, we deduced that the modification events might occur in other oxygenic organisms and might have a significant impact on the structure or function of PsbO.
Fig. 4.
Western blotting analysis of photosystem II manganese-stabilizing protein (PsbO) acetylation and amino acid sequence alignment analysis of PsbO. A, Western blotting analyses of acetylated PsbO under different light intensities, by immunoprecipitation using anti-PsbO and anti-acetyllysine antibodies. LL: low-intensity light (50 μmol photons/m2/s), NL: normal light (250 μmol photons/m2/s), and HL: high-intensity light (2000 μmol photons/m2/s). B, Three-dimensional cartoon depicting the backbone structure and localization of acetylation sites in PsbO. C, Alignment of amino acid sequences of PsbO protein from Synechococcus sp. PCC 7002 (GI:501262885), Synechocystis sp. PCC 6803 (GI:16331068), Nostoc sp. PCC 7120 (GI:499307220), Synechococcus elongatus PCC 7942 (GI:499562747), Gloeobacter violaceus PCC 7421 (GI:499456216), Chlamydomonas reinhardtii (GI:159473144), Arabidopsis thaliana (GI:15240013), Solanum tuberosum (GI:809113), Triticum aestivum (GI:21844), Spinacia oleracea (GI:21283), and Pisum sativum (GI:344004). Conserved regions (KL and DPKGR) and a unique region in cyanobacteria (cyano-loop) are indicated in red rectangles.
Site-directed Mutagenesis of PsbO Acetylation Sites
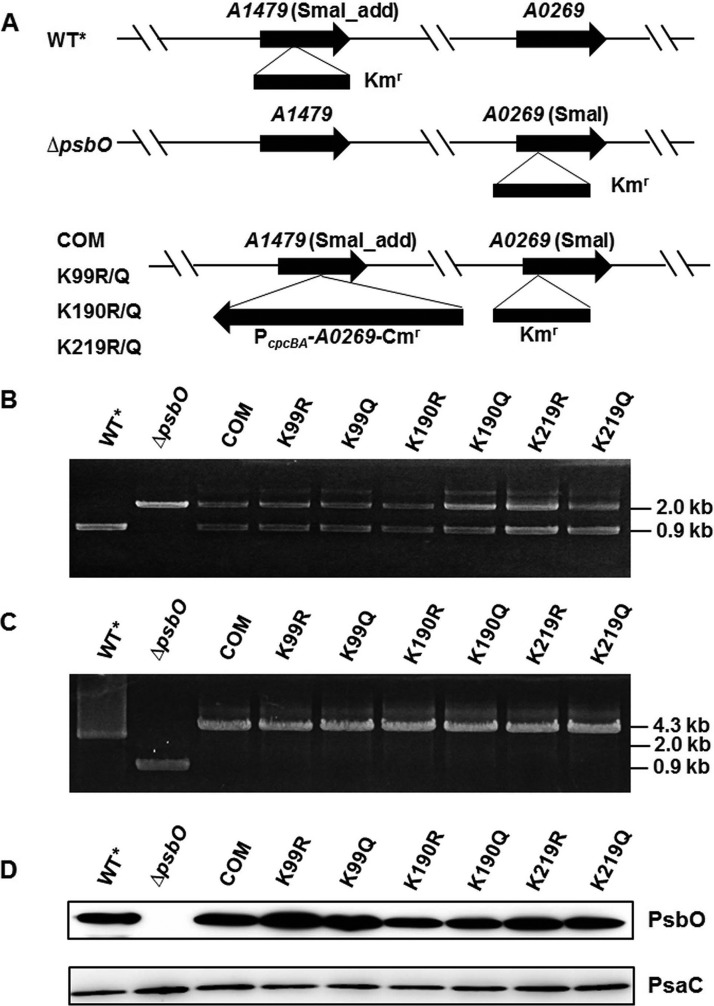
To investigate whether the acetylated sites were involved in oxygen evolution within PSII, site-directed mutagenesis of PsbO at K99 in KL region, K190 in DPKGR region, and K219 was performed. Before construction of the point mutants, a neutral site platform and a psbO null mutant were constructed as positive and negative controls, respectively. Synechococcus was adapted to grow on glycerol as an organic carbon source to favor mixotrophic growth, allowing the construction of the mutants lacking PSI and/or PSII, as previously reported (61) (Fig. 5). A neutral site platform with inactive SYNPCC7002_A1479 was obtained as the alternative wild type WT* (Fig. 5A; supplemental Fig. S4). However, the inactivation of psbO in vivo behaved distinctly from WT*. The schematic of the vector for these site-directed transformations is presented in Fig. 5A. In this study, we converted lysine to either a nonacetylated arginine to construct the mutants K99R, K190R, and K219R or to a mimic-acetylated glutamine to construct the mimic mutants K99Q, K190Q, and K219Q. All mutants were verified by PCR and direct DNA sequencing (Fig. 5B; supplemental Fig. S5).
Fig. 5.
Construction and detection of the ΔpsbO site-directed mutants. A, Schematic shows the construction of ΔpsbO site-directed mutant strains by homologous recombination. The kanamycin resistance (Kmr) cassette was inserted into the SmaI restriction sites in psbO gene and the chloramphenicol resistance (Cmr) cassette was inserted into the modified SmaI restriction sites in SYNPCC7002_A1479 gene, added to the appropriate ends of the flanking sequences (see Experimental Procedures). B, C, Agarose gel electrophoresis demonstrates complete segregation of alleles of PcpcBA-A0269::Cmr using A0269kn-F/R and A1479kn-F/R primers. D, Western blotting analyses of total cellular extracts from Synechococcus wild type and ΔpsbO site-directed mutants were performed using antibodies specific to PsbO and PsaC. PsbO, photosystem II manganese-stabilizing protein; ΔpsbO, PsbO knockout mutant; PsaC, photosystem I subunit VII; PcpcBA, cpcBA promoter derived from Synechocystis.
To validate PsbO expression level in both WT* and mutants, we analyzed PsbO abundance among these strains by Western blotting analysis (Fig. 5C). The PsbO mutant forms were expressed in each case, and the translational levels of PsbO in K99R/K99Q, K190R/K190Q, and K219R/K219Q mutants were generally the same as that in WT*. Additionally, the intrinsic core of PSI among these mutants and wild type were further analyzed using the anti-PsaC antibody. PsaC was expressed in equal amount among the wild type and mutant strains, indicating that PSI was not affected by the inactivation or site-directed mutagenesis of psbO.
K190 in the conserved DPKGR region of PsbO is present in the head domain I, as shown in Figs. 4B and 4C, and is involved in many interactions with PSII members (60). Furthermore, our results supported the speculation that lysine acetylation at K190 in PsbO might have an anciently conserved role in the maintenance of PSII system and the thylakoid membrane, since its divergent evolution from G. violaceus (62). PsbO mutations have been previously identified to be associated with a decrease in the accumulation of PSII components in C. reinhardtii (63) and A. thaliana (64–65), suggesting that deletions or amino acid substitutions might affect the intrinsic degradation of the PSII complex.
Characterization of Growth Rate of Mutant Strains
PsbO plays an essential role in the variability of both prokaryotic and eukaryotic systems (63, 66–67). Knockout and/or knockdown mutations in psbO in Synechocystis (66, 68), C. reinhardtii (63), and A. thaliana (64–65, 67, 69) have been constructed for variability investigation. Mutations involving removal of PsbO from PSII were found to have different effects in prokaryotes and eukaryotes. The Synechocystis psbO mutant is capable of slow growth, but is more vulnerable to photoinhibition (70). However, the absence of PsbO in C. reinhardtii causes a photoautotrophic growth failure and the loss of the ability to assemble functional PSII reaction centers (63). The psbo1 A. thaliana mutant exhibits retardation of photoautotrophic growth as well (67). To confirm that the growth of Synechococcus depends on the involvement of acetylated sites located on PsbO, we tested the ability of psbO point mutants to grow photoautotrophically. The different strains exhibited variation in growth in our study (supplemental Fig. S6). We failed to construct a fully segregated null mutation of psbO under photoautotrophic growth conditions, indicating the essential role of PsbO in photoautotrophic growth. Contrarily, the ΔpsbO mutant could grow under normal irradiation (∼250 μmol·m−2·s−1) when cells were supplied with 10 mm glycerol. The present experiments clearly demonstrated a decreased photoheterotrophic growth rate of all the site-directed mutants constructed under normal light compared with WT*, but an increased growth rate compared with the ΔpsbO mutant, which might be ascribed to the partial complementation of PsbO. Among these point mutants, strains mimicking constitutively acetylated states (K99Q, K190Q, and K219Q) grew under normal light irradiation, like the mutants with nonacetylated states (K99R, K190R, and K219R). Our observations indicate a minor role of PsbO acetylation in the variability of Synechococcus.
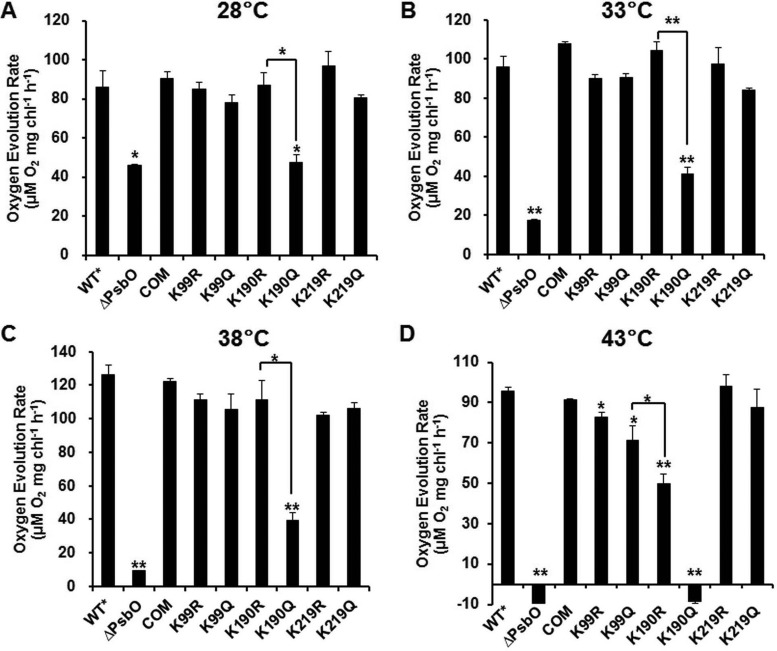
Detection of Oxygen Evolution Rate in Whole Cell under Various Temperature Conditions
PsbO stabilizes the Mn cluster that catalyzes the oxidation of water to molecular oxygen in photosynthetic organisms (59) and serves to optimize oxygen production at the physiological calcium and chloride concentrations (71). To characterize the role of PsbO acetylation in oxygen evolution in Synechococcus, the oxygen evolution rate in both WT* and mutants was examined at different temperatures. The ΔpsbO mutant has altered kinetic properties in the S-states of oxygen-evolving centers in Synechocystis (66, 72–73). Our data showed that WT*, K99R/K99Q, K190R/K190Q, and K219R/K219Q cells exhibit increased oxygen evolution with elevation in temperature and reach the maximal rate at 38 °C, after which the rate of oxygen generation remarkably reduced at 43 °C (Fig. 6). However, the mutants ΔpsbO and K190Q, with slow growth rates, exhibited dramatically decreased oxygen evolution with increase in temperature from 28 to 43 °C. On removal of PsbO, the initial oxygen evolution rate per cell was about 53.7, 18.2, 7.5, and −9.8% of the wild-type rate at 28, 33, 38, and 43 °C, respectively, whereas on substitution of K190 with uncharged residue Q, it was 55.2, 42.9, 31, and −9% at 28, 33, 38, and 43 °C, respectively. Nevertheless, K190R and other mutations had a marginal effect on this process. It has been verified that K160R mutant protein (Synechococcus K190R in PsbO) restores oxygen evolution more effectively to some extent than the K160Q mutant protein in vitro in Synechocystis, indicating participation of this residue in electrostatic interaction (74). Our findings suggested that PsbO acetylation at K190 negatively regulates oxygen evolution in Synechococcus at high temperatures.
Fig. 6.
Detection of oxygen evolution rate in alternative wild type (WT*), ΔpsbO mutant strain, and site-directed mutants based on equal cell numbers (at OD730), under 500 μmol/m2/s light at 28 (A), 33 (B), 38 (C), and 43 °C (D). Data are presented as the mean and standard deviation of three individual experiments. ΔpsbO, photosystem II manganese-stabilizing protein (PsbO) knockout mutant.
Detection of Electron Transport Rate through PSII (ETR(II)) and Transient Rise in Fluorescence
To investigate whether linear electron flow (LEF) was affected by PsbO acetylation, room temperature pulse amplitude modulated fluorescence was studied to determine the ETR(II) among the Synechococcus psbO site-directed mutants. The light response curves of ETR(II) were the same for these strains, except for the psbO null mutant, for which they not detectable, as shown in Fig. 7A. On the other hand, the K99R/K99Q, K190R/K190Q, and K219R/K219Q mutants showed a decrease in ETR(II) when compared with WT*. Nevertheless, no significant differences in ETR(II) were observed among these mutants.
Fig. 7.
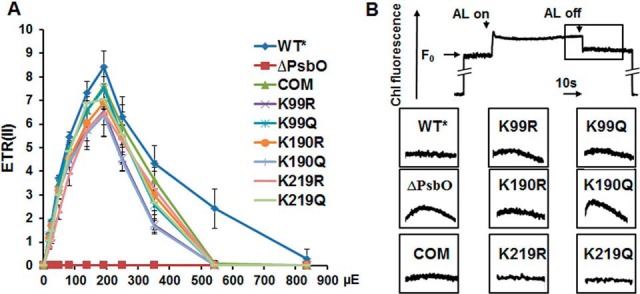
The electron transport rate through PSII (ETR(II)) and the transient increase in chlorophyll fluorescence of whole cells in the alternative wild type (WT*), ΔpsbO mutant strains, and site-directed mutants. A, ETR(II) was calculated using Dual-PAM software under different photosynthetically active radiations from 0 to 1000 μmol/m2/s (μE), and the data are presented as the mean of three independent experiments. B, The transient rise in fluorescence shown in the enlarged boxes was measured after the actinic light illumination was turned off. ΔpsbO, photosystem II manganese-stabilizing protein (PsbO) knockout mutant.
The increase in fluorescence was undetectable in WT, trans-complemented psbO, and K190R strains, whereas a trace activity was detected in K99R/K99Q (Fig. 7B). In contrast, ΔpsbO and K190Q mutants were identified by significant enhancement of postillumination increase in chlorophyll fluorescence. These results reflected the less efficient electron transfer in LEF, in contrast to the elevated cyclic electron flow (CEF) in the mimic-acetylated mutant. This enhancement of transient rise in fluorescence was because of PsbO dysfunction. Considering that post-illumination increases in chlorophyll fluorescence are thought to be involved in PSI-cyclic electron transport (75), we speculate that PsbO acetylation mediated an increase in ATP generation to compensate for the decrease in NADPH production.
DISCUSSION
Protein lysine acetylation has been extensively studied in both eukaryotes and prokaryotes. Lysine acetylation is a highly dynamic and reversible PTM. This process can be carried out by acetyltransferases and deacetylases within the cells (1). In this study, 1653 acetylated sites and several new motifs were identified on acetylated proteins in Synechococcus. Several potential acetyltransferases (SYNPCC7002_A0110, SYNPCC7002_F0063, SYNPCC7002_A2436, SYNPCC7002_A0096, and SYNPCC7002_A2015) and a deacetylase (SYNPCC7002_A2791) have also been identified among the acetylated proteins. Additionally, the motif analysis revealed several new motifs that might be recognized by protein acetyltransferases. However, no lysine acetyltransferases or deacetylases were predicted or confirmed in this model cyanobacterium. Recently, it was demonstrated that lysine acetylation can also be chemically catalyzed (76–77). Therefore, it is uncertain whether this PTM occurs enzymatically, nonenzymatically, or both ways in Synechococcus.
The proteomic profiling of lysine acetylation revealed that the acetylated proteins were involved in the regulation of many metabolic pathways (i.e. tricarboxylic acid cycle, glycolysis, and photosynthesis). Moreover, we also identified acetylated proteins involved in the two-component system and microbial metabolism in diverse environments, suggesting the evolution of adaptive responses to biotic and abiotic stresses. Thus, we speculate that acetylation is a reserve of carbon sources for cells resistant to unfavorable environment. Interestingly, our findings have indicated that protein lysine acetylation plays vital roles in photosynthesis in this model cyanobacterium, in accordance with the previous findings in Synechocystis (22) and Arabidopsis (6).
Photosynthesis serves as a system for sensing environmental stresses, besides its principal function as a solar energy converter (78–79). It has been reported that various PTMs might be regulating photosynthetic efficiency in triggering acclimation to environmental cues (80). Lately, characterization of PTMs in photosynthesis has provided a large quantity of information for the study of this process. For example, several reversible modifications, including serine/threonine/tyrosine phosphorylation (47, 81), N terminus and internal lysine acetylation (6), lysine and arginine methylation (82), and S-thiolation (83), have been well established as key regulators of Calvin cycle, ATP synthesis, electron transport, and oxygen evolution. Recently, Synechococcus proteogenomic analysis has revealed 23 different PTMs in photosynthesis, including lysine acetylation (58). Our proteomic study also suggested potential roles of lysine acetylation in photosynthesis in Synechococcus.
The precise structure of oxygen generating PSII, with a high resolution of ca. 1.9 Å, has revealed the geometric arrangement of the Mn4CaO5 cluster, which may facilitate the understanding of photolysis of water within PSII (84). However, the existence and function of PTMs, such as acetylation, in oxygen-evolving complex (OEC) have not yet been clarified with this structure. In our research, several proteins in OEC were found to be lysine acetylated, including PsbO. PsbO is responsible for maintaining optimal oxygen evolution from PSII during photosynthesis. It is poorly conserved, but has five highly conserved regions, including K99 in KL region and K190 in DPKGR region, discovered in all oxygenic photosynthetic organisms. K190 is substituted by arginine in G. violaceus, indicating the origin of this region and its role in PsbO stability. Three acetylated sites of PsbO (K99, K190, and K219) in Synechococcus were confirmed as targets for acetylation. It has been reported that PsbO in Synechocystis is acetylated at K82 (K83 in Synechococcus) and K209 (L213 in Synechococcus) (22). Nevertheless, no in-depth functional studies have been performed on PsbO acetylation in cyanobacteria.
In this study, we first focused on the role of PsbO acetylation in regulation of cellular growth under optimal growth conditions. The consequences of removal or site-directed mutagenesis of PsbO in vitro or in vivo have been analyzed in detail, to reveal its conserved role in photosynthetic organisms. Deletion or site-directed mutation of PsbO in PSII has been found to impact the photoautotrophic growth (63–66). The Synechocystis psbO mutant is capable of slow growth, but evolves oxygen at low rate, and is very sensitive to high-intensity lights under photoautotrophic conditions (66). Furthermore, the absence of PsbO in C. reinhardtii leads to the failure of photoautotrophic growth and functional PSII reaction center assembly (63). On the other hand, the psbo1 A. thaliana mutant exhibits photoautotrophic growth retardation (67). Removal of PsbO, in this research, showed similar physiological phenotypes. The growth rate of ΔpsbO mutant was partially restored by trans-complemented and site-directed mutations. Characterization of such strains showed that these mutations hindered photoheterotrophical growth when placed in a genetic background lacking PsbO. It has been studied that lack of PsbO in PSII preparations results in withdrawal of two out of the four Mn atoms comprising the water-oxidizing complex (85–86). Apart from this, PsbO has been hypothesized to be coupled with PSII turnover (87–89). In our study, PsbO acetylation was proposed to play a regulatory role in PSII and cell growth.
Next, we mainly focused on the role of PsbO acetylation in oxygen evolution under different temperature conditions. Thus, the oxygen evolution behaviors of ΔpsbO mutant and point mutant strains were further analyzed. Wild-type cells could evolve oxygen at 28 to 43 °C normally, whereas K190Q mutation in PsbO of Synechococcus suppressed oxygen evolution significantly, like ΔpsbO mutant, and the mutant could not produce oxygen as efficiently as WT* and other cells. It has been confirmed previously that oxygen evolution rate is inhibited by high temperature treatment in tobacco (90). Additionally, oxygen production was dramatically reduced on substitution of K160 in Synechocystis (K190 in Synechococcus) with uncharged residues in vitro. Contrarily, the K160R mutant protein could reconstitute oxygen evolution more effectively, implying the participation of this acetylated site in electrostatic interaction (74). In vitro reconstitution assays have shown that spinach PsbO is unique in its thermostability (91). Conversely, our data revealed that Synechococcus PsbO exhibits less thermotolerance in in vivo assays. Taken together, our observations confirmed and extended the earlier results that ΔpsbO mutant shows a decrease in oxygen evolution under high temperature conditions.
Five oxidation states of the OEC, known as S1, S2, S3, S4, and S0, have been previously characterized, with the S4 state being responsible for water oxidation and oxygen evolution (92). Many researchers have conducted experiments to investigate the molecular mechanism of regulation of S-state transitions occurring at OEC (65, 93–95). The psbo1 A. thaliana mutant exhibits longer S2 and S3 states (65). Thermoluminescence experiments reveal that the S2 state has a higher stability in the psbo1 mutant than in wild type, and electron paramagnetic resonance measurements suggest a lower yield of the S2 multiline signal in A. thaliana (94). Flash oxygen yield measurements reveal that the S2 and S3 states are more stable in the ΔpsbO mutant of Synechocystis and that there is at least 5-fold reduction in S3→[S4]→S0 transition in this mutant (96). Similar results have also been obtained for spinach PSII lacking PsbO, in vitro (97). Thus, we hypothesize that acetylation of PsbO at K190 may be one of the mechanisms that regulate S state transitions in Synechococcus.
The slow generation of oxygen in the absence of PsbO was verified to be a consequence of a decrease in the rate of electron transfer. Electron paramagnetic resonance measurement monitored the retardation in the rate of photooxidized YZ reduction in the ΔpsbO mutant from Synechocystis, indicating slower electron transfer from the Mn4CaO5 cluster to photooxidized YZ (98). On the other hand, the psbo1 A. thaliana mutant exhibits reduction in electron transfer between quinones QA and QB (65, 69). Consistent with the earlier observations, ETR(II) detected a decrease in LEF rate in the PSII of ΔpsbO and point mutants in our study. Additionally, CEF was measured, and the absence of PsbO and the substitution of K190 by glutamine resulted in an enhanced transient rise in fluorescence. These results suggested a positive correlation between acetylation and CEF.
To reveal the role of lysine acetylation in the phenotypic changes in photosynthesis, site-directed mutagenesis of psbO was performed to mimic the electrostatic charge of a nonacetylated lysine (K→R) or an acetylated lysine (K→Q) (supplemental Table S11). It is not surprising that the nonacetylated mutant has a similar phenotype to the native strain with respect to the very low acetyl stoichiometry (2, 99–100). Compared with the mutants mimicking nonacetylated states, the in vivo characterization of the K190Q mutant mimicking acetylation exhibited a decrease in oxygen evolution and an enhanced cyclic electron transport rate under normal growth conditions. Although photosynthetic performance of the regulation of acetylation at K190 in PsbO was characterized, the mechanisms that altered the ability of PsbO to reactivate oxygen evolution were not well established. To further investigate the regulatory role of acetylation in oxygen evolution, we might need to apply several strategies to study acetylation dynamics and accurate acetylation stoichiometry of PsbO, to understand the mechanisms.
DATA AVAILABILITY
The raw data and annotated peptide spectra of acetylated peptides have been deposited in the PeptideAtlas database and can be accessed with the identifier PASS00772 (http: //www.peptideatlas.org/PASS/PASS00772).
Supplementary Material
Footnotes
Author contributions: F.G. and J.Z. designed research; Z.C., G.Z., and M.Y. performed research; Z.C., M.Y., T.L., F.G., and J.Z. analyzed data; Z.C., F.G., and J.Z. wrote the paper.
* This work was supported by the National Natural Science Foundation of China (Grant No. 31570829), the Chinese Academy of Sciences Grant QYZDY-SSW-SMC004, the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB14030202).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PTM
- post-translational modification
- CEF
- cyclic electron flow
- D
- aspartate
- E
- glutamate
- EPR
- electron paramagnetic resonance
- ETR(II)
- electron transport rates through PSII
- FDR
- false discovery rate
- GO
- Gene Ontology
- K
- lysine
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- H
- histidine
- HL
- high light-intensity
- IP
- immunoprecipitation
- LEF
- linear electron flow
- Mn
- manganese
- OEC
- oxygen-evolving center
- PAM
- pulse amplitude modulated
- PsaC
- Photosystem I subunit VII
- PsbO
- photosystem II manganese-stabilizing protein
- PSI
- photosystem I
- PSII
- photosystem II
- WT*
- alternative wild type
- Y
- tyrosine.
REFERENCES
- 1. Kim G. W., and Yang X. J. (2011) Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem. Sci. 36, 211–220 [DOI] [PubMed] [Google Scholar]
- 2. Weinert B. T., Iesmantavicius V., Moustafa T., Scholz C., Wagner S. A., Magnes C., Zechner R., and Choudhary C. (2014) Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 10, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 4. Fritz K. S., Galligan J. J., Hirschey M. D., Verdin E., and Petersen D. R. (2012) Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J. Proteome Res. 11, 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weinert B. T., Wagner S. A., Horn H., Henriksen P., Liu W. R., Olsen J. V., Jensen L. J., and Choudhary C. (2011) Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci. Signal 4, ra48. [DOI] [PubMed] [Google Scholar]
- 6. Wu X., Oh M. H., Schwarz E. M., Larue C. T., Sivaguru M., Imai B. S., Yau P. M., Ort D. R., and Huber S. C. (2011) Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 155, 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henriksen P., Wagner S. A., Weinert B. T., Sharma S., Bacinskaja G., Rehman M., Juffer A. H., Walther T. C., Lisby M., and Choudhary C. (2012) Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics 11, 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaluarachchi D. S., Friesen H., Baryshnikova A., Lambert J. P., Chong Y. T., Figeys D., and Andrews B. (2012) Exploring the yeast acetylome using functional genomics. Cell 149, 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xue B., Jeffers V., Sullivan W. J., and Uversky V. N. (2013) Protein intrinsic disorder in the acetylome of intracellular and extracellular Toxoplasma gondii. Mol. Biosyst. 9, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu B. J., Kim J. A., Moon J. H., Ryu S. E., and Pan J. G. (2008) The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 18, 1529–1536 [PubMed] [Google Scholar]
- 11. Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., and Zhao Y. (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang K., Zheng S., Yang J. S., Chen Y., and Cheng Z. (2013) Comprehensive profiling of protein lysine acetylation in Escherichia coli. J. Proteome Res. 12, 844–851 [DOI] [PubMed] [Google Scholar]
- 13. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., and Zhao G. P. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee D. W., Kim D., Lee Y. J., Kim J. A., Choi J. Y., Kang S., and Pan J. G. (2013) Proteomic analysis of acetylation in thermophilic Geobacillus kaustophilus. Proteomics 13, 2278–2282 [DOI] [PubMed] [Google Scholar]
- 15. Okanishi H., Kim K., Masui R., and Kuramitsu S. (2013) Acetylome with structural mapping reveals the significance of lysine acetylation in Thermus thermophilus. J. Proteome Res. 12, 3952–3968 [DOI] [PubMed] [Google Scholar]
- 16. Kim D., Yu B. J., Kim J. A., Lee Y. J., Choi S. G., Kang S., and Pan J. G. (2013) The acetylproteome of Gram-positive model bacterium Bacillus subtilis. Proteomics 13, 1726–1736 [DOI] [PubMed] [Google Scholar]
- 17. Wu X., Vellaichamy A., Wang D., Zamdborg L., Kelleher N. L., Huber S. C., and Zhao Y. (2013) Differential lysine acetylation profiles of Erwinia amylovora strains revealed by proteomics. J. Proteomics 79, 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liao G., Xie L., Li X., Cheng Z., and Xie J. (2014) Unexpected extensive lysine acetylation in the trump-card antibiotic producer Streptomyces roseosporus revealed by proteome-wide profiling. J. Proteomics 106, 260–269 [DOI] [PubMed] [Google Scholar]
- 19. Liu F., Yang M., Wang X., Yang S., Gu J., Zhou J., Zhang X. E., Deng J., and Ge F. (2014) Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol. Cell. Proteomics 13, 3352–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xie L., Wang X., Zeng J., Zhou M., Duan X., Li Q., Zhang Z., Luo H., Pang L., Li W., Liao G., Yu X., Li Y., Huang H., and Xie J. (2015) Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int. J. Biochem. Cell Biol. 59, 193–202 [DOI] [PubMed] [Google Scholar]
- 21. Pan J., Ye Z., Cheng Z., Peng X., Wen L., and Zhao F. (2014) Systematic analysis of the lysine acetylome in Vibrio parahaemolyticus. J. Proteome Res. 13, 3294–3302 [DOI] [PubMed] [Google Scholar]
- 22. Mo R., Yang M., Chen Z., Cheng Z., Yi X., Li C., He C., Xiong Q., Chen H., Wang Q., and Ge F. (2015) Acetylome analysis reveals the involvement of lysine acetylation in photosynthesis and carbon metabolism in the model cyanobacterium Synechocystis sp. PCC 6803. J. Proteome Res. 14, 1275–1286 [DOI] [PubMed] [Google Scholar]
- 23. Liu L., Wang G. Y., Song L. M., Lv B. N., and Liang W. X. (2016) Acetylome analysis reveals the involvement of lysine acetylation in biosynthesis of antibiotics in Bacillus amyloliquefaciens. Sci. Rep. 6, 20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meng Q., Liu P., Wang J., Wang Y., Hou L., Gu W., and Wang W. (2016) Systematic analysis of the lysine acetylome of the pathogenic bacterium Spiroplasma eriocheiris reveals acetylated proteins related to metabolism and helical structure. J. Proteomics 148, 159–169 [DOI] [PubMed] [Google Scholar]
- 25. Beck C., Knoop H., Axmann I. M., and Steuer R. (2012) The diversity of cyanobacterial metabolism, genome analysis of multiple phototrophic microorganisms. BMC Genomics 13, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chellamuthu V. R., Alva V., and Forchhammer K. (2013) From cyanobacteria to plants, conservation of PII functions during plastid evolution. Planta 237, 451–462 [DOI] [PubMed] [Google Scholar]
- 27. Parmar A., Singh N. K., Pandey A., Gnansounou E., and Madamwar D. (2011) Cyanobacteria and microalgae, a positive prospect for biofuels. Bioresour. Technol. 102, 10163–10172 [DOI] [PubMed] [Google Scholar]
- 28. Xu Y., Alvey R. M., Byrne P. O., Graham J. E., Shen G., and Bryant D. A. (2011) Expression of genes in cyanobacteria, adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002. Methods Mol. Biol. 684, 273–293 [DOI] [PubMed] [Google Scholar]
- 29. Aikawa S., Nishida A., Ho S. H., Chang J. S., Hasunuma T., and Kondo A. (2014) Glycogen production for biofuels by the euryhaline cyanobacteria Synechococcus sp. strain PCC 7002 from an oceanic environment. Biotechnol. Biofuels 7, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Batterton J. C. Jr, and Van Baalen C. (1971) Growth responses of blue-green algae to sodium chloride concentration. Arch. Mikrobiol. 76, 151–165 [DOI] [PubMed] [Google Scholar]
- 31. Nomura C. T., Sakamoto T., and Bryant D. A. (2006) Roles for heme-copper oxidases in extreme high-light and oxidative stress response in the cyanobacterium Synechococcus sp. PCC 7002. Arch. Microbiol. 185, 471–479 [DOI] [PubMed] [Google Scholar]
- 32. Frigaard N. U., Sakuragi Y., and Bryant D. A. (2004) Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol. Biol. 274, 325–340 [DOI] [PubMed] [Google Scholar]
- 33. Ludwig M., and Bryant D. A. (2011) Transcription profiling of the model cyanobacterium Synechococcus sp. strain PCC 7002 by Next-Gen (SOLiD) sequencing of cDNA. Front. Microbio. 2, 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 35. Zhang K., Yau P. M., Chandrasekhar B., New R., Kondrat R., Imai B. S., and Bradbury M. E. (2004) Differentiation between peptides containing acetylated or tri-methylated lysines by mass spectrometry, an application for determining lysine 9 acetylation and methylation of histone H3. Proteomics 4, 1–10 [DOI] [PubMed] [Google Scholar]
- 36. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., and Aebersold R. (2006) The peptideatlas project. Nucleic Acids Res. 34, D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., and Robles M. (2005) Blast2GO, a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 [DOI] [PubMed] [Google Scholar]
- 38. Conesa A., and Götz S. (2008) Blast2GO, A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Götz S., García-Gómez J. M., Terol J., Williams T. D., Nagaraj S. H., Nueda M. J., Robles M., Talón M., Dopazo J., and Conesa A. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Götz S., Arnold R., Sebastián-León P., Martín-Rodríguez S., Tischler P., Jehl M. A., Dopazo J., Rattei T., and Conesa A. (2011) B2G-FAR, a species-centered GO annotation repository. Bioinformatics 27, 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., Dao P., Sahinalp S. C., Ester M., Foster L. J., and Brinkman F. S. (2010) PSORTb 3.0, improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maere S., Heymans K., and Kuiper M. (2005) BiNGO, a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 43. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape, a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 45. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villen J., Li J., Cohn M. A., Cantley L. C., and Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Treeck M., Sanders J. L., Elias J. E., and Boothroyd J. C. (2011) The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe 10, 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang M. K., Qiao Z. X., Zhang W. Y., Xiong Q., Zhang J., Li T., Ge F., and Zhao J. D. (2013) Global phosphoproteomic analysis reveals diverse functions of Serine/Threonine/Tyrosine phosphorylation in the model cyanobacterium Synechococcus sp strain PCC 7002. J. Proteome Res. 12, 1909–1923 [DOI] [PubMed] [Google Scholar]
- 48. Petersen B., Petersen T. N., Andersen P., Nielsen M., and Lundegaard C. (2009) A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., and Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 51. Kanehisa M., and Goto S. (2000) KEGG, kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., and Tanabe M. (2014) Data, information, knowledge and principle, back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nielsen M., Lundegaard C., Lund O., and Petersen T. N. (2010) CPHmodels-3.0-remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res. 38, W576–W581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., and von Mering C. (2011) The STRING database in 2011, functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bader G. D., and Hogue C. W. (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC bioinformatics 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fang X., Chen W., Zhao Y., Ruan S., Zhang H., Yan C., Jin L., Cao L., Zhu J., Ma H., and Cheng Z. (2015) Global analysis of lysine acetylation in strawberry leaves. Front. Plant Sci. 6, 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Witchell T. D., Eshghi A., Nally J. E., Hof R., Boulanger M. J., Wunder E. A. Jr, Ko A. I., Haake D. A., and Cameron C. E. (2014) Post-translational modification of LipL32 during Leptospira interrogans infection. PLoS Negl. Trop. Dis. 8, e3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang M. K., Yang Y. H., Chen Z., Zhang J., Lin Y., Wang Y., Xiong Q., Li T., Ge F., Bryant D. A., and Zhao J. D. (2014) Proteogenomic analysis and global discovery of posttranslational modifications in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 111, E5633–E5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suorsa M., and Aro E. M. (2007) Expression assembly and auxiliary functions of photosystem II oxygen-evolving proteins in higher plants. Photosynth. Res. 93, 89–100 [DOI] [PubMed] [Google Scholar]
- 60. De Las Rivas J., and Barber J. (2004) Analysis of the structure of the PsbO protein and its implications. Photosynth. Res. 81, 329–343 [DOI] [PubMed] [Google Scholar]
- 61. Shen G., and Bryant D. A. (1995) Characterization of a Synechococcus sp. Strain PCC 7002 mutant lacking Photosystem I. Protein assembly and energy distribution in the absence of the photosystem I reaction center core complex. Photosynth. Res. 44, 51–53 [DOI] [PubMed] [Google Scholar]
- 62. Rippka R., Waterbury J., and Cohen-Bazire G. A. (1974) Cyanobacterium which lacks thylakoids. Arch. Microbiol. 100, 419–436 [Google Scholar]
- 63. Mayfield S. P., Bennoun P., and Rochaix J. D. (1987) Expression of the nuclear encoded OEE-1 protein is required for oxygen evolution and stability of photosystem II particles in Chlamydomonas reinhardtii. EMBO J. 6, 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yi X., McChargue M., Laborde S., Frankel L. K., and Bricker T. M. (2005) The manganesestabilizing protein is required for photosystem II assembly/stability and photoautotrophy in higher plants. J. Biol. Chem. 280, 16170–16174 [DOI] [PubMed] [Google Scholar]
- 65. Liu H., Frankel L. K., and Bricker T. M. (2009) Functional complementation of Arabidopsis thaliana psbo1 mutant phenotype with an N-terminally His6-tagged PsbO-1 protein in photosystem II. Biochim. Biophys. Acta 1787, 1029–1038 [DOI] [PubMed] [Google Scholar]
- 66. Burnap R. L., and Sherman L. A. (1991) Deletion mutagenesis in Synechocystis sp. PCC 6803 indicates that the Mn-stabilizing protein of photosystem II is not essential for O2 evolution. Biochemistry 30, 440–446 [DOI] [PubMed] [Google Scholar]
- 67. Murakami R., Ifuku K., Takabayashi A., Shikanai T., Endo T., and Sato F. (2002) Characterization of an Arabidopsis thaliana mutant with impaired psbO, one of two genes encoding extrinsic 33-kDa proteins in photosystem II. FEBS Lett. 523, 138–142 [DOI] [PubMed] [Google Scholar]
- 68. Mayes S. R., Cook K. M., Self S. J., Zhang Z. H., and Barber J. (1991) Deletion of the gene encoding the PS II 33 kDa protein from Synechocystis PCC6803 does not inactivate water-splitting but increases vulnerability to photoinhibition. Biochim. Biophys. Acta 1060, 1–12 [Google Scholar]
- 69. Liu H., Frankel L. K., and Bricker T. M. (2007) Functional analysis of photosystem II in a PsbO-1-deficient mutant in Arabidopsis thaliana. Biochemistry 46, 7607–7613 [DOI] [PubMed] [Google Scholar]
- 70. Komenda J., and Barber J. (1995) Comparison of psbO and psbH deletion mutants of Synechocystis PCC6803 indicates that degradation of D1 protein is regulated by the QB site and is dependent on protein synthesis. Biochemistry 34, 9625–9631 [DOI] [PubMed] [Google Scholar]
- 71. Bricker T. M., Roose J. L., Fagerlund R. D., Frankel L. K., and Eaton-Rye J. J. (2012) The extrinsic proteins of Photosystem II. Biochim. Biophys. Acta 1817, 121–142 [DOI] [PubMed] [Google Scholar]
- 72. Philbrick J. B., Diner B. A., and Zilinskas B. A. (1991) Construction and characterization of cyanobacterial mutants lacking the manganese-stabilizing polypeptide of photosystem II. J. Biol. Chem. 266, 13370–13376 [PubMed] [Google Scholar]
- 73. Burnap R. L., Qian M., Shen J. R., Inoue Y., and Sherman L. A. (1994) Role of disulfide linkage and putative intermolecular binding residues in the stability and binding of the extrinsic manganese stabilizing protein to the Photosystem II reaction center. Biochemistry 33, 13712–13718 [DOI] [PubMed] [Google Scholar]
- 74. Motoki A., Usui M., Shimazu T., Hirano M., and Katoh S. (2002) A Domain of the Manganese-stabilizing Protein from Synechococcus elongatus Involved in Functional Binding to Photosystem II. J. Biol. Chem. 277, 14747–14756 [DOI] [PubMed] [Google Scholar]
- 75. Shikanai T., Endo T., Hashimoto T., Yamada Y., Asada K., and Yokota A. (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. U.S.A. 95, 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weinert B. T., Iesmantavicius V., Wagner S. A., Schölz C., Gummesson B., Beli P., Nyström T., and Choudhary C. (2013) Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51, 265–272 [DOI] [PubMed] [Google Scholar]
- 77. Kuhn M. L., Zemaitaitis B., Hu L. I., Sahu A., Sorensen D., Minasov G., Lima B. P., Scholle M., Mrksich M., Anderson W. F., Gibson B. W., Schilling B., and Wolfe A. J. (2014) Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PloS One 9, e94816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Arnon D. I. (1982) Sunlight, earth, life, the grand design of photosynthesis. Sciences 22, 22–27 [Google Scholar]
- 79. Anderson J. M., Chow W. S., and Park Y. I. (1995) The grand design of photosynthesis, acclimation of the photosynthetic apparatus to environmental cues. Photosynth. Res. 46, 129–139 [DOI] [PubMed] [Google Scholar]
- 80. Pfannschmidt T., and Yang C. (2012) The hidden function of photosynthesis, a sensing system for environmental conditions that regulates plant acclimation responses. Protoplasma 249, S125–S136 [DOI] [PubMed] [Google Scholar]
- 81. Baginsky S., and Gruissem W. (2009) The chloroplast kinase network, new insights from large-scale phosphoproteome profiling. Mol. Plant 2, 1141–1153 [DOI] [PubMed] [Google Scholar]
- 82. Alban C., Tardif M., Mininno M., Brugière S., Gilgen A., Ma S., Mazzoleni M., Gigarel O., Martin-Laffon J., Ferro M., and Ravanel S. (2014) Uncovering the protein lysine and arginine methylation network in Arabidopsis chloroplasts. PLoS ONE 9, e95512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Michelet L., Zaffagnini M., Vanacker H., Le Maréchal P., Marchand C., Schroda M., Lemaire S. D., and Decottignies P. (2008) In vivo targets of S-thiolation in Chlamydomonas reinhardtii. J. Biol. Chem. 283, 21571–21578 [DOI] [PubMed] [Google Scholar]
- 84. Umena Y., Kawakami K., Shen J. R., and Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 [DOI] [PubMed] [Google Scholar]
- 85. Ono T., and Inoue Y. (1983) Mn-preserving extraction of 33-, 24- and 16-kDa proteins from O2-evolving PS II particles by divalent salt-washing. FEBS Lett. 164, 255–260 [Google Scholar]
- 86. Miyao M., and Murata N. (1984) Role of the 33 kDa polypeptide in preserving Mn in the photosynthetic oxygen evolution. FEBS Lett. 170, 350–354 [Google Scholar]
- 87. Aro E. M., Virgin I., and Andersson B. (1993) Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143, 113–134 [DOI] [PubMed] [Google Scholar]
- 88. Aro E. M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., and Rintamäki E. (2005) Dynamics of Photosystem II, a proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56, 347–356 [DOI] [PubMed] [Google Scholar]
- 89. Lundin B., Nurmi M., Rojas-Stuetz M., Aro E. M., Adamska I., and Spetea C. (2008) Towards understanding the functional difference between the two PsbO isoforms in Arabidopsis thaliana-insights from phenotypic analyses of psbO knockout mutants. Photosynth. Res. 98, 405–414 [DOI] [PubMed] [Google Scholar]
- 90. Wang P., Duan W., Takabayashi A., Endo T., Shikanai T., Ye J. Y., and Mi H. (2006) Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol. 141, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lydakis-Simantiris N., Hutchison R. S., Betts S. D., Barry B. A., and Yocum C. F. (1999) Manganese stabilizing protein of Photosystem II is a thermostable, natively unfolded polypeptide. Biochemistry 38, 404–414 [DOI] [PubMed] [Google Scholar]
- 92. Kok B., Forbush B., and McGloin M. (1970) Cooperation of charges in photosynthetic O2 evolution-1. A linear four step mechanism. Photochem. Photobiol. 11, 457–475 [DOI] [PubMed] [Google Scholar]
- 93. Cole J., Boska M., Blough N. V., and Sauer K. (1986) Reversible and irreversible effects of alkaline pH on Photosystem II electron-transfer reactions. Biochim. Biophys. Acta 848, 41–47 [DOI] [PubMed] [Google Scholar]
- 94. Allahverdiyeva Y., Mamedov F., Holmström M., Nurmi M., Lundin B., Styring S., Spetea C., and Aro E. M. (2009) Comparison of the electron transport properties of the psbo1 and psbo2 mutants of Arabidopsis thaliana. Biochim. Biophys. Acta 1787, 1230–1237 [DOI] [PubMed] [Google Scholar]
- 95. Shoji M., Isobe H., and Yamaguchi K. (2015) QM/MM Study of the S2 to S3 transition reaction in the oxygen-evolving complex of photosystem II. Chem. Phys. Lett. 636, 172–179 [DOI] [PubMed] [Google Scholar]
- 96. Burnap R. L., Shen J. R., Jursinic P. A., Inoue Y., and Sherman L. A. (1992) Oxygen yield and thermoluminescence characteristics of a cyanobacterium lacking the manganese-stabilizing protein of Photosystem II. Biochemistry 31, 7404–7410 [DOI] [PubMed] [Google Scholar]
- 97. Miyao M., Murata N., Lavorel J., Maisonpeteri B., Boussac A., and Etienne A. L. (1987) Effect of the 33-kDa protein on the S-state transitions in photosynthetic oxygen evolution. Biochim. Biophys. Acta 890, 151–159 [Google Scholar]
- 98. Razeghifard M. R., Wydrzynski T., Pace R. J., and Burnap R. L. (1997) YZ• reduction kinetics in the absence of the manganese-stabilizing protein of Photosystem II. Biochemistry 36, 14474–14478 [DOI] [PubMed] [Google Scholar]
- 99. Choudhary C., Weinert B. T., Nishida Y., Verdin E., and Mann M. (2014) The growing landscape of lysine acetylation links metabolism and cell signaling. Mol. Syst. Biol. 15, 536–550 [DOI] [PubMed] [Google Scholar]
- 100. Baeza J., Dowell J. A., Smallegan M. J., Fan J., Amador-Noguez D., Khan Z., and Denu J. M. (2014) Stoichiometry of site-specific lysine acetylation in an entire proteome. J. Biol. Chem. 289, 21326–21338 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and annotated peptide spectra of acetylated peptides have been deposited in the PeptideAtlas database and can be accessed with the identifier PASS00772 (http: //www.peptideatlas.org/PASS/PASS00772).