Abstract
Wnt signaling pathways are of significant interest in development and oncogenesis. The first step in these pathways typically involves the binding of a Wnt protein to the cysteine-rich domain (CRD) of a Frizzled receptor. Wnt-Frizzled interactions can be antagonized by secreted Frizzled-related proteins (SFRPs), which also contain a Frizzled-like CRD. The large number of Wnts, Frizzleds, and SFRPs, as well as the hydrophobic nature of Wnt, poses challenges to laboratory-based investigations of interactions involving Wnt. Here, utilizing structural knowledge of a representative Wnt-Frizzled CRD interaction, as well as experimentally determined binding affinities for a selection of Wnt-Frizzled CRD interactions, we generated homology models of Wnt-Frizzled CRD interactions and developed a quantitative structure-activity relationship for predicting their binding affinities. The derived model incorporates a small selection of terms derived from scoring functions used in protein-protein docking, as well as an energetic term considering the contribution made by the lipid of Wnt to the Wnt-Frizzled binding affinity. Validation with an external test set suggests that the model can accurately predict binding affinity for 75% of cases and that the error associated with the predictions is comparable with the experimental error. The model was applied to predict the binding affinities of the full range of mouse and human Wnt-Frizzled and Wnt-SFRP interactions, indicating trends in Wnt binding affinity for Frizzled and SFRP CRDs. The comprehensive predictions made in this study provide the basis for laboratory-based studies of previously unexplored Wnt-Frizzled and Wnt-SFRP interactions, which, in turn, may reveal further Wnt signaling pathways.
Keywords: homology modeling, lipid-protein interaction, protein-protein interaction, structural biology, Wnt signaling, Frizzled receptor, cysteine-rich domain, structure-activity relationship
Introduction
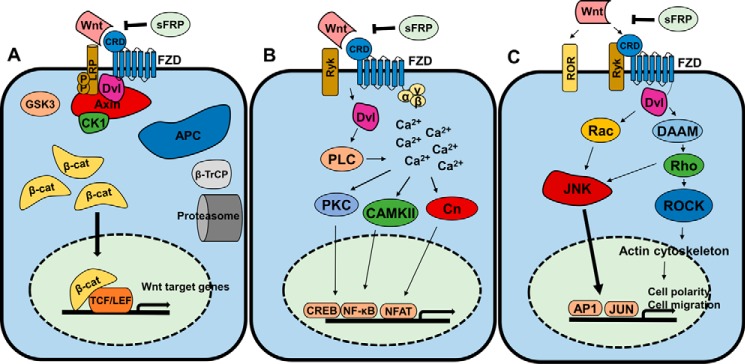
The Wnt signaling pathway is an evolutionarily conserved pathway found in vertebrates and insects (1). It functions to regulate body axis formation, cell fate, cell proliferation, and morphogenesis in embryonic development (2), whereas aberrant Wnt signaling is a hallmark of many cancers (3). Wnt signaling pathways can be divided into three separate branches: a canonical or β-catenin-dependent pathway and two non-canonical or β-catenin-independent pathways, known as the planar cell polarity (PCP)4 and Wnt/Ca2+ pathways (4, 5) (see Fig. 1). Wnt signaling is generally initiated by the binding of Wnt ligands to a Frizzled (Fzd) receptor (6). In the canonical Wnt signaling pathway (Fig. 1a), low-density lipoprotein-related protein 5/6 (LRP5/6) acts as a co-receptor for Wnt binding (7). Signal transduction by the three major Wnt signaling pathways is regulated by the interaction of Fzd with the cytoplasmic protein Dishevelled (Dvl) (8). In the canonical pathway, β-catenin-dependent signaling is mediated through the cytoplasmic “destruction complex” composed of Axin, protein phosphatase 2A, adenomatous polyposis coli protein, glycogen synthase kinase 3 (GSK3), and casein kinase 1α (CK1α) (9). In the presence of Wnt stimulation, Fzd is activated, permitting Dvl binding and resulting in the stabilization of the destruction complex and the accumulation of non-phosphorylated β-catenin, which then translocates to the nucleus and binds to T cell factor/lymphoid enhancer-binding factor transcription factors on the promoter of target genes (4). In the absence of Wnt stimulation, the destruction complex is destabilized, allowing for the phosphorylation of β-catenin by CK1α and GSK3 (10); phosphorylated β-catenin is then proteolytically degraded (11). The Wnt/Ca2+ pathway (Fig. 1b) is activated through Wnt ligands binding to Fzd receptors, resulting in an increase in intracellular calcium (12). Calcium ions are able to activate both calmodulin-dependent protein kinase II (13) and protein kinase C (PKC) (14), which subsequently activate transcription factors NFκB and cAMP-response element-binding protein. The cytosolic phosphatase calcineurin (Cn) is also activated by calcium ions. Cn-dependent dephosphorylation and activation of nuclear factor of activated T cells lead to the transcription of genes in cardiomyocytes, neuronal cells, and skeletal muscle (15). Signal transduction via the PCP pathway (Fig. 1c) is initiated through Wnt binding to Fzd and co-receptors ROR and Ryk. Fzd activation leads to Dvl-mediated activation of Rac and Rho. JNK and Rho-associated protein kinase (ROCK) are activated by Rac and Rho, respectively, which mediates actin polymerization and activates transcription factors AP-1 and JUN (16).
Figure 1.
Wnt signaling pathways. A, canonical Wnt signaling. Wnt binding to Fzd CRD initiates the destabilization of the cytoplasmic destruction complex (adenomatous polyposis coli protein (APC), Axin, GSK3, CK1, and Dvl). This allows cytosolic β-catenin (β-cat) accumulation and subsequent translocation to the nucleus where it binds to T cell factor/lymphoid enhancer-binding factor (TCF/LEF) transcription factors to transcribe Wnt target genes. SFRPs antagonize this cascade, and β-catenin is polyubiquitinated by β-transducin repeats-containing protein (β-TrCP) and degraded by proteolysis. B, the Wnt/Ca2+ pathway. Wnt binding to Fzd CRD or Ryk co-receptor activates Dvl, which stimulates calcium release. Downstream effectors PKC, calmodulin-dependent protein kinase II (CaMKII), and Cn activate transcription factors cAMP-response element-binding protein (CREB), NF-κB, and nuclear factor of activated T cells (NFAT). C, the PCP pathway. Wnt stimulation is effected initially through Fzd-Dvl interaction and co-receptors ROR/Ryk and passed through multiple effectors (Rac, phospholipase C (PLC), Disheveled-associated activator of morphogenesis (DAAM)) downstream to ROCK and JNK. ROCK regulates the actin cytoskeleton, and JNK activates AP1 and JUN transcription factors to regulate cell polarity and migration.
Wnts comprise a group of 19 proteins that are subject to numerous post-translational modifications, including the formation of a large number of characteristic disulfide bonds, glycosylation in the endoplasmic reticulum (17), and palmitoylation by Porcupine, which aids in their secretion and facilitates their interaction with Frizzled (18). Structurally, as determined by the co-crystallization of Xenopus Wnt8 (XWnt8) with the mouse Fzd8 CRD, Wnts are composed of two domains: an N-terminal domain and a C-terminal domain (19). The N-terminal domain contains 10 cysteine residues forming five disulfide bridges in a cluster of α-helices, whereas the C-terminal domain contains six disulfide bridges and a two-stranded β-sheet (19). Frizzled receptors are a group of 10 membrane-bound receptors comprising the majority of Class F G protein–coupled receptors (GPCRs). Frizzleds, like other GPCRs, consist of seven hydrophobic transmembrane helices but feature an extracellular cysteine-rich domain (CRD) in their N terminus (20). The CRD is characterized by a conserved pattern of 10 cysteines and can bind Wnt and Norrin ligands (21, 22). The five mammalian secreted Frizzled-related proteins (SFRPs) are secreted glycoproteins composed of an N-terminal CRD and a C-terminal netrin-like domain (23). These proteins function to antagonize the Wnt signaling pathway (24) through binding of either the CRD (25) or the netrin-like domain to Wnt ligands (26), thus interfering with Wnt binding to Fzd and preventing β-catenin-mediated gene transcription. The SFRPs have been studied in great detail for their potential roles as tumor suppressors and their implications in carcinogenesis (23).
Because of the large number of possible Wnt-Fzd CRD interactions (which, considering CRDs from both Fzds and SFRPs, totals 285 interactions per species), it is challenging to investigate these experimentally. A recent study utilized biolayer interferometry (BLI) to investigate a small set of mouse Wnt-Fzd CRD interactions in a combinatorial manner (27). Numerous other interactions have been identified via co-immunoprecipitation (coIP) or proposed based on co-expression of particular Wnts with particular Fzds (6). Although coIP and co-expression are valuable methods for suggesting the existence of specific protein-protein interactions, they are unable to provide an indication of the likely strength of an interaction. Computational studies provide the opportunity to complete the knowledge of interactions between Wnts and Fzd CRDs and may reveal previously unexplored high-affinity interactions.
In this study, we have generated homology models of Wnt complexes with both Fzd and SFRP CRDs and predict the likely binding affinity associated with these interactions. For a series of Wnt-Fzd CRD interactions for which dissociation constants have been reported (27), we then evaluated the interaction energy for the protein-protein and lipid-protein components of the interactions; this was achieved through scoring the interactions against the full set of functions contained in CCharPPI (28) (for the protein-protein component) and scoring using Prime MM-GB/SA (for the lipid-protein component). Strike was then used to develop and evaluate binding affinity prediction models using scores obtained from CCharPPI and Prime MM-GB/SA as descriptors for the model building. A model with high predictive performance was identified and subsequently applied to predict the binding affinities of all Wnt-Fzd and Wnt-SFRP CRD interactions in both mouse and human cases.
Results
Preparation of homology models of Wnt-Fzd CRD complexes
We prepared homology models of all mouse and human Wnts and all mouse and human Fzd and SFRP CRDs; details of UniProt accession numbers, sequence ranges, and sequence alignments used to build the models are provided in supplemental Table S1 and Figs. S2 and S3. The vast majority of proteins modeled did not feature large insertions or deletions relative to either XWnt8 or mFzd8 CRD with the exceptions of mouse and human Wnt6, Wnt10a, and Wnt10b; these Wnts feature insertions relative to XWnt8 larger than that able to be built by Prime (greater than 20 residues). To build these structures, we utilized an alternative procedure incorporating the I-TASSER server (described in detail under “Experimental procedures”), which is capable of building much longer insertions than Prime through its use of an iterative template fragment assembly approach to model building (29).
Following assembly of the complexes and refinement using a procedure automated using KNIME (supplemental Fig. S4), the MolProbity score of all models was calculated. The MolProbity score provides a single value metric of structural quality, summarizing the number of atomic clashes, percentage of backbone conformations in regions outside the Ramachandran favored regions, and the percentage of bad side-chain rotamers (30). The TM-score and the root-mean-squared deviation (RMSD) of the Cα atoms of the models with respect to the XWnt8-mFzd8 CRD complex structure (Protein Data Bank code 4F0A) (19), which was the template for all models, were also calculated. These measures assess differences in the coordinates of two structures (31). The mean value for the MolProbity scores for the mouse and human models was slightly greater than the MolProbity score obtained for the XWnt8-mFzd8 CRD complex structure (Table 1) but nonetheless comparable, indicating the generally high quality of the models. The mean values for the model TM-scores with respect to the template crystal structure were generally high, and the mean values for the model Cα RMSD values very low, further indicating the generally high quality of the models and their limited divergence from the template crystal structure. Selected complexes are shown in Fig. 2. Quality metrics are summarized in Table 1, and full details are provided in supplemental Tables S5–S10.
Table 1.
Summary of model quality metrics
| XWnt8-mFzd8 CRD complex | Mouse modelsa | Human modelsa | |
|---|---|---|---|
| MolProbity score | 1.72 | 2.13 ± 0.46 | 2.08 ± 0.42 |
| TM-scoreb | 1.00 | 0.91 ± 0.04 | 0.91 ± 0.05 |
| Cα RMSD (Å)b | 0.00 | 1.39 ± 0.39 | 1.39 ± 0.45 |
a Mean values ±2 S.D. shown; data for individual complexes shown in supplemental Tables S5–S10.
b TM-score and Cα RMSD values calculated with respect to the XWnt8-mFzd8 CRD complex (Protein Data Bank code 4F0A). By definition, the TM-score for an optimally overlaid structure compared with itself is 1; the RMSD for an optimally overlaid structure compared with itself is 0.
Figure 2.
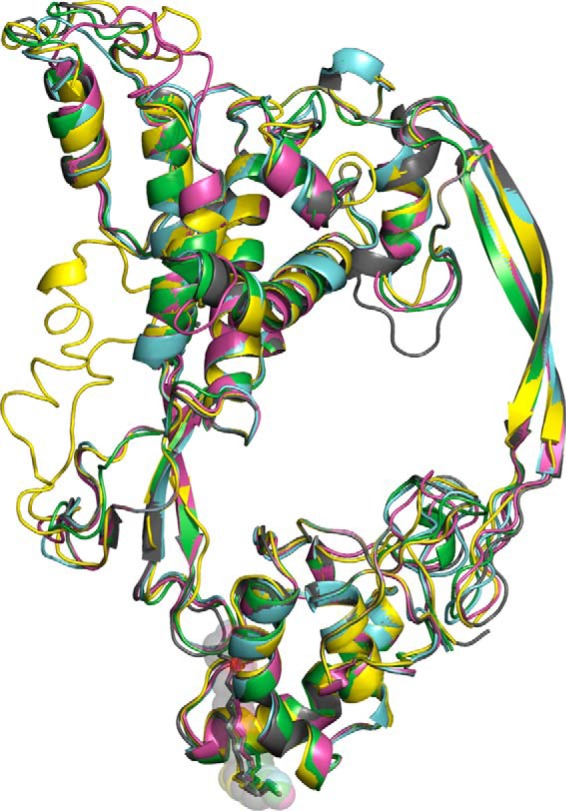
Homology models of selected Wnt-Fzd CRD complexes overlaid to the repaired XWnt8-mFzd8 CRD crystal structure (Protein Data Bank code 4F0A). Gray, repaired Protein Data Bank code 4F0A; pink, mWnt5-mFzd1 CRD complex; yellow, mWnt10a-mFzd6 complex; green, hWnt3a-hSFRP4 CRD complex; cyan, hWnt2b-hFzd9 CRD complex. Lipid is shown in all structure as sticks with transparent spheres.
Development and validation of a Wnt-Fzd CRD binding affinity prediction model
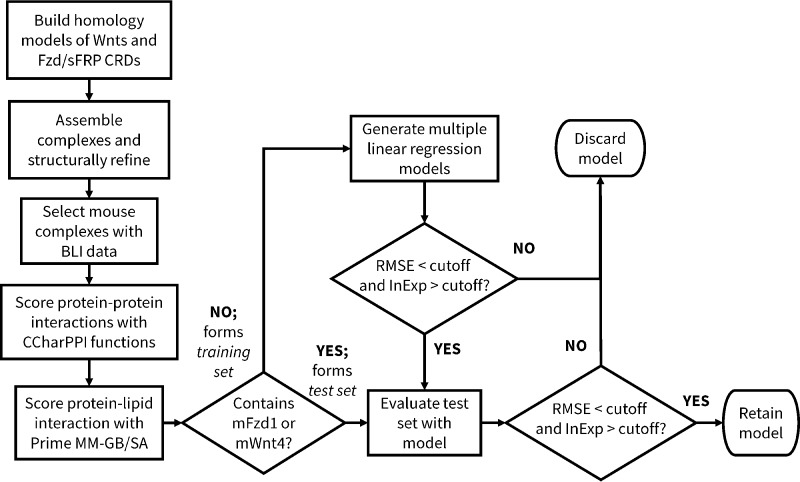
We used a set of mouse Wnt-Fzd CRD binding affinities determined by BLI (27) to develop and validate our binding affinity prediction model. The model building and evaluation procedure is summarized in Fig. 3 and herein described.
Figure 3.
Overview of the model building process. RMSE and InExp cutoffs used to select models at the relevant stages of model building are described in the Experimental Procedures.
Within the BLI data, we designated a training set, used to optimize the model, and a test set, used to demonstrate the performance of the model for data against which it had not been trained. Our training set was designated as complexes that were not part of our test set; our test set consisted of complexes involving interactions with either mFzd1 or mWnt4. The definition of the test set in this manner provided a simple means of selecting a test set covering a wide range of affinities. For all of these complexes, we then rescored, with separate procedures, the protein-protein portion and the lipid-protein portion of the interaction. The protein-protein portion was rescored against the majority of functions available within CCharPPI (28) (listed in supplemental Table S11), a server compiling a wide range of scoring functions suitable for use in protein-protein docking. The lipid-protein portion was rescored used Prime MM-GB/SA, which provides a rapid means for evaluating ligand-receptor binding energies with improved accuracy compared with typical docking scoring functions. The Prime MM-GB/SA calculation is also decomposed into its components (Coulomb/electrostatic, covalent binding, van der Waals, lipophilic, polar solvation/desolvation, hydrogen bonding, and π-π components; components used in this study listed in supplemental Table S11). The two strategies function complementarily to one another; the functions in CCharPPI are only capable of considering interactions between standard protein amino acids, whereas Prime MM-GB/SA is capable of studying interactions between small organic molecules with proteins. With this in mind, the Wnt lipid was removed from the CCharPPI calculations, and the Wnt protein was removed from the Prime MM-GB/SA calculations (that is, only the interaction between the Wnt lipid and the Fzd CRD was assessed by Prime MM-GB/SA). Multiple linear regression models combining one Prime MM-GB/SA component with one or more CCharPPI components (all herein referred to as descriptors) were then generated, thus allowing the development of a single model considering both the protein-lipid and the protein-protein portions of the interaction.
As it was computationally accessible to consider all possible three-descriptor models incorporating one Prime MM-GB/SA term and two CCharPPI-derived terms, we initially explored these. The performance of all models was evaluated using two principal metrics: 1) the root-mean-squared error (RMSE) between the predicted values and the average experimental values (RMSEtrain and RMSEtest); lower values indicate a better fit between the predictions and experimental values; and 2) the percentage of complexes for which the predicted value occurred within the experimental range reported (InExptrain and InExptest); higher values indicate a better fit between the predictions and experimental values.
High-performing three-descriptor models of Wnt-Fzd CRD binding typically incorporated the van der Waals term of the Prime MM-GB/SA calculation (supplemental Table S12). The lipophilic term of the Prime MM-GB/SA calculation also appears frequently in high-performing models. This is unsurprising considering the physicochemical properties of palmitoleic acid, which would suggest that the binding energy will likely be associated with van der Waals/non-polar interactions. The best performing three-descriptor models generally displayed RMSEs for both the training and test sets in the range of 0.3–0.4 kcal/mol, which is well outside the error range of the experiments of ∼0.2–0.3 kcal/mol (27); this indicates that three-descriptor models are insufficiently predictive.
Two models containing four descriptors were identified that were capable of high-prediction performance (Table 2). Both of these displayed RMSEs for the training and test sets less than 0.3 kcal/mol. Both included the van der Waals term of the Prime MM-GB/SA calculation, the PyRosetta hydrogen bonding potential (HBOND2) (32), and either the RW or RWplus statistical potentials (AP_calRW and AP_calRWp) (33). The fourth term in Model 1 is the antibody-antigen energy function of FireDock (FIREDOCK_AB) (34), whereas in Model 2, it is the total RosettaDock weighted energy (ROSETTADOCK). As the performance of Model 1 appeared slightly improved over Model 2, this model was selected for further study. Additionally, Model 1 was preferred over Model 2 for featuring a smaller constant term, suggesting that it may be able to predict affinities over a wider range than Model 2. The RMSE values for Model 1 suggest that the error associated with its use will be slightly larger than, but nonetheless similar to, the error range achieved by experiment.
Table 2.
Best performing four-descriptor models predicting Wnt-Fzd CRD binding energy
| Number | Model | RMSEtrain | RMSEtest | InExptrain | InExptest |
|---|---|---|---|---|---|
| kcal/mol | kcal/mol | % | % | ||
| 1 | ΔG = 0.0038165 × AP_calRW − 0.22506 × MMGBSA dG Bind vdW − 0.24626 × HBOND2 − 0.049875 × FIREDOCK_AB − 3.3475 | 0.23 | 0.27 | 80 | 75 |
| 2 | ΔG = 0.0021829 × AP_calRWp − 0.22111 × MMGBSA dG Bind vdW − 0.20861 × HBOND2 − 0.08699 × ROSETTADOCK − 7.7974 | 0.30 | 0.23 | 73 | 75 |
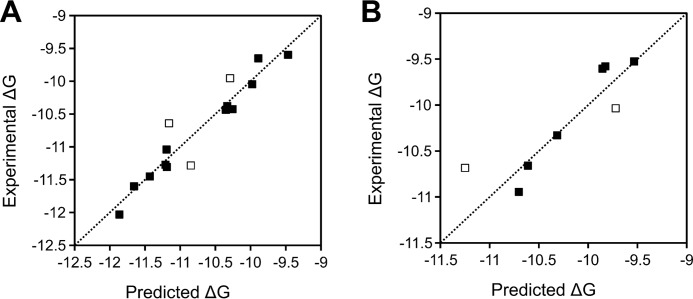
The maximum difference between any prediction made by the model, either in the training set or the test set, is ∼0.6 kcal/mol, which corresponds to a difference in Kd of approximately an order of magnitude (Fig. 4 and Table 3). Because there appears to be no particular Wnts or Fzds for which poor predictions are made, failure to make accurate predictions most likely occurs randomly and is not associated with a particular Wnt or Fzd structure; this is perhaps expected given the overall high structural quality of the models used. The binding affinities of the vast majority of cases in the training and test sets are predicted within 0.25 kcal/mol of the mean experimental values reported, which is within the experimental error range.
Figure 4.
Comparison of binding energy predictions by Model 1 in the training set (A) and test set (B). Points indicated by open squares are those where the predicted binding energy falls outside the model RMSE (0.23 kcal/mol for the training set; 0.27 kcal/mol for the test set).
Table 3.
Comparison of predictions by Model 1 with experimental data for the training and test sets
| Interaction | ΔGExpa | ΔGPreda | |ΔGExp − ΔGPred|a,b | Experimental Kdc | Predicted Kdc | Experimental range (predicted range)d | Set |
|---|---|---|---|---|---|---|---|
| mWnt3a-mFzd2 | −10.64 | −11.16 | 0.52 | 15.7 | 6.5 | +++ (++++) | Training |
| mWnt3a-mFzd4 | −11.27 | −11.21 | 0.06 | 5.4 | 6.0 | ++++ (++++) | Training |
| mWnt3a-mFzd5 | −11.60 | −11.65 | 0.05 | 3.1 | 2.9 | ++++ (++++) | Training |
| mWnt3a-mFzd7 | −11.28 | −10.85 | 0.43 | 5.3 | 11.0 | ++++ (+++) | Training |
| mWnt3a-mFzd8 | −12.03 | −11.86 | 0.17 | 1.5 | 2.0 | ++++ (++++) | Training |
| mWnt5-mFzd2 | −10.38 | −10.33 | 0.05 | 24.4 | 26.5 | +++ (+++) | Training |
| mWnt5-mFzd4 | −10.38 | −10.26 | 0.12 | 24.4 | 29.9 | +++ (+++) | Training |
| mWnt5-mFzd5 | −11.31 | −11.19 | 0.12 | 5.1 | 6.2 | ++++ (++++) | Training |
| mWnt5-mFzd7 | −10.05 | −9.98 | 0.07 | 42.6 | 47.9 | ++ (++) | Training |
| mWnt5-mFzd8 | −11.45 | −11.43 | 0.02 | 4.0 | 4.1 | ++++ (++++) | Training |
| mWnt5b-mFzd2 | −9.60 | −9.47 | 0.13 | 91.0 | 113.4 | ++ (+) | Training |
| mWnt5b-mFzd4 | −9.95 | −10.29 | 0.34 | 50.4 | 28.4 | ++ (+++) | Training |
| mWnt5b-mFzd5 | −10.44 | −10.35 | 0.09 | 22.0 | 25.7 | +++ (+++) | Training |
| mWnt5b-mFzd7 | −9.65 | −9.89 | 0.24 | 83.7 | 55.8 | ++ (++) | Training |
| mWnt5b-mFzd8 | −11.04 | −11.19 | 0.15 | 8.0 | 6.2 | ++++ (++++) | Training |
| mWnt3a-mFzd1 | −10.66 | −10.61 | 0.05 | 15.2 | 16.5 | ++++ (++++) | Test |
| mWnt4-mFzd2 | −9.53 | −9.53 | 0.00 | 102.5 | 102.5 | + (+) | Test |
| mWnt4-mFzd4 | −10.04 | −9.72 | 0.32 | 43.3 | 74.3 | ++ (++) | Test |
| mWnt4-mFzd5 | −10.68 | −11.25 | 0.57 | 14.7 | 5.6 | +++ (++++) | Test |
| mWnt4-mFzd7 | −9.58 | −9.83 | 0.25 | 94.2 | 61.7 | + (+) | Test |
| mWnt4-mFzd8 | −10.95 | −10.70 | 0.25 | 9.3 | 14.2 | ++++ (+++) | Test |
| mWnt5-mFzd1 | −10.33 | −10.31 | 0.02 | 26.5 | 27.4 | +++ (+++) | Test |
| mWnt5b-mFzd1 | −9.60 | −9.85 | 0.25 | 91.0 | 59.7 | + (+) | Test |
a ΔGexp calculated from experimental Kd values as ΔG = RT ln Kd where R is the gas constant (1.987 × 10−3 kcal K−1 mol−1) and T is the temperature at standard conditions (298 K). Predicted ΔG (ΔGpred) calculated according to Model 1. ΔG values expressed as kcal/mol.
b Absolute value of difference between experimental and predicted ΔG values.
c Kd values were obtained from Dijksterhuis et al. (27) and represent the average values reported. All Kd values expressed in nm.
d Guide to affinity range classifications: <10 nm, ++++; 10–40 nm, +++; 40–100 nm, ++; 100–400 nm, +; >400 nm, −. Cases in which the experimental and predicted Kd values occur in different ranges are underlined. Range in which value of experimental Kd occurs shown outside parentheses; range in which predicted Kd value occurs shown inside parentheses.
Further elaboration of the selected four-descriptor models into five-descriptor models was performed but did not result in models providing significant improvements in predictions (data not shown); similar RMSEs and a similar number of predictions occurring within the experimental ranges in both the training and test sets were obtained for the best four- and five-descriptor models. Thus, four-descriptor models were deemed sufficient for use in predicting binding affinities.
Prediction of binding affinities of Wnt-CRD interactions
In applying Model 1 to predict Wnt-CRD binding affinities in the mouse proteins, numerous trends are apparent (Fig. 5A and supplemental Table S13). Fzd3, Fzd5, SFRP3, and SFRP4 generally display high-affinity, nonspecific binding of Wnts, as evidenced by more than half of the interactions predicted to afford strong binding affinities (i.e. <10 nm). Fzd8 also displays nonspecific binding of Wnts; however, the majority of interactions are predicted to be of lower affinity than those with Fzd3, Fzd5, SFRP3, and SFRP4. Fzd1, Fzd4, Fzd7, and Fzd9 generally display moderate affinity for a wide variety of Wnts. Fzd1, Fzd7, and Fzd9 display high affinity for limited Wnts, indicating more selective binding compared with Fzd3, Fzd5, Fzd8, SFRP3, and SFRP4, whereas Fzd4 displays high affinity for several Wnts, indicating less selective binding. Fzd1 displays high affinity for Wnt6, Fzd7 displays high affinity for Wnt10a, and Fzd9 displays high affinity for both Wnt7a and Wnt16. Fzd2, Fzd6, Fzd10, SFRP1, SFRP2, and SFRP5 all display moderate- to high-affinity binding to less than half of the Wnts. However, this does not strictly translate to high selectivity; Fzd6 and Fzd10 bind with moderate affinity to several Wnts. Fzd2 displays high affinity for Wnt3a, Wnt7b, and Wnt10a. SFRP1, SFRP2, and SFRP5 all display high selectivity for specific Wnt ligands but retain moderate affinity for the majority of remaining Wnts. SFRP1 appears highly selective for Wnt7a, whereas SFRP2 is selective for Wnt2b and Wnt3a. SFRP5 displays moderate affinity for Wnt2b, Wnt5b, and Wnt6.
Figure 5.
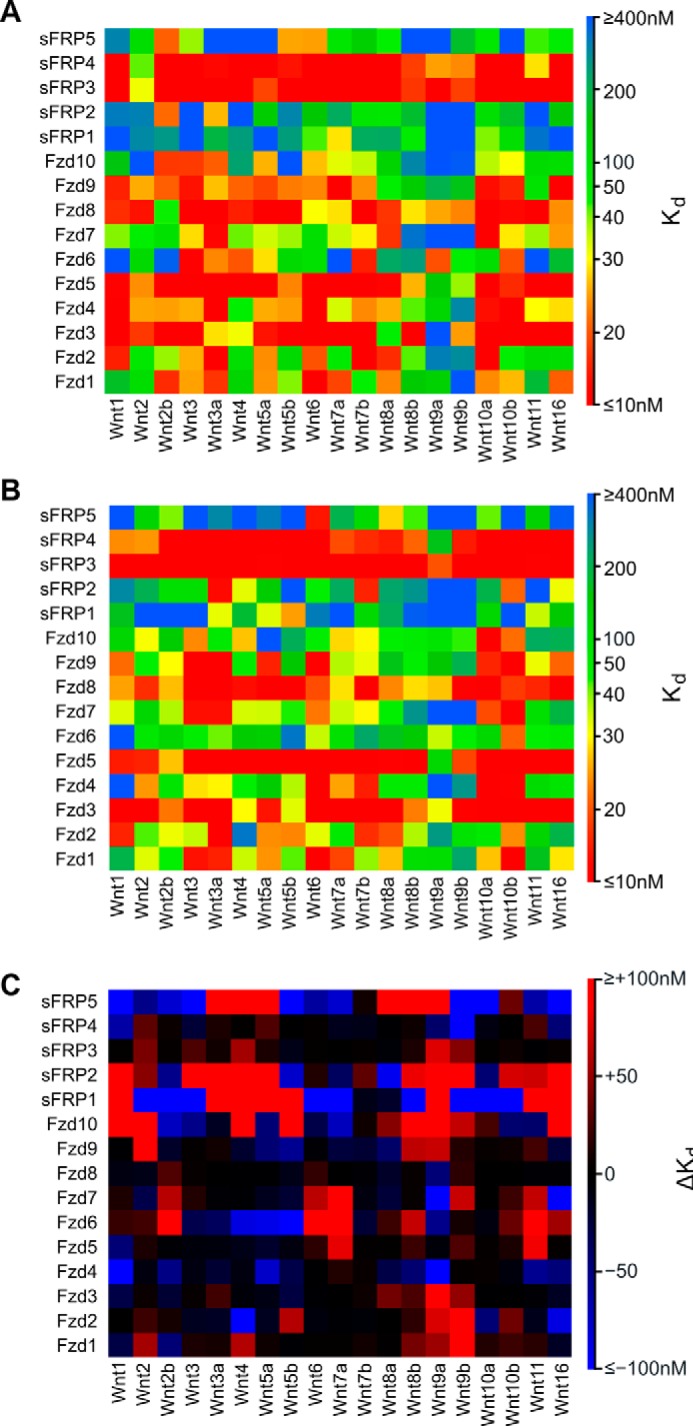
Binding affinity predictions by Model 1 for Wnt-Fzd interactions. A, mouse interactions. B, human interactions. C, binding affinity differences (ΔKd) between equivalent Wnt-Fzd interactions of mouse and human calculated as ΔKd = mouse Kd − human Kd. Positive ΔKd is indicative of a lower affinity interaction in mouse compared with human; negative ΔKd indicates a higher affinity interaction in mouse compared with human.
The human data generally display trends similar to the mouse data (Fig. 5B and supplemental Table S14). Fzd3, Fzd5, SFRP3, and SFRP4 still display generally high-affinity, nonspecific binding of Wnts; however, there are some specific points of difference. The interactions of human Fzd3 with Wnt8a and Wnt9a are predicted to be much higher affinity than in the case of the mouse, although the hFzd9-hWnt9a interaction is still predicted to be of only moderate affinity. Conversely, the interaction of human Fzd3 with Wnt5b is predicted to be of much lower affinity than the equivalent mouse interaction. The affinity of the mouse Wnt2 for SFRP3 and SFRP4 is predicted to be lower than the equivalent interactions in humans; however, Wnt9a is predicted to have increased affinity for these proteins in mouse compared with human. Significant differences in the predicted affinities of human Fzd4 for Wnt1, Wnt5a, and Wnt11 compared with the mouse interactions are observed; all of these interactions are predicted to be very low in binding affinity in humans, whereas in mice these are all predicted to be very high affinity. Large differences in the predicted affinities occur when comparing the interactions of mouse and human Fzd6, Fzd10, SFRP1, SFRP2, and SFRP5 (Fig. 5C); however, these interactions are generally predicted to be of low to moderate affinity and may not be indicative of different roles for Wnt interactions with these proteins in the two species.
Analysis of residues of functional importance to Wnt-Fzd CRD interactions
To propose residues of functional importance to Wnt-Fzd interactions, all 570 Wnt-Fzd CRD models were subject to MM-GB/SA analysis with per-residue decomposition using AMBER14 (35). This calculation allows the identification of specific residues making large contributions to the binding energy, which, in turn, can be used to suggest the most significant intermolecular contacts in the interaction. High-affinity complexes will generally have more residues making large contributions to the binding energy compared with low-affinity complexes; thus, high-affinity complexes will have greater influence on the designation of sequence positions of general importance to Wnt-Fzd CRD interactions.
Analysis of Fzd CRD-binding regions of Wnt indicates two major regions utilized by Wnt in binding Fzd CRDs (Fig. 6A). These correspond to the thumb and index finger regions of Wnt, which are already well known as Fzd CRD-binding regions (19, 36). Interestingly, Wnt residues beyond these two regions are rarely implicated in Fzd CRD binding (supplemental Fig. S15), and the majority of Wnt residues in these regions frequently implicated in Fzd CRD binding are highly (often entirely) conserved in human and mouse Wnts.
Figure 6.
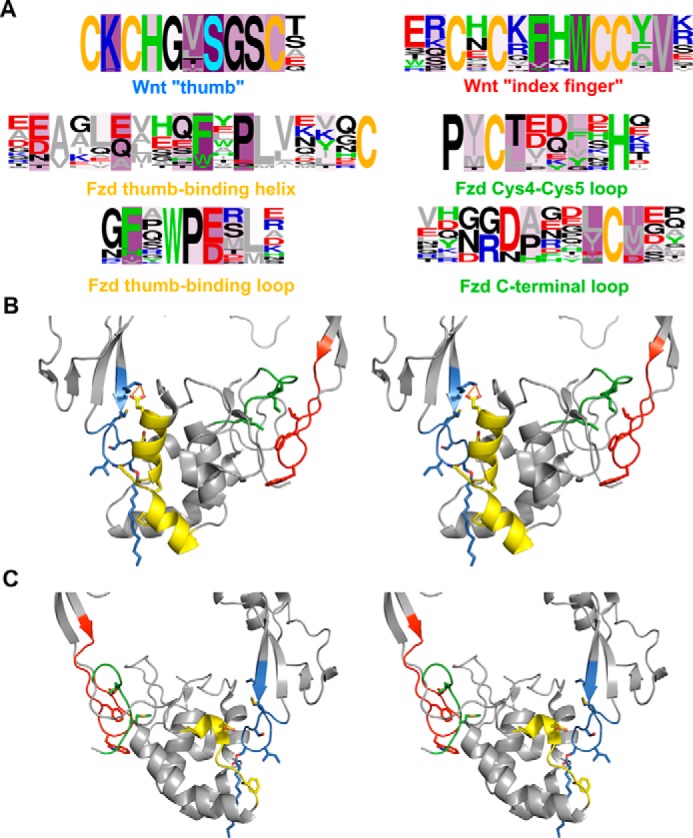
Residues making significant contributions to the binding energy in the majority of Wnt-Fzd CRD complexes. A, logo analyses of Wnts (first row) and Fzd CRDs (second two rows) highlighting the major regions involved in interactions. Fzd sequences interacting with a specific region of Wnt are shown below the sequence of Wnt corresponding to that region. Green, aromatic residues (Trp, Phe, His, and Tyr); gray, aliphatic residues (Val, Leu, Ile, Met, and Ala); blue, basic residues (Arg and Lys); red, acidic residues (Asp and Glu); yellow, cysteine; light blue, palmitoleoylserine (A only); black, all other residues (Gly, Pro, Ser, Thr, Gln, and Asn). Logos are presented as frequency plots. Intensity of purple shading indicates the number of complexes in which the residue at that position is a significant contributor to the complex binding energy. B and C, cross-eyed stereoviews of the “front” (B) and “rear” (C) of the XWnt8-mFzd8 crystal structure complex (Protein Data Bank code 4F0A) with major interacting regions highlighted. Residues corresponding to positions frequently involved in interactions across the full set of Wnt-Fzd CRD complexes are shown as sticks. Regions are colored according to the caption color in panel A. The front view displays the regions of the middle row of panel A; the rear view displays the regions of the bottom row of panel A.
In contrast to the Fzd CRD-binding regions of Wnt, which appear highly conserved and occupy relatively small sections of the Wnt sequence, the Wnt-binding regions of Fzd CRDs are distributed across several segments of the CRDs and often incorporate poorly conserved residues. Four sequences in the Fzd CRDs can be defined (Fig. 6A), two of which interact with the Wnt thumb region and two of which interact with the Wnt index finger region, with several additional residues of importance identified in specific cases (supplemental Fig. S16). Highly conserved Fzd CRD residues frequently implicated in Wnt binding are generally associated with lipid binding: the FXP motif, which frequently occurs within a helix forming one side of the lipid-binding site of the Fzd CRD, and the phenylalanine of an FXW motif in the latter part of the sequence both interact directly with the Wnt lipid (Fig. 6B). Hydrophobic residues adjacent to the final cysteine in the Fzd CRD are frequently implicated in binding the Wnt index finger, as are hydrophobic residues adjacent to the fourth cysteine of the Fzd CRD. However, the involvement of particular Fzd CRD residues in binding is often highly influenced by sequence variation, even for positions frequently implicated in Wnt binding. The greatest deviations in the utilization of Wnt-binding residues with respect to the set of Fzd CRDs occur in Fzd3, Fzd6, SFRP3, and SFRP4. The region corresponding to the FXP motif in SFRP4 occurs as YEE; the tyrosine and glutamate residues in this sequence are never implicated as strong contributors to binding any Wnt. In SFRP3, the phenylalanine of the motif is retained, but the proline is replaced by glycine; the phenylalanine is strongly implicated in binding to all Wnts, whereas this is never the case for glycine. The glutamates of a motif frequently occurring as EAGLE are often implicated in Fzd CRD binding to Wnt. In Fzd3 and Fzd6, the residue corresponding to the first glutamate is never strongly implicated in binding to any Wnt; this is replaced by a threonine in Fzd3 and an isoleucine in Fzd6. Substitution of this residue with aspartate (as occurs in several Fzds and SFRPs) or glutamine (as occurs in SFRP3 and SFRP4) does not appear to greatly influence the frequency with which this residue is involved in Wnt binding. Similarly, replacement of the second glutamate in the motif with alanine, as occurs in Fzd3, SFRP3, and SFRP4, eliminates the importance of this position to Wnt binding, whereas retaining it as a glutamate (as in Fzd6 and other Fzds and SFRPs), aspartate, or even glutamine does not seem to affect the frequency of its importance to binding.
Discussion
In this study, we have developed a model for predicting the binding affinity of Wnt-Fzd interactions. Although the model was developed against a relatively small set of data from a single study, there is nonetheless excellent agreement between affinities predicted in the current study and those experimentally determined in other studies that were not included in model building and testing here. The binding affinity of Wnt3a for the mouse SFRP3 was determined by surface plasmon resonance to be 7.9 nm (37); our model predicts this interaction to be at 0.28 nm, suggesting strong binding affinity. Binding affinities of Wnt3a, Wnt7a, Fzd10, and SFRP4 measured using ELISA (38) confirm our model's prediction that the Fzd5-Wnt3a interaction was stronger than that of Fzd10-Wnt7a and Wnt7a-SFRP4. However, direct comparisons of Kd values predicted by our model and those determined by ELISA are challenging as our model has been optimized against BLI data, where a direct interaction is measured, whereas ELISA is a coupled assay; thus, Kd values obtained from BLI are likely to indicate higher affinity than those obtained from ELISA.
As experimentally determined binding affinities of Wnt-Fzd CRD interactions are largely limited to those included in our training and test sets, it is also pertinent to investigate whether interactions demonstrated experimentally through coIP were predicted by our model to have strong binding affinities. mFzd4-mWnt2b (39), hFzd4-hWnt2 (40), mFzd4-mWnt7b (41), mFzd6-Wnt4 (42), and hWnt3a-hSFRP4 (43), which were shown by coIP to interact, are predicted by our model to bind with an affinity in the intermediate or tighter range (<40 nm). However, the interaction of SFRP1 with Wnt5a, which has been demonstrated by coIP (44), is suggested by our model to bind in the low micromolar range. Although this would be within the range detectable by coIP and is indeed a typical range for other interactions of biological relevance, particularly protein-carbohydrate interactions (45), binding affinities of functionally relevant Wnt-Fzd CRD interactions generally appear to occur in the low-to-mid nanomolar range, as evidenced in the data upon which we have based our prediction model. Therefore, it is likely that the affinity of the SFRP1-Wnt5a interaction is drastically underestimated by the model.
Despite the failure of the model in selected cases to achieve accurate predictions, the model nonetheless performs remarkably well at predicting binding affinities and likely interactors, particularly when considering that the Wnt-Fzd CRD interaction is rather complex due to involvement of both protein-protein and protein-lipid interactions at different sites. This would further suggest its usefulness in predicting the effect of Wnt/Fzd mutations to residues involved in either of the binding sites. The predictive success of the model is likely attributable to two main factors. The first is the use of a test set of cases separated from the training set to validate the model, which is not always performed in developing quantitative structure-activity relationships; even more remarkably, the use of an external data set for model validation appears to be a matter of some debate in the quantitative structure-activity relationship literature (46). The second is the incorporation of a term in the model specifically considering the contribution to binding made by the lipid. The direct involvement of Wnt lipidation in facilitating the Wnt-Fzd interaction is likely unusual among protein-protein interactions; lipidation typically appears to influence the solubility and localization of proteins, rather than directly facilitate protein-protein interactions (47). However, other post-translational modification of proteins, such as glycosylation, phosphorylation, and methylation, are very common and are often involved in facilitating protein folding and mediating protein-protein interactions (48–50). Because post-translational modifications such as these are generally not accommodated in protein-protein docking and scoring, the strategy demonstrated here is one that could be adapted to facilitate their inclusion in protein-protein docking and scoring.
This study has revealed trends with regard to the selectivity and promiscuity of Wnt ligands for Fzd CRDs. The study particularly highlights the promiscuous nature of SFRP4, a Wnt antagonist of interest to our group (51–56). SFRP3 is predicted to display similarly low selectivity for Wnt ligands, whereas SFRP1, SFRP2, and SFRP5 are predicted to display much higher selectivity. The various levels of selectivity are likely to be due to the evolutionary development of tissue expression patterns of Wnt ligands and Fzd receptors, where SFRPs can partially limit aberrant Wnt signaling for controlled tissue development (57).
This study has focused on the interactions of Wnt proteins with the Frizzled-type cysteine-rich domains of the Fzd receptors and the secreted Frizzled-related proteins. However, a variety of other proteins also contain Frizzled-type CRDs, albeit less closely sequence-related to those of the Fzds and SFRPs. These include Smoothened, atrial natriuretic peptide-converting enzyme (CORIN), the tyrosine protein kinase transmembrane receptors ROR1 and ROR2, the skeletal muscle receptor tyrosine protein kinase (MuSK, for which a structure of the Fzd CRD has been experimentally solved (58)), the collagen XVIII α-1 chain, carboxypeptidase Z, and the membrane Frizzled-related protein. With the exception of the RORs (59–62), it is unknown whether any Wnt binds to these proteins and, if so, whether such an interaction is functionally relevant in the context of Wnt signaling. The approaches utilized in the current study could be applied to investigate the binding of Wnts to the Frizzled-type CRDs of these proteins, which in turn could stimulate further research into alternative Wnt signaling pathways.
It is important to note that a high-affinity interaction between a given Wnt and a given CRD does not necessarily translate into a signal transduction event. Wnt signaling involves several additional proteins both extracellularly and intracellularly. For example, in canonical Wnt signaling, Wnt binds to a Fzd CRD as well as the co-receptor LRP5/6 (63); on the intracellular side, this likely causes a conformational change in Fzd, resulting in movement of the Fzd intracellular loop 3 and C-terminal helix, which in turn permits Dvl binding and subsequent signal transduction (64, 65). Thus, the biological relevance of given Wnt-Fzd CRD interactions will be influenced by the co-expression/co-localization of these other proteins. Recent structural data on LRP6 (66–68) and the Smoothened receptor, a Class F GPCR related to Fzd receptors (69–72), as well as the availability of Dvl domain structures (73–76) and knowledge of key residues in the Fzd-Dvl interaction (64, 77, 78) provide the opportunity to investigate more completely the structural basis of canonical Wnt signaling. Additionally, the structures of several intracellular components in non-canonical Wnt signaling pathways are known or adopt structurally characterized folds, suggesting the potential for structural investigations. The models generated in this study provide a solid basis by which to pursue further structural studies of Wnt signaling and, perhaps of greater importance, given the combinatorial nature of potential Wnt-Fzd interactions, suggest specific interacting partners on which to focus experimental and computational efforts.
Experimental procedures
Template preparation
The template structure for all models was the complex of the XWnt8 with the mouse Fzd8 CRD (Protein Data Bank code 4F0A) (19). This structure was initially processed by the Protein Preparation Wizard, with missing side chains and loops filled in by Prime. Although the identity of the lipid modification to XWnt8 in this structure could not be conclusively determined (19), we have presumed this modification to be palmitoleic acid, as indicated either by direct experimental evidence or comparison with similar sequences for which this modification has been demonstrated (18, 79–83). The lipid in the structure was manually modified using Maestro to be a palmitoleic acid modification, which involved the creation of a double bond between carbons 9 and 10 and the addition of carbons 15 and 16 to the lipid, which were missing from the structure. The lipid was subject to a Monte Carlo multiple minimum conformational search using Macromodel, with the region comprising carbons 9–16, as well as the hydrogen atoms attached to these carbons, defined as a freely moving substructure, residues within 6.0 Å of this defined as a frozen shell, and a torsional constraint to ensure cis double bond geometry about carbons 9 and 10. Automatic setup of the substructure was used to define rotatable bonds to be searched; however, all torsion check parameters were removed. Extended torsion sampling was used. A maximum of 10,000 steps was used for the search with a maximum of 2000 steps per rotatable bond. The lowest energy structure obtained from the search provided a template structure for the lipid that was used in all models.
Homology modeling
Sequences of Wnts, Frizzled, and SFRP CRDs from both mouse and human were obtained from the UniProt database (84) (accession numbers are provided in supplemental Table S1). Homology models were prepared using Prime 4.1 (85) (sequence ranges and alignments used are provided in supplemental Figs. S2 and S3). All models were prepared using knowledge-based building; however, because of the presence of large insertions in the human and mouse Wnt6, Wnt10a, and Wnt10b sequences relative to XWnt8, an alternative strategy to building these structures was performed (see below). Disulfide bonds c13–c17 and c16–c24 in Wnt (see Ref. 36 for description of cysteine numbering in Wnts) typically could not be created during the model building process due to being adjacent to insertions/gaps in the sequence alignment; these bonds were manually inserted, and the residues involved were energy-minimized. The lipid structure generated during template preparation was not included during Wnt model building but manually attached following model building.
To build the structures of the human and mouse Wnt6, Wnt10a, and Wnt10b, an initial model of the complete mouse Wnt10a was generated using the I-TASSER server (86). Structures of the remaining Wnts were then built using knowledge-based building in Prime against both the mouse Wnt10a model generated by I-TASSER (to provide the structure of the insertion) and the XWnt8 structure (to provide a template for modeling the remainder of the structure).
Complex generation and refinement
All combinations of Wnt-CRD complexes were generated by merging the structures of each of the models built in the previous step. The generated complexes were subject to refinement using Prime 4.1. The refinement process was facilitated through the use of a KNIME workflow (supplemental Fig. S4). In each complex, non-template residues and residues within 6.0 Å of the binding interface were subject to Prime Minimization and Prime Side-Chain Prediction, followed by a second Prime Minimization. The Wnt lipidation was excluded from the first minimization to allow CRD residues to relax around it but included in the second minimization. For complexes involving mouse and human Wnt6, Wnt10a, and Wnt10b, the large insertions modeled by I-TASSER were also subject to the refinement procedure. The quality of the refined models was assessed using the MolProbity score as calculated by the MolProbity module within PHENIX (87). The quality of the refined models was also assessed by calculating the RMSD of the Cα atoms and the TM-score with respect to the XWnt8-mFzd8 CRD complex. These measurements were both calculated using MM-align (88), with the option to enforce interface alignment by the default cutoff enabled.
Development and validation of the binding affinity prediction model
Complexes of mouse Wnt3a, Wnt4, Wnt5a, and Wnt5b with mouse Fzd1, Fzd2, Fzd4, Fzd5, Fzd7, and Fzd8 were rescored using all of the scoring functions contained in the CCharPPI server (28). As the scoring functions are generally only capable of considering interactions between protein residues, the lipid modification to Wnt was removed prior to rescoring. To consider contributions to the binding affinity made by the lipid, Prime MM-GB/SA calculations on the interaction between the lipid and the CRDs were performed. For these calculations, the protein component of Wnt was removed.
The scores for each complex by each scoring function in CCharPPI as well as the values of the terms provided by the Prime MM-GB/SA calculations were loaded into Maestro. A property containing the dissociation constants determined by BLI for selected mouse Wnt-Fzd CRD pairs (27) was manually created and used to define the activity property. Complexes involving interactions with either Wnt4 or Fzd1 comprised the test set, whereas all other complexes comprised the training set; the training and test sets are summarized under “Results” (Table 3). Both the training and test sets cover a diverse range of Wnts, Fzds, and binding affinities for Wnt-Fzd interactions.
Strike was used to generate affinity prediction models. Multiple linear regression was used to build models. Functions from CCharPPI and properties from Prime MM-GB/SA provided the descriptors used in model building; the full list of functions and properties considered in model building is provided in supplemental Table S5. The success of the models in predicting binding affinities for complexes in both the training and test sets was evaluated using RMSE and the percentage of complexes for which the predicted value occurred within the experimental range (RMSEtrain, RMSEtest, InExptrain, and InExptest).
All possible three-descriptor models incorporating one term from Prime MM-GB/SA and the remaining two terms from CCharPPI were investigated. Models with RMSEtrain less than 0.5 kcal/mol and InExptrain greater than 50% were selected for testing. Models performing at least as well for the test set as for the training set (i.e. RMSEtest ≤ 0.5 kcal/mol and InExptest ≥ 50%) were selected for further elaboration into four-descriptor models, which were generated by adding an additional term from CCharPPI to the best performing three-descriptor models. Four-descriptor models giving RMSEtrain and RMSEtest less than 0.3 kcal/mol and InExptrain and InExptest greater than 75% were selected as high-performing models. Elaboration of the four-descriptor models into five-descriptor models was also pursued by adding another term from CCharPPI.
As a final check of model quality, we also checked whether the approximate range of binding affinity predicted by the best models is in that expected. Dijksterhuis et al. (27) used a simplified scheme wherein Wnt-Fzd binding affinities were classified as strong (<10 nm; ++++), intermediate (10–40 nm; +++), weak (40–100 nm; ++), very weak (>100 nm; +), and non-binding (−). We have utilized this scheme with some modification; we have considered predictions of 100–400 nm to constitute the very weak (+) category and predictions greater than 400 nm to be effectively non-binding (−); the 400 nm limit was chosen in relation to the intermediate/weak affinity range defined.
Analysis of functional residues in Wnt-Fzd interactions
All 570 Wnt-Fzd CRD models were subject to MM-GB/SA analysis using AMBER14 (35). Wnt-Fzd complexes were parameterized using the ff14SB force field (89). Parameter generation for O-palmitoleoylserine was facilitated by Antechamber (90), adapting procedures described in both the AMBER14 reference manual and AMBER tutorials. MMPBSA.py facilitated MM-GB/SA calculations (91). The modified generalized Born model of Onufriev et al. (igb = 5) (92) with a salt concentration of 0.1 m was used to calculate the polar desolvation energy. The non-polar desolvation energy was calculated using surface areas derived from the linear combinations of pairwise overlaps (LCPO) method (93) multiplied by surface tension (the default of 0.0072 kcal/(mol Å2) was used). Energies calculated by MM-GB/SA were decomposed on a per-residue basis with 1–4 terms added to the internal potential terms (idecomp = 1) (94). Residues contributing greater than ±2.0 kcal/mol to the total MM-GB/SA binding energy were selected as being of functional importance to binding. Logo analysis of regions within the Wnt and Fzd sequences frequently found to contain residues of functional importance to Wnt-Fzd binding was performed using the WebLogo server (95). Sequence logos were generated as frequency plots.
Author contributions
M. A. conceived the idea for the work, conducted the experiments, and analyzed the results. M. A. and S. Ö.-G. P. prepared the manuscript. M. A., S. Ö.-G. P., and A. D. critically reviewed and revised the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We gratefully acknowledge Prof. Yvonne Jones (University of Oxford) for critical discussion in the review of the final manuscript.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1 and S5–S14 and Figs. S2–S4, S15, and S16.
- PCP
- planar cell polarity
- CRD
- cysteine-rich domain
- Fzd
- Frizzled
- SFRP
- secreted Frizzled-related protein
- LRP
- low-density lipoprotein-related protein
- Dvl
- Dishevelled
- GSK3
- glycogen synthase kinase 3
- CK1
- casein kinase 1
- Cn
- calcineurin
- ROCK
- Rho-associated protein kinase
- XWnt8
- Xenopus Wnt8
- GPCR
- G protein–coupled receptor
- BLI
- biolayer interferometry
- coIP
- co-immunoprecipitation
- mFzd
- mouse Fzd
- RMSD
- root-mean-squared deviation
- mWnt
- mouse Wnt
- hWnt
- human Wnt
- hFzd
- human Fzd
- RMSE
- root-mean-squared error
- ROR
- receptor tyrosine kinase-like orphan receptor.
References
- 1. Holstein T. W. (2012) The evolution of the Wnt pathway. Cold Spring Harb. Perspect. Biol. 4, a007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hikasa H., and Sokol S. Y. (2013) Wnt signaling in vertebrate axis specification. Cold Spring Harb. Perspect. Biol. 5, a007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polakis P. (2012) Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol. 4, a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Komiya Y., and Habas R. (2008) Wnt signal transduction pathways. Organogenesis 4, 68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pohl S. Ö.-G, Brook N., Agostino M., Arfuso F., Kumar A. P., and Dharmarajan A. (2017) Wnt signaling in triple-negative breast cancer. Oncogenesis 6, e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dijksterhuis J. P., Petersen J., and Schulte G. (2014) WNT/Frizzled signalling: receptor-ligand selectivity with focus on FZD-G protein signalling and its physiological relevance: IUPHAR Review 3. Br. J. Pharmacol. 171, 1195–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He X., Semenov M., Tamai K., and Zeng X. (2004) LDL receptor-related proteins 5 and 6 in Wnt/β-catenin signaling: arrows point the way. Development 131, 1663–1677 [DOI] [PubMed] [Google Scholar]
- 8. Wallingford J. B., and Habas R. (2005) The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development 132, 4421–4436 [DOI] [PubMed] [Google Scholar]
- 9. Clevers H. (2006) Wnt/β-catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 10. Huelsken J., and Behrens J. (2002) The Wnt signalling pathway. J. Cell Sci. 115, 3977–3978 [DOI] [PubMed] [Google Scholar]
- 11. Stamos J. L., and Weis W. I. (2013) The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 5, a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kohn A. D., and Moon R. T. (2005) Wnt and calcium signaling: β-catenin-independent pathways. Cell Calcium 38, 439–446 [DOI] [PubMed] [Google Scholar]
- 13. Kühl M., Sheldahl L. C., Malbon C. C., and Moon R. T. (2000) Ca2+/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 275, 12701–12711 [DOI] [PubMed] [Google Scholar]
- 14. Sheldahl L. C., Park M., Malbon C. C., and Moon R. T. (1999) Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr. Biol. 9, 695–698 [DOI] [PubMed] [Google Scholar]
- 15. De A. (2011) Wnt/Ca2+ signaling pathway: a brief overview. Acta Biochim. Biophys. Sin. 43, 745–756 [DOI] [PubMed] [Google Scholar]
- 16. Green J., Nusse R., and van Amerongen R. (2014) The role of Ryk and Ror receptor tyrosine kinases in Wnt signal transduction. Cold Spring Harb. Perspect. Biol. 6, a009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smolich B. D., McMahon J. A., McMahon A. P., and Papkoff J. (1993) Wnt family proteins are secreted and associated with the cell surface. Mol. Biol. Cell 4, 1267–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao X., and Hannoush R. N. (2014) Single-cell imaging of Wnt palmitoylation by the acyltransferase porcupine. Nat. Chem. Biol. 10, 61–68 [DOI] [PubMed] [Google Scholar]
- 19. Janda C. Y., Waghray D., Levin A. M., Thomas C., and Garcia K. C. (2012) Structural basis of Wnt recognition by Frizzled. Science 337, 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y., Macke J. P., Abella B. S., Andreasson K., Worley P., Gilbert D. J., Copeland N. G., Jenkins N. A., and Nathans J. (1996) A large family of putative transmembrane receptors homologous to the product of the Drosophila tissue polarity gene frizzled. J. Biol. Chem. 271, 4468–4476 [DOI] [PubMed] [Google Scholar]
- 21. Xu Q., Wang Y., Dabdoub A., Smallwood P. M., Williams J., Woods C., Kelley M. W., Jiang L., Tasman W., Zhang K., and Nathans J. (2004) Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell 116, 883–895 [DOI] [PubMed] [Google Scholar]
- 22. Ye X., Wang Y., Cahill H., Yu M., Badea T. C., Smallwood P. M., Peachey N. S., and Nathans J. (2009) Norrin, frizzled-4, and Lrp5 signaling in endothelial cells controls a genetic program for retinal vascularization. Cell 139, 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Surana R., Sikka S., Cai W., Shin E. M., Warrier S. R., Tan H. J., Arfuso F., Fox S. A., Dharmarajan A. M., and Kumar A. P. (2014) Secreted frizzled related proteins: implications in cancers. Biochim. Biophys. Acta 1845, 53–65 [DOI] [PubMed] [Google Scholar]
- 24. Cruciat C. M., and Niehrs C. (2013) Secreted and transmembrane Wnt inhibitors and activators. Cold Spring Harb. Perspect. Biol. 5, a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dann C. E., Hsieh J. C., Rattner A., Sharma D., Nathans J., and Leahy D. J. (2001) Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 412, 86–90 [DOI] [PubMed] [Google Scholar]
- 26. Lopez-Rios J., Esteve P., Ruiz J. M., and Bovolenta P. (2008) The Netrin-related domain of Sfrp1 interacts with Wnt ligands and antagonizes their activity in the anterior neural plate. Neural Dev. 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dijksterhuis J. P., Baljinnyam B., Stanger K., Sercan H. O., Ji Y., Andres O., Rubin J. S., Hannoush R. N., and Schulte G. (2015) Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J. Biol. Chem. 290, 6789–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moal I. H., Jiménez-García B., and Fernández-Recio J. (2015) CCharPPI web server: computational characterisation of protein-protein interactions from structure. Bioinformatics 31, 123–125 [DOI] [PubMed] [Google Scholar]
- 29. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y., and Skolnick J. (2004) Scoring function for automated assessment of protein structure template quality. Proteins 57, 702–710 [DOI] [PubMed] [Google Scholar]
- 32. Chaudhury S., Lyskov S., and Gray J. J. (2010) PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J., and Zhang Y. (2010) A novel side-chain orientation dependent potential derived from random-walk reference state for protein fold selection and structure prediction. PLoS One 5, e15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andrusier N., Nussinov R., and Wolfson H. J. (2007) FireDock: fast interaction refinement in molecular docking. Proteins 69, 139–159 [DOI] [PubMed] [Google Scholar]
- 35. Salomon-Ferrer R., Case D. A., and Walker R. C. (2013) An overview of the Amber biomolecular simulation package. WIREs Comput. Mol. Sci. 3, 198–210 [Google Scholar]
- 36. MacDonald B. T., Hien A., Zhang X., Iranloye O., Virshup D. M., Waterman M. L., and He X. (2014) Disulfide bond requirements for active Wnt ligands. J. Biol. Chem. 289, 18122–18136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wawrzak D., Métioui M., Willems E., Hendrickx M., de Genst E., and Leyns L. (2007) Wnt3a binds to several sFRPs in the nanomolar range. Biochem. Biophys. Res. Commun. 357, 1119–1123 [DOI] [PubMed] [Google Scholar]
- 38. Carmon K. S., and Loose D. S. (2010) Development of a bioassay for detection of Wnt-binding affinities for individual frizzled receptors. Anal. Biochem. 401, 288–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ohta K., Ito A., Kuriyama S., Lupo G., Kosaka M., Ohnuma S., Nakagawa S., and Tanaka H. (2011) Tsukushi functions as a Wnt signaling inhibitor by competing with Wnt2b for binding to transmembrane protein Frizzled4. Proc. Natl. Acad. Sci. U.S.A. 108, 14962–14967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klein D., Demory A., Peyre F., Kroll J., Augustin H. G., Helfrich W., Kzhyshkowska J., Schledzewski K., Arnold B., and Goerdt S. (2008) Wnt2 acts as a cell type-specific, autocrine growth factor in rat hepatic sinusoidal endothelial cells cross-stimulating the VEGF pathway. Hepatology 47, 1018–1031 [DOI] [PubMed] [Google Scholar]
- 41. Wang Z., Shu W., Lu M. M., and Morrisey E. E. (2005) Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol. Cell. Biol. 25, 5022–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lyons J. P., Mueller U. W., Ji H., Everett C., Fang X., Hsieh J. C., Barth A. M., and McCrea P. D. (2004) Wnt-4 activates the canonical β-catenin-mediated Wnt pathway and binds Frizzled-6 CRD: functional implications of Wnt/β-catenin activity in kidney epithelial cells. Exp. Cell Res. 298, 369–387 [DOI] [PubMed] [Google Scholar]
- 43. Constantinou T., Baumann F., Lacher M. D., Saurer S., Friis R., and Dharmarajan A. (2008) SFRP-4 abrogates Wnt-3a-induced β-catenin and Akt/PKB signalling and reverses a Wnt-3a-imposed inhibition of in vitro mammary differentiation. J. Mol. Signal. 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsuyama M., Aizawa S., and Shimono A. (2009) Sfrp controls apicobasal polarity and oriented cell division in developing gut epithelium. PLoS Genet. 5, e1000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agostino M., Velkov T., Dingjan T., Williams S. J., Yuriev E., and Ramsland P. A. (2015) The carbohydrate-binding promiscuity of Euonymus europaeus lectin is predicted to involve a single binding site. Glycobiology 25, 101–114 [DOI] [PubMed] [Google Scholar]
- 46. Veerasamy R., Rajak H., Jain A., Sivadasan S., Varghese C. P., and Agrawal R. K. (2011) Validation of QSAR models—strategies and importance. Int. J. Drug Des. Discov. 2, 511–519 [Google Scholar]
- 47. Resh M. D. (2016) Fatty acylation of proteins: the long and the short of it. Prog. Lipid Res. 63, 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu C., and Ng D. T. (2015) Glycosylation-directed quality control of protein folding. Nat. Rev. Mol. Cell Biol. 16, 742–752 [DOI] [PubMed] [Google Scholar]
- 49. Nishi H., Hashimoto K., and Panchenko A. R. (2011) Phosphorylation in protein-protein binding: effect on stability and function. Structure 19, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perumal V., Krishnan K., Gratton E., Dharmarajan A. M., and Fox S. A. (2015) Number and brightness analysis of sFRP4 domains in live cells demonstrates vesicle association signal of the NLD domain and dynamic intracellular responses to Wnt3a. Int. J. Biochem. Cell Biol. 64, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Perumal V., Pohl S., Keane K. N., Arfuso F., Newsholme P., Fox S., and Dharmarajan A. (2016) Therapeutic approach to target mesothelioma cancer cells using the Wnt antagonist, secreted frizzled-related protein 4: metabolic state of cancer cells. Exp. Cell Res. 341, 218–224 [DOI] [PubMed] [Google Scholar]
- 53. Pohl S., Scott R., Arfuso F., Perumal V., and Dharmarajan A. (2015) Secreted frizzled-related protein 4 and its implications in cancer and apoptosis. Tumour Biol. 36, 143–152 [DOI] [PubMed] [Google Scholar]
- 54. Warrier S., Balu S. K., Kumar A. P., Millward M., and Dharmarajan A. (2013) Wnt antagonist, secreted frizzled-related protein 4 (sFRP4), increases chemotherapeutic response of glioma stem-like cells. Oncol. Res. 21, 93–102 [DOI] [PubMed] [Google Scholar]
- 55. Wolf V., Ke G., Dharmarajan A. M., Bielke W., Artuso L., Saurer S., and Friis R. (1997) DDC-4, an apoptosis-associated gene, is a secreted frizzled relative. FEBS Lett. 417, 385–389 [DOI] [PubMed] [Google Scholar]
- 56. Muley A., Majumder S., Kolluru G. K., Parkinson S., Viola H., Hool L., Arfuso F., Ganss R., Dharmarajan A., and Chatterjee S. (2010) Secreted frizzled-related protein 4: an angiogenesis inhibitor. Am. J. Pathol. 176, 1505–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bovolenta P., Esteve P., Ruiz J. M., Cisneros E., and Lopez-Rios J. (2008) Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J. Cell Sci. 121, 737–746 [DOI] [PubMed] [Google Scholar]
- 58. Stiegler A. L., Burden S. J., and Hubbard S. R. (2009) Crystal structure of the frizzled-like cysteine-rich domain of the receptor tyrosine kinase MuSK. J. Mol. Biol. 393, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cui B., Zhang S., Chen L., Yu J., Widhopf G. F. 2nd, Fecteau J.-F., Rassenti L. Z., and Kipps T. J. (2013) Targeting ROR1 inhibits epithelial-mesenchymal transition and metastasis. Cancer Res. 73, 3649–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Henry C., Quadir A., Hawkins N. J., Jary E., Llamosas E., Kumar D., Daniels B., Ward R. L., and Ford C. E. (2015) Expression of the novel Wnt receptor ROR2 is increased in breast cancer and may regulate both β-catenin dependent and independent Wnt signalling. J. Cancer Res. Clin. Oncol. 141, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu Y., Rubin B., Bodine P. V., and Billiard J. (2008) Wnt5a induces homodimerization and activation of Ror2 receptor tyrosine kinase. J. Cell. Biochem. 105, 497–502 [DOI] [PubMed] [Google Scholar]
- 62. Zhang S., Chen L., Cui B., Chuang H. Y., Yu J., Wang-Rodriguez J., Tang L., Chen G., Basak G. W., and Kipps T. J. (2012) ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS One 7, e31127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. MacDonald B. T., and He X. (2012) Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb. Perspect. Biol. 4, a007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tauriello D. V., Jordens I., Kirchner K., Slootstra J. W., Kruitwagen T., Bouwman B. A., Noutsou M., Rüdiger S. G., Schwamborn K., Schambony A., and Maurice M. M. (2012) Wnt/β-catenin signaling requires interaction of the Dishevelled DEP domain and C terminus with a discontinuous motif in Frizzled. Proc. Natl. Acad. Sci. U.S.A. 109, E812–E820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cong F., Schweizer L., and Varmus H. (2004) Wnt signals across the plasma membrane to activate the β-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP. Development 131, 5103–5115 [DOI] [PubMed] [Google Scholar]
- 66. Chang T. H., Hsieh F. L., Zebisch M., Harlos K., Elegheert J., and Jones E. Y. (2015) Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan. eLife 4, e06554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cheng Z., Biechele T., Wei Z., Morrone S., Moon R. T., Wang L., and Xu W. (2011) Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat. Struct. Mol. Biol. 18, 1204–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ahn V. E., Chu M. L., Choi H. J., Tran D., Abo A., and Weis W. I. (2011) Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev. Cell 21, 862–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang C., Wu H., Evron T., Vardy E., Han G. W., Huang X. P., Hufeisen S. J., Mangano T. J., Urban D. J., Katritch V., Cherezov V., Caron M. G., Roth B. L., and Stevens R. C. (2014) Structural basis for smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun. 5, 4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang C., Wu H., Katritch V., Han G. W., Huang X. P., Liu W., Siu F. Y., Roth B. L., Cherezov V., and Stevens R. C. (2013) Structure of the human smoothened receptor bound to an antitumour agent. Nature 497, 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Weierstall U., James D., Wang C., White T. A., Wang D., Liu W., Spence J. C., Bruce Doak R., Nelson G., Fromme P., Fromme R., Grotjohann I., Kupitz C., Zatsepin N. A., Liu H., et al. (2014) Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 5, 3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Byrne E. F., Sircar R., Miller P. S., Hedger G., Luchetti G., Nachtergaele S., Tully M. D., Mydock-McGrane L., Covey D. F., Rambo R. P., Sansom M. S., Newstead S., Rohatgi R., and Siebold C. (2016) Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Madrzak J., Fiedler M., Johnson C. M., Ewan R., Knebel A., Bienz M., and Chin J. W. (2015) Ubiquitination of the Dishevelled DIX domain blocks its head-to-tail polymerization. Nat. Commun. 6, 6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wong H. C., Mao J., Nguyen J. T., Srinivas S., Zhang W., Liu B., Li L., Wu D., and Zheng J. (2000) Structural basis of the recognition of the dishevelled DEP domain in the Wnt signaling pathway. Nat. Struct. Biol. 7, 1178–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang Y., Appleton B. A., Wiesmann C., Lau T., Costa M., Hannoush R. N., and Sidhu S. S. (2009) Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat. Chem. Biol. 5, 217–219 [DOI] [PubMed] [Google Scholar]
- 76. Yu A., Xing Y., Harrison S. C., and Kirchhausen T. (2010) Structural analysis of the interaction between Dishevelled2 and clathrin AP-2 adaptor, a critical step in noncanonical Wnt signaling. Structure 18, 1311–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wong H. C., Bourdelas A., Krauss A., Lee H. J., Shao Y., Wu D., Mlodzik M., Shi D. L., and Zheng J. (2003) Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol. Cell 12, 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bertalovitz A. C., Pau M. S., Gao S., Malbon C. C., and Wang H.-Y. (2016) Frizzled-4 C-terminus distal to KTXXXW motif is essential for normal dishevelled recruitment and Norrin-stimulated activation of Lef/Tcf-dependent transcriptional activation. J. Mol. Signal. 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Coombs G. S., Yu J., Canning C. A., Veltri C. A., Covey T. M., Cheong J. K., Utomo V., Banerjee N., Zhang Z. H., Jadulco R. C., Concepcion G. P., Bugni T. S., Harper M. K., Mihalek I., Jones C. M., et al. (2010) WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J. Cell Sci. 123, 3357–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Takada R., Satomi Y., Kurata T., Ueno N., Norioka S., Kondoh H., Takao T., and Takada S. (2006) Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev. Cell 11, 791–801 [DOI] [PubMed] [Google Scholar]
- 81. Galli L. M., and Burrus L. W. (2011) Differential palmit(e)oylation of Wnt1 on C93 and S224 residues has overlapping and distinct consequences. PLoS One 6, e26636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Doubravska L., Krausova M., Gradl D., Vojtechova M., Tumova L., Lukas J., Valenta T., Pospichalova V., Fafilek B., Plachy J., Sebesta O., and Korinek V. (2011) Fatty acid modification of Wnt1 and Wnt3a at serine is prerequisite for lipidation at cysteine and is essential for Wnt signalling. Cell. Signal. 23, 837–848 [DOI] [PubMed] [Google Scholar]
- 83. Kakugawa S., Langton P. F., Zebisch M., Howell S. A., Chang T. H., Liu Y., Feizi T., Bineva G., O'Reilly N., Snijders A. P., Jones E. Y., and Vincent J. P. (2015) Notum deacylates Wnt proteins to suppress signalling activity. Nature 519, 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jacobson M. P., Friesner R. A., Xiang Z., and Honig B. (2002) On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 320, 597–608 [DOI] [PubMed] [Google Scholar]
- 86. Roy A., Kucukural A., and Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mukherjee S., and Zhang Y. (2009) MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming. Nucleic Acids Res. 37, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., and Simmerling C. (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang J., Wang W., Kollman P. A., and Case D. A. (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 [DOI] [PubMed] [Google Scholar]
- 91. Miller B. R. 3rd, McGee T. D. Jr., Swails J. M., Homeyer N., Gohlke H., and Roitberg A. E. (2012) MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 8, 3314–3321 [DOI] [PubMed] [Google Scholar]
- 92. Onufriev A., Bashford D., and Case D. A. (2004) Exploring protein native states and large-scale conformational changes with a modified generalized Born model. Proteins 55, 383–394 [DOI] [PubMed] [Google Scholar]
- 93. Weiser J., Shenkin P. S., and Still W. C. (1999) Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J. Comput. Chem. 20, 217–230 [Google Scholar]
- 94. Gohlke H., Kiel C., and Case D. A. (2003) Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 330, 891–913 [DOI] [PubMed] [Google Scholar]
- 95. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.