Abstract
The F1F0 -ATP (F-ATP) synthase is essential for growth of Mycobacterium tuberculosis, the causative agent of tuberculosis (TB). In addition to their synthase function most F-ATP synthases possess an ATP-hydrolase activity, which is coupled to proton-pumping activity. However, the mycobacterial enzyme lacks this reverse activity, but the reason for this deficiency is unclear. Here, we report that a Mycobacterium-specific, 36-amino acid long C-terminal domain in the nucleotide-binding subunit α (Mtα) of F-ATP synthase suppresses its ATPase activity and determined the mechanism of suppression. First, we employed vesicles to show that in intact membrane-embedded mycobacterial F-ATP synthases deletion of the C-terminal domain enabled ATPase and proton-pumping activity. We then generated a heterologous F-ATP synthase model system, which demonstrated that transfer of the mycobacterial C-terminal domain to a standard F-ATP synthase α subunit suppresses ATPase activity. Single-molecule rotation assays indicated that the introduction of this Mycobacterium-specific domain decreased the angular velocity of the power-stroke after ATP binding. Solution X-ray scattering data and NMR results revealed the solution shape of Mtα and the 3D structure of the subunit α C-terminal peptide 521PDEHVEALDEDKLAKEAVKV540 of M. tubercolosis (Mtα(521–540)), respectively. Together with cross-linking studies, the solution structural data lead to a model, in which Mtα(521–540) comes in close proximity with subunit γ residues 104–109, whose interaction may influence the rotation of the camshaft-like subunit γ. Finally, we propose that the unique segment Mtα(514–549), which is accessible at the C terminus of mycobacterial subunit α, is a promising drug epitope.
Keywords: ATP synthase, bioenergetics, F1FO-ATPase, membrane protein, tuberculosis, F-ATP synthase, Mycobacterium, subunit α
Introduction
The F1F0-ATP synthase has been shown to be essential for growth in Mycobacterium smegmatis and Mycobacterium tuberculosis (Mt), with the latter causing tuberculosis (TB)5 (1–3). This is different in other prokaryotes, where the enzyme is dispensable for growth on fermentable carbon sources and where increased glycolytic flux can compensate for the loss of oxidative phosphorylation (4). The difference was attributed to the extraordinarily high amount of ATP required to synthesize a mycobacterial cell (5). A special feature of the mycobacterial F-ATP synthase is its inability to establish a significant proton gradient during ATP hydrolysis, and its low or latent ATPase activity in the fast- or slow-growing form (6). Latency of ATP hydrolysis activity has also been observed for the thermoalkaliphile Bacillus sp. TA2.A1 (7–9) and the α-proteobacterium Paracoccus denitrificans F-ATP synthase (7, 10–13), which results from distortion of the asymmetrical γ-rotor within the α3β3-cylinder (13) and the ζ-inhibitor protein, respectively (14).
The mycobacterial F1F0-ATP synthase is composed of nine subunits with a stoichiometry of α3:β3:γ:δ:ϵ:a:b:b′:c9, and organized as a membrane-embedded F0 complex (abb′c9), and a water-soluble F1 complex (α3β3γδϵ). The F1 part contains three catalytic αβ-pairs that form an α3β3-hexamer, in which ATP synthesis or -hydrolysis takes place. This catalytic α3β3-headpiece is linked with the ion-pumping F0 part via the two rotating central stalk subunits γ and ϵ, as well as the peripheral stalk subunits b, b′, and δ (3). The F0 domain contains subunit a, and a ring structure consisting of 9 c subunits (15). The latter are proposed to form a helix-loop-helix structure, where the loops dock to the membrane facing, globular domain of the rotary γ subunit and enable the coupling to the F1 portion to transfer torque, derived by ion transport, to the catalytic α3β3-headpiece (16). Thus, the rotational movement of the c-ring forces the central subunits γ and ϵ to rotate (17), causing sequential conformational changes in the nucleotide-binding subunits α and β that result in the condensation of ADP + Pi to form ATP (18). In the case of ATP hydrolysis, the cleavage of ATP to ADP + Pi in the interfaces of three αβ-pairs leads to conformational changes in their C-terminal domains and subsequently to the stepwise counterclockwise 120° rotation of the central γ subunit (when viewed from the membrane side) (19). In prokaryotic F-ATP synthases described to date (20–22), each 120° rotation is divided into 40° and 80° substeps, where ATP binding to an empty αβ-pair (αβ)E induces the 80° substep, followed by ATP hydrolysis and ADP release in (αβ)TP, and inorganic phosphate (Pi) release from (αβ)DP that induces the 40° substep.
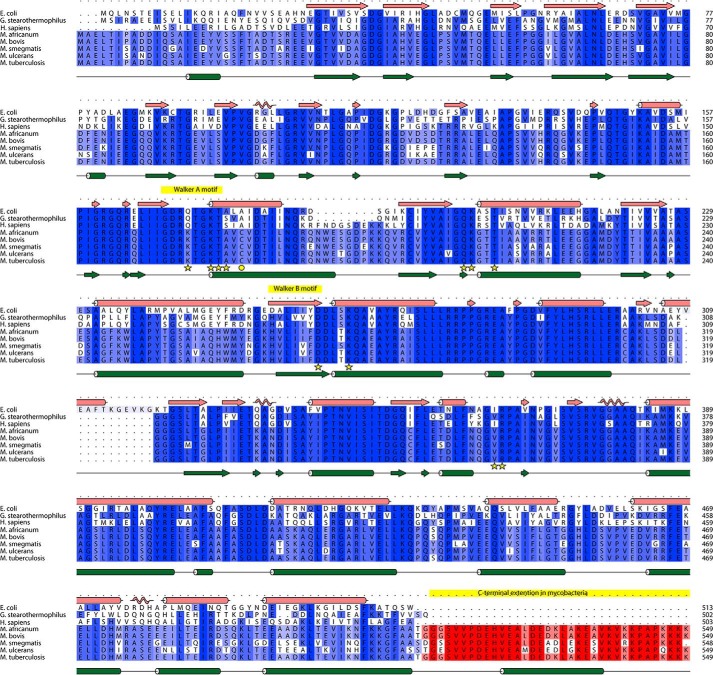
Amino acid sequence alignments of subunit α (Fig. 1) reveal that the mycobacterial F-ATP synthase subunit α has a unique extension of 36 amino acid residues at the very C terminus of subunit α, Mtα(514–549) (according to M. tuberculosis numbering), which is not present in any other known prokaryotic or eukaryotic α subunit. Questions arise whether this extension contributes to important enzymatic differences of mycobacterial F-ATP synthases such as: (i) the latency of ATP hydrolysis activity and suppression of proton pumping. Thus, activation of the latent ATP hydrolysis driven H+-pumping could reduce ATP reserves to alter the proton motive force (PMF) and to decrease the viability of the bacteria. (ii) Whether the C-terminal extension is related to possible mechanistic processes of the rotary F-ATP synthase motor, and (iii) whether this 36-amino acid extension can be employed as a potential drug target. Using a combination of recombineering, ensemble, and single molecule assays, solution X-ray scattering (SAXS), and NMR spectroscopy we demonstrated for the first time that this unique subunit α C-terminal stretch is at least one of the regulatory elements inside mycobacterial F-ATP synthases that affects ATP hydrolysis and synthesis. Removal of this C-terminal extension was found to increase the rate of ATP cleavage and therefore provides energy to enable ATP-driven H+-translocation. The ATP hydrolysis suppressing function of the mycobacterial C-terminal extension was confirmed by replacing the mycobacterial subunit α with the subunit α (502 residues) of Geobacillus stearothermophilus (formerly Bacillus PS3 or TF1), which does not contain a C-terminal extension. Furthermore, by generating an α3chiβ3γ complex, containing the chimeric α consisting of subunit α of the G. stearothermophilus F-ATP synthase (Gsα(1–502)) and Mtα(514–549), we demonstrated, that the extension alters ATPase activity also in the chimeric complex. Single molecule rotation experiments revealed that the C-terminal stretch affects ATPase activity by reducing the angular velocity of the rotating α3chiβ3γ complex after binding of ATP. In addition, solution structural and cross-linking studies lead to a model, in which the C-terminal extension of Mtα comes in close proximity with subunit γ, whose interaction may influence its rotation.
Figure 1.
Sequence alignment of F-ATP synthase subunit α from various Mycobacterium species with the ones of E. coli, G. stearothermophilus, and human. The sequences were aligned with Clustal Omega and visualized using ALINE (61).
Results
The C terminus affects enzymatic activities
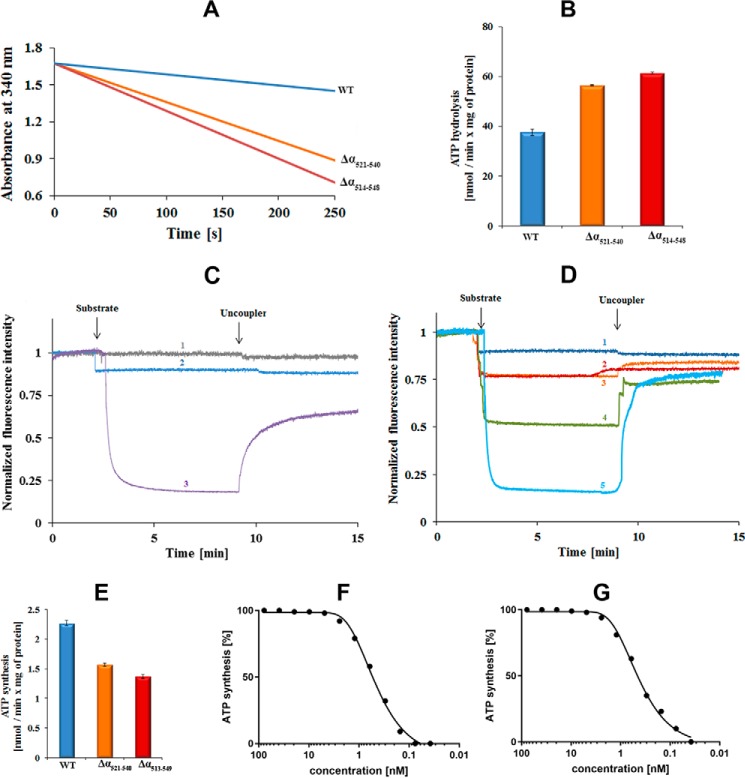
To gain insight into the enzymatic effect of the specific C-terminal domain of the mycobacterial F-ATP synthase subunit α as well as the C-terminal helix (residues 521–540) inside this extension, the M. smegmatis F-ATP synthase mutants Δα514–548 and Δα521–540 with deletion of amino acids 514–548 or 521–540 of the respective C terminus of subunit α were engineered. The effect of the C-terminal deletions of the Δα514–548 mutant on ATP hydrolysis was investigated using inverted membrane vesicles (IMVs). As demonstrated in Fig. 2, A and B, IMVs of wild type (wt) M. smegmatis revealed an ATPase activity of about 37.5 ± 1.3 nmol min−1 (mg of total protein)−1, confirming recent results that demonstrated that M. smegmatis, although at a relatively low level, hydrolyzes ATP (6, 23). In contrast, when IMVs containing the F-ATP synthase mutant Δα514–548 were used, an ATPase activity of 61.4 ± 0.4 nmol min−1 (mg of total protein)−1 was observed. Thus, deletion of the C-terminal domain caused a 64% increase in ATPase activity, which suggests that the C-terminal domain in the wt enzyme suppresses ATPase activity. To confirm that the additional C terminus causes suppression of ATPase hydrolysis, subunit α of the G. stearothermophilus F-ATP synthase, which does not include the C-terminal extension, was engineered into M. smegmatis F-ATP synthase. IMVs of the Gsα replacement mutant showed also an increase in ATP hydrolysis (80.6 ± 0.2 nmol min−1 (mg of total protein)−1) (data not shown) in comparison to the wt, supporting that the unique C-terminal extension of mycobacterial subunit α suppresses ATPase activity. To determine whether the predicted helix inside the C-terminal extension of α (Fig. 2, A and B) may be critical for this suppression, the ATPase activity of IMVs of the Δα521–540 mutant was measured and found to be 56.0 ± 0.3 nmol min−1 (mg of total protein)−1 (49% increase compared with wt activity), suggesting that the C-terminal helix contributes to this suppression.
Figure 2.
Catalytic activities of M. smegmatis F-ATP synthase wt and mutant proteins. A, continuous ATPase activity of wt (blue) M. smegmatis F-ATP synthase, Δα(514–548) (red), and Δα(521–540) mutants (orange), respectively, using IMVs measured in the presence of type II NADH dehydrogenase inhibitor thioridazine (80 μm) and 2 mm MgATP. B, specific ATPase activity of wt, Δα(514–548), and Δα(521–540) mutant IMVs. Values are the mean of six determinations with two different IMV batches of wt and mutants. C and D, substrate driven proton-pumping in IMVs. C, M. smegmatis mc2 155 wt membrane vesicles were diluted to 0.18 mg/ml. Fluorescence quenching of ACMA by wt IMVs was studied after the addition of a substrate (2 mm ATP (blue, profile 2)) or 2 mm NADH (purple, profile 3). The uncoupler (SF6847) was added at the indicated time point to collapse the proton gradient. In the control experiment, buffer was added in place for substrate (gray, profile 1). D, fluorescence quenching of ACMA by IMVs of the Δα(514–548) (red, profile 2) and Δα(521–540) mutant (orange, profile 3) after addition of ATP in comparison to the wt IMVs (blue, profile 1) and the recently described Δγ(166–179) mutant (green, profile 4) (23). Profile 5 (light blue) reveals the quenching of IMVs of the Δ(α514–548) mutant in the presence of 2 mm NADH. Fluorescence quenching of ACMA with wt- and α-mutant IMVs was performed with four and two different batches of vesicles, respectively. E, ATP synthesis measured for wt (blue), Δα(514–548) (red) and Δα(521–540) mutant (orange) IMVs of M. smegmatis. Effect of increasing concentrations of bedaquiline on ATP synthesis using the M. smegmatis Δα(514–548) mutant (F) and wt IMVs (G).
To determine whether ATP hydrolysis is coupled with proton-pumping, IMVs containing M. smegmatis wt F-ATP synthase were tested for proton pumping activity in the presence of the fluorescent dye 9-amino-6-chloro-2-methoxyacridine (ACMA). As shown in Fig. 2C, adding the substrate ATP to the assay containing IMVs resulted in no further drop of fluorescence other than the typical slight ATP-induced signal quenching. This lack of ATP-dependent fluorescence quenching was not the result of leaky IMVs, because (i) addition of NADH caused a significant and fast quenching of ACMA due to the proton-pumping of the respiratory chain complexes (Fig. 2C), and (ii) upon addition of the uncoupler SF6847 the observed fluorescence was increased. These data correlate with recent results (23, 24) that showed that ATP hydrolysis catalyzed by the M. smegmatis F-ATP synthase is not coupled to proton-pumping. Addition of ATP to IMVs containing either the M. smegmatis Δα(514–548) or Δα(521–540) mutant protein quenched ACMA fluorescence by about 10% (Fig. 2D) in a manner that was sensitive to the uncoupler SF6847. These data reveal that the increases in ATPase activity that resulted from the Δα(514–548) and Δα(521–540) deletions were sufficient to create a small proton gradient across the membrane. Because the rate of H+-conduction of both mutant proteins is similar, one can conclude that deletion of the C-terminal helix mainly causes the observed effect.
In parallel, ATP synthesis activities of the wt, the Δα(514–548) and Δα(521–540) mutants in IMVs were measured. As shown in Fig. 2E, the IMVs of wt revealed an ATP synthesis activity of 2.27 ± 0.05 nmol min−1 (mg of total protein)−1, which is similar to the value (0.96 ± 0.15 nmol min−1 (mg of total protein)−1) determined recently (6). In comparison, the Δα(514–548) and Δα(521–540) mutants were active in ATP synthesis with values of 1.55 ± 0.03 nmol min−1 (mg of total protein)−1 and 1.37 ± 0.04 nmol min−1 (mg of total protein)−1, respectively, demonstrating a decrease of about 40% in ATP synthesis activity for both mutant enzymes when compared with the wt.
In addition, the inhibitory effect of the TB drug bedaquiline (TMC207) (24) on ATP synthesis of wt and Δα(514–548) mutant IMVs was similar for both enzymes (Fig. 2F and G), resulting in comparable IC50 values of about 0.8 nmol. This is in line with those reported recently for M. smegmatis (2.5–12.9 nm) (25) and Mycobacterium phlei (20–25 nm) (15) F-ATP synthase. These data as well as Western blot analyses (data not shown) confirm that the amount of F-ATP synthases located in the Δα(514–548) mutant vesicles is comparable with that of the wt vesicles.
ATPase activity decrease in the novel α3chiβ3γ complex
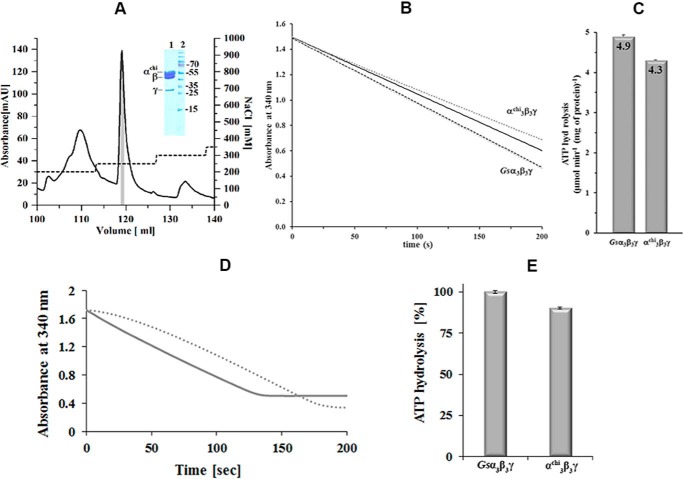
The data above raise the question, whether the apparent ATPase activity suppressing function of the Mycobacterium-specific α domain can be transplanted in F-ATP synthases of other organisms. We used the ATPase proficient and mechanistically well understood α3β3γ complex of the G. stearothermophilus F-ATP synthase (Gsα3β3γ) as a prototype with an engineered cysteine at position γ109 essential for single molecule rotation experiments (see below). An α3chiβ3γ complex with a chimeric subunit α (Gsα(1–502)), and the very C-terminal α(514–549)-segment of the M. tuberculosis F-ATP synthase (Mtα(514–549)), was compared with the Gsα3β3γ complex. Highly purified complexes were produced (Fig. 3A) to deduce the effect of subunit αchi on the ATP hydrolysis reaction (Fig. 3, B and C). At 37 °C, the Gsα3β3γ complex showed an ATPase activity of 4.9 ± 0.04 μmol min−1 (mg of protein)−1, whereas α3chiβ3γ revealed a 12% decreased ATPase activity (4.3 ± 0.01 μmol min−1 (mg of protein)−1), when the rates of the first 10 s of the reactions were compared, i.e. the time frames that were not affected by ADP inhibition of the enzyme. The effect of ADP inhibition is shown in Fig. 3D. In an additional experiment we removed all nucleotides from the enzymes prior to ATP hydrolysis measurements. A similar reduction of ATP hydrolysis (10%) was observed for the nucleotide-depleted α3chiβ3γ compared with -Gsα3β3γ (Fig. 3E). These results show that the Mycobacterium-specific C-terminal extension is indeed able to suppress ATPase activity when transplanted onto the C terminus of an ATPase proficient prokaryotic enzyme. This concurs with the complementary finding above, showing the increase of ATPase activity of IMVs from the M. smegmatis deletion mutant Δα514–548.
Figure 3.
Purification of α3chiγ and ATP hydrolytic activity of Gsα3β3γ and α3chiγ. A, the chromatogram shows an elution profile of α3chiβ3γ using a Resource-Q column (6 ml). The inset in the figure reveals a SDS gel, which corresponds to the shaded area (gray) of the elution peak. The eluted α3chiβ3γ fractions were pooled and applied on the gel. Lanes 1 and 2 reveal the purified α3chiβ3γ and protein markers, respectively. The protein size in kDa is indicated on the right. B, continuous ATPase activity of Gsα3β3γ and α3chiβ3γ measured at 2 mm MgATP, 37 °C. Decrease in NADH absorption at 340 nm is plotted against time as dotted lines. The black lines show the linear least square fit for the first 10 s. C, specific ATPase activities of Gsα3β3γ and α3chiβ3γ determined from the slope of the least square fit. Values are the mean of 10 determinations. D, ATPase profile of nucleotide-depleted Gsα3β3γ (—) in a continuous ATPase activity assay measured at 2 mm MgATP. Bound MgADP was removed prior to activity measurements by incubating the protein with 5 mm Na-P and 10 mm EDTA for 30 min at 4 °C, and passing the enzyme samples through a Superdex 300TM column (GE Healthcare). (····) ATPase profile of Gsα3β3γ, which was preincubated with 100 μm MgADP for 30 min, revealing the MgADP inhibition effect, and confirming that tightly bound MgADP was removed due to the nucleotide depletion process of nucleotide-depleted Gsα3β3γ (—) described above. E, specific ATPase activities of the nucleotide-depleted Gsα3β3γ and α3chiβ3γ. Values are the mean of three determinations.
Influence of the C terminus on ATP-dependent rotation
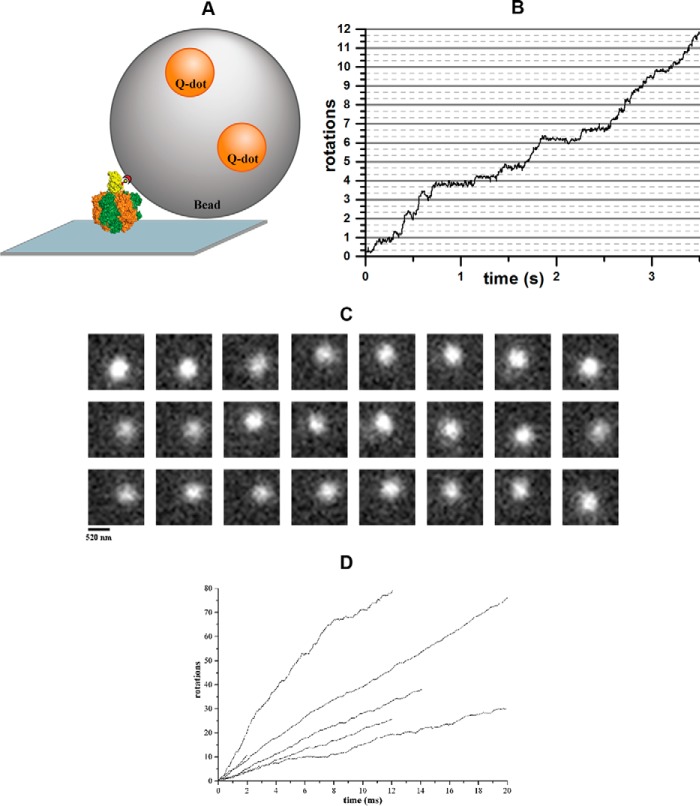
To further investigate the enzymatic influence of the C-terminal extension of mycobacterial subunit α, and to gain insight into a possible mechanistic event, we tested the Gsα3β3γ and α3chiβ3γ complexes in a single molecule rotation assay, where the rotation can be visualized directly (26). A fluorescently labeled 300-nm bead was attached to subunit γ of immobilized Gsα3β3γ or α3chiβ3γ complexes (Fig. 4A). Upon addition of saturating MgATP concentrations rotating beads were observed via TIRF microscopy. Fig. 4B shows the trajectory of a rotating Gsα3β3γ-bead complex, derived from a frame-by-frame analysis (Fig. 4C) of the recorded video (26). The average rotational rate of the Gsα3β3γ complex was 4.6 ± 1.0 s−1 (n = 9 particles). When a 600-nm bead duplex was attached to the protein, the average rotational rate under these conditions was 1.9 ± 0.5 s−1 (n = 8 particles) due to the high viscous drag. The rotation rates for this complex were similar to those reported previously (26, 27).
Figure 4.
Single-molecule measurements of rotating beads complexed with Gsα3β3γ or α3chiβ3γ. A, schematic model of the experimental setup for the single-molecule rotation assay of protein-bead complexes. The enzyme was fixed to a Ni-NTA-coated coverslide via its His tags, whereas the biotinylated cysteine at γ109 served to bind a streptavidin-coated bead (Ø = 0.3 μm) doped with biotinylated quantum dots. B, trajectory of a rotating Gsα3β3γ-bead complex with a rotational rate of 3.4 rotations per second. C, sequence of single video frames (30 ms per frame) showing the counterclockwise rotation of a single Gsα3β3γ-bead complex. Each frame has a resolution of 20 × 20 pixel with 65 nm/pixel. D, trajectories of rotating α3chiβ3γ-bead complexes, revealing that all protein-bead complexes are continuously rotating forward, i.e. counterclockwise, when active.
The question was addressed, whether the C terminus of αchi in the α3chiβ3γ complex affects the rotation and dwell times of the α3chiβ3γ complex. As demonstrated in a video (supplemental Video S1) and in Fig. 4D, all α3chiβ3γ-bead complexes rotated continuously counterclockwise without backward steps. At saturating ATP concentrations, the rotational rate measured by the bead assay was dominated by the dwell time of the catalytic dwell. A very prolonged catalytic dwell would decrease the overall rotational rate. The ATP-waiting dwell is not rate-limiting here, but again a largely increased dwell time at this position would also affect the overall rate. However, when the α3chiβ3γ complex was used in the rotation assay, we observed the same rotational rates of 4.8 ± 2.0 s−1 (n = 13 particles) and 2.1 ± 0.7 s−1 (n = 6 particles) for complexes with single beads and bead duplexes, respectively. These experiments show that ATP hydrolysis results in the unidirectional counterclockwise rotation of the chimeric complex, and that the chimeric subunit α does not alter the duration of the dwells significantly.
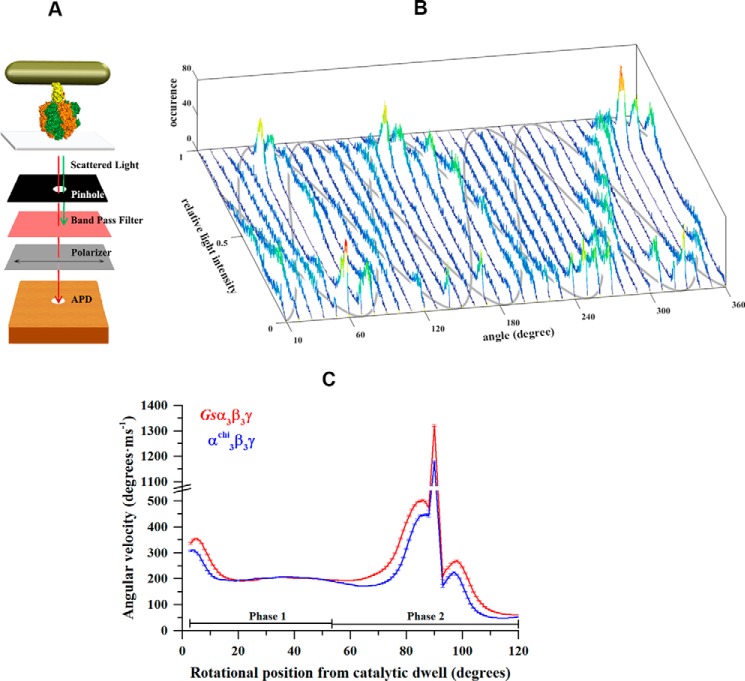
Changes in angular velocity as a function of rotational position during the power stroke cannot be investigated in detail with the bead assay because the movement of subunit γ is damped by the viscous drag of the large bead. In addition, the temporal and spatial resolution of the bead assay is too low to resolve the detailed motion during the power stroke. To overcome these limitations, we used a 40 × 76-nm gold nanorod as a probe of rotation. The viscous drag on these nanorods is not rate-limiting to the power stroke of the Escherichia coli α3β3γ (Ecα3β3γ) (28, 29), Gsα3β3γ, or the A3B3DF of the Methanosarcina mazei Gö1 A1A0-ATP synthase (MmA3B3DF) (30). The light scattered from the nanorod was detected by an avalanche photo diode (APD) after passing through a polarizing filter using dark-field microscopy (Fig. 5A). Upon addition of 1 mm MgATP, sinusoidal variations of light intensities scattered from the nanorod were observed as a function of its position relative to the axis of polarization. At a sampling rate of 1 kHz, dwells in the rotation produced a peak in the intensity distribution, as reported previously for Ecα3β3γ (28, 29), Gsα3β3γ, or MmA3B3DF (30). A series of 36 histograms of light intensities was recorded that were acquired when the rotational position of the polarizer was advanced successively by 10°. The observation of three off-set sinusoidal curves in the histograms, when the axis of rotation was close to the orthonormal position relative to the microscope slide, indicated the rotation of the ATP hydrolyzing complexes and the presence of three catalytic dwells separated by 120° steps (Fig. 5B).
Figure 5.
Single-molecule measurements of rotating gold nanorods complexed with Gsα3β3γ or α3chiβ3γ. A, schematic model of the microscope setup. Gold nanorods are illuminated by a dark-field condenser. Red polarized light scattered from a nanorod was recorded by an APD after passing through a polarizer. The model also shows the interaction of a gold nanorod with Gsα3β3γ or α3chiβ3γ. The protein Gsα3β3γ or α3chiβ3γ (subunits α, β, and γ in orange, green, and yellow, respectively) is attached via its His10 tags in subunit β to a coverslide, whereas an avidin-coated nanorod is attached to the opposing biotin modified cysteine in subunit γ. B, consecutive histograms of light intensities of a rotating nanorod attached to α3chiβ3γ upon rotating the polarizer in 10°-steps. The three peak positions in the histograms, resulting from the catalytic dwells of protein, follow a sinusoidal curve over 360° rotation of the polarizer. The approximate courses of the three sine curves are indicated in gray. C, average angular velocity over angular position at 1 mm MgATP of Gsα3β3γ (red) and α3chiβ3γ (blue). Each point represents the average over three neighboring angular positions.
The angular velocity profile for a 120° power stroke of the central stalk was calculated from the sinusoidal light intensity changes recorded with 200 kHz using the arcsin½ function (30). The three off-set sinusoidal curves like that shown in Fig. 5B was observed for each molecule used to derive the angular velocity profile. Fig. 5C shows the angular velocity of Gsα3β3γ (n = 20 molecules, average number of transitions: 2153) for a 120° power stroke between two catalytic dwells, as described recently (30). Starting from a catalytic dwell, which here was defined to be at 0°, the angular velocity reached a plateau of about 200° ms−1 between 20° and 55°. This is the range of angular positions (phase 1) where ATP binds to the catalytic site (30). Following the plateau in phase 1, the angular velocity in phase 2 increased steadily to 500° ms−1 at 85°, reached a maximum of 1300° ms−1 at 90°, and then decreased rapidly to 200° ms−1 at 93°. After another small acceleration to 270° ms−1 at 98°, the rate declined to a minimum at 120° (i.e. at the next catalytic dwell). The plot for the α3chiβ3γ complex (n = 17 molecules, average number of transitions: 1860) showed a similar pattern. However, from 55° to 120° (i.e. after the ATP-waiting dwell), the angular velocity was reduced on average by 21%. Taking into account the same dwell time for the catalytic dwell and angular velocity for the first 55° of rotation, the larger percentile decrease for the power stroke is in line with the overall 16% decrease in ATPase activity for the chimeric complex.
Purification and nucleotide-binding of Gsα and αchi
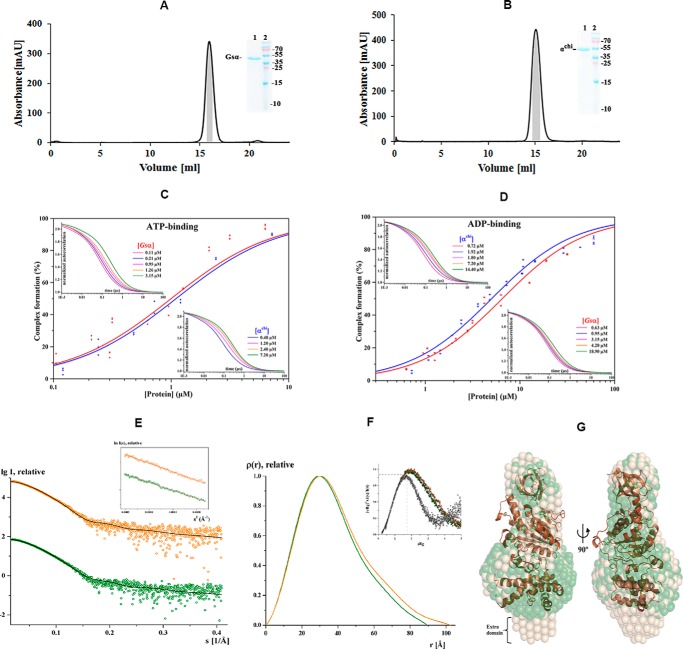
To gain insight into the location and structural traits of the extended C-terminal domain of mycobacterial subunit α, the recombinant αchi composed of Gsα(1–502) and the C-terminal Mtα(514–549) was compared with the wt Gsα. Both proteins were generated, produced, and purified in high quality (Fig. 6, A and B). The SDS gels of the purified proteins after size exclusion chromatography showed a single band that was running at a higher molecular weight in the case of αchi.
Figure 6.
Purification, FCS and solution X-ray scattering studies of Gsα and αchi. Final purification of Gsα (A) and αchi (B) with a Superdex 200 column. The inset shows an SDS gel, which corresponds to the shaded area (gray) of the elution peak, pooled, and applied on the gel. Lanes 1 and 2 reveal the purified protein and protein markers, respectively. Binding properties of subunits Gsα and αchi to fluorescently labeled nucleotides. C and D, results of FCS experiments, showing the binding of labeled nucleotides to subunit α. The upper left and lower right insets show the normalized autocorrelation curves of MgATP- (C) and MgADP-ATTO-647N (D) obtained by increasing the quantity of subunits Gsα and αchi (increased protein concentration from left to right). C, binding of subunits Gsα and αchi to MgATP-ATTO-647N and D, MgADP-ATTO-647N displayed as relative bound fraction versus protein concentration. The best fits to titration curves are shown as a non-linear, logistic curve fits. The percentage of complex formation for each concentration was calculated using a two-component fitting model. The binding constant, KD, was derived by fitting the data with the Hill equation. E, small angle X-ray scattering pattern (○) for Gsα (green) and αchi (orange). Fitting of the theoretical scattering curve (—) for Gsα computed by CRYSOL with the experimental scattering pattern (○) for Gsα (green) and αchi (orange) resulted in χ2-values of 1.44 and 1.46, respectively. The curves are displayed in logarithmic units for clarity. Inset, Guinier plots show linearity indicating no aggregation. F, pair-distance distribution function P(r) for Gsα (green) and αchi (orange). Inset, normalized Kratky plot indicating the folded nature of the protein. G, the average solution shape of Gsα (green) and αchi (wheat) as calculated by the DAMMIN program is overlapped with the crystal structure of Gsα (brown).
Fluorescence correlation spectroscopy (FCS) was used to determine the ATP- and ADP-binding properties of Gsα and αchi, and to determine whether the extension by the C-terminal Mtα(514–549) may have altered the nucleotide-binding traits of αchi (Fig. 6, C and D). The characteristic diffusion times τD of MgATP-ATTO-647N and MgADP-ATTO-647N, determined from a single component fit of the resulting autocorrelation function, were about 70 μs. After addition of proteins τD increased due to the binding of fluorescently labeled nucleotides to the proteins. The characteristic diffusion times of MgATP-ATTO-647N and MgADP-ATTO-647N bound to (i) Gsα were 250 ± 38 and 270 ± 24 μs, and (ii) αchi were 280 ± 25 and 300 ± 19 μs, respectively. Larger diffusion times of αchi are expected due to the extended C terminus. The molecular brightness was 51–52 kHz for unbound fluorescent nucleotides, and 46–54 kHz for the protein-bound fractions, whereas the number of particles remained the same after addition of the protein. This confirmed that only one nucleotide was bound to the protein. From the respective autocorrelation curves (insets in Fig. 6, C and D) the fractions of bound nucleotides were derived, plotted against the rising protein concentrations, and fitted with the Hill equation to determine the dissociation constants (KD). The KD values of Gsα and αchi for the binding of MgATP-ATTO-647N were 1.0 ± 0.1 and 1.1 ± 0.1 μm, respectively. The respective KD values for MgADP-ATTO-647N were 6.5 ± 0.3 and 5.0 ± 0.2 μm. The data indicate that the C-terminal extension of subunit α does not change the nucleotide-binding behavior of αchi, and that Gsα and αchi have a slight preference for the ATP over the ADP analogue.
Solution structure of Gsα and αchi studied by SAXS
SAXS experiments were performed to study the low resolution solution structure of Gsα and αchi. The final composite scattering curves for the SAXS pattern recorded are shown in Fig. 6E and indicate the monodispersity of the proteins. The Guinier plots at low angles were linear and revealed good data quality with no indication of protein aggregation (inset in Fig. 6E). The Kratky plots (I(s) × s2 versus s) (inset in Fig. 6F) showed a bell-shaped peak at low angles indicating a well folded protein. The radius of gyration (Rg) values determined from the Guinier approximation were 27.98 ± 0.24 and 29.56 ± 0.32 Å, for Gsα and αchi, respectively. The determined distribution functions, p(r) (Fig. 6F), showed a maximum particle dimension, Dmax, of 89.5 ± 10 and 102.5 ± 10 Å for Gsα and αchi, respectively, reflecting that αchi is more elongated (Table 1). The gross shapes of the proteins were ab initio reconstructed and had a good fit to the experimental data in the entire scattering range discrepancies of χ2 = 0.348 and 0.334 for Gsα and αchi, respectively. Ten independent reconstructions produced similar envelopes with a normalized spatial discrepancy (NSD) of 0.611 ± 0.06 and 0.603 ± 0.04 for Gsα and αchi, respectively. The average structure is shown in Fig. 6G. The theoretical scattering curve for the Gsα (PDB code 1SKY) was computed and compared with the experimental scattering curves of Gsα and αchi using CRYSOL with resulting discrepancies of χ2 = 1.44 and 1.46, respectively (Fig. 6E). Superimposition of the Gsα and αchi shapes with the crystal structure of Gsα indicated an extra domain of 15 × 22 Å, reflecting the additional 36 residues of the chimera protein. Furthermore, the comparison demonstrated that the C-terminal Mtα(514–549) of αchi is located below the very C terminus of Gsα. The NSD between the Gsα and αchi solution shapes is 0.53, whereas the NSD for the shapes of Gsα and αchi with the crystal structure of Gsα (PDB code 1SKY) are 2.12 and 2.17, respectively.
Table 1.
Data collection and scattering derived parameters for subunit Gsα and αchi using SAXS

* Reported for highest concentration measurement.
Solution structure of the C-terminal peptide Mtα(521–540)
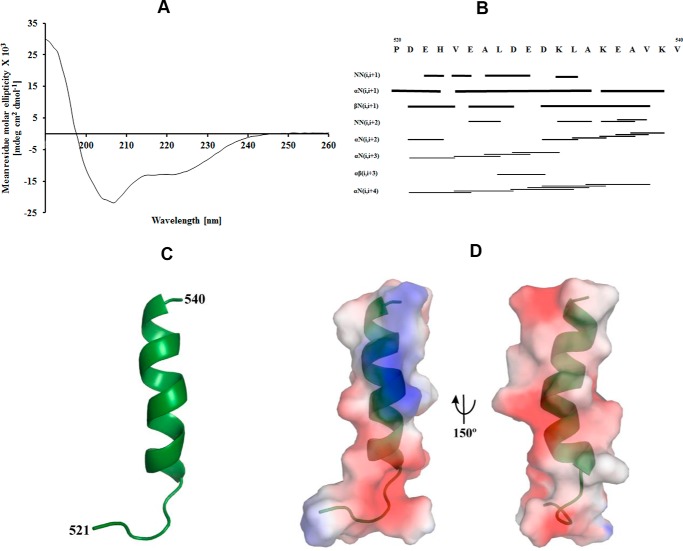
According to the secondary structure prediction in Fig. 1, the extended C-terminal region 514–549 of Mtα shows random coiled regions of the peptides 514–525 and 540–549 and a helical structure formed by the peptide 526–539. To gain structural insight about this epitope and the α-helical feature in solution, the peptide 521PDEHVEALDEDKLAKEAVKV540 of Mtα (Mtα(521–540)) was synthesized. The CD spectrum of Mtα(521–540) showed α-helical and random-coiled formation of 43 and 52%, respectively (Fig. 7A). To determine the 3D structure of Mtα(521–540), the primary sequences of α(521–540) were sequentially assigned as per standard procedures, using a combination of total correlation spectroscopy (TOCSY) and NOESY spectra. Secondary structure prediction was done using PREDITOR (31), which uses HA chemical shifts and showed the presence of an α-helical structure. The 20 lowest-energy structures of 100 were taken for further analysis. In total, an ensemble of nine calculated structures resulted in an overall root mean square deviation (r.m.s. deviation) of 0.65 Å for the backbone and 1.65 Å for heavy atoms. All the structures had energies lower than −100 kcal mol−1, with no NOE-violations and no dihedral violations (Table 2). Identified cross-peaks in the HN-HN and HA-NH regions are shown in Fig. 7B. From the assigned NOESY spectrum HN-HN, Hα-HN (i, i+3), Hα-HN (i, i+4), and Hα-Hβ (i, i+3) connectivities were plotted, which supported α-helical formation in the structure. The calculated structure has a total length of 20 Å and displays an α-helical region between residues 526 and 539 (Fig. 7C). Fig. 7D shows the molecular surface electrostatic potential of Mtα(521–540).
Figure 7.
CD and NMR studies of the peptide Mtα(521–540). A, circular dichroism (CD) spectrum of Mtα(521–540). B, the NOESY connectivity plot of Mtα(521–540). C, the average NMR structure of Mtα(521–540) is shown in schematic representation. D, molecular surface electrostatic potential of the peptide Mtα(521–540) generated by PyMOL (62), where the positive potentials are drawn in blue and the negative in red.
Table 2.
Structural statistics for Mtα(521–540)
| Statistics | |
|---|---|
| Distance restraints | |
| Total | 188 |
| Intraresidue (i–j = 0) | 83 |
| Sequential (|i-j| = 1) | 75 |
| Medium-range (2 ≤ |i−j| ≤ ) | 30 |
| Long-range (|i−j| ≥ 5) | 0 |
| Average number of violations | |
| Distance violations >5 Å | 0 |
| Ramachandran plot2 (%) | |
| Residues in most favored regions | 88 |
| Residues in additionally allowed regions | 12 |
| Residues in generously allowed regions | 0 |
| Residues in disallowed regions | 0 |
| Average root mean square deviation to mean (Å) | |
| Backbone (Cα, C′, and N) | 0.65 ± 0.25 |
| Heavy atom | 1.61 ± 0.44 |
Proximity of Mtα(521–540) to subunit γ residues 104–109
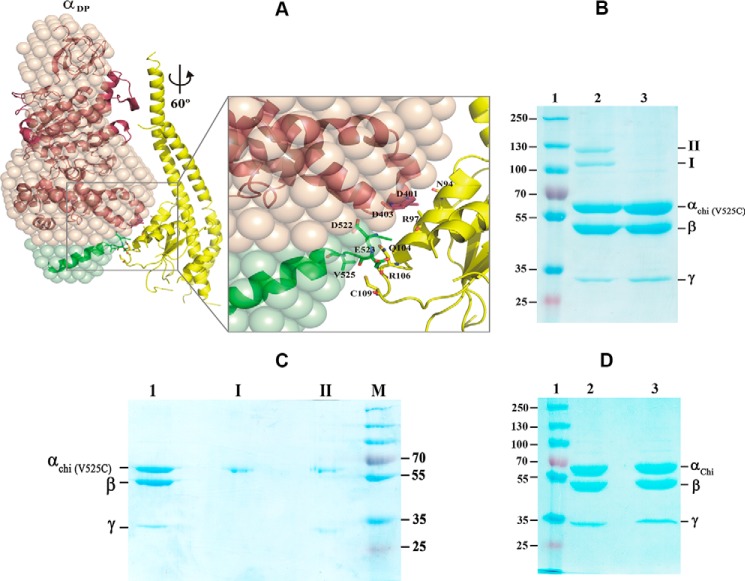
We further investigated whether the decreased hydrolysis rate correlated with a decreased angular velocity of the α3chiβ3γ complex. A structural model of the α3chiβ3γ complex was generated (Fig. 8A) based on the G. stearothermophilus F1-ATPase structure (32) (PDB code 4XD7) and the structural insights derived from the present study (see “Experimental procedures” for details). To ascertain the influence of the extended C-terminal 36 residues of αchi during hydrolysis, subunit γ was rotated counterclockwise (viewing from the membrane side) for the full 360° in steps of 20° in the α3chiβ3γ complex model. At angles of 60 ± 10° and in steps of 120°, the central globular domain of the γ subunit comes closer to the so-called “DELSEED” region, composed of residues 396AQFGSDLDK404 of subunit α, and to the extended C-terminal domain of αchi. Particularly at 60°, the α-helical residues 90AYNSNVLRLVYQT102 of the γ subunit come into the vicinity of α subunit residues 396AQFGSDLDK404. Interestingly, the polar residues Gln104 and Arg106 as well as Cys109 are in proximity to the extended C-terminal residues Asp522, Glu523, and Val525. To test their closeness, Val525 was substituted by a cysteine in a α3chiβ3γ complex mutant called (αchi-V525C)3β3γ. The isolated protein was applied on a 9% SDS gel in the absence (Fig. 8B, lane 2) or presence (lane 3) of the reductant dithiothreitol (DTT). As shown in lane 2 of Fig. 8B, two cross-linked products I and II were observed. In comparison, these products were not formed by the DTT-treated protein and the band intensities of subunits αchi-V525C and γ were stronger (lane 3). The bands of product I and II were cut out and incubated in the presence of 20 mm DTT before embedding them in a second SDS gel under reducing conditions. Fig. 8C demonstrates that product I is formed by αchi-V525C dimers and product II by a cross-linked product of αchi-V525C and γ that migrated aberrantly higher due to cross-link formation. The data revealed that C525 in αchi-V525C and Cys109 of subunit γ formed the disulfide bridge, which became cleaved after reduction. In parallel, α3chiβ3γ was treated in the same way as (αchi-V525C)3β3γ. Fig. 8D shows that no cross-link product was formed in the presence or absence of DTT. Overall, these data confirmed the proximity of the extended C-terminal helix of αchi with the subunit γ residues 104–109, whose interaction may influence rotation of the camshaft-like subunit γ.
Figure 8.
Interaction of the C-terminal stretch of α with the rotary subunit γ. A, a structural model of the α3chiβ3γ complex was generated based on the G. stearothermophilus F1-ATPase structure (32) (PDB code 4XD7). See text for details. At 60° rotation, the α-helical residues 90AYNSNVLRLVYQT102 of the central globular domain of subunit γ (yellow) come closer to the so-called DELSEED region, composed of residues 396AQFGSDLDK404 of subunit α (brown), and the extended C-terminal domain of αchi (green). The polar residues Gln104 and Arg106 as well as Cys109 are in proximity to the extended C-terminal residues Asp522, Glu523, and Val525. B, Val525 was substituted by a cysteine in the mutant α3chiβ3γ complex mutant resulting in the (αchi-V525C)3β3γ. The mutant protein was applied on a 9% SDS gel in the absence (lane 2) or presence (lane 3) of DTT. Two cross-link products of the oxidized complex are marked I and II (lane 2). Lane 1, represents molecular weight standard proteins. C, the bands of product I and II of Fig. 9B were cut out, incubated in the same buffer with 20 mm DTT, before embedding in a second 9% SDS gel under reducing condition. Lanes 1–4, represent the (αchi-V525C)3β3γ, product I, product II, and a molecular mass standard, respectively. D, the (αchi-V525C)β3γ complex in the absence (lane 2) or presence (lane 3) of DTT was applied on a 9% SDS gel. Lane 1, molecular weight standard.
Discussion
Activation of the latent ATP hydrolysis driven H+-pumping of mycobacterial F-ATP synthase may alter the magnitude of the PMF and decrease the viability of the bacteria (33). An amino acid sequence alignment of F-ATP synthase α subunits reported here (Fig. 1) revealed that the mycobacterial subunit α, important for nucleotide-binding and catalysis, as well as the coupling and rotating subunits ϵ and γ are different from any other pro- and eukaryotic counterparts (34, 35). Here, the deletion of the mycobacterial C terminus of subunit α increased ATP hydrolysis and H+-pumping activity of about 63 and 10%, respectively, compared with the wt enzyme (Fig. 2B, 2D). These increases of ATP hydrolysis and H+-pumping that were comparable for both the Δα(514–548) and Δα(521–540) mutants demonstrated that the C-terminal helix plays a role in the regulation of catalytic and coupling events. In contrast, when this C-terminal extension was inserted into Gsα3β3γ to form a chimeric protein, the ATPase rate was reduced by about 10%. These data reveal that the C-terminal extension of subunit α contributes significantly to the reduced (inhibited) ATPase activity that does not provide sufficient energy to couple proton-conduction in mycobacteria.
The extent of the reduced ATPase activity provided by the unique α subunit extension occurs in addition to similar effects, like those reported in Fig. 2D, which result from the unique insertion of 14 amino acid residues of the M. tuberculosis subunit γ (165TDNGEDQRSDSGEG178) (23). This subunit γ insertion that forms a loop of polar residues has been proposed to be in vicinity to the polar residues Arg41, Gln42, Glu44, and Gln46 of the c-ring (23, 24) (Fig. 9, A and B). Deletion of this loop in IMVs, containing the M. smegmatis Δγ(166–179) mutant protein, led to ATP-driven H+-pumping (about 61%) (23), as shown in Fig. 2D. It has been proposed that this unique polar loop of the mycobacterial subunit γ interacts with the c-ring such that it affects rotation and thereby inhibits proton pumping. Thus, the results reported here for the unique extension of subunit α, in addition to those reported previously for the unique insertion of subunit γ, provide a novel inhibitory mechanism to regulate the PMF such that it enables the pathogenic bacterium to survive.
Figure 9.
Model of the interaction between subunits γ and αchi in α3chiβ3γ during rotation. A, the unique mycobacterial γ-loop (red), which is in the vicinity to the M. phlei c-ring (brown, PDB code 4V1G) (15), was inserted in the Gsγ (yellow). The arrow indicates the interaction between the 396AQFGSDLDK404 peptide (green) and the extra C-terminal domain of αchi (light green) with the α-helical peptide 90AYNSNVLRLVYQT102 of Gsγ. B, the proximity of the C-terminal helix of Mtϵ with the extended C-terminal stretch of αchi. Subunit ϵ (blue) inside the α3chiβ3γ complex was modeled based on the recently determined solution structure of Mtϵ (PDB code 5WY7), which was then superimposed onto the δ subunit of the bovine F1-ATPase. At 80° rotation of the central stalk, the compact C-terminal helix of Mtϵ might interact with the extended C-terminal domain of αchi on the way to form an extended conformation. C, subunits αchi, β with nucleotide occupancy, and γ with rotational position are shown in green, orange, and yellow, respectively, and labeled according to the native crystal structure (60). The extended C terminus of αchi is shown in dark green. Catalytic events are based on the model by Watanabe et al. (63). The state on the left shows the catalytic dwell. During phase 1 (see Fig. 5C) Pi release from βE triggers a 40° rotation of subunit γ, which leads the enzyme to adopt the ATP-waiting dwell. During phase 2, ATP binding and ADP release, accompanied by a change in the conformational states of the αchi and β subunits, causes subunit γ to further rotate by 80°. At a rotational position of 90° subunit γ is in close proximity to the extended C terminus of αchi.
The structural model described here (Fig. 8A), which is confirmed by the formation of a cross-link between residues αCys525 and γCys109 of (αchi-V525C)3β3γ, clearly shows that the extended C-terminal stretch of mycobacterial subunit α with its α-helical region between residues 526 and 539 and the rotating γ are in close proximity. Particularly at 60°, the α-helical residues 90AYNSNVLRLVYQT102 of subunit γ come into the vicinity of subunit α residues 396AQFGSDLDK404, and the polar residues Gln104, Arg106, and Cys109 of subunit γ are in proximity to the extended C-terminal α residues Asp522, Glu523, and Val525. The possible occurrence of additional interactions due to the extended α subunit may also influence the rate of catalysis, which may explain the 12% reduction of the overall ATPase activity rate in bulk ATP hydrolysis of α3chiβ3γ when compared with wt Gsα3β3γ.
The effect of the C-terminal extension of subunit α unique to mycobacteria on ATPase-dependent rotation is summarized in Fig. 9C. Hydrolysis of ATP occurs at the (αβ)DP catalytic site during the catalytic dwell that ends upon release of phosphate (Pi) from the (αβ)E catalytic site to initiate rotation (36). This initiates phase 1 of the power stroke (29) that rotates subunit γ by ∼40°. If ATP has not become bound to (αβ)E by the time that subunit γ reaches that position, an ATP-binding dwell is observed. Binding of ATP then induces phase 2 in which subunit γ rotates for 80° to complete the 120° power stroke and form the next catalytic dwell (29).
The single-molecule rotation assays with a fluorescent bead (Fig. 4B) indicate that ATPase-dependent rotation occurred in the counterclockwise direction without significant back-steps, and that the chimeric subunit α does not alter the duration of the dwells significantly. This was corroborated by our FCS studies that showed that the binding affinities for ATP or ADP remained the same in the chimeric protein (Fig. 6, C and D). Using the gold nanorod assay capable of resolving the angular velocity of the power stroke under conditions in which not the viscosity but rather by the internal mechanism of the motor is rate-limiting, we found that the presence of the C-terminal extension of subunit α decreased the angular velocity of the power stroke of the motor (Fig. 5C). Although the angular velocity profile of phase 1 remained unchanged, the final 65° of the power stroke (phase 2) was on average 21% slower with the chimeric subunit α. These results were consistent with the structural and cross-linking results presented here that showed an interaction between the C-terminal extension of subunit α and subunit γ.
The motion of the DELSEED sequence in subunit β has been shown to contribute significantly to angular velocity of ATPase-dependent subunit γ rotation (29). We now show for the first time that the extension of the C terminus of mycobacterial subunit α significantly limits the angular velocity of the rotor that is likely to result from steric hindrance between one of the αchi subunits and subunit γ during the power stroke. An effect of steric interference due to electrostatic forces between subunit γ and the C-terminal domain of the catalytic βE subunit of the thermoalkaliphilic Bacillus sp. TA2.A1 F-ATP synthase has been described to prevent the enzyme from rotating in the ATP hydrolysis direction (23). Our hypothesis is further strengthened by the observation that the ATP hydrolysis rate of the M. smegmatis ATP synthase deletion mutant Δα(514–548) is enhanced when compared with its wt form (Fig. 2, A and B). This underlines that the C-terminal extension of the mycobacterial subunit α contributes to the regulation of ATPase hydrolysis.
It is noteworthy that mycobacterial subunit ϵ, which also forms part of the drive shaft of the motor in the intact F-ATP synthase, has a truncated C-terminal domain compared with other organisms (Fig. 9B) and was predicted to be too short to reach into the upper region of the α-β interface close to the adenine-binding pocket (34). A recent study (34) extended our structural model to M. tuberculosis F-ATP synthase subunit ϵ based on the recently determined NMR structure of Mtϵ (PDB code 5WY7) (Fig. 9B). Application of similar rotational simulations applied to subunits γ and Mtϵ in this model revealed that the C-terminal helix of Mtϵ lies closer to the αchi (Fig. 9B). At a rotational angle of 80 ± 10° (in hydrolysis direction from a catalytic dwell position) the C terminus of Mtϵ is in close proximity to the extended C-terminal domain of αchi when the compact Mtϵ is on the way to form an extended conformation. It is for future studies to reveal how these proposed interactions might influence the activity of the holoenzyme.
In summary, a novel combination of complementary approaches of genetic engineering, protein chemistry, ensemble enzymatic and single-molecule rotation assays, and structural biology has provided new insights into the unique mycobacterial C-terminal 36-amino acid residues extension of subunit α. The C-terminal α-helical extension of one α subunit is suggested to interact with polar residues of subunit γ, whereas a second α (αTP) is suggested to interact with the C-terminal helix of Mtϵ to generate an inhibited state that reduces ATP hydrolysis. The low ATPase activity and the interaction of the γ(166–179)-loop with the c-ring significantly limit the extent of ATPase-driven H+-pumping such that they alter the magnitude of PMF and decreases the viability of bacteria.
Experimental procedures
Genetic manipulation
DNA manipulations were done according to standard protocols (37). Plasmid and DNA isolations were performed according to Qiagen protocols. Mycobacterial DNA manipulations were done with some minor modifications according to published protocols (38). Primers and dsDNA oligonucleotides were synthesized by Integrated DNA Technologies (USA) and sequencing of DNA was done by AIT Biotech (Singapore).
Cultivation and preparation of electro-competent cells of M. smegmatis mc2 155 was done according to the protocol described by Goude and Parish (39). For recombineering, M. smegmatis mc2 155 cells containing the episomal plasmid pJV53, with the inducible proviral Che9c genes gp60 and gp61 under control of the acetamide promoter were used. Electrocompetent cells were prepared (39), washed, and cells after overnight growth were expanded in 7H9 medium supplemented with succinate (0.2% v/v) to an A600 = 0.6. Prior to cooling the cells on ice, recombineering genes were induced with 0.2% (v/v) acetamide (3 h, 37 °C).
Recombineering assay with dsDNA in M. smegmatis mc2 155
Recombineering was done according to van Kessel and Hatfull (40, 41). To generate a M. smegmatis mc2 155 F-ATP synthase mutant form with a deletion of amino acids 514–548 of the C terminus of subunit α (Δα(514–548)), a stop codon TAG at position 514 in the double-stranded 100-bp DNA oligonucleotide of the gene atpA was synthesized by Integrated DNA Technologies, USA. This oligonucleotide was further expanded by 80 bp using PCR with the forward (forward ext Δα1) and reverse extending primers (reverse ext Δα1) as described in supplemental Table S1, which resulted in a 200-bp dsDNA oligonucleotide AES (allelic exchange substrate). In case of the M. smegmatis F-ATP synthase mutant Δα(521–540), the deletion of the C-terminal helix was achieved by synthesizing a 90-bp junction dsDNA containing 45-bp DNA sequences upstream and downstream of the DNA coding for deleted helix 521–540 (dsDNA Δα(521–540)). The junction dsDNA sequence was further extended to 210 bp by PCR using 90-bp extended primers (forward ext Δα2/reverse ext Δα2) (supplemental Table S1). The M. smegmatis F-ATP synthase recombinant strain with the gene atpA replaced by the gene atpA from G. stearothermophilus (GsatpA) was constructed using a 2.5-kb dsDNA AES and plasmid pSJ25 as a co-transformation vehicle. The 2.5-kb dsDNA AES contained the 1.47-kb GsatpA gene flanked at both 5′ and 3′ ends by a 500-bp DNA, homologous to the DNA upstream and downstream of atpA from M. smegmatis. The AES was generated by several PCRs. First, the upstream and downstream 500-bp homologous DNA was generated by separate PCR, using the primers Forward_Gs_A/Reverse_Gs_A (upstream homologous DNA) and Forward_Gs _B/Reverse_Gs_B (downstream homologous DNA). The GsatpA gene was amplified using primers Forward_ext_Gs/Reverse_ext_Gs (see supplemental Table S1). Finally, the 2.5-kb AES was obtained by PCR using amplified DNA: 500-bp upstream/downstream homologous DNA and 1.47-kb GsatpA as templates and the pair of primers Forward_Gs_A/Reverse_Gs_B. The DNA sequence of the 2.5-kb AES was confirmed by sequencing.
Electrocompetent M. smegmatis mc2 155 cells containing the plasmid pJV53 (KanR) were mixed with 200 ng of double-stranded DNA (210 bp dsDNA) and 100 ng of co-transforming plasmid pSJ25. Transformation was done using the following parameters: single pulse 2.5 kV, conductivity 25 μF, resistance 1000 Ω, and Gene pulser Xcell (Bio-Rad) as previously described (39–41). Transformed cells were recovered (4 h, 37 °C) in 7H9 supplemented with 10% ADC and 0.05% Tween 80, and plated on 7H10 agar supplemented with 10% oleic albumin dextrose catalase, kanamycin, and hygromycin. About 200 hygromycin-resistant transformants were streaked on master 7H10 agar plates and tested by colony PCR. Colony PCR was done as follows: cells from master agar plates were resuspended in 200 μl of H2O, boiled (95 °C, 20 min), and immediately cooled on ice. An aliquot a 1/10 of the reaction mixture was used as a template in PCR detection using forward atpA/Reverse atpA-mama primers (supplemental Table S1). PCR parameters used in detecting recombinants were as follows: Taq DNA polymerase (Thermo Fischer), initial denaturation (95 °C, 3 min), cycle denaturation (95 °C, 30 s), annealing (65 °C, 30 s), extension (72 °C, 30 s), final extension (72 °C, 5 min), and 30 cycles in total. MAMA-PCR (Mismatch Amplification Mutation Assay) of recombinants carrying the mutation resulted in a PCR product, whereas unchanged DNA of the wt failed to amplify DNA. Detected recombinants were re-streaked several times on non-selective 7H10 agar plates to lose the recombineering plasmid pJV53 (KanR). The targeted deletion of the DNA sequence in the atpA gene and inserted stop-codon of M. smegmatis of both mutants was confirmed by sequencing the DNA (AIT Singapore).
Preparation of inverted membrane vesicles from M. smegmatis
Inverted vesicles of M. smegmatis for ATP synthesis, hydrolysis, and proton-pumping assays, were generated (23, 24).
Cloning, production, and purification of mutants
To amplify subunit α of the G. stearothermophilus F-ATP synthase, the atpA gene was amplified using the plasmid DNA encoding for subcomplex Gsα3β3γ of G. stearothermophilus as a template. NcoI and BamHI restriction sites were introduced in the forward and reverse primers, respectively: forward, 5′-GAGACCATGGCAATGAGCATTCGAGCGGAAGAA-3′ and reverse, 5′-CCCGGGATCCTTATTGAGAAACGACAAACGTTTTCTT-3′.
To generate an α3chiβ3γ complex, overlap extension PCR was performed, using templates of plasmid DNA for Gsαγβ and Mtα. The plasmid DNA of Gsαγβ encodes for the α3β3γ complex, including mutations in αC193S and γS109C. The presence of a single cysteine residue enabled the selective biotin labeling for single molecule rotation studies. Restriction sites BamHI and HindIII were incorporated in the forward and reverse primers, respectively: forward, 5′-AGATGGATCCAATGAGCATTCGAGCGGA-3′ and reverse, 5′-GAATAAGCTTTCACACTTCGACACCCATCGC-3′. These served as the flanking primers (primer a and primer f). Primers b, c, d, and e were 5′-AGAGCCGCCTTGAGAAACGACAAACGT-3′, 5′-GTTTCTCAAGGCGGCTCTGTGGTGCCC-3′, 5′-CCGCCGGCCTTATTTCTTCTTCTTCGG-3′, and 5′-AAGAAATAAGGCCGGCGGGGGCATATC-3′, respectively. These were the chimeric primers, 27 nucleotides in length containing anchoring segments from the template and the adjoining anchoring segment. To amplify the individual fragments, overlap extension PCR was performed to allow insertion of the fragment encoding for the C terminus of Mtα at the end of the Gsα in the complex to yield αchiγβ.
The plasmid containing αchiγβ genes was used as a template for site-directed mutagenesis and to generate the mutant (αchi-V525C)3β3γ. The entire plasmid (pET-24b vector) was amplified by KAPA HiFi DNA polymerase (KAPA BIOSYSTEMS) using forward, 5′-CGCCAAGGAAGCCTGCAAGGTCAAAAAGCC-3′ and the reverse, primer 5′-GGCTTTTTGACCTTGCAGGCTTCCTTGGCG-3′. The amplified PCR product was purified and DpnI digestion was performed to remove the methylated parental plasmid. After DpnI digestion the PCR product of mutant (αchi-V525C)3β3γ was purified and transformed into E. coli TOP10 cells. The DNA coding for the chimeric αchi was amplified using the primers 5′-GAGACCATGGCAATGAGCATTCGAGCGGAAGAA-3′ and 5′-CCGGGATCCTTATTTCTTCTTCTTCGGCGC-3′ as forward and reverse, respectively, which include the NcoI and BamHI restriction sites.
The amplified gene products for Gsα and αchi were ligated into the pET-9d(+) vector (42), and αchiγβ into the pET-24b vector, and transformed into E. coli DH5α cells for plasmid amplification. The generated plasmid with the Gsα and αchi insert, respectively, was transformed into E. coli BL21(DE3) cells (Stratagene), and plasmids carrying the DNA for αchiγβ and mutant αchi-V525Cγβ were transformed into E. coli DK8 cells for protein production. Gsα and αchi cells were grown in kanamycin-containing (30 μg ml−1) LB-medium (37 °C). The cells producing the mutants αchiγβ and (αchi-V525C)3β3γ were grown in 2YT medium, supplemented with 30 μg ml−1 of kanamycin. In all cases, protein production was induced by the addition of isopropyl β-d-thiogalactopyranoside (1 mm) at an A600 = 0.6, and cultivated at 37 °C. Cells were harvested after overnight induction, frozen by liquid nitrogen, and stored at −80 °C.
Cells containing recombinant subunits Gsα and αchi were lysed on ice by sonication for 3× 1 min in buffer A (50 mm Tris/HCl, pH 7.5, 200 mm NaCl, 2 mm PMSF, 0.8 mm DTT, and 2 mm PefablocSC (4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride) (BIOMOL)). Cells debris were pelleted by centrifugation (10,000 × g, 35 min). The supernatant was filtered (0.45 μm; Millipore). Proteins were allowed to bind to the Ni2+-NTA matrix, and eluted with an imidazole gradient (20–400 mm) in buffer A. Fractions containing Gsα and αchi were identified by SDS-PAGE (43), pooled, diluted with buffer B (50 mm Tris/HCl, pH 7.5) to reduce the salt concentration to 50 mm, and subsequently applied onto an anion-exchange column (Mono Q, 8 ml, GE-Healthcare). Gsα and αchi eluted at 100 mm NaCl. Fractions were concentrated using Centricon YM-30 (30-kDa molecular mass cut off) spin concentrators (Millipore) and applied onto a size exclusion column (Superdex 200 HR 10/30 column (GE Healthcare)). The purified protein was eluted with buffer B containing 200 mm NaCl and 0.8 mm DTT. The purity of the samples was ascertained via an SDS-PAGE (43).
The protocol for the purification of Gsα3β3γ, α3chiβ3γ, or (αchi-V525C)3β3γ mutant was similar to the one above, except PefablocSC (BIOMOL) was used in Ni2+-NTA affinity and anion-exchange chromatography. Ni2+-NTA affinity chromatography was performed with an imidazole gradient (20–600 mm). Fractions containing the protein were pooled and applied onto an anion-exchange column (Resource Q, 6 ml, GE Healthcare) with buffer B containing 50 mm NaCl. α3chiβ3γ eluted at 250 mm NaCl. Fractions were concentrated using Centricon YM-100 (100-kDa molecular mass cut off) spin concentrators (Millipore).
ATP hydrolysis activity measurements
A continuous ATP hydrolysis assay was applied to measure the ATPase activity of wt IMVs of M. smegmatis mc2 155, Δα(514–548), and Δα(521–540) mutants, and of the recombinant Gsα3β3γ and α3chiβ3γ complexes (26, 44). In the case of IMVs the consumption of NADH by the type II NADH dehydrogenase was inhibited by thioridazine. In addition, ATPase activity of IMVs was also measured in the absence of ATP and presence of thioridazine to identify any background NADH oxidation and MgATP hydrolysis.
Assay for ATP-driven proton translocation
ATP-driven proton translocation into IMVs of M. smegmatis with wt F-ATP synthase, Δα(514–548), or Δα(521–540) mutant was measured by a decrease of ACMA fluorescence using a Cary Eclipse Fluorescence spectrophotometer (Varian Inc., Palo Alto, CA) (6, 23).
ATP synthesis assay
ATP synthesis was measured as described recently (23).
Biotinylation of protein complexes
The cysteine at γ109 in the Gsα3β3γ and α3chiβ3γ complexes was biotinylated for the attachment of beads or nanorods coated with streptavidin or neutravidin, respectively. Freshly prepared proteins in buffer C (50 mm Tris/HCl, pH 7.5, 250 mm NaCl) were incubated for 5 min on ice with a 1.2-fold molar excess of biotin-PEAC5-maleimide (Dojindo, Japan). The reaction was stopped by adding 1 mm acetylcysteine, and unbound biotin-maleimide was removed by adding buffer C, followed by filtration using Centricon YM-100 (100 kDa molecular mass cut off) spin concentrators (Millipore). Samples were frozen in liquid N2 and stored at −80 °C.
Single molecule rotation assay with beads
Gsα3β3γ or α3chiβ3γ complexes were fixed on a Ni2+-NTA-coated coverslide with their His10 tag at the N termini of each β subunit. On the opposing end the biotinylated cysteine at γ109 was bound to a streptavidin-coated bead (Ø = 300 nm) doped with biotinylated quantum dots 605 (Q-dots). After addition of 1 to 4 mm MgATP, videos of rotating protein-bead complexes were recorded with an inverted fluorescence microscope (IX83/Cell∧TIRF) and analyzed to determine the rotational rates as described before (26).
Single molecule rotation assay with nanorods
Biotinylated Gsα3β3γ or α3chiβ3γ complexes were fixed on a Ni2+-NTA-coated coverslide. The biotinylated cysteine at γ109 was bound to a gold nanorods (40 × 76 nm, Nanopartz) that was prepared by mixing 9 μl of nanorods with 1 μl of Neutravidin (5 mg/ml in H2O, Molecular Probes) for 1 h before adding 1 μl of BSA (10% solution, Aurion, The Netherlands) for 1 min and 89 μl of buffer RB (50 mm Tris, pH 8.0, 10 mm KCl). Solutions were infused in a flow cell (20 μl, 5 min incubation) in the following order, with washing steps of 60 μl of buffer RB in between: 1) Ni2+-NTA-horseradish peroxidase conjugate (Qiagen, Germany) in deionized H2O; 2) 1 nm biotinylated protein in buffer RB; 3) gold nanorods in buffer RB; 4) 2 mm ATP, 1 mm MgCl2 in buffer RB. Rotation of protein-nanorod complexes via polarized darkfield microscopy and analysis of the data via customized software to obtain intensities and velocities at each angular position were performed as described before (28–30). In brief, samples were illuminated by a mercury lamp through a dark field condenser in a confocal microscope (Axiovert, Zeiss, Germany). When observed through a band pass filter and a polarizer the intensity of red light scattered from the nanorod, detected by an APD, changed sinusoidal from a maximum to a minimum when the long axis of the nanorod was parallel and perpendicular, respectively, to the axis of polarization. Data were collected for 10 and 30 s at 1 and 200 kHz, respectively. The obtained signals were evaluated with customized software (Matlab, MathWorks) to obtain intensities and velocities at each angular position of the rotating protein-nanorod complex (28–30). From the light intensity fluctuations recorded by the APD, power strokes were identified by intensity changes from a minimum taken from the lowest 10 percentile of peaks to a maximum taken from the highest 5 percentile of peak, for which a linear least square fit gave an R-value of at least 0.9.
Synthesis of the peptide α(521–540) from M. tuberculosis
Peptide Mtα(521–540) of the C-terminal amino acids 521–540 of Mtα (strain H37RV) was synthesized at the peptide core facility, School of Biological Sciences, NTU on Liberty Automatic. The final product with at least 99% purity was confirmed by analytical high pressure liquid chromatography.
Circular dichroism spectroscopy of Mtα(521–540)
Circular dichroism (CD) spectra of the peptide Mtα (521–540) were recorded with a CHIRASCAN spectrometer (Applied Photo-physics) using a 60-μl quartz cell (Hellma, Germany) with 1-mm path length. Light of 190–260 nm was used to record the far UV-spectra at 20 °C with 1-nm resolution. The measurement was done three times for each sample. The CD-spectra were acquired in a buffer of 25 mm phosphate (pH 6.5), and 30% 2,2,2-trifluoroethanol with a peptide concentration of 2.0 mg/ml.
Solution structure determination of Mtα(521–540)
For NMR spectroscopy experiments 2 mm Mtα(521–540) were dissolved in 25 mm phosphate buffer (pH 6.5) and 30% deuterated 2,2,2-trifluoroethanol. All spectra were obtained at 298 K on a 700-MHz Avance Bruker NMR spectrometer, equipped with actively shielded cryoprobe and pulse field gradients. TOCSY and nuclear Overhauser effect spectroscopy (NOESY) spectra of the peptide were recorded with mixing times of 80 and 200 ms, respectively. 2,2-Dimethyl-2-silapentane-5-sulfonate sodium salt was used as an internal reference. Data acquisition and processing were done with the Topspin (Bruker Biospin) program. The Sparky suite was used for spectrum visualization and peak picking. Standard procedures based on spin system identification and sequential assignments were adopted to identify the resonances. Interproton distances were obtained from the NOESY spectra by using the caliba script included in the CYANA 2.1 package (45). Dihedral angle constraints were derived from TALOS. The predicted dihedral angle restraints were used for the structure calculation, with a variation of ±30° from the average values. The CYANA 2.1 package (45) was used to generate the three-dimensional (3D) structure of the peptide. Several rounds of structural calculation were performed. Angle and distance constraints were adjusted until no NOE violations occurred. In total, 100 structures were calculated and an ensemble of 8 structures with lowest total energy was chosen for structural analysis.
Solution small angle X-ray scattering of Gsα and αchi
SAXS data of recombinant Gsα and αchi (2.5–5 mg ml−1) were measured with the BRUKER NANOSTAR SAXS instrument as described recently (46–49). Data processing was performed automatically using the program package PRIMUS (50). The experimental data obtained for different concentrations were analyzed for aggregation and folding state using Guinier (51) and Kartky plots (52), respectively. The forward scattering I(0) and the radius of gyration Rg were evaluated using the Guinier approximation (50). These parameters were also computed from the entire scattering patterns using the indirect transform package GNOM (53), which also provided the distance distribution function p(r), which gives the maximal particle diameter, Dmax. Low-resolution models of Gs3α and αchi were built using DAMMIF (54). Ab initio solution shapes were obtained by superposition of 10 independent model reconstructions with the program package SUBCOMP (55) and building an averaged model from the most probable one using DAMAVER (56). Comparison of the experimental scattering curves with the theoretical scattering curve calculated from high resolution structures were performed with CRYSOL (57).
FCS studies of Gsα and αchi
The binding affinities of Gsα and αchi toward ATP or ADP were examined by FCS. Measurements were performed on an LSM510 Meta/ConfoCor 3 microscope (Carl Zeiss, Germany) (58) and data were fitted with a two-component fit from 1 μs to 3.36 s where the diffusion time of the smaller component was fixed to 70 μs (26).
Models on the interaction of αchi with subunits γ and Mtϵ
The structure of Gsα (PDB code 4XD7) (32) was superimposed on to the α-subunits of the bovine mitochondrial F1-ATPase structure (PDB code 1H8E) (59) in all three states, with r.m.s. deviation being 1.55 Å for αE, 1.38 Å for αDP, and 1.01 Å for αTP. Subunit γ of Gs (PDB code 4XD7) (32) was superimposed onto the bovine γ subunit with an r.m.s. deviation of 1.57. The orientation of subunit γ was adjusted manually such as to have 0° rotation in the ground state based on the native structure (PDB code 1BMF) (60). The solution shape of αchi was superimposed onto the Gsα structure in all sites with an NSD of 1.56. The NMR solution structure of the C-terminal peptide Mtα(521–540) was placed in the extra density of the SAXS shape. Similar orientations were maintained for all the three sites of αchi. The Mtϵ was modeled based on the recently determined solution structure of the M. tuberculosis ϵ (Mtϵ) subunit (PDB code 5WY7). Mtϵ was superimposed on to the homologue δ subunit of the mitochondrial bovine F1-ATPase with an r.m.s. deviation of 1.62 Å.
Author contributions
P. R., H. S., L. S., G. B., T. W., W. F., T. D., and G. G. conceived and designed the experiments. P. R., H. S., L. S., G. B., M. S. S. M., D. S., S. K., and G. G. performed the experiments. P. R., H. S., L. S., G. B., and M. S. S. M. analyzed the data. P. R., H. S., L. S., G. B., M. S. S. M., T. W., W. F., T. D., and G. G. wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. J. Martin (School of Life Sciences, AZ State University) and Dr. R. Machan (Centre for Bioimaging Sciences, NUS, Singapore) for support in the single molecule experiments. We thank Dr. Wuan Geok Saw (School of Biological Sciences, NTU, Singapore) for support with the FCS experiments. We thank Prof. Dr. M. Futai (Iwate University, Japan) for kindly providing the antibody against subunit β.
This work was supported in part by the Ministry of Education, Singapore Grants MOE2011-T2-2-156 and ARC 18/12, Ministry of Health, Singapore, NMRC Grant CBRG12nov049 (to G. G.), Ministry of Health, Singapore Grant MOE2012-T2-1-101 (to T. W.), and National Institute of Health Grant R01GM097510 (to W. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Video S1 and Table S1.
The atomic coordinates and structure factors (code 5WY7) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TB
- tuberculosis
- ACMA
- 9-amino-6-chloro-2-methoxyacridine
- Gsα
- G. stearothermophilus subunit α
- Mtα
- M. tuberculosis F-ATP synthase subunit α
- TOCSY
- total correlation spectroscopy
- PDB
- Protein Data Bank
- PMF
- proton motive force
- IMV
- inverted membrane vesicle
- APD
- avalanche photo diode
- FCS
- fluorescence correlation spectroscopy
- NSD
- normalized spatial discrepancy
- AES
- allelic exchange substrate
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- r.m.s. deviation
- root mean square deviation
- chi
- chimeric.
References
- 1. Sassetti C. M., Boyd D. H., and Rubin E. J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 2. Cook G. M., Hards K., Vilcheze C., Hartman T., and Berney M. (2014) Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol. Spectr. 2, 10.1128/microbiolspec.MGM2-0015-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu P., Lill H., and Bald D. (2014) ATP synthase in mycobacteria: special features and implications for a function as drug target. Biochim. Biophys. Acta 1837, 1208–1218 [DOI] [PubMed] [Google Scholar]
- 4. Santana M., Ionescu M. S., Vertes A., Longin R., Kunst F., Danchin A., and Glaser P. (1994) Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J. Bacteriol. 176, 6802–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cox R. A., and Cook G. M. (2007) Growth regulation in the mycobacterial cell. Curr. Mol. Med. 7, 231–245 [DOI] [PubMed] [Google Scholar]
- 6. Haagsma A. C., Driessen N. N., Hahn M. M., Lill H., and Bald D. (2010) ATP synthase in slow- and fast-growing mycobacteria is active in ATP synthesis and blocked in ATP hydrolysis direction. FEMS Microbiol. Lett. 313, 68–74 [DOI] [PubMed] [Google Scholar]
- 7. Cook G. M., Keis S., Morgan H. W., von Ballmoos C., Matthey U., Kaim G., and Dimroth P. (2003) Purification and biochemical characterization of the F1Fo-ATP synthase from thermoalkaliphilic Bacillus sp. strain TA2.A1. J. Bacteriol. 185, 4442–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hicks D. B., and Krulwich T. A. (1990) Purification and reconstitution of the F1F0-ATP synthase from alkaliphilic Bacillus firmus OF4: evidence that the enzyme translocates H+ but not Na+. J. Biol. Chem. 265, 20547–20554 [PubMed] [Google Scholar]
- 9. Hoffmann A., and Dimroth P. (1990) The ATPase of Bacillus alcalophilus: purification and properties of the enzyme. Eur. J. Biochem. 194, 423–430 [DOI] [PubMed] [Google Scholar]
- 10. Pérez J. A., and Ferguson S. J. (1990) Kinetics of oxidative phosphorylation in Paracoccus denitrificans: 1. mechanism of ATP synthesis at the active site(s) of F0F1-ATPase. Biochemistry 29, 10503–10518 [DOI] [PubMed] [Google Scholar]
- 11. Pacheco-Moisés F., Minauro-Sanmiguel F., Bravo C., and García J. J. (2002) Sulfite inhibits the F1F0-ATP synthase and activates the F1F0-ATPase of Paracoccus denitrificans. J. Bioenerg. Biomembr. 34, 269–278 [DOI] [PubMed] [Google Scholar]
- 12. Zharova T. V., and Vinogradov A. D. (2003) Proton-translocating ATP-synthase of Paracoccus denitrificans: ATP-hydrolytic activity. Biochemistry 68, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 13. Stocker A., Keis S., Vonck J., Cook G. M., and Dimroth P. (2007) The structural basis for unidirectional rotation of thermoalkaliphilic F1-ATPase. Structure 15, 904–914 [DOI] [PubMed] [Google Scholar]
- 14. Zarco-Zavala M., Morales-Ríos E., Mendoza-Hernández G., Ramírez-Silva L., Pérez-Hernández G., and García-Trejo J. J. (2014) The ζ subunit of the F1FO-ATP synthase of α-proteobacteria controls rotation of the nanomotor with a different structure. FASEB J. 28, 2146–2157 [DOI] [PubMed] [Google Scholar]
- 15. Preiss L., Langer J. D., Yildiz Ö., Eckhardt-Strelau L., Guillemont J. E., Koul A., and Meier T. (2015) Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline. Sci. Adv. 1, e1500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okuno D., Iino R., and Noji H. (2011) Rotation and structure of FoF1-ATP synthase. J. Biochem. 149, 655–664 [DOI] [PubMed] [Google Scholar]
- 17. Pänke O., Gumbiowski K., Junge W., and Engelbrecht S. (2000) F-ATPase: specific observation of the rotating c subunit oligomer of EF(o) EF(1). FEBS Lett. 472, 34–38 [DOI] [PubMed] [Google Scholar]
- 18. von Ballmoos C., Wiedenmann A., and Dimroth P. (2009) Essentials for ATP synthesis by F1F0 ATP synthases. Annu. Rev. Biochem. 78, 649–672 [DOI] [PubMed] [Google Scholar]
- 19. Adachi K., Yasuda R., Noji H., Itoh H., Harada Y., Yoshida M., and Kinosita K. (2000) Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl. Acad. Sci. U.S.A. 97, 7243–7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yasuda R., Noji H., Yoshida M., Kinosita K. Jr., and Itoh H. (2001) Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 [DOI] [PubMed] [Google Scholar]
- 21. Nishizaka T., Oiwa K., Noji H., Kimura S., Muneyuki E., Yoshida M., and Kinosita K. Jr. (2004) Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 11, 142–148 [DOI] [PubMed] [Google Scholar]
- 22. Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K. Jr., and Yoshida M. (2003) Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40 degree substep rotation. Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hotra A., Suter M., Biuković G., Ragunathan P., Kundu S., Dick T., and Grüber G. (2016) Deletion of a unique loop in the mycobacterial F-ATP synthase γ subunit sheds light on its inhibitory role in ATP hydrolysis-driven H+ pumping. FEBS J. 283, 1947–1961 [DOI] [PubMed] [Google Scholar]
- 24. Andries K., Verhasselt P., Guillemont J., Göhlmann H. W., Neefs J. M., Winkler H., Van Gestel J., Timmerman P., Zhu M., Lee E., Williams P., de Chaffoy D., Huitric E., Hoffner S., Cambau E., Truffot-Pernot C., Lounis N., and Jarlier V. (2005) A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227 [DOI] [PubMed] [Google Scholar]
- 25. Haagsma A. C., Abdillahi-Ibrahim R., Wagner M. J., Krab K., Vergauwen K., Guillemont J., Andries K., Lill H., Koul A., and Bald D. (2009) Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob. Agents Chemother. 53, 1290–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ho J., Sielaff H., Nadeem A., Svanborg C., and Grüber G. (2015) The molecular motor F-ATP synthase is targeted by the tumoricidal protein HAMLET. J. Mol. Biol. 427, 1866–1874 [DOI] [PubMed] [Google Scholar]
- 27. Sakaki N., Shimo-Kon R., Adachi K., Itoh H., Furuike S., Muneyuki E., Yoshida M., and Kinosita K. Jr. (2005) One rotary mechanism for F1-ATPase over ATP concentrations from millimolar down to nanomolar. Biophys. J. 88, 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spetzler D., York J., Daniel D., Fromme R., Lowry D., and Frasch W. (2006) Microsecond time scale rotation measurements of single F1-ATPase molecules. Biochemistry 45, 3117–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin J. L., Ishmukhametov R., Hornung T., Ahmad Z., and Frasch W. D. (2014) Anatomy of F1-ATPase powered rotation. Proc. Natl. Acad. Sci. U.S.A. 111, 3715–372024567403 [Google Scholar]
- 30. Sielaff H., Martin J., Singh D., Biuković G., Grüber G., and Frasch W. D. (2016) Power stroke angular velocity profiles of archaeal A-ATP synthase versus thermophilic and mesophilic F-ATP synthase molecular motors. J. Biol. Chem. 291, 25351–25363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berjanskii M. V., Neal S., and Wishart D. S. (2006) PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 34, W63–W69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shirakihara Y., Shiratori A., Tanikawa H., Nakasako M., Yoshida M., and Suzuki T. (2015) Structure of a thermophilic F1-ATPase inhibited by an ϵ-subunit: deeper insight into the ϵ-inhibition mechanism. FEBS J. 282, 2895–2913 [DOI] [PubMed] [Google Scholar]
- 33. Rao S. P., Alonso S., Rand L., Dick T., and Pethe K. (2008) The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 105, 11945–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Biukovic G., Basak S., Manimekalai M. S., Rishikesan S., Roessle M., Dick T., Rao S. P., Hunke C., and Grüber G. (2013) Variations of subunit epsilon of the Mycobacterium tuberculosis F1Fo ATP synthase and a novel model for mechanism of action of the tuberculosis drug TMC207. Antimicrob. Agents Chemother. 57, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Priya R., Biuković G., Manimekalai M. S., Lim J., Rao S. P., and Grüber G. (2013) Solution structure of subunit γ (γ(1–204)) of the Mycobacterium tuberculosis F-ATP synthase and the unique loop of γ(165–178), representing a novel TB drug target. J. Bioenerg. Biomembr. 45, 121–129 [DOI] [PubMed] [Google Scholar]
- 36. Adachi K., Oiwa K., Nishizaka T., Furuike S., Noji H., Itoh H., Yoshida M., and Kinosita K. Jr. (2007) Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 [DOI] [PubMed] [Google Scholar]
- 37. Sambrook J., Fritsch E. F., and Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Parish T., and Brown A. C. (2008) Methods in Molecular Biology. Mycobacteria protocols. 2 Ed., Vol. 465, Humana Press, New York [Google Scholar]
- 39. Goude R., and Parish T. (2008) Electroporation of Mycobacteria. in Methods in Molecular Biology. Mycobacteria protocols. Vol. 564, 203–215, Humana Press, New York: [DOI] [PubMed] [Google Scholar]
- 40. van Kessel J. C., and Hatfull G. F. (2008) Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol. Microbiol. 67, 1094–1107 [DOI] [PubMed] [Google Scholar]
- 41. van Kessel J. C., Marinelli L. J., and Hatfull G. F. (2008) Recombineering mycobacteria and their phages. Nat. Rev. Microbiol. 6, 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grüber G., Godovac-Zimmermann J., Link T. A., Coskun U., Rizzo V. F., Betz C., and Bailer S. M. (2002) Expression, purification, and characterization of subunit E, an essential subunit of the vacuolar ATPase. Biochem. Biophys. Res. Commun. 298, 383–391 [DOI] [PubMed] [Google Scholar]
- 43. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 44. Lötscher H. R., deJong C., and Capaldi R. A. (1984) Interconversion of high and low adenosinetriphosphatase activity forms of Escherichia coli F1 by the detergent lauryldimethylamine oxide. Biochemistry 23, 4140–4143 [DOI] [PubMed] [Google Scholar]
- 45. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 46. Balakrishna A. M., Basak S., Manimekalai M. S., and Grüber G. (2015) Crystal structure of subunits D and F in complex gives insight into energy transmission of the eukaryotic V-ATPase from Saccharomyces cerevisiae. J. Biol. Chem. 290, 3183–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dip P. V., Kamariah N., Subramanian Manimekalai M. S., Nartey W., Balakrishna A. M., Eisenhaber F., Eisenhaber B., and Grüber G. (2014) Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 70, 2848–2862 [DOI] [PubMed] [Google Scholar]
- 48. Tay M. Y., Saw W. G., Zhao Y., Chan K. W., Singh D., Chong Y., Forwood J. K., Ooi E. E., Grüber G., Lescar J., Luo D., and Vasudevan S. G. (2015) The C-terminal 50 amino acid residues of dengue NS3 protein are important for NS3-NS5 interaction and viral replication. J. Biol. Chem. 290, 2379–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kamariah N., Nartey W., Eisenhaber B., Eisenhaber F., and Grüber G. (2016) Low resolution solution structure of an enzymatic active AhpC10:AhpF2 ensemble of the Escherichia coli alkyl hydroperoxide reductase. J. Struct. Biol. 193, 13–22 [DOI] [PubMed] [Google Scholar]
- 50. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 51. Guinier A., and Fournet G. (1955) Small-angle Scattering of X-rays. Wiley, New York [Google Scholar]
- 52. Glatter O., and Kratky O. (1982) Small-angle X-ray Scattering. Academic Press, London, UK: [DOI] [PubMed] [Google Scholar]
- 53. Svergun D. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 54. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kozin M. B., and Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 56. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL, a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 58. Hunke C., Chen W. J., Schäfer H. J., and Grüber G. (2007) Cloning, purification, and nucleotide-binding traits of the catalytic subunit A of the V1VO ATPase from Aedes albopictus. Protein Expr. Purif. 53, 378–383 [DOI] [PubMed] [Google Scholar]
- 59. Menz R. I., Walker J. E., and Leslie A. G. (2001) Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell 106, 331–341 [DOI] [PubMed] [Google Scholar]
- 60. Abrahams J. P., Leslie A. G., Lutter R., and Walker J. E. (1994) Structure at 2.8-Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 61. Bond C. S., and Schüttelkopf A. W. (2009) ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D Biol. Crystallogr. 65, 510–512 [DOI] [PubMed] [Google Scholar]
- 62. DeLano W. (2002) The PyMOL Molecular Graphics System, Schroedinger, LLC, New York [Google Scholar]
- 63. Watanabe R., and Noji H. (2014) Timing of inorganic phosphate release modulates the catalytic activity of ATP-driven rotary motor protein. Nat. Commun. 5, 3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.