Abstract
The let-7/Lin28 axis is associated with the regulation of key cellular regulatory genes known as microRNAs in various human disorders and cancer development. This study evaluated the role of the let-7/Lin28 axis in regulating a mesenchymal phenotype of hepatic stellate cells in alcoholic liver injury. We identified that ethanol feeding significantly down-regulated several members of the let-7 family in mouse liver, including let-7a and let-7b. Similarly, the treatment of human hepatic stellate cells (HSCs) with lipopolysaccharide (LPS) and transforming growth factor-β (TGF-β) significantly decreased the expressions of let-7a and let-7b. Conversely, overexpression of let-7a and let-7b suppressed the myofibroblastic activation of cultured human HSCs induced by LPS and TGF-β, as evidenced by repressed ACTA2 (α-actin 2), COL1A1 (collagen 1A1), TIMP1 (TIMP metallopeptidase inhibitor 1), and FN1 (fibronectin 1); this supports the notion that HSC activation is controlled by let-7. A combination of bioinformatics, dual-luciferase reporter assay, and Western blot analysis revealed that Lin28B and high-mobility group AT-hook (HMGA2) were the direct targets of let-7a and let-7b. Furthermore, Lin28B deficiency increased the expression of let-7a/let-7b as well as reduced HSC activation and liver fibrosis in mice with alcoholic liver injury. This feedback regulation of let-7 by Lin28B is verified in hepatic stellate cells isolated by laser capture microdissection from the model. The identification of the let-7/Lin28 axis as an important regulator of HSC activation as well as its upstream modulators and down-stream targets will provide insights into the involvement of altered microRNA expression in contributing to the pathogenesis of alcoholic liver fibrosis and novel therapeutic approaches for human alcoholic liver diseases.
Keywords: fibroblast, gene knockout, hepatic stellate cell (HSC), liver injury, microRNA (miRNA), HMGA2, alcoholic liver diseases, let-7, lin28
Introduction
Alcoholic steatohepatitis with steatosis (fatty liver) and perisinusoidal fibrosis represents a crucial step in progression of alcoholic liver disease (ALD)4 (1, 2). Despite the importance of liver fibrosis in the disease process, the causes of fibrosis and diminished regeneration, especially in liver cirrhosis, remain poorly understood. Enhanced mesenchymal reactions are associated with liver fibrosis (3) and are characterized as generation of myofibroblasts (4–6).
Repair of the liver occurs after injury through a coordinated interplay of resident and infiltrating cells. However, if the injury is too great to repair at a similar or greater rate than the injury, scarring ensues in the form of fibrosis (7). The main cell types that are involved in liver fibrosis are hepatic stellate cell (HSCs) (8). HSCs are liver-specific mesenchymal cells that play a critical role in liver injury and fibrosis. HSCs are typically quiescent, vitamin A-storing pericytes (9). However, when the liver is injured, HSCs are activated to become myofibroblastic cells (10). Activated HSCs travel to the site of an injury to secrete extracellular matrix (ECM) for dividing cells to attach (11). If the liver is unable to replace the damaged cells, HSCs deposit an excessive amount of ECM causing a fibrotic scar. The mechanisms by which the liver reacts to alcohol are being studied extensively. One such mechanism is the activity of the small regulatory non-coding RNAs, microRNAs (miRNAs). These small non-coding RNAs can regulate genes by direct binding to DNA or mRNA (12). We previously showed that let-7 is important in alcoholic and chronic liver injury; we also showed that the reduction of let-7 increases the severity of fibrosis and other injuries (13–15). Several variants of let-7, let-7a, let-7b, and let-7g are down-regulated in the serum of ethanol-fed mice (16). The potential target genes of let-7 family include Lin28, a key promoter of mesenchymal reaction, cell migration, and generation of induced pluripotent stem cells (17). Recent studies show that miR-200c, another miRNA that is regulated by Lin28, may regulate the mesenchymal reaction in cancer. Overexpression of miR-200c inhibits mesenchymal reaction and supports termination of cancer progression (18, 19). It has been hypothesized that the mesenchymal phenotypes regulate the ability to either recover from injury or progress to a cirrhotic condition. More mesenchymal reactions promote liver fibrosis, whereas inhibiting them triggers liver healing and regeneration (20).
The let-7/Lin28 axis regulates hepatocellular carcinoma development (16, 21) and is associated with an increased risk of type 2 diabetes mellitus (22). However, the functional role of let-7/Lin28 in alcoholic liver injury remains unclear. Thus, we assessed the role of aberrant expression of the let-7/Lin28 axis in HSCs in ALD by posing the following four questions. (i) Is let-7/Lin28 expression altered in ethanol-exposed mice and ALD human liver tissues? (ii) Does modulation of let-7/Lin28 alter activation of HSCs in vitro and in animals with ALD? (iii) What is the upstream regulator of let-7/Lin28 in HSCs in ALD? (iv) What are downstream targets of let-7/Lin28 involved in HSC activation in ALD?
Results
Presence of enhanced mesenchymal phenotypes due to long-term ethanol exposure in mice liver
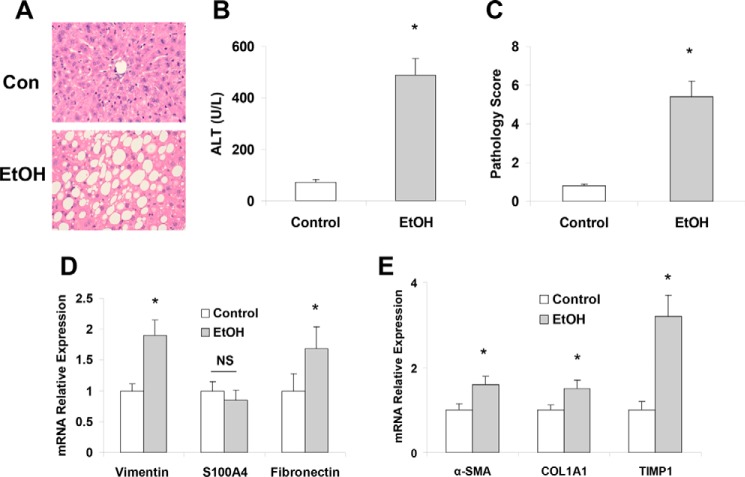
Recent studies from the chronic-plus-binge ethanol feeding model suggest that binge drinking induces significant activation of HSCs, leading to liver fibrosis in chronic ethanol-fed mice (25). To test the impact of chronic alcohol drinking on hepatic mesenchymal phenotypes and liver injury, we analyzed the role of the mesenchymal markers vimentin, S100A4, and fibronectin in mouse livers with chronic alcoholic liver injury. Eight weeks of intragastric feeding of ethanol (∼27 g/kg/day) induced significant liver damage (Fig. 1A) and increased in serum alanine aminotransferase levels compared with control mice (Fig. 1B) along with significantly enhanced expression of fibrosis markers such as Vim (vimentin), Fn1 Acta2, Col1a1, and Timp1 (Fig. 1, D and E). These results along with liver histologic findings (Fig. 1C) indicate that long-term ethanol feeding induced liver fibrotic responses.
Figure 1.
Intragastric ethanol feeding induced liver injury and increased expression of mesenchymal and fibrotic markers. A, C57Bl/6J mice were fed either a control (Con) or ethanol diet for 8 weeks (n = 5). Livers were harvested, fixed in formalin, and embedded in paraffin. A number of 6-mm sliced liver sections were affixed to glass slides, rehydrated, and stained with hematoxylin and eosin H&E to evaluate gross histology. The H&E histology of control (upper) and ethanol fed mice (lower) showed increased steatosis and liver injury in ethanol-fed mice. B, serum was extracted from both control-fed and ethanol-fed mice, and ALT levels were measured. The ALT levels were shown to be increased in ethanol-fed mice, correlating the increased steatosis seen in panel A. C, the H&E histology was scored by three separate observers from one (no damage) to 10 (severe damage) to quantify the level of liver damage based on the levels of steatosis, hepatic ballooning, loss of liver structure, etc. It was found after combining the scores from the three observers that the histopathology scores were significantly higher in the ethanol-fed mice. D, the levels of the mesenchymal markers (vimentin, S100A4, and fibronectin) in total liver samples were measured with real-time PCR control and ethanol-fed mice. Mesenchymal markers were shown to be elevated in ethanol-fed mice compared with control. E, the fibrosis markers (α-SMA, Col1A1, and TIMP1) were evaluated in total liver samples derived from control and ethanol-fed mice. Ethanol-fed mice were shown to have significantly increased levels of these markers compared with control-fed mouse livers. Data are presented as ± S.E. (n = 5). *, p < 0.05 relative to controls. NS, no significant difference.
Altered let-7/Lin28 axis in ethanol-exposed mice liver tissues
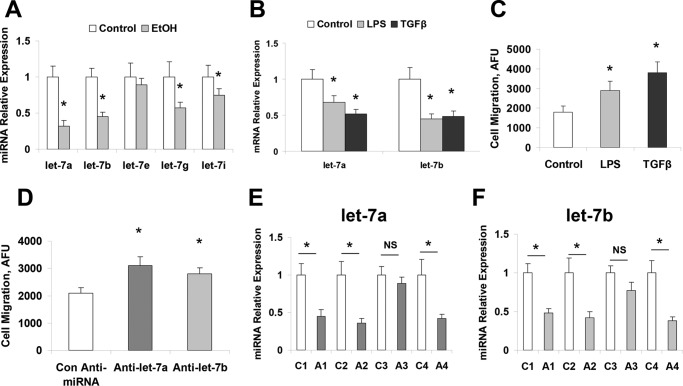
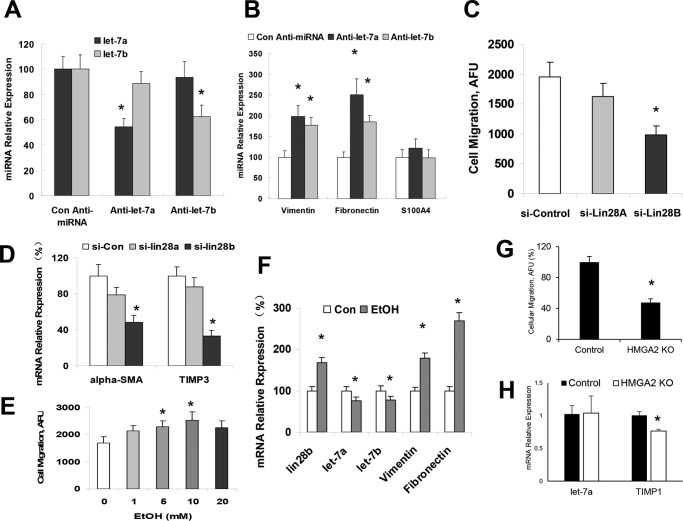
We have demonstrated the aberrant expression of selected miRNAs after alcoholic liver injury, including the significant reduction of let-7 family members (2, 13, 26). To further extend these observations in alcoholic liver injury, we analyzed the expression of let-7 family members in five pairs of ethanol-treated mouse and control liver tissues using the Taqman real-time PCR assay. The expressions of several let-7 miRNAs (including let-7a, let-7b, and let-7g) were markedly decreased (more than half) in ethanol-exposed liver tissues compared with control liver tissues (Fig. 2A). The expression of let-7a and let-7b was also decreased in lipopolysaccharide (LPS) and transforming growth factor β (TGF-β)-treated hHSCs with a higher migration rate compared with controls (Fig. 2, B and C). Inhibition of let-7a and let-7b in hHSCs also significantly increased cell migration rate (Fig. 2D). To further verify the expression of let-7 in human ALD, real-time PCR analysis was performed for total RNA from patient ALD and normal human liver tissues obtained from XenoTech (Kansas City, KS). The expression of let-7a and let-7b was decreased by 50% or more in 3 of the 4 ALD samples compared with the control liver tissues (Fig. 2, E and F). These results indicate that reduced expression of let-7 is a common event in clinical and experimental ALD.
Figure 2.
let-7 levels affected migration and levels were decreased upon exposure to LPS, TGF-β, or EtOH. These hHSCs were cultured as recommended by the manufacturer on poly-l-lysine-coated plates. All hHSCs at 70–80% confluency were treated with 10 mm ethanol, 1 ng/ml TGF-β, or LPS (1 μg/ml) for 72 h. Cells were collected, and total RNA was extracted. Real-time PCR was performed to evaluate the levels of let-7 after treatment with ethanol (A), LPS, or TGF-β (B). Cellular migration was analyzed in hHSCs with the Oris Cell Migration assay kit after LPS or TGF-β treatment (C) or let-7 knockdown (D). Migrating cells were positively stained by calcein AM, and levels of migrating cells were quantified with a spectrophotometer. The presence of LPS, TGF-β, and let-7 inhibition increased the migration ability of hHSCs. E and F, total liver samples from human alcoholic liver disease or control patients were analyzed for let-7 levels with real-time PCR (E and F). Alcoholic patient liver samples showed significant reductions in both let-7a and let-7b. Data are presented as ± S.E. (n = 5). *, p < 0.05 relative to specific control group. NS, no significant difference.
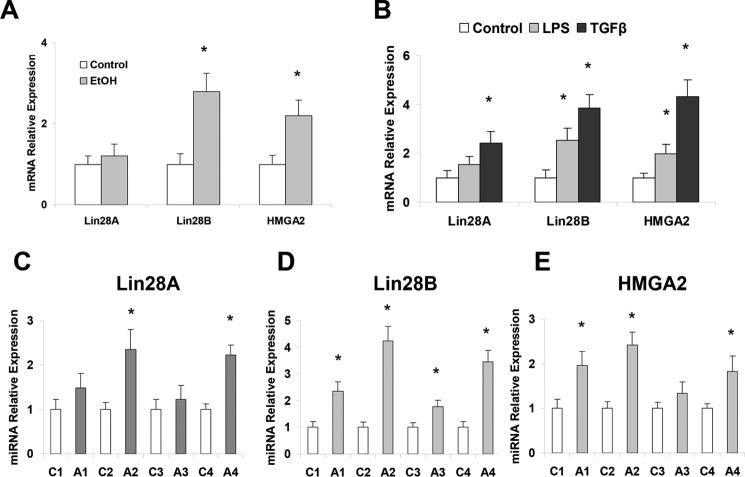
A main target gene of let-7 is an RNA-binding protein and stem cell marker, Lin28. Elevation of Lin28 and decrease of let-7 are linked with highly aggressive diseases in other organs, and their importance in liver diseases is also suggested (16). Significantly increased expression of Lin28B (but not Lin28A) was observed in ethanol-fed mouse livers relative to controls along with the enhanced expression of another let-7 target gene, HMGA2 (27) (Fig. 3A). Additionally, both Lin28B and HMGA2 are significantly increased in LPS- and TGFβ-treated hHSCs (Fig. 3B) as well as in human ALD samples, suggesting the potential role of the let-7/Lin28B axis in human ALD and liver fibrosis (Fig. 3, C–E).
Figure 3.
Ethanol, LPS or TGF-β exposure increased Lin28 and HMGA2 levels. These hHSCs were cultured as recommended by the manufacturer on poly-l-lysine-coated plates. The HSCs at 70–80% confluency were treated with 10 mm ethanol as well as 1 ng/ml TGF-β or LPS (1 μg/ml) for 72 h. Cells were collected, and total RNA was extracted. The Lin28A/B and HMGA2 levels were measured by real-time PCR in hHSCs treated with ethanol (A) and LPS or TGF-β (B). The Lin28A/B and HMGA2 levels were all elevated in hHSCs treated with LPS, TGF-β, and ethanol. C–E, total liver samples from human alcoholic liver disease or control patients were analyzed for Lin28A/B and HMGA2 levels with real-time PCR. Data are presented as ±S.E. (n = 5). *, p < 0.05 relative to specific control group.
Regulation of Lin28B and HMGA2 by let-7 in hHSCs
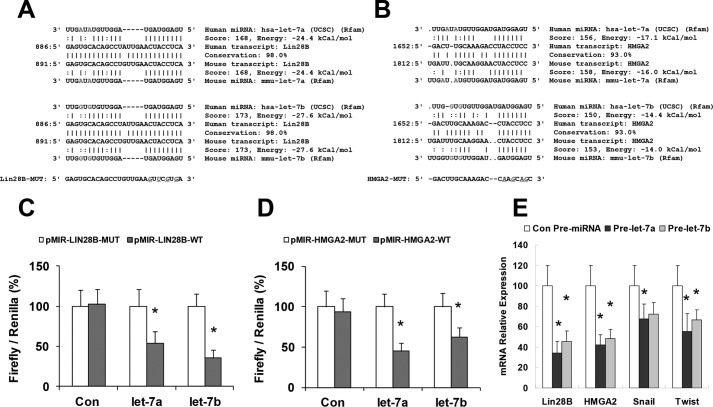
To verify that Lin28B and HMGA2 are bona fide targets of translational regulation by let-7 in hHSCs, we performed studies using luciferase reporter constructs containing the let-7a/let-7b recognition sequence from the 3′-UTR of Lin28B and HMGA2 inserted downstream of the luciferase gene (Fig. 4, A and B). Transfection with let-7a/let-7b precursor in hHSCs decreased reporter activity of both Lin28B and HMGA2 in normal hHSCs. However, when these studies were repeated with reporter constructs that contained random mutations in the recognition sequence, the effects of reporter deactivation by let-7a/let-7b precursor were abolished (Fig. 4, C and D). Moreover, reductions in Lin28B and HMGA2 expression occurred in hHSCs incubated with pre-let-7a/let-7b for 3 days (Fig. 4E). Western blot analysis also demonstrated significant reduction of Lin28B (0.40 ± 0.08 (Pre-let-7a, p = 0.01), 0.62 ± 0.10 (Pre-let-7b, p = 0.03)) and HMGA2 (0.55 ± 0.09 (Pre-let-7a, p = 0.03), 0.44 ± 0.07 (Pre-let-7b, p = 0.02)) relative to pre-miRNA controls, respectively (n = 6). Concomitant with reduced HMGA2 expression, there were significant repressions of the snail family transcriptional repressor (Snail) and the twist family basic helix-loop-helix transcription factor (Twist), which are the known downstream mediators of HMGA2 mesenchymal signaling (Fig. 4E). In contrast, transfection with a precursor to miR-21 (which can also modulate mesenchymal phenotypes in normal hHSCs) did not alter the expression of Lin28B and HMGA2 with a relative expression of 1.2 ± 0.2-fold and 0.9 ± 0.4-fold of controls, respectively. Taken together, these findings confirm that Lin28B and HMGA2 are the biologically relevant targets of let-7a/let-7b in hHSCs.
Figure 4.
let-7 was increased upon blockage of Lin28B or HMAG2 and blockage of let-7 can alter mesenchymal mediators. The intact let-7a and let-7b recognition sequences from the 3′-UTR of Lin28B and HMGA2 or with random mutations were cloned downstream of the firefly luciferase reporter gene (A and B). Luciferase assays were performed 72 h after transfection of pre-let-7a, pre-let-7b, and control pre-miRNA. The let-7 levels were analyzed with real-time PCR after these constructs were used to block Lin28B (C) and HMGA2 (D). The let-7 levels were increased after knockdown of either Lin28B or HMGA2. E, overexpression plasmids for let-7a/b were transfected into hHSCs by electrophoresis, cells were harvested after 48 h, total RNA was extracted, and Lin28 and HMGA2 (as well as the mesenchymal mediators Snail and Twist) were evaluated with real-time PCR. Overexpression of let-7 induced decreases in both isoforms of Lin28 as well as HMGA2, Snail, and Twist. Data are presented as ± S.E. (n = 5). *, p < 0.05 relative to specific control group.
Regulation of let-7/Lin28-associated mesenchymal phenotypes in hHSCs
We previously demonstrated a role for miR-34a in alcoholic liver injury. However, the role of let-7 in ALD remains unknown. Thus, we performed studies aimed to explore the possible biological significance of aberrant let-7 by using an antisense oligo inhibitor specific to let-7a and let-7b. First, we verified the efficacy of transfection and target effects by assessing the expression of mature let-7a and let-7b by real-time PCR in hHSCs transfected with anti-let-7a and let-7b inhibitors (Fig. 5A). Significant up-regulation of vimentin and fibronectin was seen after anti-let-7a and anti-let-7b treatment (Fig. 5B). We also quantified let-7a/let-7b expression in let-7a/let-7b-transfected cells before and after alcohol exposure and demonstrated that alcohol significantly decreased let-7a and let-7b by 0.6-fold and 0.5-fold in hHSCs, respectively. A prolonged ethanol treatment (1 week) without let-7a/let-7b transfection also reduced let-7a/let-7b expression by 0.4-fold and 0.3-fold, respectively. In these cells the expression of vimentin and fibronectin was up-regulated. These observations indicate a role for let7 in the regulation of mesenchymal phenotypes of hHSCs after alcoholic exposure.
Figure 5.
The Lin28/let-7 axis controlled cellular migration and mesenchymal marker expression. The hHSCs were transiently transfected for inhibitors of let-7 via electrophoresis. Cells were harvested 48 h after transfection, RNA was extracted, and real-time PCR was performed for levels of let-7 (A) and mesenchymal markers (B). Knockdown of let-7 was confirmed in panel A. The silencing of let-7 induced these cells to increase levels of the mesenchymal markers, vimentin, S100A4, and fibronectin. C, the hHSCs were transiently transfected for siRNAs to Lin28A or Lin28B via electrophoresis. Cellular migration was analyzed in Lin28A/B repressed hHSCs with the Oris Cell Migration assay kit. D, the Lin28A/B-repressed cells were harvested 48 h after transfection, RNA was extracted, and real-time PCR was performed for the fibrosis markers α-SMA and TIMP3. Silencing of Lin28B reduced the levels of fibrotic markers. E, the hHSCs were treated with 10 mm ethanol for 72 h, and cellular migration was measured as above. Treatment with ethanol increased cellular migration in a dose-dependent manner. F, the hHSCs treated with ethanol were harvested after 72 h, RNA was extracted, and real-time PCR was performed for Lin28, let-7, and the mesenchymal markers vimentin and fibronectin. Lin28B levels were elevated, let-7 levels were decreased, and vimentin and fibronectin levels were increased after treatment with ethanol. G, HSCs were transfected with siRNA against HMGA2, and cellular migration was measured as above. Migration is presented as a percentage of si-control migration. H, mRNA was extracted from HSCs transfected for si-HMGA2 and let-7a, and TIMP1 levels were measured by qPCR. Data are presented as ±S.E. (n = 5). *, p < 0.05 relative to specific control groups.
The ability to migrate into injured areas of tissues is a phenotypic characteristic of HSC activation and a key determinant of hepatic fibrogenetic process. To assess the effect of Lin28B on cell motility, hHSCs were transfected with either control, Lin28B or HMGA2 siRNAs, and vertical cell migration was assessed. Silencing Lin28B or HMAG2 decreased the cell migration index (Fig. 5, C and G). In addition, silencing of Lin28B significantly decreased the fibrogenic markers ACTA2 and TIMP3 (Fig. 5D). Furthermore, cell motility was significantly increased after ethanol treatment (Fig. 5E). This effect was accompanied by increased Lin28B, decreased let-7a/let-7b, and up-regulated vimentin and fibronectin (Fig. 5F). Silencing of HMGA2 in stellate cells did not alter let-7a levels but decreased TIMP1 expression in HSCs (Fig. 5H). Collectively, these results support a functional role for ethanol-induced Lin28B/let-7 axis in activating hHSC and stimulating their migration.
Reduced mesenchymal phenotypes and liver fibrosis in Lin28B-deficient mice
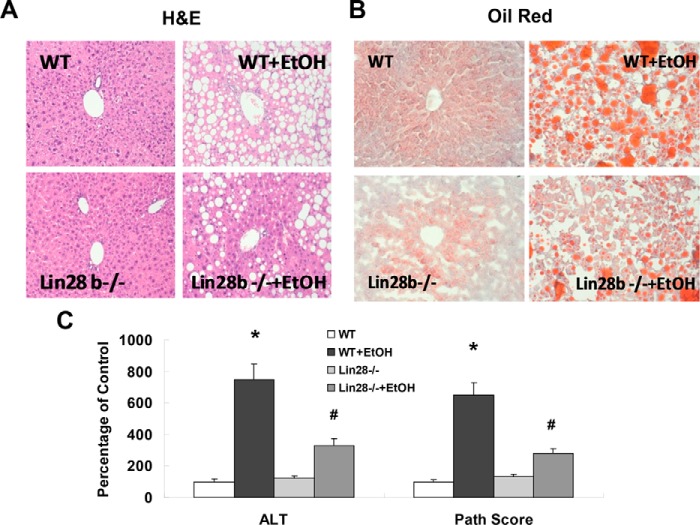
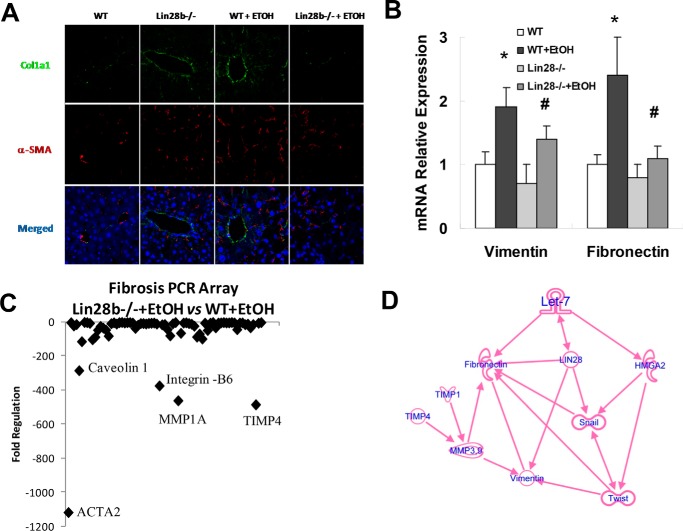
To extend our study on Lin28B to in vivo, we carried out intragastric ethanol feeding for 8 weeks in Lin28B-deficient and WT control mice. Intragastric ethanol feeding induced severe steatosis and changes in liver structure. Alternatively, Lin28B-deficient mice fed an intragastric ethanol diet showed a reduction in the degree of steatosis and liver structure alterations, suggesting that Lin28B promotes during alcoholic liver injury (Fig. 6, A and B). Plasma levels of the liver enzyme alanine aminotransferase and total liver pathology scores were decreased in Lin28B-deficient mice with EtOH compared with WT mice with EtOH (Fig. 6C). Significant reductions in type I collagen and ACTA2 were noted in Lin28B deficient-EtOH mice compared with WT-ETOH mice (Fig. 7A). Furthermore, significant reductions in vimentin and fibronectin mRNA were noted in Lin28B-deficient EtOH mice (Fig. 7B). A fibrosis PCR array also identified obvious reduction of liver fibrosis-related genes such as α-SMA (ACTA2), TIMP-4, MMP-1, intergrin-B6, and caveolin (Fig. 7C). Ingenuity pathway analysis (IPA) was performed to ascertain the cellular context of the differentially expressed genes related to Lin28B deficiency-mediated liver repair. Pathway analysis indicated that the hepatic mesenchymal/fibrotic pathway was most inhibited through let-7/Lin28-related signaling mechanisms (Fig. 7D). Further analysis with IPA uncovered several differentially regulated mesenchymal/fibrotic gene alterations after Lin28B silencing in ethanol-feeding mice liver. Several of these genes are regulated by let-7/Lin28 axis, including vimentin, fibronectin, HMGA2, Twist, Snail, TIMP-1, TIMP-4, MMP-3, and MMP-9. These studies indicate that the loss of Lin28B ameliorates alcoholic liver injury and fibrosis through the inhibition of mesenchymal signaling pathways.
Figure 6.
The recovery effects of Lin28B deficiency in ethanol-feeding mice. The H&E (A) and Oil Red O (B) staining was performed in FFPE and frozen sections, respectively, in wild-type and Lin28B-deficient mice with or without ethanol feeding. Red staining in oil red sections is positive for lipid. The H&E staining showed reduced steatosis and other liver damage in Lin28B-deficient mice. Additionally, oil red staining showed reduced fat deposition in Lin28B-deficient mice. Serum ALT levels and histopathological scores were evaluated in wild-type and Lin28B-deficient mice with or without ethanol feeding (C). The ALT levels and overall histopathology scores were shown to be reduced in Lin28B-deficient mice. Data are presented as ± S.E. (n = 5). *, p < 0.05 relative to WT control group; #, p < 0.05 relative to WT+EtOH group.
Figure 7.
Liver fibrosis and mesenchymal markers are decreased in Lin28B-deficient mice with alcoholic liver injury. Immunofluorescence was used to identify the location of collagen 1A1 (green) and α-SMA (red) in liver sections (A). Real-time PCR was performed in total liver isolated for the mesenchymal markers vimentin and fibronectin (B). These mesenchymal markers were reduced in Lin28B-deficient mice. A fibrosis PCR array (SABiosciences) was used to evaluate levels of fibrotic markers in Lin28B-deficient ethanol-fed mice compared with wild-type ethanol-fed mice (C). Lower levels of markers indicate that these markers were decreased in Lin28B-deficient ethanol-fed mice compared with wild-type ethanol-fed mice. The markers the most down-regulated in Lin28B-deficient mice were ACTA2 (α-SMA), TIMP-4, and MMP-1. Panel D presents IPA of differentially regulated gene network after Lin28B deficiency in ethanol feeding mice. IPA was performed to understand the cellular context of the differentially expressed genes related to the recovery effects of Lin28B silencing during alcoholic liver injury. Several mesenchymal and fibrotic genes implicated in HSC activations are regulated by Lin28B deficiency in vivo, including vimentin, fibronectin, HMGA2, Twist, Snail, TIMP-1, TIMP-4, MMP-3, and MMP-9. Data are presented as ±S.E. (n = 5). *, p < 0.05 relative to WT control group; #, p < 0.05 relative to WT+EtOH group.
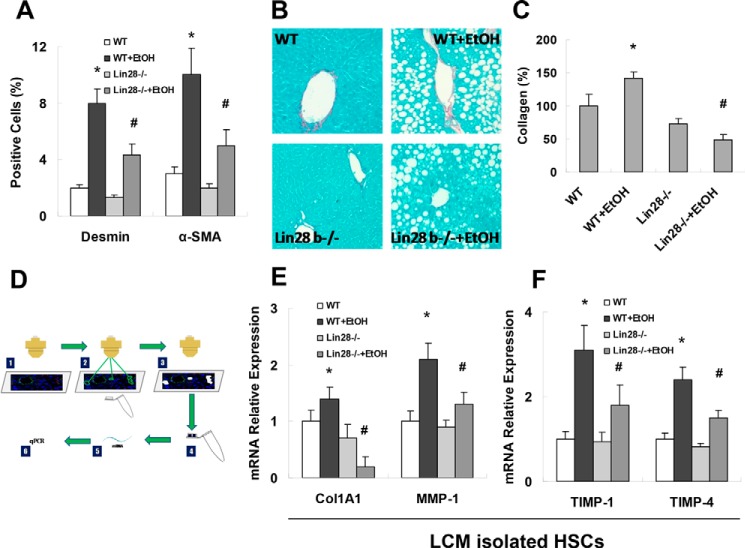
Because activation of HSCs is central in the development of liver fibrosis, we analyzed HSC activation in vivo by staining liver sections for desmin (HSC marker) or α-SMA (marker of activated HSCs) and quantified the percentage of positive cells. Ethanol feeding increased the number of desmin-positive and α-SMA-positive cells in WT mice; these findings were accompanied by increased Sirius Red staining and collagen deposition (Fig. 8, A–C). In Lin28B-deficient mice, EtOH inductions of desmin-positive or α-SMA-positive cells were suppressed compared with WT mice with EtOH; these findings were also accompanied by decreased Sirius Red staining and collagen deposition (Fig. 8, A–C). In HSCs that were isolated from WT-EtOH mice by laser capture microdissection (LCM) (Fig. 8D), we detected increased mRNA expression of Col1a1, MMP-13 (mouse MMP-1 is MMP-13), Timp-1, and Timp-4 when compared with WT without EtOH (Fig. 8, E–F). In contrast, HSCs that were isolated from Lin28B-deficient mice with EtOH had reduced inductions of these parameters compared with WT mice with EtOH (Fig. 8, E and F). No significant differences of fibrosis markers were noted between HSCs isolated from WT controls and Lin28B-deficient controls. These data further support our notion that increased Lin28B contributes to HSC activation and alcoholic liver fibrosis.
Figure 8.
Reduced hepatic stellate cell activation in Lin28B-deficient mice with alcoholic liver injury. Frozen liver sections were stained for desmin with immunofluorescence (A). Quantitative analysis of desmin and α-SMA staining for hepatic stellate cells in wild-type and Lin28B-deficient mice with and without ethanol feeding are displayed. Desmin-positive cells were reduced in Lin28B-deficient mice with or without ethanol feeding compared with their respective controls. Sirius Red staining for collagen deposition was performed in liver sections (B) and quantified with ImageJ software (C). Collagen deposition was shown to be significantly reduced in Lin28B ethanol-fed mice compared with wild-type ethanol-fed mice. The LCM procedure was used to isolate desmin positive hepatic stellate cells. RNA was extracted from hepatic stellate cells, and real-time PCR was performed (D). The fibrosis markers, collagen 1A1, MMP1 (E) TIMP1, and TIMP4 (F) were evaluated with real-time PCR after LCM of desmin-positive cells in liver sections. Collagen 1A1, MMP1, TIMP1, and TIMP4 that were elevated in wild-type ethanol-fed mice was reduced in Lin28B ethanol-fed mice. Data are presented as ±S.E. (n = 6). *, p < 0.05 relative to WT control group; #, p < 0.05 relative to WT+EtOH group.
Identification of feedback loop in HSCs isolated from Lin28B-deficient mice
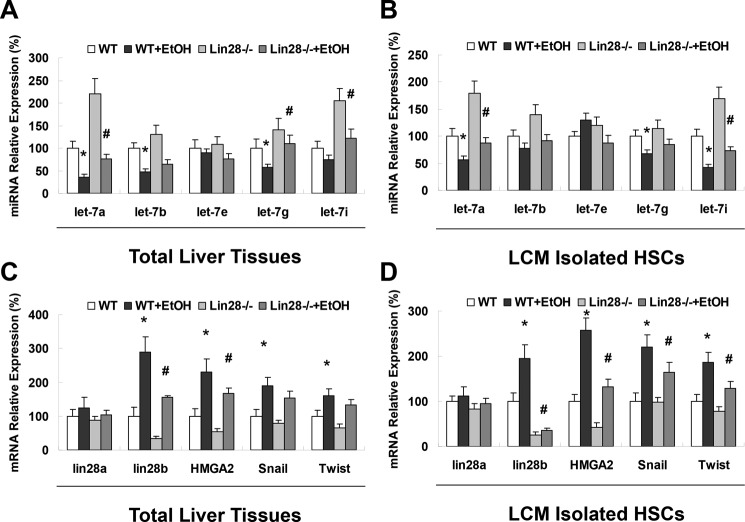
let-7 miRNA can activate TRIM71, which targets Lin28 for ubiquitin-mediated proteasomal degradation, thus inhibiting Lin28 expression at both posttranscriptional and posttranslational levels (28). The Lin28B-let-7-positive feedback loop is discovered in human cancer development and progression and established that Lin28 (which is targeted by let-7) inhibits the biogenesis of let-7 family miRNAs to support Lin28 expression (29, 30). To confirm the feedback loop in HSC in alcoholic liver injury, total liver tissues and LCM-isolated HSCs from WT and Lin28B0-deficient mice (with or without ethanol treatment) were analyzed for the expression of let-7 family members by Taqman real-time PCR assay. Indeed, the expression of let-7a and let-7b was significantly up-regulated in the total liver from Lin28B-deficient mice when compared with WT mice (with or without EtOH respectively) (Fig. 9A). However, only let-7a was significantly up-regulated in LCM-isolated HSCs from Lin28B-deficient mice when compared with WT mice (with or without EtOH, respectively), suggesting the pathologic role of the Lin28B-let-7a-positive feedback in HSC activation and liver fibrosis (Fig. 9B). Furthermore, the expression of let-7 target gene HMGA2 along with its downstream mediators of mesenchymal transition, Snail and Twist, were all significantly down-regulated in Lin28B-deficient mice with EtOH when compared with WT mice with EtOH, further supporting a critical role of a Lin28-let-7a-HMGA2 feedback loop in activation of HSCs in alcoholic liver fibrogenesis (Fig. 9, C and D).
Figure 9.
Lin28B-deficient mice showed increased let-7 levels, decreased HMGA2 levels, and decreased levels of mesenchymal mediators in total liver and isolated hepatic stellate cells. The levels of let-7 (A and B), Lin28, HMGA2, and the mesenchymal mediators Snail and Twist (C and D) were evaluated with real-time PCR in total liver (A and C) and after laser capture microdissection of desmin-positive cells in liver sections (B and D). Data are presented as ±S.E. (n = 6). *, p < 0.05 relative to WT control group; #, p < 0.05 relative to WT+EtOH group.
Discussion
In this study we described the role of altered expression of let-7/Lin28 axis by ethanol in the contribution to mesenchymal phenotypic changes that are associated with ALD progression. The results suggest that decreased let-7a and let-7b is associated with HSC activation in ALD mouse livers and in LPS/TGFβ-treated HSCs compared with controls. We propose that let-7 contributes to anti-alcoholic liver injury and tissue reparation through modulating HSC activation by inhibition of HSC activation markers. Some of these effects are mediated through let-7 targets, Lin28B, and HMGA2, which are all known to be involved in mesenchymal reactions and ALD. Altered expression of let-7/Lin28 was shown by in situ hybridization during hepatocellular carcinoma (HCC) development, and a similar role for let-7/Lin28 has been postulated in the development of type 2 diabetes mellitus. The silencing of Lin28 in Lin28B-deficient mice can inhibit activation of HSCs and subsequently facilitate tissue recovery in ethanol-feeding mice liver. These findings taken together support a functional role for let-7/anti-Lin28 modulation in promoting liver tissue repair and against liver fibrosis during the development of ALD.
The miRNA, let-7, is a vital mediator of liver damage in many liver diseases. It has been shown to be decreased in several forms of acute and chronic liver diseases, but it has not yet been analyzed in ALD. We have shown that administration of ethanol both in vitro and in vivo decreases let-7 levels and increases fibrosis markers and the stem cell marker, Lin28B. In vitro, HSCs treated with the inflammatory proteins LPS or TGF-β exhibited decreased levels of let-7 as well as increased levels of migration, Lin28A/B, HMGA2, and mesenchymal markers. Knock-out of let-7 in HSCs led to increased migration, indicating that a lack of let-7 allows HSCs to become more activated. In human ALD tissues, let-7 levels were decreased, whereas Lin28A/B levels and HMAG2 levels were increased, which indicates that these factors are involved in the severe injury to the liver induced by alcohol consumption. Knock-out of Lin28B or HMGA2 in HSCs decreased cell migration and fibrosis levels. However, silencing of HMGA2 in HSCs failed to change the expression of let-7a, suggesting that HMGA2 is only the direct target of let-7 but not inside the loop of let-7/Lin28. Lin28B-deficient mice showed decreased levels of fibrosis and mesenchymal markers in addition to decreased histopathology scores and decreased alanine aminotransferase levels compared with wild-type ethanol-fed mice. Isolated HSCs from these mice showed decreased levels of fibrosis markers and mesenchymal mediators as well as increased levels of let-7.
It has been noted that ALD is characterized by a typical disease progression that begins at alcoholic fatty liver and eventually progresses to cirrhosis and possible hepatocellular carcinoma. It is unclear what controls the progression of the disease, but it may be tied to the livers of certain patients that have the inability to repair the damage caused by alcohol in a timely manner. The let-7/Lin28 axis is vitally important in the differentiation of cells during development. Cells that have more Lin28 activation have more of a stem cell or mesenchymal phenotype, whereas cells with high levels of let-7 are well-differentiated cells. This is well displayed in HCC. The Lin28B antibody has been shown to be up-regulated in HCC samples, whereas let-7 is down-regulated (31). This reactivation of Lin28B in HCC samples indicates an attempt by the liver to repair the extensive damage caused by cirrhosis that continued uncontrolled.
The let-7/Lin28 axis appears to be an important tool that is utilized by the liver to repair injury and control the transformation of cells from the epithelial to mesenchymal phenotype. Because HSCs need to undergo a mesenchymal transition to become activated and move to the area of injury where they lay down the ECM for dividing cells, the let-7/Lin28 axis could be an important key to this activation. As we have shown, down-regulation of Lin28B may decrease overall liver damage such as steatosis and fibrosis. This fibrosis reduction appears to be a result of blockage of HSC activation through modulation of the mesenchymal mediators Snail and Twist. Therefore, inhibition of Lin28B in patients could be an important therapeutic target for halting the progression of ALD and saving the lives of these patients as well as reducing the cost of care for these patients and the cost of liver transplantation.
Our findings identify a previously unrecognized mechanism for direct regulation of Lin28 and HMGA2 and mesenchymal reactions in HSCs; this mechanism involves non-coding RNA (ncRNA) in ALD. Aberrant mesenchymal reactions have been implicated in many human diseases including ALDs. The abusive consumption of alcohol can cause serious cellular injuries that often lead to activation of mesenchymal transition. Although let-7/Lin28 signaling has been tightly linked to liver injuries and disease outcome in many hepatic disorders, including human ALDs, its application to ethanol-dependent ncRNA expression is novel. A better understanding of how ethanol interacts with specific cytokines to contribute to aberrant ncRNA expression in HSCs will clearly advance the field and increase our understanding of the mechanisms involved in the development of ALDs. Employing LCM and single cell analysis approaches to identify transcriptional and translational factors and their modified targets in HSCs during ALD development and progression are lacking, but such strategies could identify other novel targets that could be genetically or epigenetically modified in ALD.
With chronic alcohol abuse, the early and reversible liver damage occurs in HSCs and other hepatic cell types long before the onset of clinically symptomatic and irreversible form of hepatitis, fibrosis, and cirrhosis. Currently, the mechanisms and regulation of ALDs at single cell levels are poorly understood, and the ALD-associated putative biomarkers and regulatory molecules in HSCs remain tenuous and unproven. In this study we characterized the role of dysregulated let-7/Lin28 axis in ethanol-induced altered mesenchymal signaling in HSCs and Lin28 deficiency to ALD progression. The data presented here may have direct application to the future translational research targeting HSC activation and mesenchymal reactions for an improved diagnosis and treatment of ALD patients.
Experimental procedures
Evaluation of the role of the let-7/Lin28 axis in the mesenchymal reaction in hHSCs
Primary hHSCs were purchased from Sciencell (Carlsbad, CA). Cells were cultured as recommended by the manufacturer. Cells were treated with 10 ml of ethanol (EtOH; Sigma) for 72 h. Cells were harvested with a cell scraper, and RNA was extracted with the mirVana isolation kit (ThermoFisher Scientific, Waltham, MA). The cDNA was made with the iScript reverse transcriptase kit (Bio-Rad). Real-time PCR was performed using the SYBR Green Supermix (Bio-Rad) and premade PCR primers (Qiagen, Germantown, MD) on an Applied Biosystems (Foster City, CA) ViiA 7 PCR machine. For miRNA real-time PCR, cDNA was made with the RT2 reverse transcriptase kit (ThermoFisher) with premade primers (ThermoFisher) and Taqman reagents (ThermoFisher). The HSCs were also treated with 5 ng of TGF-β (R&D Biosystems, Minneapolis, MN) or LPS (Sigma). Cells were harvested, RNA was extracted, and real-time PCR was performed as above.
Transfections
Transfections were performed by nuclear electroporation or Lipofectamine 3000 (ThermoFisher) using the Nucleofector system (Amaxa Biosystems, Koln, Germany). For electroporation, 50 μl of 100 nm miRNA precursor, antisense inhibitor, or controls (Ambion, Austin, TX) were added to 1 × 106 cells suspended in 50 μl of Nucleofector solution at room temperature. The sequences of the miRNA precursors and inhibitors used can be obtained from Ambion. After electroporation, transfected cells were resuspended in culture medium containing 10% fetal bovine serum for 48–72 h before study. Lipofectamine transfections were performed according to the manufacturer's instructions using siRNA obtained from ThermoFisher. All studies were performed in quadruplicate unless otherwise specified.
Real-time PCR for mature miRNA
The expression of mature miRNAs in human hepatic cell lines was analyzed by TaqMan miRNA Assay (Applied Biosystems). Briefly, single-stranded cDNA was synthesized from 10 ng of total RNA in a 15-μl reaction volume by using the TaqMan MiRNA reverse transcription kit (Applied Biosystems). The reactions were initially incubated at 16 °C for 30 min and then at 42 °C for 30 min. The reactions were inactivated by incubation at 85 °C for 5 min. Each cDNA generated was amplified by quantitative PCR via sequence-specific primers from the TaqMan miRNA Assays on an MX 3000P PCR Instrument (Stratagene, San Diego, CA). The 20-μl PCR included 10 μl of 2 × Universal PCR Master Mix (No AmpErase UNG), 2 μl of each 10× TaqMan MiRNA Assay Mix, and 1.5 μl of reverse transcription product. The reactions were incubated in a 96-well plate at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The threshold cycle (CT) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold.
SuperArray real-time PCR array and real-time PCR analysis
The RNA was isolated from liver tissues or cell lysates using TRIzol (Invitrogen) according to the manufacturer's protocol; after isolation, the RNA was subsequently cleaned using the Qiagen RNeasy kit, again according to the manufacturer's protocol. The optional on-column DNase treatment was performed. Reverse transcription was done using 1 μg of RNA with the SABiosciences RT2 First Strand kit (SABiosciences, Frederick, MD) according to the manufacturer's protocol. Mouse liver tissue or normal cDNA from hHSCs was analyzed using SuperArray plates Mouse Fibrosis PCR array (#PAMM-120A; SABiosciences). To validate the translational significance of these gene expression findings, mice liver and human HSC samples were analyzed using real-time PCR. The SABiosciences RT2 Real-Time PCR Primer Assays or Taqman miRNA PCR assay were used. Real-time PCR was performed using the SABiosciences RT2 SYBR Green/ROX Real-Time PCR Master Mix for a Stratagene Mx3005P Real-time PCR System according to the manufacturer's protocol. ROX was used as an endogenous reference, and data were analyzed using the SABiosciences PCR Array Data Analysis Template. The comparative CT method was used for quantification of gene expression. All samples were tested in triplicate, and average values were used for quantification.
Animal studies
All animal studies were reviewed and approved by the Animal Care and Use Committees of Baylor Scott & White and the University of Southern California according to the National Research Council's Guide for the Care and Use of Laboratory Animals. Lin28atm1.2Gqda/J (Lin28B KO) and C57BL/6J (wild type) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Animals were treated with the intragastric EtOH infusion model as previously described (23). After euthanasia, livers were harvested, part of the liver was fixed with formalin and embossed in paraffin, and the remaining liver was flash-frozen and stored at −80 °C. Evaluation of Lin28A/B and let-7 levels was performed by extracting RNA with the miRVana kit, and real-time PCR was performed as above.
Western blotting
Cells grown in 100-mm dishes were lysed, and protein content was quantitated using the Bradford protein assay. Equivalent amounts of protein were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked and incubated with the specific primary antibody (rabbit anti-Lin28B and goat anti-high-mobility group AT-hook (HMGA2); Santa Cruz Biotechnology, Dallas, TX) overnight at 4 °C, washed, and incubated with the appropriate IRDye700- and IRDye800-labeled secondary antibodies (Rockland, Gilbertsville, PA) (1:1000) for 1 h. Blots were stripped and reprobed with mouse monoclonal antibodies for β-actin (Sigma) (1:1000) or total glyceraldehyde-3-phosphate dehydrogenase antibody (1:1000), which was used for normalization. Protein expression was visualized and quantified using the LI-COR Odyssey Infrared Imaging System (LI-COR Bioscience, Lincoln, NE).
Luciferase reporter assay
Intact putative let-7a and let-7b recognition sequence from the 3′-UTR of Lin28B and HMGA2 (pMIR-Lin28B/HMGA2-wt-3′-UTR) or with random mutations (pMIR- Lin28B/HMGA2-mut-3′-UTR) were cloned downstream of the firefly luciferase reporter gene. Cells were co-transfected with 1 μg of pMIR- Lin28B/HMGA2-wt or mut-3′-UTR construct and 1 μg of pRL-TK Renilla luciferase expression construct with or without precursor miR-34a using TransIT-siQUEST transfection reagent (Mirus, Madison, WI). Luciferase assays were performed 72 h after transfection using the Dual Luciferase Reporter Assay system (Promega, Madison, WI).
Cell migration assays
Cell migration was assessed by using the Oris Cell Migration assay kit (Platypus Technologies, Madison, WI). Briefly, cells were stained with calcein AM (Calbiochem) and plated (10,000 cells/well) in a 96-well plate provided with an insert that prevented the cells from attaching in a central analytic zone. After overnight incubation at 37 °C, medium was changed to serum-free medium containing the compounds, and the insert was removed to allow cells to migrate into the central zone. The cells that had migrated into the central zone were labeled by calcein AM and quantified fluorometrically by using SpectraMax M5 Plate Reader from Molecular Devices (Sunnyvale, CA).
Immunohistochemistry analysis
Lobular necrosis was evaluated in liver sections stained with hematoxylin-eosin. Liver sections were incubated overnight at 4 °C with specific antibody (1:50), washed in phosphate buffered saline (PBS), incubated for 20 min at room temperature with a secondary biotinylated antibody (Dako Cytomation LSAB Plus System-HRP, Glostrup, Denmark), and further incubated with Dako ABC for 20 min and developed with 3–3′ diaminobenzidine (Dako Cytomation Liquid DAB Plus Substrate Chromogen System). For all immunoreactions, negative controls were included. Immunohistochemical observations were taken by BX-51 light microscopy (Olympus, Tokyo, Japan) with a Videocam (Spot Insight; Diagnostic Instrument, Sterling Heights, MI) and processed with an image analysis system.
Evaluation of liver steatosis and fat deposition
Formalin fixed paraffin-embedded (FFPE) sections were sectioned at 6 μm and affixed to charged glass slides. Slides were rehydrated in xylenes followed by decreasing percentages of ethanol. Hematoxylin and eosin (H&E) staining was performed by immersing slides in gill's hematoxylin (Vector Laboratories, Burlingame, CA) and subsequently eosin before dehydration slides through elevating alcohol washes and xylenes. The Oil Red O kit (IHC World, Woodstock, MD) was used to stain slides for fat deposition following directions from the manufacturer and dehydrated as above.
Evaluation of liver fibrosis
The FFPE sections were sectioned at 6 μm and affixed to charged glass slides. Slides were rehydrated in xylenes followed by decreasing percentages of ethanol. The Sirius Red kit (IHC World, Woodstock, MD) was used to stain slides for collagen deposition following directions from the manufacturer and dehydrated as above. Sirius Red staining was quantified with ImageJ software (24). Immunofluorescence was performed on FFPE liver sections using antibodies from ABCam (San Francisco, CA). Real-time PCR was performed on total liver RNA isolates as described above.
Isolation of HSCs and evaluation of mesenchymal and fibrotic markers
Frozen liver tissue was sectioned at 10 μm and stained with immunofluorescence for desmin (Abcam). Desmin-positive cells were isolated with LCM using a Leica LMD7 microscope. The RNA was isolated from desmin-positive cells using the PicoPure RNA Isolation Kit (ThermoFisher), cDNA was made, and real-time PCR was performed as above.
Human tissues
Human liver tissue from normal and ALD patients was purchased from XenoTech (Table 1). The RNA was isolated, cDNA was made, and real-time PCR was performed as above.
Table 1.
Characteristics of liver donors from control and ALD patients
| Tissue Type | Age | Gender | Pathological diagnosis | Alcohol frequency |
|---|---|---|---|---|
| ALD Patient 1 | 48 | Male | Steatohepatitis, ballooned hepatocytes, lobular inflammation, moderate portal inflammation (40% fat) | Heavy |
| ALD Patient 2 | 43 | Male | Steatohepatitis (80% fat) | Heavy |
| ALD Patient 3 | 64 | Male | Steatohepatitis, scattered ballooned hepatocytes (20% fat) | Heavy |
| ALD Patient 4 | 51 | Female | Steatohepatitis (70% fat) | Heavy |
| Control 1 | 67 | Female | None | Occasional |
| Control 2 | 68 | Male | None | N/A |
| Control 3 | 51 | Male | None | Occasional |
| Control 4 | 63 | Female | None | Occasional |
Statistical analysis
Data are expressed as the means ± S.E. from at least three separate experiments performed in triplicate unless otherwise noted. The differences between groups were analyzed using a double-sided Student's t test when only two groups were present and analysis of variance when there were more than two groups. The null hypothesis was rejected at the 0.05 level unless otherwise specified.
Author contributions
K. M. and L. H. designed, performed, and analyzed most of the experiments and wrote the manuscript. K. S., N. W., T. A., T. Z., S.-R. L., and Y. W. performed and analyzed some experiments. Q. H., H. F., and S. G. designed and modified the manuscript. H.T., G. A., and F. M. designed and finalized manuscript.
Acknowledgments
We express our deepest appreciation for the animal core and its tissue sharing program of the Southern California Research Center for ALPD and Cirrhosis (National Institutes of Health Grant P50AA011999) for assistance with the animal studies for this project. We also acknowledge Adam C. Stephens in the Publication Department of Baylor Scott & White Health for editing assistance. This material is the result of work supported by resources at the Central Texas Veterans Health Care System.
This work was supported, in whole or in part, by National Institutes of Health Grants P50AA011999 and U01AA018663 (to H. T.), DK058411, DK107310, DK076898, DK095291, and DK062975 (to G. A., F. M., and S. G.), and DK108959 (to H. F.). This work was also supported by Veterans Affairs Merit Awards 1I01BX001724 (to F. M.), 5I01BX000574 (to G. A.), 5I01BX002192 (to S. G.), 1I01BX003031 (to H. F.), and 5I01BX001991 (to H. T.) and by the Dr. Nicholas C. Hightower Centennial Chair of Gastroenterology from Baylor Scott & White. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs, the United States government, or the National Institutes of Heath. The authors declare that they have no conflicts of interest with the contents of this article.
- ALD
- alcoholic liver disease
- miRNA
- microRNA
- HMGA2
- high-mobility group AT-hook 2
- Snail
- snail family transcriptional repressor
- Twist
- twist family basic helix-loop-helix transcription factor
- LCM
- laser capture microdissection
- let-7
- lethal-7
- ACTA2
- α-actin 2
- Col1A1
- collagen 1A1
- TIMP1
- TIMP metallopeptidase inhibitor 1
- TIMP3
- TIMP metallopeptidase inhibitor 3
- FN1
- fibronectin 1
- Lin28A/B
- Lin-28 homolog A/B
- ALT
- alanine aminotransferase
- (h)HSC
- (human) hepatic stellate cell
- FFPE
- formalin fixed paraffin embedded
- ECM
- extracellular matrix
- IPA
- Ingenuity pathway analysis
- HCC
- hepatocellular carcinoma
- ncRNA
- non-coding RNA.
References
- 1. Louvet A., and Mathurin P. (2015) Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat. Rev. Gastroenterol. Hepatol. 12, 231–242 [DOI] [PubMed] [Google Scholar]
- 2. McDaniel K., Herrera L., Zhou T., Francis H., Han Y., Levine P., Lin E., Glaser S., Alpini G., and Meng F. (2014) The functional role of microRNAs in alcoholic liver injury. J. Cell Mol. Med. 18, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taura K., Iwaisako K., Hatano E., and Uemoto S. (2016) Controversies over the epithelial-to-mesenchymal transition in liver fibrosis. J. Clin. Med. 5, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Y. P., Yu G. R., Lee M. J., Lee S. Y., Chu I. S., Leem S. H., and Kim D. G. (2013) Lipocalin-2 negatively modulates the epithelial-to-mesenchymal transition in hepatocellular carcinoma through the epidermal growth factor (TGF-β1)/Lcn2/Twist1 pathway. Hepatology 58, 1349–1361 [DOI] [PubMed] [Google Scholar]
- 5. Mavila N., James D., Shivakumar P., Nguyen M. V., Utley S., Mak K., Wu A., Zhou S., Wang L., Vendyres C., Groff M., Asahina K., and Wang K. S. (2014) Expansion of prominin-1-expressing cells in association with fibrosis of biliary atresia. Hepatology 60, 941–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee S. J., Kim K. H., and Park K. K. (2014) Mechanisms of fibrogenesis in liver cirrhosis: the molecular aspects of epithelial-mesenchymal transition. World J. Hepatol. 6, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pellicoro A., Ramachandran P., Iredale J. P., and Fallowfield J. A. (2014) Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat. Rev. Immunol. 14, 181–194 [DOI] [PubMed] [Google Scholar]
- 8. Bansal M. B. (2016) Hepatic stellate cells: fibrogenic, regenerative or both? Heterogeneity and context are key. Hepatol. Int. 10, 902–908 [DOI] [PubMed] [Google Scholar]
- 9. Senoo H., Kojima N., and Sato M. (2007) Vitamin A-storing cells (stellate cells). Vitam. Horm. 75, 131–159 [DOI] [PubMed] [Google Scholar]
- 10. Karin D., Koyama Y., Brenner D., and Kisseleva T. (2016) The characteristics of activated portal fibroblasts/myofibroblasts in liver fibrosis. Differentiation 92, 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Omar R., Yang J., Liu H., Davies N. M., and Gong Y. (2016) Hepatic stellate cells in liver fibrosis and siRNA-based therapy. Rev. Physiol. Biochem. Pharmacol. 172, 1–37 [DOI] [PubMed] [Google Scholar]
- 12. Chen Y., Xiao J., Zhang X., and Bian X. (2016) MicroRNAs as key mediators of hepatic detoxification. Toxicology 368, 80–90 [DOI] [PubMed] [Google Scholar]
- 13. Meng F., Glaser S. S., Francis H., Yang F., Han Y., Stokes A., Staloch D., McCarra J., Liu J., Venter J., Zhao H., Liu X., Francis T., Swendsen S., Liu C. G., Tsukamoto H., and Alpini G. (2012) Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am. J. Pathol. 181, 804–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glaser S., Meng F., Han Y., Onori P., Chow B. K., Francis H., Venter J., McDaniel K., Marzioni M., Invernizzi P., Ueno Y., Lai J. M., Huang L., Standeford H., Alvaro D., Gaudio E., Franchitto A., and Alpini G. (2014) Secretin stimulates biliary cell proliferation by regulating expression of microRNA 125b and microRNA let7a in mice. Gastroenterology 146, 1795–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han Y., Meng F., Venter J., Wu N., Wan Y., Standeford H., Francis H., Meininger C., Greene J. Jr., Trzeciakowski J. P., Ehrlich L., Glaser S., and Alpini G. (2016) miR-34a-dependent overexpression of Per1 decreases cholangiocarcinoma growth. J. Hepatol. 64, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McDaniel K., Hall C., Sato K., Lairmore T., Marzioni M., Glaser S., Meng F., and Alpini G. (2016) Lin28 and let-7: roles and regulation in liver diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G757–G765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., and Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 18. Hurteau G. J., Carlson J. A., Roos E., and Brock G. J. (2009) Stable expression of miR-200c alone is sufficient to regulate TCF8 (ZEB1) and restore E-cadherin expression. Cell Cycle 8, 2064–2069 [DOI] [PubMed] [Google Scholar]
- 19. Hurteau G. J., Carlson J. A., Spivack S. D., and Brock G. J. (2007) Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 67, 7972–7976 [DOI] [PubMed] [Google Scholar]
- 20. Choi S. S., and Diehl A. M. (2009) Epithelial-to-mesenchymal transitions in the liver. Hepatology 50, 2007–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Viswanathan S. R., Powers J. T., Einhorn W., Hoshida Y., Ng T. L., Toffanin S., O'Sullivan M., Lu J., Phillips L. A., Lockhart V. L., Shah S. P., Tanwar P. S., Mermel C. H., Beroukhim R., Azam M., et al. (2009) Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J., Zhang L., Fan R., Guo N., Xiong C., Wang L., Jin S., Li W., and Lu J. (2013) The polymorphism in the let-7 targeted region of the Lin28 gene is associated with increased risk of type 2 diabetes mellitus. Mol. Cell Endocrinol. 375, 53–57 [DOI] [PubMed] [Google Scholar]
- 23. Lazaro R., Wu R., Lee S., Zhu N. L., Chen C. L., French S. W., Xu J., Machida K., and Tsukamoto H. (2015) Osteopontin deficiency does not prevent but promotes alcoholic neutrophilic hepatitis in mice. Hepatology 61, 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mathews S., Xu M., Wang H., Bertola A., and Gao B. (2014) Animals models of gastrointestinal and liver diseases. Animal models of alcohol-induced liver disease: pathophysiology, translational relevance, and challenges. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G819–G823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Francis H., McDaniel K., Han Y., Liu X., Kennedy L., Yang F., McCarra J., Zhou T., Glaser S., Venter J., Huang L., Levine P., Lai J. M., Liu C. G., Alpini G., and Meng F. (2014) Regulation of the extrinsic apoptotic pathway by microRNA-21 in alcoholic liver injury. J. Biol. Chem. 289, 27526–27539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shell S., Park S. M., Radjabi A. R., Schickel R., Kistner E. O., Jewell D. A., Feig C., Lengyel E., and Peter M. E. (2007) Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 11400–11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee S. H., Cho S., Kim M. S., Choi K., Cho J. Y., Gwak H. S., Kim Y. J., Yoo H., Lee S. H., Park J. B., and Kim J. H. (2014) The ubiquitin ligase human TRIM71 regulates let-7 microRNA biogenesis via modulation of Lin28b protein. Biochim. Biophys. Acta 1839, 374–386 [DOI] [PubMed] [Google Scholar]
- 29. Viswanathan S. R., Daley G. Q., and Gregory R. I. (2008) Selective blockade of microRNA processing by Lin28. Science 320, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Wynsberghe P. M., Kai Z. S., Massirer K. B., Burton V. H., Yeo G. W., and Pasquinelli A. E. (2011) LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 18, 302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng S. W., Tsai H. W., Lin Y. J., Cheng P. N., Chang Y. C., Yen C. J., Huang H. P., Chuang Y. P., Chang T. T., Lee C. T., Chao A., Chou C. Y., Chan S. H., Chow N. H., and Ho C. L. (2013) Lin28b is an oncofetal circulating cancer stem cell-like marker associated with recurrence of hepatocellular carcinoma. PLoS ONE 8, e80053. [DOI] [PMC free article] [PubMed] [Google Scholar]