Abstract
How each metalloprotein assembles the correct metal at the proper binding site presents challenges to the cell. The di-iron enzyme ribonucleotide reductase (RNR) uses a diferric-tyrosyl radical (FeIII2-Y•) cofactor to initiate nucleotide reduction. Assembly of this cofactor requires O2, FeII, and a reducing equivalent. Recent studies show that RNR cofactor biosynthesis shares the same source of iron, in the form of [2Fe-2S]-GSH2 from the monothiol glutaredoxin Grx3/4, and the same electron source, in the form of the Dre2-Tah18 electron transfer chain, with the cytosolic iron-sulfur protein assembly (CIA) machinery required for maturation of [4Fe-4S] clusters in cytosolic and nuclear proteins. Here, we further investigated the interplay between the formation of the FeIII2-Y• cofactor in RNR and the cellular iron-sulfur (Fe-S) protein biogenesis pathways by examining both the iron loading into the RNR β subunit and the RNR catalytic activity in yeast mutants depleted of individual components of the mitochondrial iron-sulfur cluster assembly (ISC) and the CIA machineries. We found that both iron loading and cofactor assembly in RNR are dependent on the ISC machinery. We also found that Dre2 is required for RNR cofactor formation but appears to be dispensable for iron loading. None of the CIA components downstream of Dre2 was required for RNR cofactor formation. Thus, the pathways for RNR and Fe-S cluster biogenesis bifurcate after the Dre2-Tah18 step. We conclude that RNR cofactor biogenesis requires the ISC machinery to mature the Grx3/4 and Dre2 Fe-S proteins, which then function in iron and electron delivery to RNR, respectively.
Keywords: electron transfer, iron metabolism, iron-sulfur protein, metalloprotein, ribonucleotide reductase, iron-sulfur cluster, metallocofactor, deoxyribonucleotide biosynthesis
Introduction
Iron is an essential element for all eukaryotes because of its role as a redox-active cofactor in many fundamental biological reactions including respiration, ribosomal biogenesis, DNA replication, and repair. Many cytosolic and nuclear enzymes use iron as a cofactor in the form of iron-sulfur (Fe-S)4 clusters, heme, mono-iron, and di-iron. Given the abundance of metalloproteins in the prokaryotic and eukaryotic proteomes, a main challenge for the cell is to load the Fe2+ onto the proper binding site and convert it to into the active forms (1).
One family of di-iron enzymes is the class Ia ribonucleotide reductases (RNRs). RNRs catalyze the conversion of ribonucleoside 5′-diphosphates (NDPs) to the corresponding deoxyribonucleotides in some prokaryotes and all eukaryotes, providing the monomeric building blocks for DNA replication and repair (2, 3). All class Ia RNRs utilize a diferric-tyrosyl radical (FeIII2-Y•) metallocofactor to catalyze the nucleotide reduction reaction. The level of the tyrosyl radical (Y•) of the FeIII2-Y• cofactor is directly correlated with the catalytic activity of the RNR enzyme (4, 5). The RNR holoenzyme consists of α and β subunits with a likely active quaternary structure of (α2)m(β2)n (6). Subunit α contains the active site where NDPs are reduced and the allosteric effector-binding sites that control substrate specificity and overall activity. Subunit β houses the FeIII2-Y• cofactor that is assembled from FeII, O2, and a reducing equivalent. Structural and biochemical studies of Escherichia coli α2 and β2 support a mechanism in which the Y• in β is required for the reversible oxidation, over a distance of 35 Å, of a conserved cysteine within the active site of α into a transient thiyl radical, which initiates the nucleotide reduction reaction (7).
Previous biochemical studies of the E. coli NrdB (β2) as well as other eukaryotic β2s have demonstrated that the FeIII2-Y• cofactor can be generated in vitro by self-assembly from apo-β2, FeII, and O2, with FeII serving as both the metal ligand and the source of the obligatory reducing equivalent (Fig. 1A) (4, 8). However, FeII is unlikely the electron donor in vivo because it is highly redox-reactive and has to be chaperoned inside the cell (9). Moreover, the self-assembly process is difficult to control and of varying efficiency, pointing to the importance of a regulated and catalyzed biosynthesis pathway in vivo.
Figure 1.
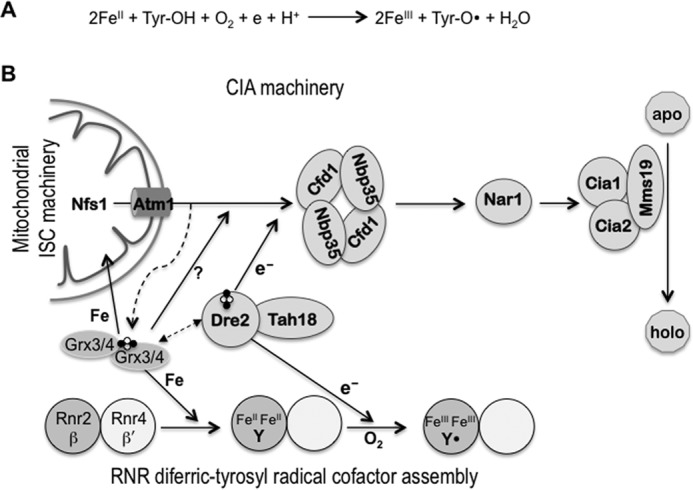
The relationship between the cofactor assembly in RNR and the pathways of iron-sulfur protein biogenesis in the mitochondria (ISC) and the cytosol (CIA) is shown. A, self-assembly of the RNR metallocofactor. The FeIII2-Y• cofactor can self-assemble in vitro from apo-β, FeII, and O2 in a four-electron reduction of O2 with the indicated stoichiometry. Of the four electrons required, two are from the two FeII ions serving as the metal ligand and one from a conserved tyrosine residue that is converted into a tyrosyl radical (Tyr-O•). The fourth electron, which can come from the excess of FeII in vitro, has to be supplied by an electron donor in vivo. B, connection between RNR radical cofactor assembly and cellular Fe-S protein biogenesis. Assembly of the FeIII2-Y• cofactor in Rnr2 (β) is facilitated by Rnr4 (β′), which stabilizes β in a conformation that allows iron binding. The monothiol glutaredoxins Grx3/4 function as [2Fe-2S]-GSH2-bridged dimers, and are required for several aspects of intracellular iron metabolism. They are involved in intracellular iron regulation (not depicted here), cytosolic Fe-S protein assembly, and in the formation of the FeIII2-Y• cofactor in RNR. The CIA pathway and RNR cofactor assembly share the electron donor complex Tah18-Dre2, which delivers electrons from NADPH to the scaffold protein complex Cfd1-Nbp35 in the CIA and to β in RNR. Genetic and physical interactions have been observed between Dre2 and Grx3/4 in both yeast and human cells (14, 29, 30), although the mechanistic basis of this interaction is unclear.
The RNR of budding yeast Saccharomyces cerevisiae has been an excellent model system for studying the proteins involved in the RNR metallocofactor biosynthesis. S. cerevisiae RNR has two β isoforms, Rnr2 (β) and Rnr4 (β′), which readily form the active ββ′ heterodimer from apo-β2 and apo-β′2 (5, 10). Only β is capable of iron binding and cofactor assembly and thus there is a maximum of one Y• radical per ββ′ (11). Subunit β′ can neither bind iron nor form its own cofactor because of multiple substitutions of the conserved iron-binding residues (12, 13). However, β′ is required to maintain β in a conformation competent for iron binding and cofactor assembly both in vivo and in vitro (12, 14). The two key issues of in vivo RNR cofactor assembly are the sources of iron and reducing equivalent. Our recent studies have demonstrated that the paralogous monothiol glutaredoxins Grx3 and Grx4 are required for iron loading into β (15), whereas the electron donor complex Dre2-Tah18 is required for RNR cofactor formation in yeast cells (16).
Both Grx3/4 and Dre2-Tah18 are evolutionarily conserved in eukaryotes; the human homologs can substitute the essential function of their respective yeast counterparts (17–20). Grx3 and Grx4 form a heterodimer bridged by a [2Fe-2S] cluster ligated to glutathione that is involved in the trafficking and delivery of iron to almost all iron-requiring proteins inside the cell (15). Dre2 contains a [2Fe-2S] cluster and a [4Fe-4S] cluster. In vivo, Dre2 forms a complex with the diflavin reductase Tah18, thus constituting an electron-transfer chain in which electrons are relayed from NADPH via the FAD- and FMN-containing Tah18 to the [2Fe-2S] cluster of Dre2 (19). In addition to being essential to RNR cofactor formation, Grx3/4 and Dre2-Tah18 are also required for cytosolic Fe-S protein assembly pathway (CIA). The CIA pathway is responsible for biogenesis of the [4Fe-4S] cluster in many cytosolic and nuclear proteins that function in divergent processes such as ribosome maturation, tRNA modification, and DNA replication and repair (21, 22). The first step of CIA involves formation of a transient [4Fe-4S] cluster on the scaffold protein complex Cfd1-Nbp35, which requires a sulfur-containing compound synthesized by the mitochondrial Fe-S cluster assembly (ISC) machinery and exported via the mitochondrial inner membrane ABC transporter Atm1 (Fig. 1B). In addition, CIA requires Grx3/4 function and reducing equivalents from Dre2-Tah18 (21). The Dre2-Tah18 electron donor complex acts at an early step in CIA, and is required for the generation of the N-terminal [4Fe-4S] cluster of Nbp35 (19).
Our previous studies have established that the formations of two different iron-containing cofactors, the FeIII2-Y• of RNR and the [4Fe-4S] cluster in cytosolic and nuclear proteins, share the requirement of Grx3/4, likely as an iron donor, and of Dre2-Tah18 as the source of electrons. In this work, we further investigate the connection between the two processes. In particular, we asked whether iron loading and/or radical cofactor formation in RNR is dependent on specific components of the ISC or CIA machinery. Our results indicate that the ISC machinery, together with Grx3/4, is essential for both iron loading and radical cofactor formation in RNR. In contrast, the CIA components are dispensable for these steps with the exception of Dre2 that supplies electrons for RNR cofactor formation, downstream of the iron-loading step.
Results
Depletion of ISC and CIA components under iron-replete conditions decreases both RNR ββ′ activity and the β protein level
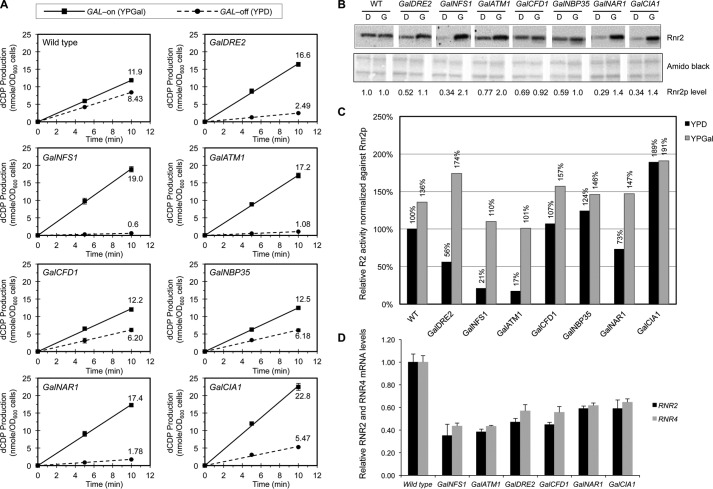
To determine the contribution of the mitochondrial and cytosolic Fe-S biogenesis machineries to the radical cofactor assembly process in RNR, we examined the effects of depletion of individual components of the ISC and CIA on the catalytic activity of ββ′ by a previously developed permeabilized cell-based RNR activity assay (14). Toward this end, individual ISC and CIA genes were placed under the control of the glucose-repressible GAL1 promoter by homologous recombination-based replacement of the native promoters at their respective chromosomal loci (23). These strains were grown in rich media containing either galactose (YPGal, GAL promoter-on) or glucose (YPD, GAL promoter-off) for 40 h before being harvested for assays of RNR ββ′ catalytic activity in the presence of an excess of the α subunit. Wild-type cells grown in YPD exhibited 70% of the RNR activity compared with growth in YPGal (Fig. 2A). Consistent with our previous report (14), GalDRE2 cells grown in YPD only had 15% of the RNR activity as those grown in YPGal. Under the same growth conditions, all six Gal-ISC and Gal-CIA mutants characterized exhibited various degrees of reduction in RNR activity (Fig. 2A).
Figure 2.
Comparison of the RNR ββ′ activity in the Gal-ISC and Gal-CIA mutants grown in iron-replete rich media under GAL-on and GAL-off conditions is shown. A, measurement of the ββ′ catalytic activity in permeabilized spheroplasts of wild-type, GalNFS1, GalATM1, GalDRE2, GalCFD1, GalNBP35, GalNAR1, and GalCIA1 strains grown in rich media containing galactose (YPGal, GAL-on, solid lines) or glucose (YPD, GAL-off, dashed lines). Cells from log-phase cultures were harvested and converted into spheroplasts, and the catalytic activity of ββ′ was assayed in the presence of an excess of α2 as described previously (14). Values are given as means ± S.D. (n = 4). OD, optical desity. B, comparison of the protein levels of Rnr2 (β) in the wild-type (WT) and Gal strains grown in rich media containing glucose (D, GAL-off) or galactose (G, GAL-on). Total protein extracts from an equal number of log-phase cells were loaded for each sample. The protein blot was probed with anti-Rnr2 polyclonal antibodies (top panel) and stained with Amido Black (bottom panel) as a control for loading. Relative Rnr2 signals were normalized against that of the WT in YPD, which was set to 1. The Western blotting was done three times from independently grown cultures; one representative blot was shown with quantitation. C, relative specific activities of ββ′ after being normalized against the levels of the Rnr2 (β) protein, with the activity of WT cells in YPD arbitrarily defined as 100%. D, comparison of mRNA levels of RNR2 and RNR4 in wild-type, GalNFS1, GalATM1, GalDRE2, GalCFD1, GalNBP35, and GalCIA1 cells by RT-qPCR. The signals of RNR2 and RNR4 were normalized against that of ACT1 in each strain, and the resulting ratios in the wild-type cells were arbitrarily defined as 1-fold. Values are given as means ± S.D. (n = 3).
The differences in the ββ′ activity may reflect the severely decreased β and β′ protein levels in the Gal-ISC and Gal-CIA mutants (Fig. 2B). As observed previously Dre2-depleted cells had a significantly reduced β protein level (16). We therefore corrected the Rnr2 activity results for β protein levels. The resulting relative Rnr2 activities fell into two groups: those for GalNFS1 and GalATM1 cells were lower than those for GalDRE2 cells, whereas those for GalCFD1, GalNBP35, GalNAR1, and GalCIA1 cells were much higher (Fig. 2C). The correction for Rnr2 protein levels seemed justified because its decrease was mainly because of a lower transcription of the RNR2 gene and thus lower mRNA levels (Fig. 2D). Our recent study had shown that the decrease in Rnr2 and Rnr4 mRNA levels in the GalDRE2 mutant is partially because of Cth1/2-mediated mRNA degradation (16). Taken together, these results provided a first hint that the later CIA components may not play a major role in RNR cofactor biogenesis.
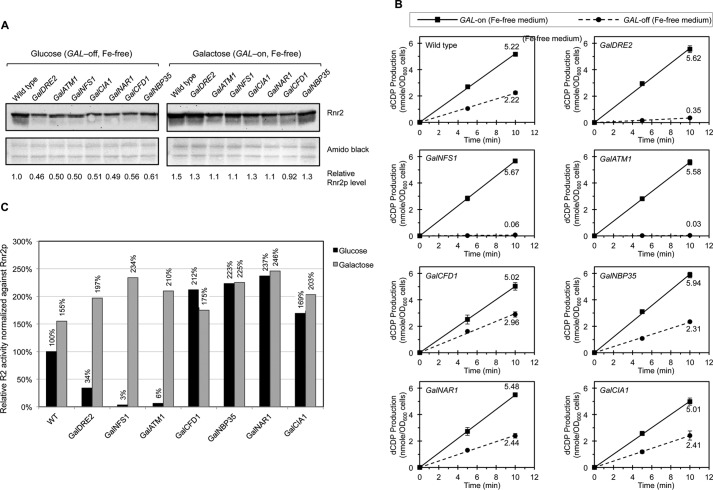
Activity assay of RNR ββ′ in the ISC- and CIA-depletion mutants under iron-deficient conditions revealed a requirement for ISC and Dre2 but for none of the other CIA components
We searched for an experimental condition under which the Rnr2/4 levels do not alter strongly. From previous work, we knew that under iron-limiting conditions the Rnr2 levels hardly change (15). We therefore repeated the experiment of Fig, 2 and grew the cells in the synthetic iron-free instead of rich medium, with either galactose (GAL-on) or glucose (GAL-off) as the sole carbon source, for 40 h before harvesting them for RNR activity assay and protein analysis. Protein blotting revealed more consistent Rnr2 protein levels across the strains under both GAL-on and GAL-off conditions when cells were iron-starved, although the levels were still slightly lower in the Gal mutants relative to the wild-type under GAL-off conditions (Fig. 3A). Hardly any variation in RNR activity (5.01–5.67 nmol dCDP/A600 cells) was found among the wild-type and Gal strains under iron-free and GAL-on conditions (Fig. 3B), as opposed to the high variation (11.9–22.8 nmol dCDP/A600 cells) in the iron-rich and GAL-on (YPGal) medium (Fig. 2A). Under iron-free and GAL-off conditions, RNR activity of these strains clearly fell into two categories. All the Gal-CIA strains with the exception of GalDRE2 had an RNR activity comparable with that of the wild-type. On the contrary, GalDRE2 and the two ISC mutants GalNFS1 and GalATM1 had a drastically diminished RNR activity relative to the wild-type (Fig. 3B). After correction for Rnr2 protein levels, the relative specific activities of ββ′ were drastically lower in cells depleted of Nfs1, Atm1, or Dre2, ranging from 3 to 34% of the level of the wild-type under Gal-off conditions (Fig. 3C). By contrast, depletion of the CIA components Cfd1, Nbp35, Nar1, and Cia1 did not cause any decrease in the RNR specific activity. (Fig. 3C). Taken together, these results strongly indicated that the assembly of the metallocofactor in RNR, as measured by the catalytic activity of ββ′, requires the mitochondrial ISC system and Dre2, but not the rest of the CIA components downstream of Dre2.
Figure 3.
Comparison of the RNR ββ′ activity in the Gal-ISC and Gal-CIA mutants grown in iron-free media under GAL-on and GAL-off conditions is shown. A, comparison of the Rnr2 (β) protein levels in the wild-type and the indicated Gal mutants grown in synthetic and iron-free media containing glucose or galactose as described in Fig. 2B. The Western blotting was done three times from independently grown cultures; one representative blot was shown with quantitation. B, measurement of the ββ′ activity in permeabilized spheroplasts of the indicated strains grown in synthetic and iron-free media containing either galactose (GAL-on, solid lines) or glucose (GAL-off, dashed lines) as described in Fig. 2A. Values are given as means ± S.D. (n = 4). C, comparison of the relative specific activities of ββ′ as described in Fig. 2C.
Iron loading into β requires ISC but not Dre2 and the remainder of the CIA components
Our recent studies have demonstrated that in vivo assembly of the FeIII2-Y• cofactor in β (Rnr2) of the yeast RNR requires Grx3/4 as the source of iron, Dre2-Tah18 as the source of reducing equivalents, as well as β′ (Rnr4) that stabilizes β in a conformation to allow stable iron binding (14–16). One unresolved question was whether the iron loading into β occurs independently of the electron delivery step. Another important issue was the potential involvement of the ISC and CIA machineries in these two steps. Maturation of the electron-transferring [2Fe-2S] cluster in Dre2 is dependent on the ISC and Grx3/4, whereas iron binding of Grx3/4 is independent of Dre2 (15, 19). To address which components of the ISC and CIA are required for iron loading into β of the RNR, we used an established 55Fe radiolabeling and immunoprecipitation assay in wild-type, Gal-ISC, and Gal-CIA mutant cells (24). 55Fe loading into β was decreased 2.5- to 5-fold in the GalNFS1, GalATM1, and GalGRX3/grx4Δ mutants, but was unaffected in the GalDRE2, GalCFD1, GalNBP35, and GalNAR1 mutants relative to the wild-type control (Fig. 4A). To monitor the efficiency of protein depletion in the Gal-ISC and Gal-CIA mutants, we measured in parallel the 55Fe incorporation into Leu1, the cytosolic Fe-S cluster-containing isopropylmalate isomerase. 55Fe loading into Leu1 was greatly diminished in all ISC- and CIA-depleted mutant cells, consistent with the notion that Fe-S cluster assembly in this cytosolic protein depends on both the ISC and CIA machineries (Fig. 4B). The protein levels of Rnr2, Leu1, as well as individual ISC and CIA components being depleted were monitored by Western blotting. The levels of Rnr2 and Leu1 were comparable across the strains except for the marked decrease of Leu1 in the GalGRX3/grx4Δ mutant (Fig. 4C), an observation consistent with earlier findings (15). Together, our results indicated that iron loading into RNR was dependent on the ISC but none of the CIA components. We conclude that iron loading into β can occur in the absence of electron delivery from Dre2.
Figure 4.
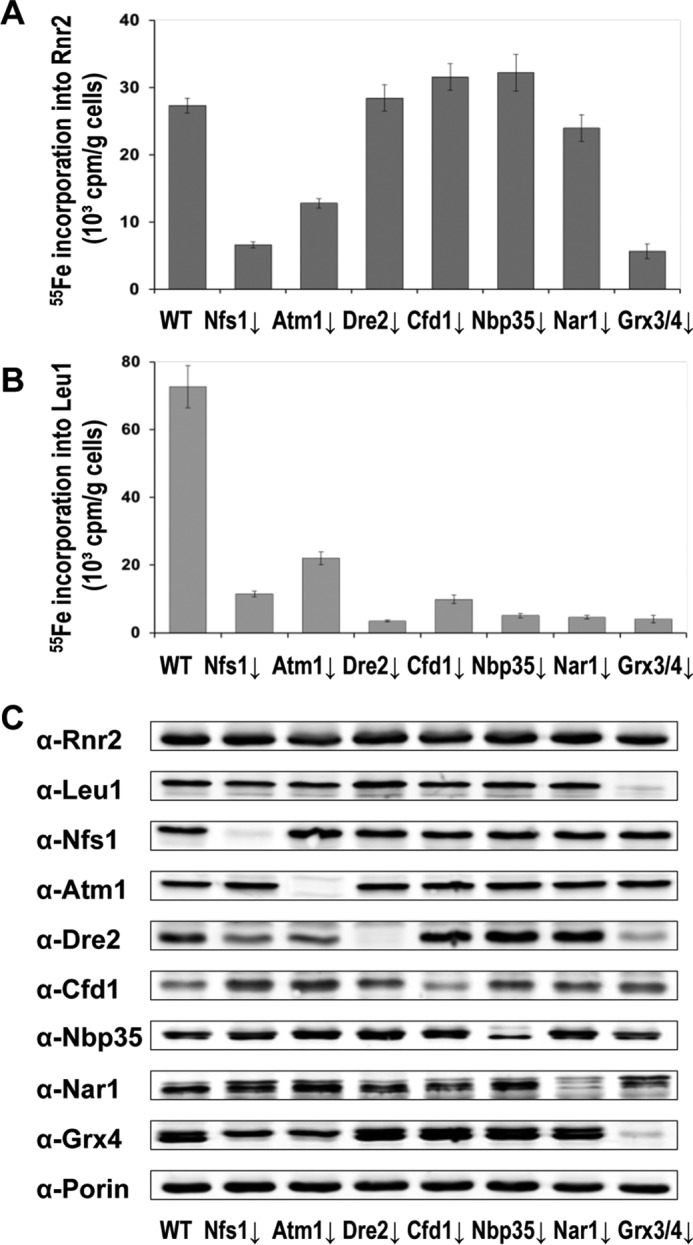
Iron loading into Rnr2 protein depends on the mitochondrial ISC system and cytosolic Grx3/4 proteins but not the CIA machinery. A and B, wild-type (WT), GalNFS1, GalATM1, GalDRE2, GalCFD1, GalNBP35, GalNAR1, and Gal-GRX4grx3Δ cells were grown in a synthetic iron-free medium containing glucose (GAL-off) for 40 h or 64 h (26) before being radiolabeled with 55FeCl3 for 2 h. After 55Fe incorporation, the Rnr2 (A) and Leu1 (B) proteins were immunoprecipitated with specific polyclonal antibodies. Incorporation of 55Fe into Rnr2 and Leu1 was quantified by scintillation counting. Values are given as means ± S.D. (n ≥ 3). C, comparison of protein levels of Rnr2, Leu1, as well as individual ISC and CIA components in wild-type (WT), GalNFS1, GalATM1, GalDRE2, GalCFD1, GalNBP35, GalNAR1, and Gal-GRX4grx3Δ mutant cells by Western blot. The protein blot was also probed with anti-Porin as a loading control.
Discussion
The molecular requirements for correct metalation of many metalloenzymes are poorly understood. In this study, we dissected the key steps involved in the assembly of the FeIII2-Y• metallocofactor of the yeast RNR. Based on our previous observation that RNR cofactor assembly and the CIA share the same sources of iron and electron supply, we further examined the requirement of individual ISC and CIA components for RNR iron acquisition and cofactor formation. In particular, we asked whether the steps of iron loading and electron delivery are interdependent during cofactor assembly in RNR or can occur in a stepwise fashion. Several conclusions can be drawn from this study. First, iron loading into RNR is dependent on the mitochondrial ISC system and the previously identified cytosolic Grx3/4, but not on the CIA machinery. Second, the electron donor Dre2 is dispensable for the iron-loading step in RNR cofactor assembly, suggesting that iron can assemble with β relatively stably without being converted to the active cofactor FeIII2-Y• by further electron supply (Fig. 1B). Third, none of the CIA components downstream of Dre2 participates directly in the RNR radical cofactor formation.
The requirement of the mitochondrial ISC machinery for iron loading into RNR is likely because of the essential role of ISC in the biogenesis of the cytosolic Fe-S proteins Grx3/4 and Dre2. Their maturation requires the ISC but not the CIA machinery (15, 19). Accordingly, we observed a severe diminution of the RNR activity in ISC- and Dre2-depletion mutants but not in any of the other CIA mutants downstream of Dre2. The increase of RNR ββ′ catalytic activity in CIA-depletion mutants relative to wild-type cells (Fig. 3C) suggests that under iron-limiting conditions, blocking of the CIA pathway may allow for more bioavailable iron to be funneled into RNR, thereby enhancing its cofactor assembly process. This notion was consistent with the observed moderate increase of 55Fe incorporation into the RNR β (15)(Fig. 4A).
Dre2 (together with its electron supply partner Tah18) (Fig. 1B) stands unique among all the CIA components identified to date because of its dual role as an electron donor both in cytosolic Fe-S protein maturation and in RNR cofactor assembly. It is evident from our studies that Dre2 is essential for assembly of the active FeIII2-Y• cofactor of RNR but dispensable for the initial step of iron loading into β. Iron loading also appears to occur independently of the Dre2-mediated electron-delivery step during the maturation of [4Fe-4S] in cytosolic and nuclear proteins, as depletion of Dre2 leads to an increase in 55Fe incorporation into the CIA scaffold protein Cfd1 but abolishes 55Fe-S insertion in all the rest of the CIA components and downstream substrate proteins (19). Although Dre2 is unlikely directly involved in the iron loading step mediated by Grx3/4, we cannot rule out the possibility that Dre2 can become critical for loading of the correct metal (e.g. iron versus manganese) into RNR under conditions of skewed cellular metal pools.
Experimental procedures
Strains and media
All yeast strains used for RNR activity assay in this study were of the S288C strain background. GalNFS1, GalATM1, GalCFD1, GalNBP35, GalNAR1, and GalCIA1 strains were constructed in the BY4741 strain (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and GalDRE2 in BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) by replacing the endogenous promoters with the galactose-inducible GAL1 promoter that was marked by HIS3MX6 as described (23). Transformants were selected on synthetic complete histidine dropout agar plates (SC-His) containing 1% raffinose and 1% galactose, and confirmed by PCR diagnosis as well as Western blot using specific antibodies recognizing individual CIA and ISC components. The GalGRX4grx3Δ strain was described previously (15). The strains used for 55Fe incorporation analysis were of the W303 background.
Rich YP medium contained 1% yeast extract, 2% peptone, and 2% of one of the following three carbohydrates as the sole carbon source: glucose (YPD) that represses the GAL1 promoter (GAL-off), raffinose (YP-Raffinose) that keeps the GAL1 promoter at the basal level of transcription activity (uninduced state), or galactose (YPGal) that activates the GAL1 promoter (induced state or GAL-on). Synthetic complete (SC) medium contained 0.17% yeast nitrogen base (YNB) (MP Biomedicals), 0.5% (NH4)2SO4, 20 amino acids (Sigma) as described (25), and the indicated carbon source. The iron-free medium was based on the SC medium except for the use of an iron-free YNB (Sunrise Science Products). All solid media contained 2% Bacto Agar that was added before autoclaving.
55Fe incorporation analysis
Radiolabeling of yeast cells with 55FeCl3 and measurement of 55Fe incorporation into RNR and Leu1 by immunoprecipitation were performed as described (26).
Protein analysis
Yeast cells were harvested from early- to mid-log phase cultures (A600 0.5–1.0). Protein extracts were prepared by trichloroacetic acid precipitation (10) and subjected to Western blotting. Antibodies used for Western blotting were anti-G6PDH (Sigma-Aldrich); anti-Rnr2 and anti-Rnr4 (13); anti-Leu1, anti-Nfs1, anti-Atm1, anti-Dre2, anti-Cfd1, anti-Nbp35, anti-Nar1, anti-Grx4, and anti-Porin as described previously (15). Signals from protein blots were recorded and quantitated by using ChemiDoc MP Imaging System (Bio-Rad) or enhanced chemiluminescence imaging system (Intas, Gottingen, Germany).
RNA extraction, reverse transcription, and quantitative real-time PCR (RT-qPCR)
Yeast strains were grown in YPD medium with starting A600 = 0.002 for 16 h. Cells were harvested from early- to mid-log phase cultures and total RNA was extracted by using a hot-phenol method (27) Ten μg of total RNA were treated with 10 units of RNase-free DNase I (New England Biolabs) for 30 min at 37 °C to remove contaminating DNA. First-strand cDNA synthesis was carried out by M-MuLV Reverse Transcriptase (New England Biolabs) on aliquots of 1 μg RNA with a random primer mix. The single-stranded cDNA products were used in quantitative PCR on a Bio-Rad CFX96 Real-Time PCR Detection System based on SYBR Green fluorescence. Sequences of oligonucleotide pairs used were 5′-CCTAAAGAGACCCCTTCCAAAG-3′ and 5′-GCCTTGTGATTTTCAGCGTC-3′ for RNR2; 5′-CATAAGGCTGCTTTCATCGAG-3′ and 5′-CTGTTGGCCATTGCTAAACC-3′ for RNR4; 5′-GTATGTGTAAAGCCGGTTTTG-3′ and 5′-CATGATACCTTGGTGTCTTGG-3′ for ACT1.
RNR ββ′ activity assays in permeabilized spheroplasts
Preparation of permeabilized yeast spheroplasts and measurement of RNR activity were performed as described previously (14). Assay of ββ′ activity in permeabilized spheroplasts was performed in the presence of an excess of α2 (4.4 μm) using DTT as a reductant. Production of [3H]dCDP at 0-, 5-, and 10-min time points was calculated and normalized against cell number using A600 (28).
Author contributions
M. H., R. L., and J. S. designed the study and wrote the paper. H. L. performed RNR activity using permeabilized yeast cells. M. S. characterized 55Fe incorporation in GAL depleted strains. C. Z. determined RNR mRNA and protein levels. X. A. constructed the GAL promoter replacement strains in the S288C background. All authors analyzed the results and approved the final version of the manuscript.
This work was supported by National Institutes of Health Grants GM81393 (to J. S. and M. H), and CA125574 (to M. H.) and by Deutsche Forschungsgemeinschaft (SFB 987, SPP 1710, LI 415–5), LOEWE program of the state of Hesse, and the Max-Planck Gesellschaft (to R. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Fe-S
- iron-sulfur cluster
- RNR
- ribonucleotide reductase
- ISC
- iron-sulfur cluster assembly
- CIA
- cytosolic iron-sulfur protein assembly
- RT-qPCR
- reverse transcription and quantitative real-time PCR
- FeIII2-Y•
- diferric-tyrosyl radical.
References
- 1. Waldron K. J., Rutherford J. C., Ford D., and Robinson N. J. (2009) Metalloproteins and metal sensing. Nature 460, 823–830 [DOI] [PubMed] [Google Scholar]
- 2. Nordlund P., and Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 [DOI] [PubMed] [Google Scholar]
- 3. Hofer A., Crona M., Logan D. T., and Sjöberg B. M. (2012) DNA building blocks: Keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang M., Parker M. J., and Stubbe J. (2014) Choosing the right metal: Case studies of class I ribonucleotide reductases. J. Biol. Chem. 289, 28104–28111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perlstein D. L., Ge J., Ortigosa A. D., Robblee J. H., Zhang Z., Huang M., and Stubbe J. (2005) The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry 44, 15366–15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cotruvo J. A., and Stubbe J. (2011) Class I ribonucleotide reductases: Metallocofactor assembly and repair in vitro and in vivo. Annu. Rev. Biochem. 80, 733–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Minnihan E. C., Nocera D. G., and Stubbe J. (2013) Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase. Acc. Chem. Res. 46, 2524–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Atkin C. L., Thelander L., Reichard P., and Lang G. (1973) Iron and free radical in ribonucleotide reductase. Exchange of iron and Mössbauer spectroscopy of the protein B2 subunit of the Escherichia coli enzyme. J. Biol. Chem. 248, 7464–7472 [PubMed] [Google Scholar]
- 9. Kaplan C. D., and Kaplan J. (2009) Iron acquisition and transcriptional regulation. Chem. Rev. 109, 4536–4552 [DOI] [PubMed] [Google Scholar]
- 10. An X., Zhang Z., Yang K., and Huang M. (2006) Cotransport of the heterodimeric small subunit of the Saccharomyces cerevisiae ribonucleotide reductase between the nucleus and the cytoplasm. Genetics 173, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ortigosa A. D., Hristova D., Perlstein D. L., Zhang Z., Huang M., and Stubbe J. (2006) Determination of the in vivo stoichiometry of tyrosyl radical per ββ' in Saccharomyces cerevisiae ribonucleotide reductase. Biochemistry 45, 12282–12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang M. X., and Elledge S. J. (1997) Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 6105–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nguyen H. H., Ge J., Perlstein D. L., and Stubbe J. (1999) Purification of ribonucleotide reductase subunits Y1, Y2, Y3, and Y4 from yeast: Y4 plays a key role in diiron cluster assembly. Proc. Natl. Acad. Sci. U.S.A. 96, 12339–12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y., Liu L., Wu X., An X., Stubbe J., and Huang M. (2011) Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: Requirement of Rnr4 and contribution of Grx3/4 AND Dre2 proteins. J. Biol. Chem. 286, 41499–41509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mühlenhoff U., Molik S., Godoy J. R., Uzarska M. A., Richter N., Seubert A., Zhang Y., Stubbe J., Pierrel F., Herrero E., Lillig C. H., and Lill R. (2010) Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12, 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y., Li H. R., Zhang C. G., An X. X., Liu L. L., Stubbe J., and Huang M. X. (2014) Conserved electron donor complex Dre2-Tah18 is required for ribonucleotide reductase metallocofactor assembly and DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 111, E1695–E1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y., Lyver E. R., Nakamaru-Ogiso E., Yoon H., Amutha B., Lee D. W., Bi E., Ohnishi T., Daldal F., Pain D., and Dancis A. (2008) Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 28, 5569–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng N. H., Zhang W., Chen W. Q., Jin J., Cui X., Butte N. F., Chan L., and Hirschi K. D. (2011) A mammalian monothiol glutaredoxin, Grx3, is critical for cell cycle progression during embryogenesis. FEBS. J. 278, 2525–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Netz D. J., Stümpfig M., Doré C., Mühlenhoff U., Pierik A. J., and Lill R. (2010) Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6, 758–765 [DOI] [PubMed] [Google Scholar]
- 20. Haunhorst P., Hanschmann E. M., Bräutigam L., Stehling O., Hoffmann B., Mühlenhoff U., Lill R., Berndt C., and Lillig C. H. (2013) Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol. Biol. Cell 24, 1895–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Netz D. J., Mascarenhas J., Stehling O., Pierik A. J., and Lill R. (2014) Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol. 24, 303–312 [DOI] [PubMed] [Google Scholar]
- 22. Paul V. D., and Lill R. (2015) Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim. Biophys. Acta 1853, 1528–1539 [DOI] [PubMed] [Google Scholar]
- 23. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 24. Stehling O., Smith P. M., Biederbick A., Balk J., Lill R., and Mühlenhoff U. (2007) Investigation of iron-sulfur protein maturation in eukaryotes. Methods Mol. Biol. 372, 325–342 [DOI] [PubMed] [Google Scholar]
- 25. Burke D., Sawson D., and Stearns T. (2000) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Springs Harbor Laboratory Press, Long Island, NY [Google Scholar]
- 26. Pierik A. J., Netz D. J., and Lill R. (2009) Analysis of iron-sulfur protein maturation in eukaryotes. Nat. Protoc. 4, 753–766 [DOI] [PubMed] [Google Scholar]
- 27. Köhrer K., and Domdey H. (1991) Preparation of high molecular weight RNA. Methods Enzymol. 194, 398–405 [DOI] [PubMed] [Google Scholar]
- 28. Steeper J. R., and Steuart C. D. (1970) A rapid assay for CDP reductase activity in mammalian cell extracts. Anal. Biochem. 34, 123–130 [DOI] [PubMed] [Google Scholar]
- 29. Tarassov K., Messier V., Landry C. R., Radinovic S., Molina M. M. S., Shames I., Malitskaya Y., Vogel J., Bussey H., and Michnick S. W. (2008) An in vivo map of the yeast protein interactome. Science 320, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 30. Saito Y., Shibayama H., Tanaka H., Tanimura A., Matsumura I., and Kanakura Y. (2011) PICOT is a molecule which binds to anamorsin. Biochem. Biophys. Res. Commun. 408, 329–333 [DOI] [PubMed] [Google Scholar]