Abstract
Cystatin C (CysC) is a versatile and ubiquitously-expressed member of the cysteine protease inhibitor family that is present at notably high concentrations in cerebrospinal fluid. Under mildly denaturing conditions, CysC forms inactive domain-swapped dimers. A destabilizing mutation, L68Q, increases the rate of domain-swapping and causes a fatal amyloid disease, hereditary cystatin C amyloid angiopathy. Wild-type (wt) CysC will also aggregate into amyloid fibrils under some conditions. Propagated domain-swapping has been proposed as the mechanism by which CysC fibrils grow. We present evidence that a CysC mutant, V57N, stabilized against domain-swapping, readily forms fibrils, contradicting the propagated domain-swapping hypothesis. Furthermore, in physiological buffer, wt CysC can form oligomers without undergoing domain-swapping. These non-swapped oligomers are identical in secondary structure to CysC monomers and completely retain protease inhibitory activity. However, unlike monomers or dimers, the oligomers bind fluorescent dyes that indicate they have characteristics of pre-amyloid aggregates. Although these oligomers appear to be a pre-amyloid assembly, they are slower than CysC monomers to form fibrils. Fibrillation of CysC therefore likely initiates from the monomer and does not require domain-swapping. The non-swapped oligomers likely represent a dead-end offshoot of the amyloid pathway and must dissociate to monomers prior to rearranging to amyloid fibrils. These prefibrillar CysC oligomers were potent inhibitors of aggregation of the Alzheimer's-related peptide, β-amyloid. This result illustrates an example where heterotypic interactions between pre-amyloid oligomers prevent the homotypic interactions that would lead to mature amyloid fibrils.
Keywords: amyloid, amyloid-β (AB), cysteine protease, protein aggregation, protein misfolding, cystatin, domain swapping, oligomers
Introduction
Cystatin C (CysC)2 is a soluble basic protein belonging to the cysteine protease inhibitor family. The protein is at high concentrations in the cerebrospinal fluid (CSF) relative to its concentration in blood (∼5:1 ratio, 3–7 mg/liter in CSF versus 0.6–1.8 mg/liter in plasma), an unusually high ratio given that the total protein content of plasma is ∼300-fold higher than that of CSF (1–3). High CSF and brain tissue content (4) is a consequence of endogeneous synthesis of CysC in the choroid plexus, and by neurons, astrocytes, and neural progenitor cells (5). CysC is normally secreted and is therefore thought of as acting extracellularly. However, it can also be re-internalized, where it may localize to endosomes or lysosomes (6).
CysC is a potent inhibitor of cysteine proteases such as the cathepsins. These proteases degrade intracellular and endocytosed proteins, but also can be secreted to serve a role in remodeling and degrading extracellular matrix (7). Leakage of cathepsin B (CatB) to the cytosol leads to caspase activation (8), so CysC participates in regulating autophagy by inhibiting CatB. In the brain, CysC-CatB interactions are believed to play a role in regulating neuronal apoptosis (9). A high level of cathepsin activity has been linked to a number of neurological disorders (8), supporting a role for CysC in maintaining healthy neurons. CysC also inhibits asparginyl proteases such as legumain, which is involved with antigen processing (10). In addition, CysC serves as a regulatory factor in neural stem cell growth and glial development, and may be involved in induction of the unique properties of the blood-brain barrier (11–13).
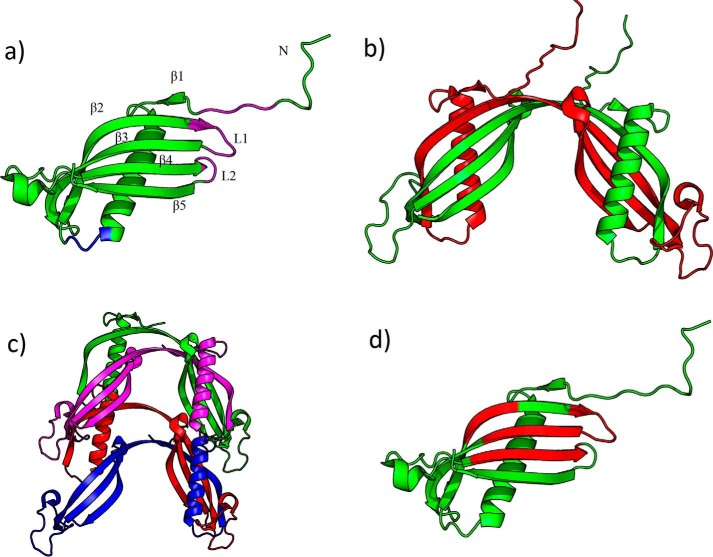
CysC is a small (13.3 kDa) protein that contains two disulfide bonds and is usually non-glycosylated. In solution, the native protein is monomeric; each monomer contains a single five-stranded β-sheet with a highly curved β-bulge encircling the lone α-helix, along with a large disordered loop (Fig. 1a). The monomer is meta-stable: when crystallized (14) or when subjected to mildly destabilizing solvent conditions (15), the protein readily forms domain-swapped dimers, in which the N-terminal β-strands and α-helices are swapped between subunits, and the L1 turn is lost (Fig. 1b). Once formed, domain-swapped dimers are fairly stable under physiological conditions (16). Domain-swapping results in a complete loss of inhibitory activity against cysteine proteases, although legumain inhibition is retained. Newly synthesized CysC leaves the ribosome as roughly half-monomer and half-dimer, but only the monomer is secreted in quantity (17).
Figure 1.
Crystal structure of stabilized monomeric human cystatin C (PDB code 3NX0, modified to include flexible N-terminal residues that were not resolvable in the original structure). a, annotated structure with substrate-binding residues emphasized. Papain, cathepsin-binding site: magenta. Legumain-binding site: blue. b, domain-swapped (PDB code 1R4C). c, propagated domain swapped (hypothesized, adapted from Ref. 21). d, amyloid-prone regions identified by AMYLPRED algorithm (68) highlighted in red.
Besides its normal biological functions, CysC is linked to neurodegenerative amyloid diseases. Wild-type (wt) CysC is a component in the amyloid deposits found in the cerebral vasculature of patients (mostly elderly) with sporadic cerebral amyloid angiopathy (18–20). A mutation, L68Q, is responsible for all known cases of inherited cerebral amyloid angiopathy (Icelandic type), a lethal disease that causes brain hemorrhaging and strikes young adults (21, 22). This mutation destabilizes CysC, making it prone to aggregate into amyloid fibrils. It has been proposed that CysC amyloid fibrils form via propagated domain-swapping (Fig. 1c) (15, 21, 23, 24), although this view has been challenged recently (25, 26).
CysC is found co-deposited with β-amyloid (Aβ) in cerebrovascular and senile plaques of patients with Alzheimer's disease (27, 28). Soluble CysC-Aβ complexes were detected in CSF of Alzheimer's patients (29). Some studies suggest that CysC binds to Aβ, prevents its aggregation, and partially protects neurons from Aβ toxicity (30). Maintenance of the balance between CysC and CatB may be crucial for controlling Aβ deposition and clearance of amyloid deposits (31), and studies with transgenic animals indicate that increasing CysC expression diminishes Aβ deposition (32, 33). The role of CysC in Alzheimer's disease is complex, because of the multiple functions of the protein as a protease inhibitor as well as an inhibitor of Aβ fibrillogenesis (34–36).
Recently, we reported on the production and purification of recombinant human CysC. We discovered that a fraction of the protein readily associates into small soluble oligomers (37), raising the question as to whether these oligomers are intermediates to amyloid fibrils. In this report, we test the proposal that CysC forms fibrils by propagated domain-swapping, we investigate the structure of oligomeric CysC, and we probe the role of oligomers in the CysC amyloidogenesis pathway. Additionally, we demonstrate that these oligomers inhibit Aβ aggregation better than monomeric or domain-swapped dimeric CysC.
Results
V57N CysC does not undergo domain-swapping but still forms fibrils
Wt CysC forms amyloid fibrils under partially denaturing conditions, whereas the disease-related L68Q mutant forms dimers and fibrils more readily than wt (24, 38). When disulfide bonds were inserted to link strands 2 and 3, thereby preventing domain-swapping and dimerization, the stabilized monomer was resistant to dimerization or fibril formation (24). These and similar observations led to the proposal that CysC fibril formation proceeds via propagated domain-swapping (Fig. 1c) (14, 21).
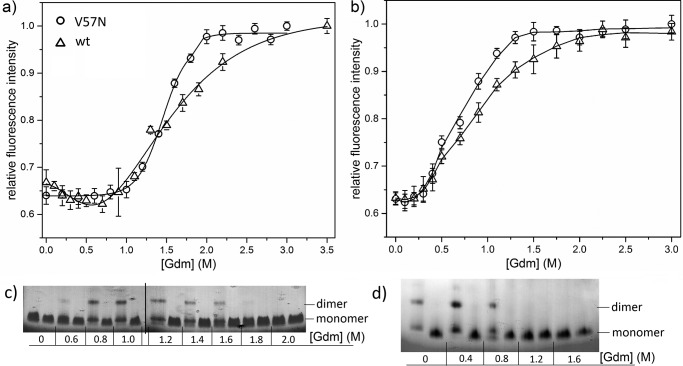
Given the propagated domain-swapping hypothesis, we speculated that a single-point mutation that inhibits domain-swapping should also reduce its amyloid-forming capability. To test, we chose V57N, an engineered mutant designed to prevent domain-swapping by relieving the high strain of the hinge-loop region associated with the distorted Val57 backbone angle (39). Wt and V57N were incubated in 0–3.5 m GdmHCl at physiological pH and temperature (pH 7.4, 37 °C), conditions known to lead to domain-swapping and dimerization in wt CysC. The experiment was repeated at acidic pH and high temperature (pH 4.0, 45 °C), conditions that are known to induce fibrillogenesis of wt (21). Denaturation was monitored by an increase in Trp106 fluorescence emission intensity. Data were fit to a two-state unfolding model to determine apparent [GdmHCl]50, the concentration of GdmHCl at which half the protein has unfolded. Samples were separated by native gel electrophoresis to visualize dimer formation.
The unfolding curve for wt CysC at pH 7.4 was broad, with [GdmHCl]50 of 1.61 ± 0.08 m and complete unfolding at ∼2.75 m (Fig. 2a). CysC formed dimers over a range of 0.6–1.6 m GdmHCl, with the densest dimer bands appearing around 1.0 m (Fig. 2c). Of note, there is a slight dip in fluorescence intensity at 0.6 m GdmHCl that coincides with the appearance of dimers (Fig. 2c). Others have shown that low concentrations of GdmHCl will facilitate disruption of hydrogen bonding between β2 and β3 strands in the monomer and subsequent re-association as a domain-swapped dimer (Fig. 1) (15, 40). Because the lone Trp, Trp106, natively contacts the loop between β2 and β3, we attribute the dip in fluorescence intensity at 0.6 m GdmHCl to the rearrangement of the β2 and β3 strands during the domain-swapping process. At pH 4.0, [GdmHCl]50 of wt was reduced to 0.78 ± 0.04 m (Fig. 2b), indicating a destabilization of the native-fold relative to physiological conditions. At this acidic pH and higher temperature, wt formed a mixture of dimers and monomers even at 0 m GdmHCl, and dimers were present up to 0.8 m GdmHCl (Fig. 2d).
Figure 2.
Comparison of denaturation of wt to V57N CysC. a and b, CysC V57N mutant and wt stability at pH 7.4, 37 °C (a), and pH 4.0, 45 °C (b), monitored by Trp106 fluorescence (excitation, 295 nm; emission, 350 nm) with increasing GdmHCl concentration. Error bars are standard deviation for 3 independent samples. Lines shown are smooth curves. c and d, dimerization tendency of wt (left lanes) compared with V57N (right lanes) in various concentrations of GdmHCl at pH 7.4, 37 °C (c), or pH 4.0, 45 °C (d).
The V57N unfolding curve at pH 7.4 lacked the dip at 0.6 m GdmHCl and had a single sharp transition, indicative of simple cooperative two-state unfolding. [GdmHCl]50 at pH 7.4 was 1.47 ± 0.01 m, declining to 0.68 ± 0.03 m at pH 4.0 (Fig. 2, a and b). Thus, by this measure V57N is slightly less stable than wt at both tested pH conditions. V57N did not produce dimers at any GdmHCl concentration or pH (Fig. 2, c and d), consistent with previous reports (39, 40).
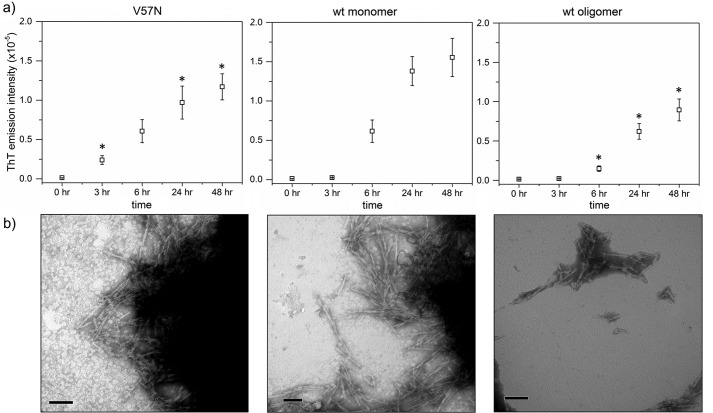
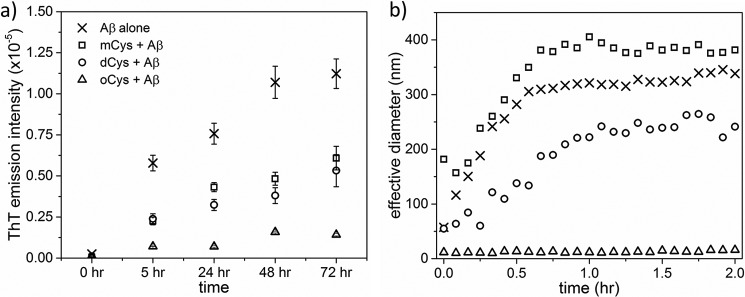
To produce fibrils, we incubated wt and V57N in acidic buffer at elevated temperature and agitation, following literature protocols (21). Fibril growth was tracked using ThT fluorescence and TEM. With wt, ThT-positive fibrils were first detected at 6 h; fibril formation was faster for V57N, with a ThT-positive signal at only 3 h (Fig. 3a). For both samples, ThT intensity continued to increase over 1–2 days. Samples taken at 6 h were imaged by TEM (Fig. 3b). The total fibril content was similar between wt and V57N samples, although V57N fibrils tended to cluster more into larger aggregates. Taken together, these data contradict the hypothesis that CysC fibrils must grow by propagated domain-swapping, because V57N is resistant to dimerization and domain-swapping but readily forms amyloid fibrils. Due to clustering effects, high resolution images of individual fibrils could not be taken. We attempted to form fibrils without agitation to discourage clustering, but found no fibrils after 2 weeks of incubation. Agitation appears to be necessary for fibrillation of both wt and V57N.
Figure 3.
Amyloid fibril growth of CysC. Fibrils were formed by incubation of protein at 60 μm in 50 mm acetate, 5 mm phosphate, 100 mm NaCl buffered to pH 4.0. Samples were continuously agitated at 45 °C. a, ThT fluorescence intensity (excitation, 440 nm; emission, 480 nm). Error bars are the standard deviation of 6 independent measurements. Statistically relevant difference (p < 0.05) of V57N or oCys compared with mCys for a given time point is indicated by an asterisk (*). b, TEM images of the 6-h time point of each sample. Scale bars are 100 nm.
oCys forms fibrils more slowly than mCys
In previous work, we developed a simplified affinity chromatography-based protocol for production of recombinant CysC (37). During the routine protein concentration, we discovered CysC contained some oligomers. As shown previously, oligomer formation is not a consequence of Escherichia coliexpression but rather arises naturally during processes that lead to locally high concentrations (37). Once formed, oligomers do not readily dissociate back to monomers with dilution (37) and so can be considered kinetically trapped. Oligomers (oCys) were fractionated from monomers (mCys) using membrane filters of the appropriate molecular weight cutoff size. Speculating that the oligomers might be intermediates on the path toward amyloid fibrils, we tested the capacity of oCys to form fibrils under the same conditions as previously described. In contrast to our initial expectation, fibril onset was considerably later for oCys compared with mCys, and ThT fluorescence increased at a slower rate (Fig. 3a). A TEM sample confirmed that the fibril content was significantly lower for oCys than either mCys or V57N (Fig. 3b). Rather than being amyloid intermediates, oCys proved to be resistant to amyloidogenesis.
Size characteristics of mCys, dCys, and oCys
To uncover reasons why oCys was less prone to amyloid fibril formation, we searched for differences in its structural characteristics compared with mCys and the domain-swapped dimeric CysC (dCys). We first confirmed the monomeric or oligomeric status of each of the three preparations. Weight-averaged molecular weights (M̄W) were obtained by static light scattering (SLS) (Table 1). M̄W of mCys was ∼8% higher than the known molecular weight of the monomer, indicative of a small (<10%) fraction of dimer or oligomer. M̄W of dCys was smaller than that expected for a pure dimer (26,600 g/mol); from the measurement we estimated that dCys contained ∼30–35% (by mass) monomer and 65–70% dimer. This is consistent with other reports of incomplete domain-swapping by CysC, presumably due to establishment of an equilibrium between monomers and dimers (16, 41). M̄W for oCys was approximately that of a trimer.
Table 1.
Size characterization of CysC
| CysC preparation | M̄Wa | dhb | dspherec | dh/dsphered |
|---|---|---|---|---|
| g/mol | nm | |||
| mCys | 14,500 ± 300 | 5.4 ± 0.1 | 3.7 | 1.46 |
| dCys | 21,800 ± 400 | 5.9 ± 0.2 | 4.2 | 1.40 |
| oCys | 42,300 ± 300 | 8.3 ± 1.6 | 5.3 | 1.57 |
a M̄W for CysC monomers is 13,300 g/mol. Error in M̄W determination was found from the zero-angle extrapolation error during least squares analysis of SLS data.
b Z-averaged hydrodynamic diameter and variance, determined from cumulants analysis.
c dsphere is calculated based on the diameter of a spherical protein occupying the same volume as a protein with the molecular weight of each fraction shown, assuming an average density of 0.73 g/ml and average 0.35 g water/g dry protein (43, 78).
d dh/dsphere is a measure of the non-sphericity or excess hydration of the protein (78).
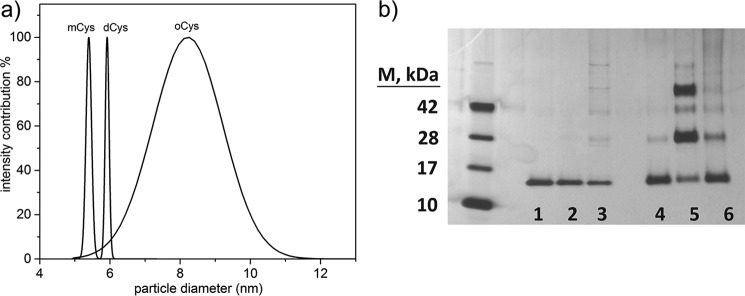
Z-averaged hydrodynamic diameter dh was obtained by cumulants analysis of dynamic light scattering (DLS) data (Table 1). As expected, dh increased in the order mCys < dCys < oCys. The monomer diameter is consistent with other reports (42). We compared the measured dh to that expected for a hydrated spherical protein of the same molecular weight, dsphere (43). A large ratio dh/dsphere is indicative of an elongated shape and/or a highly hydrated molecule. As shown in Table 1, dCys was more compact than either mCys or oCys. DLS data were further analyzed to ascertain the polydispersity of the solutions. Although mCys and dCys size distributions were very narrow, a relatively broad distribution was obtained for oCys (Fig. 4a), indicating that oCys is a population ensemble with an average M̄W of a trimer, rather than a single species.
Figure 4.
a, DLS analysis of mCys, dCys, and oCys fractions. Fits obtained by CONTIN. b, glutaraldehyde cross-linking used to stabilize oligomer complexes and separate by SDS-PAGE. Lanes 1–3 are not cross-linked, whereas lanes 4–6 were cross-linked using 0.5% (w/v) glutaraldehyde for 1 min. Lanes 1, 4-mCys; 2, 5-dCys; 3, 6-oCys.
We next used cross-linking plus gel electrophoresis to obtain further evidence of stable protein complexes, and to search for indirect evidence of structural differences. In the absence of cross-linker, each fraction ran as a monomer, with a small fraction of SDS-resistant species in oCys (Fig. 4b, lanes 1–3). After cross-linking, mCys was largely monomeric but with a small dimer band, consistent with SLS results. dCys contained a roughly equal mixture of dimers and tetramers, with ∼15% monomer (based on image analysis of band density). Existence of a significant stable tetramer population, although inconsistent with our SLS data (Table 1), is attributable to glutaraldehyde-mediated capture of transient tetramers formed during cross-linking.
Cross-linked oCys resolved to monomer (52%), dimer (23%), trimer (10%), and tetramer (8%). This exponential decay in concentration suggests that oligomers assemble non-cooperatively. M̄W calculated from glutaraldehyde cross-linking is 30 kDa, versus 42 kDa by SLS. This suggests that glutaraldehyde is not as efficient at trapping dimers and other oligomers in the oCys population compared with dCys, and may indicate that the monomers are not as tightly compacted in the oligomer as in the dimer, consistent with the larger dh/dsphere of the oligomer.
Secondary and tertiary structural characterization
We collected and analyzed CD spectra for dCys and compared it to mCys and oCys (supplemental Fig. S1 and Table 2). Secondary structure analysis of mCys is consistent with that reported by others (38). Although mCys and oCys have nearly identical spectra, dCys is shifted with a slight increase in β-sheet content. Our spectra for dCys is consistent with previously reported CD spectra of domain-swapped CysC at low pH (41).
Table 2.
Structural characterization of CysC
| CysC preparation | α (%) | β (%) | Turn (%) | Random (%) | λmax | τa | kqb |
|---|---|---|---|---|---|---|---|
| nm | ns | m−1 ns−1 | |||||
| mCys | 15 | 31 | 21 | 34 | 349 | 1.6 ± 0.1 | 4.6 ± 0.3 |
| dCys | 11 | 37 | 22 | 31 | 347 | 2.6 ± 0.2 | NDc |
| oCys | 11 | 32 | 22 | 34 | 343 | 2.5 ± 0.2 | 2.4 ± 0.2 |
a Mean ± S.D. are given. τ of dCys and oCys are not significantly different, but mCys is different (p = 0.05).
b Mean ± S.D. are given. kq of mCys and oCys were found to be significantly different (p = 0.01). kq of dCys was not determined due to nonlinearity.
c ND, not determined.
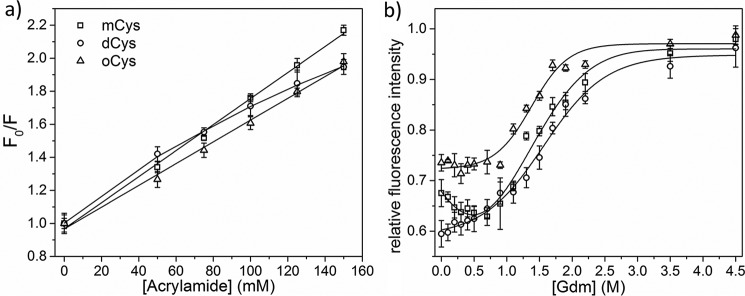
Acrylamide quenching was used to determine the solvent accessibility of the lone Trp, Trp106 (44). The data were plotted according to the Stern-Volmer relationship (Fig. 5a). mCys and oCys yielded linear plots, from which kq was calculated (Table 2). kq for mCys was approximately twice as large as oCys, indicating greater burial of Trp106 in oCys compared with mCys. The plot for dCys was nonlinear, possibly due to occurrence of homoFRET between the dimer Trp groups (supplemental Figs. S2 and S3), so kq was not determined. Given that the oCys plot is linear, whereas dCys is not, it is likely that the geometric arrangement of the monomeric subunits in oCys is different from that in dCys. Specifically, the relative distances between Trps from different monomeric subunits in the oligomer is larger than 16 Å (supplemental Fig. S2).
Figure 5.
a, acrylamide quenching in PBS, pH 7. 4, 22 °C, monitored at 350 nm emission. Lines are given by Stern-Volmer fits. b, denaturation of mCys, dCys, and oCys by GdmHCl addition. Lines are smooth fits. Average of 3 samples per data point is shown. Error bars represent standard deviation of samples.
We compared Trp106 fluorescence of dCys and oCys to mCys, as a function of denaturant concentration (GdmHCl). With no GdmHCl, maximum fluorescence for mCys occurs at 349 nm (supplemental Fig. S4), due to high solvent accessibility of Trp106 in the native-fold, and consistent with other reports (38). dCys was slightly blue-shifted (347 nm) compared with mCys and the emission intensity was smaller. Maximum emission for oCys was further blue-shifted (343 nm) and emission intensity was higher than mCys, indicative of Trp burial, consistent with acrylamide quenching results.
At low GdmHCl (∼0.6 m), fluorescence intensity of mCys decreased (Fig. 5b), a decrease that we previously ascribed to disruption of the β2/β3 strand association in the monomer and rearrangement into domain-swapped dimers (Fig. 2). Fluorescence intensity of dCys was lower than mCys at 0 m GdmHCl, but the two curves converged at moderate GdmHCl, presumably because dCys is in the domain-swapped arrangement even at 0 m GdmHCl. In contrast to mCys, there was no decrease in fluorescence for oCys at low GdmHCl, and the denaturation curve follows a simple two-state transition.
Functional characterization
Natively folded monomeric CysC is a potent inhibitor of cysteine-type proteases, whereas domain-swapping destroys inhibition because of the loss of the L1 loop (15). We evaluated the enzyme inhibitory properties of each of our three preparations, to confirm native-fold in mCys and domain-swapping in dCys, and to determine whether there is evidence that oCys is also domain-swapped.
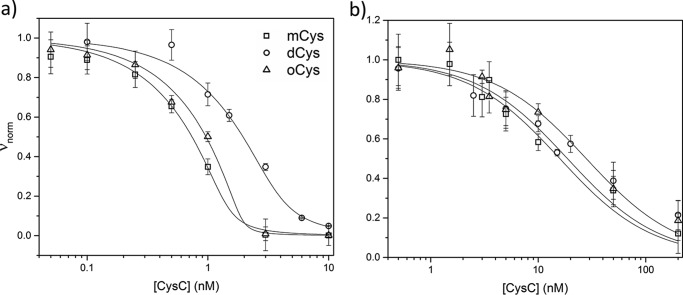
Inhibition of papain activity was detected by monitoring cleavage of a fluorogenic substrate (Z-FR-AMC) as a function of CysC concentration (Fig. 6a). Inhibition constants derived from fitting the data to the Morrison equation for tight-binding enzyme inhibition (45) are found in Table 3. mCys inhibited papain with low picomolar affinity, as expected (46). dCys was much less effective as an inhibitor, supporting assignment as domain-swapped; residual activity could be completely accounted for by considering that dCys contained 30% monomer and 70% dimer, based on M̄W (Table 1). oCys was just as effective as mCys at papain inhibition. CysC also inhibits asparaginyl proteases such as legumain through a binding site between the helix and β2 strand involving Asn39 (10); inhibition is not affected by domain-swapping (10, 23). We added CysC to legumain and measured enzyme cleavage of a fluorogenic substrate (Z-AAN-AMC). Although Ki for oCys is slightly weaker than for dCys or mCys, the difference is not statistically significant (Fig. 6b, Table 3). Taken together, we conclude that oCys retains the secondary and tertiary structure of mCys and, importantly, oCys is not domain-swapped.
Figure 6.
Enzyme inhibition assays. mCys, dCys, and oCys activity against papain (a) and legumain (b). Morrison equation best fit lines are drawn for each sample. Error bars are the standard deviation of 3 independent measurements.
Table 3.
Functional characterization of CysC
Mean ± S.D. in Ki values was found from weighted fitting of enzyme activity data to Morrison equation for tight-binding inhibitors.
| CysC preparation | Papain Ki | Legumain Ki |
|---|---|---|
| pm | nm | |
| mCys | < 15 ± 10 | 4 ± 2 |
| dCys | 3100 ± 200 | 5 ± 2 |
| oCys | < 25 ± 12 | 7 ± 3 |
N terminus is protected in oCys
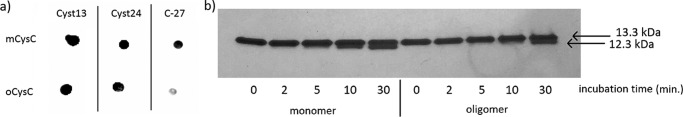
CysC has a Pro-rich, unstructured N-terminal domain (47). Because Pro-rich sequences near protein termini can project outward as stiff arms from the protein body, and form the contact area for protein-protein interactions (48), we examined the solvent accessibility of the N terminus on mCys and oCys using three monoclonal antibodies: C-27, which recognizes the N terminus (49); Cyst13, which targets the C terminus (50); and Cyst24, with an unknown epitope that does not overlap with Cyst13 (51). Cyst13 and Cyst24 bound to both preparations, whereas C-27 bound mCys but not oCys (Fig. 7a, supplemental Fig. S5).
Figure 7.
Characterization of N-terminal solvent accessibility. a, dot blot for determination of free CysC epitope. b, digestion of mCys and oCys in human neutrophil elastase and separation by SDS-PAGE. The 12.3-kDa band represents proteolytically digested CysC.
In addition, we digested CysC with human neutrophil elastase, an enzyme known to specifically cleave the 10Val-Gly11 peptide bond and release the N-terminal decapeptide (52). The proteolytic digestion products were separated by SDS-PAGE and visualized by silver stain (Fig. 7b). With mCys, a 12.3-kDa band, corresponding to N terminally-truncated CysC, appeared within 5–10 min. oCys was degraded more slowly than the monomer (Fig. 7b). Together, these results indicate that the N-terminal segment is less accessible in oCys than in the monomer.
oCys has pre-amyloid characteristics
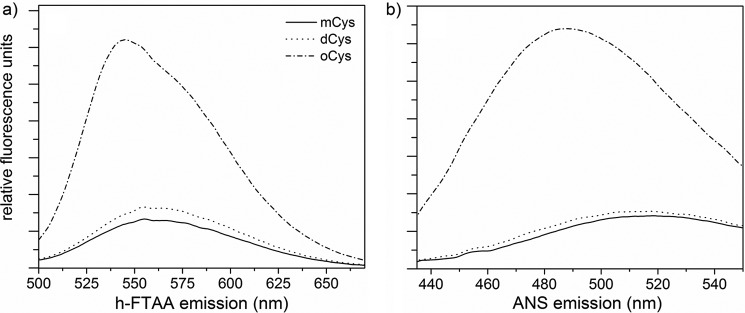
With some amyloidogenic proteins, oligomeric intermediates appear prior to maturation into fibrils (25). To determine whether mCys, dCys, and/or oCys preparations contained any amyloid-like oligomers, we tested for binding to fluorescent dyes that are used to characterize misfolded or aggregated proteins. 8-Anilinonapthalene-1-sulfonic acid (ANS), a dye that binds to hydrophobic patches or cavities on proteins or protein complexes, has frequently been used to detect misfolding intermediates and molten globules (53) and to monitor formation of pre-fibrillar amyloid intermediates (54, 55).
We observed no ANS fluorescence enhancement with mCys or dCys. In contrast, ANS fluorescence intensity was dramatically increased with oCys (Fig. 8a), showing that oligomer formation creates a hydrophobic cavity or surface. Hepta-formylthiophene acetic acid (h-FTAA), a conjugated thiophene, is believed to bind to pre-fibrillar aggregates or early amyloid structures, causing conformational constraints on its backbone planarity that increase its fluorescence quantum yield (56). Co-incubation with mCys and dCys did not cause fluorescence enhancement of h-FTAA, whereas co-incubation with oCys strongly increased its fluorescence intensity (Fig. 8b). None of the CysC preparations resulted in enhancement of ThT fluorescence, demonstrating that oCys does not form mature amyloid fibrils at physiological pH and temperature (data not shown).
Figure 8.
Structure-specific dye binding. Binding and fluorescence of h-FTAA (a) and ANS (b) to each of the 3 fractions of CysC. Proteins at 2 μm were incubated with dye in PBS, pH 7.4, at 22 °C. Data shown are the average of 3 independent scans.
oCys is a potent inhibitor of Aβ aggregation
CysC has been reported to inhibit Aβ fibril formation (57, 58), although the interaction has not been characterized in much detail. We added mCys, dCys, or oCys to freshly-prepared monomeric Aβ and followed aggregation by DLS and ThT fluorescence. In physiological buffer, and in the absence of Aβ, none of the CysC samples were ThT positive and no aggregate growth was measured by DLS (data not shown). mCys added to Aβ did not alter the rate of early aggregate development (Fig. 9b), but did modestly reduce the total fibril mass (Fig. 9a). dCys addition to Aβ resulted in a minor reduction of early aggregate growth rate compared with mCys, but the rate of fibril formation by ThT was not significantly different from the mCys sample. In sharp contrast, oCys addition to Aβ almost completely inhibited aggregation. There was no growth in aggregate size measured by DLS, and very little development of ThT fluorescence.
Figure 9.
Interaction of CysC fractions with Aβ peptide. Aggregation of Aβ (28 μm) in the presence of CysC at 2:1 (Aβ:CysC) molar ratio was monitored. a, ThT fluorescence emission. All co-incubation samples after the 0-h time point are statistically different from Aβ alone (p < 0.05). oCys co-incubation samples after 0 h are statistically different from other co-incubation samples (p < 0.05). Error bars are the standard deviation of 4 independent measurements. b, DLS mean particle size. Samples shown are representative of at least 3 independent samples.
Discussion
The underlying cause of several neurodegenerative diseases, including Alzheimer's, Parkinson's, and Huntington's, is attributed to aggregation of functional proteins into misfolded amyloid fibrils, motivating numerous investigations into the kinetics and mechanism of amyloidogenesis. Commonly observed mechanisms for amyloid aggregation include the nucleation-elongation model, where a rare event provides a nucleus for further growth by monomer addition, and the aggregation-conformational conversion model, where association of natively folded or intrinsically disordered proteins into soluble oligomers is followed by subsequent rearrangement within the hydrophobic core of the oligomer to the cross-β-sheet structure typical of amyloid fibrils (59–62). CysC has been proposed to aggregate via a third mechanism, propagated domain-swapping (15, 21, 23, 24), a mechanism suggested for a few other proteins such as β2-microglobulin (β2m) (63).
The propagated domain-swapping hypothesis arose from the observation that a disease-associated mutant form of CysC, L68Q, readily undergoes unfolding at the L1 loop and also forms amyloid fibrils more readily than wt (64). However, direct evidence of L68Q aggregation by domain-swapping is lacking. Although wt CysC crystallizes as domain-swapped dimers, the L68Q crystal structure is not available due to the instability of the protein. L68Q aggregation has been monitored by agarose gel electrophoresis and size-exclusion chromatography (24, 40), but these methods only identify size, not structure.
Additional support for propagated domain-swapping was found from a study of disulfide-stabilized CysC (21, 24). In this mutant (HCC-stab1, L47C/G69C), a disulfide bond was engineered to restrict movement of the L1 region and impede domain-swapping. HCC-stab1 did not form fibrils under conditions where wt CysC does; fibril formation occurred only after reduction of the disulfide bond (21, 23).
However, the propagated domain-swapping mechanism has been challenged recently (25, 65). For example, cystatin B, a close relative of CysC, is known to dimerize by domain-swapping but forms fibrils that are structurally inconsistent with propagated domain-swapping (26).
We designed a test of the hypothesis by comparing the aggregation rate of wt CysC with a mutant, V57N, which has previously been shown to resist domain-swapping and dimerization, because the mutation stabilizes the L1 loop (39). We first confirmed that V57N, unlike wt, did not undergo dimerization at low GdmHCl concentrations or in acidic buffer. Next, we showed that both wt and V57N readily formed fibrils if mechanically agitated at pH 4 and 45 °C, with a somewhat shorter lag time for V57N than for wt. We conclude that domain-swapping is not necessary for CysC amyloid fibril formation, and that it is not necessary to invoke a propagated domain-swapping mechanism for this protein. Rather, fibril formation propensity is likely due to destabilization of the native protein fold. This is supported by our finding that V57N is both less stable than wt and forms fibrils with a shorter lag period.
The secondary structure of L68Q contains less helix and more disorder than wt, Trp106 is more solvent-exposed (38), and L68Q is more readily denatured than wt (41). The lower stability in L68Q could explain why the mutant is prone to fibril formation, without the need to invoke susceptibility to domain-swapping per se. Conversely, the disulfide bond in HCC-stab1 may increase the stability of that mutant relative to wt and explain its resistance to fibrillogenesis. An additional factor may arise because the HCC-stab1 mutation flattens and shortens the β-strand around the 47–51 region, whereas the V57N mutation does not (39), one of three regions that is predicted to be an aggregation-prone region by amyloid predictor algorithms (Fig. 1d) (66, 67). Consistent with this prediction, the pentapeptide LQVVR has been shown to spontaneously form amyloid fibrils in vitro (68). The observed reduction in amyloid propensity of HCC-stab1 might be explained by the structural change in this region.
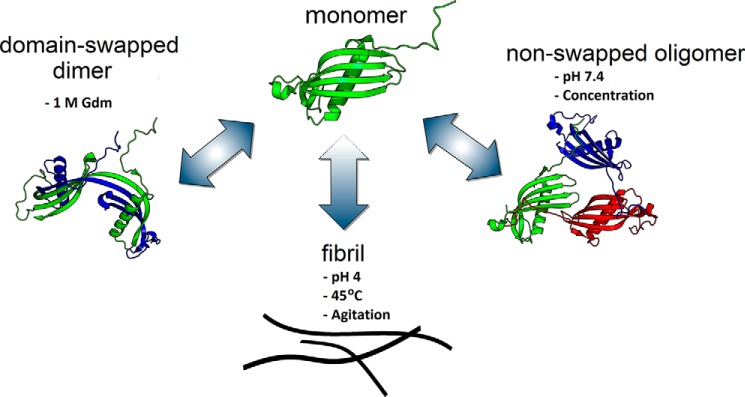
Soluble oligomers of CysC were resistant to fibril formation, with a significantly longer lag time and significantly lower fibril content than either monomeric wt or V57N. We propose that CysC oligomers must dissociate to monomer prior to fibril initiation and growth, in other words, oligomers are “off-pathway” for amyloid fibril formation (Fig. 10). A similar mechanism was shown for β2m, specifically, β2m self-assembled by two competing pathways, one of which led to formation of oligomers and wormlike linear aggregates, whereas the other led to classical rigid amyloid fibrils (69). β2m oligomers and wormlike aggregates were required to dissociate to monomers before re-associating into amyloid fibrils.
Figure 10.
Conceptual diagram of CysC aggregation pathways. CysC monomers self-associate by 3 distinct pathways. Mild chemical denaturation (1 m GdmHCl) supports rearrangement into domain-swapped dimers that are stable and no longer inhibit cysteine proteases. Mild concentrations during ultrafiltration or adsorption steps produces oligomers with similar secondary structure and enzyme inhibition properties as monomers. Agitation at acid pH and high temperature induces amyloid fibril formation. All 3 self-associated forms are fairly stable when returned to physiological solvent, which suggests that the monomer is a metastable state. Fibril growth proceeds through the monomer and does not require propagated domain-swapping.
Because oCys was resistant to fibril growth, we undertook further characterization of oligomer size and structure. Briefly, the oligomers are small clusters of natively folded monomers. They retain the native secondary structure and enzyme inhibitory function of the monomer and are not domain-swapped, but differ from monomers in reduced solvent accessibility of Trp106. The blue-shift in Trp106 fluorescence in oCys (relative to wt) is in contrast to the red-shift in L68Q (38), suggesting that the relative burial or exposure of Trp106 correlates with relative resistance or susceptibility to amyloidogenesis, respectively. Another structural distinction is the protection of the Pro-rich N terminus of oCys compared with mCys. Involvement of stiff Pro-rich “arms” in protein self-association has been observed in other systems (48). Building on this, we speculate that oligomer formation may be mediated through interactions of the extended Pro-rich N-terminal arms (Fig. 10).
Although oCys are “dead ends” for mature amyloid fibrils, the oligomers exhibit some structural features of pre-amyloid intermediates. Specifically, oCys are positive for both ANS and h-FTAA, two fluorescent dyes that are useful markers of amyloid intermediates (54–56). This behavior contrasts with that of β-amyloid (Aβ), the Alzheimer-related peptide that is one of the best known examples of amyloidogenesis. Aβ amyloid fibrils arise directly via maturation of soluble oligomers that are ANS- and h-FTAA-positive (70, 71). The soluble oligomers of Aβ are believed to be more toxic than the mature fibrils (70).
A few previous studies have demonstrated that CysC affects Aβ aggregation in vitro. Sastre et al. (57) co-incubated CysC and Aβ in PBS (pH 7, 37 °C) and observed a decrease in fibril formation by TEM, with a concomitant increase in amorphous aggregates. Similar amorphous aggregates were observed with CysC alone, which somewhat confounds interpretation of the data with Aβ. Using gel electrophoresis and Western blotting, Selenica and co-workers (58) observed a decrease in soluble Aβ monomers and oligomers with the addition of CysC as well as an increase in precipitable protein. These two studies both indicate that CysC changes Aβ aggregates from fibrils to amorphous, but does not inhibit Aβ aggregation overall. In another study, binding of the conformation-dependent antibody A11 to Aβ oligomers decreased with increasing CysC concentration (30), which could be due either to a competition for binding to Aβ between A11 and CysC, or a CysC-mediated shift in Aβ aggregation pathway.
In our work, we used two well known assays to detect the effect of CysC on Aβ aggregation kinetics: DLS, where the average size of aggregates is measured in solution, and ThT fluorescence, which is proportional to the mass of amyloid fibrils. At physiological pH, CysC alone did not aggregate by DLS and contained no amyloid fibrils by ThT. By DLS we observed that monomeric and dimeric CysC have only a minor effect at delaying Aβ aggregation. With both mCys and dCys, we observed little reduction in Aβ aggregation as measured by DLS and a modest inhibition of Aβ fibril growth as measured by ThT, suggesting that CysC monomers and dimers may slightly re-direct Aβ away from amyloid fibril formation and toward non-amyloid aggregates, but the effect is minor.
In sharp contrast, CysC oligomers were remarkably potent at arresting aggregation of Aβ, as measured by either fibril mass (ThT) or light scattering. Combined with the observation that oCys (but not mCys or dCys) have characteristics of pre-amyloid intermediates (e.g. ANS and h-FTAA fluorescence), we hypothesize that this strong hetero-interaction arises because oCys shares some structural features of Aβ oligomers. We envision the CysC oligomers as “frustrated amyloid,” resistant to full growth into fibrils but with some structural characteristics of prefibrillar intermediates. The interaction between oCys and Aβ could arise from their structural similarities, as an example of heterotypic binding of one pre-amyloid aggregate to another (72). However, the result of this cross-amyloid interaction is prevention of full maturation into amyloid fibrils, possibly because of partial structural mismatch between the different oligomers. This could be an example of an amyloid regulatory network, where homotypic interactions lead to amyloid fibrils but heterotypic interactions lead to off-pathway complexes, preventing amyloid formation.
Experimental procedures
Cystatin C preparation
Recombinant human CysC was produced and purified as previously described (37). Purified CysC in PBSA-E buffer (PBSA-E: 10 mm phosphate, 150 mm NaCl, 0.02% (w/v) azide, 1 mm EDTA, pH 7.4) was concentrated by ultrafiltration in a 3-kDa centrifugal membrane (Amicon Ultra-15) from 20 to 120 μm. Concentrated protein was fractionated by centrifugal ultrafiltration through a 30-kDa membrane. The filtrate, “mCys,” was stored at 4 °C. The retained volume, enriched in oligomeric cystatin C, “oCys,” was diluted to 45 μm with PBSA-E and stored at 4 °C. Oligomer stability under these conditions was confirmed by ANS-binding assays and light scattering up to 2 months after the initial preparation.
To produce domain-swapped dimers, mCys was concentrated to 100 μm by centrifugal ultrafiltration through a 3-kDa membrane. GdmHCl (8 m in PBSA-E) was added to a final concentration of 1.0 m (70 μm CysC). This solution was incubated for 1 day at 37 °C, quickly diluted in PBSA-E to 0.1 m GdmHCl, extensively dialyzed against PBSA-E and concentrated via ultrafiltration to 20 μm. The domain-swapped dimer “dCys” was stored without further purification at 4 °C. The protein concentration was determined by measurement of absorbance at 280 nm, using an extinction coefficient of 11,100 m−1 cm −1 (21). All reported protein molar concentrations are given on the basis of a CysC monomer, regardless of oligomer state.
The V57N CysC mutant was constructed by site-directed mutagenesis of the wt plasmid. Plasmid sequence was confirmed following transformation into the BL21(DE3)pLysS expression cell line. Expression, collection, and purification of V57N was identical to wt.
Chemical denaturation
Spectroscopic grade GdmHCl (Calbiochem) was dissolved to saturation (8.4 m at room temperature) in PBSA-E or sodium acetate buffer (50 mm NaAc, 100 mm NaCl, pH 4.0) and filtered through a 0.22-μm PVDF syringe filter (Millex). GdmHCl concentration was confirmed by measurement of refractive index relative to buffer. Samples were prepared to final concentrations of 0–3.5 m GdmHCl and 9 μm CysC in either PBSA-E or sodium acetate buffer. Samples were stored at 4 °C (PBSA-E) or 45 °C (sodium acetate) for 2 days, then either warmed to room temperature for 30 min (PBSA-E) or maintained at 45 °C (sodium acetate) prior to collection of Trp106 fluorescence emission spectra on a PTI QuantaMaster 40 fluorimeter. Samples were excited at 295 nm and emission spectra were collected from 320–380 nm. Spectra were measured at least 3 times and averaged, with background subtracted. Unfolding was monitored by an increase of intensity at 350 nm.
Data were fit to
| (Eq. 1) |
where F is the measured fluorescence intensity at [Gdm], FF and FU are fluorescence intensities at 0 and 100% unfolding, respectively, m is related to the slope of the transition, and [Gdm]50 is the GdmHCl concentration at 50% unfolding. In the case of simple two-state unfolding, [Gdm]50 = ΔGunf/m, where ΔGunf is the Gibbs energy of unfolding at 0 GdmHCl.
Characterization of dimerization by native PAGE
Wt and V57N were concentrated to 85 μm via ultrafiltration. GdmHCl (8.0 m in PBSA-E) was added to 20 μl of either protein and diluted with PBSA-E to final concentrations of 70 μm protein and 0–2.0 M GdmHCl. Samples were incubated for 48 h at 37 °C. Because domain-swapped dimers may dissociate in denaturing conditions, we applied a non-denaturing gel electrophoresis technique for resolving basic proteins (73). Protein samples were diluted to 15 μm in PBSA-E, then mixed with native PAGE sample buffer (×4, Novex) containing Coomassie dye and loaded onto a 4–16% bis-Tris gradient gel (Novex). The cathode was filled with dark blue running buffer, pH 7.0 (50 mm Tricine, 7.5 mm imidazole, 0.02% Coomassie G-250; Novex). The anode was filled with native PAGE running buffer, pH 7.0 (25 mm imidazole; Novex). Proteins were separated at 150 V for 2 h and visualized by destaining in 35% methanol, 7% acetic acid in water overnight.
Fibril formation
V57N, mCys, and oCys in PBSA-E were concentrated to 120 μm (1.6 mg/ml) by ultrafiltration, then combined in equal volumes with sodium acetate buffer (100 mm NaAc, 50 mm NaCl, pH 2.85) in a 96-well plate to a final volume of 200 μl. Final solution conditions were: 60 μm protein, 50 mm NaAc, 5 mm phosphate, 100 mm NaCl, pH 4.0. Samples were agitated by micro-stir bar and heated to 45 °C. Wells were sealed to prevent evaporative loss. At various time intervals, an aliquot was diluted into ThT stock solution (50 μm ThT in PBSA-E), to final concentrations of 2.5 μm protein and 10 μm ThT. Fluorescence measurements were taken with a PTI QuantaMaster 40 fluorimeter at an excitation wavelength of 440 nm and emission of 460–550 nm. Samples at the 6-h time point were taken for TEM imaging. Samples were diluted to 10 μm in PBSA-E, fixed on a pioloform-coated grid, and stained with methylamine tungstate. Images were taken with a Philips CM120 scanning transmission electron microscope (FEI Corp.). Samples were prepared using the same method, but incubated at pH 4.0 and 45 °C but without agitation for 12 days, then imaged by TEM.
Dynamic and static light scattering
mCys, dCys, or oCys (20–50 μm, in PBSA-E) were filtered through a Millipore HV 0.22-μm syringe filter directly into a scrupulously cleaned cylindrical quartz cuvette. The cuvette was inserted into a sample bath containing refractive index-matching fluid (decahydronaphthalene, cis-trans mix, >99% purity, Sigma) and equilibrated to 23 °C. Laser source was an Innova 90C (Coherent, Inc.) 488 nm, 150 milliwatt argon laser. The sample bath was mounted on a turntable goniometer (Brookhaven BI-200SM). Measurement angles were 90° (DLS) or 30–130° (SLS). DLS measurements were collected for 30 min. The method of cumulants was used to determine a mean hydrodynamic diameter, whereas CONTIN was used to determine an intensity-weighted size distribution (74).
SLS data were taken at 23 angles, along with PBSA-E buffer and a toluene standard. Prior to each experiment, the concentration of the protein sample was determined by A280 measurement. The Rayleigh ratio of each sample was calculated as described previously (75), and the weight-averaged molecular weight, M̄W, was determined by weighted linear regression. A more detailed description of DLS and SLS analysis is included in the supplemental information.
Glutaraldehyde cross-linking
Glutaraldehyde is a well known, indiscriminant cross-linker (76, 77), which primarily targets ϵ-amino groups on lysines. Protein was diluted to 20 μm in PBSA-E. Glutaraldehyde (50% (v/v) in water, Sigma) was added to each protein sample to a final concentration of 0.5% (19 μm protein). After 1 min, cross-linking reactions were quenched by addition of NaBH4 to a final concentration of 0.5% (w/v). Reducing SDS sample buffer (10% (w/v) SDS, 10 mm DTT, 80% (v/v) glycerol, 0.5% (w/v) bromphenol blue in 400 mm Tris-HCl) was added to each cross-linked protein sample in 1:10 volume ratio. Samples were heated to 85 °C for 5 min, then loaded onto a 4–20% gradient Tris-HEPES polyacrylamide gel (Thermo) and separated at 125 V for 40 min. Protein bands were visualized by silver stain (Pierce).
Acrylamide quenching
Acrylamide (electrophoresis grade, >99%, Sigma) was dissolved in PBSA-E buffer to 4.5 m and filtered through a 0.22-μm PVDF syringe filter (Millex), then added to protein samples to final concentrations of 25–150 mm acrylamide and 9 μm mCys, dCys, or oCys. Samples were incubated at room temperature for 1 h prior to measurement of steady state Trp106 fluorescence. Sample data were fit to the Stern-Volmer equation to determine the quenching constant, KSV, and the bimolecular quenching rate constant, kq.
| (Eq. 2) |
F and F0 are the fluorescence intensity in the presence and absence of quencher, respectively, [Q] is the molar concentration of quencher, and τ̄0 is the mean fluorescence lifetime. Trp106 fluorescence lifetime was measured by time-correlated single photon counting (TCSPC) by a PTI QuantaMaster 40 with 280-nm LED excitation source. Photon emission events were monitored at 350 nm. An instrument response function was measured using a 1 wt % suspension of Ludox particles in water. Data collection and analysis were performed through FelixGX software (PTI, Inc.). Sample curves were fit to model exponential decay functions using a nonlinear least squares analysis to find the mean lifetime.
Enzyme inhibition assay
Inhibition of papain was measured as described previously (37). Briefly, cleavage of fluorogenic substrate Z-FR-AMC by papain was monitored in the presence of increasing concentrations of mCys, dCys, or oCys. Final reaction conditions were: 2.5 nm activated papain, 0–10 nm CysC, 100 μm Z-FR-AMC in Tris buffer (50 mm Tris-HCl, pH 7.0, 0.02% Tween 20).
Inhibition of legumain was measured similarly to papain, with minor modifications (10). Human legumain (R&D Systems) was activated by dilution into acetate buffer (50 mm acetate, 100 mm NaCl, pH 4) to 100 μg/ml and incubation at 37 °C for 2 h. Activated legumain was diluted into MES buffer (50 mm MES, 250 mm NaCl, pH 5) to 2 μg/ml. CysC was diluted from stock to 0.2–4 μm in MES buffer, mixed in equal volume with legumain, and incubated at 37 °C for 10 min. The fluorogenic substrate Z-AAN-AMC (Bachem) in N,N-dimethylformamide was diluted to 200 μm using MES buffer immediately before use. The CysC/legumain mixtures were diluted 5-fold with MES buffer and mixed in an equal volume with Z-AAN-AMC (200 μm) in a black polystyrene 96-well plate. The final concentrations of substrate, legumain, and CysC were: 100 μm, 2 nm, and 0–200 nm, respectively.
For both enzyme assays, samples were excited at 360 nm and fluorescence was measured at 460 nm by a Biotek FLX800 fluorescence plate reader for 5 min. The rate of fluorescence increase was measured and normalized to the zero-inhibition sample. Fractional inhibition of active enzyme was plotted against CysC concentration and fit to the Morrison equation (37, 45) to obtain the inhibition constant, Ki.
To find Km, substrate (0–300 μm) was mixed with a fixed enzyme concentration (5 nm) and initial rates were measured as described, then fit to the Michaelis-Menten equation. Km for papain/Z-FR-AMC was 110 ± 10 μm and legumain/Z-AAN-AMC was 30 ± 3 μm.
Neutrophil elastase digestion
Human neutrophil elastase (Athens Research & Technology) was reconstituted in acetate buffer (50 mm acetate, 150 mm NaCl, pH 5.5) and stored at 58 μm. Elastase was added to either mCys or oCys to a final concentration of 150 nm elastase and 15 μm CysC and incubated at 37 °C for 2–30 min. To quench the elastase activity, 20 μl of sample was combined with 20 μl of reducing SDS buffer (4% SDS, 4 mm DTT, 2 mm PMSF, 16% glycerol, 0.1% bromphenol blue in 400 mm Tris-HCl). Samples were heated, separated by SDS-PAGE, and silver stained as described earlier. Elastase cleavage of the N-terminal segment of CysC was monitored by the appearance of a smaller molecular weight band.
Antibody presentation by dot blot
mCys and oCys were diluted to 10 μm and 2 μl was blotted onto a nitrocellulose membrane and dried completely. Each membrane was blocked by incubation overnight at 4 °C in 5% nonfat dry milk. After thorough washing in TBS-T (20 mm Tris, 150 mm NaCl, 0.05% Tween 20, pH 7.5), each membrane was incubated with one of three mouse monoclonal anti-CysC antibodies: Cyst13 (Novus, NB100-62492, 1:3000 dilution in TBS-T), Cyst24 (Novus, NB100-62127, 1:3000 dilution in TBS-T), or C-27 (Santa Cruz Biotechnology, sc-73878, 1:500 dilution in TBS-T) for 1 h at room temperature. After washing in TBS-T, each membrane was incubated with goat anti-mouse poly-HRP-conjugated antibody (Pierce, 32230, 1:3000 dilution in TBS-T) at room temperature for 1 h. Membranes were washed in TBS-T and antibody binding was visualized by chemiluminescence (Amersham Biosciences ECL Western blot Analysis system, protocol and kit by GE Healthcare Life Sciences).
Dye binding assays
mCys, dCys, or oCys (2 μm) were incubated with either ANS (28 μm, TCI Chemicals) or h-FTAA (0.5 μm) at room temperature for 10 min. h-FTAA was a generous gift from Drs. Peter Nilsson and Per Hammarström (Linköping University, Sweden). Fluorescence emission spectra were taken on a PTI QuantaMaster40 fluorimeter. Excitation wavelengths were 370 nm for ANS and 480 nm for h-FTAA. Emission was recorded over 440–550 nm for ANS and 500–670 nm for h-FTAA.
β-Amyloid (Aβ) interaction studies
Lyophilized Aβ(1–40) (Bachem) was reconstituted to 2.8 mm in 100 mm glycine, 8 m urea, pH 10, buffer to completely disaggregate the peptide. Aggregation of Aβ was initiated by rapid dilution into PBSA-E to 28 μm. In some experiments, stock mCys, dCys, or oCys was added to a final concentration of 14 μm. For DLS studies, the protein mixture was filtered through a 0.22-μm syringe filter directly into a clean cuvette. The cuvette was placed into a solvent bath maintained at 37 °C. Sample autocorrelation functions were averaged over a 5-min period and recorded for up to 2 h. For ThT measurements, protein mixtures were incubated at 37 °C in sealed tubes with no agitation. Over the course of 72 h, samples were taken and ThT fluorescence spectra were determined as described earlier.
Author contributions
T. J. P. and R. M. M. designed the study and wrote the paper. J. D. M. produced and purified CysC and V57N, and performed fibrillation studies described by Fig. 3. T. J. P. carried out all other experiments. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
CD data were obtained at the University of Wisconsin-Madison Biophysics Instrumentation Facility, which was established with support from the University of Wisconsin-Madison and National Science Foundation Grant BIR-9512577 and National Institutes of Health Grant S10 RR13790, with technical assistance provided by Dr. Darrell McCaslin. Randall Massey provided technical assistance with TEM. h-FTAA pre-amyloid dye was a generous gift from Drs. Peter Nilsson and Per Hammarström (Linköping University, Sweden).
This work was supported by National Science Foundation Grant CBET-1262729. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S5.
- CysC
- cystatin C
- CSF
- cerebrospinal fluid
- CatB
- cathepsin B
- SLS
- static light scattering
- DLS
- dynamic light scattering
- ThT
- thioflavin T
- TEM
- transmission electron microscopy
- wt
- wild-type
- dCys
- dimeric CysC
- oCys
- oligomeric cystatin C
- mCys
- monomeric cystatin C
- β2m
- β2-microglobulin
- GdmHCl
- guanidinium hydrochloride
- bisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- ANS
- 8-anilinonapthalene-1-sulfonic acid
- h-FTAA
- hepta-formylthiophene acetic acid
- PDB
- Protein Data Bank
- Z
- benzyloxycarbonyl.
References
- 1. Spector R., Snodgrass S., and Johanson C. E. (2015) A balanced view of the cerebrospinal fluid composition and functions: focus on adult humans. Exp. Neurol. 273, 57–68 [DOI] [PubMed] [Google Scholar]
- 2. Filler G., Bökenkamp A., Hofmann W., Le Bricon T., Martínez-Brú C., and Grubb A. (2005) Cystatin C as a marker of GFR: history, indications, and future research. Clin. Biochem. 38, 1–8 [DOI] [PubMed] [Google Scholar]
- 3. Pate K. M., and Murphy R. M. (2017) Cerebrospinal fluid proteins as regulators of β-amyloid aggregation and toxicity. Isr. J. Chem. 10.1002/ijch.201600078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Håkansson K., Huh C., Grubb A., Karlsson S., and Abrahamson M. (1996) Mouse and rat cystatin C: Escherichia coli production, characterization and tissue distribution. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 114, 303–311 [DOI] [PubMed] [Google Scholar]
- 5. Mussap M., and Plebani M. (2004) Biochemistry and clinical role of human cystatin C. Crit. Rev. Clin. Lab. Sci. 41, 467–550 [DOI] [PubMed] [Google Scholar]
- 6. Wallin H., Bjarnadottir M., Vogel L. K., Wassélius J., Ekström U., and Abrahamson M. (2010) Cystatins, extra- and intracellular cysteine protease inhibitors: high-level secretion and uptake of cystatin C in human neuroblastoma cells. Biochimie 92, 1625–1634 [DOI] [PubMed] [Google Scholar]
- 7. Lutgens S. P., Cleutjens K. B., Daemen M. J., and Heeneman S. (2007) Cathepsin cysteine proteases in cardiovascular disease. FASEB J. 21, 3029–3041 [DOI] [PubMed] [Google Scholar]
- 8. Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., and Turk D. (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta 1824, 68–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu Y., Ding Y., Li X., and Wu X. (2015) Cystatin C is a disease-associated protein subject to multiple regulation. Immunol. Cell Biol. 93, 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alvarez-Fernandez M., Barrett A. J., Gerhartz B., Dando P. M., Ni J., and Abrahamson M. (1999) Inhibition of mammalian legumain by some cystatins is due to a novel second reactive site. J. Biol. Chem. 274, 19195–19203 [DOI] [PubMed] [Google Scholar]
- 11. Taupin P., Ray J., Fischer W. H., Suhr S. T., Hakansson K., Grubb A., and Gage F. H. (2000) FGF-2-responsive neural stem cell proliferation requires CCg, a novel autocrine/paracrine cofactor. Neuron 28, 385–397 [DOI] [PubMed] [Google Scholar]
- 12. Hasegawa A., Naruse M., Hitoshi S., Iwasaki Y., Takebayashi H., and Ikenaka K. (2007) Regulation of glial development by cystatin C. J. Neurochem. 100, 12–22 [DOI] [PubMed] [Google Scholar]
- 13. Enerson B. E., and Drewes L. R. (2006) The rat blood-brain barrier transcriptome. J. Cereb. Blood Flow Metab. 26, 959–973 [DOI] [PubMed] [Google Scholar]
- 14. Janowski R., Kozak M., Jankowska E., Grzonka Z., Grubb A., Abrahamson M., and Jaskolski M. (2001) Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Biol. 8, 316–320 [DOI] [PubMed] [Google Scholar]
- 15. Ekiel I., and Abrahamson M. (1996) Folding-related dimerization of human cystatin C. J. Biol. Chem. 271, 1314–1321 [DOI] [PubMed] [Google Scholar]
- 16. Szymańska A., Jankowska E., Orlikowska M., Behrendt I., Czaplewska P., and Rodziewicz-Motowidło S. (2012) Influence of point mutations on the stability, dimerization, and oligomerization of human cystatin C and its L68Q variant. Front. Mol. Neurosci. 5, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu Y., Lindemann P., Vega-Ramos J., Zhang J. G., and Villadangos J. A. (2014) Developmental regulation of synthesis and dimerization of the amyloidogenic protease inhibitor cystatin C in the hematopoietic system. J. Biol. Chem. 289, 9730–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levy E., Jaskolski M., and Grubb A. (2006) The role of cystatin C in cerebral amyloid angiopathy and stroke: cell biology and animal models. Brain Pathol. 16, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McCarron M. O., Nicoll J. A., Stewart J., Ironside J. W., Mann D. M., Love S., Graham D. I., and Grubb A. (2000) Absence of cystatin C mutation in sporadic cerebral amyloid angiopathy-related hemorrhage. Neurology 54, 242–244 [DOI] [PubMed] [Google Scholar]
- 20. Yamada M. (2000) Cerebral amyloid angiopathy: an overview. Neuropathology 20, 8–22 [DOI] [PubMed] [Google Scholar]
- 21. Wahlbom M., Wang X., Lindström V., Carlemalm E., Jaskolski M., and Grubb A. (2007) Fibrillogenic oligomers of human cystatin C are formed by propagated domain swapping. J. Biol. Chem. 282, 18318–18326 [DOI] [PubMed] [Google Scholar]
- 22. Palsdottir A., Snorradottir A. O., and Thorsteinsson L. (2006) Hereditary cystatin C amyloid angiopathy: genetic, clinical, and pathological aspects. Brain Pathol. 16, 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Östner G., Lindström V., Hjort Christensen P., Kozak M., Abrahamson M., and Grubb A. (2013) Stabilization, characterization, and selective removal of cystatin C amyloid oligomers. J. Biol. Chem. 288, 16438–16450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nilsson M., Wang X., Rodziewicz-Motowidlo S., Janowski R., Lindström V., Onnerfjord P., Westermark G., Grzonka Z., Jaskolski M., and Grubb A. (2004) Prevention of domain swapping inhibits dimerization and amyloid fibril formation of cystatin C: use of engineered disulfide bridges, antibodies, and carboxymethylpapain to stabilize the monomeric form of cystatin C. J. Biol. Chem. 279, 24236–24245 [DOI] [PubMed] [Google Scholar]
- 25. van der Wel P. C. (2012) Domain swapping and amyloid fibril conformation. Prion 6, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davis P. J., Holmes D., Waltho J. P., and Staniforth R. A. (2015) Limited proteolysis reveals that amyloids from the 3D domain-swapping cystatin B have a non-native β-sheet topology. J. Mol. Biol. 427, 2418–2434 [DOI] [PubMed] [Google Scholar]
- 27. Levy E., Sastre M., Kumar A., Gallo G., Piccardo P., Ghetti B., and Tagliavini F. (2001) Codeposition of cystatin C with amyloid-β protein in the brain of Alzheimer disease patients. J. Neuropathol. Exp. Neurol. 60, 94–104 [DOI] [PubMed] [Google Scholar]
- 28. Coria F., Castaño E. M., and Frangione B. (1987) Brain amyloid in normal aging and cerebral amyloid angiopathy is antigenically related to Alzheimer's disease beta-protein. Am. J. Pathol. 129, 422–428 [PMC free article] [PubMed] [Google Scholar]
- 29. Mi W., Jung S. S., Yu H., Schmidt S. D., Nixon R. A., Mathews P. M., Tagliavini F., and Levy E. (2009) Complexes of amyloid-β and cystatin C in the human central nervous system. J. Alzheimers Dis. 18, 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tizon B., Ribe E. M., Mi W., Troy C. M., and Levy E. (2010) Cystatin C protects neuronal cells from amyloid-β-induced toxicity. J. Alzheimers Dis. 19, 885–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zerovnik E. (2009) The emerging role of cystatins in Alzheimer's disease. BioEssays 31, 597–599 [DOI] [PubMed] [Google Scholar]
- 32. Kaeser S. A., Herzig M. C., Coomaraswamy J., Kilger E., Selenica M.-L., Winkler D. T., Staufenbiel M., Levy E., Grubb A., and Jucker M. (2007) Cystatin C modulates cerebral β-amyloidosis. Nat. Genet. 39, 1437–1439 [DOI] [PubMed] [Google Scholar]
- 33. Mi W., Pawlik M., Sastre M., Jung S. S., Radvinsky D. S., Klein A. M., Sommer J., Schmidt S. D., Nixon R. A., Mathews P. M., and Levy E. (2007) Cystatin C inhibits amyloid-β deposition in Alzheimer's disease mouse models. Nat. Genet. 39, 1440–1442 [DOI] [PubMed] [Google Scholar]
- 34. Sun B., Zhou Y., Halabisky B., Lo I., Cho S. H., Mueller-Steiner S., Devidze N., Wang X., Grubb A., and Gan L. (2008) Cystatin C-cathepsin B axis regulates amyloid β levels and associated neuronal deficits in an animal model of Alzheimer's disease. Neuron 60, 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang C., Sun B., Zhou Y., Grubb A., and Gan L. (2012) Cathepsin B degrades amyloid-β in mice expressing wild-type human amyloid precursor protein. J. Biol. Chem. 287, 39834–39841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mathews P. M., and Levy E. (2016) Cystatin C in aging and in Alzheimer's disease. Ageing Res. Rev. 32, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perlenfein T. J., and Murphy R. M. (2016) Expression, purification, and characterization of human cystatin C monomers and oligomers. Protein Expr. Purif. 117, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Calero M., Pawlik M., Soto C., Castaño E. M., Sigurdsson E. M., Kumar A., Gallo G., Frangione B., and Levy E. (2001) Distinct properties of wild-type and the amyloidogenic human cystatin C variant of hereditary cerebral hemorrhage with amyloidosis, Icelandic type. J. Neurochem. 77, 628–637 [DOI] [PubMed] [Google Scholar]
- 39. Orlikowska M., Jankowska E., Kołodziejczyk R., Jaskólski M., and Szymańska A. (2011) Hinge-loop mutation can be used to control 3D domain swapping and amyloidogenesis of human cystatin C. J. Struct. Biol. 173, 406–413 [DOI] [PubMed] [Google Scholar]
- 40. Szymańska A., Radulska A., Czaplewska P., Grubb A., Grzonka Z., and Rodziewicz-Motowidło S. (2009) Governing the monomer-dimer ratio of human cystatin C by single amino acid substitution in the hinge region. Acta Biochim. Pol. 56, 455–463 [PubMed] [Google Scholar]
- 41. Gerhartz B., Ekiel I., and Abrahamson M. (1998) Two stable unfolding intermediates of the disease-causing L68Q variant of human cystatin C. Biochemistry 37, 17309–17317 [DOI] [PubMed] [Google Scholar]
- 42. Zhou Y., Zhou Y., Li J., Chen J., Yao Y., Yu L., Peng D., Wang M., Su D., He Y., and Gou L. (2015) Efficient expression, purification and characterization of native human cystatin C in Escherichia coli periplasm. Protein Expr. Purif. 111, 18–22 [DOI] [PubMed] [Google Scholar]
- 43. Erickson H. P. (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online. 11, 32–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kayser V., Turton D. A., Aggeli A., Beevers A., Reid G. D., and Beddard G. S. (2004) Energy migration in novel pH-triggered self-assembled β-sheet ribbons. J. Am. Chem. Soc. 126, 336–343 [DOI] [PubMed] [Google Scholar]
- 45. Williams J. W., and Morrison J. F. (1979) The kinetics of reversible tight-binding inhibition. Methods Enzymol. 63, 437–467 [DOI] [PubMed] [Google Scholar]
- 46. Björk I., Pol E., Raub-Segall E., Abrahamson M., Rowan A. D., and Mort J. S. (1994) Differential changes in the association and dissociation rate constants for binding of cystatins to target proteinases occurring on N-terminal truncation of the inhibitors indicate that the interaction mechanism varies with different enzymes. Biochem. J. 299, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ekiel I., Abrahamson M., Fulton D. B., Lindahl P., Storer A. C., Levadoux W., Lafrance M., Labelle S., Pomerleau Y., Groleau D., LeSauteur L., and Gehring K. (1997) NMR structural studies of human cystatin C dimers and monomers. J. Mol. Biol. 271, 266–277 [DOI] [PubMed] [Google Scholar]
- 48. Bergdoll M., Remy M. H., Cagnon C., Masson J. M., and Dumas P. (1997) Proline-dependent oligomerization with arm exchange. Structure 5, 391–401 [DOI] [PubMed] [Google Scholar]
- 49. Guo S.-L., Han C.-T., Jung J.-L., Chen W.-J., Mei J. J., Lee H.-C., and Cheng Y.-C. (2014) Cystatin C in cerebrospinal fluid is upregulated in elderly patients with chronic osteoarthritis pain and modulated through matrix metalloproteinase 9-specific pathway. Clin. J. Pain. 30, 331–339 [DOI] [PubMed] [Google Scholar]
- 50. Sladewska A., Szymańska A., Kordalska M., Lewandowska A., Kołodziejczyk A. S., Paraschiv G., Przybylski M., and Czaplewska P. (2011) Identification of the epitope for anti-cystatin C antibody. J. Mol. Recognit. 24, 687–699 [DOI] [PubMed] [Google Scholar]
- 51. Ristiniemi N., Qin Q. P., Postnikov A., Grubb A., and Pettersson K. (2010) Dry-reagent double-monoclonal assay for cystatin C. Clin. Chem. 56, 1424–1431 [DOI] [PubMed] [Google Scholar]
- 52. Abrahamson M., Mason R. W., Hansson H., Buttle D. J., Grubb A., and Ohlsson K. (1991) Human cystatin C: role of the N-terminal segment in the inhibition of human cysteine proteinases and in its inactivation by leucocyte elastase. Biochem. J. 273, 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hawe A., Sutter M., and Jiskoot W. (2008) Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 25, 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kremer J. J., Pallitto M. M., Sklansky D. J., and Murphy R. M. (2000) Correlation of β-amyloid aggregate size and hydrophobicity with decreased bilayer fluidity of model membranes. Biochemistry 39, 10309–10318 [DOI] [PubMed] [Google Scholar]
- 55. Ladiwala A. R., Litt J., Kane R. S., Aucoin D. S., Smith S. O., Ranjan S., Davis J., Van Nostrand W. E., and Tessier P. M. (2012) Conformational differences between two amyloid β oligomers of similar size and dissimilar toxicity. J. Biol. Chem. 287, 24765–24773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sjöqvist J., Maria J., Simon R. A., Linares M., Norman P., Nilsson K. P., and Lindgren M. (2014) Toward a molecular understanding of the detection of amyloid proteins with flexible conjugated oligothiophenes. J. Phys. Chem. A. 118, 9820–9827 [DOI] [PubMed] [Google Scholar]
- 57. Sastre M., Calero M., Pawlik M., Mathews P. M., Kumar A., Danilov V., Schmidt S. D., Nixon R. A., Frangione B., and Levy E. (2004) Binding of cystatin C to Alzheimer's amyloid β inhibits in vitro amyloid fibril formation. Neurobiol. Aging 25, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 58. Selenica M. L., Wang X., Ostergaard-Pedersen L., Westlind-Danielsson A., and Grubb A. (2007) Cystatin C reduces the in vitro formation of soluble A β1–42 oligomers and protofibrils. Scand. J. Clin. Lab. Invest. 67, 179–190 [DOI] [PubMed] [Google Scholar]
- 59. Bleiholder C., Dupuis N. F., Wyttenbach T., and Bowers M. T. (2011) Ion mobility-mass spectrometry reveals a conformational conversion from random assembly to β-sheet in amyloid fibril formation. Nat. Chem. 3, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Harper J. D., and Lansbury P. T. (1997) Models of amyloid seeding in Alzheimer's disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 66, 385–407 [DOI] [PubMed] [Google Scholar]
- 61. Kelly J. W. (1998) The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 8, 101–106 [DOI] [PubMed] [Google Scholar]
- 62. Kodali R., and Wetzel R. (2007) Polymorphism in the intermediates and products of amyloid assembly. Curr. Opin. Struct. Biol. 17, 48–57 [DOI] [PubMed] [Google Scholar]
- 63. Bennett M. J., Sawaya M. R., and Eisenberg D. (2006) Deposition diseases and 3D domain swapping. Structure 14, 811–824 [DOI] [PubMed] [Google Scholar]
- 64. Ghiso J., Jensson O., and Frangione B. (1986) Amyloid fibrils in hereditary cerebral hemorrhage with amyloidosis of Icelandic type is a variant of γ-trace basic protein (cystatin C). Proc. Natl. Acad. Sci U.S.A. 83, 2974–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li J., Hoop C. L., Kodali R., Sivanandam V. N., and van der Wel P. C. (2011) Amyloid-like fibrils from a domain-swapping protein feature a parallel, in-register conformation without native-like interactions. J. Biol. Chem. 286, 28988–28995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tsolis A. C., Papandreou N. C., Iconomidou V. A., and Hamodrakas S. J. (2013) A consensus method for the prediction of “aggregation-prone” peptides in globular proteins. PLoS ONE 8, e54175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tsiolaki P. L., Louros N. N., Hamodrakas S. J., and Iconomidou V. A. (2015) Exploring the “aggregation-prone” core of human Cystatin C: a structural study. J. Struct. Biol. 191, 272–280 [DOI] [PubMed] [Google Scholar]
- 68. Tsiolaki P. L., Hamodrakas S. J., and Iconomidou V. A. (2015) The pentapeptide LQVVR plays a pivotal role in human cystatin C fibrillization. FEBS Lett. 589, 159–164 [DOI] [PubMed] [Google Scholar]
- 69. Gosal W. S., Morten I. J., Hewitt E. W., Smith D. A., Thomson N. H., and Radford S. E. (2005) Competing pathways determine fibril morphology in the self-assembly of β-2-microglobulin into amyloid. J. Mol. Biol. 351, 850–864 [DOI] [PubMed] [Google Scholar]
- 70. Fändrich M. (2012) Oligomeric intermediates in amyloid formation: structure determination and mechanisms of toxicity. J. Mol. Biol. 421, 427–440 [DOI] [PubMed] [Google Scholar]
- 71. Klingstedt T., Aslund A., Simon R. A., Johansson L. B., Mason J. J., Nyström S., Hammarström P., and Nilsson K. P. (2011) Synthesis of a library of oligothiophenes and their utilization as fluorescent ligands for spectral assignment of protein aggregates. Org. Biomol. Chem. 9, 8356–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Luo J., Wärmländer S. K., Gräslund A., and Abrahams J. P. (2016) Cross-interactions between the Alzheimer disease amyloid-β peptide and other amyloid proteins: a further aspect of the amyloid cascade hypothesis. J. Biol. Chem. 291, 16485–16493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schägger H. (2001) Blue-Native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65, 231–244 [DOI] [PubMed] [Google Scholar]
- 74. Pecora R. (2000) Dynamic light scattering measurement of nanometer particles in liquids. J. Nanoparticle Res. 2, 123–131 [Google Scholar]
- 75. Murphy R. M., and Pallitto M. M. (2000) Probing the kinetics of β-amyloid self-association. J. Struct. Biol. 130, 109–122 [DOI] [PubMed] [Google Scholar]
- 76. Wine Y., Cohen-Hadar N., Freeman A., and Frolow F. (2007) Elucidation of the mechanism and end products of glutaraldehyde cross-linking reaction by X-ray structure analysis. Biotechnol. Bioeng. 98, 711–718 [DOI] [PubMed] [Google Scholar]
- 77. Migneault I., Dartiguenave C., Bertrand M. J., and Waldron K. C. (2004) Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme cross-linking. BioTechniques 37, 790–796, 798–802 [DOI] [PubMed] [Google Scholar]
- 78. Cantor C. R., and Schimmel P. R. (1980) Biophysical chemistry: Part II: Techniques for the study of biological structure and function, pp. 549–562, W. H. Freeman and Company, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.