Abstract
The peroxins Pex19 and Pex3 play an indispensable role in peroxisomal membrane protein (PMP) biogenesis, peroxisome division, and inheritance. Pex19 plays multiple roles in these processes, but how these functions relate to the structural organization of the Pex19 domains is unresolved. To this end, using deletion mutants, we mapped the Pex19 regions required for peroxisome biogenesis in the yeast Pichia pastoris. Surprisingly, import-competent peroxisomes still formed when Pex19 domains previously believed to be required for biogenesis were deleted, although the peroxisome size was larger than that in wild-type cells. Moreover, these mutants exhibited a delay of 14–24 h in peroxisome biogenesis. The shortest functional N-terminal (NTCs) and C-terminal constructs (CTCs) were Pex19 (aa 1–150) and Pex19 (aa 89–300), respectively. Deletions of the N-terminal Pex3-binding site disrupted the direct interactions of Pex19 with Pex3, but preserved interactions with a membrane peroxisomal targeting signal (mPTS)-containing PMP, Pex10. In contrast, deletion of the C-terminal mPTS-binding domain of Pex19 disrupted its interaction with Pex10 while leaving the Pex19-Pex3 interactions intact. However, Pex11 and Pex25 retained their interactions with both N- and C-terminal deletion mutants. NTC-CTC co-expression improved growth and reversed the larger-than-normal peroxisome size observed with the single deletions. Pex25 was critical for peroxisome formation with the CTC variants, and its overexpression enhanced their interactions with Pex3 and aided the growth of both NTC and CTC Pex19 variants. In conclusion, physical segregation of the Pex3- and PMP-binding domains of Pex19 has provided novel insights into the modular architecture of Pex19. We define the minimum region of Pex19 required for peroxisome biogenesis and a unique role for Pex25 in this process.
Keywords: membrane biogenesis, organelle, peroxisome, protein assembly, trafficking
Introduction
Peroxisomes are single membrane-enclosed organelles that are vital in the maintenance of cellular metabolic homeostasis (1). In response to the metabolic needs of the cell, peroxisomes can alter their enzyme content and modulate their number and size (2). Proteins involved in peroxisome assembly, division, and inheritance are known as peroxins and encoded by PEX genes. Over a dozen peroxins conserved from yeasts to mammals are essential for normal human development (3). A failure in peroxisome biogenesis can have detrimental, even lethal consequences in humans as exemplified by the Zellweger syndrome spectrum, a series of genetic disorders leading to craniofacial and ocular abnormalities (4). Future improvements in the quality of life for those affected by peroxisomal metabolism defects rely on a more thorough understanding of peroxisome biogenesis, turnover, and homeostasis.
Peroxisomes are replenished by growth and division of pre-existing peroxisomes (5, 6) and are also formed de novo from the endoplasmic reticulum (ER)3 (7, 8). Peroxins Pex3 and Pex19 are essential for both processes, because in their absence, mature peroxisomes are not formed (9–12). Pex19 is a hydrophilic and acidic protein with an intrinsically disordered region and characteristic N- and C-terminal domains (13, 14). Although a predominant amount of Pex19 is cytosolic, a small but significant portion is also associated with the peroxisome membrane through the farnesylation of its C-terminal end and association with Pex3 (15, 16). The C-terminal domain of Pex19 (aa ∼160–300) participates in the recognition and binding of putative membrane peroxisomal targeting signal (mPTS) motifs on peroxisomal membrane proteins (PMPs) (17–19). Currently, many mPTSs have been identified, and it is clear that these signals show significant variability regarding their length and sequence (20). The N-terminal region of Pex19 (aa 1–44) contains a high-affinity Pex3-binding site (17, 21–23).
Different models of peroxisome biogenesis ascribe Pex3 and Pex19 with different biological roles. In the growth and division model, it has been suggested that Pex19 functions as a chaperone protein that post-translationally shuttles cytoplasmic mPTS-containing cargoes to the peroxisomal membrane (9, 22, 24, 25). In this scenario, the N-terminal domain of Pex19 is anchored to the peroxisomal membrane by its interaction with Pex3, while inserting other PMPs bound to the C-terminal part of Pex19 into the peroxisomal membrane (22). Consistent with its chaperone function, PMPs synthesized in the absence of Pex19 form aggregates during translation (26). Pex19 also plays an important part in delivering the tail-anchored (TA) proteins to the peroxisomal membrane, through Pex3 (27, 28). Recently, a unique structural region of Pex19 was identified that is critical for TA protein insertion (28). Mutants that affect the interaction of Pex3 and Pex19 mostly have a negative impact on peroxisome biogenesis (13, 18, 19). Thus the interactions of Pex19 with the mPTSs of PMPs and with Pex3 are critical for peroxisome biogenesis (29). In addition, during de novo peroxisome biogenesis, roles for Pex3 and Pex19 are well demonstrated in the intra-ER sorting and budding of pre-peroxisomal vesicles (ppVs) from the ER (11, 30–32). Additionally, Pex19 is necessary for proper inheritance of peroxisomes between the dividing cells (33) and it is also involved in peroxisome division by virtue of its association with the organelle division machinery (34, 35). These observations point to Pex3 and Pex19 as versatile, multifunctional proteins.
The multiple roles of Pex19 raise the question as to how these functions correlate with the structural organization of the domains of this protein. In this study, we have identified the critical regions of Pex19 required for peroxisome biogenesis by segregating its N- and C-terminal domains. Several different constructions of the N- or C-terminal domains of Pex19 were expressed in Pichia pastoris and analyzed by fluorescence microscopy and biochemical studies to characterize their individual contributions in promoting peroxisome biogenesis and proper PMP localization. The physical segregation of the Pex3- and mPTS-binding domains of Pex19 has provided novel insights into the role of Pex19. Our findings demonstrate novel functions of Pex25 in peroxisome biogenesis and successfully integrate the classical role of Pex25 in promoting cell growth by regulating peroxisome size and number.
Results
Construction of Pex19 N-and C-terminal deletion
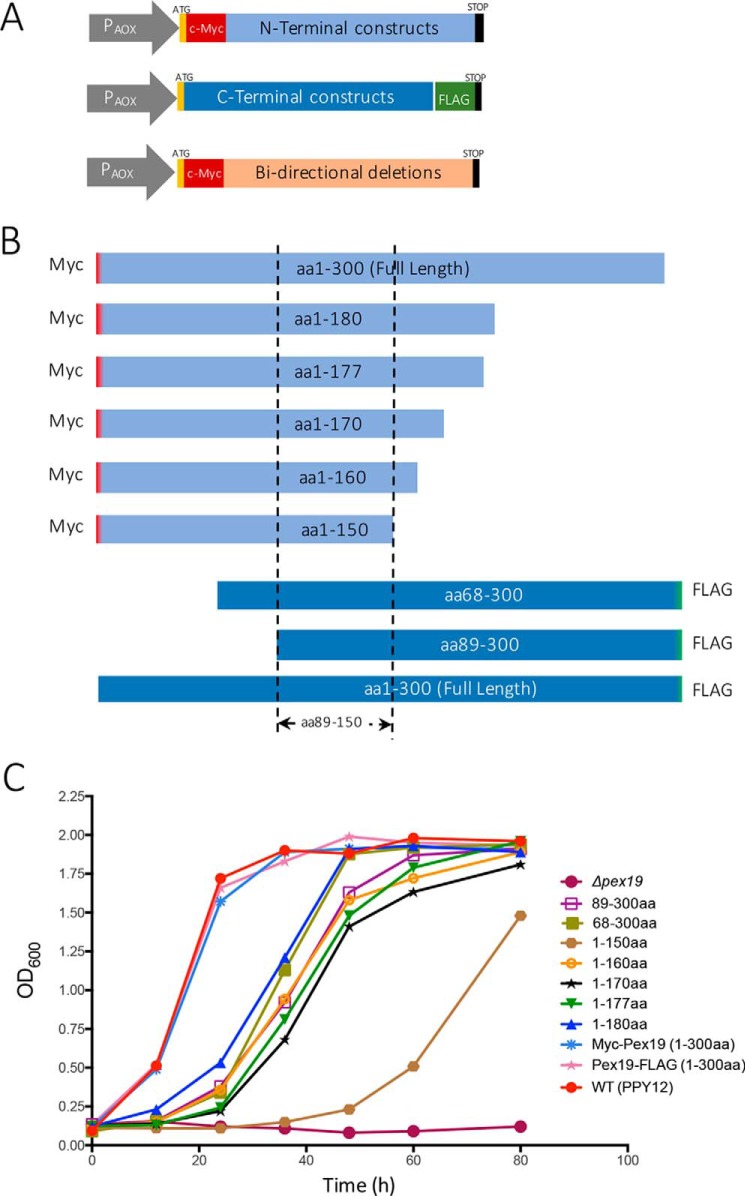
To study the essential structural domains of Pex19 necessary for supporting peroxisome biogenesis, we created several N- and C-terminal deletion constructs. To achieve proper folding, the length of the constructs was determined using several open source domain prediction tools (DomPred, DoBo, DOMpro, and Globplot 2) (36–39). The N-terminal constructs (defined as Pex19 variants with deletions from the C terminus), are termed NTCs, and the C-terminal constructs (Pex19 variants with deletions from the N terminus), are termed CTCs. All Pex19 constructs were expressed from the inducible alcohol oxidase promoter (PAOX) in pex19Δ cells. FLAG and c-myc tags were attached to all the CTCs and NTCs, respectively. The bidirectional deletions that preserved the central domains had the c-myc tag at the N-terminal end (Fig. 1A). The start (ATG) and stop codons (TAA) were present at the 5′ and the 3′ ends of the PEX19 ORFs for all constructs (Fig. 1A). Full-length Pex19 with an N-terminal c-myc tag and another one with a C-terminal FLAG tag were used as wild-type (WT) controls.
Figure 1.
Schematic representations of Pex19 deletion constructs. A, all the deletion constructs were expressed from the inducible alcohol oxidase promoter (PAOX) in pex19Δ cells. The NTCs and the bidirectional deletion constructs each had an N-terminal c-myc tag, whereas the CTCs each had a C-terminal FLAG tag. All the constructs had a start (ATG) and a stop codon (TAA), at the beginning and end of the ORF, respectively. B, schematic representation of the functional Pex19 deletion constructs. All the functional deletion constructs have an overlap between aa 89 and 150. C, the pex19Δ cells transformed with plasmids expressing specified deletion constructs from chromosomally-integrated plasmid DNA, were assayed for growth on methanol medium. Cells were grown overnight in YPD and ∼0.1 A600/ml was further inoculated into methanol medium. Cell growth was measured at specific times. The experiment was repeated three times with similar results. P. pastoris PPY12 cells were used as WT control.
Growth of pex19 deletion mutants under peroxisome proliferation conditions
P. pastoris is a methylotropic yeast that utilizes methanol as a sole carbon and energy source (1, 40). Primarily localized to peroxisomes, alcohol oxidase catalyzes a series of reduction-oxidation reactions in which methanol and its subsequent derivatives are oxidized and energy is stored in the form of NADH (41). Thus, under methanol conditions, to meet cellular growth and maintenance demands, peroxisomes proliferate to metabolize methanol. Mutants such as pex19Δ, which lack Pex19 completely and are deficient in peroxisome assembly, do not grow in methanol. For this reason, strains expressing various NTC and CTC variants of Pex19 were examined for their growth characteristics in methanol medium for up to 80 h. Fig. 1, B and C, show the constructs used and the growth characteristics for only the Pex19 deletion mutants that supported peroxisome biogenesis in methanol medium, in comparison with the pex19Δ control.
As expected, the pex19Δ strain showed no significant growth in methanol medium. The WT PPY12 cells and the strains expressing full-length Pex19 with either c-myc or FLAG tags exhibited similar growth profiles and normal peroxisome size (Figs. 1C and 2, A and B), suggesting that the fusion of these tags to Pex19 did not affect its function. Strains expressing the Pex19 NTCs, Pex19(1–180), Pex19(1–177), Pex19(1–170), Pex19(1–160), and the CTCs Pex19(68–300) and Pex19(89–300) displayed logarithmic growth in methanol media after an initial lag of nearly 14–24 h. However, the strain expressing the Pex19(1–150) showed a longer delay of nearly 30 h (Fig. 1C). The bidirectional deletion mutants, Pex19(89–230) and Pex19(89–220), grew very poorly with more than a 40–50-h delay (Table 1). This suggests that although specific regions in the N- and C-terminal domains of Pex19 are dispensable, a common central domain likely contains the minimum structural requirement for supporting peroxisome biogenesis. From these results, we identified Pex19(1–150) as containing the minimal region of Pex19 that supports peroxisome biogenesis. In addition, the longest constructs, Pex19(1–180) and Pex19(68–300) grew better than other mutants, whereas the shortest construct, Pex19(1–150) was the slowest with more than a 30-h delay (Fig. 1C). The common overlapping region between the functional Pex19 mutants was aa 89–150 (Fig. 1B), although this particular construct, Pex19(89–150), was non-functional when expressed alone (Table 1).
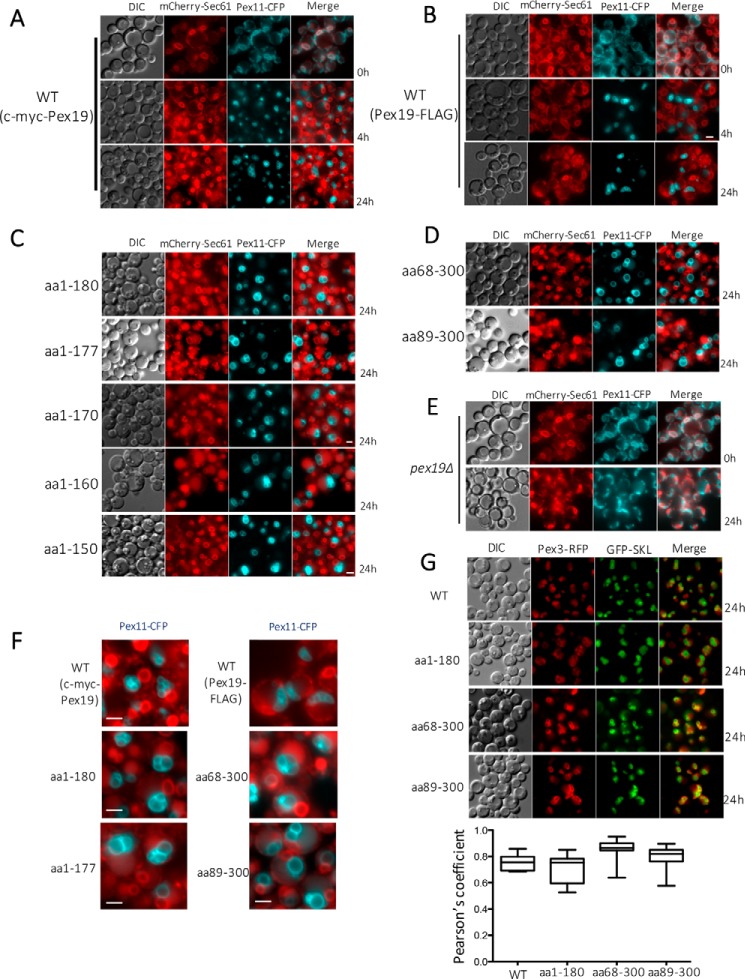
Figure 2.
Fluorescence microscopy analysis of methanol-grown cells expressing the specified fluorescently-tagged proteins. All the constructs were expressed in the pex19Δ cells. Cells were grown in YPD and switched during exponential phase to methanol medium. DIC, differential interference contrast. Scale bar, 2 μm. A and B, localization of Pex11-CFP and Sec61-mCherry, the ER marker, in pex19Δ cells expressing full-length Pex19 constructs at specified times. Pex11-CFP colocalized with the ER marker at 0 h only in the absence of Pex19, but it did not colocalize at 4 or 24 h as it was peroxisomal. C and D, absence of co-localization of Pex11-CFP with Sec61-mCherry in NTC and CTC variants capable of growing on methanol. Pex11-CFP is peroxisomal in these strains because the Pex19 constructs retain their ability to facilitate peroxisome biogenesis. E, co-localization of Pex11-CFP and mCherry-Sec61 in methanol-grown pex19Δ cells after 24 h. F, larger than normal peroxisomes labeled with Pex11-CFP, relative to those in WT cells, in NTC and CTC variants competent to form peroxisomes. G, evidence of bona fide peroxisome formation in WT and specified NTC and CTC variants of Pex19, as judged by the colocalization of peroxisomal membrane marker, Pex3-RFP, and the peroxisomal matrix marker, GFP-SKL. Also shown is colocalization of Pex3-RFP and GFP-SKL in Pex19 variants. Pearson's correlation coefficients were calculated using the coloc2 plug-in for ImageJ, and the data are displayed as interquartile boxes and whisker plots. Pex3-RFP was expressed from the PAOX promoter, and GFP-SKL was expressed from the PGAP promoter.
Table 1.
Summary of Pex19 N- and C-terminal constructs
| Constructs | Peroxisome reintroduction | Growth (methanol) | Peroxisome size | |
|---|---|---|---|---|
| WT | Myc-Pex19 (aa 1–300) | + | ++++ | Normal |
| Pex19-FLAG (aa 1–300) | + | ++++ | Normal | |
| pex19Δ | − | − | No peroxisomes | |
| N-terminal constructs (NTCs) | aa 1–180 | + | +++ | Enlarged |
| aa 1–177 | + | ++ | Enlarged | |
| aa 1–170 | + | ++ | Enlarged | |
| aa 1–160 | + | ++ | Enlarged | |
| aa 1–150 | + | + | Enlarged | |
| aa 1–146 | − | − | No peroxisomes | |
| aa 1–140 | − | − | No peroxisomes | |
| aa 1–128 | − | − | No peroxisomes | |
| aa 1–110 | − | − | No peroxisomes | |
| aa 1–88 | − | − | No peroxisomes | |
| aa 1–67 | − | − | No peroxisomes | |
| C-terminal constructs (CTCs) | aa 68–300 | + | ++ | Enlarged |
| aa 89–300 | + | ++ | Enlarged | |
| aa 101–300 | − | − | No peroxisomes | |
| aa 111–300 | − | − | No peroxisomes | |
| aa 121–300 | − | − | No peroxisomes | |
| aa 125–300 | − | − | No peroxisomes | |
| aa 131–300 | − | − | No peroxisomes | |
| aa 147–300 | − | − | No peroxisomes | |
| aa 151–300 | − | − | No peroxisomes | |
| aa 178–300 | − | − | No peroxisomes | |
| Bi-directional deletions | aa 89–230 | +/− | +/− | Enlarged |
| aa 89–220 | +/− | +/− | Enlarged | |
| aa 89–210 | − | − | No peroxisomes | |
| aa 89–200 | − | − | No peroxisomes | |
| aa 89–190 | − | − | No peroxisomes | |
| aa 89–180 | − | − | No peroxisomes | |
| aa 89–170 | − | − | No peroxisomes | |
| aa 89–160 | − | − | No peroxisomes | |
| aa 89–150 | − | − | No peroxisomes |
Localization of PMPs in cells expressing Pex19 constructs under peroxisome proliferation conditions
We monitored peroxisome formation and the localization of PMPs in our Pex19 mutants for over 70 h under peroxisome proliferation conditions. Previously, we observed that Pex11-CFP was relocated to the de novo formed peroxisomes when the PEX19 gene was reintroduced and expressed from the inducible alcohol oxidase promoter in pex19Δ cells (11). In a similar manner, Pex19 deletion mutants were also co-expressed with Pex11-CFP and mCherry-Sec61, an ER-marker, in pex19Δ cells. Pex11-CFP was expressed from the constitutive glyceraldehyde-3-phosphate dehydrogenase (PGAP) promoter, whereas the mCherry-Sec61 was expressed from its endogenous promoter.
Under peroxisome proliferation conditions, in pex19Δ cells expressing the full-length Pex19 with either c-myc or FLAG tag, peroxisomes appeared within 4 h as characteristic punctate structures labeled with Pex11-CFP, depicting proper peroxisomal localization of this PMP (Fig. 2, A and B). This also affirmed that the fusion of a c-myc or FLAG tag to Pex19 did not affect normal peroxisome formation. All the Pex19 constructs that supported growth on methanol medium also formed peroxisomes. However, the peroxisomes formation was delayed by nearly 14–24 h and peroxisome size was remarkably larger as compared with that in WT cells (Figs. 2, C, D, and F). In addition, as expected the peroxisomes formed in the NTC and CTC variants were import competent, as seen from the significant co-localization of GFP-SKL (an artificial peroxisomal matrix protein marker) and Pex3-RFP (Fig. 2G). These large peroxisomal structures are reminiscent of a Δpex11 phenotype, suggesting that peroxisomal division may be impaired in these strains (42, 43). However, strains expressing the other Pex19 deletion mutants did not grow in methanol medium, had no detectable peroxisomal structures, and appeared similar to the pex19Δ cells (Fig. 2E).
Co-expression of N- and C-terminal domains of Pex19 promotes peroxisome biogenesis
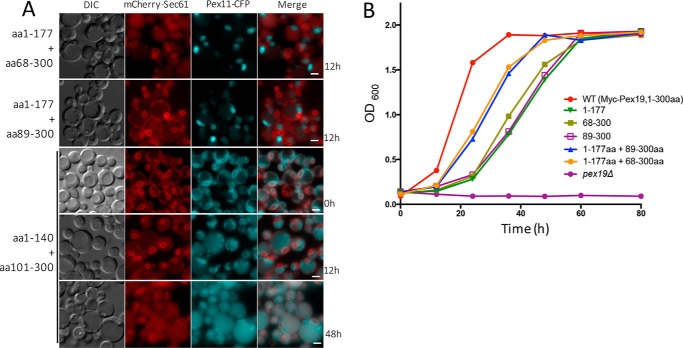
We identified six NTCs and eight CTCs that were non-functional and did not support peroxisome biogenesis (Table 1). We investigated whether the pairwise co-expression of any two of these non-functional constructs would restore peroxisome biogenesis. We constructed strains co-expressing various combinations of N- and C-terminal constructs, and analyzed them for growth on methanol medium and observed the localization of Pex11-CFP. However, none of the combinations disrupting the central domain from aa 89–150, including the two longest N- and C-terminal constructs (Pex19(1–140) with Pex19(101–300)), restored peroxisome biogenesis in these strains (Fig. 3A).
Figure 3.
Fluorescence microscopy and growth analysis of P. pastoris strains co-expressing the specified NTC and CTC variants in pex19Δ cells. Cells were grown in YPD and switched during exponential phase to methanol medium. A, fluorescence microscopy analysis of methanol-grown cells co-expressing the specified NTC and CTC variants. Pex11-CFP and Sec61-mCherry co-localized only at 0 h, when Pex19 was absent, but this co-location was lost as peroxisomes formed and Pex11-CFP became peroxisomal. Scale bar: 2 μm. B, cells co-expressing the specified NTC and CTC variants were assayed for growth on methanol medium as described in the legend to Fig. 1B. The experiment was repeated three times with similar results.
The most pronounced growth advantage was observed when the C-terminal constructs were co-expressed with Pex19(1–177) (Fig. 3B), although the growth was not completely restored to the same level seen for the WT cells. Notably, the onset of the exponential phase was less visibly affected, regardless of the combination of constructs. In addition, the delay in peroxisome formation was reduced to 9–12 h as compared with 20–28 h, and the size of peroxisomes was also restored very close to that of the WT cells (Fig. 3A). Thus, by co-expressing the N-terminal Pex3-binding domain with the C-terminal mPTS-binding domain of Pex19, a synergistic effect on peroxisome biogenesis was observed, as long as the essential central domain between aa 89 and 150 was intact.
Characterizing the interactions of Pex19 domains with different PMPs
Previous studies on the role of Pex19 as a chaperone and co-receptor for PMP insertion into the peroxisome membrane suggested that Pex19 should simultaneously interact with Pex3 and insert the bound PMP in the peroxisomal membrane and thus, have non-overlapping binding sites for both Pex3 and PMPs.
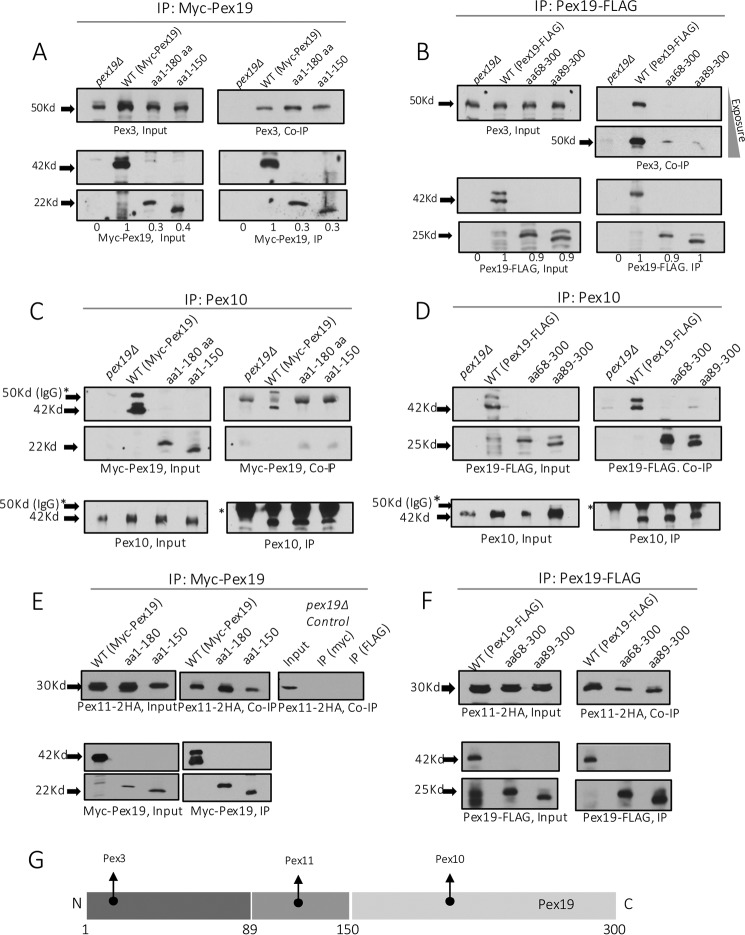
To verify this, we analyzed the functional NTCs and CTCs for their interactions with Pex3 and other PMPs. Several co-immunoprecipitations (co-IP) were performed to determine whether the functional Pex19 constructs maintained interactions with Pex3, Pex10, and Pex11. Pex3 was chosen due to the proposed importance of Pex19-Pex3 interactions for peroxisome biogenesis, whereas Pex10 and Pex11 were chosen due to their different binding sites on Pex19; Pex11 is suggested to bind within the internal domain, between aa 135 and 150, whereas Pex10 binds at the C-terminal mPTS-binding domain, between aa 180 and 300 (26).
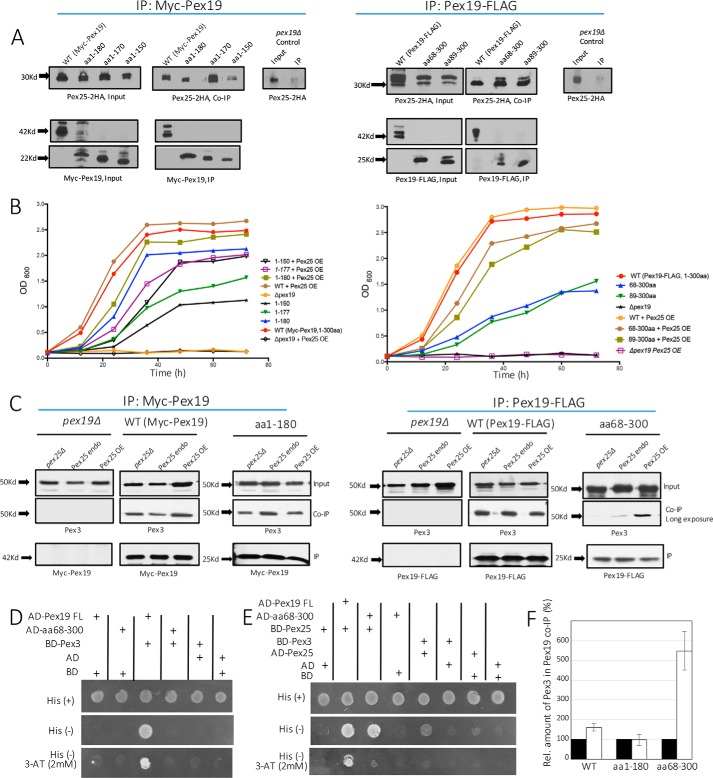
As expected, all three proteins were detected in the input and co-IP fractions in strains expressing WT c-myc or FLAG-tagged Pex19, suggesting that these PMPs were present and interacted with Pex19 (Fig. 4). The binding of Pex3 with the NTC mutants, Pex19(1–180) and Pex19(1–150), was preserved (Fig. 4A) as expected, because aa 1–44 form a high-affinity Pex3-binding site (13, 44, 45). However, both the functional C-terminal constructs, Pex19(68–300) and Pex19(89–300), were drastically reduced in their interactions with Pex3 to nearly undetectable levels (a very weak interaction was noted in a longer exposition of the film) (Fig. 4B). Although this was expected, given that these constructs do not encompass the canonical Pex3-binding site of the Pex19, it was difficult to reconcile the growth of strains expressing these two CTCs in methanol and the presence of peroxisomes (Figs. 1C and 2D) with previous studies, which suggested that the interaction between Pex3 and Pex19 is critical for peroxisome biogenesis (10, 18, 23, 29, 31). However, it is also suggestive of another parallel, perhaps redundant mechanism, which might compensate by anchoring these CTC variants of Pex19 with Pex3, albeit with a lower affinity (see later).
Figure 4.
Interaction of WT Pex19 and its NTC and CTC variants with different PMPs. All the constructs were expressed in pex19Δ cells. Co-immunoprecipitations were performed with appropriate antibodies against c-myc-Pex19 (A and E), Pex19-FLAG (B and F), or Pex10 (C and D) as described under ”Materials and methods.“ The immunoprecipitates were then immunoblotted with appropriate antibodies to detect Pex3 (A and B), Myc-Pex19 (C), Pex19-FLAG (D), and Pex11–2HA (E and F). The proteins detected in the Input, IP, and co-IP are shown. The relative protein abundances of WT Pex19 and its variants, as detected by quantitation of the protein band intensities, are shown by numbers in A and B. The WT Pex19 level was set to 1 and Pex19 in pex19Δ cells to 0. The co-IP was repeated three times with similar qualitative results regarding the presence or absence of the proteins that co-immunoprecipitated with Pex19 or its variants. G, schematic representation of the binding sites on Pex19 for different PMPs.
The interactions between Pex10 and the C-terminal constructs remained intact (Fig. 4D); however, the interaction with the NTC mutants, Pex19(1–180) and Pex19(1–150), was nearly abolished (Fig. 4C). This observation further confirmed that the binding site for Pex10 in Pex19 was toward the C terminus of Pex19 (aa 180–300). In contrast, Pex11 showed a strong interaction with both N- and C-terminal constructs (Fig. 4, E and F). Together, these results show a clear segregation of various PMP-binding sites on Pex19 (Fig. 4G).
Conditional requirement of Pex25 in peroxisome biogenesis
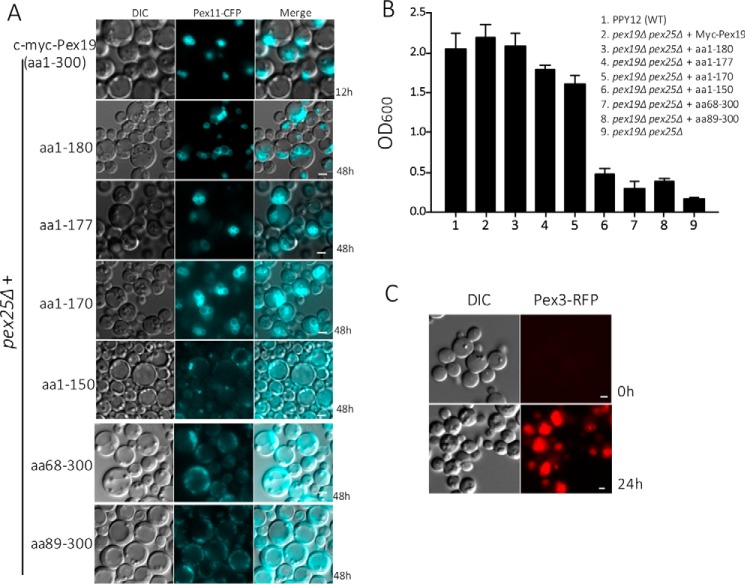
Previous studies have demonstrated that Pex25 plays an important role in peroxisome biogenesis by regulating peroxisome size and number (46–48). Although yeast cells lacking Pex25 alone are still capable of supporting peroxisome biogenesis, in the double deletion mutant, pex3Δ pex25Δ, where peroxisomes are absent, the formation of new peroxisomes de novo occurred only upon the reintroduction of both Pex3 and Pex25 (47). Based on these findings, it was suggested that ScPex25 participates in membrane elongation of existing peroxisomes and acts in concert with Pex3 at the ER to initiate de novo peroxisome biogenesis. We wondered if in P. pastoris the double mutants, pex3Δ pex25Δ and pex19Δ pex25Δ, would also require the reintroduction of both Pex25 in addition to Pex3 or Pex19 for restoring de novo peroxisome biogenesis.
Contrary to studies in Saccharomyces cerevisiae, in P. pastoris, we discovered that for pex3Δ pex25Δ, or pex19Δ pex25Δ cells, the reintroduction of Pex3 or Pex19 alone was capable of restoring peroxisome biogenesis. In pex3Δ pex25Δ cells, when Pex3-RFP was reintroduced by expression from the PAOX promoter (in methanol medium), peroxisome biogenesis was completely restored (Fig. 5C). We employed the Pex11-CFP localization assay to visualize the formation of peroxisomes upon the reintroduction of Pex19. Pex19 initiated the proper localization of Pex11-CFP at the peroxisomal membrane within 12 h in methanol (Fig. 5A). These results suggest that for P. pastoris, the absence of Pex25 alone is not sufficient to block de novo peroxisome formation.
Figure 5.
Pex25 is required for peroxisome biogenesis in the CTC variants of Pex19. A, localization of Pex11-CFP in methanol-grown pex19Δ pex25Δ cells expressing either the WT Pex19 or the specified NTCs or CTCs (scale bar: 2 μm). Note that only one NTC (aa 1–150) and two CTC variants (aa 68–300 and aa 89–300 of Pex19) expressing Pex11-CFP did not show punctate, peroxisome-like structures. B, methanol-grown pex19Δ pex25Δ cells expressing constructs described in A were assayed for growth in methanol. Note that only those Pex19 variants that failed to localize Pex11-CFP to punctate peroxisomes in A failed to grow in methanol. The experiment was repeated three times with similar results. C, localization of Pex3-RFP, expressed from the inducible PAOX promoter, to punctate, peroxisome-like structures in methanol-grown pex3Δ pex25Δ cells. Cells were grown in YPD and switched during exponential phase to methanol medium.
We also evaluated the ability of functional N- and C-terminal constructs of Pex19 to support peroxisome biogenesis in pex19Δ pex25Δ cells. In the absence of Pex25, for all the NTC variants except the Pex19(1–150), Pex11-CFP was localized at the mature peroxisomal clusters (Fig. 5A). Surprisingly, the CTC mutants failed to restore peroxisome biogenesis in the absence of Pex25. In these cells, Pex11-CFP was mislocalized along the peripheral ER (Fig. 5A). These results were further confirmed for these deletion mutants with growth in methanol medium. The NTC, except for Pex19(1–150), but not the CTC, mutants rescued the growth of pex19Δ pex25Δ cells (Fig. 5B). These results suggest that Pex25 is necessary for restoring peroxisome biogenesis in the CTC mutants, perhaps by restoring a weak interaction between Pex3 and the CTC mutants (see below), which lack the Pex3-binding site.
The Pex25-binding site on Pex19 is between amino acids 89 and 150
Pex25 interacts with Pex19 in yeast two-hybrid studies in S. cerevisiae (49). In addition, using synthetic peptide scanning data, the binding site of Pex25 on Pex19 was predicted between aa 135 and 150 (49). To establish the binding site of Pex25 on Pex19 in P. pastoris, we performed co-IPs with cells expressing the functional Pex19 constructs with an HA-tagged Pex25. Although the interaction with Pex19(1–150) was weak, Pex25 interacted strongly with Pex19(1–170) and Pex19(89–300), suggesting that Pex25 interacts with Pex19 within aa 89–150 (Fig. 6A).
Figure 6.
Pex25 is required for restoring the interaction between CTC variants and Pex3. A, co-immunoprecipitation was performed by immunoprecipitating either Myc-Pex19 (for NTCs, left panel) or Pex19-FLAG (for the CTCs, right panel) using appropriate antibodies, as described under ”Materials and methods.“ Immunoblotted proteins detected in the Input, IP, and co-IP samples are shown. The co-IP was repeated three times with similar results. B, methanol-grown pex19Δ cells, either expressing Pex25 from the constitutive GAP promoter (Pex25 OE) or from the endogenous (endo) promoter, with the WT Pex19 or the specified NTCs or the CTCs were assayed for growth in methanol medium. The experiment was repeated three times with similar results. C, co-immunoprecipitation was performed by immunoprecipitating either Myc-Pex19 (for NTCs, left panel) or Pex19-FLAG (for the CTCs, right panel) in strains expressing different Pex25 levels. Immunoblots were then done to detect Pex3 levels in the co-IP. The co-IP was repeated three times with similar results. D and E, yeast two-hybrid assay of Pex19 full-length (FL), Pex19(68–300), Pex3, and Pex25. F, effect of Pex25 overexpression on Pex3-Pex19 co-immunoprecipitation. The relative abundance of Pex3 in the Pex19 co-immunoprecipitates in strains expressing either endogenous (black bars) or overexpressed (white bars) levels of Pex25 for Pex19 variants depicted in C was quantitated. The Pex3 levels of cells expressing endogenous levels of Pex25 were set to 100%. Error bars represent mean ± S.E. of three independent experiments.
Overexpression of Pex25 provides a growth advantage for the N- and C-terminal constructs
The functional CTC mutants, Pex19 (68–300) and Pex19(89–300), are the most interesting because they support peroxisome biogenesis even without the high-affinity Pex3-binding site in the N-terminal domain of Pex19. This was not in agreement with the accepted dogma that proposes that the Pex3-Pex19 interaction is central to peroxisome biogenesis. Nonetheless, we detected a very weak interaction with Pex3 with these C-terminal constructs (long exposure in Fig. 4B), which could be an indirect interaction. We suspected that Pex25 might bridge the C-terminal constructs and Pex3. Supporting this idea was the observation that otherwise functional CTC mutants failed to restore peroxisome biogenesis without Pex25 (Fig. 5B).
Thus, we overexpressed Pex25 in functional NTC and CTC mutants and analyzed the effects on peroxisome biogenesis. Pex25 was overexpressed from the constitutively active promoter, PGAP, and expressed as an HA fusion protein (2HA-Pex25), for all the following assays. Growth analyses of the mutants under peroxisome proliferation conditions confirmed that the overexpression of Pex25 promotes peroxisome biogenesis. For all functional NTCs and CTCs of Pex19 analyzed, the overexpression of Pex25 provided a distinct kinetic advantage for cellular growth in methanol medium (Fig. 6B). The lag in cell growth during the exponential phase was visibly reduced and the maximum growth reached approached that for wild-type controls. These results are consistent with previous studies showing that overexpression of Pex25 promotes peroxisome biogenesis in oleate for S. cerevisiae (47). The positive effect of Pex25 overexpression on the CTC variants, which lack the Pex3-binding site, could be due to increased binding between Pex3 and Pex19; however, the reason for the growth advantage on the NTC variants was not apparent. A plausible explanation could be that the binding of the functional NTC variants (1–177 and 1–150) with the PMPs binding toward the C terminus of Pex19 was restored when Pex25 was overexpressed. Thus, we investigated the binding of both various PMPs and Pex3 to NTC and CTC variants, respectively, upon Pex25 overexpression.
Pex25 strengthens Pex19 and specific PMP interactions to promote peroxisome biogenesis
First, we investigated whether the levels of Pex25 affected the Pex19-Pex3 interaction in the absence of the N-terminal Pex3-binding domain in Pex19. We also included the NTC Pex19(1–180) for these studies to determine whether this novel function of Pex25 was active only when the direct Pex19-Pex3 interaction is compromised. For these studies, individual interactions between Pex3 and mutants Pex19(1–180) and Pex19(68–300) were analyzed when Pex25 was either absent (pex25Δ), or overexpressed (PGAP-2HA-Pex25) or at the endogenous levels (Fig. 6C).
The levels of expression of the Pex19 variants in the input and their ability to be immunoprecipitated was relatively constant (Figs. 6C and 7A). Co-IP results revealed that in the absence of Pex25, in the CTC variant Pex19(68–300), a negligible amount of Pex3 was detected in the eluate (Fig. 6C), suggesting the absence of a significant Pex3 interaction, as noted earlier. When Pex25 was endogenously expressed with Pex19(68–300), a very weak interaction was detected. However, upon the overexpression of Pex25, the level of Pex3 interaction was visibly increased (Fig. 6, C and F). As expected, the interaction of Pex3 with Pex19(1–180) was relatively constant (Fig. 6F), as seen for WT Pex19, irrespective of Pex25 expression levels, suggesting that when the N-terminal Pex3-binding site in Pex19 is intact, the indirect binding of Pex3 to Pex19 via Pex25 is masked.
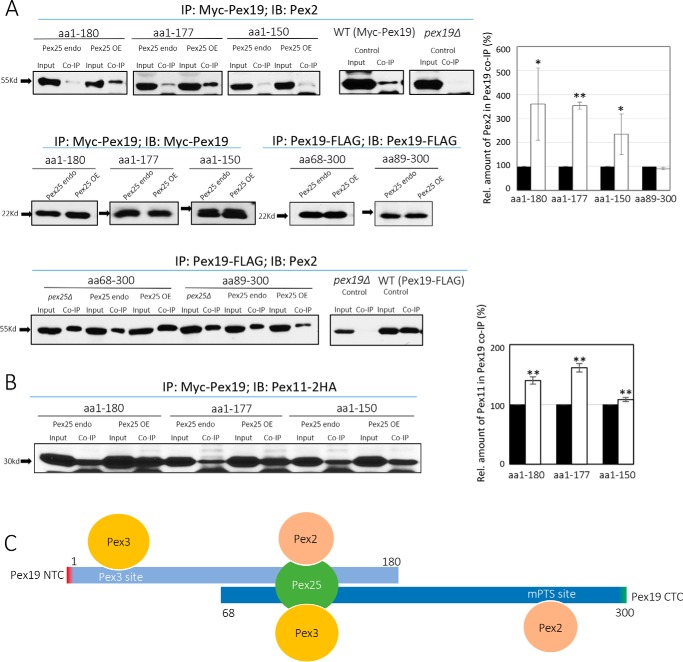
Figure 7.
Effect of Pex25 on the ability of Pex19 variants to bind other PMPs. A, co-immunoprecipitation of Myc-Pex19 or Pex19-FLAG with Pex2, in strains expressing different levels of Pex25. The bar graph depicts the levels of Pex2 in the co-IP of Pex19 variants, in the presence of either endogenous (black bars) or overexpressed (OE) levels (white bars) of Pex25. *, p < 0.05; **, p < 0.01. B, co-immunoprecipitation of Myc-Pex19 NTCs with Pex11-2HA, in strains expressing different levels of Pex25. The bar graph depicts the levels of Pex11-2HA in the co-IP of Pex19 variants, in the presence of either endogenous (black bars) or overexpressed levels (white bars) of Pex25. *, p < 0.05; **, p < 0.01. The co-immunoprecipitations were repeated twice with similar results. C, schematic representation of key binding sites missing in the Pex19 NTC and CTC variants. Pex25 binding to the central domain of Pex19 bridges the missing PMP (e.g. Pex2) and Pex3 interactions in the NTC and CTC, respectively, and explains why Pex25 overexpression stimulates growth of cells expressing these constructs, relative to strains expressing endogenous levels of Pex25. The direct binding of Pex3 and Pex2/Pex10 to Pex19 through the sites shown is the preferred mode of binding, because pex25Δ cells show no defect in peroxisome biogenesis, but the bridging interactions mediated by Pex25 become prominent only when the Pex3 site and/or the PMP site in Pex19 is missing.
To further confirm the interaction of Pex25 with the CTC variant Pex19(68–300) or Pex3, we used a yeast two-hybrid assay. First, Pex3 and the full-length Pex19 or Pex19(68–300) were fused to GAL4-binding domain (BD) and activation domain (AD), respectively. Only cells co-transformed with BD-Pex3 and AD-full-length Pex19 survived in the His- and His-(3AT) selection plates (Fig. 6D), indicating their direct interaction. As expected, Pex19(68–300) failed to interact with Pex3 because this variant lacked the complete Pex3-binding domain (Δ1–67). We also tested if Pex19(68–300) could still bind to Pex25. Consistent with our co-IP data as shown above (Fig. 6A), Pex19(68–300) retained a robust interaction with Pex25, comparable with that seen between the full-length Pex19 and Pex25 (Fig. 6E). Pex25 also manifested a moderate interaction with Pex3 (Fig. 6E), consistent with the co-IP experiments (Fig. 6C), confirming that it bridges the Pex3 association with Pex19(68–300) (Fig. 7C).
Additionally, we investigated whether Pex25 also strengthens the compromised Pex19 interaction with the PMPs that interact normally with the C-terminal region of Pex19. The interactions of the NTC variants with Pex2 in different Pex25 backgrounds were then analyzed. Interestingly, we found that the binding of Pex2 was indeed stronger for the NTC variants, but not the CTC mutants, when Pex25 was overexpressed (Fig. 7A). This role of Pex25 in strengthening the interactions between the NTC variants, Pex19(1–180) and Pex19(1–177), and Pex2, explains why these Pex19 variants showed a significant advantage in growth with the overexpression of Pex25 (Fig. 7C). In addition, we also found that the interactions of some of the NTC variants with Pex11 were stronger when Pex25 was overexpressed (Fig. 7B).
Discussion
The crystal structure of Pex19 has identified a Pex3-binding domain (aa 1–44) at its N-terminal region and an mPTS-binding domain (∼aa 160–300) at its C-terminal region (17, 19, 21). To understand the molecular mechanism of Pex19 function, we segregated and studied these two domains independently of each other using N- and C-terminal deletions of Pex19 in P. pastoris.
Interesting insights were obtained with the characterization of these Pex19 variants. First, several Pex19 variants restored peroxisome biogenesis in pex19Δ cells and utilized methanol as a sole carbon source, but grew with a lag of 14–24 h in methanol medium. Proper peroxisomal localization of Pex11-CFP (Fig. 2D) and Pex3-RFP (Fig. 2G) was observed in cells lacking the N-terminal Pex3-binding domain of Pex19. However, the size of peroxisomes reintroduced by these variants was very large (Fig. 2F). This was surprising because previous studies characterizing mammalian PEX19 deletions either in the N- or C-terminal domains showed a complete loss of interaction with different PMPs (26) and were unable to restore peroxisome biogenesis (18). The minimum region of mammalian PEX19, which was able to restore peroxisome biogenesis, although with a reduced efficiency, in pex19Δ cells was between aa 12 and 261 (full-length: aa 1–299) (18). We believe that P. pastoris has redundant systems to back up the protein-protein interactions lost in those Pex19 deletions that still function in peroxisome biogenesis and have uncovered these in this study.
Second, the NTC variants that contain the minimum stretch of aa 1–150 restored peroxisome biogenesis, albeit with a lag of 14–24 h after methanol induction. These variants lacked most of the mPTS-binding domain (∼aa 160–300) and were unable to retain interaction with Pex10, a PMP that is known to interact with the mPTS-binding region of Pex19 (aa 233–300 of P. pastoris) (17). Previous studies have reported that the farnesylation of Pex19 at the C-terminal end is critical for matrix protein import (15, 16) and structural integrity of the protein (16). Other reports also suggested an important role of Pex19 farnesylation in peroxisome inheritance (33, 50) and its interaction with various PMPs (51). However, the role of Pex19 farnesylation is controversial because other studies show that Pex19 farnesylation is not essential for peroxisome biogenesis in mammalian, and also in yeast, cells (52). Our results with the functional NTC variants demonstrates that the farnesylation is not important for restoring peroxisome biogenesis (Figs. 1C and 2, C and G) and are quite consistent with our earlier results showing dispensability of Pex19 farnesylation in P. pastoris (17). Given the varying opinions on the role of Pex19 farnesylation in S. cerevisiae (15, 16, 51), we cannot rule out that this role of Pex19 farnesylation in peroxisome biogenesis is slightly different in P. pastoris and mammalian cells.
Mammalian PEX19 is involved in the early events of PMP biogenesis, functioning as a chaperone for newly-synthesized PMPs (Type I) and targeting them to the peroxisomal membrane (9, 26). PMPs form aggregates when translated in the absence of Pex19 (26, 53). In this chaperone role, mammalian PEX3 functions as an anchor for PEX19 at the peroxisomal membrane and assists in the membrane insertion of PMPs (22, 24, 28, 29, 44). To accomplish this function, PEX19 contains distinct and non-overlapping PEX3- and PMP-binding domains. Critical mutations (13) or deletions in the PEX3-binding domain of mammalian PEX19 (18, 19) result either in a non-functional protein or severely delayed peroxisome reintroduction (13).
In view of these studies with domains of mammalian PEX19, our observations with the functional CTC variants of P. pastoris Pex19 were highly unexpected. These variants (Δ1–67 or Δ1–88) lacked the complete Pex3-binding domain, and as expected, their interaction with Pex3 was nearly abolished (Fig. 4B). However, they still supported peroxisome biogenesis (Figs. 1 and 2), challenging the dogma that a direct interaction between Pex3-Pex19 is essential for peroxisome biogenesis. This is the first observation where Pex19 lacking the Pex3-binding site is capable of restoring peroxisome biogenesis in pex19Δ cells. As discussed below, we uncovered a role for Pex25 in enabling these CTC variants to restore peroxisome biogenesis, by restoring their interaction with Pex3. In contrast to P. pastoris, mammalian cells likely lack such a mechanism, explaining why the N-terminal PEX3-binding site of mammalian PEX19 is essential.
The functional NTCs and CTCs of Pex19 contain the consensus central region between aa 89 and 150. Deletions from either end that encroach into this central domain resulted in a pex19Δ phenotype. Interestingly, residues 89–150 lie within an internally disordered region (13, 54) of Pex19, rather than within its Pex3- or the predicted mPTS-binding domains. A recent study in Neurospora crassa found certain amphipathic segments in Pex19 whose hydrophobicity is required for peroxisomal membrane insertion of TA proteins (28). The amphipathic segment of Pex19 was not involved in its PMP-chaperone activity or in Pex3 binding. Interestingly, this amphipathic segment aligns along aa 96–107 with P. pastoris Pex19 and is present within the essential domain of aa 89–150. Thus, the aa 89–150 domain of Pex19 may also be involved in post-translational insertion of TA proteins into the peroxisomal membrane, but this hypothesis remains to be tested.
We also paired and analyzed the various functional N- and C-terminal variants and found a synergistic effect in restoring the growth of cells in methanol and in suppressing the enlarged size of peroxisome phenotype but still not comparable with the properties of WT Pex19. These results point to a modular nature of Pex19 domains where the Pex3-binding domain and the mPTS-binding domains, although they have unique characteristics, work synergistically even when present, in trans, on different molecules.
It was intriguing to note that the NTC and CTC variants of Pex19 showed an initial lag in resuming exponential growth and displayed large peroxisomes. Previous studies with Pex19 have demonstrated its essential role in peroxisomal inheritance (33, 50), division (34, 35), and more recently, in the budding of ppVs from the ER (11, 30, 55). Upon careful analysis we concluded that the inheritance process was not affected because every dividing cell showed a clear peroxisome signal in the bud. In some instances, peroxisomes were also observed to segregate between mother and daughter cells. Our recent study demonstrated that the interaction of Pex3 and Pex19 in P. pastoris is required for intra-ER sorting of the RING subcomplex proteins (31). Proper sorting and budding of the RING subcomplex is critical for de novo peroxisome biogenesis (31, 56). Although the functional CTC variants analyzed in this study have compromised interaction with Pex3 (Fig. 4B), they still supported peroxisome biogenesis (Figs. 2 and 3), thus further ruling out the possibility of a budding defect. In addition, we noted the import of the peroxisomal matrix protein marker, GFP-SKL, into the peroxisomal structures as soon as they appeared, in both functional NTC and CTC variants (Fig. 2G), further excluding a defect in the peroxisomal matrix protein import pathway. However, the cells expressing the deletion constructs have enlarged peroxisomes and their growth was affected on methanol medium, suggesting a defect in peroxisome division. Previous studies have demonstrated that Pex19 is required for recruiting the components of the organelle division machinery on the peroxisomal membrane (34, 35). From these observations, we conclude that the delay in growth and the enlarged size of peroxisomes are likely due to a defect in peroxisome division in both NTC and CTC variants.
Several studies in yeast have implicated Pex25, a member of Pex11-family of proteins, in peroxisome membrane elongation, division, and de novo biogenesis (46, 47). In pex25Δ cells there is a growth delay with reduced number of peroxisomes (often enlarged) per cell, but peroxisome biogenesis is still supported (47, 48). However, Pex25 was reported in S. cerevisiae to be indispensable for restoring peroxisome biogenesis in pex3Δ cells (47). In contrast, we did not observe a similar requirement of Pex25 in restoring peroxisomes in pex3Δ cells in P. pastoris (Fig. 5C). Like the restoration of peroxisome biogenesis in pex3Δ pex25Δ cells by Pex3 expression, the pex19Δ pex25Δ cells regenerated peroxisomes when a WT copy of Pex19 was reintroduced (Fig. 5). These results suggest that unlike S. cerevisiae, the Pex11 protein family has greater redundancy in P. pastoris. Thus, surprisingly, when the NTC or CTC variants were reintroduced in pex19Δ pex25Δ cells, instead of the full-length WT Pex19, only the NTC variants (except for the aa 1–150) were capable of supporting peroxisome biogenesis, as detected through proper Pex11-CFP localization in these strains (Fig. 5). Because the CTC variants mainly differ in lacking the Pex3-binding, we presumed that Pex25 could be involved in enabling the CTC variants in establishing an indirect interaction with Pex3. In support of this concept, Pex25 binds Pex19 within the essential domain (aa 89–150) (Fig. 6A) and also interacts with Pex3 (Fig. 6E) and thus was able to bridge the interaction between the CTC variants and Pex3 (Fig. 7C). We also found that the binding of Pex2 to Pex19 was indeed stronger for the NTC variants lacking the PMP-binding site, when Pex25 was overexpressed (Fig. 7A). These results explain why Pex25 overexpression stimulates growth of cells expressing Pex19 NTCs and CTCs, relative to strains expressing endogenous levels of Pex25 (Fig. 6B).
Materials and methods
Cloning of N- and C-terminal deletion mutants
The coding sequence for the Pex19 constructs were amplified from a plasmid containing the PEX19 coding sequence. For NTCs, we used a forward primer that included the c-myc tag sequence in-frame with the PEX19 5′ codon. The reverse primer was designed to bind to the 3′-region of the PEX19 coding sequence. The amplicon was subsequently cloned into plasmid pJCF230 between XhoI and SphI sites. For CTCs, genomic DNA from a strain expressing Pex19 with a FLAG tag was used. The forward primer was designed to bind the 5′-region of the PEX19 coding sequence, whereas the reverse primer was designed to bind within the AOX terminator that followed the FLAG sequence in the genomic DNA. The amplicon was subsequently cloned into plasmid pIB4 between XhoI and AgeI sites. The primers for the CTCs included a start codon in the forward primer, whereas the reverse primer for the NTCs contained a stop codon at the 3′ end. The full-length Pex19 constructs either containing an N-terminal c-myc tag or a C-terminal FLAG tag were similarly constructed but were cloned into pIB4 plasmid between XhoI-AgeI sites.
Yeast strains and growth conditions
Yeast cells were grown at 30 °C in YPD medium (1% yeast extract, 2% peptone, 2% glucose) for the preparation of S1 fractions to an OD 1.2–2.0 and were transferred to methanol medium (0.67% yeast nitrogen base (YNB) without amino acids, 0.02 g of l-histidine/liter, 0.02 g of l-arginine/liter, 0.1% yeast extract, 0.5% (v/v) methanol) for 6 h.
Fluorescence microscopy
Cells were grown on YPD and switched to methanol medium during the exponential phase. Images were captured using a Plan Apochromat ×100 1.40 NA oil immersion objective on a motorized fluorescence microscope (Axioskop 2 MOT plus; Carl Zeiss, Inc.) coupled to a monochrome digital camera (AxioCam MRm; Carl Zeiss, Inc.) and processed using AxioVision software (version 4.5; Carl Zeiss, Inc.).
Co-immunoprecipitations
Cells were grown on YPD to an A600 of 1.0–1.2 and switched to methanol media during exponential phase. A 150 A600 sample of methanol-grown cells were resuspended in 2 ml of immunoprecipitation lysis buffer (50 mm HEPES-NaOH, pH 7.4), 0.15 m NaCl, 1% CHAPS, 10% glycerol, 5 mm NaF, 1 mm PMSF, and yeast protease inhibitor mixture). Cells were lysed by vortexing with acid-washed glass beads 4 times for 1 min. Lysate was then solubilized for 1 h at 4 °C with rotation with a pre-clearing step using 100 μl of GammaBind G-Sepharose beads (GE Healthcare) prewashed in immunoprecipitation lysis buffer. The lysate was then centrifuged at 20,000 × g for 10 min. A 1-ml amount of lysate was incubated with 10 μl of anti-myc mouse monoclonal antibody overnight at 4 °C. 100 μl of GammaBind-G-Sepharose beads (GE Healthcare) (prewashed in immunoprecipitation lysis buffer) was added to the lysate and incubated for 1 h at 4 °C. The beads were then washed three times with 2 ml of IP lysis buffer for 10 min each and eluted in 2× SDS loading buffer with boiling for 5 min. Eluted protein was resolved by SDS-PAGE. 7.5 A600 equivalents of the eluate and 0.2 A600 equivalents of the input were then loaded and analyzed by Western blotting using the specified antibodies.
Yeast two-hybrid assay
The GAL4-based Matchmaker yeast two-hybrid system (Clontech Laboratories Inc.) was performed according to the standard user manual. PEX3 and PEX19 or its variants were cloned to pGBKT7 (BD) and pGADT7 (AD) vectors, respectively. PEX25 was cloned to each vector. The BD and AD plasmids were co-transformed into AH109 strains using SD (Leu-, Trp-) plates. Transformants were then plated on SD (Leu-, Trp-), SD (Leu-, Trp-, His-), and SD (Leu-, Trp-, His-) with 2 mm 3-amino-1,2,4-triazole, respectively. The strains were incubated at 30 °C until visible colonies appeared. Two transformants from each strain were tested.
Author contributions
G. A. and S. S. conceived the experiments. G. A., H. H. S., and Z. J. X. performed the experiments. G. A., Z. J. X., and S. S. analyzed and interpreted the data and wrote the manuscript.
Acknowledgments
We thank members of the Subramani lab for their critical inputs. G. A. thanks Dr. Nidhi Vashistha for incisive discussions and tenacious support.
This work was supported by National Institutes of Health Grant RO1DK41737 (to S. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ER
- endoplasmic reticulum
- mPTS
- membrane peroxisomal targeting signal
- PMP
- peroxisomal membrane protein
- TA
- tail-anchored
- NTC
- N-terminal construct
- CTC
- C-terminal construct
- co-IP
- co-immunoprecipitation
- BD
- binding domain
- AD
- activation domain.
References
- 1. Subramani S. (1998) Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol. Rev. 78, 171–188 [DOI] [PubMed] [Google Scholar]
- 2. Wanders R. J., and Waterham H. R. (2006) Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295–332 [DOI] [PubMed] [Google Scholar]
- 3. Schrader M., and Fahimi H. D. (2008) The peroxisome: still a mysterious organelle. Histochem. Cell Biol. 129, 421–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klouwer F. C., Berendse K., Ferdinandusse S., Wanders R. J., Engelen M., and Poll-The B. T. (2015) Zellweger spectrum disorders: clinical overview and management approach. Orphanet J. Rare Dis. 10, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schrader M., and Fahimi H. D. (2006) Growth and division of peroxisomes. Int. Rev. Cytol. 255, 237–290 [DOI] [PubMed] [Google Scholar]
- 6. Motley A. M., and Hettema E. H. (2007) Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178, 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tabak H. F., Braakman I., and van der Zand A. (2013) Peroxisome formation and maintenance are dependent on the endoplasmic reticulum. Annu. Rev. Biochem. 82, 723–744 [DOI] [PubMed] [Google Scholar]
- 8. Agrawal G., and Subramani S. (2016) De novo peroxisome biogenesis: evolving concepts and conundrums. Biochim. Biophys. Acta 1863, 892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones J. M., Morrell J. C., and Gould S. J. (2004) PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164, 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoepfner D., Schildknegt D., Braakman I., Philippsen P., and Tabak H. F. (2005) Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95 [DOI] [PubMed] [Google Scholar]
- 11. Agrawal G., Joshi S., and Subramani S. (2011) Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 108, 9113–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hettema E. H., and Motley A. M. (2009) How peroxisomes multiply. J. Cell Sci. 122, 2331–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sato Y., Shibata H., Nakatsu T., Nakano H., Kashiwayama Y., Imanaka T., and Kato H. (2010) Structural basis for docking of peroxisomal membrane protein carrier Pex19p onto its receptor Pex3p. EMBO J. 29, 4083–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giannopoulou E.-A., Emmanouilidis L., Sattler M., Dodt G., and Wilmanns M. (2016) Towards the molecular mechanism of the integration of peroxisomal membrane proteins. Biochim. Biophys. Acta 1863, 863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Götte K., Girzalsky W., Linkert M., Baumgart E., Kammerer S., Kunau W. H., and Erdmann R. (1998) Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol. Cell. Biol. 18, 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rucktäschel R., Thoms S., Sidorovitch V., Halbach A., Pechlivanis M., Volkmer R., Alexandrov K., Kuhlmann J., Rottensteiner H., and Erdmann R. (2009) Farnesylation of Pex19p is required for its structural integrity and function in peroxisome biogenesis. J. Biol. Chem. 284, 20885–20896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Snyder W. B., Faber K. N., Wenzel T. J., Koller A., Lüers G. H., Rangell L., Keller G. A., and Subramani S. (1999) Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol. Biol. Cell 10, 1745–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsuzono Y., Matsuzaki T., and Fujiki Y. (2006) Functional domain mapping of peroxin Pex19p: interaction with Pex3p is essential for function and translocation. J. Cell Sci. 119, 3539–3550 [DOI] [PubMed] [Google Scholar]
- 19. Schueller N., Holton S. J., Fodor K., Milewski M., Konarev P., Stanley W. A., Wolf J., Erdmann R., Schliebs W., Song Y. H., and Wilmanns M. (2010) The peroxisomal receptor Pex19p forms a helical mPTS recognition domain. EMBO J. 29, 2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sacksteder K. A., Jones J. M., South S. T., Li X., Liu Y., and Gould S. J. (2000) PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 148, 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt F., Treiber N., Zocher G., Bjelic S., Steinmetz M. O., Kalbacher H., Stehle T., and Dodt G. (2010) Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19. J. Biol. Chem. 285, 25410–25417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang Y., Morrell J. C., Jones J. M., and Gould S. J. (2004) PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sato Y., Shibata H., Nakano H., Matsuzono Y., Kashiwayama Y., Kobayashi Y., Fujiki Y., Imanaka T., and Kato H. (2008) Characterization of the interaction between recombinant human peroxin Pex3p and Pex19p: identification of TRP-104 IN Pex3p as a critical residue for the interaction. J. Biol. Chem. 283, 6136–6144 [DOI] [PubMed] [Google Scholar]
- 24. Matsuzono Y., and Fujiki Y. (2006) In vitro transport of membrane proteins to peroxisomes by shuttling receptor Pex19p. J. Biol. Chem. 281, 36–42 [DOI] [PubMed] [Google Scholar]
- 25. Matsuzaki T., and Fujiki Y. (2008) The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183, 1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shibata H., Kashiwayama Y., Imanaka T., and Kato H. (2004) Domain architecture and activity of human Pex19p, a chaperone-like protein for intracellular trafficking of peroxisomal membrane proteins. J. Biol. Chem. 279, 38486–38494 [DOI] [PubMed] [Google Scholar]
- 27. Yagita Y., Hiromasa T., and Fujiki Y. (2013) Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J. Cell Biol. 200, 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Y., Pieuchot L., Loh R. A., Yang J., Kari T. M., Wong J. Y., and Jedd G. (2014) Hydrophobic handoff for direct delivery of peroxisome tail-anchored proteins. Nat. Commun. 5, 5790. [DOI] [PubMed] [Google Scholar]
- 29. Fujiki Y., Matsuzono Y., Matsuzaki T., and Fransen M. (2006) Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions. Biochim. Biophys. Acta. 1763, 1639–1646 [DOI] [PubMed] [Google Scholar]
- 30. Lam S. K., Yoda N., and Schekman R. (2010) A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 107, 21523–21528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Agrawal G., Fassas S. N., Xia Z.-J., and Subramani S. (2016) Distinct requirements for intra-ER sorting and budding of peroxisomal membrane proteins from the ER. J. Cell Biol. 212, 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mast F. D., Jamakhandi A., Saleem R. A., Dilworth D. J., Rogers R. S., Rachubinski R. A., and Aitchison J. D. (2016) Peroxins Pex30 and Pex29 dynamically associate with reticulons to regulate peroxisome biogenesis from the endoplasmic reticulum. J. Biol. Chem. 291, 15408–15427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otzen M., Rucktäschel R., Thoms S., Emmrich K., Krikken A. M., Erdmann R., and van der Klei I. J. (2012) Pex19p contributes to peroxisome inheritance in the association of peroxisomes to Myo2p. Traffic 13, 947–959 [DOI] [PubMed] [Google Scholar]
- 34. Joshi S., Agrawal G., and Subramani S. (2012) Phosphorylation-dependent Pex11p and Fis1p interaction regulates peroxisome division. Mol. Biol. Cell 23, 1307–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delille H. K., and Schrader M. (2008) Targeting of hFis1 to peroxisomes is mediated by Pex19p. J. Biol. Chem. 283, 31107–31115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bryson K., Cozzetto D., and Jones D. T. (2007) Computer-assisted protein domain boundary prediction using the DomPred server. Curr. Protein Pept. Sci. 8, 181–188 [DOI] [PubMed] [Google Scholar]
- 37. Eickholt J., Deng X., and Cheng J. (2011) DoBo: protein domain boundary prediction by integrating evolutionary signals and machine learning. BMC Bioinformatics 12, 43–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheng J., Sweredoski M. J., and Baldi P. (2006) DOMpro: protein domain prediction using profiles, secondary structure, relative solvent accessibility, and recursive neural networks. Data Min. Knowl. Disc. 13, 1–10 [Google Scholar]
- 39. Linding R., Russell R. B., Neduva V., and Gibson T. J. (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31, 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Veenhuis M., Van Dijken J. P., and Harder W. (1983) The significance of peroxisomes in the metabolism of one-carbon compounds in yeasts. Adv. Microb. Physiol. 24, 1–82 [DOI] [PubMed] [Google Scholar]
- 41. Sibirny A. A., Titorenko V. I., Gonchar M. V., Ubiyvovk V. M., Ksheminskaya G. P., and Vitvitskaya O. P. (1988) Genetic control of methanol utilization in yeasts. J. Basic Microbiol. 28, 293–319 [DOI] [PubMed] [Google Scholar]
- 42. Erdmann R., and Blobel G. (1995) Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128, 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marshall P. A., Dyer J. M., Quick M. E., and Goodman J. M. (1996) Redox-sensitive homodimerization of Pex11p: a proposed mechanism to regulate peroxisomal division. J. Cell Biol. 135, 123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmidt F., Dietrich D., Eylenstein R., Groemping Y., Stehle T., and Dodt G. (2012) The role of conserved PEX3 regions in PEX19-binding and peroxisome biogenesis. Traffic 13, 1244–1260 [DOI] [PubMed] [Google Scholar]
- 45. Fransen M., Vastiau I., Brees C., Brys V., Mannaerts G. P., and Van Veldhoven P. P. (2005) Analysis of human Pex19p's domain structure by pentapeptide scanning mutagenesis. J. Mol. Biol. 346, 1275–1286 [DOI] [PubMed] [Google Scholar]
- 46. Rottensteiner H., Stein K., Sonnenhol E., and Erdmann R. (2003) Conserved function of Pex11p and the novel Pex25p and Pex27p in peroxisome biogenesis. Mol. Biol. Cell 14, 4316–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huber A., Koch J., Kragler F., Brocard C., and Hartig A. (2012) A subtle interplay between three Pex11 proteins shapes de novo formation and fission of peroxisomes. Traffic 13, 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tower R. J., Fagarasanu A., Aitchison J. D., and Rachubinski R. A. (2011) The peroxin Pex34p functions with the Pex11 family of peroxisomal divisional proteins to regulate the peroxisome population in yeast. Mol. Biol. Cell 22, 1727–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rottensteiner H., Kramer A., Lorenzen S., Stein K., Landgraf C., Volkmer-Engert R., and Erdmann R. (2004) Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol. Biol. Cell 15, 3406–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Otzen M., Krikken A. M., Ozimek P. Z., Kurbatova E., Nagotu S., Veenhuis M., and van der Klei I. J. (2006) In the yeast Hansenula polymorpha, peroxisome formation from the ER is independent of Pex19p, but involves the function of p24 proteins. FEMS Yeast Res. 6, 1157–1166 [DOI] [PubMed] [Google Scholar]
- 51. Fransen M., Wylin T., Brees C., Mannaerts G. P., and Van Veldhoven P. P. (2001) Human Pex19p binds peroxisomal integral membrane proteins at regions distinct from their sorting sequences. Mol. Cell. Biol. 21, 4413–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vastiau I. M., Anthonio E. A., Brams M., Brees C., Young S. G., Van de Velde S., Wanders R. J., Mannaerts G. P., Baes M., Van Veldhoven P. P., and Fransen M. (2006) Farnesylation of Pex19p is not essential for peroxisome biogenesis in yeast and mammalian cells. Cell Mol. Life Sci. 63, 1686–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kashiwayama Y., Asahina K., Shibata H., Morita M., Muntau A. C., Roscher A. A., Wanders R. J., Shimozawa N., Sakaguchi M., Kato H., and Imanaka T. (2005) Role of Pex19p in the targeting of PMP70 to peroxisome. Biochim. Biophys. Acta 1746, 116–128 [DOI] [PubMed] [Google Scholar]
- 54. Theodoulou F. L., Bernhardt K., Linka N., and Baker A. (2013) Peroxisome membrane proteins: multiple trafficking routes and multiple functions? Biochem. J. 451, 345–352 [DOI] [PubMed] [Google Scholar]
- 55. Agrawal G., and Subramani S. (2013) Emerging role of the endoplasmic reticulum in peroxisome biogenesis. Front. Physiol. 4, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van der Zand A., Gent J., Braakman I., and Tabak H. F. (2012) Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149, 397–409 [DOI] [PubMed] [Google Scholar]