Abstract
The presence of minimal residual disease (MRD) is widely recognized as a powerful predictor of therapeutic outcome in acute myeloid leukemia (AML), but methods of measurement and quantification of MRD in AML are not yet standardized in clinical practice. There is an urgent, unmet need for robust and sensitive assays that can be readily adopted as real-time tools for disease monitoring. NPM1 frameshift mutations are an established MRD marker present in half of patients with cytogenetically normal AML. However, detection is complicated by the existence of hundreds of potential frameshift insertions, clonal heterogeneity, and absence of sequence information when the NPM1 mutation is identified using capillary electrophoresis. Thus, some patients are ineligible for NPM1 MRD monitoring. Furthermore, a subset of patients with NPM1-mutated AML will have false-negative MRD results because of clonal evolution. To simplify and improve MRD testing for NPM1, we present a novel digital PCR technique composed of massively multiplex pools of insertion-specific primers that selectively detect mutated but not wild-type NPM1. By measuring reaction end points using digital PCR technology, the resulting single assay enables sensitive and specific quantification of most NPM1 exon 12 mutations in a manner that is robust to clonal heterogeneity, does not require NPM1 sequence information, and obviates the need for maintenance of hundreds of type-specific assays and associated plasmid standards.
CME Accreditation Statement: This activity (“The JMD 2017 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint providership of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“The JMD 2017 CME Program in Molecular Diagnostics”) for a maximum of 36 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Acute myeloid leukemia (AML) is a fatal disease with dismal outcomes. Even after achieving initial remission, most patients relapse and ultimately succumb to their disease. The presence of minimal residual disease (MRD) using a variety of molecular and immunophenotypic criteria is predictive of outcome in patients with AML.1, 2, 3, 4 Flow cytometric methods of MRD detection are effective but require multiantibody panels and high levels of user expertise in the flow cytometric identification of rare cell populations.5, 6, 7 Worldwide, there are no accepted standards for the identification and quantification of MRD in AML. PCR-based detection of MRD, in the form of fusion transcripts, can be accomplished in approximately 30% of patients with AML with abnormal cytogenetics, for example, inv(16), t(8;21), and t(15;17).8 More than half of patients with AML have normal cytogenetics, and 40% to 50% of these patients, in turn, have NPM1 mutations (NPM1muts).9, 10 Posttherapy monitoring of MRD in patients with NPM1mut AML using real-time quantitative PCR (qPCR) with allele-specific oligonucleotide primers has been evaluated in several clinical trials.1, 11 In all these studies, the levels of NPM1mut transcript were associated with patient outcome that pointed to NPM1 as a stable marker for disease progression. The presence of the NPM1mut transcript was associated with high risk of relapse compared with patients with MRS and a lower survival rate, with detection after the second chemotherapy cycle the most discriminatory time point.10 Conversely, a decrease in NPM1mut transcript copies correlated with hematologic remission, and half of the patients achieving complete remission presented <100 copies of the mutant transcript.11 This value agrees with other studies in which NPM1mut levels >1% after treatment and >10% after allogeneic transplantation were associated with poor overall and disease-free survival.12 Similarly, a threshold of 200 NPM1mut per 104 ABL1 (NPM1mut/104 ABL1) copies was proposed for early detection of relapse after therapy by Krönke et al.13
Approximately 95% of NPM1muts consist of four nucleotide insertions in exon 12 at the 863 position, the most common of which is type A (c.860_863dupTCTG), found in approximately 75% of patients with NPM1mutAML and an additional 15% comprising both type B (c.863_ins864insCATG) and type D (c.863_864insCCTG) mutations.3, 9, 14, 15, 16, 17 The remaining patients often have rarer subtypes, with differing and sometimes highly patient-specific insertion sequences. More than 50 such frameshift insertion mutations have been reported,18 and at least hundreds are theoretically possible. MRD testing in patients with NPM1mut AML currently requires prior DNA sequencing to identify the specific insertion sequence to match the patient to an appropriate allele-specific qPCR test. Thus, quantitative NPM1 MRD testing requires maintenance of qPCR assays and plasmid standards for each mutation, with commercial plasmid standards being widely available for only the top three mutations. Digital PCR (dPCR) assays in which the standard type-specific qPCR assays for NPM1mut are directly adapted individually to dPCR format have been reported to circumvent the need for plasmid standards while demonstrating excellent agreement with qPCR for the detection of rare NPM1muts on clinical validation.18 Despite this concordance, custom tests are still required for every new NPM1mut that is encountered.
The need to develop patient-specific MRD tests currently poses regulatory challenges. In the United States, for instance, each patient-specific MRD test for rare NPM1mut would comprise a separate laboratory developed test and, as such, would require submission of a new or amended application with subsequent approval by regulatory agencies before the laboratory developed test is clinically reportable. In addition, a practical problem encountered by oncologists is that NPM1mut diagnosis is sometimes made without sequencing using capillary electrophoresis19 with no widely used, clinically actionable sequencing technology sensitive enough to sequence NPM1 during remission. In such cases, reporting of MRD as mutated NPM1 transcripts is precluded using any current approach. Therefore, a substantial number of patients with NPM1mut AML would be ineligible for MRD assessment because they lack NPM1 sequence information and/or the availability of an appropriate test. Finally, although NPM1muts are generally stable over time,20, 21, 22 there have been reports of intrapatient heterogeneity23 and type switching,24 both of which could result in false-negative determinations of MRD status in individual cases. Next-generation sequencing–based approaches can circumvent these challenges because they do not require prior sequence information for detection of NPM1muts in the context of MRD and have sensitivities that exceed flow cytometry.23, 25 However, this technology currently does not express MRD in NPM1mut/104 ABL1 copies used by much of the concurrent and historical work. Thus, MRD testing for NPM1 in AML would be significantly enhanced by a more robust assay that is usable on more patients, simplifies the testing process for practitioners, and better addresses the possibility of intrapatient heterogeneity or clonal evolution. We present a new single dPCR-based test that detects 95% of NPM1mut types in AML using a single multiplex assay pool that enables quantitative assessments in a manner that is robust to NPM1 evolution, mixture, and subtype even when sequence information is missing.
Materials and Methods
Primary Sample Isolation and Cell Culture
Mononuclear cell isolation was performed using Ficoll-Paque (Pharmacia Biotech, Piscataway, NJ) density gradient centrifugation. GM12878 cell line (Coriell, Camden, NJ) was cultured in RPMI 1640 medium supplemented with 15% fetal bovine serum and 2 mmol/L l-glutamine (ThermoFisher, Pittsburgh, PA). OCI-AML3 cells were cultured in α–minimal essential medium supplemented with 20% heat-inactivated fetal bovine serum (ThermoFisher). Patient samples and healthy donor samples were acquired with patient consent in accordance with the Declaration of Helsinki and following Weill Cornell Medicine Institutional Review Board–approved regulations and protocols.
RNA Extraction
Cells were washed with Dulbecco's phosphate-buffered saline, and total RNA was isolated using the RNeasy extraction kit (Qiagen, Valencia, CA) as per the manufacturer's specifications.
Primers, Probes, Synthetic Targets, and Plasmid Standards
NPM1mut detection was performed using primers and probes described by Gorello et al,11 with modifications when performing dPCR. The ABL1 gene was used as endogenous control.26 Multiplex dPCR reactions for NPM1 detection consisted of a common forward primer (5′-GAAGAATTGCTTCCGGATGACT-3′) and probe (5′FAM-ACCAAGAGGCTATTCAA-MGB-3′) as previously described11 combined with a degenerate reverse primer (5′-CTTCCTCCACTGCNNNNCAGA-3′) adapted from the study by Gorello et al11 to generate a multiplex reaction pool (where N is a mixture of A, C, G, and T). A VIC (4,7,2′-trichloro-7′-phenyl-6-carboxyfluorescein)–labeled ABL1 probe was used for dPCR.27 Additional primers are indicated in Table 1. Probes were synthesized by ThermoFisher and primers by ThermoFisher or Integrated DNA Technologies (Coralville, IA). Synthetic targets for rare NPM1 subtypes were generated as gBlocks (Integrated DNA Technologies). A synthetic NPM1mut target pool and wild-type NPM1 (GeneArt fragments) were obtained from ThermoFisher. Sequences for all synthetic DNA targets are listed in Supplemental Table S1. Plasmid reference standards were used for quantification by qPCR (Qiagen).
Table 1.
Primers and Probes Used in This Study
| Target | Reverse primer sequence | Annealing temperature, °C |
|---|---|---|
| NPM1 multiplex | 5′-CTTCCTCCACTGCNNNNCAGA-3′ | 58 |
| NPM1 deoxyinosine (dI) | 5′-CTTCCTCCACTGC(dI)4CAGA-3′ | 58 |
| NPM1 5-nitroindole (5NI) | 5′-CTTCCTCCACTGC(5NI)4CAGA-3′ | 58 |
| NPM1 universal | 5′-GCCAGATATCAACTGTTACAGAAATG-3′ | 58 |
| c.860_863dupTCTG (type A) | 5′-CTTCCTCCACTGCCAGACAGA-3′ | 62 |
| c.863_864insCATG (type B) | 5′-CCTCCACTGCCATGCAGAG-3′ | 60 |
| c.863_864insCCTG (type D) | 5′-CCTCCACTGCCAGGCAGA-3′ | 61 |
| c.863_864insCTTG (type DD1) | 5′-CCTCCACTGCCAAGCAGAG-3′ | 62 |
| c.863_864insTATG | 5′-CTTCCTCCACTGCCATACAGA-3′ | 60 |
| c.863_864insTCGG | 5′-CTCCACTGCCCGACAGAGA-3′ | 60 |
| c.863_864insTAAG | 5′-CTTCCTCCACTGCCTTACAGAGA-3′ | 63 |
| c.863_864insCGTG | 5′-CCTCCACTGCCACGCAG-3′ | 60 |
| c.863_864insTTTG | 5′-TCCTCCACTGCCAAACAGA-3′ | 60 |
| c.863_864insCAAA | 5′-TCCTCCACTGCTTTGCAGA-3′ | 60 |
| c.863_864insTAGG | 5′-CTTCCTCCACTGCCCTACAGAG-3′ | 60 |
| c.863_864insCTCG | 5′-CCTCCACTGCCGAGCAGA-3′ | 60 |
| c.864_864delinsCCGTT | 5′-TCCTCCACTGAACGGCAGA-3′ | 60 |
| c.865_866insCAGC | 5′-CTTCCTCCACTGCCTGGCAGA-3′ | 60 |
| c.863_864insGCCG | 5′-CTTCCTCCACTGCCCGGCAGA-3′ | 61 |
Sequences for reverse primers are listed for all NPM1 mutation (NPM1mut) subtypes tested and massively multiplex NPM1mut and NPM1 universal assays, which amplifies both wild-type and mutant NPM1. Annealing temperature is indicated for universal, multiplex, NPM1 c.863_864insCTTG, NPM1 c.865_866insCAGC, and NPM1 c.863_864insGCCG assays.
Reverse Transcription
cDNA was synthesized from 500 ng of total RNA with SuperScript VILO cDNA Synthesis (Life Technologies, Carlsbad, CA). Reactions were incubated at 25°C for 10 minutes, 42°C for 60 minutes, and at 85°C for 5 minutes.
qPCR
qPCR reactions were performed on a QuantStudio 5K platform (Applied Biosystems, Carlsbad, CA) as described in Gorello et al.11 Annealing temperature varied according to target; individual values are specified for each assay in Table 1.
Droplet dPCR
Droplet dPCR was performed using the RainDrop platform (RainDance Technologies, Billerica, MA) or QX-200 platform (BioRad, Hercules, CA). For the RainDrop platform, reaction mixtures consisted of 1× Taqman Genotyping Master Mix (Life Technologies), 1× Droplet Stabilizer (RainDance Technologies), 500 nmol/L of each forward and reverse primer for NPM1 and ABL1, and 250 nmol/L of each probe. After generation of droplet partitions, 10 minutes of polymerase activation (95°C) were followed by 45 cycles of denaturation (95°C) and annealing and extension (58°C for multiplex assay) using BioMetra TAdvanced Thermocycler (Göttingen, Germany). Other temperatures are indicated in Table 1. Reactions were terminated with a 98°C incubation and held at 10°C until analysis. For the QX-200 platform, reactions were performed using the droplet dPCR Supermix for Probes (no dUTP) (BioRad) and processed according to the manufacturer's protocols on a C1000 Touch Thermal Cycler (BioRad) using the amplification protocol and primer and probe concentrations as above. Droplet positivity was quantified using the manufacturer's software: RainDrop Analyst II version 1.1 (RainDance Technologies) or QuantaSoft version 1.0 (BioRad).
Limit of Detection
Limit of detection was determined as the mean NPM1mut/104 ABL1 ratio for 12 blank samples ± 3 SDs according to International Union of Pure and Applied Chemistry recommendations.28, 29
RNA Sequencing
Total RNA was used to generate sequencing libraries using the Kapa Biosystems Stranded mRNA Sequencing Kit with an mRNA capture beads protocol according to the manufacturer instructions (Kapa Biosystems, Wilmington, MA). Targeted enrichment for ultradeep RNA sequencing was performed using Integrated DNA Technologies Lockdown probes. Hybridization and washing were performed as per the company's specifications. Paired end 150-bp sequencing data were generated on an Illumina HiSeq 2500. Reads were aligned to the human reference genome (GRCh38) using the STAR aligner version 2.5.2a.30 Postalignment processing included duplicate marking using SamBlaster version 0.1.2131 and splice junction splitting using splitNreads commit a84339a (splitNreads, https://github.com/mjafin/splitNreads, last accessed August 31, 2015). Variant calling was performed using FreeBayes version 0.9.21.32
Targeted DNA Sequencing
Genomic DNA was extracted from frozen pellets using the Qiagen QiaAmp DNA Micro Kit. Libraries for DNA sequencing were prepared using 250 ng of material with the Kapa Biosystems HyperPrep Kit following the manufacturer's instructions. Libraries were hybridized for targeted enrichment with a custom-designed panel specific for hematologic malignant neoplasms. Capture probes were synthesized by Nimblegen (Roche Diagnostics, Indianapolis, IN); hybridization, washing, and purification steps were performed following their protocol. Paired-end sequencing (150 bp) was performed on an Illumina Hiseq 2500. Reads were aligned to the human reference genome (hg37d5) using BWA-MEM version 0.7.10 (http://bio-bwa.sourceforge.net).33 Variant calling was performed using VarDict version 1.4.6 (https://github.com/AstraZeneca-NGS/VarDictJava).34
Results
Cross-Detection between Existing qPCR-Based NPM1 Allele-Specific Assays
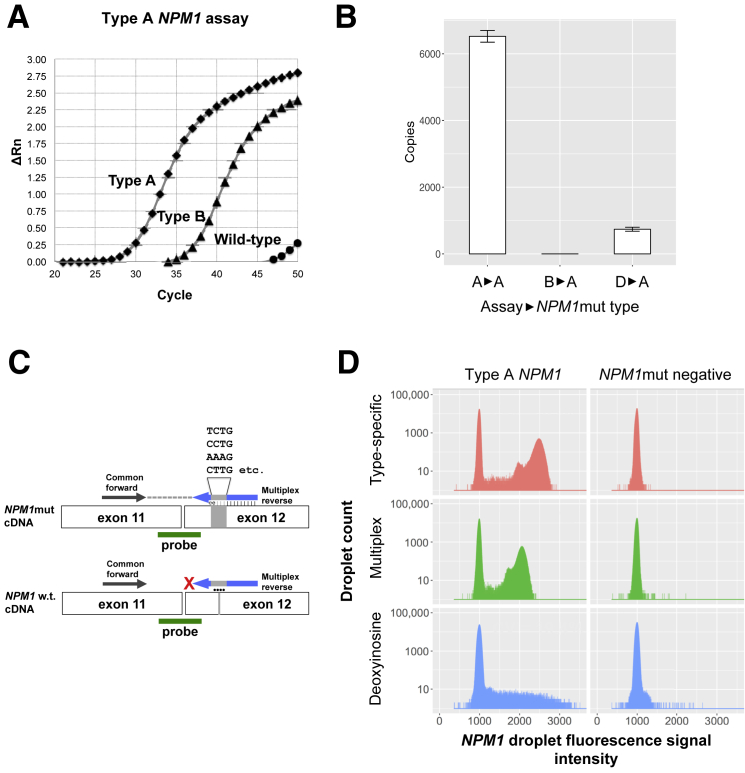
NPM1 allele-specific assays are able to cross-detect other NPM1 subtypes but not wild-type NPM1. Representative qPCR amplification curves (Figure 1A) reveal the ability of the established type A NPM1 assay11 to detect 2000 copies of reference plasmid for type A or type B NPM1 insertions (Ipsogen) alongside wild-type NPM1. The type A assay effectively discriminated between the type A NPM1mut and the wild-type NPM1, reproducing the known specificity of this assay for NPM1mut. However, the same type A–specific assay was notably less selective between type A and type B NPM1muts, where type B mutations had delayed reaction kinetics and approached a reaction plateau above 45 cycles that was not achieved by wild-type NPM1. Next, we determined the extent to which the established NPM1 type-specific assays for the most common mutations (types A, B, and D) could cross-detect the type A NPM1mut by testing each of the three assays against the same amount of type A plasmid (Figure 1B). When the type A–specific assay was correctly matched to the type A NPM1mut, means ± SEM of 6524 ± 176 NPM1mut copies were detected. However, when the type B–specific and type D–specific assays were used against the same amount of type A target, means ± SEM of only 0.3 ± 0.3 and 740 ± 60 copies were detected, respectively. The resulting underestimation of NPM1mut copies arising from the mismatch of assays to mutations was significant (P < 0.001; one-way analysis of variance). Despite these observations, the apparent ability to achieve a reaction plateau in the case of NPM1mut but not wild-type NPM1 irrespective of mismatch between assay and mutation suggested the possibility that an end point–sensitive technology, such as dPCR, could demonstrate quantitative characteristics in these scenarios. We hypothesized that the observed specificity for NPM1muts versus wild-type was likely because NPM1mut primers need to loop out their insertion sequence in an energetically unfavorable manner to successfully amplify wild-type NPM1, with the insertion sequence itself being a significant but lesser contributor to specificity. We further hypothesized that most NPM1mut AMLs would be detectable with a single assay if it were possible to develop a multiplex pool of assays against all known and theoretical insertion NPM1muts with subsequent quantification of reaction end points by dPCR.
Figure 1.
Cross-detection between NPM1 assays motivates design of a digital PCR-based multiplex NPM1 assay. A: Real-time quantitative PCR (qPCR) amplification curves indicating normalized reporter value (ΔRn) as a function of the PCR cycle. The type A NPM1 assay is able to amplify 2000 copies of Ipsogen reference plasmids for type A and type B NPM1 mutations (NPM1muts) but not wild-type NPM1. B:NPM1mut copies detected by qPCR with type A, type B, or type D assays when type A plasmid standard was used as template. C: Schematic of massively multiplex assay indicating relative positions of the common forward primer, multiplex reverse primer, and common probe. Productive amplification occurs with NPM1mut templates (top row) but not wild-type NPM1 templates (bottom row). D: Histograms indicate the distribution of positive droplets of type-specific, multiplex, and deoxyinosine-based NPM1 mutation assays in type A NPM1 patient samples (NPM1mut acute myeloid leukemia; type A) alongside NPM1-negative mononuclear cells (negative). The vertical axis indicates log10 (absolute counts) with the horizontal axis indicating droplet signal intensity. The CV of positive droplets is indicated. Data are expressed as means ± SEM (B). n = 3 (B).
Development of Massively Multiplex dPCR Assay for NPM1muts
Multiplex insertion-specific assays covering all known and theoretical four nucleotide insertions were synthesized for the most common insertion site (position 863) (Table 1). The relative position of primers and probes is indicated schematically in Figure 1C. In parallel, we synthesized assays in which the common insertion site was spanned by universal bases deoxyinosine35 and 5-nitroindole,36 which have relaxed base-pairing properties relative to the four standard nucleotides, thus permitting base pairing across a range of insertion sequences. As a baseline, the established qPCR–based assay for type A NPM1muts11 was adapted to dPCR. The performance of these assays was initially compared using cDNA from a patient with AML who tested positive for a type A NPM1mut by clinical sequencing (Genoptix Medical Laboratory, Carlsbad, CA). Both the type-specific assay and the multiplex assay produced distinct positive droplet populations indicative of successful amplification of NPM1mut with CV values of 5.5% and 5.1%, respectively, among positive droplets (Figure 1D). In contrast, dPCR reactions conducted with deoxyinosine-containing primers produced a nondistinct smear of positive droplets (CV = 19.4%) (Figure 1D), rendering it less useful for MRD detection. 5-Nitroindole–containing primers failed to produce amplified product. Given that the multiplex assay was both the most successful and cost-effective option tested, we continued to evaluate the multiplex assay on other NPM1mut and in serial AML cases.
Agreement with Established Assays
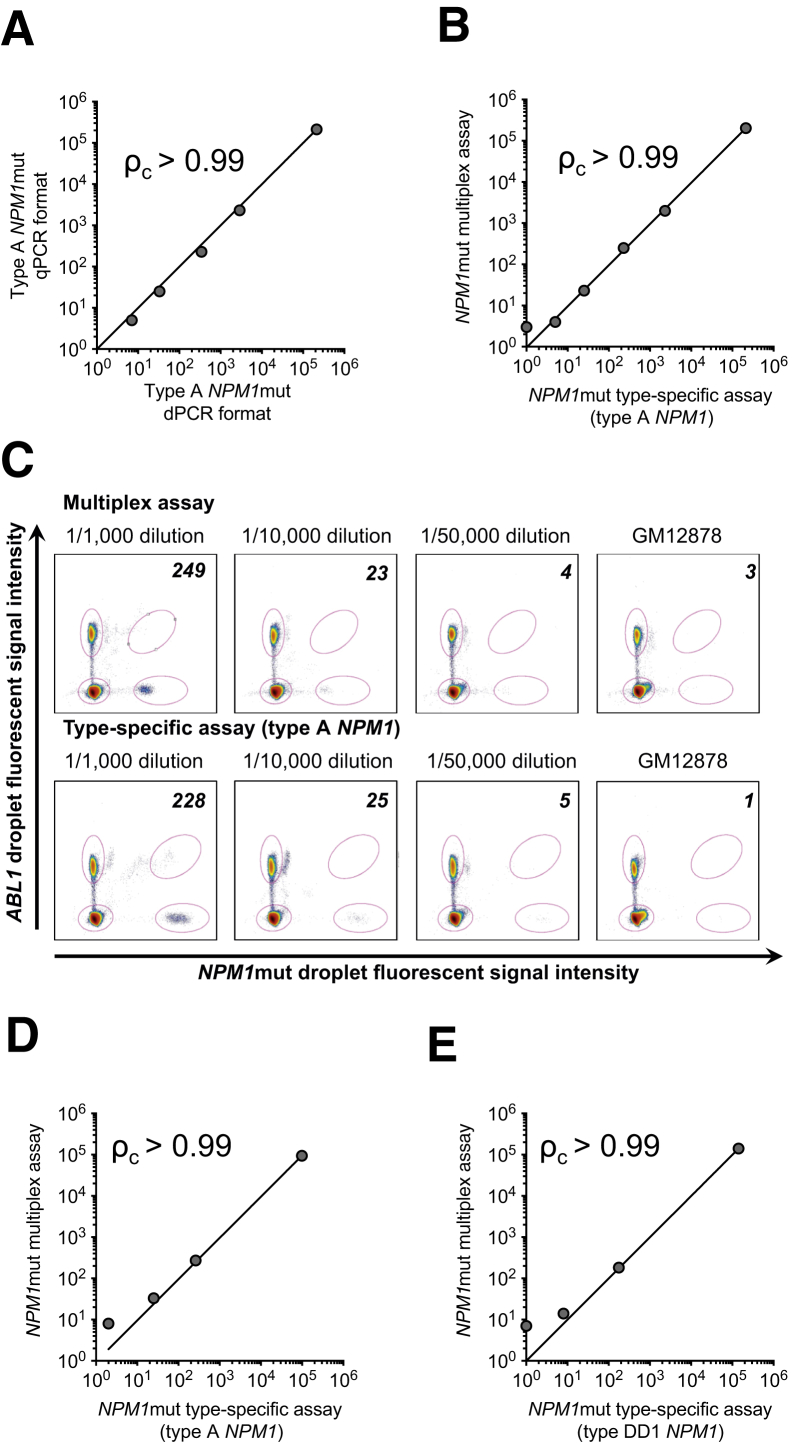
To test whether the NPM1 multiplex assay agrees with established subtype-specific qPCR and dPCR assays already used in MRD assessment, we generated a spike-in dilution series of the cell line OCI-AML3 (type A NPM1) into GM12878 (wild-type NPM1), ranging from 1:1000 cells to 1:50,000 cells (OCI-AML3:GM12878). NPM1mut subtype and variant allele fraction were verified by next-generation sequencing. For each dilution, we quantified NPM1mut/104 ABL1 transcript copies according to methods and reporting convention established by Gorello et al.11 The established type A allele-specific assays in qPCR were then compared with type-specific and multiplex assays in dPCR. For each comparison, we computed Lin's concordance correlation coefficient (ρc),37 which evaluates the extent to which pairwise comparisons agree (ie, fit to a diagonal line with a slope of 1 and y-intercept of 0). Previous studies noted general concordance between MRD measurements obtained by qPCR and dPCR using allele-specific primers across a range of common and rare NPM1muts.18 Indeed, we similarly noted excellent concordance between qPCR and dPCR across the range of cell dilutions tested (ρc = 0.998; 95% CI, 0.994–0.999; qPCR versus dPCR) (Figure 2A). NPM1mut/104 ABL1 ratios were further concordant when comparing the new NPM1 multiplex assay to the type-specific assay in dPCR (ρc = 0.999; 95% CI, 0.997–0.999; multiplex versus type specific) (Figure 2B), producing essentially identical NPM1mut/104 ABL1 ratios (Figure 2C). The 1:50,000 dilution demonstrated NPM1mut/104 ABL1 ratios close to double the background seen in negative control GM12878. However, increasing cDNA input 10-fold improved the signal/noise ratio for 1:50,000 cell dilution (Supplemental Figure S1C). OCI-AML3 serial dilutions in MV411, a myelomonocytic cell line, produced similar results (data not shown). This concordance was preserved when using an alternative dPCR technology (BioRad QX200) (Supplemental Figure S1, A and B), indicating that results across platforms are generally comparable. To transition from cell lines to primary samples, we further tested the NPM1 multiplex assay against type-specific assays in primary AML samples identified as positive for common, type A NPM1 (Figure 2D), or rare naturally occurring mutation types in patient samples, such as DD1 (c.863_864insCTTG) (Figure 2E), c.863_864insTATG, c.865_866insCAGC, and c.863_864insGCCG (Supplemental Figure S2). The cDNA from primary samples was diluted into healthy cord blood cDNA at 1:1000 and 1:10,000 (w/w). Concordance was noted between massively multiplex assay and type-specific assays for all mutation subtypes tested (ρc = 0.995 to 1.000).
Figure 2.
Performance of multiplex NPM1 minimal residual disease assay on spike-in dilution series. A: Correlation of real-time quantitative PCR (qPCR)–based type A–specific assay (vertical axis) to digital PCR (dPCR)–based adaptation of the type A–specific assay (horizontal axis) when using cDNA derived from undiluted OCI-AML3 or OCI-AML3 spiked into GM12878 at 1:1000, 1:10,000, or 1:50,000 cells or GM12878 alone. Axes indicate NPM1 mutation (NPM1mut)/104ABL1. B: Correlation of the massively multiplex NPM1 assay (vertical axis) to dPCR-based adaptation of the type A–specific assay (horizontal axis) on the same dilution series as A. Axes indicate NPM1mut/104ABL1. C: Dot plot of dPCR data for the multiplex assay (top row) and type A–specific assay adapted to dPCR (bottom row) on the same spike-in dilution series as A and B. Dilutions are indicated. The vertical axis indicates the signal intensity of droplets in the ABL1 channel (VIC), and the horizontal axis indicates positive droplets in the NPM1 mutant channel (FAM). NPM1mut/104ABL1 copies are indicated in the upper right hand corner of each scatterplot. D: Correlation of the massively multiplex NPM1 assay (vertical axis) to dPCR-based adaptation of the type A–specific assay (horizontal axis) on cDNA from NPM1mut acute myeloid leukemia (AML; type A) undiluted or diluted into cDNA from healthy cord blood at 1:1000 or 1:10,000 (w/w) alongside cord blood cDNA alone. Axes indicate NPM1mut/104ABL1. E: Same as D except using a type DD1–specific dPCR assay on cDNA from a rare NPM1mut AML (type DD1). Lin's concordance correlation coefficient is indicated (ρc).
To determine the lower limit of detection of multiplex assay, background signal was measured in a panel of 12 NPM1mut-negative samples. The panel included primary AML samples (bearing rearrangements rarely found to co-occur with NPM1, such as CBFβ-MYH11 or AML1/ETO),15 bone marrow from healthy donor, cord blood, and the GM12878 and MV411 cell lines. Total NPM1 transcripts were also quantified in the same set of samples using an NPM1-universal primer with binding site downstream of the 863 position in exon 12 of NPM1 transcript. NPM1-universal primer nonselectively amplifies all NPM1 transcripts irrespective of the presence or absence of a 4-nucleotide insertion (ie, the number of NPM1 copies counted consist of wild-type and, if present, mutant transcripts). Background levels in 12 NPM1mut wild-type normal primary samples and cell lines tested ranged from 0 to 7 NPM1mut/104 ABL1 (median, 1 NPM1mut/104 ABL1) for the multiplex assay compared with 0 to 1 NPM1mut (median, 0 NPM1mut/104 ABL1) for the type-specific assay (NPM1mut type A) (Supplemental Figure S3). Thus, the limit of detection for the multiplex and type A–specific assays under dPCR conditions are estimated at 2.4 and 1.0, respectively, for normal primary blood samples, well below reported decision cutoffs of 100 to 1000 NPM1mut/104 ABL1 copies.38 Both allele-specific and massively multiplex assays produced a single amplicon as determined by gel electrophoresis thus further corroborating specificity (Supplemental Figure S4).
The multiplex dPCR assay for detection of NPM1mut demonstrated overall agreement with established type-specific assays in the spike-in dilution series and the primary AML cells for common and rare NPM1 subtypes. Moreover, NPM1mut/104 ABL1 ratios produced by the multiplex assay agreed with established qPCR–based assays. We thus proceeded to determine its robustness against a variety of rare NPM1muts and its ability to monitor MRD in actual patients.
Other Rare NPM1mut Types
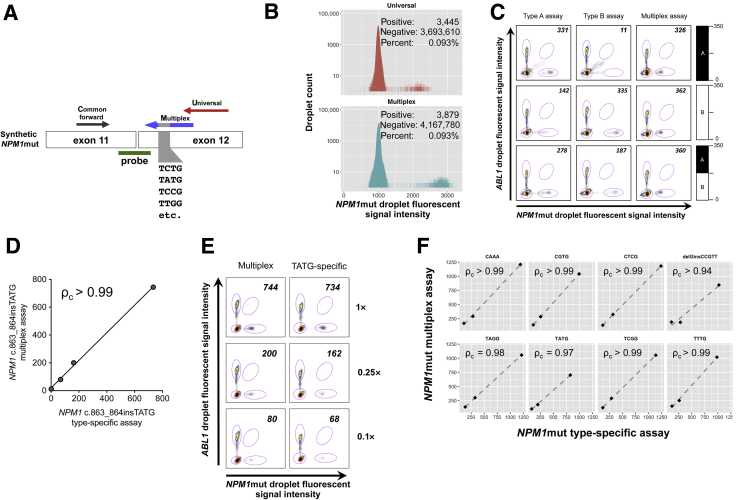
To further assess the general robustness of the multiplex assay and its ability to detect rare NPM1muts, we generated a synthetic NPM1mut target pool that contained all possible four nucleotide insertions at the c.863 position of exon 12 such that 100% of DNA fragments contain NPM1 insertions. The sequence of the synthetic target was verified by Sanger sequencing (Table 2). The synthetic fragment and primers are indicated schematically in Figure 3A. NPM1 copies were quantified using a multiplex assay or NPM1 universal assay. If the multiplex NPM1mut assay can detect all NPM1 c.863 insertions, the expectation is that the number of NPM1-positive partitions will not differ statistically if the synthetic target is amplified by NPM1 multiplex (mutant specific) or the NPM1 universal primer set. Indeed, we found that the universal and mutant-specific multiplex primer sets resulted in 0.093% positive partitions, indicating that counting was accurate irrespective of subtype (odds ratio, 1.002; 95% CI, 0.956–1.049; P = 0.934) (Figure 3B). Given the reported instances of clonal heterogeneity23 and type switching24 in selected AML cases and success with the multiplex target pool in Figure 3B, we next sought to determine the ability of the massively multiplex assay to quantify NPM1muts in the setting of clonal heterogeneity (Figure 3C). To this end, plasmids harboring type A and/or type B NPM1muts were spiked into NPM1 wild-type GM12878 cDNA alone or combined as a 50:50 mixture of type A and type B (A:B) to target a total of approximately 350 NPM1mut/104 ABL1 copies in all cases. Each of the targets was tested with allele-specific assays for type A or type B NPM1muts alongside the massively multiplex assay. As expected, the type-specific assays produced accurate measurements, within approximately 5% of the targeted 350 NPM1mut/104 ABL1 ratios ranging from 331 to 335 NPM1mut/104 ABL1 copies when each assay was correctly matched to the correct NPM1mut. Notably, the massively multiplex assay demonstrated similar accuracy against all targets, including the mixture detecting 326, 362, and 360 NPM1mut/104 ABL1 copies for the type A, type B, and A:B (50:50) mixture, respectively. However, in sharp contrast, the type-specific assays variably underestimated the NPM1mut/104 ABL1 ratio when either used with the wrong NPM1mut or challenged with the A:B mixture, producing unreliable measurements ranging from 11 to 278 NPM1mut/104 ABL1 copies.
Table 2.
Sequence of Synthetic NPM1 Mutant Transcripts
| Insertion | Synthetic sequence |
|---|---|
| NPM1-c863del864G_insCCGTT | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGCCGTTCAGTGGAGGAAGTCTCTTTAAG-3′ |
| NPM1-c863_864insCAAA | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGCAAAGCAGTGGAGGAAGTCTCTTTAAG-3′ |
| NPM1-c863_864insCGTG | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGCGTGGCAGTGGAGGAAGTCTCTTTAAG-3′ |
| NPM1-c863_864insCTCG | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGCTCGGCAGTGGAGGAAGTCTCTTTAAG-3′ |
| NPM1-c863_864insTAGG | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGTAGGGCAGTGGAGGAAGTCTCTTTAAG-3′ |
| NPM1-c863_864insTATG | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGTATGGCAGTGGAGGAAGTCTCTTTAAG-3′ |
| NPM1-c863_864insTCGG | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGTCGGGCAGTGGAGGAAGTCTCTTTAAG-3′ |
| NPM1-c863_864insTTTG | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGTTTGGCAGTGGAGGAAGTCTCTTTAAG-3′ |
| NPM1-c863ins4N | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGNNNNGCAGTGGAGGAAGTCTCTTTAAGAAAATAGTTTAAACAATTTGTTAAAAAATTTTCCGTCTTATTTCATTTCTGTAACAGTTGATATCTGGCTGTCCTTTTTATAATGCAGAGTGAGAACTTTCCCTACCGTGTTTGATAAATGTTGTCCAGGTTCTATTGCCAAGAATGTGTTGT-3′ |
| NPM1-wt | 5′-GACCTAGTTCTGTAGAAGACATTAAAGCAAAAATGCAAGCAAGTATAGAAAAAGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGTGAAGAATTGCTTCCGGATGACTGACCAAGAGGCTATTCAAGATCTCTGGCAGTGGAGGAAGTCTCTTTAAGAAAATAGTTTAAACAATTTGTTAAAAAATTTTCCGTCTTATTTCATTTCTGTAACAGTTGATATCTGGCTGTCCTTTTTATAATGCAGAGTGAGAACTTTCCCTACCGTGTTTGATAAATGTTGTCCAGGTTCTATTGCCAAGAATGTGTTGT-3′ |
Sequences for mutant and wild-type NPM1 synthetic cDNA fragments.
Figure 3.
Multiplex NPM1 assay accurately detects rare NPM1 mutation (NPM1mut) types. A: Schematic representation of synthetic NPM1mut target consisting of a pool of degenerate NPM1mut insertion sequences. Amplification is performed using common forward primer with multiplex reverse primer pool (mutant specific) or the universal reverse primer (amplifies both mutant and wild-type). Common probe is used for all reactions. B: Histogram indicating counts and distribution of fluorescence signal intensity of NPM1-positive and -negative droplets. C: Plasmids harboring type A (top row), type B (middle row), or a 50:50 mixture of type A and B (bottom row) NPM1 mutations were spiked into GM12878 background cDNA. A target of 350 NPM1mut/104ABL1 copies was used in all cases and the expected content indicated graphically by the bar plot on each row. Detection was attempted by digital PCR (dPCR) using the specific assays for the type A or type B mutation alongside the massively multiplex assay. The vertical axis of each dot plot indicates the signal intensity of ABL1-positive droplets, and the horizontal axis indicates NPM1mut-positive droplets. NPM1mut/104ABL1 ratios are indicated in the upper right corner. D: Synthetic template for NPM1 c.863_864insTATG is spiked into GM12878 cDNA targeting approximately 1000 NPM1mut/104ABL1 copies and left undiluted (1×) or diluted fourfold (0.25×) or 10-fold (0.1×) alongside negative control cell line GM12878. The axes indicate NPM1mut/104ABL1 as determined by the NPM1 multiplex assay (vertical axis) versus type-specific assay (horizontal axis). E: Scatterplot of dPCR data for the multiplex assay and c.863_864insTATG-specific assay adapted to dPCR for the dilution series from A with dilutions 1×, 0.25×, and 0.1× shown. F: Correlation between NPM1mut/104ABL1 ratios for a different synthetic rare NPM1 insertion mutations spiked into GM12878 cDNA targeting an approximately 1000 NPM1mut/104ABL1 starting ratio. Undiluted spike-in mixture (1×) is then compared with fourfold (0.25×) and 10-fold (0.1×). The position 863 insertion sequence is indicated over each plot except for delGinsCCGTT, which is NPM1 c.864_865delGinsCCGTT. The axes indicate NPM1mut/104ABL1 ratios for the multiplex assay (vertical axis) versus type-specific assays (horizontal axis). Lin's concordance correlation coefficient is indicated (ρc).
Next, to further explore the ability of multiplex assay to detect NPM1mut transcripts independently of the subtype sequence, we proceeded to test our NPM1mut-specific multiplex assay on individual rare insertion sequences reported recently by Ivey et al10 along with the published type-specific assay for those mutations (Table 1). NPM1 copies were determined using both the NPM1 multiplex assay and type-specific assay, testing a subset of rare NPM1 insertions on dual dPCR platforms. First, NPM1 c.863_864insTATG synthetic targets in a dilution series were spiked into GM12878 alongside nonspiked GM12878 controls. NPM1mut/104 ABL1 ratios obtained by type-specific and multiplex assay were concordant (ρc = 0.998; 95% CI, 0.981–1.000) (Figure 3D), producing similar values and distinct clusters of positive dPCR partitions/droplets (Figure 3E). Measurements of NPM1mut copies were also consistent across dPCR platforms using the NPM1 multiplex assay (ρc = 0.985; 95% CI, 0.861–0.999) (Supplemental Figure S5 and Supplemental Table S1). We additionally generated synthetic NPM1mut spike-in dilution series for eight rare NPM1muts and compared NPM1mut/104 ABL1 ratios derived from multiplex and type-specific assays (Figure 3F). All the NPM1muts demonstrated substantial concordance between type-specific and multiplex assays (ρc = 0.97 to 0.99), with the notable exception of NPM1 c.864_865delGinsCCGTT, which contains an extra mismatch, likely leading to reduced PCR priming efficiency. The multiplex assay detected NPM1 insertions in all cases tested.
Serial Monitoring of MRD in Patients with AML
To ascertain the potential utility of the multiplex assay in patient care, we determined molecular MRD levels sequentially in three patients with AML undergoing treatment at the Leukemia Program of Weill Cornell Medicine and New York Presbyterian Hospital. Patients had confirmation of NPM1mut by an independent diagnostic laboratory, and all clinical decision making was based on the results from the Clinical Laboratory Improvement Amendments–certified laboratory. Bone marrow aspirates (BMAs) and/or peripheral blood (PB) were collected periodically from patients with AML seen during their care. The patients signed informed consent documents approved by the institutional review board and agreed to participate in a research protocol for serial sampling of PB and bone marrow. One of these patients presented to our clinic in remission without prior sequencing having been diagnosed for NPM1mut AML only by capillary electrophoresis.
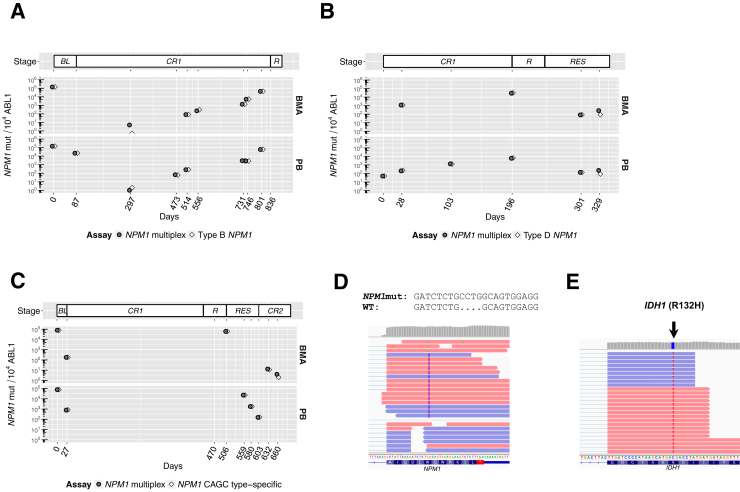
Patient 1 was confirmed positive for type B NPM1mut (c.863_864insCATG) by routine clinical sequencing performed as standard care (Genoptix). NPM1 MRD levels were assessed during a period of 801 days (Figure 4A). PB and BMAs demonstrated significant decreased levels of NPM1mut (<100 NPM1mut/104 ABL1) after therapy measured at day 297 as determined by the type-specific assay (type B) and NPM1 multiplex assay, with both assays demonstrating overall agreement and trending upward over time. NPM1mut levels are expected to be higher in BMA versus PB based on previous reports.10, 15 However, the present case demonstrated higher PB mutant NPM1 percentages compared with BMA at day 514 and 731. The patient eventually evolved to relapsed disease after 836 days.
Figure 4.
Monitoring of minimal residual disease in serial acute myeloid leukemia (AML) cases. A: Patient 1 [type B NPM1 mutated (NPM1mut)]: NPM1mut/104ABL1 as a function of time (top panel). The vertical axis indicates NPM1mut/104ABL1, and the horizontal axis indicates time since initial monitoring in days. Diamond points indicate results with the type B–specific assay. Circular points indicate results with the NPM1 multiplex assay. Peripheral blood (PB) and bone marrow aspirate (BMA) samples are shown in the bottom and middle panels, respectively. Disease status is indicated in the top panel. B: Patient 2 (type D NPM1mut): NPM1mut/104ABL1 as a function of time (top panel). The vertical axis indicates NPM1mut/104ABL1, and the horizontal axis indicates time since initial monitoring in days. Diamond points indicate results with the type D–specific assay. Circular points indicate results with the NPM1 multiplex assay. PB and BM samples are shown in the bottom and middle panels, respectively. Disease status is indicated in the top panel. C: Patient 3 [NPM1 c.865_866ins(CAGC)]: NPM1mut/104ABL1 as a function of time (top panel). The vertical axis indicates NPM1mut/104ABL1, and the horizontal axis indicates time since initial monitoring in days. Diamond points indicate results with the CAGC type-specific assay. Circular points indicate results with the NPM1 multiplex assay. PB and BM samples are shown in the bottom and middle panels, respectively. D: The insertion sequence identified by deep targeted mRNA sequencing for patient 2 above the alignment of reads supporting the type D NPM1 insertion. E: Reads supporting IDH1 (p.ArgR132His) mutation identified by deep targeted mRNA sequencing for patient 2. BL, baseline; CR1, complete remission 1; CR2, complete remission 2; R, relapse; RES, resistant disease; WT, wild-type.
Patient 2 is a 62-year-old woman with a history of breast cancer treated with multiagent chemotherapy and radiation who presented for care at our center after having been diagnosed with AML with normal cytogenetics and treated elsewhere. She received a standard cytarabine and anthracycline-based induction, followed by four cycles of high-dose cytarabine consolidation. The patient had an NPM1mut identified at the time of diagnosis using capillary electrophoresis by a clinical diagnostic laboratory. However, a diagnostic specimen was not available to determine the patient's NPM1 sequence. We thus followed up NPM1mut MRD levels for 329 days (Figure 4B) using the NPM1 multiplex assay. A surge in NPM1mut transcripts was noted in BM from day 28, reaching 1012 NPM1mut/104 ABL1 copies. Leftover RNA initially isolated for MRD assessment was submitted for ultradeep targeted mRNA-sequencing hybrid capture sequencing to determine the NPM1 subtype. This analysis revealed the NPM1 subtype as type D (c.863_864insCCTG) supported by 48 (0.34%) of 14,271 reads (Figure 4D) and incidentally identified an IDH1 (p.Arg132His; R132H) unknown in this patient supported by 32 (0.9%) of 3679 reads (Figure 4E). The type-specific and multiplex assay closely tracked each other, with levels of NPM1mut gradually increasing in the PB and BMA during MRD monitoring through this patient's eventual clinical and morphologic relapse 6 months later. On relapse, clinical DNA sequencing (Genoptix) identified the IDH1 R132H mutation at 6% variant allele fraction but not the type D NPM1mut. Targeted deep DNA sequencing then confirmed the presence of the type D NPM1mut at 3% variant allele fraction. The patient is currently undergoing treatment with an investigational isocitrate dehydrogenase (IDH1) inhibitor, which was associated with an observed reduction in NPM1mut transcripts during the resistant disease stage.
Patient 3 presented with a rare NPM1mut subtype (c.865_866insCAGC) confirmed by routine clinical sequencing. A type-specific assay was designed to retrospectively monitor NPM1 status alongside multiplex assay during a period of 660 days (Figure 4C). NPM1mut copies drastically decreased after standard induction therapy after day 27, consistent with treatment response and progression into remission status. The patient received a stem cell transplant and remained in remission for >1 year, after which the patient relapsed as indicated by the increase in NPM1mut copies with levels comparable to those observed at baseline. Reemergence of an IDH1 mutation present at baseline was observed by next-generation sequencing, and at this point treatment was started with an IDH1 inhibitor under clinical trial. Finally, the patient evolved to resistant disease and secondary remission, with NPM1mut levels trending downward. The patient is currently in second remission. Type-specific and multiplex assays showed close agreement at all times in a case that is not otherwise tractable for clinically reportable molecular MRD evaluation by the most common qPCR tests.
Discussion
Historically, the most important prognostic criteria in AML have been age and cytogenetics. Recently, molecular genetics are also emerging as important prognostic criteria and, in some cases, a means to identify potential targets for treatment. Despite these advances, outcomes remain poor, especially for older patients with AML and for those of any age with unfavorable cytogenetics. Recent studies suggest that assessment of NPM1mut transcript copies as a marker of MRD improves risk stratification over established cytogenetic and molecular criteria.2, 3, 4, 10, 39 Indeed, a recent prospective study of 223 patients revealed subgroups of patients with NPM1muts with improved survival when the PCR results were NPM1 negative, despite having prognostically adverse co-occurring mutations, such as FLT3-ITD and/or DNMT3A.10 In addition, several studies have found that patients with AML who are treated with allogeneic stem cell transplantation in first remission have distinctly more favorable outcomes if undergo transplantation without detectable MRD, as measured by immunophenotype.40 Thus, MRD monitoring in general, and MRD monitoring of patients with NPM1mut AML in particular need to become more widely accessible tools for prognostication and treatment planning in general practice not only for patients in clinical trials.
Selected patients with NPM1mut AML are currently ineligible for quantitative MRD monitoring because their NPM1mut is rare or their insertion sequence is unknown because detection was performed using capillary electrophoresis. It is possible to use qPCR difference-in-cycle thresholds1 as an alternative for rare NPM1muts, but this approach is sensitive to input RNA and does not express MRD in NPM1mut per 10,000 ABL1 copies, making cross-study comparisons challenging, especially when patient materials are limiting. The development of many individual patient-tailored tests is also a possibility but may pose challenges in the clinic, where each type-specific PCR assay and its corresponding quantitative standard require rigorous quality control and regulatory approval that may not be feasible in agency-regulated clinical laboratories, such as those encountered in the United States. Moreover, site-to-site differences in tests developed at different laboratories for rare mutations may further fragment studies across sites and raise the complexity of interpreting results across centers. Finally, AML is a dynamic and heterogeneous disease that can evolve. Reports demonstrating intrapatient NPM1 heterogeneity and type switching indicate the possibility of MRD underestimation or misdiagnosis in some patients with type-specific assays, which can result in adverse patient outcomes when used to guide patient care.
The multiplex assay described in this work was effective across a range of diverse common and rare NPM1muts, with overall concordance with type-specific assays. The efficacy of the new multiplex assay was demonstrated in a total of 14 common and rare subtypes and accurately measured NPM1mut even in an extremely challenging heterogeneous pool of 256 subtypes, illustrating the extreme robustness of the approach. The observed background was higher compared with type-specific assays but still well below proposed cutoffs for treatment decisions.38 In addition, the assay successfully quantified NPM1mut in a mixture of different mutant subtypes, illustrating its utility in cases of clonal heterogeneity. NPM1 levels are expressed by the new assay in the same units as previous NPM1 studies, thus enabling comparison to concurrent and historical work.10, 11, 15 Importantly, the multiplex test remained effective and quantitative in the absence of sequence information at diagnosis and matched the upward and downward MRD trend in serially monitored patients seen with NPM1mut MRD assays. Overall, these data indicate that a single, easily deployed NPM1mut test can effectively simplify NPM1mut MRD testing capabilities for laboratories, while reducing the potential complexities associated with NPM1 quantification.
Acknowledgments
We thank Drs. Michael J. Kluk and Wayne Tam for critical evaluation of this manuscript and Leukemia Fighters for funding.
N.M.-T., M.L.G., G.J.R., and D.C.H. designed research, analyzed data, and wrote the manuscript; F.A. coordinated sample and clinical data; B.J.W. provided clinical samples; E.K.R., S.L., P.D., and G.J.R. provided clinical samples and designed research; N.M.-T., Y.H., and M.A.A. performed experiments and analyzed data.
Footnotes
See related Commentary on page 498
Supported by Leukemia Fighters (D.C.H.).
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2017.03.005.
Supplemental Data
Performance of multiplex NPM1 minimal residual disease assay on spike-in dilution series on alternative digital PCR (dPCR) platform and with higher cDNA input. A: Concordance of real-time quantitative PCR (RQ-PCR) (qPCR)–based type A–specific assay (vertical axis) to dPCR–based adaptation of the standard type A–specific assay on the BioRad QX200 (horizontal axis) when using cDNA derived from either undiluted OCI-AML3 or OCI-AML3 spiked into GM12878 at 1:1000, 1:10,000, or 1:50,000 cells or GM12878 alone. Axes indicate NPM1 mutations (NPM1muts) per 104ABL1 (NPM1mut/104ABL1). Lin's concordance correlation coefficient is indicated (ρc). B: Concordance of the massively multiplex NPM1 assay on RainDance RainDrops (vertical axis) to dPCR–based adaptation of the type A–specific assay on BioRad QX200 (horizontal axis) on the same dilution series as A. Axes indicate NPM1mut/104ABL1. Lin's concordance correlation coefficient is indicated (ρc). C: Scatterplot of dPCR data for the multiplex assay (top row) and type A–specific assay adapted to dPCR (bottom row) on the same spike-in dilution series as A and B but with 10× as much input to ascertain improved sensitivity. Dilutions are indicated. The vertical axis indicates the signal intensity of droplets in the ABL1 channel (VIC) and the horizontal axis indicates positive droplets in the NPM1mut channel (FAM). NPM1mut/104ABL1 copies are indicated in the upper right hand corner of each scatterplot. BioRad QX200 could not be tested in C because the number of ABL1 copies saturated the instruments so that it was no longer quantitative with respect to ABL1.
The type-specific and massively multiplex assays agree in rare NPM1 mutations observed in patients. cDNA from NPM1-mutated acute myeloid leukemia was diluted into cDNA from healthy cord blood at 1:1000 and 1:10,000 (w/w) alongside cord blood controls. Detection was performed with the multiplex assay or the corresponding type-specific assay. Concordance of the massively multiplex NPM1 assay (vertical axis) to type-specific assays (horizontal axis) is shown for NPM1 c.863_864insTATG (A), NPM1 c.865_866insCAGC (B), and NPM1 c.863_864insGCGG (C). The axes indicate detected NPM1mut/104ABL1 ratios. Lin's concordance correlation coefficient (ρc) is indicated.
Agreement for NPM1 c.863_864insTATG on digital PCR platforms using multiplex assay. The horizontal axis indicates NPM1 mutation/104ABL1 ratios obtained using the BioRad QX200. The vertical axis indicates NPM1mut/104ABL1 ratios obtained using the RainDance RainDrops. Lin's concordance correlation coefficient is indicated (ρc).
Massively multiplex assay background in NPM1 mutation (NPM1mut)–negative samples. Representative scatter plot of digital PCR data in a set of NPM1mut-negative samples for multiplex assay (top row) or type A NPM1mut–specific assay (middle row) and universal assay (bottom row). The vertical axis indicates the signal intensity of ABL1-positive droplets, and the horizontal axis indicates NPM1mut-positive droplets. NPM1mut/104ABL1 ratios are indicated in the upper right corner.
The massively multiplex NPM1 mutation (NPM1mut) PCR produces a single product. NPM1 was quantified by real-time quantitative PCR in a panel of wild-type (GM12878, MV411, and synthetic wild-type NPM1 cDNA) and NPM1mut samples (OCI-AML3 and synthetic pool of transcripts NPM1mut c863_864insNNNN). DNA gel electrophoresis demonstrating specificity of PCR product for multiplex (lanes 2, 5, 8, 11, 14, 17, and 20), type A NPM1 (lanes 3, 6, 9, 12, 18, and 21), and universal (lanes 1, 4, 7, 10, 13, 16, and 19) assays. Amplicon size is 61 bp for multiplex and type A NPM1mut assays and 140 bp for universal assay. No template control (NTC) is shown. L, DNA ladder.
References
- 1.Grimwade D., Jovanovic J.V., Hills R.K., Nugent E.A., Patel Y., Flora R., Diverio D., Jones K., Aslett H., Batson E., Rennie K., Angell R., Clark R.E., Solomon E., Lo-Coco F., Wheatley K., Burnett A.K. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol. 2009;27:3650–3658. doi: 10.1200/JCO.2008.20.1533. [DOI] [PubMed] [Google Scholar]
- 2.Hourigan C.S., Karp J.E. Minimal residual disease in acute myeloid leukaemia. Nat Rev Clin Oncol. 2013;10:460–471. doi: 10.1038/nrclinonc.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perea G., Lasa A., Aventín A., Domingo A., Villamor N., Queipo de Llano M.P., Llorente A., Juncà J., Palacios C., Fernández C., Gallart M., Font L., Tormo M., Florensa L., Bargay J., Martí J.M., Vivancos P., Torres P., Berlanga J.J., Badell I., Brunet S., Sierra J., Nomdedéu J.F. Prognostic value of minimal residual disease (MRD) in acute myeloid leukemia (AML) with favorable cytogenetics [t(8;21) and inv(16)] Leukemia. 2006;20:87–94. doi: 10.1038/sj.leu.2404015. [DOI] [PubMed] [Google Scholar]
- 4.Paietta E. Should minimal residual disease guide therapy in AML? Best Pract Res Clin Haematol. 2015;28:98–105. doi: 10.1016/j.beha.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Vidriales M.-B., Pérez-López E., Pegenaute C., Castellanos M., Pérez J.-J., Chandía M., Díaz-Mediavilla J., Rayón C., de las Heras N., Fernández-Abellán P., Cabezudo M., de Coca A.G., Alonso J.M., Olivier C., Hernández-Rivas J.M., Montesinos P., Fernández R., García- Suárez J., García M., Sayas M.-J., Paiva B., González M., Orfao A., San Miguel J.F. Minimal residual disease evaluation by flow cytometry is a complementary tool to cytogenetics for treatment decisions in acute myeloid leukaemia. Leuk Res. 2016;40:1–9. doi: 10.1016/j.leukres.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Kern W., Bacher U., Haferlach C., Schnittger S., Haferlach T. The role of multiparameter flow cytometry for disease monitoring in AML. Best Pract Res Clin Haematol. 2010;23:379–390. doi: 10.1016/j.beha.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Herzenberg L.A., Tung J., Moore W.A., Herzenberg L.A., Parks D.R. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 8.van der Velden V.H.J., Hochhaus A., Cazzaniga G., Szczepanski T., Gabert J., van Dongen J.J.M. Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia. 2003;17:1013–1034. doi: 10.1038/sj.leu.2402922. [DOI] [PubMed] [Google Scholar]
- 9.Verhaak R.G.W., Goudswaard C.S., Van Putten W., Bijl M.A., Sanders M.A., Hugens W., Uitterlinden A.G., Erpelinck C.A.J., Delwel R., Lowenberg B., Valk P.J.M. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 10.Ivey A., Hills R.K., Simpson M.A., Jovanovic J.V., Gilkes A., Grech A., Patel Y., Bhudia N., Farah H., Mason J., Wall K., Akiki S., Griffiths M., Solomon E., McCaughan F., Linch D.C., Gale R.E., Vyas P., Freeman S.D., Russell N., Burnett A.K., Grimwade D. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374:422–433. doi: 10.1056/NEJMoa1507471. [DOI] [PubMed] [Google Scholar]
- 11.Gorello P., Cazzaniga G., Alberti F., Dell'Oro M.G., Gottardi E., Specchia G., Roti G., Rosati R., Martelli M.F., Diverio D., Lo Coco F., Biondi A., Saglio G., Mecucci C., Falini B. Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia. 2006;20:1103–1108. doi: 10.1038/sj.leu.2404149. [DOI] [PubMed] [Google Scholar]
- 12.Cherian J., Nacro K., Poh Z.Y., Guo S., Jeyaraj D.A., Wong Y.X., Ho M., Yang H.Y., Joy J.K., Kwek Z.P., Liu B., Wee J.L.K., Ong E.H., Choong M.L., Poulsen A., Lee M.A., Pendharkar V., Ding L.J., Manoharan V., Chew Y.S., Sangthongpitag K., Lim S., Ong S.T., Hill J., Keller T.H. Structure-activity relationship studies of mitogen activated protein kinase interacting kinase (MNK) 1 & 2 and BCR-ABL1 inhibitors targeting chronic myeloid leukemic cells. J Med Chem. 2016;59:3063–3078. doi: 10.1021/acs.jmedchem.5b01712. [DOI] [PubMed] [Google Scholar]
- 13.Krönke J., Schlenk R.F., Jensen K.-O., Tschürtz F., Corbacioglu A., Gaidzik V.I., Paschka P., Onken S., Eiwen K., Habdank M., Späth D., Lübbert M., Wattad M., Kindler T., Salih H.R., Held G., Nachbaur D., von Lilienfeld-Toal M., Germing U., Haase D., Mergenthaler H.-G., Krauter J., Ganser A., Göhring G., Schlegelberger B., Döhner H., Döhner K. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: a study from the German-Austrian acute myeloid leukemia study group. J Clin Oncol. 2011;29:2709–2716. doi: 10.1200/JCO.2011.35.0371. [DOI] [PubMed] [Google Scholar]
- 14.Schnittger S., Schoch C., Kern W., Mecucci C., Tschulik C., Martelli M.F., Haferlach T., Hiddemann W., Falini B. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 15.Thiede C., Koch S., Creutzig E., Steudel C., Illmer T., Schaich M., Ehninger G. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 16.Andersen M.T., Andersen M.K., Christiansen D.H., Pedersen-Bjergaard J. NPM1 mutations in therapy-related acute myeloid leukemia with uncharacteristic features. Leukemia. 2008;22:951–955. doi: 10.1038/leu.2008.17. [DOI] [PubMed] [Google Scholar]
- 17.Falini B., Nicoletti I., Bolli N., Martelli M.P., Liso A., Gorello P., Mandelli F., Mecucci C., Martelli M.F. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica. 2007;92:519–532. doi: 10.3324/haematol.11007. [DOI] [PubMed] [Google Scholar]
- 18.Bacher U., Dicker F., Haferlach C., Alpermann T., Rose D., Kern W., Haferlach T., Schnittger S. Quantification of rare NPM1 mutation subtypes by digital PCR. Br J Haematol. 2014;167:710–714. doi: 10.1111/bjh.13038. [DOI] [PubMed] [Google Scholar]
- 19.Szankasi P., Jama M., Bahler D.W. A new DNA-based test for detection of nucleophosmin exon 12 mutations by capillary electrophoresis. J Mol Diagn. 2008;10:236–241. doi: 10.2353/jmoldx.2008.070167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmisano M., Grafone T., Ottaviani E., Testoni N., Baccarani M., Martinelli G. NPM1 mutations are more stable than FLT3 mutations during the course of disease in patients with acute myeloid leukemia. Haematologica. 2007;92:1268–1269. doi: 10.3324/haematol.11202. [DOI] [PubMed] [Google Scholar]
- 21.Kristensen T., Møller M.B., Friis L., Bergmann O.J., Preiss B. NPM1 mutation is a stable marker for minimal residual disease monitoring in acute myeloid leukaemia patients with increased sensitivity compared to WT1 expression. Eur J Haematol. 2011;87:400–408. doi: 10.1111/j.1600-0609.2011.01673.x. [DOI] [PubMed] [Google Scholar]
- 22.Meloni G., Mancini M., Gianfelici V., Martelli M.P., Foa R., Falini B. Late relapse of acute myeloid leukemia with mutated NPM1 after eight years: evidence of NPM1 mutation stability. Haematologica. 2009;94:298–300. doi: 10.3324/haematol.2008.000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salipante S.J., Fromm J.R., Shendure J., Wood B.L., Wu D. Detection of minimal residual disease in NPM1-mutated acute myeloid leukemia by next-generation sequencing. Mod Pathol. 2014;27:1438–1446. doi: 10.1038/modpathol.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webersinke G., Kranewitter W., Deutschbauer S., Zach O., Hasenschwandtner S., Wiesinger K., Erdel M., Marschon R., Böhm A., Tschurtschenthaler G. Switch of the mutation type of the NPM1 gene in acute myeloid leukemia (AML): relapse or secondary AML? Blood Cancer J. 2014;4:e221. doi: 10.1038/bcj.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malmberg E.B.R., Ståhlman S., Rehammar A., Samuelsson T., Alm S.J., Kristiansson E., Abrahamsson J., Garelius H., Pettersson L., Ehinger M., Palmqvist L., Fogelstrand L. Patient-tailored analysis of minimal residual disease in acute myeloid leukemia using next-generation sequencing. Eur J Haematol. 2017;98:26–37. doi: 10.1111/ejh.12780. [DOI] [PubMed] [Google Scholar]
- 26.Gabert J., Beillard E., van der Velden V.H.J., Bi W., Grimwade D., Pallisgaard N., Barbany G., Cazzaniga G., Cayuela J.M., Cavé H., Pane F., Aerts J.L.E., De Micheli D., Thirion X., Pradel V., González M., Viehmann S., Malec M., Saglio G., van Dongen J.J.M. Standardization and quality control studies of “real-time” quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia – A Europe Against Cancer Program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 27.Gerrard G., Mudge K., Foskett P., Stevens D., Alikian M., White H.E., Cross N.C.P., Apperley J., Foroni L. Fast-mode duplex qPCR for BCR-ABL1 molecular monitoring: innovation, automation, and harmonization. Am J Hematol. 2012;87:717–720. doi: 10.1002/ajh.23212. [DOI] [PubMed] [Google Scholar]
- 28.Shrivastava A., Gupta V., Article R. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron Young Sci. 2011;2:21–25. [Google Scholar]
- 29.Analytical Methods Committee Recommendations for the definition, estimation and use of the detection limit. Analyst. 1987;112:199–204. [Google Scholar]
- 30.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faust G.G., Hall I.M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. Genomics. 2012;1207:9. [Google Scholar]
- 33.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Genomics. 2013;1303:3. [Google Scholar]
- 34.Lai Z., Markovets A., Ahdesmaki M., Chapman B., Hofmann O., McEwen R., Johnson J., Dougherty B., Barrett J.C., Dry J.R. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watkins N.E., SantaLucia J. Nearest-neighbor thermodynamics of deoxyinosine pairs in DNA duplexes. Nucleic Acids Res. 2005;33:6258–6267. doi: 10.1093/nar/gki918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loakes D., Brown D.M., Linde S., Hill F. 3-Nitropyrrole and 5-nitroindole as universal bases in primers for DNA sequencing and PCR. Nucleic Acids Res. 1995;23:2361–2366. doi: 10.1093/nar/23.13.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin L.I.-K. Assay validation using the concordance correlation coefficient. Biometrics. 1992;48:599–604. [Google Scholar]
- 38.Shayegi N., Kramer M., Bornhäuser M., Schaich M., Schetelig J., Platzbecker U., Röllig C., Heiderich C., Landt O., Ehninger G., Thiede C. The level of residual disease based on mutant NPM1 is an independent prognostic factor for relapse and survival in AML. Blood. 2013;122:83–92. doi: 10.1182/blood-2012-10-461749. [DOI] [PubMed] [Google Scholar]
- 39.Kern W., Haferlach C., Haferlach T., Schnittger S. Monitoring of minimal residual disease in acute myeloid leukemia. Cancer. 2008;112:4–16. doi: 10.1002/cncr.23128. [DOI] [PubMed] [Google Scholar]
- 40.Walter R.B., Gyurkocza B., Storer B.E., Godwin C.D., Pagel J.M., Buckley S.A., Sorror M.L., Wood B.L., Storb R., Appelbaum F.R., Sandmaier B.M. Comparison of minimal residual disease as outcome predictor for AML patients in first complete remission undergoing myeloablative or nonmyeloablative allogeneic hematopoietic cell transplantation. Leukemia. 2015;29:137–144. doi: 10.1038/leu.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Performance of multiplex NPM1 minimal residual disease assay on spike-in dilution series on alternative digital PCR (dPCR) platform and with higher cDNA input. A: Concordance of real-time quantitative PCR (RQ-PCR) (qPCR)–based type A–specific assay (vertical axis) to dPCR–based adaptation of the standard type A–specific assay on the BioRad QX200 (horizontal axis) when using cDNA derived from either undiluted OCI-AML3 or OCI-AML3 spiked into GM12878 at 1:1000, 1:10,000, or 1:50,000 cells or GM12878 alone. Axes indicate NPM1 mutations (NPM1muts) per 104ABL1 (NPM1mut/104ABL1). Lin's concordance correlation coefficient is indicated (ρc). B: Concordance of the massively multiplex NPM1 assay on RainDance RainDrops (vertical axis) to dPCR–based adaptation of the type A–specific assay on BioRad QX200 (horizontal axis) on the same dilution series as A. Axes indicate NPM1mut/104ABL1. Lin's concordance correlation coefficient is indicated (ρc). C: Scatterplot of dPCR data for the multiplex assay (top row) and type A–specific assay adapted to dPCR (bottom row) on the same spike-in dilution series as A and B but with 10× as much input to ascertain improved sensitivity. Dilutions are indicated. The vertical axis indicates the signal intensity of droplets in the ABL1 channel (VIC) and the horizontal axis indicates positive droplets in the NPM1mut channel (FAM). NPM1mut/104ABL1 copies are indicated in the upper right hand corner of each scatterplot. BioRad QX200 could not be tested in C because the number of ABL1 copies saturated the instruments so that it was no longer quantitative with respect to ABL1.
The type-specific and massively multiplex assays agree in rare NPM1 mutations observed in patients. cDNA from NPM1-mutated acute myeloid leukemia was diluted into cDNA from healthy cord blood at 1:1000 and 1:10,000 (w/w) alongside cord blood controls. Detection was performed with the multiplex assay or the corresponding type-specific assay. Concordance of the massively multiplex NPM1 assay (vertical axis) to type-specific assays (horizontal axis) is shown for NPM1 c.863_864insTATG (A), NPM1 c.865_866insCAGC (B), and NPM1 c.863_864insGCGG (C). The axes indicate detected NPM1mut/104ABL1 ratios. Lin's concordance correlation coefficient (ρc) is indicated.
Agreement for NPM1 c.863_864insTATG on digital PCR platforms using multiplex assay. The horizontal axis indicates NPM1 mutation/104ABL1 ratios obtained using the BioRad QX200. The vertical axis indicates NPM1mut/104ABL1 ratios obtained using the RainDance RainDrops. Lin's concordance correlation coefficient is indicated (ρc).
Massively multiplex assay background in NPM1 mutation (NPM1mut)–negative samples. Representative scatter plot of digital PCR data in a set of NPM1mut-negative samples for multiplex assay (top row) or type A NPM1mut–specific assay (middle row) and universal assay (bottom row). The vertical axis indicates the signal intensity of ABL1-positive droplets, and the horizontal axis indicates NPM1mut-positive droplets. NPM1mut/104ABL1 ratios are indicated in the upper right corner.
The massively multiplex NPM1 mutation (NPM1mut) PCR produces a single product. NPM1 was quantified by real-time quantitative PCR in a panel of wild-type (GM12878, MV411, and synthetic wild-type NPM1 cDNA) and NPM1mut samples (OCI-AML3 and synthetic pool of transcripts NPM1mut c863_864insNNNN). DNA gel electrophoresis demonstrating specificity of PCR product for multiplex (lanes 2, 5, 8, 11, 14, 17, and 20), type A NPM1 (lanes 3, 6, 9, 12, 18, and 21), and universal (lanes 1, 4, 7, 10, 13, 16, and 19) assays. Amplicon size is 61 bp for multiplex and type A NPM1mut assays and 140 bp for universal assay. No template control (NTC) is shown. L, DNA ladder.