Abstract
Invasion is a hallmark of advanced head and neck squamous cell carcinoma (HNSCC). We previously determined that low relative miR-375 expression was associated with poor patient prognosis. HNSCC cells with increased miR-375 expression have lower invasive properties and impaired invadopodium activity. Using stable isotope labeling with amino acids in cell culture and reverse-phase liquid chromatography mass spectrometry, we assessed the impact of miR-375 expression on protein levels in UM-SCC-1 cells. Increased miR-375 expression was associated with down-regulation of proteins involved in cellular assembly and organization, death and survival, and movement. Two invasion-associated proteins, vimentin and L-plastin, were strongly down-regulated by miR-375. Luciferase reporter assays demonstrated that high miR-375 expression reduced vimentin promoter activity, suggesting that vimentin is an indirect target of miR-375. Runt-related transcription factor 1 (RUNX1) is a potential miR-375 direct target, and its knockdown reduced vimentin and L-plastin expression. Data in The Cancer Genome Atlas HNSCC database showed a significant inverse correlation between miR-375 expression and RUNX1, vimentin, and L-plastin RNA expression. These clinical correlations validate our in vitro model findings and support a mechanism in which miR-375 suppresses RUNX1 levels, resulting in reduced vimentin and L-plastin expression. Furthermore, knockdown of RUNX1, L-plastin, and vimentin resulted in significant reductions in cell invasion in vitro, indicating the functional significance of miR-375 regulation of specific proteins involved in HNSCC invasion.
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide.1, 2 HNSCC develops in the mucosal layer of the upper aerodigestive tract, arising in the squamous epithelium of the oral cavity, oropharynx, larynx, hypopharynx, and nasopharynx.3, 4 Despite advances in multimodal treatment strategies, including surgery, radiotherapy, and chemotherapy, locoregional recurrence, lymph node metastases, and second primary tumors continue to contribute significantly to morbidity and mortality among HNSCC patients.5, 6
miRNAs are small noncoding single-stranded RNA molecules (approximately 22 nucleotides in length), which are post-transcriptional gene expression regulators.7, 8, 9 Mature miRNAs usually bind to the 3′-untranslated region of the target mRNAs, leading to either mRNA degradation or translational repression.8, 9, 10 Each miRNA targets hundreds of mRNA transcripts, and each mRNA transcript may possess multiple different miRNA binding sites.11, 12 The ability of miRNAs to target diverse mRNAs accounts for their ability to control many cancer phenotypes, including cellular proliferation, differentiation, apoptosis, invasion, metastasis, and angiogenesis.9, 13, 14, 15 Some miRNAs, including miR-375, have been identified as diagnostic and prognostic markers in human cancers.9, 14, 16, 17 We previously determined that low relative miR-375 expression in HNSCC tumors is associated with poor patient prognosis.17 We also identified that when the level of miR-375 is stably increased in HNSCC cells, these cells exhibit impaired cell invasion and invadopodial function in vitro.17, 18
miRNAs have the ability to regulate their mRNA targets by translational repression without necessarily causing mRNA degradation/cleavage.9, 19 Stable isotope labeling of amino acids in cell culture (SILAC)–based proteomic analysis has emerged as a useful, unbiased method for the identification of miRNA-regulated proteins.11, 20, 21, 22, 23, 24 The incorporation of nonradioactive, stable isotopes of amino acids (ie, lysine and arginine) into proteins22, 25 in combination with quantitative mass spectrometry allows for the detection of differences in protein levels between biological samples.20, 26
This study focuses on the identification of miR-375–regulated proteins using a SILAC-based proteomic strategy. We quantitated 2188 proteins in four subcellular fractions, subsets of which were down-regulated or up-regulated in UM-SCC-1 cells with increased miR-375 expression. Among the down-regulated proteins were two previously published miR-375 targets, metadherin and lactate dehydrogenase B.8 Vimentin and L-plastin emerged as the most down-regulated proteins. As vimentin and L-plastin are not predicted to be direct targets of miR-375, we went on to identify runt-related transcription factor 1 (RUNX1) as a potential target of miR-375 that regulates vimentin and L-plastin expression. These results are supported by negative correlations between miR-375 and RUNX1, vimentin, and L-plastin mRNA levels in the HNSCC database from The Cancer Genome Atlas (TCGA).
Materials and Methods
Cell Culture and SILAC Labeling of UM-SCC-1
The generation of stable UM-SCC-1 empty vector control (Control) and precursor miR-375–expressing (Pre375) lentiviral transductant cell lines was previously described.17, 18 For the SILAC labeling, the SILAC Protein Identification and Quantitation Media Kit (catalog number MS10030; Invitrogen, Waltham, MA) was used. UM-SCC-1 transductant lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% dialyzed fetal bovine serum, penicillin-streptomycin (100 U/mL penicillin, 100 μg/mL streptomycin) (catalog number 15140-122; Gibco/Life Technologies, Waltham, MA), and 2 mmol/L l-glutamine (catalog number 25030-081; Gibco/Life Technologies). The heavy medium was supplemented with 13C6-lysine and 13C6, 15N4-arginine. The light medium was supplemented with 12C6-lysine and 12C6, 14N4-arginine. To prevent the conversion of heavy arginine to heavy proline, 200 mg/mL l-proline was also added to the heavy SILAC growth media.27 The UM-SCC-1 Control and Pre375 cells were grown for 7 days in the heavy and light SILAC growth medium, respectively. Cell lines were maintained in a 37°C incubator with 5% CO2.
Subcellular Fractionation for Proteomic Analysis
A total of 5 × 106 cells each of the light-grown and heavy-grown UM-SCC-1 cells were combined, and subcellular fractionation was performed to improve the dynamic range of detection. Subcellular fractionation of the combined cells was conducted with the QProteome Cell Compartment kit (catalog number 37502; Qiagen, Germantown, MD), according to the manufacturer's suggested protocol. In brief, the combined cells were centrifuged at 500 × g for 10 minutes at 4°C. The combined cells were washed twice with 2 mL ice-cold phosphate-buffered saline and centrifuged after each wash. The pellet was resuspended in 1 mL ice-cold Extraction Buffer CE1 (containing 1× protease inhibitor) and incubated for 10 minutes at 4°C on an end-over-end shaker. The lysate was centrifuged at 1000 × g for 10 minutes at 4°C. The supernatant was designated the cytosolic fraction. The pellet was resuspended in 1 mL ice-cold Extraction Buffer CE2 (containing 1× protease inhibitor) and incubated for 30 minutes at 4°C on an end-over-end shaker. The suspension was centrifuged at 6000 × g for 10 minutes at 4°C. The supernatant was designated the membrane fraction. To the pellet, 7 μL of benzonase nuclease and 13 μL of distilled water were added and resuspended by gently flicking the bottom of the tube. The suspension was incubated for 15 minutes at room temperature. Ice-cold Extraction Buffer CE3 (500 μL; containing 1× protease inhibitor) was added into the tube and incubated for 10 minutes at 4°C on an end-over-end shaker. The suspension was centrifuged at 6800 × g for 10 minutes at 4°C. The supernatant was designated the nuclear fraction. The pellet was resuspended in 500 μL of Extraction Buffer CE4 and designated the cytoskeletal fraction.
To concentrate the subcellular fractions, the protein concentrations of the fractions were quantitated with the Pierce BCA Protein Assay (catalog number 23225; Thermo Scientific, Waltham, MA). To 70 μg of cytosol, nucleus, membrane, or cytoskeleton fractions, four times the volume of ice-cold acetone was added. After a 20-minute incubation on ice, each suspension was centrifuged at 12,000 × g for 10 minutes at 4°C. After removing the supernatant, each pellet was air dried and resuspended with 35 μL of Laemmli buffer containing 5 mmol/L of Tris (2-carboxyethyl) phosphine and incubated at 40°C for 30 minutes with 600 rpm agitation. The samples were placed in a prechilled TLA100 rotor and spun down at 70,000 × g for 30 minutes at 4°C in the Beckman TL-100 centrifuge and supernatants (60 μg in 30 μL) loaded on a 4% to 20% gradient gel with protein ladders. The gel was stained with GelCode Blue Stain Reagent (catalog number 24590; Thermo Scientific) for 30 minutes and destained overnight.
Mass Spectrometry Analysis
Complete lanes were excised and sliced to produce 24 bands sized 10 × 1 × 1 mm3 from each subcellular fraction. Band slices were placed in wells of 96-well plates and in-gel digested using an Ettan Digester (GE Healthcare, Chicago, IL). Gel pieces were reduced with dithiothreitol and alkylated with iodoacetamide, then digested with trypsin, as described previously.28 A nano Acquity ultra performance liquid chromatograph interfaced to an Orbitrap Velos mass spectrometer (Thermo Scientific) was used for analysis. Digested peptides were loaded onto a Symmetry C18 nanoACQUITY trap column (100 Å, 5 μm, 180 μm × 20 mm) at a flow rate of 8 μL/minute with 0.1% formic acid in water and delivered to an ACQUITY UPLC PST C18 nanoACQUITY Column (300 Å, 1.7 μm, 75 μm × 150 mm; Waters, Milford, MA). Peptides were eluted from the nanocolumn at a flow rate of 270 nL/minute with 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) gradient. The gradient used was as follows: 0 minute, 1% B; 1 minute, 2% B; 2 minutes, 5% B; 42 minutes, 42% B; 52 minutes, 65% B; 57 minutes, 70% B; 60 minutes, 80% B; 65 minutes, 85% B; 70 minutes, 85% B; 72 minutes, 1% B; 90 minutes, 1% B. The 20 most intense ions with charge states between 2 and 4 from an initial survey scan between 300 and 1650 m/z, were selected for tandem mass spectrometry (MS/MS). MS/MS was performed using an isolation width of 2 m/z, normalized collision energy of 35%, and a minimum signal intensity of 1000 counts. Dynamic exclusion was enabled, so once a certain ion was selected twice for MS/MS within 30 seconds, the ion was excluded from being selected again for MS/MS during the next 120 seconds.
Protein Identification
Once obtained, peak lists were generated from MS/MS spectra using Proteome Discoverer (version 1.3.0.339; Thermo Scientific) and searched against the IPI-Human database (version 3.73) concatenated with a reverse decoy, using Mascot (version 2.3; Matrix Science, Boston, MA). Fixed modification of cysteines to S-carbamidomethyl derivatives and variable ones of methionine oxidation, 13C6-lysine, and 13C615N4-arginine were defined for the database search. Mass tolerance was set to 5 ppm for precursor ions and 0.6 Da for fragment ions. Search results were exported as DAT files and grouped to protein matches using ProteoIQ (version 2.6.03; Nusep, Sydney, Australia). Protein hits were filtered to include only those that were identified with <1% false discovery rate and two peptides detection.
Protein Quantitation from MS Data
Raw files were converted to mzXML with Trans Proteomic Pipeline (version 4.4).29 Intensities of peptide peaks were extracted from mzXML using deisotoped accurate mass retention time pairs, which were replicated throughout the experiments. Windows of the mass tolerance and retention time were 5 ppm and 0 seconds each. Intensity measurements were matched to the redundant, tryptic peptides identified from each protein to determine an average, relative protein fold change using ProteoIQ. Nonproteotypic peptides with significantly different ratios from proteotypic peptides of the same protein were excluded from quantitation. Systematic bias was corrected using intensity normalization. Presence of an interfering precursor that overlaps with the distribution of interest was removed from quantitation using the correlation coefficient measured between the theoretical and experimental isotopic distributions for the quantitative precursors.
Proteomic Analysis
We screened all of the SILAC-identified proteins for those that had at least a 20% reduction or increase in protein levels in UM-SCC-1 Pre375 (high miR-375) compared to the UM-SCC-1 control (low miR-375). The lists of down-regulated and up-regulated proteins were compared to the theoretical direct target lists based on seed matches in miRWalk.30 miRWalk has a combined list of the putative target lists from 10 different prediction algorithms, including miRWalk,30 TargetScan,31 DIANA-mT,32 miRanda,33 RNA22,34 and RNAhybrid.35 We confirmed some of the down-regulated and up-regulated proteins by microarray analysis. We compared the average UM-SCC-1 Pre375/Control ratios of mRNA abundance in three independent replicates (the procedure for the microarray analysis is described below).
Ingenuity Pathway Analysis
The lists of SILAC-identified down-regulated and up-regulated proteins and the SILAC Pre375/Control log ratio values of the proteins were uploaded into Ingenuity Pathway Analysis (IPA; Qiagen, Redwood City, CA). For the data set, a Core Analysis was run in IPA. The top molecular and cellular functions that the down-regulated and up-regulated proteins were associated with were identified. The list of down-regulated and up-regulated proteins that were recognized in IPA analysis as associated with cellular movement was plugged to STRING version 10 (http://string-db.org, last accessed November 2015).36 The confidence view was selected to show the STRING network view of these dysregulated proteins.
Expression Analysis of RNA Samples
Total RNA from three biological replicates from the UM-SCC-1 transductant cells was isolated using TRIzol (catalog number 15596-026; Life Technologies), according to the manufacturer's protocol. The quality of the RNA samples was monitored using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). For each RNA sample (total of six samples), linear amplification and biotin labeling of 500 ng of total RNA were performed using the Illumina TotalPrep RNA Amplification Kit (Ambion, Waltham, MA), according to the suggested manufacturer's protocol. The amplified RNA was hybridized to Illumina HumanHT-12 v3 Expression BeadChips (Illumina, San Diego, CA). BeadChips were scanned using an Illumina BeadArray Reader and the Bead Scan software GenomeStudio version 2011.1 (Illumina). The raw data were quantile normalized and uploaded to ArrayExpress. Microarray data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress; accession number E-MTAB-5405). The average signal of each mRNA from the independent replicates of UM-SCC-1 Control and Pre375 cells was determined. Unpaired t-tests were calculated for each of the mRNAs comparing the signals from the three replicates of the UM-SCC-1 Control and Pre375 cells.
Western Blotting
Vimentin (catalog number sc-6260) and zinc finger E-box binding homeobox (ZEB) 2 (catalog number sc-271984) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). L-plastin (catalog number 5350), metadherin (catalog number 9596), RUNX1/AML1 (catalog number 4334), JunD (catalog number 5000), c-Jun (catalog number 9165), and phospho-c-Jun (Ser 63) (catalog number 9261) antibodies were purchased from Cell Signaling Technology (Danvers, MA). Lactate dehydrogenase B (catalog number 14824-1-AP) was purchased from ProteinTech (Rosemont, IL). B-tubulin (catalog number T4026) and B-actin (catalog number A5441) antibodies were purchased from Sigma-Aldrich (St. Louis, MO). IRDye 680RD anti-mouse (catalog number 926-68072) and IRDye 800CW anti-rabbit (catalog number 926-32213) secondary antibodies were purchased from LI-COR Biosciences (Lincoln, NE).
The UM-SCC-1 cell lysates were prepared in two ways: the cells were lysed in radioimmunoprecipitation assay buffer containing 1× protease inhibitors (catalog number 11836170-001; Roche, New York, NY). Cell lysates were sonicated for 15 seconds at 4°C. Quantitation of total protein concentration was performed with the Pierce BCA Protein Assay (catalog number 23225; Thermo Scientific). SDS-PAGE sample loading buffer was added to 40 μg of total protein and heated at 95°C for 5 minutes, and the cells were lysed in 2× sample buffer with 2× protease inhibitors. The cell lysates were diluted down to 1× and sheared with a 28-gauge syringe needle. The sheared lysates were heated at 95°C for 5 minutes.
The prepared lysates were loaded in polyacrylamide gels and run at 120 V for approximately 1.5 hours. Gels were transferred to nitrocellulose membranes at 100 V for 1 hour. Membranes were blocked in LI-COR Odyssey blocking buffer (catalog number 927-40000; LI-COR Biosciences) for 1 hour at room temperature and incubated with primary antibodies overnight at 4°C. The membranes were washed three times with 1× Tris-buffered saline–Tween and incubated with secondary antibodies (IRDye 680RD anti-mouse and IRDye 800CW anti-rabbit; 1:5000) for 1 hour at room temperature. The membranes were then washed again three times with 1× Tris-buffered saline–Tween and scanned using the LI-COR Odyssey Infrared Imaging System. The Western blot images were analyzed using ImageJ software version 1.50i (NIH, Bethesda, MD; http://imagej.nih.gov/ij).37, 38 β-Tubulin or β-actin was used as loading control. The mean intensities of the protein bands were measured, and the corresponding background mean intensities were subtracted. The background subtracted mean intensities of the protein bands of interest were normalized to the background subtracted mean intensity of the β-actin or β-tubulin band. The ratio of the value for the miR-375–overexpressing line to its respective control line was calculated. In the case of the siRNA knockdown experiments, the ratio of the value for the targeted siRNA–treated cells to the nontargeting control siRNA cells was determined.
RT-qPCR
The previously described protocol for reverse transcription and quantitative real-time RT-PCRs (RT-qPCRs) for miRNA quantitation in the HNSCC transductant lines17 was used to determine miR-375 expression in HNSCC patient tumor and normal samples using RNU48 as a loading control.
For quantitating mRNA, total RNA was extracted using the Qiagen RNeasy Mini Kit (catalog number 74104) following the manufacturer's protocol. Two hundred nanograms of RNA from each cell line was analyzed using the TaqMan RNA to Ct One Step Kit (catalog number 4392938) from Life Technologies, using their recommended parameters. TaqMan Gene Expression assays used were Hs00185584_m1 (vimentin), Hs00158701_m1 (L-plastin), Hs00757841_m1 (metadherin), Hs00929956_m1 (lactate dehydrogenase B), Hs01021970_m1 (RUNX1), Hs00231692_m1 (RUNX2), Hs00232783_m1 (ZEB1), Hs00207691_m1 (ZEB2), Hs00195591_m1 (zinc finger protein SNAI1), Hs00950344_m1 (zinc finger protein SNAI2), Hs00916521_m1 (transcription factor Sp1), and Hs99999905_m1 (glyceraldehyde-3-phosphate dehydrogenase). For each cell line, the mean CT value was calculated from three technical replicates, and glyceraldehyde-3-phosphate dehydrogenase was used for normalization with fold changes calculated as 2−ΔΔCT.
Luciferase Reporter Assays
The vimentin promoter firefly luciferase reporter construct was graciously provided by Dr. Christine Gilles (Liège University, Liège, Wallonia, Belgium). The activator protein 1 (AP-1) firefly luciferase reporter constructs were graciously provided by Dr. Hong Wan (Queen Mary University, London, UK). The UM-SCC-1 transductant cells were plated in duplicate for each transfection experiment. Vimentin promoter and AP-1 luciferase reporter construct (1980 ng) were cotransfected with 20 ng of Renilla luciferase construct using Fugene HD. Twenty-four hours after transfection, the luciferase activity was measured using the Dual Luciferase Reporter Assay, according to the manufacturer's instructions (catalog number E1910; Promega, Madison, WI). Luciferase signals were measured with a FluoStar Optima. Firefly luciferase activity was normalized to the Renilla luciferase activity. Means and SEM of three independent experiments were calculated.
siRNA Transfection
UM-SCC-1 cells were plated at a concentration of 50,000 cells in 6-well plates the day before the transfection. siGENOME SMARTpool siRNAs (50 nmol/L; Dharmacon, Lafayette, CO) specifically targeting vimentin (catalog number M-003551-02-0005), L-plastin (catalog number M-011716-00-0005), RUNX1 (catalog number M-003926-02-0005), and nontargeting siRNA pools (catalog number D-001206-14-05) were transfected into UM-SCC-1 cells with Dharmafect 4 transfection reagent (1 μL per well; catalog number T-2004-01). The siRNA transfectant cells were harvested 72 hours after transfection for RT-qPCR, Western blot analyses, and in vitro invasion assays.
In Vitro Invasion
For the in vitro invasion assessments, Matrigel Invasion Chambers (catalog number 354480; BD Biosciences) were used. Before adding the cells to the assays, the invasion chambers were rehydrated for 2 hours in Dulbecco's modified Eagle's medium. Cells were detached using Accutase and resuspended in Dulbecco's modified Eagle's medium + 0.7% bovine serum albumin (catalog number BP1605-100; Fisher Scientific, Waltham, MA) at a density of 2 × 105 cells/mL. The rehydrated invasion chambers were submerged in wells filled with 0.750 mL 0.1 nmol/L epidermal growth factor in Dulbecco's modified Eagle's medium + 0.7% bovine serum albumin in a 24-well plate. A total of 1 × 105 cells in 0.5 mL Dulbecco's modified Eagle's medium + 0.7% bovine serum albumin were added to the inside of each chamber. The chambers were incubated for 24 hours at 37°C in a humidified incubator with 5% CO2. After 24 hours, the chambers were fixed in 10% phosphate-buffered formalin for 15 minutes and stained with 0.2% crystal violet for 10 minutes. The cells that had not invaded were removed by scraping the upper surface of the membrane. An Epson 11000XL Scanner was used to image the cells that had invaded the lower surface. The percentage of the filter covered with cells that invaded was calculated using ImageJ software.37, 38
Statistical Analysis
With the exception of the protein analysis using SILAC and mass spectrometry, all of the results shown are based on at least three independent experiments. Statistical analyses were assessed using two-tailed unpaired t tests with Welch corrections in Graphpad Prism software version 6 (GraphPad Software Inc., La Jolla, CA) or z tests in Microsoft Excel 2010 (Microsoft, Redmond, WA). The P values are shown as stars based on the degree of significance. Data are shown as the means ± SEM from at least three independent experiments.
For analysis of TCGA data, the following HNSCC data files were downloaded from the Firehose website (https://gdac.broadinstitute.org,39 last accessed January 28, 2016: gdac.broadinstitute.org_HNSC.Merge_mirnaseq__illuminahiseq_mirnaseq__bcgsc_ca__Level_3__miR_gene_expression__data.Level_3.2016012800.0.0.tar and gdac.broadinstitute.org_HNSC.Merge_rnaseqv2__illuminahiseq_rnaseqv2__unc_edu__Level_3__RSEM_genes_normalized__data.Level_3.2016012800.0.0.tar). Data were used from primary tumors that had both miRNA and mRNA quantitation. The data were log-normalized and then Pearson correlations calculated using SPSS (IBM, Armonk, NY), with two-tailed significance tests.
Results
miR-375–Regulated Proteins Identified by SILAC and MS
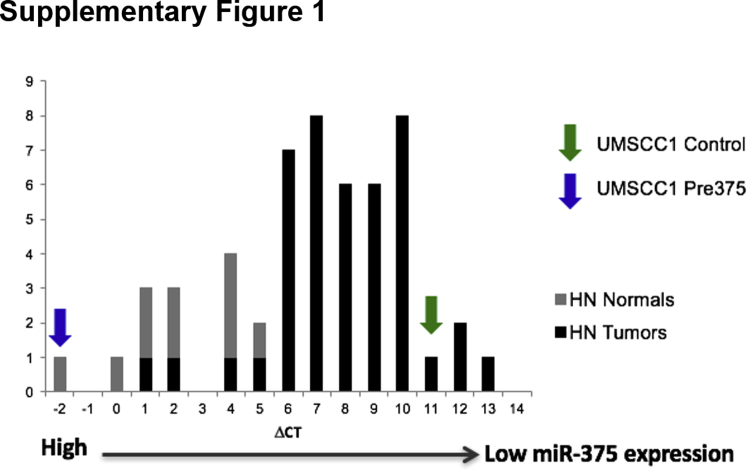
We previously described the generation of stable transductants17, 18 of the oral cavity UM-SCC-1 HNSCC cell line40, 41 with increased miR-375 expression. The empty vector control cells (Control) exhibit low levels of miR-375, consistent with primary HNSCC patient tumors. The Pre375-expressing cells exhibit high levels of mature miR-375, at levels similar to what is seen in normal tissue neighboring HNSCC tumors (Supplemental Figure S1).
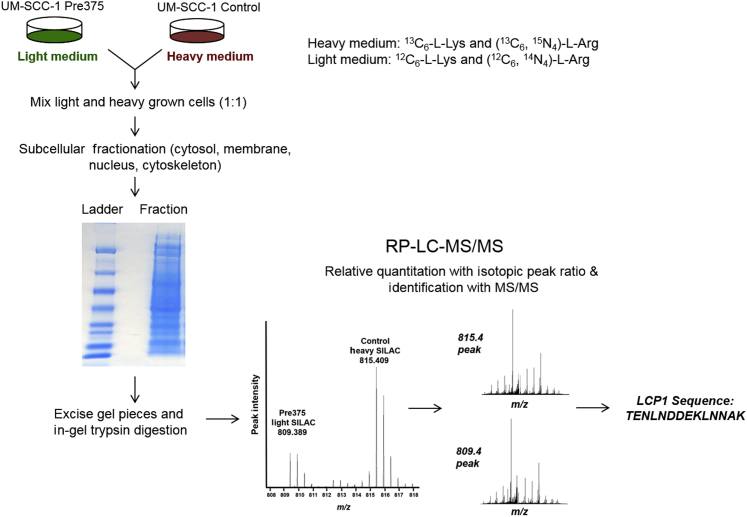
We conducted quantitative proteomic comparisons of the Control and Pre375 lines using SILAC to identify differentially expressed miR-375–regulated proteins. The intensity peak ratios between heavy- and light-labeled peptides were used to indicate the changes in the amount of protein as a result of miR-375 expression (Figure 1). We identified a total of 19,179 peptides in the four subcellular fractions, which corresponded to 2692 proteins (Table 1). There were 14,948 peptides used for quantitation, which corresponded to 2188 proteins (Table 1 and Supplemental Tables S1, S2, S3, and S4). The protein lists from the different subcellular fractions used for quantitation were filtered for P value in ProteoIQ; the P value is determined as an approximation of how likely the assigned protein is correct, which relies on the probabilities that peptides for the identified protein have been correctly assigned. Of the proteins used for quantitation, there were 761 in the four subcellular fractions with significant quantitative precision (P ≤ 0.05) (Table 1 and Supplemental Tables S5, S6, S7, and S8). Using a 20% reduction in the Pre375/Control ratio as the cutoff, 125 proteins were down-regulated (Table 1 and Supplemental Table S9). In contrast, 121 proteins showed at least a 20% increase in the Pre375 cells compared to the Control cells (Table 1 and Supplemental Table S10).
Figure 1.
Flow diagram of the SILAC MS/MS experiment. The UM-SCC-1 control and Pre375 cells were grown in medium supplemented with heavy (13C6-lysine and 13C6, 15N4-arginine) or light (12C6-lysine and 12C6, 14N4-arginine) amino acids, respectively. Equal amounts of heavy grown control cells and light grown Pre375 cells were combined and subcellular fractionation was conducted. The fractions were run on SDS-PAGE. The gel pieces were excised and subjected to in-gel tryptic digestion. The tryptic peptides from digested bands were analyzed by mass spectrometry. Alternating MS (left) and tandem MS (MS/MS; right) provided the intensity peak ratios between heavy- and light-labeled peptides and the identity of peptides acquired (data for a peptide from L-plastin are shown). LC, liquid chromatography; MS, mass spectrometry; RP, reverse phase.
Table 1.
Number of Peptides and Proteins Identified, Quantitated, and Differentially Expressed from the SILAC MS/MS Analysis
| Cell fraction | Peptides identified | Proteins identified | Peptides used for quantitation | Proteins used for quantitation | Peptides used for quantitation (P < 0.05)∗ | Proteins used for quantitation (P < 0.05)∗ | Proteins down-regulated at least 20% | Proteins up-regulated at least 20% |
|---|---|---|---|---|---|---|---|---|
| Cytosol | 6489 | 842 | 6025 | 797 | 2384 | 223 | 36 | 31 |
| Nucleus | 5210 | 818 | 3217 | 564 | 1607 | 218 | 58 | 16 |
| Membrane | 3983 | 609 | 2478 | 430 | 960 | 124 | 9 | 11 |
| Cytoskeleton | 3497 | 423 | 3228 | 397 | 2085 | 196 | 22 | 63 |
| Total | 19,179 | 2692 | 14,948 | 2188 | 7036 | 761 | 125 | 121 |
MS/MS, tandem mass spectrometry; SILAC, stable isotope labeling of amino acids in cell culture.
Differentially regulated proteins were selected only from proteins with significant quantitative precision (P ≤ 0.05). P values were calculated using peptide ratios using t-test.
We evaluated the list of altered proteins by the comparative analysis search program in miRWalk30 using a database that has a compiled list of putative miRNA targets from 10 different prediction algorithms, including miRWalk,30 TargetScan,31 DIANA-mT,32 miRanda,33 RNA22,34 and RNAhybrid.35 Thirty-three of the down-regulated proteins and 22 of the up-regulated proteins are predicted by at least one algorithm to be theoretical miR-375 targets based on seed matches between miR-375 and the mRNA encoding each protein (Supplemental Tables S9 and S10).
Because miRNAs can also regulate gene expression through mRNA degradation, we compared the gene expression profiles of the UM-SCC-1 transductant cells using Illumina HumanHT-12 v3 Expression BeadChip arrays with the SILAC results. Thirty-two of the down-regulated proteins had at least a 20% reduction in RNA levels (Supplemental Table S9). Of the down-regulated proteins that showed reduced RNA levels, 12 were potential miR-375 targets. Comparing the RNA expression data to the list of the SILAC-identified up-regulated proteins revealed that 28 up-regulated proteins also had increased RNA expression in the microarray analysis (Supplemental Table S10). Of the up-regulated proteins that showed increased RNA expression, seven of them were potential miR-375 targets.
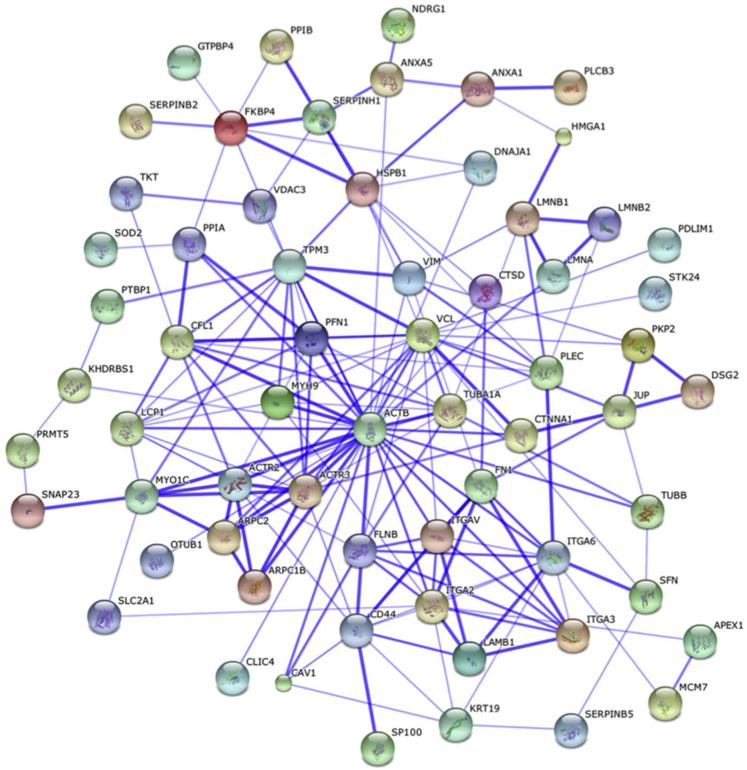
To identify the cellular functions affected by miR-375 expression, an IPA was performed with the combined list of the down-regulated and up-regulated proteins. The core analysis in IPA identified that the top five cellular functions affected by miR-375 expression were as follows: i) cellular death and survival, ii) cellular growth and proliferation, iii) cellular movement, iv) cellular assembly and organization, and v) cellular function and maintenance (Supplemental Table S11). The interactions of the down-regulated and up-regulated proteins associated with cellular movement are shown in Figure 2.
Figure 2.
Interactions of dysregulated proteins recognized in Ingenuity Pathway Analysis as associated with cellular movement. STRING10 was used to show the known and predicted protein-protein interactions of these differentially regulated proteins. The thickness of the lines between the proteins indicates the strength of the association.
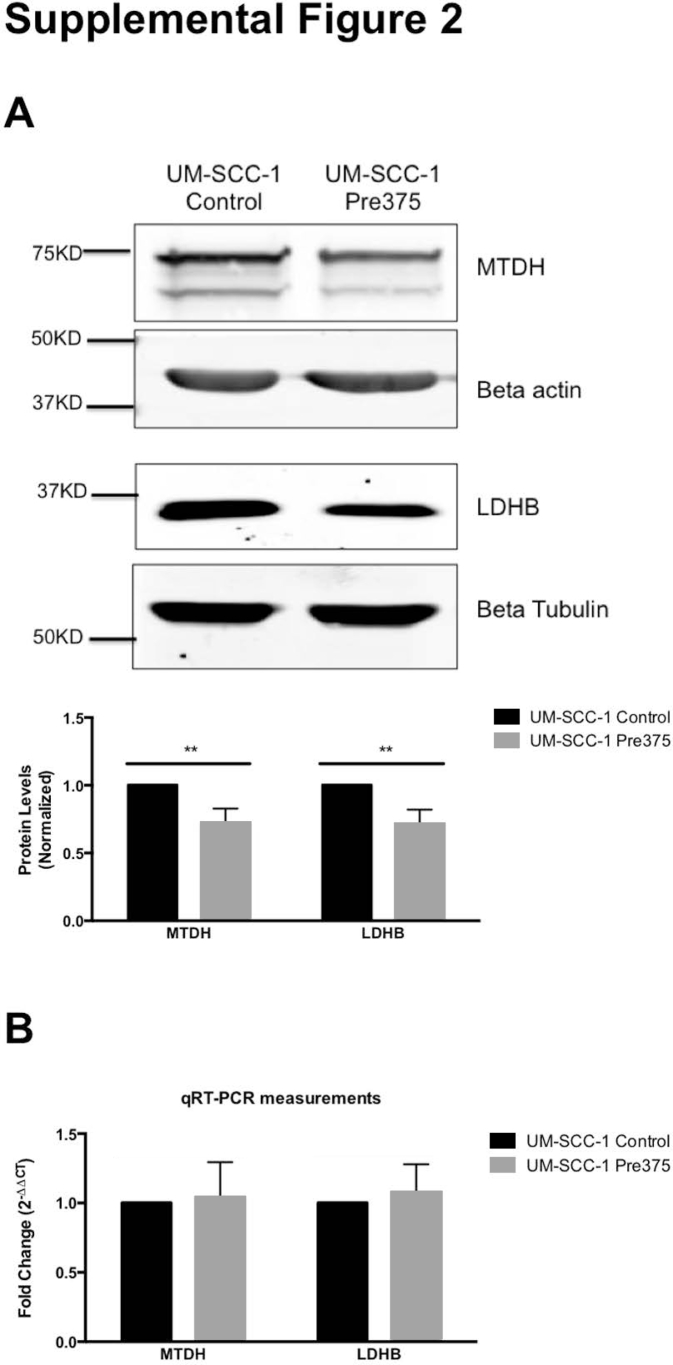
Among the down-regulated proteins, two previously published experimentally validated miR-375 targets were identified: metadherin and lactate dehydrogenase B (Supplemental Table S9).8 We confirmed these results at the protein level by Western blotting, although there did not seem to be a change at the mRNA level (Supplemental Figure S2).
L-Plastin and Vimentin Identified as the Most Down-Regulated Proteins
There were two proteins that were identified as substantially down-regulated in both the SILAC proteomic and microarray analyses: L-plastin and vimentin. L-plastin is an actin-bundling protein that promotes cell invasion in epithelial cells.42, 43 Vimentin is a cytoskeletal intermediate filament that is commonly expressed in mesenchymal cells.44, 45, 46 In the SILAC analysis, L-plastin was identified in the cytosolic fraction, whereas vimentin was identified in the cytoskeleton fraction. There was a 65% reduction in L-plastin and 62% reduction in vimentin protein levels with increased miR-375 expression (Supplemental Table S1). In the microarray analysis, L-plastin mRNA was reduced by 54% and vimentin mRNA was reduced by 66% with increased miR-375 expression (Supplemental Table S9).
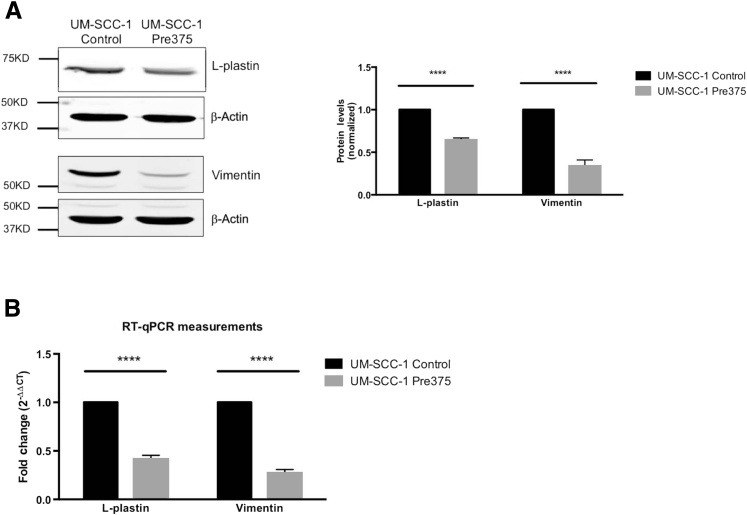
We performed whole cell Western blot and RT-qPCR analyses to confirm the SILAC analysis. In the Pre375 cells, L-plastin and vimentin protein levels were reduced by 36% and 64%, respectively, relative to the Control cells (Figure 3A). At the mRNA level, increased miR-375 expression resulted in 59% and 73% reductions in the mRNA abundance of L-plastin and vimentin, respectively (Figure 3B). Thus, we validated that L-plastin and vimentin were altered at both the mRNA and protein levels with increased miR-375 expression.
Figure 3.
Increased miR-375 expression in HNSCC cells results in reduced protein levels and mRNA expression levels of L-plastin and vimentin. A: Representative whole cell Western blots of L-plastin and vimentin in UM-SCC-1 transductant lines and its quantification. The L-plastin and vimentin bands were normalized to the loading control (β-actin). B: Quantitative real-time RT-PCR (RT-qPCR) analysis of the mRNA expression of L-plastin and vimentin in UM-SCC-1. ∗∗∗∗P < 0.0001.
Vimentin Promoter Activity Is Reduced by miR-375
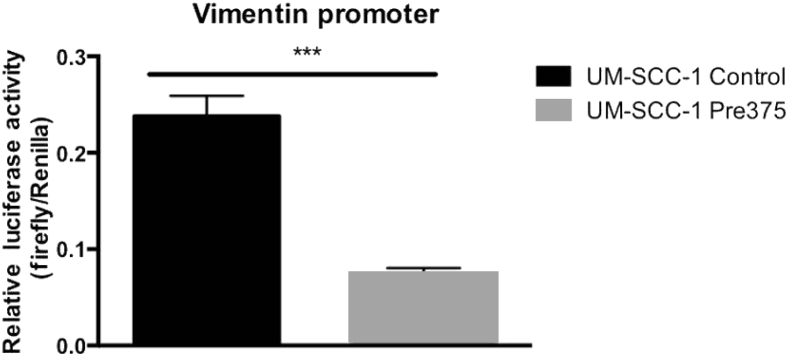
Although high miR-375 expression correlated with reduced vimentin mRNA and protein abundance, vimentin is not a theoretical miR-375 target. Transcriptional regulation is an alternative to direct post-transcriptional miRNA interaction; therefore, we asked if miR-375 expression reduces vimentin promoter activity using a luciferase reporter assay. We transiently transfected a firefly luciferase reporter construct expressing the vimentin promoter47, 48 into the UM-SCC-1 transductant lines using a Renilla luciferase construct as a transfection normalization control. We observed a 69% reduction in relative firefly luciferase activity in the Pre375 cells (Figure 4), suggesting that miR-375 regulates a transcription factor that drives the vimentin promoter.
Figure 4.
HNSCC cells with increased miR-375 expression show reduced vimentin promoter luciferase activity. The vimentin promoter firefly luciferase construct was cotransfected with a Renilla luciferase construct into the UM-SCC-1 lines. The luciferase signals were measured with the dual luciferase reporter assay. The firefly luciferase signal of the vimentin promoter construct was normalized to the Renilla luciferase signal. ∗∗∗P < 0.001.
Transcriptional Regulation of L-Plastin and Vimentin
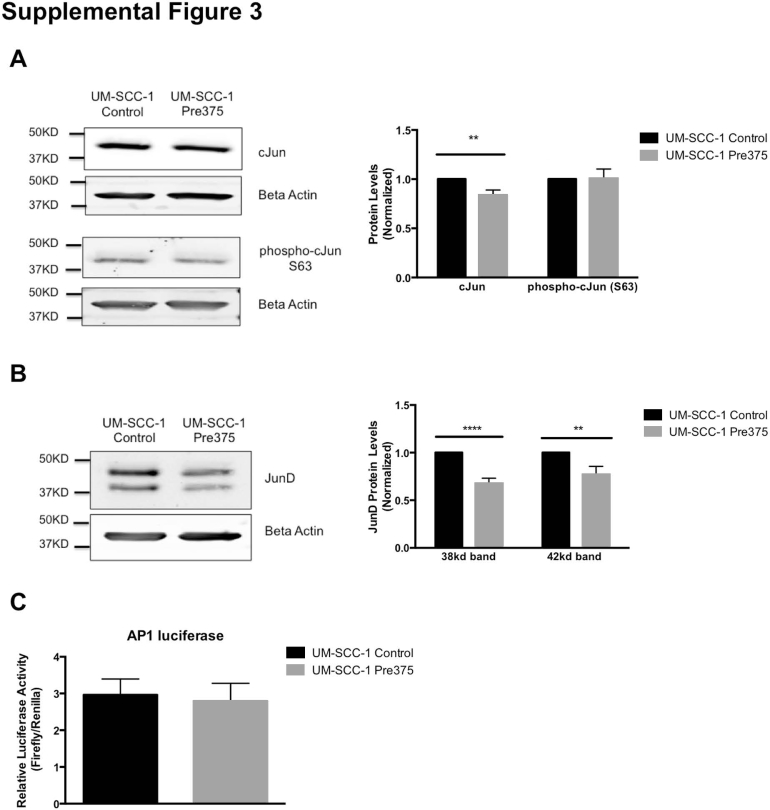
Because L-plastin and vimentin expression can be positively regulated by the AP-1 transcription factor,49, 50, 51, 52 we studied expression of AP-1 proteins (Supplemental Figure S3, A and B) and AP-1 activity using a luciferase reporter assay (Supplemental Figure S3C). We conclude that the diminished vimentin and L-plastin expression is not because of the regulation of AP-1 transcription factor activity by miR-375.
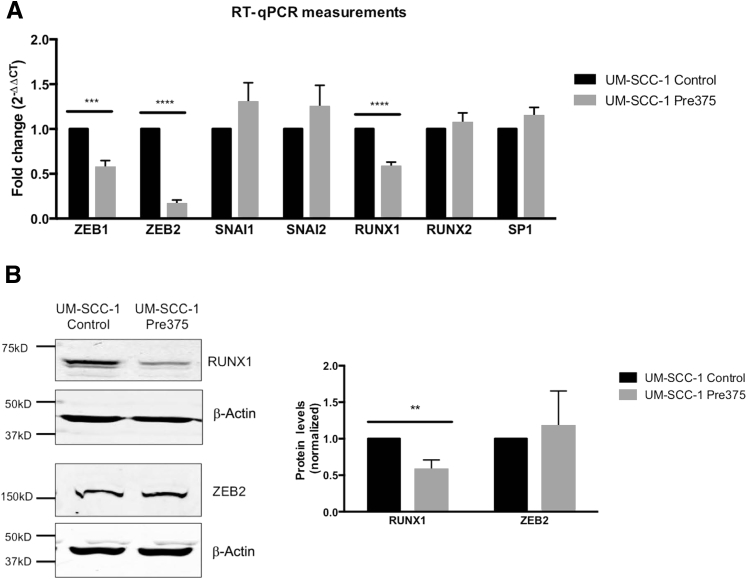
We then evaluated epithelial-mesenchymal transition–associated transcription factors that could potentially regulate L-plastin and/or vimentin expression: ZEB1, ZEB2, zinc finger protein SNAI1, zinc finger protein SNAI2, RUNX1, and RUNX2. All of these transcription factors are potential direct miR-375 targets, except Snail (SNAI1). We first assessed the mRNA abundance of these transcription factors by RT-qPCR. ZEB1, ZEB2, and RUNX1 RNA levels were reduced with increased miR-375 expression (Figure 5A). ZEB2 mRNA expression levels were reduced by 84%, whereas RUNX1 mRNA expression was reduced by 42%. Compared to RUNX1 (average ΔCT, 7.2), the mRNA levels of ZEB1 and ZEB2 were significantly lower (ΔCT values of 15.7 and 14.4, respectively), but because of the strong reduction of ZEB2 mRNA we included it along with RUNX1 for evaluation of protein levels.
Figure 5.
UM-SCC-1 cells with increased miR-375 expression show reduction in both the mRNA expression and protein levels of RUNX1. A: The mRNA abundance of selected transcription factors in the UM-SCC-1 transductant lines was analyzed by quantitative real-time RT-PCR (RT-qPCR). B: Representative Western blot image of RUNX1 protein levels and its quantification. ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
To assess the impact of miR-375 expression on ZEB2 and RUNX1 protein levels, Western blots were performed. There was a 42% reduction in RUNX1 protein similar to the reduction in RUNX1 RNA (Figure 5B). In contrast, we did not observe reductions in the ZEB2 protein levels in these cells (Figure 5B). These findings suggest that RUNX1 could be a direct miR-375 target.
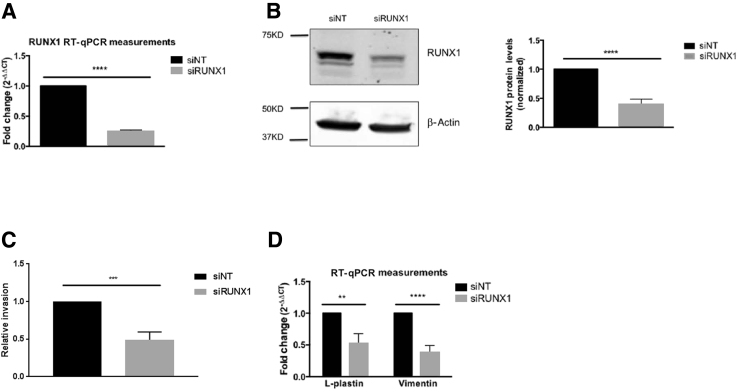
Knockdown of RUNX1 Diminishes Expression of Vimentin and L-Plastin, and Invasion
Because both the vimentin and L-plastin promoters have putative RUNX1 binding sites, we tested if vimentin and L-plastin expression would be suppressed when RUNX1 is transiently knocked down. Application of RUNX1 siRNA resulted in a 76% reduction in RUNX1 mRNA (Figure 6A) and a 61% reduction in RUNX1 protein (Figure 6B) as well as a 51% reduction in epidermal growth factor–stimulated invasion (Figure 6C). Knockdown of RUNX1 resulted in a 48% reduction in L-plastin mRNA expression and a 62% reduction in vimentin mRNA abundance in UM-SCC-1 cells (Figure 6D).
Figure 6.
Transient siRNA knockdown of RUNX1 suppresses invasion and vimentin and L-plastin mRNA expression. A: Quantitative real-time RT-PCR (RT-qPCR) analysis of the mRNA abundance of RUNX1 in UM-SCC-1 siRNA transfectants. B: Representative Western blots of RUNX1 with β-actin as loading control and its quantification. C: Epidermal growth factor–stimulated invasion of cells treated with nontargeting (siNT) and RUNX1 (siRUNX1) siRNA. D: Quantitative real-time RT-PCR (RT-qPCR) analysis of the mRNA expression of L-plastin and vimentin. ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
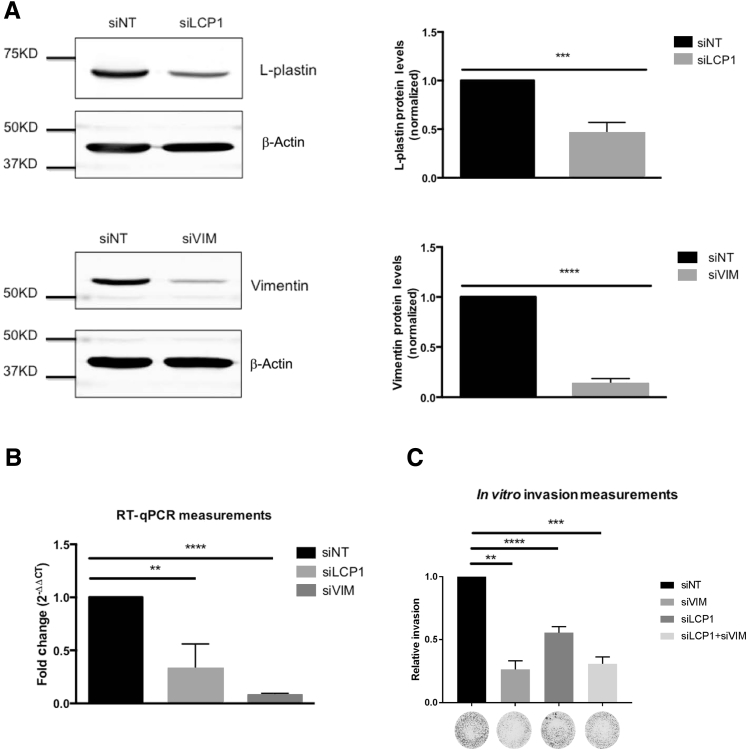
Knockdown of L-Plastin and Vimentin Reduces Cell Invasion
L-plastin is an actin bundling protein and is involved in cell invasion.42, 53, 54 Vimentin is a marker of mesenchymal cells, and cells with mesenchymal-like properties tend to be more motile and invasive.55, 56 To evaluate the roles of L-plastin and vimentin in HNSCC cell invasion, cells were treated with siRNAs against L-plastin and vimentin. The Western blot assessment of the L-plastin siRNA knockdowns revealed that the L-plastin protein level was reduced by 54% (Figure 7A). The Western blot assessments of the vimentin siRNA knockdowns revealed that the vimentin protein levels were reduced by 87% (Figure 7A). Application of L-plastin and vimentin siRNAs resulted in corresponding 68% and 93% reductions in L-plastin and vimentin mRNA levels, respectively (Figure 7B).
Figure 7.
Transient knockdown of L-plastin and vimentin inhibits invasion in vitro. A: Representative Western blots of L-plastin and vimentin and β-actin loading control and their quantification. B: Quantitative real-time RT-PCR (RT-qPCR) analysis of the mRNA abundance of L-plastin and vimentin in UM-SCC-1 siRNA transfections. C: Epidermal growth factor (EGF)–stimulated (0.1 nmol/L EGF) in vitro invasion was evaluated with representative examples of invasion filters provided underneath. ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
To test the effect of L-plastin and vimentin knockdown on cell invasion, the invasion of the siRNA transfectants was measured in response to 0.1 nmol/L epidermal growth factor using Matrigel invasion chambers. Knockdown of either L-plastin or vimentin showed significant reduction in epidermal growth factor–induced invasion compared to nontargeting siRNA. Invasion was reduced by 45% and 74% by siRNAs against L-plastin and vimentin, respectively (Figure 7C). Simultaneous knockdown of both L-plastin and vimentin did not reduce invasion further.
Evaluation of the TCGA HNSCC Data Set
We evaluated the relationships between miR-375, RUNX, vimentin, and L-plastin RNA expression in the TCGA HNSCC data set consisting of 478 tumor samples (Table 2). miR-375 RNA abundance was negatively correlated with RUNX1, vimentin, and L-plastin RNA abundance, consistent with miR-375 suppressing their expression. In addition, there was a positive correlation between RUNX1 and vimentin and L-plastin, consistent with the possibility that RUNX1 can stimulate transcription of vimentin and L-plastin. Vimentin and L-plastin also positively correlated with each other, supporting a common mechanism of regulation, such as through RUNX1 transcription factor activation. In summary, the TCGA RNA expression data from a large number of clinical samples are consistent with data generated using the UM-SCC-1 cell line in which miR-375 suppresses RUNX1 levels, and RUNX1 in turn contributes to the transcription of vimentin and L-plastin. Taken together, the model and clinical data support a role for miR-375 in regulating HNSCC invasion.
Table 2.
Analysis of TCGA HNSCC Primary Tumors
| Variable | miR-375 | RUNX1 | Vimentin | L-plastin |
|---|---|---|---|---|
| miR-375 | ||||
| Pearson correlation | 1 | −0.116∗ | −0.291∗∗ | −0.109∗ |
| Significance (two-tailed) | 0.011 | 0.000 | 0.017 | |
| N | 478 | 478 | 478 | 478 |
| RUNX1 | ||||
| Pearson correlation | −0.116∗ | 1 | 0.499∗∗ | 0.337∗∗ |
| Significance (two-tailed) | 0.011 | 0 | 0 | |
| N | 478 | 478 | 478 | 478 |
| Vimentin | ||||
| Pearson correlation | −0.291∗∗ | 0.499∗∗ | 1 | 0.491∗∗ |
| Significance (two-tailed) | 0 | 0 | 0 | |
| N | 478 | 478 | 478 | 478 |
| L-plastin | ||||
| Pearson correlation | −0.109∗ | 0.337∗∗ | 0.491∗∗ | 1 |
| Significance (two-tailed) | 0.017 | 0.000 | 0.000 | |
| N | 478 | 478 | 478 | 478 |
∗P < 0.05, ∗∗P < 0.01 using two-tailed t-test.
HNSCC, head and neck squamous cell carcinoma; RUNX1, Runt-related transcription factor 1; TCGA, The Cancer Genome Atlas.
Discussion
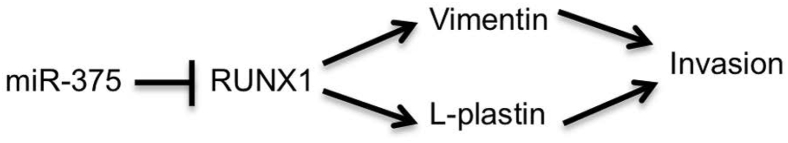
In this study, we used a SILAC-based proteomic strategy to survey the proteome to identify proteins whose expression changed with miR-375 abundance in HNSCC cells. There were 125 down-regulated proteins and 121 up-regulated proteins associated with increased miR-375 expression. We compared the lists of altered proteins to microarray data collected from the same lines and identified a subset of the proteins that were similarly correlated at the mRNA level. The most down-regulated proteins attributed to miR-375 expression were vimentin and L-plastin. RUNX1 emerged as a transcriptional regulator sensitive to miR-375 that contributes significantly to vimentin and L-plastin transcription. Reduction in levels of L-plastin or vimentin using siRNA significantly reduced cell invasion. Analysis of the TCGA HNSCC database revealed a negative correlation between miR-375 and RUNX, vimentin, and L-plastin, supporting a model in which miR-375 suppresses RUNX1 expression, resulting in turn in reduced transcription of vimentin and L-plastin (Figure 8).
Figure 8.
Our proposed mechanism by which miR-375 regulates invasion. miR-375 suppresses the expression of RUNX1. Because RUNX1 positively drives expression of L-plastin and vimentin, the reduction in RUNX1 results in reduced L-plastin and vimentin expression. Vimentin and L-plastin are important for cell invasion, and therefore reductions in L-plastin and vimentin lead to a reduction in invasion.
Our research group previously determined that increasing miR-375 levels in HNSCC cells resulted in decreased extracellular matrix degradation and invadopodial activity.18 Both vimentin and L-plastin have been reported to play a role in invasion and invadopodial activity. Vimentin is activated in cells undergoing epithelial-mesenchymal transition.46 High vimentin expression is associated with elevated invasion and metastatic potential of tumor cells, including in HNSCC.44, 55 In breast cancer cells, vimentin has been shown to be indispensable for the elongation of invadopodia, but is not essential for invadopodium formation.57 In kidney cancer cells, it has been determined that there is a complex formed at the base of invadopodia that has vimentin, plectin, and F-actin.58 Vimentin is bound to F-actin through its interaction with plectin and is believed to serve as the scaffold for invadopodium formation.58
L-plastin was originally described as being exclusively in hematopoietic cell lineages, but many epithelial carcinomas also express L-plastin.42, 51 L-plastin is an actin bundling protein, and is reported to promote cell invasion in cholangiocarcinoma and colon cancer43, 53 and in vivo metastasis of human prostate and melanoma cancer cells.42, 59 L-plastin promotes vinculin-containing podosome formation in melanoma cells; however, the phosphorylation of L-plastin is not necessary for podosome formation.42 Phosphorylation of L-plastin on serine-5 increases its ability to bind F-actin and promotes its targeting to actin assembly in cells.60 L-plastin and phosphorylated L-plastin colocalize with cortactin in phorbol-12-myristate-13-acetate–treated MCF-7 cells and control actin turnover in focal adhesions.61 In summary, down-regulation of vimentin and/or L-plastin could mediate the inhibition of invadopod formation caused by increased expression of miR-375. Future studies will test if restoration of vimentin and L-plastin in HNSCC cells with more normal miR-375 levels would promote the invadopodial matrix degradation phenotype we previously observed in aggressive cells.18
We observed regulation of the vimentin promoter associated with varied miR-375 abundance, suggesting that miR-375 could be altering transcription factors driving vimentin expression. Because AP-1 has been previously reported to increase the expression of L-plastin and vimentin expression by binding to their promoters,49, 50, 51 we evaluated the expression of AP-1 components and AP-1 activity. The AP-1 luciferase reporter did not show significant reductions in AP-1 activity. Thus, in our cell type, AP-1 is unlikely to be a major component in the observed changes in vimentin and L-plastin expression, though under some conditions it may contribute to the phenotype.
We then explored if changes in the abundance of other transcription factors that potentially activate L-plastin and/or vimentin expression could be responsible for the invasion phenotype. The top candidate that emerged was RUNX1. RUNX1 is predicted to be a miR-375 target by several miRNA target prediction databases, including TargetScan,31 DIANA-mT,32 and miRanda.33 RUNX1 has putative consensus DNA binding sites in both the vimentin and L-plastin promoters. Our observation that tumor cells with elevated miR-375 levels exhibit suppressed RUNX1 protein and mRNA expression levels suggests that RUNX1 could be a miR-375 target and prove that RUNX1 is regulated at some level by miR-375. Suppressing RUNX1 expression by siRNA treatment led to diminished L-plastin and vimentin mRNA expression, consistent with a transcriptional role for this factor on those promoters. This result is consistent with other studies showing that RUNX1 can be involved in cell invasion and metastasis.62, 63, 64 RUNX1 promotes ovarian cancer proliferation, migration, and invasion.62 In the MMTV-PyMT model of breast cancer, RUNX1 acts as an oncogene, promoting tumor growth and metastasis.63 RUNX1 also promotes metastasis of endometrial cancer cells in vivo.64 Planaguma et al65 reported that RUNX1 colocalized with matrix metalloproteinases (2 and 9) at the invasive front of endometrioid endometrial carcinoma and ovarian endometrioid carcinoma. In our squamous carcinoma cells, RUNX1 expression could promote invasion by increasing expression of L-plastin and vimentin.
Evaluation of the RNA expression data from 478 head and neck cancer clinical samples from the TCGA revealed the same relationships between miR-375, RUNX1, vimentin, and L-plastin expression that we observed in our model. miR-375 expression is negatively correlated with RUNX1, vimentin, and L-plastin mRNA levels, consistent with it negatively regulating them. Conversely, RUNX1, vimentin, and L-plastin mRNAs are positively correlated, as predicted if RUNX1 can drive expression of vimentin and L-plastin. Thus, data obtained from clinical samples support the model developed from our studies with the UM-SCC-1 cell line.
In summary, we used a SILAC-based proteomic technique to identify miR-375–regulated proteins. L-plastin and vimentin emerged as the most down-regulated proteins in the SILAC analysis. We confirmed that miR-375 expression in HNSCC cells indirectly regulates the mRNA levels of L-plastin and vimentin and that L-plastin and vimentin are important for HNSCC invasion. RUNX1 emerged as a candidate miR-375 target that could subsequently regulate L-plastin and vimentin expression (Figure 8). Inhibition of RUNX1 activity could provide a new opportunity for inhibiting invasion in HNSCC.
Acknowledgments
We thank members of the Condeelis, Cox, and Hodgson laboratories, as well as the Einstein/Montefiore Head and Neck Cancer Multidisciplinary Research Group for helpful discussions; Jennifer T. Aguilan for assistance with mass spectrometry data; Dr. Christine Gilles (Liège University, Liège, Wallonia, Belgium) for the vimentin promoter firefly luciferase; and Dr. Hong Wan (Queen Mary University, London, UK) for the AP-1 firefly luciferase reporter construct.
Footnotes
Supported by NIH predoctoral fellowship F31CA168337 (L.J.); NIH research grants R01CA77522 (J.E.S.), P01CA100324 (J.E.S.), 2K12 CA132783-06 (T.J.O.), and UL1TR001073 (T.J.O.); the NIH Albert Einstein Cancer Center Support grant P30CA013330; and the Department of Pathology, Albert Einstein College of Medicine/Montefiore Medical Center. J.E.S. is the Betty and Sheldon Feinberg Senior Faculty Scholar in Cancer Research.
Disclosures: None declared.
Current address of L.J., Department of Cancer Biology, Vanderbilt University, Nashville, TN; of J.L., Janssen Research & Development, LLC, Large Molecule Analytical Development, Malvern, PA.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2017.02.019.
Supplemental Data
Supplemental Figure S1.
Comparison of relative miR-375 expression in the transductant lines and HNSCC patient samples. Quantitative real-time RT-PCR analysis of the miR-375 expression in the UM-SCC-1 transductant cell lines and HNSCC patient tumor (HN Tumors) and normal (HN Normals) samples. The histogram of the ΔCT values of the HNSCC patient samples is shown with miR-375 expression decreasing from left to right. The arrows show the ΔCT values of the UM-SCC-1 control vector (green) and pre-375 (blue) transductant cell lines.
Supplemental Figure S2.
UM-SCC-1 cells with increased miR-375 expression show reductions in protein levels of metadherin (MTDH) and lactate dehydrogenase B (LDHB). A: Representative Western blots of MTDH and LDHB and quantification of Western blot measurements. The MTDH and LDHB bands were normalized to the loading controls (β-actin or β-tubulin). B: Quantitative real-time RT-PCR (RT-qPCR) analysis of the mRNA expression of MTDH and LDHB in UM-SCC-1. Data are shown as the means and SEM from at least three independent experiments (A). ∗∗P < 0.01.
Supplemental Figure S3.
Evaluation of miR-375 effects on AP-1 activity. A: Western blot analyses of cJun and phospho-cJun (Ser63) protein levels in the UM-SCC-1 transductant lines. Representative Western blot images and their quantitation. The cJun and phospho-cJun bands were normalized to the loading controls (β-actin or β-tubulin). B: Western blot analyses of JunD protein levels in the UM-SCC-1 transductant lines. Representative Western blot images and their quantitation. Each of the JunD bands was normalized to the loading controls (β-actin or β-tubulin). C: AP-1 promoter activity in the UM-SCC-1 transductant lines. The AP-1 firefly luciferase reporter constructs were cotransfected with a Renilla luciferase construct into the UM-SCC-1 transductant cells. The luciferase signals were measured with the dual luciferase reporter assay. The firefly luciferase signal of the AP-1 reporter construct was normalized to the Renilla luciferase signal. ∗∗P < 0.01, ∗∗∗∗P < 0.0001.
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–316. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4.Pai S.I., Westra W.H. Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Annu Rev Pathol. 2009;4:49–70. doi: 10.1146/annurev.pathol.4.110807.092158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganci F., Sacconi A., Manciocco V., Covello R., Spriano G., Fontemaggi G., Blandino G. Molecular genetics and biology of head and neck squamous cell carcinoma: implications for diagnosis, prognosis and treatment. Head and Neck Cancer. In: Agulnik D.M., editor. InTech. 2012. pp. 73–122. Available at https://www.intechopen.com/books/head-and-neck-cancer/molecular-genetics-and-biology-of-head-and-neck-squamous-cell-carcinoma-implications-for-diagnosis-p. [Google Scholar]
- 6.Nisa L., Aebersold D.M., Giger R., Caversaccio M.D., Borner U., Medova M., Zimmer Y. Profiling invasiveness in head and neck cancer: recent contributions of genomic and transcriptomic approaches. Cancers (Basel) 2015;7:585–597. doi: 10.3390/cancers7020585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Yan J.W., Lin J.S., He X.X. The emerging role of miR-375 in cancer. Int J Cancer. 2014;135:1011–1018. doi: 10.1002/ijc.28563. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y.S., Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pencheva N., Tavazoie S.F. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bargaje R., Gupta S., Sarkeshik A., Park R., Xu T., Sarkar M., Halimani M., Roy S.S., Yates J., Pillai B. Identification of novel targets for miR-29a using miRNA proteomics. PLoS One. 2012;7:e43243. doi: 10.1371/journal.pone.0043243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh R., Mo Y.Y. Role of microRNAs in breast cancer. Cancer Biol Ther. 2013;14:201–212. doi: 10.4161/cbt.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu H.F., Lin S.C., Chang K.W. MicroRNA aberrances in head and neck cancer: pathogenetic and clinical significance. Curr Opin Otolaryngol Head Neck Surg. 2013;21:104–111. doi: 10.1097/MOO.0b013e32835e1d6e. [DOI] [PubMed] [Google Scholar]
- 15.Farazi T.A., Hoell J.I., Morozov P., Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Childs G., Fazzari M., Kung G., Kawachi N., Brandwein-Gensler M., McLemore M., Chen Q., Burk R.D., Smith R.V., Prystowsky M.B., Belbin T.J., Schlecht N.F. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris T., Jimenez L., Kawachi N., Fan J.B., Chen J., Belbin T., Ramnauth A., Loudig O., Keller C.E., Smith R., Prystowsky M.B., Schlecht N.F., Segall J.E., Childs G. Low-level expression of miR-375 correlates with poor outcome and metastasis while altering the invasive properties of head and neck squamous cell carcinomas. Am J Pathol. 2012;180:917–928. doi: 10.1016/j.ajpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez L., Sharma V.P., Condeelis J., Harris T., Ow T.J., Prystowsky M.B., Childs G., Segall J.E. MicroRNA-375 suppresses extracellular matrix degradation and invadopodial activity in head and neck squamous cell carcinoma. Arch Pathol Lab Med. 2015;139:1349–1361. doi: 10.5858/arpa.2014-0471-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iorio M.V., Croce C.M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics: a comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lossner C., Meier J., Warnken U., Rogers M.A., Lichter P., Pscherer A., Schnolzer M. Quantitative proteomics identify novel miR-155 target proteins. PLoS One. 2011;6:e22146. doi: 10.1371/journal.pone.0022146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong Q., Zhong Q., Zhang J., Yang M., Li C., Zheng P., Bi L.J., Ge F. Identification of novel miR-21 target proteins in multiple myeloma cells by quantitative proteomics. J Proteome Res. 2012;11:2078–2090. doi: 10.1021/pr201079y. [DOI] [PubMed] [Google Scholar]
- 22.Yan G.R., Xu S.H., Tan Z.L., Liu L., He Q.Y. Global identification of miR-373-regulated genes in breast cancer by quantitative proteomics. Proteomics. 2011;11:912–920. doi: 10.1002/pmic.201000539. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y., Chaerkady R., Kandasamy K., Huang T.C., Selvan L.D., Dwivedi S.B., Kent O.A., Mendell J.T., Pandey A. Identifying targets of miR-143 using a SILAC-based proteomic approach. Mol Biosyst. 2010;6:1873–1882. doi: 10.1039/c004401f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang T.C., Renuse S., Pinto S., Kumar P., Yang Y., Chaerkady R., Godsey B., Mendell J.T., Halushka M.K., Civin C.I., Marchionni L., Pandey A. Identification of miR-145 targets through an integrated omics analysis. Mol Biosyst. 2015;11:197–207. doi: 10.1039/c4mb00585f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer K.M., Hummon A.B. Effects of the miR-143/-145 microRNA cluster on the colon cancer proteome and transcriptome. J Proteome Res. 2012;11:4744–4754. doi: 10.1021/pr300600r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 27.Bendall S.C., Hughes C., Stewart M.H., Doble B., Bhatia M., Lajoie G.A. Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol Cell Proteomics. 2008;7:1587–1597. doi: 10.1074/mcp.M800113-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 29.Deutsch E.W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J.K., Martin D.B., Nesvizhskii A.I., Aebersold R. A guided tour of the trans-proteomic pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dweep H., Sticht C., Pandey P., Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Vlachos I.S., Kostoulas N., Vergoulis T., Georgakilas G., Reczko M., Maragkakis M., Paraskevopoulou M.D., Prionidis K., Dalamagas T., Hatzigeorgiou A.G. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miranda K.C., Huynh T., Tay Y., Ang Y.S., Tam W.L., Thomson A.M., Lim B., Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 35.Kruger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasband W.S. US NIH; Bethesda, MD: 1997-2014. ImageJ. [Google Scholar]
- 39.Broad Institute TCGA Genome Data Analysis Center (2016): Analysis-ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. Broad Institute of MIT and Harvard. Dataset. https://doi.org/10.7908/C11G0KM9
- 40.Lin C.J., Grandis J.R., Carey T.E., Gollin S.M., Whiteside T.L., Koch W.M., Ferris R.L., Lai S.Y. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29:163–188. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 41.Yokoi T.Y.A., Odajima T., Furukawa K. Establishment and characterization of a human cell line derived from a squamous cell carcinoma of the tongue. Tumor Res. 1988;23:43–57. [Google Scholar]
- 42.Klemke M., Rafael M.T., Wabnitz G.H., Weschenfelder T., Konstandin M.H., Garbi N., Autschbach F., Hartschuh W., Samstag Y. Phosphorylation of ectopically expressed L-plastin enhances invasiveness of human melanoma cells. Int J Cancer. 2007;120:2590–2599. doi: 10.1002/ijc.22589. [DOI] [PubMed] [Google Scholar]
- 43.Foran E., McWilliam P., Kelleher D., Croke D.T., Long A. The leukocyte protein L-plastin induces proliferation, invasion and loss of E-cadherin expression in colon cancer cells. Int J Cancer. 2006;118:2098–2104. doi: 10.1002/ijc.21593. [DOI] [PubMed] [Google Scholar]
- 44.Chaw S.Y., Majeed A.A., Dalley A.J., Chan A., Stein S., Farah C.S. Epithelial to mesenchymal transition (EMT) biomarkers–E-cadherin, beta-catenin, APC and vimentin–in oral squamous cell carcinogenesis and transformation. Oral Oncol. 2012;48:997–1006. doi: 10.1016/j.oraloncology.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 45.Kim K.H., Kim L., Choi S.J., Han J.Y., Kim J.M., Chu Y.C., Kim Y.M., Park I.S., Lim J.H. The clinicopathological significance of epithelial mesenchymal transition associated protein expression in head and neck squamous cell carcinoma. Korean J Pathol. 2014;48:263–269. doi: 10.4132/KoreanJPathol.2014.48.4.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scanlon C.S., Van Tubergen E.A., Inglehart R.C., D'Silva N.J. Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J Dent Res. 2013;92:114–121. doi: 10.1177/0022034512467352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bindels S., Mestdagt M., Vandewalle C., Jacobs N., Volders L., Noel A., van Roy F., Berx G., Foidart J.M., Gilles C. Regulation of vimentin by SIP1 in human epithelial breast tumor cells. Oncogene. 2006;25:4975–4985. doi: 10.1038/sj.onc.1209511. [DOI] [PubMed] [Google Scholar]
- 48.Gilles C., Polette M., Mestdagt M., Nawrocki-Raby B., Ruggeri P., Birembaut P., Foidart J.M. Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res. 2003;63:2658–2664. [PubMed] [Google Scholar]
- 49.Sommers C.L., Skerker J.M., Chrysogelos S.A., Bosseler M., Gelmann E.P. Regulation of vimentin gene transcription in human breast cancer cell lines. Cell Growth Differ. 1994;5:839–846. [PubMed] [Google Scholar]
- 50.Rittling S.R., Coutinho L., Amram T., Kolbe M. AP-1/jun binding sites mediate serum inducibility of the human vimentin promoter. Nucleic Acids Res. 1989;17:1619–1633. doi: 10.1093/nar/17.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin C.S., Chen Z.P., Park T., Ghosh K., Leavitt J. Characterization of the human L-plastin gene promoter in normal and neoplastic cells. J Biol Chem. 1993;268:2793–2801. [PubMed] [Google Scholar]
- 52.Karin M., Liu Z., Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 53.Chaijan S., Roytrakul S., Mutirangura A., Leelawat K. Matrigel induces L-plastin expression and promotes L-plastin-dependent invasion in human cholangiocarcinoma cells. Oncol Lett. 2014;8:993–1000. doi: 10.3892/ol.2014.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng J., Rudra-Ganguly N., Powell W.C., Roy-Burman P. Suppression of prostate carcinoma cell invasion by expression of antisense L-plastin gene. Am J Pathol. 1999;155:115–122. doi: 10.1016/S0002-9440(10)65106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dal Vechio A.M., Giudice F.S., Sperandio F.F., Mantesso A., Pinto Junior Ddos S. Vimentin expression and the influence of Matrigel in cell lines of head and neck squamous cell carcinoma. Braz Oral Res. 2011;25:235–240. doi: 10.1590/s1806-83242011000300008. [DOI] [PubMed] [Google Scholar]
- 56.Nijkamp M.M., Span P.N., Hoogsteen I.J., van der Kogel A.J., Kaanders J.H., Bussink J. Expression of E-cadherin and vimentin correlates with metastasis formation in head and neck squamous cell carcinoma patients. Radiother Oncol. 2011;99:344–348. doi: 10.1016/j.radonc.2011.05.066. [DOI] [PubMed] [Google Scholar]
- 57.Schoumacher M., Goldman R.D., Louvard D., Vignjevic D.M. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol. 2010;189:541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutoh Yoneyama M., Hatakeyama S., Habuchi T., Inoue T., Nakamura T., Funyu T., Wiche G., Ohyama C., Tsuboi S. Vimentin intermediate filament and plectin provide a scaffold for invadopodia, facilitating cancer cell invasion and extravasation for metastasis. Eur J Cell Biol. 2014;93:157–169. doi: 10.1016/j.ejcb.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 59.Riplinger S.M., Wabnitz G.H., Kirchgessner H., Jahraus B., Lasitschka F., Schulte B., van der Pluijm G., van der Horst G., Hammerling G.J., Nakchbandi I., Samstag Y. Metastasis of prostate cancer and melanoma cells in a preclinical in vivo mouse model is enhanced by L-plastin expression and phosphorylation. Mol Cancer. 2014;13:10. doi: 10.1186/1476-4598-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Janji B., Giganti A., De Corte V., Catillon M., Bruyneel E., Lentz D., Plastino J., Gettemans J., Friederich E. Phosphorylation on Ser5 increases the F-actin-binding activity of L-plastin and promotes its targeting to sites of actin assembly in cells. J Cell Sci. 2006;119:1947–1960. doi: 10.1242/jcs.02874. [DOI] [PubMed] [Google Scholar]
- 61.Al Tanoury Z., Schaffner-Reckinger E., Halavatyi A., Hoffmann C., Moes M., Hadzic E., Catillon M., Yatskou M., Friederich E. Quantitative kinetic study of the actin-bundling protein L-plastin and of its impact on actin turn-over. PLoS One. 2010;5:e9210. doi: 10.1371/journal.pone.0009210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keita M., Bachvarova M., Morin C., Plante M., Gregoire J., Renaud M.C., Sebastianelli A., Trinh X.B., Bachvarov D. The RUNX1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle. 2013;12:972–986. doi: 10.4161/cc.23963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Browne G., Taipaleenmaki H., Bishop N.M., Madasu S.C., Shaw L.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Runx1 is associated with breast cancer progression in MMTV-PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. J Cell Physiol. 2015;230:2522–2532. doi: 10.1002/jcp.24989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doll A., Gonzalez M., Abal M., Llaurado M., Rigau M., Colas E., Monge M., Xercavins J., Capella G., Diaz B., Gil-Moreno A., Alameda F., Reventos J. An orthotopic endometrial cancer mouse model demonstrates a role for RUNX1 in distant metastasis. Int J Cancer. 2009;125:257–263. doi: 10.1002/ijc.24330. [DOI] [PubMed] [Google Scholar]
- 65.Planaguma J., Liljestrom M., Alameda F., Butzow R., Virtanen I., Reventos J., Hukkanen M. Matrix metalloproteinase-2 and matrix metalloproteinase-9 codistribute with transcription factors RUNX1/AML1 and ETV5/ERM at the invasive front of endometrial and ovarian carcinoma. Hum Pathol. 2011;42:57–67. doi: 10.1016/j.humpath.2010.01.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.