SUMMARY
Umbilical cord blood (CB) is a convenient and broadly used source of hematopoietic stem cells (HSCs) for allogeneic stem cell transplantation. However, limiting numbers of HSCs remain a major constraint for its clinical application. Although one feasible option would be to expand HSCs to improve therapeutic outcome, available protocols and the molecular mechanisms governing the self-renewal of HSCs are unclear. Here, we show that ectopic expression of a single microRNA (miRNA), miR-125a, in purified murine and human multipotent progenitors (MPPs) resulted in increased self-renewal and robust long-term multi-lineage repopulation in transplanted recipient mice. Using quantitative proteomics and western blot analysis, we identified a restricted set of miR-125a targets involved in conferring long-term repopulating capacity to MPPs in humans and mice. Our findings offer the innovative potential to use MPPs with enhanced self-renewal activity to augment limited sources of HSCs to improve clinical protocols.
In Brief
Wojtowicz et al. report that enforced expression of miR-125a confers enhanced long-term self-renewal potential to multipotent hematopoietic progenitors (MPPs). Quantitative proteomics reveals a miR-125a target network that normally functions to restrain self-renewal in more committed progenitors. These results suggest that enhanced MPPs may be used to augment limited HSC sources.

INTRODUCTION
Allogeneic stem cell transplantation is an established treatment for malignant diseases such as leukemia, myelodysplasia, and myelofibrosis as well as non-malignant disorders such as severe aplastic anemia and congenital immune or enzyme deficiencies, with >40,000 transplants undertaken worldwide (Aljurf et al., 2014; Pineault and Abu-Khader, 2015). Despite this success, ~67% of patients who require allogeneic transplantation are not able to undergo this procedure due to the lack of matched donors (Oran and Shpall, 2012). Umbilical cord blood (CB) banks have the potential to alleviate these obstacles due to broader representation of histocompatibility leukocyte antigen (HLA) types, less restriction on HLA matching, reduced incidence of graft versus host disease (GvHD), and rapid access compared to registries. However, widespread use of CB is impaired due to insufficient hematopoietic stem cell (HSC) numbers in single CB units, resulting in extended times to full engraftment (Ballen et al., 2013). Transplants combining two unrelated CB units have attempted to overcome the shortfall in HSC number, although improvements are modest (Barker et al., 2001; Wagner et al., 2014). Multiple efforts have been undertaken to attempt HSC expansion, but these have largely been empirical and rarely successful (Metcalf, 2008; Ogawa, 1993; Sauvageau et al., 2004). Recently, promising HSC expansion protocols have employed small molecules such as prostaglandin E2, SR1, UM171, or UM729, although clinical efficacy remains elusive (Boitano et al., 2010; Fares et al., 2014; North et al., 2007).
The unique self-renewal properties of HSCs are thought to be controlled by a combination of intrinsic and extrinsic signaling networks that finely balance the equilibrium among quiescence, proliferation, and differentiation. Generating increased numbers of HSC could come from controlling self-renewal or inducing self-renewal in the more numerous progenitor compartment. To achieve the latter, numerous signaling pathways would have to be appropriately fine-tuned. Small, non-coding RNAs such as microRNAs (miRNAs) act as post-transcriptional regulators of gene expression, and one miRNA can target hundreds of transcripts comprising an entire network of regulated genes, positioning miRNAs as pivotal regulators of cellular homeostasis (Bartel, 2009; Filipowicz et al., 2008). miRNA profiling of hematopoietic stem and progenitor cells (HSPCs) has identified several miRNAs as differentially expressed and functionally able to govern HSC properties (Gerrits et al., 2012; Lechman et al., 2012; O’Connell et al., 2008, 2010). The family of miR-125 is particularly interesting, as they are highly expressed in murine HSCs (Gerrits et al., 2012; Guo and Scadden, 2010; O’Connell et al., 2010). This family consists of three paralogs (miR-125a, miR-125b1, and miR-125b2) sharing an identical seed sequence and exerting similar effects on HSPCs (Wojtowicz et al., 2014). MicroRNA-125a expression levels significantly decrease upon hematopoietic differentiation in mice (Gerrits et al., 2012; O’Connell et al., 2010), implying a correlation between the level of miR-125 activity and differentiation status. Ectopic expression of miR-125 (a, b1, or b2) in murine bone marrow (BM) cells results in an increased self-renewal, a proliferative advantage to HSCs and the initiation of myeloproliferation or leukemia, depending on miR-125 expression level (O’Connell et al., 2010; Wojtowicz et al., 2014). Targeted downregulation of miR-125a and miR-125b caused reduced HSPC self-renewal in vitro (Guo and Scadden, 2010) and reduced proliferation and blood cell output in vivo (Ooi et al., 2010; Wojtowicz et al., 2014), confirming a pivotal role for miR-125 family members in the molecular control of HSC self-renewal.
In the current study, we explored the potential of a single miRNA, miR-125a, to confer long-term repopulating ability to non-stem cell populations. Here, we report that enforced expression of miR-125a endows multipotent progenitors (MPPs) with enhanced self-renewal potential, offering an innovative means to use MPPs to augment limited sources of HSCs to improve clinical outcomes.
RESULTS
Enforced Expression of miR-125a Increases Human HSC Self-Renewal
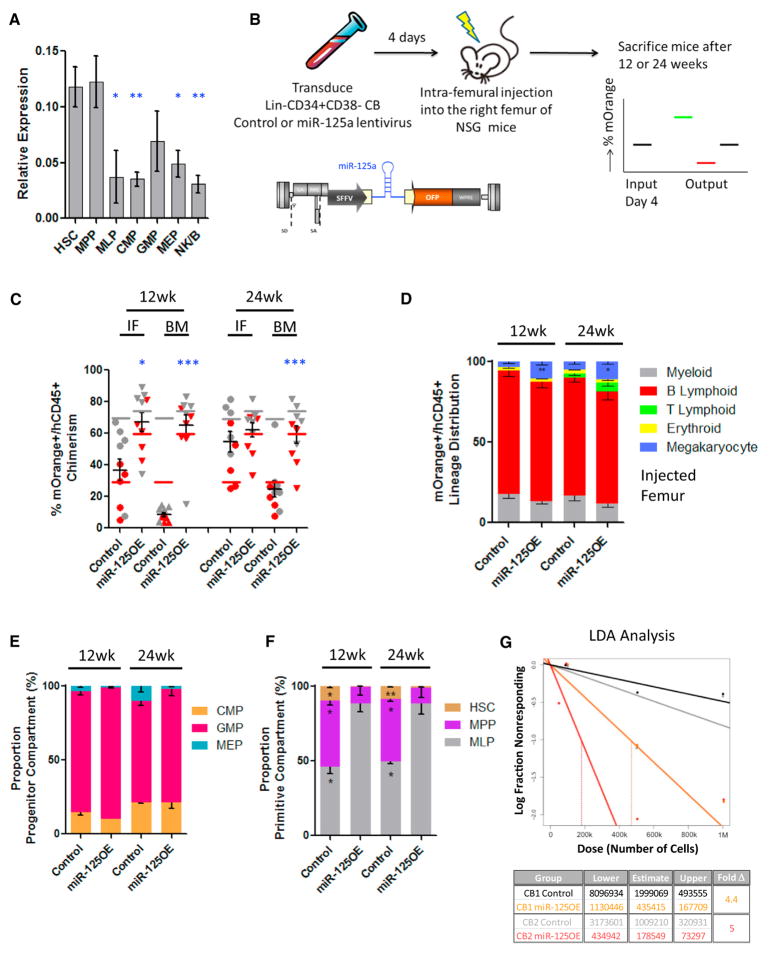
Since miR-125a is highly expressed within mouse long-term HSCs (LT-HSCs) and has been shown to increase their self-renewal activity (Gerrits et al., 2012; Guo and Scadden, 2010; O’Connell et al., 2010; Ooi et al., 2010; Wojtowicz et al., 2014), we wanted to evaluate if these attributes are evolutionarily conserved within human hematopoiesis. We sorted immunophenotypically defined HSC and progenitor cell subsets from human-lineage-depleted CB (Figure S1A) (Doulatov et al., 2010; Notta et al., 2011) to obtain a detailed expression profile of mature miR-125a. We observed that the highest expression levels of miR-125a were restricted to the HSC-enriched CD34+CD38− CD45RA−CD90+CD49f+ and CD34+CD38−CD45RA−CD90− CD49f− (MPP) fractions and become significantly downregulated in multi-lymphoid progenitors (MLPs) and committed CD34+ CD38+ progenitor fractions (common myeloid progenitor [CMP], granulocyte monocyte progenitor [GMP], and megakaryocytic-erythroid progenitor [MEP]) (Figure 1A). These data demonstrate an inverse correlation between differentiation state and miR-125a expression levels.
Figure 1. Enforced Expression of miR-125a Gives a Proliferative Advantage to HSCs.
(A) RT-PCR expression levels of miR-125a in flow-sorted populations from the human Lin− CB hierarchy. Cells were sorted to high purity according to the scheme outlined in Figure S1A. RNU48 was used as an endogenous control, and miR-125a levels were normalized to RNU48 levels.
(B) The scheme describing the experimental system.
(C) Human mOrange+ (mO+) chimerism at 12 and 24 weeks post-transplantation with pre-sorted CD34+CD38− umbilical cord blood cells transduced with a miR-125 lentiviral vector (miR-125OE) (n = 10) or an empty control vector (n = 10) in injected femur (IF) and distant bone marrow (BM) sites. Red and gray symbols represent replicate experiments using two distinct human CB pools. Color-matched bars represent input levels of mOrange+ cells at time of transplant for each experiment.
(D) Lineage distribution of mO+ BM grafts 12 and 24 weeks post-transplantation.
(E) Frequency of CMP, GMP, and MEP populations recovered from total BM within the CD34+CD38+ committed progenitor compartment.
(F) Proportion of HSC, MPP, and MLP from total BM within the CD34+CD38− primitive compartment 12 and 24 weeks post-transplant.
(G) Secondary transplantations. BM cells were harvested from two primary recipients and flow sorted for hCD45+mO+ cells. Stem cell frequencies were evaluated by limiting dilution transplantation into secondary mice. Confidence intervals were calculated using ELDA software.
All data reflect mean values ± SEM. Statistical significance was assessed using a non-parametric Mann-Whitney test, where *p < 0.05, **p < 0.01, and ***p < 0.001.
To study the function of miR-125a in human HSPCs, we generated a lentiviral vector with miR-125a (miR-125OE) under a constitutive SFFV promoter using an orange fluorescent protein (OFP) reporter (Figure 1B). To determine the impact of increased miR-125a expression (Figure S1B) on human HSPC function, CD34+CD38− human CB HSPCs transduced with miR-125OE and control vector transduced cells were analyzed in xenotransplantation assays (Figure 1B). At 12 and 24 weeks after transplantation, we observed significant increases in human chimerism within distant BM sites in the miR-125aOE group compared to controls; chimerism levels in the injected femur (IF) were significantly increased only at 12 weeks (Figure 1C). Mice transplanted with miR-125OE cells also exhibited white BM and splenomegaly (Figure S1C). Furthermore, within enlarged spleens, we found increased proportions of CD19+ CD10+CD20− B lymphoid cells, suggesting a partial B cell differentiation block at the pro-B cell stage (Figure S1D). Examination of the lineage distribution of OFP+ cells in the BM revealed that all lineages were present, with significant expansion of the CD41+ megakaryocyte population (Figure 1D) and an increased CD3+ T cell population. At the progenitor level (Figures 1E and S1E), we observed a trend for increased GMPs and decreased MEP frequency in the BM, whereas the frequency of the multi-lymphoid progenitor fraction (MLP) was significantly increased at the expense of immunophenotypic HSCs and MPPs in the primitive compartment (Figures 1F and S1E). These observations are in line with previous reports in murine HSPCs pointing to conservation of function across species (Gerrits et al., 2012; Guo and Scadden, 2010; O’Connell et al., 2010).
The self-renewal potential of transduced human HSC within primary engrafted mice was assessed using quantitative secondary transplantation assays. Recipients transplanted with miR-125OE cells showed higher chimerism levels compared to controls, and the HSC frequency increased by 4.4- and 5-fold in miR-125OE recipients in independent experiments (Figures 1G and S1F), similar to published murine studies (Gerrits et al., 2012; Guo and Scadden, 2010; O’Connell et al., 2010).
Enforced Expression of miR-125a Confers a Stem Cell-like Program to Human CD34+CD38+ Committed Progenitors and MPPs
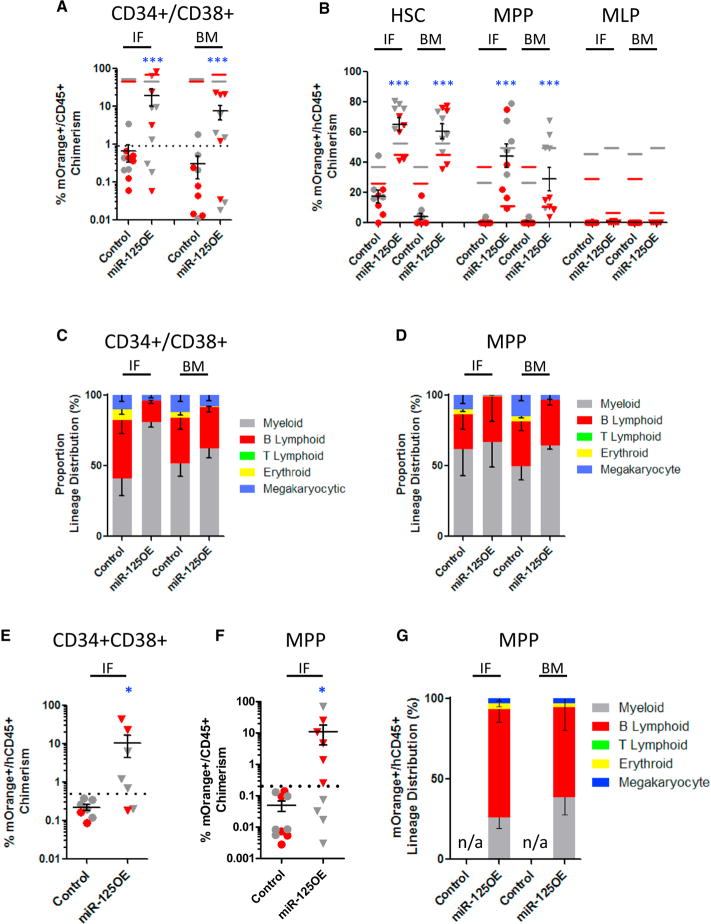
Given the robust increase in HSC frequency upon miR-125a expression, we asked whether more committed cell populations might also be endowed with enhanced self-renewal. We flow sorted CD34+CD38− compartment sub-populations (HSC, MPP, and MLP) and CD34+CD38+ committed progenitors, transduced them with miR-125OE or control vectors, and transplanted cells into immuno-deficient mice measuring repopulation after 12 weeks. miR-125a overexpression in CD34+ CD38+ progenitors produced a substantial graft that disseminated to distant BM locations after 12 weeks, whereas control transduced CD34+CD38+ cells generated a barely discernable graft (in one mouse recipient only) in the IF site (Figure 2A). As expected, only control transduced cells from sorted HSC (CD34+ CD38−CD45RA−CD90+CD49f+) generated a disseminating graft in recipient mice (Figure 2B). Similarly, sorted HSCs and MPPs (CD34+CD38−CD45RA−CD90−CD49f−) transduced with miR-125OE generated robust engraftment within the IFs and distant BM locations (Figure 2B). Sorted and transduced MLP (CD34+ CD38−CD45RA+) were unable to generate any detectable graft (Figure 2B). In all cases, xenografts generated by CD34+CD38+ and MPP transduced with miR-125OE showed multi-lineage repopulation (Figures 2C and 2D). To evaluate if the grafts derived from CD34+CD38+ committed progenitors and MPP were durable, we transplanted these cells into secondary recipients. Whereas control transduced progenitors did not generate a graft in secondary recipient mice, the CD34+CD38+ and MPP cells that overexpressed miR-125a generated a multi-lineage graft for 20 weeks (five out of seven and six out of ten mice, respectively; Figures 2E and 2F). Furthermore, MPP secondary grafts remained multi-lineage (Figure 2G), and in no case did we observe myeloproliferative disease or leukemia in mice engrafted with miR-125OE human CB cells. Finally, to evaluate the long-term self-renewal potential of miR-125a expressing MPP, we transplanted the secondary MPP grafts into tertiary recipients for another 10 weeks. After a total of 30 weeks in mice, we observed small but measurable multi-lineage grafts both from CD34+CD38− CB progenitors and MPP-expressing miR-125a (Figures S2A–S2D).
Figure 2. miR-125a-Enforced Expression Improves the Repopulating Abilities of MPPs and CD34+CD38+ Committed Progenitors.
(A) Human mO+ chimerism at 12 weeks post-transplantation with pre-sorted CD34+CD38+ committed progenitor compartment from human cord blood transduced with control or miR-125a expressing lentivectors. Red and gray symbols represent replicate experiments using two distinct human CB pools. Color-matched bars represent input levels of mOrange+ cells at time of transplant for each experiment.
(B) Human mO+ chimerism at 12 weeks post-transplantation with pre-sorted HSC, MPP, and MLP human cord blood populations transduced with control or miR-125a expressing lentivectors.
(C) Lineage distribution of mO+ BM grafts from transplanted CD34+CD38+ CB cells 12 weeks post-transplantation. (D) Lineage distribution of mO+ BM grafts from transplanted MPP 12 weeks post-transplantation.
(E) Human mO+ chimerism at 8 weeks post-secondary transplant from CD34+CD38+ progenitors from primary mouse recipients.
(F) Human mO+ chimerism at 8 weeks post-secondary transplant from MPP from primary mouse recipients.
(G) Lineage distribution of mO+ BM grafts from MPP secondary recipient mice.
To distinguish between rare contaminating HSCs or enhanced self-renewal of MPPs as the primary mechanism for the observed MPP phenotype in secondary and tertiary mice, we used a modified splinkerette PCR approach to assess the clonality of secondary miR-125OE MPP grafts. Our data demonstrate that miR-125OE MPP BM grafts were oligoclonal, with no lentivirus insertions near or within strong oncogenes (Table S1), suggesting that the enhanced self-renewal observed in miR-125OE MPP mice was not attributable to rare contaminating HSCs. Given that our data strongly suggests that MPPs are frequently endowed with enhanced self-renewal potential, we should be able to model this phenomenon at the clonal level. Using in vitro long-term culture initiating cell (LTC-IC) assays that were established with presorted transduced MPP (Table S2), we found that miR-125OE increased the LTC-IC frequency of transduced MPPs by ~4-fold compared to controls (1/11.9 versus 1/44.7, respectively, p = 0.0000000975), with a concomitant ~9-fold increase in colonies/LTC-IC (6.6 ± 2.1 versus 54.4 ± 11.6, p = 0.0041). Together, these data strongly suggest that the enhancement of self-renewal by enforced expression of miR-125 occurs not only in HSCs but also in MPPs and in an as-yet-unidentified population within the CD34+38+ committed progenitor compartment.
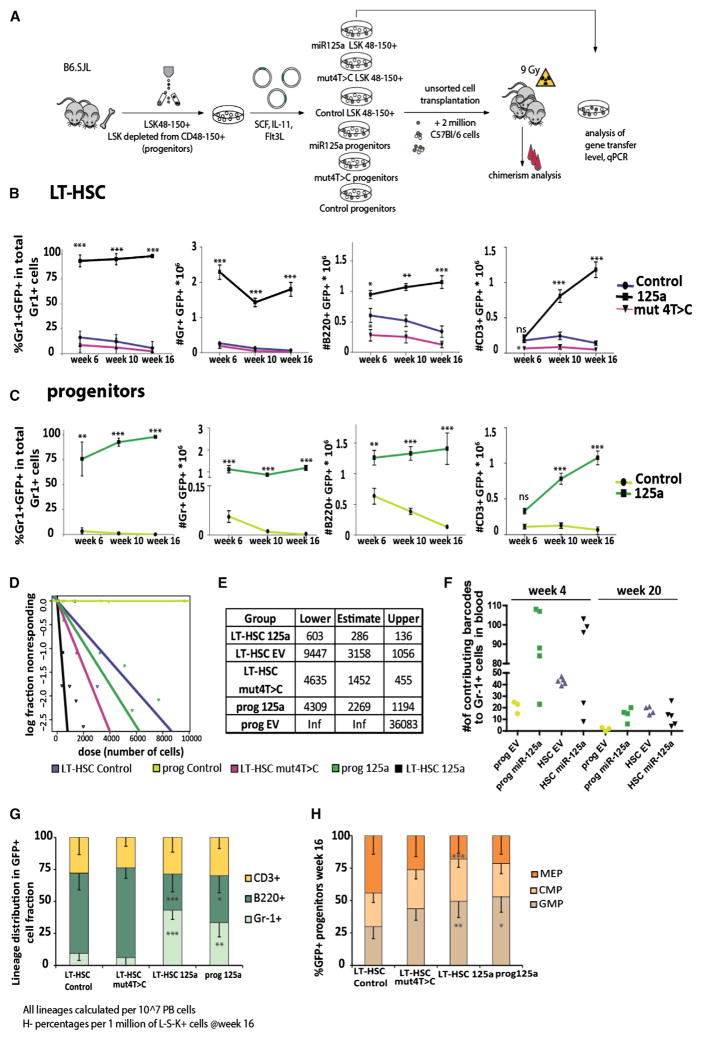
miR-125a Expression Prolongs Self-Renewal Activity in Murine MPPs
To assess whether this enhanced self-renewal induced by miR-125a is conserved in mice, we overexpressed miR-125a (Figure 3A) in Lin−Sca-1+c-Kit+ cells, depleted from CD150+CD48− cells (hereafter referred as progenitors) and in LT-HSCs (Lin− Sca-1+c-Kit+CD150+CD48−) (Figure S3A). We confirmed enforced overexpression by qPCR (Figure S3B) and performed a series of in vivo HSC assays. As controls, we included cells transduced with a vector expressing only GFP and a vector expressing a mutant form of miR-125a, where the fourth base of the seed sequence was modified (for a seed 5′-CCCTGAC-3′ miR-125mut4T→C) (Figure 3A). The mutant construct was as effectively expressed and processed as non-mutated miR-125a (Figure S3B). We competitively transplanted transduced cells in limiting dilution to lethally irradiated recipients together with a high dose of 2 × 106 fresh BM cells (Figure 3A) and monitored GFP+ chimerism over time. As expected, miR-125OE in LT-HSCs strongly increased their competitive reconstitution activity compared to controls, similar to what we and others have reported (Figure 3B) (Gerrits et al., 2012; Guo and Scadden, 2010; O’Connell et al., 2010; Ooi et al., 2010; Wojtowicz et al., 2014) and similar to our results using human cells. Control transduced progenitors (and the mutant construct; data not shown) were unable to provide long-term repopulation (Figure 3C). By contrast, miR-125OE progenitors generated robust, high levels of multi-lineage reconstitution up to 16 weeks post-transplantation (Figure 3C). Limiting dilution experiments demonstrated a 10-fold expansion of stem cell pool size (Figures 3D, 3E, and S4A). Importantly, miR-125OE progenitors sustained long-term blood cell production with an estimated frequency similar to control LT-HSCs (Figures 3D, 3E, and S4A). Using cellular barcoding (for gene transfer efficiency and cell doses, see Figure S4B), we observed a significant increase in the number of clones contributing to myeloid engraftment upon miR-125a overexpression, indicating that induction of stem cell activity in progenitors was not a rare event, and likely not from contamination of rare LT-HSC (Figure 3F). At week 20 post-transplantation, we observed a reduced number of clones compared to week 4, but overall, the numbers of contributing clones were comparable in all groups (Figure 3F). Similar to our human CB data, mouse 125OE progenitors were able to sustain multi-lineage potential with myeloid skewing represented by increased numbers of Gr-1+ cells (Figure 3G) and increased numbers of GMPs, at the expense of MEPs (Figure 3H).
Figure 3. miR-125a Overexpression in Mouse LT-HSC and Progenitors Increases the Self-Renewal Potential.
(A) Experimental setup.
(B) Chimerism levels in Gr-1+ cells in primary recipients transplanted with LT-HSCs transduced with miR-125mut4T→C (n = 7), LT-HSCs with control vector (n = 11), or LT-HSCs with miR-125a (n = 16). Panels also show the absolute cell numbers of B220+, CD3ε+, or Gr-1+ cells in 106 viable peripheral blood cells, assessed by FACS at indicated time points.
(C) Chimerism levels in recipients transplanted with progenitors overexpressing control vector (n = 17) or miR-125a (n = 9), measured by FACS at indicated time points.
(D) Stem cell frequencies in mice transplanted with virally transduced LT-HSCs or progenitor cell populations, measured by limiting dilution. Confidence intervals were calculated using ELDA software. The colors indicate the various experimental groups and are the same throughout all figures.
(E) The table containing the upper and lower confidence interval and estimated frequency of repopulating cells, calculated using ELDA.
(F) The number of detected barcodes in peripheral blood in Gr-1+ cells at 4 or 20 weeks post-transplantation.
(G) Lineage distribution in GFP+ peripheral blood cells.
(H) Progenitor (L−S−K+) distribution Lin−GFP+ BM cells.
Data in (E)–(H) reflect mean values ± SD. Statistical significance was assessed using a non-parametric Mann-Whitney test, where *p < 0.05, **p < 0.01, and ***p < 0.001.
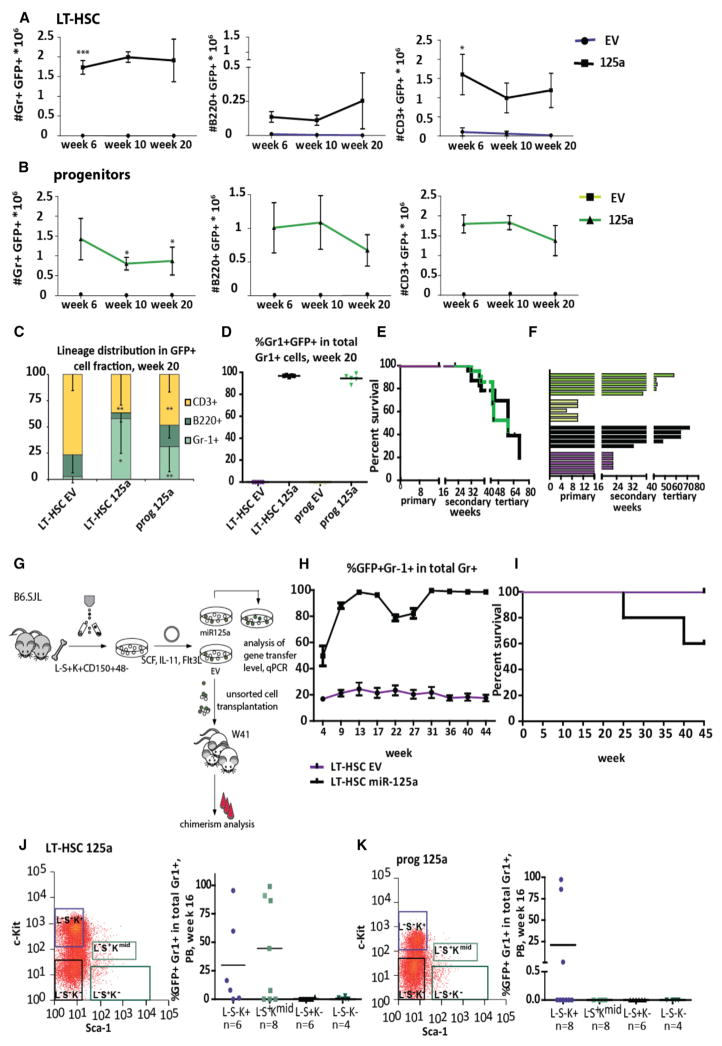
To assess the potential of miR-125a to induce self-renewal, we serially transplanted 107 BM cells from primary recipients into lethally irradiated secondary recipients. While the repopulating potential of control LT-HSCs in secondary recipients was restricted to short-term engraftment (up to 6 weeks, most probably due to the high dose of competitors used in the first round of transplantation), the secondary recipients transplanted with miR-125OE LT-HSCs or progenitors were highly reconstituted, with an increased frequency of myeloid cells 20 weeks post-transplantation in peripheral blood (Figures 4C and 4D). No lethality was observed in primary recipients. However, in secondary mice, we observed ~20% mortality, starting at week 32, in mice transplanted with either miR-125OE LT-HSC or progenitors. Necropsy revealed increased white blood cell (WBC) counts, splenomegaly, pale bones, and blast-like cells in the blood. Fluorescence-activated cell sorting (FACS) analysis of BM and spleen revealed dominance of EGFP+ cells of myeloid origin (Gr-1+ and/or Mac-1+, a MPN-like phenotype, as previously described; Gerrits et al., 2012; data not shown). In tertiary recipients, mortality further increased to 40% for 125OE progenitors, and 60% for 125OE LT-HSCs. Surviving mice in the tertiary transplantation showed no engraftment (Figure 4E). In addition, mice transplanted with 125OE progenitors exhibited an increased fraction of erythroid progenitors (Figures S5A and S5B). These data suggest that both LT-HSCs and progenitors overexpressing miR-125a remained functionally active for significantly longer periods of time compared to control cells (Figure 4F), and the cell type distribution of GFP+ cells in secondary recipients 20 weeks post-transplantation showed myeloid skewing in mice transplanted with LT-HSC overexpressing miR-125a.
Figure 4. Enforced miR-125a Ectopic Expression Sustains Repopulation Potential to MPPs in Serial Transplantation.
(A) The number of donor-derived GFP+ B220+, CD3ε+, or Gr-1+ cells per 106 peripheral blood cells at indicated time points in secondary recipients transplanted with LT-HSCs transduced with control vector (n = 5) or miR-125a (n = 5).
(B) The number of donor-derived GFP+ B220+, CD3ε+, or Gr-1+ cells per 106 peripheral blood cells at indicated time points in secondary recipients transplanted with progenitors transduced with control vector (n = 3) or miR-125a (n = 5). Data reflect mean values ± SEM. Statistical significance assessed using non-parametric Mann-Whitney test in case of Gr-1+ cells.
(C) Peripheral blood lineage distribution of GFP+ cells in secondary recipients.
(D) Peripheral blood Gr-1+ chimerism levels in secondary recipients 20 weeks post-transplantation.
(E) The lifespan of transduced cells (plotted mice were considered positive if the percentage of Gr-1+ was >1%; brakes indicate serial transplantations).
(F) The survival statistics of primary, secondary, and tertiary recipients transplanted with LT-HSCs or progenitors overexpressing miR-125a. Interruptions of the x axis indicate time points of serial transplantations.
(G) Representative FACS plot of the Lin−Sca-1+ c-kit+ compartment in BM cells from primary recipients transplanted with LT-HSCs overexpressing miR-125a. Right panel shows repopulation potential of indicated cell population assessed in secondary recipients. Donor cells were obtained from three mice.
(H) Same as (G), but for progenitors overexpressing miR-125a.
(I) Experimental setup for the second model, non-conditioned W41 mice used as recipients.
(J) The kinetics of chimerism changes in Gr-1+ cells in the course of experiment.
(K) The survival curve of non-irradiated W41 recipients transplanted with control or miR-125a-overexpressing LT-HSCs.
All data reflect mean values ± SD, except for (K), which shows mean values ± SEM. Statistical significance was assessed using a non-parametric Mann-Whitney test, where *p < 0.05, **p < 0.01, and ***p < 0.001.
We postulated that the myeloproliferative disease observed within 125OE mice could be due to a combination of miR-125-induced intrinsic changes and replicative stress (Figure 4E). To explore the role of replicative stress, we transplanted control and miR-125OE LT-HSCs into non-irradiated W41 recipients (Figure 4G). During the 11-month follow-up period, chimerism levels in 125OE LT-HSCs increased from 26% at transplantation reaching upward of 90% after 9 weeks post transplantation and remained very high. In contrast, the contribution of control LT-HSCs was stable over time, starting at 30% and slightly decreasing to 20% at week 45 (Figure 4H). In this non-irradiated model, we observed only ~40% mortality in mice transplanted with miR-125OE HSC, a 50% reduction compared to serial transplantation, where replicative stress is augmented (Figure 4I). These data suggest that replicative stress, due to irradiation and accompanying cytokine storm, contributes to the development of myeloproliferative disease in 125OE murine cells.
As recently reported, miR-125a overexpression leads to changes in expression of several HSC markers, including Sca-1, c-Kit, and CD150, precluding standard LT-HSC isolation from repopulated mice (O’Connell et al., 2010; Wojtowicz et al., 2014). To determine which cell population contained secondary repopulating potential, we harvested BM cells from reconstituted primary recipients and sub-fractionated GFP+ Lin− cells into four populations (Figures 4J and 4K). Sub-fractionated cells from primary recipients were transplanted with competitor cells into secondary recipients (see Figures 4J, 4K, and S5C for transplanted cell doses). Repopulating activity in miR-125-overexpressing cells was exclusively observed in L−S−K+ and L−S+Kmid cells derived from LT-HSC and in L−S−K+ cells if progenitors were used (Figures 4J and 4K). Taken together, these data show that miR-125a overexpression interferes with markers expression and cells with the repopulating potential are present in L−S−K+ and/or L−S+Kmid cell populations upon miR-125a overexpression.
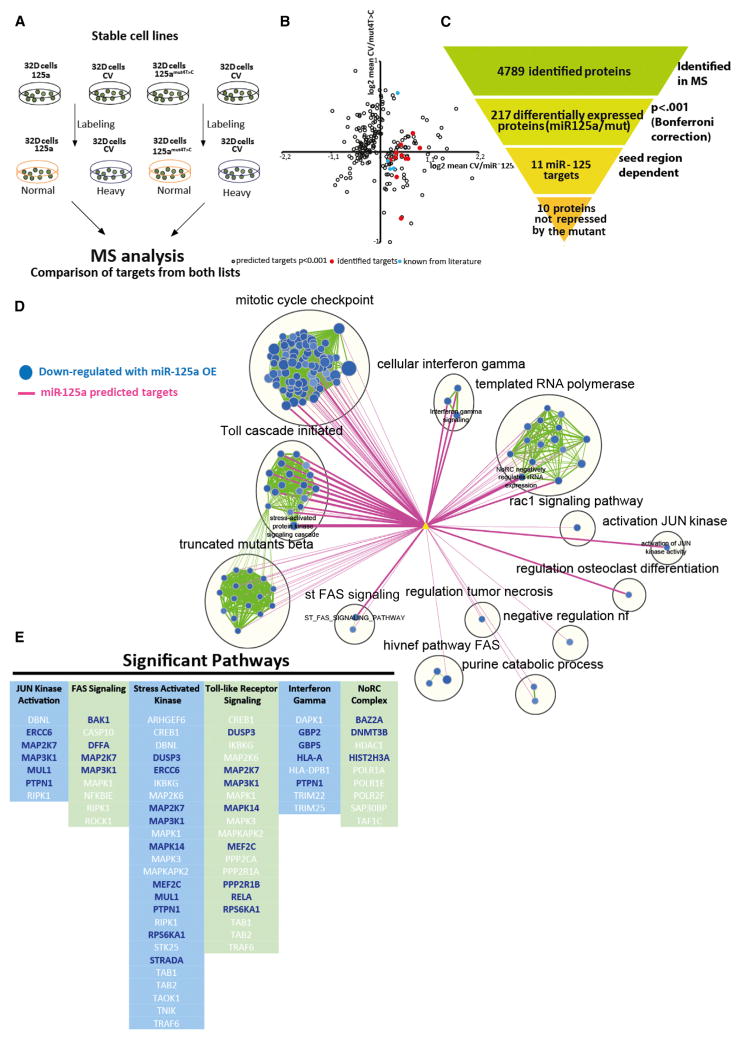
SILAC Reveals a miR-125a-Specific Network of Targets
To gain more insight in the molecular network of miR-125a targets, we performed stable isotope labeling by amino acids in cell culture (SILAC) proteomics in the 32D murine hematopoietic progenitor cell line (Ong et al., 2002; Ward et al., 1999). To do this, we expressed either miR-125a, miR-125mut4T→C or a control vector and cultured cells in media with isotope-labeled amino acids (Figure 5A). We detected a total of 4,789 expressed proteins in duplicate experiments when analyzing fold-change expression ratios, comparing cells expressing the control vector with those expressing miR-125a and control vector with 125mut4T→C samples. As the induction of a long-term repopulating program in HSPCs was largely seed sequence dependent, we excluded proteins that were similarly affected by both miR-125a and 125mut4T→C. This filtered out previously reported miR-125 targets such as Bak 1, Traf 6, Klf13, and Bmf (Guo and Scadden, 2010; O’Connell et al., 2010; Ooi et al., 2010) (Figure 5B). We identified 217 proteins that were differentially expressed upon miR-125a overexpression. Of these, 11 genes contained a 3′ UTR target sequence that is perfectly complementary to the seed sequence of miR-125 (Figure 5C); ten targets showed decreased protein levels upon miR-125a overexpression in our cell system and were therefore considered to be direct miR-125 targets. These targets include p38 kinase, a negative HSC self-renewal regulator (Ito et al., 2006), and its inhibition leads to ex vivo HSC expansion (Wang et al., 2011), three protein tyrosine phosphatases (Ptpn1, Ptpn7, and Smek1, all known to suppress cytokine signaling and negatively regulate MAP kinases), a ubiquitin ligase (Syvn1, shown to block differentiation of embryonic stem cells; Yagishita et al., 2005), and Vps4b and M6pr, which are involved in transport of protein to vesicles and their lysosomal degradation. In addition, we identified interleukin-16 (IL-16) and Txnrd1, the latter gene playing a role in protection against oxidative stress (Peng et al., 2014).
Figure 5. Molecular Mechanism of Sustained Self-Renewal Potential in Human and Mouse MPPs.
(A) SILAC experimental setup. 32D cells were transduced with control vector, miR-125a, or 125mut4T →C. Cells were cultured in medium containing normal (miR-125a OE or miR-125mut4T→C samples) or heavy labeled amino acids (empty vector [EV]), as indicated.
(B) Graphical representation of 217 differentially expressed proteins. Blue symbols indicate previously reported targets of the miR-125 family. Red symbols indicate the ten differentially expressed seed-sequence-containing proteins in miR-125a and 125mut4T→C samples.
(C) Overview of filtering criteria.
(D) Functional enrichment map for protein MS-based expression revealing miR-125a modulated pathways in human CD34+ CB cells. Only the subset of pathways that were found to have a significant overlap with miR-125a targets is shown. Blue nodes (circles) represent gene sets enriched in proteins downregulated in CD34+ human cord blood cells overexpressing miR-125a. Green line (edge) width between nodes corresponds to the number of shared proteins. Predicted miR-125a targets (yellow triangle) are connected to enriched pathways by pink edges and edge width is proportional to the overlap significance (Mann-Whitney [lesser] proteomics p = < 0.01).
(E) Top six pathways significantly downregulated by miR-125a expression. Columns represent individual pathways, with the protein names listed vertically underneath. Protein names in blue are predicted miR-125a targets.
To gain further insight into the mechanism whereby miR-125a was exerting its biological effects on human HSPCs, we performed label-free protein mass spectrometry on 100,000 sorted mO+CD34+ human cord blood (huCB) HSPCs transduced with miR-125a or control vector. We were able to reliably quantify 6,687 protein groups (Figure S5D), and 517 of them showed a significant change (p < 0.05) between 125OE and control samples (Table S3). Gene set enrichment analysis (GSEA) of the proteomics dataset identified pathways and leading edge genes directly targeted by miR-125a. In post-analysis, predicted miR-125a targets were correlated with proteomic-modulated pathways (Figure 5D). The most significant pathways centered on JUN kinase activation, FAS signaling, stress-activated kinase signaling, Toll-like receptor signaling, interferon gamma signaling, and the NoRC complex, an epigenetic pathway involved in heterochromatin formation (Figures 5D and 5E). These data suggest that miR-125 exerts its effects on HSPCs by downregulating several genes within multiple pathways. Our data also confirm that P38MAPK14 and PTPN1 are miR-125a targets in both murine and human HSPCs.
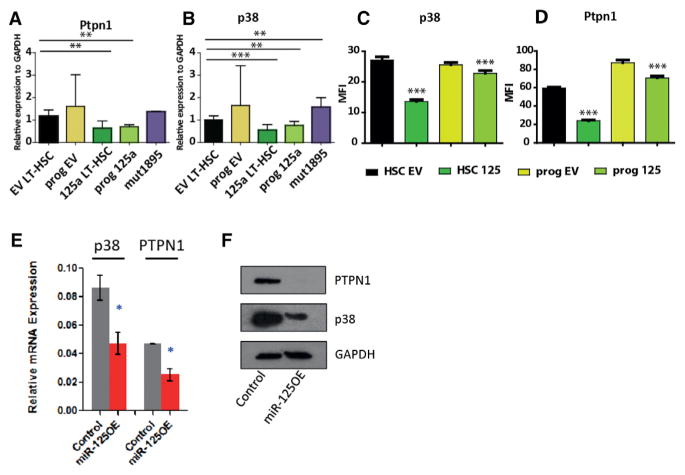
To validate identified targets, we performed nanofluidic proteomic analysis and confirmed the suppression of p38, Ptpn1 and Ptpn7, and Vps4b in cell lines used for SILAC (Figure S6A). To assess the downregulation of miR-125a targets in human CD34+ and mouse LT-HSCs and progenitors overexpressing miR-125a, we measured the expression of p38, Ptpn1, and Ptpn7, as well as Smek1 transcripts, by qPCR. We confirmed that all tested targets showed decreased expression compared to controls in both mouse and human cells (Figures 6A, 6B, and 6E). We were also able to verify the repression of these targets at the protein level by western blot or immunofluorescence in transduced human CD34+ CB cells or sorted and transduced mouse LT-HSCs and progenitors (Figures 6C, 6D, and 6F). Together, these data suggest that the endowment of an HSC-like self-renewal program in hematopoietic progenitors that normally lack self-renewal capacity upon miR-125a overexpression is linked to repression of genes controlling the p38/mitogen-activated protein kinase (MAPK) pathway as well as several other pathways. Furthermore, our results reinforce the notion that miR-125a exerts similar functions in mouse and human cells.
Figure 6. Target Validation.
(A–D) Validation of selected targets in mouse transduced LT-HSC or progenitors at the transcript level (A and B) and protein level (C and D).
(E and F) Repression of miR-125a targets in human CD34+ cells at the transcript level (E) and protein level (F).
All data reflect mean values ± SEM. Statistical significance was assessed using a non-parametric Mann-Whitney test, where *p < 0.05, **p < 0.01, and ***p < 0.001.
DISCUSSION
Our study provides key insights into the powerful role that miR-NAs can play in the regulation of HSC expansion/self-renewal properties. Previous work demonstrated expansion of the HSC pool upon miR-125 expression (Ooi et al., 2010). Here, we establish that miR-125a can endow repopulation potential to progenitor populations and potently increase their self-renewal capacity. Our findings highlight the possibility of generating MPPs with enhanced self-renewal to augment limited sources of HLA-matched CB-derived HSCs to improve transplant outcomes. As CB-transplant-related mortality is influenced by low numbers of infused HSCs, rapid in vitro expansion of CB progenitors could increase the transplanted dose of stem cells, thus accelerating engraftment and reducing complications. Although it may be possible to therapeutically employ miR-125a to confer MPPs with self-renewal capacity, it is more likely that small-molecule regulation of several critical miR-125a targets will be required to fully translate our work. In the future, combinatorial approaches combining miR-125a modified MPPs with existing small-molecule compounds like PGE2, SR1, and UM171 (Boitano et al., 2010; Fares et al., 2014; North et al., 2007) may support expansion of undersized HLA-matched CB units, allowing them to become a selected source of cells for transplantation.
The development of myeloproliferative disease/leukemia upon miR-125a overexpression in murine cells in secondary and tertiary recipients is in agreement with previous reports (Bousquet et al., 2010; Gerrits et al., 2012; O’Connell et al., 2010) and has been shown to be dependent upon sustained expression of miR-125a (Guo et al., 2012) and dosage (O’Connell et al., 2008, 2010). Our data suggest the miR-125a-induced myeloproliferative disease occurs, in part, as a function of replicative stress. We surmise that with careful titration of miR-125 levels, it may be possible to exploit the beneficial self-renewal effects of miR-125 without incurring the malignant side effects. Interestingly, overt malignancy was never observed in human xenografts through three serial transplantations (30 weeks total), suggesting that the inherent susceptibility of murine cells to oncogenic transformation may contribute to the development of (pre-) malignancy (Balmain and Harris, 2000; Rangarajan et al., 2004). We cannot completely discount the possibility that vector integration contributed to malignant transformation. However, we deem this unlikely, considering the reproducibility of the acquired phenotypes and the absence of any known strong oncogenes in insertion site analysis. In addition, our data on cellular barcoding confirm that mice transplanted with LT-HSCs or progenitors overexpressing miR-125a had polyclonal repopulation similar to control LT-HSC, excluding the possibility that engraftment results from clonal dominance of a single clone.
We found similar behavior between control HSCs and miR-125a-transduced MPPs regarding their long-term multi-lineage repopulating ability, self-renewal potential, and lineage distribution in transplanted recipients, bothinmouse and human systems. This similarity points to a common mechanism whereby miR-125a functions to govern stem cell properties. Using proteomic analysis, we uncovered a subset of the relevant miR-125a-targeted pathways. Our study predicts that several of our identified targets should normally act to repress self-renewal in cells downstream of HSCs. Of the ten identified targets in murine cells, four are involved in the MAPK signaling pathway. The best-known and most-studied candidate protein is p38. Chemical inhibitors of p38 activity have previously shown efficacy in ex vivo mouse and human HSC expansion protocols (Baudet et al., 2012; Wang et al., 2011). Another candidate target is PTPN1, of which little is known in the hematopoietic system. Loss of PTPN1 leads to increased numbers of pro- and pre-B cells in the BM (Dubé et al., 2005), similar to previous reports showing increased numbers of pro-B cells upon miR-125b overexpression (Chaudhuri et al., 2012) and correlating with our human CB xenotransplantation data. Moreover, deletion of PTPN1 in fibroblasts leads to suppression of Erk signaling (Dubé et al., 2004) and increased insulin growth factor-1 (IGF-1)-induced AKT protein kinase B activity (AKT/PKB) (Buckley et al., 2002). Constitutive activation of AKT in HSPCs has been shown to deplete B lymphocytes and induce myeloid skewing and the development of myeloproliferative disorders that progressed to leukemia (Helgason et al., 1998; Zhang et al., 2006). We speculate that PTPN1 downregulation may similarly activate AKT in HSPCs, resulting in myeloid skewing and a block in B cell differentiation. However, the exact direct and indirect contribution of each individual miR-125a target in moderating self-renewal activity within progenitors requires additional studies. Multiple targets within several pathways are probably required to fully recapitulate the effects of miR125a on MPPs.
Irrespective of the precise mechanism of action, our findings provide clear evidence that miR-125a confers hematopoietic stem cell potential to cells that are normally devoid of this activity. Our results open the prospect of searching for other regulators that have this capability laying the foundation for utilizing progenitors in clinically relevant therapeutics. Molecular identification and functional evaluation of the full complement of miR-125a targets may allow the dissection of targets inhibiting self-renewal and distinguishing them from targets that may be closely aligned with malignancy. Furthermore, small-molecule screening and candidate identification on hematopoietic progenitor populations using induction of miR-125a as an operational readout could circumvent the requirement for virally enforced miRNA expression and accelerate the clinical application of miR-125a for CB transplantation.
EXPERIMENTAL PROCEDURES
Cell Sorting
Human Lin− CB cells were thawed and stained with CD34-APC-Cy7 and CD38-PE-Cy7 (BD). For HSC, MPP, and MLP sorts, Lin− CB cells were stained with CD34, CD38, CD49f, CD45RA, CD90, CD7, and CD10. Murine BM cells were isolated by crushing and then stained with a cocktail of antibodies against lineage markers (Ter119, Gr-1, Mac-1, CD3, and B220), c-Kit, Sca-1, CD48, and CD150 and sorted for LT-HSCs (Lin−Sca-1+c-Kit+CD150+48−) and progenitors (Lin−Sca-1+c-Kit+ depleted from CD150+48− cells).
Lentiviral and Retroviral Constructs and Transduction
For the human part of the study, the lentiviral vector for ectopic miRNA expression (gain of function) has been described previously (Gentner et al., 2010). In the murine part of the study, we used a retroviral vector, miR-125a, and mut 4T→C cells were cloned as previously described (Gerrits et al., 2012; Wojtowicz et al., 2014).
NSG Repopulation Assay
Male NSG mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ; Jackson Laboratory) were sublethally irradiated (225 cGy) 24 hr before intrafemoral injection. Competitive transplantation was performed using the equivalent of 400 HSCs, 400 MPPs, and 1,000 MLP cells after overnight culture in low-cytokine conditions. Limiting dilution analysis of human CD34+/CD38− CB cell was performed a secondary mouse was scored as positive if it had >0.5% hCD45+, mOrange+ BM engraftment 8 weeks post transplantation. HSC frequency was estimated using ELDA software (http://bioinf.wehi.edu.au/software/elda/; Hu and Smyth, 2009).
C57BL/6 (B6) mice were purchased from Harlan Laboratories. C57BL/6.SJL mice were bred at the Central Animal Facility of University Medical Centre of Groningen (Groningen, the Netherlands). All animal experiments were approved by the University of Groningen Animal Care Committee.
Lentiviral Insertion Site Analysis by Modified Splinkerette PCR
To determine the lentiviral integration sites we used a splinkerette PCR approach (Uren et al., 2009) that we modified to be non-restrictive and have increased sensitivity for low DNA input amounts (see Supplemental Experimental Procedures). For identified integration sites in MPP-engrafted mice, see Table S1.
Cellular Barcoding
Cells were isolated and transduced as described above (see also Verovskaya et al., 2013). We transplanted 15,760 GFP+ progenitor cells per mouse and 4,300 GFP+ LT-HSCs overexpressing miR-125a, 39,800 GFP+ progenitors, or 16,200 GFP+ LT-HSCs for controls.
Hematopoietic Cell Transplantation and Blood Analysis
For limiting dilution experiments, cells were injected in given doses (Figure S3A) into lethally irradiated (9 Gy) female B6 recipients together with 2 × 106 fresh BM B6 cells. Blood samples were taken every 4–6 weeks and stained with antibodies against CD45.1 and CD45.2 to determine the donor chimerism and CD3ε, Gr-1, and B220 for FACS analysis. HSC frequency was estimated using ELDA software (http://bioinf.wehi.edu.au/software/elda/).
Serial BM Transplantation
Primary recipients were sacrificed at 16 weeks post-transplantation for BM collection prior to secondary transplantation. 107 whole BM cells were transplanted into lethally irradiated secondary recipients. For tertiary recipients, donors were sacrificed 24 weeks post-secondary transplantation, and 5 × 106 whole BM cells were injected together with 5 × 105 fresh competitor cells from B6.
qPCR
RNA was extracted from 25,000 to 30,000 GFP+ cells using the miRNeasy Kit (QIAGEN), and commercially available TaqMan probes for miRNA qPCR or primers (Table S4) were used according to the manufacturer’s protocol.
Label-free Quantitative Proteomics Analysis
The raw files were analyzed using MaxQuant version 1.5.2.8 (Cox and Mann, 2008) and standard settings. For differentially expressed proteins, see Table S3. For sample preparation details, see Supplemental Experimental Procedures.
Quantitative Mass Spectrometry: SILAC
The murine 32D cell line was used for to identify miR-125a targets. After transduction, GFP+ cells were FACS sorted and expanded prior to stable isotope labeling as previously reported (Meenhuis et al., 2011). A nano-high performance liquid chromatography (HPLC) system consisting of Easy-nLC 1000 and Q-Exactive (Thermo Fisher Scientific) was used.
Statistical Analysis
Unless otherwise stated, mean ± SEM values are shown in the graphs. For pairwise comparison, a Mann-Whitney U test was applied. Statistical analysis was performed using PRISM 6 (GraphPad Software). For proteomics data analysis, Student’s t test with Bonferroni correction was applied.
Western Blot Analysis
For immunoblotting, proteins were transferred to PVDF membranes, incubated with the specific antibody (anti-p38, Cell Signaling #9212 1:1,000; anti-PTPN1, LifeSpan BioSciences #LS-C61924 1:500; and mouse anti-GAPDH, 1:10,000, Sigma) followed by peroxidase-conjugated secondary antibodies. Bands were visualized on Amersham Hyperfilm (GE Healthcare).
Nanofluidic Proteomic Analysis
The expression levels of p38 in transduced GFP+ 32D cells used for SILAC or in LT-HSCs and progenitors overexpressing miR-125a were investigated by a size-based Simple western immunoassay, using a WES device (Protein Simple).
Immunofluorescence
To measure protein expression levels in transduced GFP+ LT-HSC or progenitors (Lin-Sca-1+c-Kit+ depleted from CD150+ 48- cells), cells were stained with antibodies directed against p38 (1:50, Cell Signaling, #9212) and Ptpn1 (1:100, Millipore, clone# ABS40). We used rabbit-specific IgG conjugated with Alexa-633 (Molecular Probes, Invitrogen) as a secondary antibody. For nuclear staining we treated cells with DAPI (Sigma).
Supplementary Material
Highlights.
Enforced expression of miR-125 increases hematopoietic stem frequency in vivo
miR-125 induces stem cell potential in murine and human progenitor cells
miR-125 represses, among others, targets of the MAP kinase signaling pathway
miR-125 function and targets are conserved in human and mouse
Acknowledgments
The authors thank H. Moes, G. Mesander, and R. J. van der Lei for expert cell-sorting assistance; Klaas Sjollema from the UMCG Microscopy and Imaging Center (UMIC) for advice; and E. Verovskaya and R.P. van Os for discussions and assistance in the laboratory. The authors also thank the obstetrics units of Trillium Health Partners (Missisauga and Credit Valley sites) for the cord blood units and the UHN/Sick Kids and PMH Flow Cytometry Facilities for cell sorting. We would also like to thank Dr. K. Itoh (Kyoto University, Japan) for kindly providing MS-5 stromal cells.
This work was supported by a Rubicon fellowship from the Netherlands Organization for Scientific Research (to K.G.H.), a Canadian Institutes for Health Research (CIHR) fellowship (to K.G.H.), and grants from Netherlands Organization for Scientific Research (to G.d.H), EuroCSCTraining Eurocancer Stem-cell Training Network ITN-FP7-Marie Curie Action 264361 (to E.E.W.), and the Netherlands Institute for Regenerative Medicine. E.M.S. is an EMBO Postdoctoral Fellow (ALTF 1595-2014) and is co-funded by the European Commission (LTFCOFUND2013, GA-2013-609409) and Marie Curie Actions. The work in J.E.D.’s lab is funded by the CIHR, Canadian Cancer Society Research Institute, Terry Fox Foundation, Genome Canada through the Ontario Genomics Institute, Ontario Institute for Cancer Research, a Canada Research Chair, the Princess Margaret Hospital Foundation, and the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed in this manuscript do not necessarily reflect those of the OMOHLTC.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2016.06.008.
AUTHOR CONTRIBUTIONS
E.E.W., K.G.H., E.R.L., L.V.B., J.E.D., and G.d.H. designed research; E.E.W., M.J.C.B., E.W., and M.R. performed mouse experiments; K.G.H., E.R.L., A.T.-G., S.M.D., G.K., J.E., J.K., E.W., and O.I.G. performed human experiments; P.A.v.V., G.M.C.J., M.F.A., H.W.J.d.L., S.J.E., G.K., R.I., E.M.S., and G.D.B. performed proteomics and analyzed MS data; E.E.W., E.R.L., and L.V.B. analyzed and interpreted data; and E.E.W., E.R.L., L.V.B, J.E.D., and G.d.H. wrote the manuscript.
References
- Aljurf M, Rizzo JD, Mohty M, Hussain F, Madrigal A, Pasquini MC, Passweg J, Chaudhri N, Ghavamzadeh A, Solh HE, et al. Challenges and opportunities for HSCT outcome registries: perspective from international HSCT registries experts. Bone Marrow Transplant. 2014;49:1016–1021. doi: 10.1038/bmt.2014.78. [DOI] [PubMed] [Google Scholar]
- Ballen KK, Gluckman E, Broxmeyer HE. Umbilical cord blood transplantation: the first 25 years and beyond. Blood. 2013;122:491–498. doi: 10.1182/blood-2013-02-453175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmain A, Harris CC. Carcinogenesis in mouse and human cells: parallels and paradoxes. Carcinogenesis. 2000;21:371–377. doi: 10.1093/carcin/21.3.371. [DOI] [PubMed] [Google Scholar]
- Barker JN, Weisdorf DJ, Wagner JE. Creation of a double chimera after the transplantation of umbilical-cord blood from two partially matched unrelated donors. N Engl J Med. 2001;344:1870–1871. doi: 10.1056/NEJM200106143442417. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet A, Karlsson C, Safaee Talkhoncheh M, Galeev R, Magnusson M, Larsson J. RNAi screen identifies MAPK14 as a druggable suppressor of human hematopoietic stem cell expansion. Blood. 2012;119:6255–6258. doi: 10.1182/blood-2012-01-403949. [DOI] [PubMed] [Google Scholar]
- Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DA, Cheng A, Kiely PA, Tremblay ML, O’Connor R. Regulation of insulin-like growth factor type I (IGF-I) receptor kinase activity by protein tyrosine phosphatase 1B (PTP-1B) and enhanced IGF-I-mediated suppression of apoptosis and motility in PTP-1B-deficient fibroblasts. Mol Cell Biol. 2002;22:1998–2010. doi: 10.1128/MCB.22.7.1998-2010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri AA, So AY, Mehta A, Minisandram A, Sinha N, Jonsson VD, Rao DS, O’Connell RM, Baltimore D. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc Natl Acad Sci USA. 2012;109:4233–4238. doi: 10.1073/pnas.1200677109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- Dubé N, Cheng A, Tremblay ML. The role of protein tyrosine phosphatase 1B in Ras signaling. Proc Natl Acad Sci USA. 2004;101:1834–1839. doi: 10.1073/pnas.0304242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé N, Bourdeau A, Heinonen KM, Cheng A, Loy AL, Tremblay ML. Genetic ablation of protein tyrosine phosphatase 1B accelerates lymphomagenesis of p53-null mice through the regulation of B-cell development. Cancer Res. 2005;65:10088–10095. doi: 10.1158/0008-5472.CAN-05-1353. [DOI] [PubMed] [Google Scholar]
- Fares I, Chagraoui J, Gareau Y, Gingras S, Ruel R, Mayotte N, Csaszar E, Knapp DJ, Miller P, Ngom M, et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science. 2014;345:1509–1512. doi: 10.1126/science.1256337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gentner B, Visigalli I, Hiramatsu H, Lechman E, Ungari S, Giustacchini A, Schira G, Amendola M, Quattrini A, Martino S, et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci Transl Med. 2010;2:58ra84. doi: 10.1126/scitranslmed.3001522. [DOI] [PubMed] [Google Scholar]
- Gerrits A, Walasek MA, Olthof S, Weersing E, Ritsema M, Zwart E, van Os R, Bystrykh LV, de Haan G. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood. 2012;119:377–387. doi: 10.1182/blood-2011-01-331686. [DOI] [PubMed] [Google Scholar]
- Guo S, Scadden DT. A microRNA regulating adult hematopoietic stem cells. Cell Cycle. 2010;9:3637–3638. doi: 10.4161/cc.9.18.13174. [DOI] [PubMed] [Google Scholar]
- Guo S, Bai H, Megyola CM, Halene S, Krause DS, Scadden DT, Lu J. Complex oncogene dependence in microRNA-125a-induced myeloproliferative neoplasms. Proc Natl Acad Sci USA. 2012;109:16636–16641. doi: 10.1073/pnas.1213196109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason CD, Damen JE, Rosten P, Grewal R, Sorensen P, Chappel SM, Borowski A, Jirik F, Krystal G, Humphries RK. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, Ohmura M, Naka K, Hosokawa K, Ikeda Y, Suda T. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, Hiramatsu H, Restuccia U, Bachi A, Voisin V, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11:799–811. doi: 10.1016/j.stem.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, Sun SM, Taskesen E, Stern P, de Ru AH, van Adrichem AJ, et al. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 2011;118:916–925. doi: 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf D. Hematopoietic cytokines. Blood. 2008;111:485–491. doi: 10.1182/blood-2007-03-079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci USA. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M. Differentiation and proliferation of hematopoietic stem cells. Blood. 1993;81:2844–2853. [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci USA. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oran B, Shpall E. Umbilical cord blood transplantation: a maturing technology. Hematology (Am Soc Hematol Educ Program) 2012;2012:215–222. doi: 10.1182/asheducation-2012.1.215. [DOI] [PubMed] [Google Scholar]
- Peng X, Mandal PK, Kaminskyy VO, Lindqvist A, Conrad M, Arnér ES. Sec-containing TrxR1 is essential for self-sufficiency of cells by control of glucose-derived H2O2. Cell Death Dis. 2014;5:e1235. doi: 10.1038/cddis.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineault N, Abu-Khader A. Advances in umbilical cord blood stem cell expansion and clinical translation. Exp Hematol. 2015;43:498–513. doi: 10.1016/j.exphem.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species-and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Sauvageau G, Iscove NN, Humphries RK. In vitro and in vivo expansion of hematopoietic stem cells. Oncogene. 2004;23:7223–7232. doi: 10.1038/sj.onc.1207942. [DOI] [PubMed] [Google Scholar]
- Uren AG, Mikkers H, Kool J, van der Weyden L, Lund AH, Wilson CH, Rance R, Jonkers J, van Lohuizen M, Berns A, Adams DJ. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat Protoc. 2009;4:789–798. doi: 10.1038/nprot.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verovskaya E, Broekhuis MJ, Zwart E, Ritsema M, van Os R, de Haan G, Bystrykh LV. Heterogeneity of young and aged murine hematopoietic stem cells revealed by quantitative clonal analysis using cellular barcoding. Blood. 2013;122:523–532. doi: 10.1182/blood-2013-01-481135. [DOI] [PubMed] [Google Scholar]
- Wagner JE, Jr, Eapen M, Carter S, Wang Y, Schultz KR, Wall DA, Bunin N, Delaney C, Haut P, Margolis D, et al. Marrow Transplant Clinical Trials Network. One-unit versus two-unit cord-blood transplantation for hematologic cancers. N Engl J Med. 2014;371:1685–1694. doi: 10.1056/NEJMoa1405584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kellner J, Liu L, Zhou D. Inhibition of p38 mitogen-activated protein kinase promotes ex vivo hematopoietic stem cell expansion. Stem Cells Dev. 2011;20:1143–1152. doi: 10.1089/scd.2010.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AC, Smith L, de Koning JP, van Aesch Y, Touw IP. Multiple signals mediate proliferation, differentiation, and survival from the granulocyte colony-stimulating factor receptor in myeloid 32D cells. J Biol Chem. 1999;274:14956–14962. doi: 10.1074/jbc.274.21.14956. [DOI] [PubMed] [Google Scholar]
- Wojtowicz EE, Walasek MA, Broekhuis MJ, Weersing E, Ritsema M, Ausema A, Bystrykh LV, de Haan G. MicroRNA-125 family members exert a similar role in the regulation of murine hematopoiesis. Exp Hematol. 2014;42:909–18. e1. doi: 10.1016/j.exphem.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Yagishita N, Ohneda K, Amano T, Yamasaki S, Sugiura A, Tsuchimochi K, Shin H, Kawahara K, Ohneda O, Ohta T, et al. Essential role of synoviolin in embryogenesis. J Biol Chem. 2005;280:7909–7916. doi: 10.1074/jbc.M410863200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.