Abstract
Protein phosphorylation is an important cellular regulatory mechanism as many enzymes and receptors are activated/deactivated by phosphorylation and dephosphorylation events, by means of kinases and phosphatases. In particular, the protein kinases are responsible for cellular transduction signaling and their hyperactivity, malfunction or overexpression can be found in several diseases, mostly tumors. Therefore, it is evident that the use of kinase inhibitors can be valuable for the treatment of cancer. In this review, we discuss the mechanism of action of phosphorylation, with particular attention to the importance of phosphorylation under physiological and pathological conditions. We also discuss the possibility of using kinase inhibitors in the treatment of tumors.
Keywords: protein phosphorylation, kinase, phosphatase, phospho-signaling networks, cancer, drug target
1. Introduction
Protein phosphorylation is one of the most common and important post-translational modifications (PTMs) (1,2). This reversible mechanism occurs through protein kinases and consists of the addition of a phosphate group (PO4) to the polar group R of various amino acids. Consequently, this addition modifies the protein from hydrophobic apolar to hydrophilic polar, allowing the protein to change conformation when interacting with other molecules. A phosphorylated amino acid can bind molecules able to interact with other proteins and consequently assemble and detach proteic complexes (3).
The interactive capacity of the phosphate group is mainly due to its components. One of its main elements is phosphorus. It has five outer electrons able to form a maximum of five covalent bonds, has three pKas, high water solubility and it can form, for its versatility, mono, di and trialkyl and aryl esters with hydroxyl groups, but also acid anhydrides (4).
In particular, many cellular phosphate esters are phosphoproteins that form, via a catalytic enzyme and adenosine triphosphate (ATP), a phosphate anhydride, acting as a donor of a phosphate group.
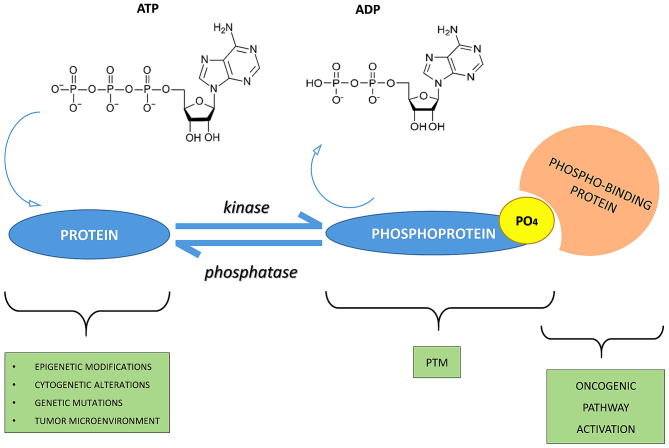
A good energy balance also favors phosphorylation. Indeed, there is a constant balance between phosphorylation and dephosphorylation events mediated by kinases, phosphatases, ATP and/or ADP (protein + ATP ⇄ phosphoprotein + ADP) (5,6) (Fig. 1).
Figure 1.
Phospho-signaling networks. The mechanism of phosphorylation regulation consists of kinases, phosphatases and their substrates phospho-binding proteins. For example, phosphorylation is activated by stimuli such as epigenetic modifications, cytogenetic alterations, genetic mutations or the tumor micro-environment. Consequently, the protein receives a phosphate group by adenosine triphosphate (ATP) hydrolysis and due to enzymatic activity of kinase. This is the mechanism for the basis of post-translational modification (PTM) formation. In addition, phosphorylation is a reversible process due to activity of phosphatase. Phosphorylation and dephosphorylation are a molecular switch and, in particular, a PTM can cause oncogenic pathway activation by a phospho-binding protein that bind to the phosphate group of a phosphoprotein.
The Cell Signaling Technology PhosphoSitePlus (www.phosphosite.org) and the Kinexus PhosphoNET (www.phosphonet.ca) websites both list over 200,000 known human phosphosites, and the Kinexus website predicts another 760,000 additional sites that are likely to be phosphorylated. More than two-thirds of the 21,000 proteins encoded by the human genome has been shown to be phosphorylated, and it is likely that more than 90% are actually subjected to this type of PTM.
More than one-third of the protein phosphorylation events occurs on serine (Ser or S), threonine (Thr or T), and tyrosine residues (Tyr or Y) (O-phosphorylation) (7). In particular, the phosphorylated residues of serine are 86.4%, followed by residues of threonine 11.8% whereas only 1.8% of tyrosine residues are phosphorylated (8,9). Tyrosine phosphorylation is relatively rare compared to the other PTMs and is typical of the epidermal growth factor receptor (EGFR) family, which owns a domain called, precisely, tyrosine kinase. Sometimes, phosphorylation of histidine (His or H) and aspartate residues (Asp or D) (N-phosphorylation) also occurs, but, in both cases, this phosphorylation is less stable than others.
Protein phosphorylation is a mechanism of regulation that is extremely important in most cellular processes such as protein synthesis, cell division, signal transduction, cell growth, development and aging as many enzymes and receptors are activated and deactivated via phosphorylation/dephosphorylation events due to specific kinases and phosphatases (10).
The human genome, in fact, includes approximately 568 protein kinases and 156 protein phosphatases that regulate phosphorylation events and, therefore, play an important role in the control of biological processes such as proliferation, differentiation and apoptosis.
For instance, p53 protein is activated by phosphorylation and is then able to stimulate transcription of genes to inhibit the cell cycle, activate DNA repair and in some cases lead to apoptosis (13). An imbalance in the mechanism of phosphorylation/dephosphorylation of the p53 protein can lead to a chronic inactivation of the protein itself, which in turn can transform the cell into a cancer cell.
2. Protein kinases
The protein kinases belong to the great family of kinases and are responsible for the mechanism of phosphorylation. They are activated by phosphorylation which in turn activates a cascade of events leading to the phosphorylation of different amino acids (3). Activation or deactivation of kinase occurs in different ways: through the kinase itself with a cis-phosphorylation/autophosphorylation, by binding with activator or inhibitor proteins or checking their localization in the cell in relation to their substrate (7).
The catalytic domain of protein kinase has 2 subdomains, N- and C-terminal (8). Both are connected by a peptidic stand, which forms an active site with a front pocket (catalytic residues) and a back pocket. Access to the rear pocket is controlled by a conserved lysine residue and a residue 'gatekeeper'. The catalytic domain is unavailable when it is active because propellers of the N- and C-terminal subdomains rotate inward. The activation of the catalytic domain occurs through phosphorylation of the activation loop or through an allosteric mechanism (8). Moreover, the kinases also have non-catalytic domains allowing the attachment of substrates and the recruitment of other signaling proteins (9).
Up to 30% of all human proteins may be modified by kinase activity, and kinases are known to regulate the majority of cellular pathways, especially those involved in signal transduction (10).
In the last few years, kinases have been considered important not only for their crucial role in signaling but also for the transduction of the signal, controlling its amplitude (11–13). To facilitate the study of phospho-signaling networks, different databases have been designed (2,14,15).
The 518 human protein kinases are classified according to the amino acid residue that it phosphorylates. Most kinases act on both serine and threonine (serine/threonine kinases; STKs), others act on tyrosine (tyrosine kinases; TKs), and a number act on all three (dual-specificity kinases; DSKs) (16). The latter can phosphorylate STKs and TKs (17); at least 125 of the human protein kinases are STKs (18).
The STKs are enzymes that phosphorylate the OH group of serine or threonine, and are activated by different events such as DNA damage or chemical signals mediated for instance by Ca2+/calmodulin, cyclic-adenosine monophosphate/cyclic-guanosine monophosphate (cAMP/cGMP) and diacylglycerol.
There are also the following subfamilies of protein kinases: AGC, CaMK, CK1, CMGC, STE, TK and TKL (19–27) (Table I).
Table I.
Subfamilies of protein kinases.
| Protein kinase family | Origin of the name | Description | Refs. |
|---|---|---|---|
| AGC | Named after the protein kinase A, G and C families (PKA, PKC, PKG) | Subgroup of Ser/Thr protein kinases that, based on sequence alignments of their catalytic kinase domain, are related to cAMP-dependent protein kinase 1 (PKA; also known as PKAC), cGMP-dependent protein kinase (PKG; also known as CGK1α) and protein kinase C (PKC) | (20) |
| CaMK | Ca2+/calmodulin-dependent protein kinases | CaMKs transfer phosphates from ATP to serine or threonine residues in proteins in response to increase in concentration of intracellular calcium ions. They are important for expression of various genes because after activation, CAMKs phosphorylate several transcription factors. Members of this enzyme class include: CaMK I, CaMK II, CaMK III, CaMK IV and CaMK V | (21) |
| CK1 | Originally known as casein kinase 1 and now renamed cell kinase 1 | CK1 family of monomeric serine-threonine protein kinases. This family has seven members and are serine/threonine-selective enzymes that function as regulators of signal transduction pathways. CK1 isoforms are involved in Wnt signaling, circadian rhythms, nucleocytoplasmic shuttling of transcription factors, DNA repair and DNA transcription | (22) |
| CMGC | Named after another set of families (CDK, MAPK, GSK3 and CLK) | CDKs regulate cell progression through the different phases of the cell cycle | |
| MAP kinases are signal transduction molecules and they play a key role in the regulation of many cellular processes such as proliferation, differentiation and death. Abnormalities in MAP kinase cascades are tightly linked to oncogenic transformation | (23) | ||
| GSK3, initially described as a key enzyme involved in glycogen metabolism, is now known to regulate a diverse array of functions. GSK3 is a well-established component of the Wnt pathway, which is essential for establishing the entire body pattern during embryonic development | (28) | ||
| CLK encodes phosphorylation of serine/arginine-rich proteins involved in pre-mRNA processing, releasing them into the nucleoplasm and it may play an indirect role in governing splice site selection | (25) | ||
| STE | Sterile kinase | The STE group consists of three main families, which sequentially activate each other to then activate the MAPK family. The Ste7 family directly phosphorylates MAPKs, while many Ste20 members (MAP4K) act on Ste11 kinases. The Ste20 (MAP4K) family is the largest of the three and is divided into many subfamilies. Some are implicated in MAPK cascades, while others are not and may have completely distinct functions | (26) |
| TK | Tyrosine kinase | Members of the TK group specifically phosphorylate tyrosine residues and are therefore distinct from dual specificity kinases, which phosphorylate serine/threonine in addition to tyrosine. TKs are cell surface receptors (RTKs) and many of the others function close to the surface of the cell | (19) |
| TKL | Tyrosine kinase-like | Tyrosine kinase-like kinases are serine-threonine protein kinases named so because of their close sequence similarity to tyrosine kinases. Members of this family include MLK, RAF, STKR, LRRK, LISK, IRAK and RIPK | (27) |
In the column 'Origin of the name' some letters are underlined and in bold as they generate the acronym of the corresponding protein kinase. ATP, adenosine triphosphate; MAPK, mitogen-activated protein kinase.
3. Protein phosphatases
Phosphatases have the opposite function of kinases. They remove the phosphate group from phosphoproteins by hydrolyzing phosphoric acid monoesters into a phosphate group and a molecule with a free hydroxyl group (28,29).
Enzymatic removal reverts the protein to a non-phosphorylated state with a kinetics more rapid than kinases (30). When working with proteins in the laboratory, phosphatases are inactivated using denaturation or inhibitors so phosphorylation inside of a sample is not lost (31).
The protein phosphatases are considered passive housekeeping enzymes compared with protein kinases; their different structure makes them harder to identify and less important than the protein kinases (32).
Currently, there are approximately 226 known protein phosphatases (33) which are classified into 3 families: phosphoprotein phosphatase (PPP) family, metallo-dependent protein phosphatase (PPM) family and protein-tyrosine phosphatase (PTP) family (19).
The PPP family includes PP1, PP2A, PP2B and PP4–7 responsible for many dephosphorylation reactions (1) whereas PP2C is one of the most important members of the PPM family (34). PPP and PMP groups are responsible for most dephosphorylation reactions of phosphoserine and phosphotreonine (pSer/pThr) and they can also dephosphorylate phosphotyrosine (pTyr) (35,36). However, pSer/pThr have different domain sequences compared with pTyr (37).
In contrast, the phosphatases that belong to the PTP family have the same catalytic domain but different selectivity for phosphorylated proteins (38,39). Of all the phosphatases, at least 100 belong to those that dephosphorylate tyrosine residues, such as the Tyr-specific phosphatase subfamily, the Cdc25 family and myotubularin-related phosphatase and low molecular weight Tyr phosphatase (12,14,32,33,39,40). Aspartate-based phosphatases, such as FCP/SCP (small CTD phosphatase) (41–43) and TAD (haloacid dehalogenase) family enzymes are part of the PTP group (44).
PTPs are well-known as they can also dephosphorylate non-protein targets such as carbohydrates, mRNA and phosphoinositides (12,45–47).
4. Activities and role of protein phosphorylation under physiological conditions
Protein phosphorylation is one of the initial steps that is vital for the coordination of cellular and organic functions such as the regulation of metabolism, proliferation, apoptosis, subcellular trafficking, inflammation, and other important physiological processes.
There are several ways in which the phosphorylation acts to fulfill its role. First, the activity of phosphorylation/dephosphorylation acts as a molecular switch (Fig. 1).
For instance, protein kinase B is only activated follo wing phosphorylation of its Ser and Thr residues and, thus, is able to regulate cell survival (48); on the other hand, when proto-oncogene tyrosine-protein kinase (c-Src) is dephosphorylated, it is turned off causing a block in the regulation of cell growth (49).
Another mode of phosphorylation is temporary protein-protein interaction, which allows the adjustment of many signaling pathways (50,51). An example is glomerular podocyte protein nephrin 1 (Neph1), an important protein of renal cells, which once phosphorylated by Src Fyn, interacts with Grb2, an adapter protein involved in signal transduction and cell communication (52).
In addition, the phosphorylation of a protein can regulate the process of signal transduction since it is able to trigger the subcellular translocation of the protein phosphorylated by the mechanism itself.
Moreover, phosphorylation on serine/threonine-protein kinase (Ser350) residue of the death-associated protein (DAP) leads to the translocation from the cytoplasm to the nucleus of apoptosis-inducing kinase 2 (DRAK2) which is able to induce apoptosis in T and B cells (53).
Another example is the membrane translocation of synaptosomal-associated protein 25 (SNAP25). After being phosphorylated, SNAP25 has a reduced binding affinity for syntaxin-1A and thus changes location (54,55).
Furthermore, phosphorylation is mainly involved in the production and recycling of ATP and, therefore, is important in biological reactions that require energy (5,56).
Sometimes, protein phosphorylation may promote the formation or removal of a second PTM (4,57) (Fig. 1). An example is the phosphorylation of insulin receptor substrate-1 (IRS-1), a mediator of insulin signaling pathway, by the ribosomal protein S6 kinase β1 that induces polyubiquitination of IRS-1 due to E3 ligase-CUL7 and its successive proteasomal degradation (58).
The processes of phosphorylation and dephosphorylation can be etremely complex, since a single kinase or phosphatase may simultaneously have more substrates and may function in various cell signaling pathways. One of the signaling pathways known for this function is mitogen activated protein kinase (MAPK/ERK). It is activated through phosphorylation of MAPK which in turn phosphorylates many substrates, including 40S ribosomal protein S6 kinase (RSK, which then phosphorylates ribosomal protein S6), c-Myc and MNK (which then phosphorylates CREB in this cascade) (59,60).
MAPK is a known protein involved in a signaling pathway activated by a cascade effect of phosphorylation events (61). The binding of interferon-γ (IFN-γ) to its receptor induces phosphorylation of the Tyr-440 receptor, which promotes the formation of a complex with the tyrosine kinase JAK1 and JAK2. This complex phosphorylates Stat1, leading to its dimerization and nuclear translocation, where it regulates gene transcription (62–65).
Therefore, phospho-signaling networks represent the basis of many cellular processes. They consist mainly of protein kinases, phosphatases, and their respective substrates phospho-binding proteins (66) (Fig. 1).
There is also a mechanism of competition for kinases and phosphatases at the level of protein sites to adjust the states of phosphorylation of common substrates (67,68).
PhosphoNET and PhosphoSitePlus websites document the inhibition or activation of human protein with more than 850 different phosphosites with predictions for over 1,000 additional sites.
Therefore, it is customary to classify phosphorylation into two categories: one refers to functional changes (stable) and the other one, transitory, has no effect on regulatory functions. For this reason, it is thought that all stable phosphosites are functional and those not stable, are not functional (69–71).
In addition, the functional effects of phosphosites within a protein are site-dependent (72), and this means that they are functional only if phosphorylation takes place on a specific site and not random. This endorses the view that the detailed study of phosphorylation networks may help to understand the physiological and pathological mechanisms (72–76).
5. Protein phosphorylation and cancer
Phosphorylation is one of the most common PTMs involved in the regulation of multiple biological processes and overexpression of kinase. Mutations or defects in regulatory mechanisms can lead to aberrant activation or dysregulation of kinase signaling pathways (77) and this is the basis of oncogenesis for multiple tumors (78–80).
Cancer is not only considered a disease that arises from genetic mutations, but also a disease that results from epigenetic changes (81–83) that mainly lead to a deregulation of signal transduction pathways with subsequent changes in normal cellular mechanisms (84).
Many key regulatory proteins controlling gene expression are targets of kinases. The addition of a phosphate group to a protein by a kinase can alter the activity of the protein and this action is often exploited as a switch on or off (85,86).
In chronic myeloid leukemia, a particular chromosomal translocation (Philadelphia chromosome) was identified that generates a novel kinase that is always active, the retinoblastoma, pRb. The process normally controlled by this kinase is stuck in the 'on' position. This leads to the proliferation of tumor cells (87).
D. Stehelin was one of the first researchers to understand the direct involvement of protein kinases in tumors, with the study of the oncogene v-SRC (88). This tyrosine kinase with the phosphate group of Tyr-527 has a key role in tumor cell proliferation, and has been studied extensively in Rous sarcoma virus (RSV) as the main cause of sarcoma in chickens (89–92). Its carcinogenic action is due to a mutation of the carboxyl terminal of the molecule able to eliminate the tyrosine residue, which causes conformational changes and also an irregular unregulated autophosphorylation, leading to a signal of increased growth (93,94).
Aberrations of kinases have been reported in different types of cancer. An example is the amplification of Her2/neu observed in tumor cells of invasive breast cancer (95,96).
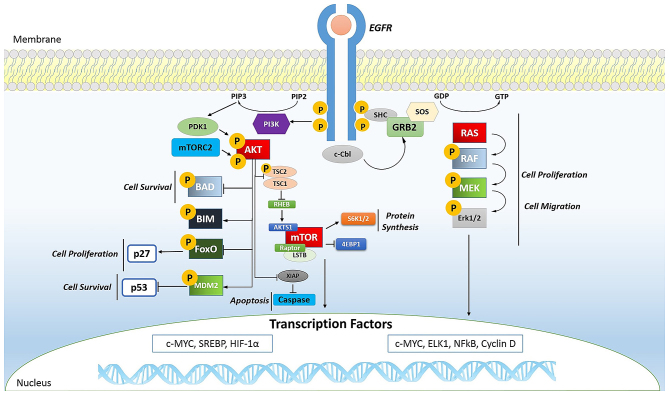
Phosphorylation plays a key role even in oral cancer. In fact, the phosphorylation of EGFR (Fig. 2) is important for transactivation of interleukin (IL)-1β through the CXCL1 and CXCR2 axis (97).
Figure 2.
Endothelial growth factor receptor (EGFR) signaling. Ligand binding to the EGFR activates its intrinsic tyrosine kinase activity. The autophosphorylation of EGFR causes the activation of several signaling pathways such as PI3K/AKT and RAS/mitogen-activated protein kinase (MAPK). In both, the phosphorylation is a predominant event and plays an important role in cell survival (AKT activation phosphorylates BAD and MDM2), cell proliferation (AKT activation phosphorylates FoxO, RAS phosphorylates RAF and it phosphorylates MEK that phosphorylates Erk1/2), cell migration (phosphorylation cascade of RAS), apoptosis (AKT phosphorylation is able to activate caspase). However, the phosphorylation of AKT causes mTOR activation, important in protein synthesis.
Alteration of the phosphoproteome also affects gastrointestinal stromal tumors (GISTs) (98,99), lung cancer (73,100), hematologic malignancies (101,102), breast cancer (103,104), pancreatic cancer (105,106) and prostate cancer (103,107).
To date, more than 1,000 variations in the expression of protein kinases have been detected in human tumors (108–111). Many of these kinases are now considered cancer biomarkers, such as EGFR for colon cancer, cKIT for GISTs, and human EGFR-related gene (Her2) for breast cancer (112).
In tumors, mTOR (Fig. 2) is a kinase and, when activated, is able to induce activation of its downstream effectors and consequently increases the synthesis of cell cycle proteins such as hypoxia-inducible factor-1 (HIF-1) (cyclin D and HIF-1α) which in turn stimulates vascular endothelial growth factor (VEGF) (113,114). mTOR is particularly active in renal cancer by promoting angiogenesis (115,116).
The Ras oncogene (Fig. 2) is the most common in human tumors (117) and its activation is very complex, characterized by countless phosphorylation events. It begins with the binding of a ligand to a receptor tyrosine kinase (RTK) loca ted on the plasma membrane. This receptor is activated only if it dimerizes with another RTK. They then phosphorylate each other and become activated. The activated receptor binds to the SH2 domain of the adapter protein Grb2, which plays its role without being phosphorylated. In fact, its SH3 domain binds to the protein activating SOS, without any phosphorylation reactions. SOS then moves close to the plasma membrane where it can bind Ras, replacing GDP with GTP and then becomes activated; SOS therefore acts as a nucleotide exchange factor (GEF). It is known that activated Ras binds to the N-terminus of a protein Ser/Thr kinase called c-Raf, activating them (118).
6. Protein kinases as drug targets
The signaling pathways regulated by protein kinases contribute to the onset and progression of almost all types of cancer. Consequently, research of the signaling pathways mediated by kinase and therefore the possibility of blocking them with targeted treatment could have major clinical-therapeutic utility especially since many of these proteins act as oncogenes (78,119,120).
Considerable advances have led to the identification of inhibitors directed against activated tyrosine kinases in cancer, 17 of which are already used for the treatment of several cancers and more than 390 molecules are being tested (121).
Imatinib (Glivec®) is a known inhibitor that blocks the action of the tyrosine kinase BCR-Abl in patients with chronic myeloid leukemia (CML) (122,123). This drug also targets against PI3K in solid tumors (124,125), serine/threonine kinase BRAF to treat melanoma (126–128), the receptor tyrosine kinase EGFR for lung cancer (129,130), and serine/threonine kinase mTOR for the treatment of renal tumors (131).
Gefitinib/erlotinib (Tarceva®) is targeted against EGFR in lung tumors (132) with a success rate of 71.2% (129), whereas crizotinib (Xalkori®) acts in the same tumor against EML4-ALK (133).
Vemurafenib (Zelboraf®) in melanoma is directed against mutations of BRAF V600E (134) with a successful response of 48% during treatment (127).
If overexpressed, HER2 is a protein tyrosine kinase which enhances the proliferation of cancer cells, and enhances the formation of blood vessels thereby increasing the invasiveness of breast cancer. Currently, improvements in the prognosis of this cancer are due to the use of trastuzumab (Herceptin®) a monoclonal antibody targeted against this protein (135).
Sorafenib (Nexavar®) is another kinase inhibitor which blocks the action of Raf kinase in kidney and liver tumors (136).
Sunitinib (Sutent®) is a multi-targeted receptor tyrosine kinase inhibitor that acts against platelet-derived growth factor receptors (PDGFRs) and VEGF receptors (VEGFRs) (137). The simultaneous inhibition of these targets induces a reduction in tumor vascularization and triggers cancer cell apoptosis. It has been recommended as a drug in renal cell carcinoma and in GISTs (138,139). Furthermore, since sunitinib targets many different receptors, it leads to dermatologic toxic side effects such as hand-foot syndrome (140).
Temsirolimus (Torisel®) is a drug used for the treatment of renal cell carcinoma and it is a specific inhibitor of mTOR (141), a cellular kinase enzyme that may favor tumor growth. Temsirolimus leads to cell cycle arrest in the G1 phase, and also inhibits tumor angiogenesis by reducing synthesis of VEGF (142).
The success of therapies based on kinase inhibitors relies on different aspects: the clinical targeted kinase, the structure of the signaling network and the mechanisms of innate or acquired resistance.
First of all, both the patients and the therapeutic approach functions must be appropriately selected (119). For instance, in CML, this therapy only works with BCR-Abl-positive patients as the main targets of therapy are BCR and ABL (Philadelphia chromosome genes) fused together by means of an activated protein tyrosine kinase.
In the same way, the response rate of Herceptin was 34% in patients whose tumors had amplified HER2 compared with 7% in those whose tumors did not (143).
However, not all tumors respond to inhibitors of kinases and often patients with the same cancer respond differently to the same therapy. For this reason, patients should be further stratified using biomarkers (144) and further studies are warranted to investigate the signaling pathways (145–147).
In this respect, we know that changes in the signaling pathways, caused by several factors (genetic and epigenetic mutations, alterations of the microenvironment), lead to the formation of oncogenes and, very often, there is a release of tumoral molecules that can be tracked and used as biomarkers. For example, hepatocyte growth factor (HGF) released in stromal cells of melanoma, influences the way BRAF inhibitors act (148).
The signaling networks of cancer cells can also develop innate or acquired resistance, since they are able to create the most common or rare oncogenic mutations different from tumor to tumor (the so-called polygenic tumor biology) (149).
There are two main types of resistance to a drug treatment based on kinase inhibitors. Intrinsic or innate resistance (on target) occurs when the drug target protein has changed due to steric hindrance to inhibitor binding (150), altered active site topography (151), disruption of favorable inhibitor interactions (152), altered protein dynamics (153), increased oncogenicity (151), and alteration of ATP affinity (154). In this way, this resistance is not inhibited by the drug and continues to perform its normal activity in the tumor cell.
In extrinsic or acquired resistance (off target), the signaling network is able to restore the function of oncogenic mutation in the signaling network (155,156) or via bypass/compensatory signaling or even through feedback loops to adjust the signal. The cancer cells are able in fact to exploit and reactivate the mechanism of signaling that the drug would inhibit (157). In addition, during treatment acquired resistance can occur and the tumors can develop subclones which foster even relapse.
New studies of the signaling network of tumors with particular attention to the mechanism of action of drug inhibitors of protein kinases are therefore needed.
7. Conclusions
Phosphoproteomics has a critical relevance for many aspects of biology and has a significant role for understanding the molecular mechanisms, especially those that lead to the genesis and growth of tumors (77–79). Signaling networks in which protein kinases operate are highly complex, but we believe that understanding the regulatory functions of kinases may be a valid means to identify more effective therapies against cancer (146,147). Many drug kinase inhibitors are already on the market (122,133–137) but, often, their effectiveness is reduced due to the development of complex mechanisms of drug resistance (149).
However, great progress has been made in recent years thanks to the numerous techniques of proteomics. Proteomics is the most important way by which to study the sites and behavior of phosphoprotein and phosphosite in tumor biology. The identification of biomarkers that aid in the selection of the most appropriate therapy for individual patients remains a major challenge (144).
Glossary
Abbreviations
- ADP
adenosine diphosphate
- AGC kinases
protein kinase A, G and C families
- Asp or D
aspartate residues
- ATP
adenosine triphosphate
- CaMK
Ca2+/calmodulin-dependent protein kinase class
- cAMP/cGMP
cyclic-adenosine monophosphate/cyclic-guanosine monophosphate
- Cdc25
cell division cycle 25 phosphatase
- CK1
casein kinase 1
- CML
chronic myeloid leukemia
- c-Src
proto-oncogene tyrosine-protein kinase
- DAP protein
death-associated protein
- DRAK2
apoptosis-inducing kinase 2
- DSKs
dual-specificity kinases
- EGF
epidermal growth factor
- GISTs
gatrointestinal stromal tumors
- Grb2
growth factor receptor-bound protein 2
- HGF
hepatocyte growth factor
- HIF-1α
hypoxia-inducible factor-1α
- His or H
histidine residues and N-phosphorylation
- IFN-γ
interferon-γ
- IRS-1
insulin receptor substrate-1
- mRNA
messenger ribonucleic acid
- Neph1
glomerular podocyte protein nephrin 1
- PDGFRs
platelet-derived growth factor receptors
- PO4
phosphate group
- PP1
phosphoprotein phosphatase 1
- PP2A
phosphoprotein phosphatase 2A
- PP2B
phosphoprotein phosphatase 2B
- PP2C
phosphoprotein phosphatase 2C
- PP4–7
phosphoprotein phosphatase 4–7
- PPM
metallo-dependent protein phosphatase family
- PPP
phosphoprotein phosphatase family
- pSer/pThr
phosphoserine and phosphothreonine
- PTMs
post-translational modifications
- PTP
protein-tyrosine phosphatase family
- pTyr
phosphotyrosine
- Ser or S
serine residues
- Ser350
serine/threonine-protein kinase
- SNAP25
synaptosomal-associated protein 25
- STKs
serine/threonine kinases
- Thr or T
threonine residues
- TKs
tyrosine kinases
- TKLs
tyrosine kinase-like kinases
- Tyr or Y
tyrosine residues
- VEGFRs
vascular endothelial growth factor receptors
References
- 1.Li X, Wilmanns M, Thornton J, Köhn M. Elucidating human phosphatase-substrate networks. Sci Signal. 2013;6:rs10. doi: 10.1126/scisignal.2003203. [DOI] [PubMed] [Google Scholar]
- 2.Sacco F, Perfetto L, Castagnoli L, Cesareni G. The human phosphatase interactome: an intricate family portrait. FEBS Lett. 2012;586:2732–2739. doi: 10.1016/j.febslet.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. In: Molecular Biology of the Cell. 5th edition. Anderson M, Granum S, editors. Garland Science; New York, NY: 2007. p. 175. [Google Scholar]
- 4.Hunter T. Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci. 2009;367:2513–2516. doi: 10.1098/rstb.2012.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukami Y, Lipmann F. Reversal of Rous sarcoma-specific immunoglobulin phosphorylation on tyrosine (ADP as phosphate acceptor) catalyzed by the src gene kinase. Proc Natl Acad Sci USA. 1983;80:1872–1876. doi: 10.1073/pnas.80.7.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kole HK, Abdel-Ghany M, Racker E. Specific dephosphorylation of phosphoproteins by protein-serine and -tyrosine kinases. Proc Natl Acad Sci USA. 1988;85:5849–5853. doi: 10.1073/pnas.85.16.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roskoski R., Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz PA, Murray BW. Protein kinase biochemistry and drug discovery. Bioorg Chem. 2011;39:192–210. doi: 10.1016/j.bioorg.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Nishi H, Shaytan A, Panchenko AR. Physicochemical mechanisms of protein regulation by phosphorylation. Front Genet. 2014;5:270. doi: 10.3389/fgene.2014.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCance KL, Huether SE. In: Pathophysiology: The Biologic Basis for Disease in Adults and Children. 7th edition. Brashers VL, Rote NS, editors. Elsevier; 2014. [Google Scholar]
- 11.Hornberg JJ, Bruggeman FJ, Binder B, Geest CR, de Vaate AJ, Lankelma J, Heinrich R, Westerhoff HV. Principles behind the multifarious control of signal transduction. ERK phosphorylation and kinase/phosphatase control. FEBS J. 2005;272:244–258. doi: 10.1111/j.1432-1033.2004.04404.x. [DOI] [PubMed] [Google Scholar]
- 12.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 13.Heinrich R, Neel BG, Rapoport TA. Mathematical models of protein kinase signal transduction. Mol Cell. 2002;9:957–970. doi: 10.1016/S1097-2765(02)00528-2. [DOI] [PubMed] [Google Scholar]
- 14.Liberti S, Sacco F, Calderone A, Perfetto L, Iannuccelli M, Panni S, Santonico E, Palma A, Nardozza AP, Castagnoli L, et al. HuPho: the human phosphatase portal. FEBS J. 2013;280:379–387. doi: 10.1111/j.1742-4658.2012.08712.x. [DOI] [PubMed] [Google Scholar]
- 15.Hatzihristidis T, Liu S, Pryszcz L, Hutchins AP, Gabaldón T, Tremblay ML, Miranda-Saavedra D. PTP-central: a comprehensive resource of protein tyrosine phosphatases in eukaryotic genomes. Methods. 2014;65:156–164. doi: 10.1016/j.ymeth.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 16.Miller ML, Jensen LJ, Diella F, Jørgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1 doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capra M, Nuciforo PG, Confalonieri S, Quarto M, Bianchi M, Nebuloni M, Boldorini R, Pallotti F, Viale G, Gishizky ML, et al. Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res. 2006;66:8147–8154. doi: 10.1158/0008-5472.CAN-05-3489. [DOI] [PubMed] [Google Scholar]
- 19.Jin J, Pawson T. Modular evolution of phosphorylation-based signalling systems. Philos Trans R Soc Lond B Biol Sci. 2012;367:2540–2555. doi: 10.1098/rstb.2012.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 21.Wayman GA, Tokumitsu H, Davare MA, Soderling TR. Analysis of CaM-kinase signaling in cells. Cell Calcium. 2011;50:1–8. doi: 10.1016/j.ceca.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eide EJ, Virshup DM. Casein kinase I: another cog in the circadian clockworks. Chronobiol Int. 2001;18:389–398. doi: 10.1081/CBI-100103963. [DOI] [PubMed] [Google Scholar]
- 23.Sundaram MV. RTK/Ras/MAPK signaling. WormBook: 2006. pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen P, Goedert M. GSK3 inhibitors: development and therapeutic potential. Nat Rev Drug Discov. 2004;3:479–487. doi: 10.1038/nrd1415. [DOI] [PubMed] [Google Scholar]
- 25.Moeslein FM, Myers MP, Landreth GE. The CLK family kinases, CLK1 and CLK2, phosphorylate and activate the tyrosine phosphatase, PTP-1B. J Biol Chem. 1999;274:26697–26704. doi: 10.1074/jbc.274.38.26697. [DOI] [PubMed] [Google Scholar]
- 26.Müller-Taubenberger A, Ishikawa-Ankerhold HC, Kastner PM, Burghardt E, Gerisch G. The STE group kinase SepA controls cleavage furrow formation in Dictyostelium. Cell Motil Cytoskeleton. 2009;66:929–939. doi: 10.1002/cm.20386. [DOI] [PubMed] [Google Scholar]
- 27.Abdi AI, Carvalho TG, Wilkes JM, Doerig C. A secreted Plasmodium falciparum kinase reveals a signature motif for classification of tyrosine kinase-like kinases. Microbiology. 2013;159:2533–2547. doi: 10.1099/mic.0.070409-0. [DOI] [PubMed] [Google Scholar]
- 28.Barford D. Molecular mechanisms of the protein serine/thre-onine phosphatases. Trends Biochem Sci. 1996;21:407–412. doi: 10.1016/S0968-0004(96)10060-8. [DOI] [PubMed] [Google Scholar]
- 29.Zhang ZY. Protein tyrosine phosphatases: structure and function, substrate specificity, and inhibitor development. Annu Rev Pharmacol Toxicol. 2002;42:209–234. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 30.Mackintosh C. In: Protein Phosphorylation: A Practical Approach. Hardie GD, editor. IRL Press; New York, NY: 1993. p. 328. [Google Scholar]
- 31.Thingholm TE, Larsen MR, Ingrell CR, Kassem M, Jensen ON. TiO(2)-based phosphoproteomic analysis of the plasma membrane and the effects of phosphatase inhibitor treatment. J Proteome Res. 2008;7:3304–3313. doi: 10.1021/pr800099y. [DOI] [PubMed] [Google Scholar]
- 32.Stern DF. Phosphoproteomics for oncology discovery and treatment. Expert Opin Ther Targets. 2005;9:851–860. doi: 10.1517/14728222.9.4.851. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Chance MR. Integrating phosphoproteomics in systems biology. Comput Struct Biotechnol J. 2014;10:90–97. doi: 10.1016/j.csbj.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 35.Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- 36.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Guan KL, Dixon JE. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. J Biol Chem. 1991;266:17026–17030. [PubMed] [Google Scholar]
- 39.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 40.Tonks NK, Neel BG. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr Opin Cell Biol. 2001;13:182–195. doi: 10.1016/S0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 41.Wrighton KH, Willis D, Long J, Liu F, Lin X, Feng XH. Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-beta signaling. J Biol Chem. 2006;281:38365–38375. doi: 10.1074/jbc.M607246200. [DOI] [PubMed] [Google Scholar]
- 42.Archambault J, Pan G, Dahmus GK, Cartier M, Marshall N, Zhang S, Dahmus ME, Greenblatt J. FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J Biol Chem. 1998;273:27593–27601. doi: 10.1074/jbc.273.42.27593. [DOI] [PubMed] [Google Scholar]
- 43.Salton SR. Teaching resources. Protein phosphatases. Sci STKE. 2005;2005:tr8. doi: 10.1126/stke.2732005tr8. [DOI] [PubMed] [Google Scholar]
- 44.Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- 45.Gentry MS, Dixon JE, Worby CA. Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem Sci. 2009;34:628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gentry MS, Dowen RH, III, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niittylä T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MD, Gatehouse JA, Villadsen D, Smith SM, Chen J, et al. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem. 2006;281:11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 48.Case N, Thomas J, Sen B, Styner M, Xie Z, Galior K, Rubin J. Mechanical regulation of glycogen synthase kinase 3β (GSK3β) in mesenchymal stem cells is dependent on Akt protein serine 473 phosphorylation via mTORC2 protein. J Biol Chem. 2011;286:39450–39456. doi: 10.1074/jbc.M111.265330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cole PA, Shen K, Qiao Y, Wang D. Protein tyrosine kinases Src and Csk: a tail's tale. Curr Opin Chem Biol. 2003;7:580–585. doi: 10.1016/j.cbpa.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Nishi H, Hashimoto K, Panchenko AR. Phosphorylation in protein-protein binding: effect on stability and function. Structure. 2011;19:1807–1815. doi: 10.1016/j.str.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishi H, Fong JH, Chang C, Teichmann SA, Panchenko AR. Regulation of protein-protein binding by coupling between phosphorylation and intrinsic disorder: analysis of human protein complexes. Mol Biosyst. 2013;9:1620–1626. doi: 10.1039/c3mb25514j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harita Y, Kurihara H, Kosako H, Tezuka T, Sekine T, Igarashi T, Hattori S. Neph1, a component of the kidney slit diaphragm, is tyrosine-phosphorylated by the Src family tyrosine kinase and modulates intracellular signaling by binding to Grb2. J Biol Chem. 2008;283:9177–9186. doi: 10.1074/jbc.M707247200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuwahara H, Nishizaki M, Kanazawa H. Nuclear localization signal and phosphorylation of Serine350 specify intracellular localization of DRAK2. J Biochem. 2008;143:349–358. doi: 10.1093/jb/mvm236. [DOI] [PubMed] [Google Scholar]
- 54.Shimazaki Y, Nishiki T, Omori A, Sekiguchi M, Kamata Y, Kozaki S, Takahashi M. Phosphorylation of 25-kDa synaptosome-associated protein. Possible involvement in protein kinase C-mediated regulation of neurotransmitter release. J Biol Chem. 1996;271:14548–14553. doi: 10.1074/jbc.271.24.14548. [DOI] [PubMed] [Google Scholar]
- 55.Kataoka M, Kuwahara R, Iwasaki S, Shoji-Kasai Y, Takahashi M. Nerve growth factor-induced phosphorylation of SNAP-25 in PC12 cells: a possible involvement in the regulation of SNAP-25 localization. J Neurochem. 2000;74:2058–2066. doi: 10.1046/j.1471-4159.2000.0742058.x. [DOI] [PubMed] [Google Scholar]
- 56.Rosen OM, Erlichman J. Reversible autophosphorylation of a cyclic 3′:5′-AMP-dependent protein kinase from bovine cardiac muscle. J Biol Chem. 1975;250:7788–7794. [PubMed] [Google Scholar]
- 57.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 58.Xu X, Sarikas A, Dias-Santagata DC, Dolios G, Lafontant PJ, Tsai SC, Zhu W, Nakajima H, Nakajima HO, Field LJ, et al. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol Cell. 2008;30:403–414. doi: 10.1016/j.molcel.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferrer I, Blanco R, Carmona M, Puig B, Domínguez I, Viñals F. Active, phosphorylation-dependent MAP kinases, MAPK/ERK, SAPK/JNK and p38, and specific transcription factor substrates are differentially expressed following systemic administration of kainic acid to the adult rat. Acta Neuropathol. 2002;103:391–407. doi: 10.1007/s00401-001-0481-9. [DOI] [PubMed] [Google Scholar]
- 61.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 62.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 63.Greenlund AC, Farrar MA, Viviano BL, Schreiber RD. Ligand-induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91) EMBO J. 1994;13:1591–1600. doi: 10.1002/j.1460-2075.1994.tb06422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Igarashi K, Garotta G, Ozmen L, Ziemiecki A, Wilks AF, Harpur AG, Larner AC, Finbloom DS. Interferon-gamma induces tyrosine phosphorylation of interferon-gamma receptor and regulated association of protein tyrosine kinases, Jak1 and Jak2, with its receptor. J Biol Chem. 1994;269:14333–14336. [PubMed] [Google Scholar]
- 65.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 66.Liu Z, Wang Y, Xue Y. Phosphoproteomics-based network medicine. FEBS J. 2013;280:5696–5704. doi: 10.1111/febs.12380. [DOI] [PubMed] [Google Scholar]
- 67.Hirschi A, Cecchini M, Steinhardt RC, Schamber MR, Dick FA, Rubin SM. An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat Struct Mol Biol. 2010;17:1051–1057. doi: 10.1038/nsmb.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salazar C, Höfer T. Competition effects shape the response sensitivity and kinetics of phosphorylation cycles in cell signaling. Ann NY Acad Sci. 2006;1091:517–530. doi: 10.1196/annals.1378.093. [DOI] [PubMed] [Google Scholar]
- 69.Lienhard GE. Non-functional phosphorylations? Trends Biochem Sci. 2008;33:351–352. doi: 10.1016/j.tibs.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Landry CR, Levy ED, Michnick SW. Weak functional constraints on phosphoproteomes. Trends Genet. 2009;25:193–197. doi: 10.1016/j.tig.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 71.Levy ED, Michnick SW, Landry CR. Protein abundance is key to distinguish promiscuous from functional phosphorylation based on evolutionary information. Philos Trans R Soc Lond B Biol Sci. 2012;367:2594–2606. doi: 10.1098/rstb.2012.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 73.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 74.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jørgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Newman RH, Hu J, Rho HS, Xie Z, Woodard C, Neiswinger J, Cooper C, Shirley M, Clark HM, Hu S, et al. Construction of human activity-based phosphorylation networks. Mol Syst Biol. 2013;9:655. doi: 10.1038/msb.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Drake JM, Graham NA, Stoyanova T, Sedghi A, Goldstein AS, Cai H, Smith DA, Zhang H, Komisopoulou E, Huang J, et al. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc Natl Acad Sci USA. 2012;109:1643–1648. doi: 10.1073/pnas.1120985109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harsha HC, Pandey A. Phosphoproteomics in cancer. Mol Oncol. 2010;4:482–495. doi: 10.1016/j.molonc.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 79.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 80.Sharma A, Tan TH, Cheetham G, Scott HS, Brown MP. Rare and novel epidermal growth factor receptor mutations in non-small-cell lung cancer and lack of clinical response to gefitinib in two cases. J Thorac Oncol. 2012;7:941–942. doi: 10.1097/JTO.0b013e31825134ac. [DOI] [PubMed] [Google Scholar]
- 81.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 82.Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA. Clinical proteomics: translating benchside promise into bedside reality. Nat Rev Drug Discov. 2002;1:683–695. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 83.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 84.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 85.Tarrant MK, Cole PA. The chemical biology of protein phosphorylation. Annu Rev Biochem. 2009;78:797–825. doi: 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paul MK, Mukhopadhyay AK. Tyrosine kinase - role and significance in cancer. Int J Med Sci. 2004;1:101–115. doi: 10.7150/ijms.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murphree AL, Benedict WF. Retinoblastoma: clues to human oncogenesis. Science. 1984;223:1028–1033. doi: 10.1126/science.6320372. [DOI] [PubMed] [Google Scholar]
- 88.Stehelin D, Guntaka RV, Varmus HE, Bishop JM. Purification of DNA complementary to nucleotide sequences required for neoplastic transformation of fibroblasts by avian sarcoma viruses. J Mol Biol. 1976;101:349–365. doi: 10.1016/0022-2836(76)90152-2. [DOI] [PubMed] [Google Scholar]
- 89.Hunter T, Cooper JA. Protein-tyrosine kinases. Annu Rev Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 90.Sefton BM, Hunter T. From c-src to v-src, or the case of the missing C terminus. Cancer Surv. 1986;5:159–172. [PubMed] [Google Scholar]
- 91.Sefton BM, Hunter T, Raschke WC. Evidence that the Abelson virus protein functions in vivo as a protein kinase that phosphorylates tyrosine. Proc Natl Acad Sci USA. 1981;78:1552–1556. doi: 10.1073/pnas.78.3.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sefton BM, Hunter T, Beemon K, Eckhart W. Evidence that the phosphorylation of tyrosine is essential for cellular transformation by Rous sarcoma virus. Cell. 1980;20:807–816. doi: 10.1016/0092-8674(80)90327-X. [DOI] [PubMed] [Google Scholar]
- 93.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/S1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 94.Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell. 2001;105:115–126. doi: 10.1016/S0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 95.Shawver LK, Slamon D, Ullrich A. Smart drugs: tyrosine kinase inhibitors in cancer therapy. Cancer Cell. 2002;1:117–123. doi: 10.1016/S1535-6108(02)00039-9. [DOI] [PubMed] [Google Scholar]
- 96.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 97.Lee CH, Syu SH, Liu KJ, Chu PY, Yang WC, Lin P, Shieh WY. Interleukin-1 beta transactivates epidermal growth factor receptor via the CXCL1-CXCR2 axis in oral cancer. Oncotarget. 2015;6:38866–38880. doi: 10.18632/oncotarget.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22:3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 99.Javidi-Sharifi N, Traer E, Martinez J, Gupta A, Taguchi T, Dunlap J, Heinrich MC, Corless CL, Rubin BP, Druker BJ, et al. Crosstalk between KIT and FGFR3 promotes gastrointestinal stromal tumor cell growth and drug resistance. Cancer Res. 2015;75:880–891. doi: 10.1158/0008-5472.CAN-14-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 101.Zhu N, Xiao H, Wang LM, Fu S, Zhao C, Huang H. Mutations in tyrosine kinase and tyrosine phosphatase and their relevance to the target therapy in hematologic malignancies. Future Oncol. 2015;11:659–673. doi: 10.2217/fon.14.280. [DOI] [PubMed] [Google Scholar]
- 102.Kraus J, Kraus M, Liu N, Besse L, Bader J, Geurink PP, de Bruin G, Kisselev AF, Overkleeft H, Driessen C. The novel β2-selective proteasome inhibitor LU-102 decreases phosphorylation of I kappa B and induces highly synergistic cytotoxicity in combination with ibrutinib in multiple myeloma cells. Cancer Chemother Pharmacol. 2015;76:383–396. doi: 10.1007/s00280-015-2801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jagarlamudi KK, Hansson LO, Eriksson S. Breast and prostate cancer patients differ significantly in their serum thymidine kinase 1 (TK1) specific activities compared with those hematological malignancies and blood donors: implications of using serum TK1 as a biomarker. BMC Cancer. 2015;15:66. doi: 10.1186/s12885-015-1073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hou S, Isaji T, Hang Q, Im S, Fukuda T, Gu J. Distinct effects of β1 integrin on cell proliferation and cellular signaling in MDA-MB-231 breast cancer cells. Sci Rep. 2016;6:18430. doi: 10.1038/srep18430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Paladino D, Yue P, Furuya H, Acoba J, Rosser CJ, Turkson J. A novel nuclear Src and p300 signaling axis controls migratory and invasive behavior in pancreatic cancer. Oncotarget. 2016;7:7253–7267. doi: 10.18632/oncotarget.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Z, Lin P, Gao C, Peng C, Liu S, Gao H, Wang B, Wang J, Niu J, Niu W. Integrin β6 acts as an unfavorable prognostic indicator and promotes cellular malignant behaviors via ERK-ETS1 pathway in pancreatic ductal adenocarcinoma (PDAC) Tumour Biol. 2016;37:5117–5131. doi: 10.1007/s13277-015-4353-7. [DOI] [PubMed] [Google Scholar]
- 107.Meh raein-Ghom i F, Chu rch DR, Sch reiber CL, Weichmann AM, Basu HS, Wilding G. Inhibitor of p52 NF-κB subunit and androgen receptor (AR) interaction reduces growth of human prostate cancer cells by abrogating nuclear translocation of p52 and phosphorylated AR(ser81) Genes Cancer. 2015;6:428–444. doi: 10.18632/genesandcancer.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med. 2004;351:2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]
- 109.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 110.Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, Silliman N, Ptak J, Szabo S, Willson JK, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 111.Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 112.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 113.Rubio-Viqueira B, Hidalgo M. Targeting mTOR for cancer treatment. Adv Exp Med Biol. 2006;587:309–327. doi: 10.1007/978-1-4020-5133-3_24. [DOI] [PubMed] [Google Scholar]
- 114.Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dancey JE. Therapeutic targets: MTOR and related pathways. Cancer Biol Ther. 2006;5:1065–1073. doi: 10.4161/cbt.5.9.3175. [DOI] [PubMed] [Google Scholar]
- 116.Thomas GV, Tran C, Mellinghoff IK, Welsbie DS, Chan E, Fueger B, Czernin J, Sawyers CL. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- 117.Ahmadian MR. Prospects for anti-ras drugs. Br J Haematol. 2002;116:511–518. doi: 10.1046/j.0007-1048.2001.03314.x. [DOI] [PubMed] [Google Scholar]
- 118.Goodsell DS. The molecular perspective: the ras oncogene. Oncologist. 1999;4:263–264. [PubMed] [Google Scholar]
- 119.Sawyers CL. Shifting paradigms: the seeds of oncogene addiction. Nat Med. 2009;15:1158–1161. doi: 10.1038/nm1009-1158. [DOI] [PubMed] [Google Scholar]
- 120.Hainaut P, Plymoth A. Targeting the hallmarks of cancer: towards a rational approach to next-generation cancer therapy. Curr Opin Oncol. 2013;25:50–51. doi: 10.1097/CCO.0b013e32835b651e. [DOI] [PubMed] [Google Scholar]
- 121.Gonzalez de Castro D, Clarke PA, Al-Lazikani B, Workman P. Personalized cancer medicine: molecular diagnostics, predictive biomarkers, and drug resistance. Clin Pharmacol Ther. 2013;93:252–259. doi: 10.1038/clpt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Druker BJ. Imatinib mesylate in the treatment of chronic myeloid leukaemia. Expert Opin Pharmacother. 2003;4:963–971. doi: 10.1517/14656566.4.6.963. [DOI] [PubMed] [Google Scholar]
- 123.Stegmeier F, Warmuth M, Sellers WR, Dorsch M. Targeted cancer therapies in the twenty-first century: lessons from imatinib. Clin Pharmacol Ther. 2010;87:543–552. doi: 10.1038/clpt.2009.297. [DOI] [PubMed] [Google Scholar]
- 124.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, et al. The IK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 125.Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 126.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 127.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Young K, Minchom A, Larkin J. BRIM-1, -2 and -3 trials: improved survival with vemurafenib in metastatic melanoma patients with a BRAF(V600E) mutation. Future Oncol. 2012;8:499–507. doi: 10.2217/fon.12.43. [DOI] [PubMed] [Google Scholar]
- 129.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 130.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 131.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, Grünwald V, Thompson JA, Figlin RA, Hollaender N, et al. RECORD-1 Study Group Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 132.Abdurahman A, Anwar J, Turghun A, Niyaz M, Zhang L, Awut I. Epidermal growth factor receptor gene mutation status and its association with clinical characteristics and tumor markers in non-small-cell lung cancer patients in Northwest China. Mol Clin Oncol. 2015;3:847–850. doi: 10.3892/mco.2015.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ulivi P, Chiadini E, Dazzi C, Dubini A, Costantini M, Medri L, Puccetti M, Capelli L, Calistri D, Verlicchi A, et al. Nonsquamous, non-small-cell lung cancer patients who carry a double mutation of EGFR, EML4-ALK or KRAS: frequency, clinical-pathological characteristics, and response to therapy. Clin Lung Cancer. 2016;17:384–390. doi: 10.1016/j.cllc.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 134.Larkin J, Ascierto PA, Dréno B, Atkinson V, Liszkay G, Maio M, Mandalà M, Demidov L, Stroyakovskiy D, Thomas L, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 135.Carvajal-Hausdorf DE, Schalper KA, Pusztai L, Psyrri A, Kalogeras KT, Kotoula V, Fountzilas G, Rimm DL. Measurement of domain-specific HER2 (ERBB2) expression may classify benefit from trastuzumab in breast cancer. J Natl Cancer Inst. 2015;107:djv136. doi: 10.1093/jnci/djv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hasskarl J. Sorafenib: targeting multiple tyrosine kinases in cancer. Recent Results Cancer Res. 2014;201:145–164. doi: 10.1007/978-3-642-54490-3_8. [DOI] [PubMed] [Google Scholar]
- 137.Aita Y, Ishii KA, Saito Y, Ikeda T, Kawakami Y, Shimano H, Hara H, Takekoshi K. Sunitinib inhibits catecholamine synthesis and secretion in pheochromocytoma tumor cells by blocking VEGF receptor 2 via PLC-γ-related pathways. Am J Physiol Endocrinol Metab. 2012;303:E1006–E1014. doi: 10.1152/ajpendo.00156.2012. [DOI] [PubMed] [Google Scholar]
- 138.Demetri GD, van Oosterom AT, Garrett CR, Blackstein ME, Shah MH, Verweij J, McArthur G, Judson IR, Heinrich MC, Morgan JA, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet. 2006;368:1329–1338. doi: 10.1016/S0140-6736(06)69446-4. [DOI] [PubMed] [Google Scholar]
- 139.Czarnecka AM, Solarek W, Kornakiewicz A, Szczylik C. Tyrosine kinase inhibitors target cancer stem cells in renal cell cancer. Oncol Rep. 2016;35:1433–1442. doi: 10.3892/or.2015.4514. [DOI] [PubMed] [Google Scholar]
- 140.Lankheet NA, Huitema AD, Mallo H, Adriaansz S, Haanen JB, Schellens JH, Beijnen JH, Blank CU. The effect of seasonal variation and secretion of sunitinib in sweat on the development of hand-foot syndrome. Eur J Clin Pharmacol. 2013;69:2065–2072. doi: 10.1007/s00228-013-1579-4. [DOI] [PubMed] [Google Scholar]
- 141.Axelsson J, Rippe A, Rippe B. mTOR inhibition with temsirolimus causes acute increases in glomerular permeability, but inhibits the dynamic permeability actions of puromycin aminonucleoside. Am J Physiol Renal Physiol. 2015;308:F1056–F1064. doi: 10.1152/ajprenal.00632.2014. [DOI] [PubMed] [Google Scholar]
- 142.Wan X, Shen N, Mendoza A, Khanna C, Helman LJ. CCI-779 inhibits rhabdomyosarcoma xenograft growth by an antiangiogenic mechanism linked to the targeting of mTOR/Hif-1alpha/VEGF signaling. Neoplasia. 2006;8:394–401. doi: 10.1593/neo.05820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 144.Cutillas PR. Role of phosphoproteomics in the development of personalized cancer therapies. Proteomics Clin Appl. 2015;9:383–395. doi: 10.1002/prca.201400104. [DOI] [PubMed] [Google Scholar]
- 145.Klempner SJ, Myers AP, Cantley LC. What a tangled web we weave: emerging resistance mechanisms to inhibition of the phosphoinositide 3-kinase pathway. Cancer Discov. 2013;3:1345–1354. doi: 10.1158/2159-8290.CD-13-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Robin X, Creixell P, Radetskaya O, Santini CC, Longden J, Linding R. Personalized network-based treatments in oncology. Clin Pharmacol Ther. 2013;94:646–650. doi: 10.1038/clpt.2013.171. [DOI] [PubMed] [Google Scholar]
- 147.Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, Tao J, Campos AB, Rodon J, Ibrahim YH, et al. mTORC1 inhibition is required for sensitivity to PI3K110α inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med. 2013;5:196ra99. doi: 10.1126/scitranslmed.3005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Murray BW, Miller N. Durability of kinase-directed therapies - a network perspective on response and resistance. Mol Cancer Ther. 2015;14:1975–1984. doi: 10.1158/1535-7163.MCT-15-0088. [DOI] [PubMed] [Google Scholar]
- 150.Daub H, Specht K, Ullrich A. Strategies to overcome resistance to targeted protein kinase inhibitors. Nat Rev Drug Discov. 2004;3:1001–1010. doi: 10.1038/nrd1579. [DOI] [PubMed] [Google Scholar]
- 151.Yoshida T, Zhang G, Haura EB. Targeting epidermal growth factor receptor: central signaling kinase in lung cancer. Biochem Pharmacol. 2010;80:613–623. doi: 10.1016/j.bcp.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 152.Balius TE, Rizzo RC. Quantitative prediction of fold resistance for inhibitors of EGFR. Biochemistry. 2009;48:8435–8448. doi: 10.1021/bi900729a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dixit A, Verkhivker GM. Hierarchical modeling of activation mechanisms in the ABL and EGFR kinase domains: thermodynamic and mechanistic catalysts of kinase activation by cancer mutations. PLOS Comput Biol. 2009;5:e1000487. doi: 10.1371/journal.pcbi.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Denis MG, Vallée A, Théoleyre S. EGFR T790M resistance mutation in non small-cell lung carcinoma. Clin Chim Acta. 2015;444:81–85. doi: 10.1016/j.cca.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 157.Niederst MJ, Engelman JA. Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci Signal. 2013;6:re6. doi: 10.1126/scisignal.2004652. [DOI] [PMC free article] [PubMed] [Google Scholar]