Abstract
Q-ball imaging (QBI) is a popular high angular resolution diffusion imaging (HARDI) technique used to study brain architecture in vivo. Simulation and phantom-based studies suggest that QBI results are affected by the b-value, the number of diffusion weighting directions, and the signal-to-noise ratio (SNR). However, optimal acquisition schemes for QBI in clinical settings are largely undetermined given empirical (observed) imaging considerations. In this study, we acquire a HARDI dataset at five b-values with 11 repetitions on a single subject to investigate the effects of acquisition scheme and subsequent analysis models on the accuracy and precision of measures of tissue composition and fiber orientation derived from clinically feasible QBI at 3T. Clinical feasibility entails short scan protocols - less than five minutes in the current study - resulting in lower SNR, lower b-values, and fewer diffusion directions than are typical in most QBI protocols with research applications, where time constraints are less prevalent. In agreement with previous studies, we find that the b-value and number of diffusion directions impact the magnitude and variation of QBI indices in both white matter and gray matter regions; however, QBI indices are most heavily dependent on the maximum order of the spherical harmonic (SH) series used to represent the diffusion orientation distribution function (ODF). Specifically, to ensure numerical stability and reduce the occurrence of false peaks and inflated anisotropy, we recommend oversampling by at least 8–12 more diffusion directions than the number of estimated coefficients for a given SH order. In addition, in an equal scan time comparison of QBI accuracy, we find that increasing the directional resolution of the HARDI dataset is preferable to repeating observations; however, our results indicate that as few as 32 directions at a low b-value (1,000 s/mm2) captures most of the angular information in the q-ball ODF. Our findings provide guidance for determining an optimal acquisition scheme for QBI in the low SNR and low scan time regime, and suggest that care must be taken when choosing the basis functions used to represent the QBI ODF.
Introduction
Diffusion-weighted magnetic resonance imaging (DW-MRI) is a medical imaging technique sensitive to the random Brownian motion of water, which can provide contrasts that give insight into the tissue architecture of the brain. There exists a wide variety of methods that attempt to model the underlying three-dimensional displacement of water molecules (i.e., the diffusion propagator) from a series of “diffusion-weighted” (DW) images in order to make inferences regarding tissue microstructure dimensions, organization, and orientation. Diffusion tensor imaging (DTI) has proven useful in providing indices related to white matter integrity and estimates of the primary direction of diffusion (1), which are assumed to lie parallel to the axonal tract orientation within each MRI voxel. The diffusion directional preference has been widely applied to map the white matter fiber pathways of the brain in a process called fiber tractography (2–4).
Despite its successes, DTI can only recover a single fiber orientation in each voxel, which leads to ambiguous orientation estimates in regions of complex fiber geometries and subsequent failure of tractography (5,6). These limitations have led to the development of alternative models and algorithms that aim to recover more detailed information about the fiber geometry - collectively referred to as high angular resolution diffusion imaging (HARDI) techniques (7–11). Q-ball imaging (QBI), one of the most commonly applied HARDI methods, aims to extract an estimate of a function called the diffusion orientation distribution function (ODF) using the Funk-Radon transform of the DW signal (10). The diffusion ODF is a continuous function over a sphere that describes the probability that a water molecule diffuses in a given direction, and it is generally assumed that the ODF has its maxima aligned with the underlying fiber directions in every voxel. Measures of anisotropy and orientation can be extracted from the ODF and used for subsequent tractography. QBI and tractography have been shown to resolve complex fiber geometries in both white matter (WM) and gray matter (GM) tissue and successfully track through crossing-fiber regions (12,13). While cutting edge research in the field of diffusion MRI now largely focuses on microstructural quantification, the ability to accurately and reliably estimate orientation information is essential for subsequent fiber tractography and the study of the human connectome. Studies utilizing QBI remain pervasive in recent literature (14–20), yet there is little consensus on how best to implement this algorithm, particularly in clinical (or low SNR) settings.
The QBI acquisition is characterized by the amount of diffusion weighting (b-value) and the number of DW-directions sensitized to diffusion. Current practice is to acquire as many directions, at as high a b-value, as time will allow. For example, Tuch et al. acquire 492 gradient directions at a b-value of 12,000s/mm2 (21), and later collect 252 directions at b=3,000s/mm2 (10), while Kuo et al (22) study schemes consisting of b-values ranging from 1,000 to 6,000s/mm2 with 253+ gradient directions. However, the increased angular resolution and large diffusion-weightings come at the expense of increased scan times and decreased signal-to-noise ratios (SNR), making these schemes impractical in clinical applications or neuroscience investigations where scan time constraints exist due to cost, patient comfort, or the need for additional neuroimaging sequences. Because of the complex tradeoffs between these acquisition parameters, the optimal acquisition scheme in terms of b-value and number of DW-directions remains unclear. This problem has been partially tackled by imaging physical phantoms in order to assess the effects of acquisition schemes on the accuracy and precision of the q-ball ODF, or ODF derived metrics (23,24). However, these studies, too, have little to no constraints on acquisition time, leading to high b-values, large number of DW-directions, and favorable SNR. This problem has also been addressed with the use of synthetic data. For example, simulations (25–27) at realistic SNRs suggest QBI can adequately resolve complex fiber configurations at moderate b-values (~2000–2500s/mm2) and a relatively low number of DW-directions (48–100); however, it is unclear how well the simulated data reflects the complexities of live tissue. To date, there has been no attempt to address the effects of acquisition scheme on the reproducibility and performance of QBI using empirical, clinically feasible datasets. The current study focuses on short time acquisition paradigms, specifically scan times less than 4–5 minutes in duration. As a consequence of the short duration, the number of diffusion weighting directions remains low (less than 96), the b-value remains low (less than a maximum b-value of 3000 s/mm2), and the resulting images have SNRs much lower than is typical in research QBI applications (in this study, an SNR of ~10–12 in WM of b0 images), all of which are expected to impact the fidelity of the estimated diffusion ODF.
In addition to the QBI acquisition, there is also the question of appropriate QBI reconstruction and implementation. The q-ball ODF is typically represented using basis functions on a sphere. Original work used spherical radial basis functions (10,21) to represent the ODF. Later work uses spherical harmonics (SH) (8,28,29), which have become popular because they give a more compact representation of the ODF and reduce numerical computations. Because the SH are a complete set of orthogonal functions on a sphere, they are capable of describing any bounded function defined on the surface of a sphere (i.e., any ODF) given a sufficiently large harmonic series order. However, the number of DW-images acquired limits the maximum order that can be applied to QBI. Truncating the SH order is equivalent to apodizing the ideal ODF with a windowing function (30) which can smooth the ODF and impact both the angular resolution and measures of anisotropy. Thus, while it is desirable to reconstruct the ODF using high order SH for the highest angular resolution, the higher frequency harmonics are less reliably estimated, and, in the presence of low SNR data, can lead to false peaks in the ODF. Conversely, a smaller SH series ensures numerical stability in generating the q-ball ODF with low SNR data while potentially blurring out informative features of the ODF. While these properties are well-known, and have been characterized both theoretically (28) and experimentally using high SNR data (31), it is unknown how the maximum order SH fit of QBI impacts measures derived from the ODF on low-SNR data, and what the appropriate numerical computation should be for empirical datasets with varying numbers of DW-directions.
The aim of this study is to investigate how the acquisition and reconstruction of clinically feasible q-ball data affects the derived contrasts through a direct in vivo analysis of experimental data acquired in a short acquisition time - less than five minutes. This is done using repeated scans on a single healthy subject, at a range of b-values. The study is divided into two parts. In part one, we determine how the magnitude and reproducibility of the Q-ball ODF (and subsequent contrasts) vary as a function of the number of gradient directions, the b-value, and the maximum SH order. In the second half of the study, we ask whether increasing directional resolution is preferable to repeating observations by characterizing the effects of different scan-time-equivalent (STE) schemes on the accuracy and precision of QBI contrasts. This work extends (32) by considering the effects of SH order, different DW-schemes, and additional metrics of accuracy and reproducibility. Overall, this study provides guidance for both clinicians and researchers for determining an optimal acquisition scheme for QBI in a clinically feasible scan time.
2. Methods
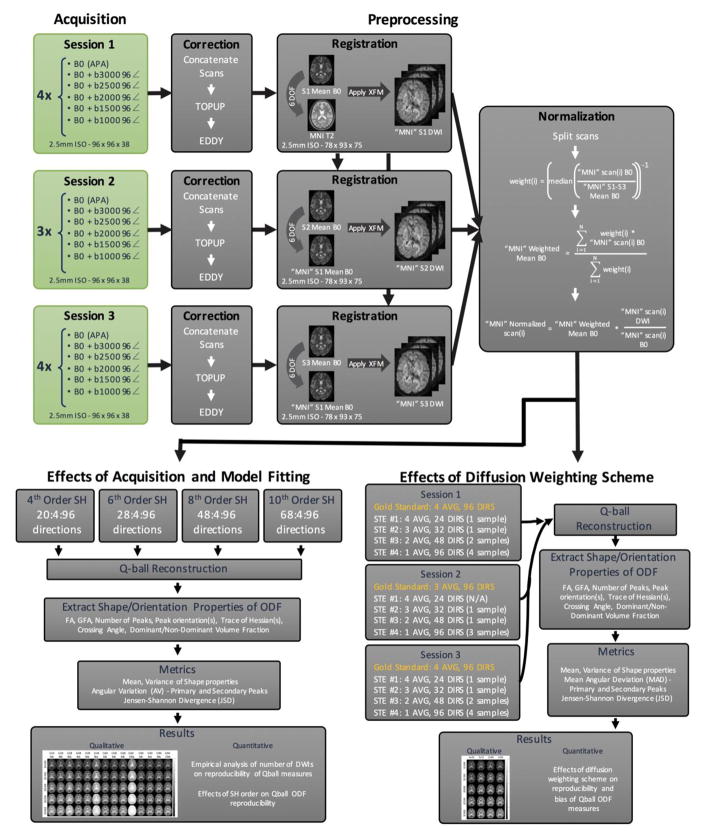
2.1 Acquisition
All data were acquired in accordance with the Vanderbilt University Institution Institutional Review Board (IRB) guidelines and with signed consent from the volunteer. Diffusion MRI was performed with a 32-channel head coil on a single healthy subject scanned on a 3T Philips Achieva Scanner (Philips Medical Systems, Best, The Netherlands), in three two-hour long sessions, on three consecutive days. The scan protocol consisted of five diffusion shells (b-values of 1000, 1500, 2000, 2500, and 3000 s/mm2) with 96 diffusion-weighted directions each (Figure 1). Each was preceded by a non-diffusion weighted (b0) volume, formed by 10 repeats averaged on the scanner. The data were acquired at 2.5mm isotropic resolution, with a matrix of 96 × 96 × 38 slices, using a DW Stejskal-Tanner monopolar PGSE-EPI sequence (multi-band=2; SENSE=2.2; TR=2650 ms; TE=101 ms; Partial Fourier=0.7, receiver bandwidth=318 kHz), giving a total scan time for a diffusion shell of 106 volumes (96 DWIs + 10 b0 images) of just over 4.5 minutes. Fold-over direction was A-P with a P fat shift. For each set of 5 shells, an additional “b0” was acquired with reverse phase encoded volumes (i.e., fold over direction A-P with A fat shift). This protocol was performed four times on days 1 and 3, and three times on day 2, resulting in a total of 11 sets of data for each diffusion shell.
Figure 1.
Standard pre-processing routines were performed within each session and within-scan normalization was performed. The effects of acquisition and model fitting were examined through subsampling the number of gradient directions used for the fits. Scan equivalent sessions were constructed to understand the tradeoffs between SNR and angular sampling.
2.2 Data Pre-Processing
All volumes from each scan session were concatenated and corrected for movement, susceptibility induced distortions, and eddy currents using FSLs topup and eddy algorithms (33) (Figure 1). The gradient table was rotated based on the transformations obtained from the corrections. Data were then registered to a common space to facilitate voxel-wise comparisons (Figure 1). First, for each scan session, all b0 images were combined to obtain the mean b0 for individual sessions. The mean b0 from session 1 (S1 Mean b0) was registered to the MNI T2 template using a 6 degrees-of-freedom (DOF) rigid registration, resulting in the “MNI” S1 Mean b0. The mean b0 from the other two sessions were then registered to “MNI” S1 Mean b0 with 6 DOF. Finally, all b0’s and diffusion weighted images from all sessions are moved to target space by applying the appropriate transform. All registration was implemented using FSLs FLIRT command (34). The images were then in MNI space, at 2.5mm isotropic resolution, resulting in an image matrix of 78X93X75.
Because of potential signal drift over time within sessions, and differences in gain and signal magnitudes between sessions, global intensity normalization was needed (Figure 1). To do this, all b0’s from all sessions (in common space) were averaged to form the overall mean b0 (MNI S1–S3 Mean b0). The individual b0 images were then divided by the overall mean b0 and the median intensity (within a brain mask) was calculated. The inverse of this value was the normalization weight used to create a weighted arithmetic mean of the b0s (“MNI” Weighted Mean b0). Finally, all DWIs were divided by their corresponding b0, and multiplied by the weighted average. This results in all DWIs normalized to the weighted b0, and corrected for scanner gains across sessions, and signal drifts within sessions.
The full processed dataset is made available through the NITRC Project: “multimodal”.
2.3 Effects of acquisition and model fitting
The first aim is to perform an empirical analysis of the number of diffusion weighted images and maximum SH order on the reproducibility of QBI measures. To assess the effects of varying the number of sampled diffusion weighting directions (Figure 1), the full 96 gradient directions were re-ordered to minimize the electrostatic potential energy of any partial set of the first “N” directions, ensuring that the first “N” directions are maximally uniformly distributed (35). From this, for each diffusion shell (b-value), subsets of DWIs from 20 to 96 directions (in increments of 4) were created for all 11 scans.
The q-ball ODF was stored in the form of spherical harmonic basis functions, which allows a simplified representation of the ODF in terms of a small number of basis coefficients. Because dMRI is incapable of distinguishing diffusion along opposite directions (i.e., ODF is antipodally symmetric), only even order SH coefficients need to be calculated. Second, because both the ODF and diffusion-weighted signal are real, the SH basis functions exhibit conjugate symmetry, further reducing the number of estimated coefficients. This means that for a SH representation of maximum order L, the number of coefficients to be estimated is reduced from (L+1)2 to (L+1)×(L+2)/2. Thus, representing the ODF using 4th, 6th, 8th, and 10th order SH necessitates acquisition of at least 15, 28, 45, and 66 diffusion weighted directions, respectively. To assess the effects of varying orders of the SH fit, all datasets were fit to 4th order SH coefficients, datasets containing 28 to 96 directions were fit to 6th order SH coefficients, those containing 48 to 96 directions were fit to 8th order, and those containing greater than 68 were fit to 10th order (Figure 1). Note that these correspond to the first multiple of 4 that meets the requirements for model fitting. A maximum order of 10 was chosen as it is the highest observed in literature (while fits of 4, 6, and 8 are by far more common).
Next, for all combinations of gradient directions and SH fits (for 11 scans), Q-ball reconstruction of the ODF was performed following the analytic (non-regularized) solution proposed in (29) using UCL’s Camino Diffusion MRI Toolkit (36). From this, shape and orientation measures were extracted (Figure 1). First, peaks of the ODF were extracted by searching for local maxima in the ODF that are larger than a threshold defined as 20% the magnitude of the largest peak (this is common in diffusion literature (37–39) to reduce the presence of false positive peaks). For each voxel, the peak orientation(s) and number of peaks were stored, and if multiple peaks were detected, the crossing angle is calculated in addition to a volume fraction for each peak. Next, the fractional anisotropy (FA), a scalar value between 0 and 1 derived from DTI that describes the anisotropy of diffusion, was calculated, as well as the generalized fractional anisotropy (GFA), an analogue of FA that describes the variation of the ODF magnitude over the sphere. Finally, for each peak, the Hessian, or second derivative matrix of the ODF, was derived and its trace was calculated. The trace of the Hessian (TH) describes the sharpness of each peak, and has been used in fiber tracking algorithms, where the log of the TH is used to find the uncertainty in peak orientation estimation (40).
From these measures, metrics of precision and reproducibility were calculated. The voxel-wise mean and variance across 11 observations were calculated for FA, GFA, log of TH, number of peaks, and crossing angle (if >1 peak). To describe precision of orientation measures, the angular variability for the primary (AV1) and secondary (AV2) peaks were computed using a method similar to that described in (41,42), although adapted to accommodate more than one peak. Briefly, the ODFs from all 11 sessions were averaged to create the mean ODF in each voxel. Local maxima of the mean ODF were extracted and designated as the mean primary peak ( ), and (if it exists) the mean secondary peak ( ). The angular variability of the primary fiber orientation was computed as the mean angle between each observation and the primary peak of the mean ODF:
where N=11, and PEAKi is the peak from the observed data that minimizes the angular difference from the mean peak. Similarly, the angular variability of the second fiber orientation (if it exists) was computed as:
where N is less than or equal to 11, and PEAKi is the peak from the observed data that minimizes the angular difference from the mean peak. Note that this can be the same peak used when calculating AV1.
Finally, to have a quantitative assessment of the reproducibility of the ODF (from which all other parameters are derived), the Jensen-Shannon divergence (JSD) was calculated. The JSD measures the distance between multiple probability distributions (in this case, the ODF over a sphere) and has been used to compare ODFs from different reconstruction methods (43) and in bootstrap analysis to quantify reproducibility of the Q-ball ODF (44). Here, we used JSD to quantify the similarity between the observed ODFs and the mean ODF. Similar to (44), we project both ODFs onto 724 values distributed equally over a sphere. Using the same notation as in (44) the JSD is defined as
with
and P(i) and Q(i) are the ODF magnitudes of the observed and mean ODFs along index i (i=1…724), and DKL is the Kullback-Leibler divergence:
The JSD is calculated for all combinations of directions, SH order, and b-values, across all 11 sessions.
All results regarding acquisition and model fit are displayed as qualitative images of a single axial slice for selected acquisition schemes and SH fits, as well as quantitative plots in select regions of interest (ROIs) (defined below).
2.4 Effects of diffusion weighting schemes
The second aim is to study the effects of diffusion weighting schemes on the reproducibility and accuracy of QBI measures. Specifically, we ask, given a specific scan time, is it better to improve angular resolution by acquiring more unique DWIs, or to improve SNR by averaging a smaller set of DWIs? We chose to define a scan time equivalent (STE) as 96 DWIs (approximately 4.5 minutes with our protocol). We grouped data from DWIs within sessions to form 4 “composite acquisitions” (Figure 1). STE#1 is an acquisition of 4 averages of 24 DW-directions (resulting in 2 total samples), STE#2 is 3 averages of 32 directions (3 samples), STE#3 is 2 averages of 48 directions (5 samples), and STE#4 is 1 average of 96 directions (11 samples). Although scans have been normalized across sessions to remove potential changes in scanner calibration, we have chosen not to directly combine diffusion data from different scan sessions in order to mitigate any potential inaccuracies in registration. Finally, all schemes are compared to the “gold standard” contrasts derived from the concatenation of all DWIs in an imaging session. It is important to remember that the SNR of the DWIs is expected to increase proportional to the square-root of the number of averages. For example, compared to the 1X96 acquisition, we expect SNR to increase by a factor of 2, , and for the 4X24, 3X32, and 2X48 schemes, respectively.
Q-ball reconstruction and extraction of shape and orientation properties of the ODF are performed as described previously. Again, metrics of accuracy and precision were calculated, including the mean and variance across all samples in each STE scheme for GFA, log of TH, number of peaks, and crossing angle. To describe errors associated with orientation estimates, the mean angular difference for the primary (MAD1) and secondary (MAD2) peaks were computed. The mean angular difference describes the averaged angular error between each observation and the gold standard primary peak (PEAK1gs) and (if it exists) the gold standard secondary peak (PEAK2gs), and is calculated as:
where N=2, 3, 5, and 11 for STE#1, 2, 3, and 4, respectively. PEAKi is the peak from the observed data that minimizes the angular difference from the gold standard peak. Finally, the JSD is calculated as described previously; however, in this case, we are measuring the similarity of each observed ODF with the gold standard ODF. Again, results are displayed as qualitative images of a single axial slice in the 4 STE schemes, as well as quantitative plots in select ROIs (defined below).
2.5 Region of interest definition
Five regions of interest (ROIs) were chosen to examine the effects of acquisition and analysis in both WM and GM structures. ROIs were manually defined in order to minimize partial volume contamination from adjacent structures. For analysis of WM structures, ROIs were chosen in the splenium of the corpus callosum (SCC), the internal capsule (IC), and the centrum semiovale (CS). These regions are representative of the spectrum of WM complexity expected in the brain, ranging from dense, coherent fiber bundles of the SCC to the complex CS which contains multiple fiber populations of varying volume fractions. The FA in the areas of the SCC, IC, and CS, are approximately 0.85, 0.62, and 0.27, respectively (Note that the results for FA are not shown - FA is derived from the diffusion tensor, and there is considerable literature on the effects of b-value, DW-scheme, and SNR on this measure). For analysis of GM structures, ROIs were chosen in the globus pallidus (GP) a sub-cortical structure (FA ~0.25) and in the precentral gyrus (PCG) of the cerebral cortex (FA ~0.12).
3. Results
3.1 Qualitative Results
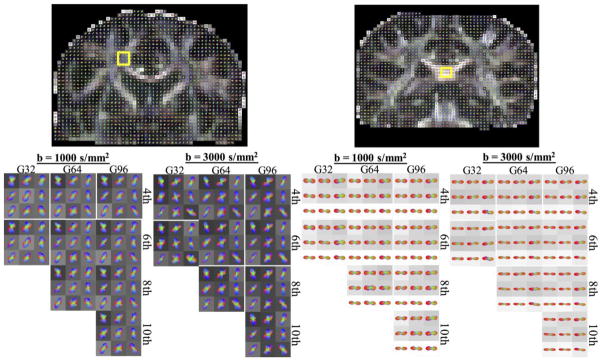
QBI glyphs for two anatomical locations are shown in Figure 2 for various combinations of diffusion weightings, number of diffusion directions, and different maximum SH orders. The glyphs in a complex fiber crossing region (Figure 2, left) are generally able to resolve the intersection of the callosal fibers and cortical spinal fibers, with some exhibiting the presence of a third peak oriented anterior/posterior (association fibers). However, the most striking feature of these glyphs is the presence of an increased number of peaks (often more than three), and the sharpness of the peaks, when fitting with 8th and 10th order SH, for both low and high b-values. In addition, there is little difference in the 4th and 6th order fits between the lowest 32 directions and the full set of 96 directions.
Figure 2.
QBI glyphs in crossing fiber (left) and single fiber (right) regions, for various b-values, number of gradient directions, and maximum SH order fit. Glyphs are shown min-max normalized and are displayed on top of fractional anisotropy maps.
For the single fiber region of the SCC (Figure 2, right), there is little noticeable difference across various fitting orders, and across gradient directions. However, the ODF lobes are noticeably sharper at a b-value of 3,000 s/mm2 than at 1,000 s/mm2. Small, spurious peaks are sometimes noticeable, which justifies the use of a threshold (relative to the maximum peak amplitude) when counting the number of fiber populations in a voxel.
3.2 Effects of acquisition and model fitting
GFA and crossing angle
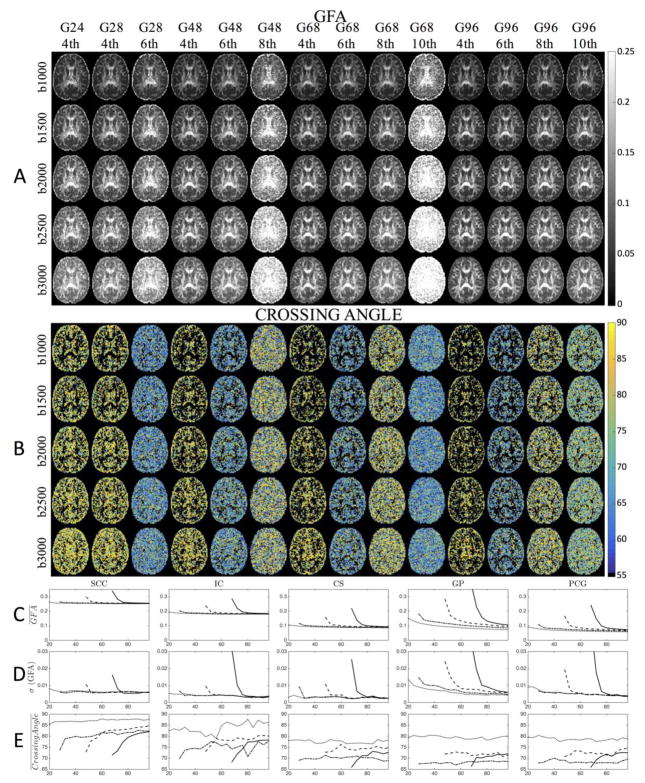
Representative maps of GFA and crossing angle are shown for select combinations of DW-directions and maximum SH order in Figure 3 (a and b), for the middle axial slice. The directions (for all qualitative results) are chosen to span a wide range of possible acquisitions schemes and to highlight the “transition zones” where the next highest SH order can be fit. This includes: 24 directions, from which only 4th order SH can be fit; 28 directions, from which 4th order can be fit, and is the minimum sampling requirement for 6th order fit; 48 directions, from which 4th and 6th order can be fit, and is the minimum sampling requirement (which is a multiple of 4) for 8th order fit; 68 directions, from which 4th, 6th, and 8th, order can be fit, and is the minimum sampling requirement (which is a multiple of 4) for 10th order fit; and 96 directions, which is the maximum acquired in this study.
Figure 3.
The effect of the number of gradient directions, and maximum SH order on the GFA and Crossing Angle are shown in representative axial slices (A and B) and ROI-based analysis (C–E) for a b-value of 2,000 s/mm2. Maximum SH order fit is indicated by line style (dotted line=4th order; dash-dot=6th order; dashed=8th order; solid=10th order). Note, in the Crossing Angle figures (B), voxels with only one resolved fiber population are shown as black. For results at all b-values, see Supplementary Figure 1.
Qualitatively, GFA appears higher in WM than in GM, and the contrast between the two tissues is decreased at larger b-values (Figure 3a). Most striking is the loss of GM/WM contrast and increased GFA across the entire brain when first transitioning towards higher SH orders. However, in areas not in these transition zones, the GFA maps appear to be quite similar and of comparable quality.
These trends are in agreement with quantitative ROI analysis. Here, the mean (Figure 3c) and standard deviation (Figure 3d) of the GFA across all 11 sessions for a b-value of 2,000 s/mm2 are shown, across all ROIs (for results at all b-values, see Supplementary Figure 1). In descending order, the GFA for ROIs closely follow that of FA (i.e. SCC, IC, GP, CS, PCG), and there is an increase in GFA as b-value increases (Supplementary 1a). Here, it is clear that the GFA, for all regions, is overestimated when switching to a higher order SH fit. The anisotropy is regularized with increasing angular samples, approaching that of the 4th order fit after approximately 32, 52, and 80 samples for 6th, 8th, and 10th orders, respectively (with the exception of GP which takes slightly longer to regularize). However, in all cases, a higher-order fit always results in a larger GFA. The variation across 11 sessions also shows that these transition zones result in less reproducible measurements. In addition, the results in GM regions show more variance than those of the WM ROIs.
Maps of crossing angles (Figure 3b) show that resolved crossing occurs at more acute angles at the “transition zones”. It is also interesting that the crossing angle appears somewhat homogenous across the entire brain in most cases, suggesting that the resolved intravoxel crossing is influenced more heavily by the number of DW-directions and q-ball model rather than the underlying anatomical geometry. Plots of the crossing angle (Figure 3) show that the SCC and IC tend towards orthogonal crossing as directional resolution increases; however very few crossings are expected in these regions, so they are likely false positive peaks. Crossings in GP, CS, and PCG are heavily dependent on SH order, and tend to remain fairly constant for all direction numbers (except for 10th order, where there is again a trend towards orthogonality). It is expected that 4th order SH resolve mostly orthogonal crossings (78–90°), however it is unexpected that the 6th order SH fits result in the most acute crossings (for GP, CS, PCG), with 8th and 10th order in the intermediate range. Crossing angles become slightly more acute at higher b-values (see Supplementary Figure 1c); however, the average crossing angle in any ROI never reached below 65 degrees, and very few crossings were observed at less than 60 degrees.
Trace of Hessian (log) and Number of Peaks
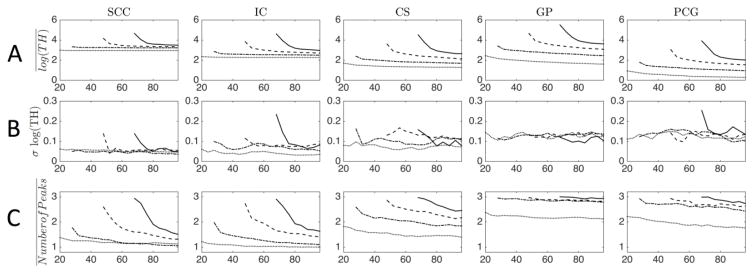
Maps of the log(TH) and the number of peaks are shown in Supplementary Figure 2 (a and b), again for an axial slice of the brain, and for the same acquisition and processing schemes as in Figure 3. Qualitatively, the TH shows strong contrast between GM and WM (Supplementary Figure 2a), with very little dependence on b-value. Similar to GFA, contrast is lost at transition zones with resulting overestimation of peak sharpness throughout the entire brain. Plots of the log(TH) across gradient directions (Figure 4a) show that the highest TH belong to the primary peaks of the GP and the SCC, followed by IC, CS, and PCG. Again, regularization occurs after oversampling by approximately 4, 7, and 14 samples for 6th, 8th, and 10th order SH, although the log(TH) never approaches that of the 4th order fit, and is continually decreasing as the number of DW-directions increases. Finally, the variation of log(TH) (Figure 4b) shows slight decreases as the number of directions increase for higher order fits, although it remains fairly constant for 4th and 6th orders.
Figure 4.
The effect of the number of gradient directions, and maximum SH order on the log(TH) (A, B) and Number of Peaks (C) are shown as an ROI-based analysis for a b-value of 2,000 s/mm2. Maximum SH order fit is indicated by line style (dotted line=4th order; dash-dot=6th order; dashed=8th order; solid=10th order). For results at all b-values, see Supplementary Figure 3.
The number of extracted peaks is also heavily dependent on the SH order (see Supplementary Figure 2b for qualitative results). Fitting the data exactly in the low-SNR clinical scans results in 3 peaks consistently, in both WM and GM, even when no crossing fibers are expected. Overall, there are fewer peaks identified with a lower order fit. A 4th order SH fit consistently shows one peak throughout most of WM with one or two peaks in GM, while all other orders tend to show one or two peaks in WM, with 3 identified in GM. These results are verified in plots (Figure 4c) for a b-value of 2,000 s/mm2 (plots for all b-values, and all ROIs are provided in Supplementary Figure 3). The GP and PCG consistently show the presence of 3 peaks (except for 4th order fit, where 2–2.5 peaks are identified on average), the SCC and IC trend towards 1 peak, while the CS is intermediate, ranging from as many as 3 to as few as 1.2 average peaks depending on the scheme. Three trends are apparent in all regions. First, for a given number of directions, a lower order SH fit always results in a smaller number of extracted peaks. Second, the number of local maxima decreases as the number of directions increase. Finally, a higher b-value results in an elevated number of peaks (Supplementary Figure 3).
JSD, Angular Variation 1, Angular Variation 2
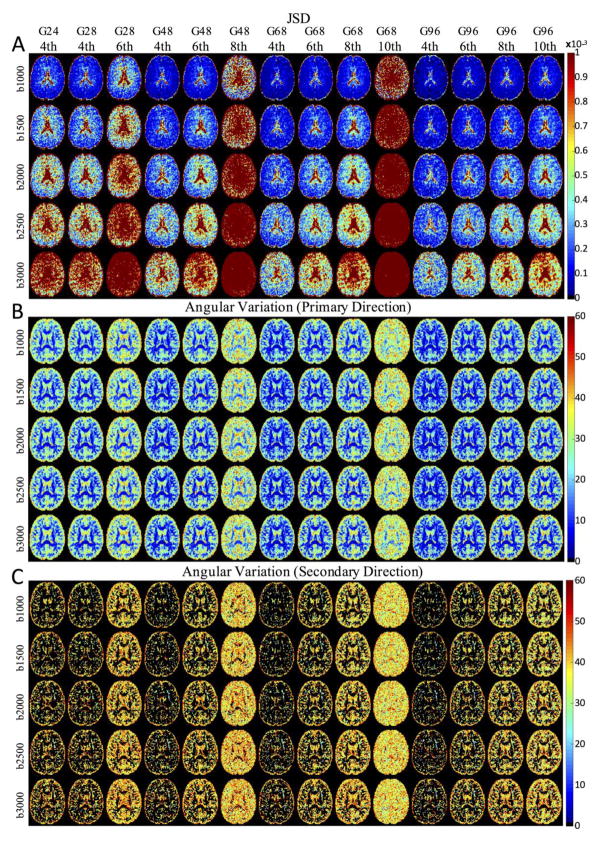
To describe the reproducibility of the entire ODF, as well as the spread of the estimated orientations about the observation mean, maps of JSD, AV1, and AV2 are shown in Figure 5. In most cases, the JSD is higher in the ventricles, and there is no clear contrast between GM and WM (Figure 5a). The findings of (44) are reproduced, where the JSD increases towards the center of the brain, likely reflecting the decreased sensitivity of the array coils in this area and suggesting a direct relationship between SNR and reproducibility of the ODF reconstruction. Similarly, the ODFs reconstructed at higher b-values (lower SNR in DWIs) show larger JSD. As in all previous measures (all of which are derived from the ODF), the reproducibility is significantly reduced at the transition zones, but quickly overcome by compensating with greater angular oversampling.
Figure 5.
The effect of b-value, number of gradient directions, and maximum SH order on JSD, AV1, and AV2 are presented in representative axial slices.
The angular variation around the primary diffusion direction (Figure 5b) is typically less than 10° throughout much of the WM, but approaches 30° in GM regions. However, the angular variation about the secondary direction (Figure 5c) is much higher, typically greater than 30° throughout the entire brain. For both AV1 and AV2, there is little difference across b-value, and the only schemes where differences can be appreciated are 8th order SH fit at 48 directions, and 10th order SH fit at 68 directions (the transition zones).
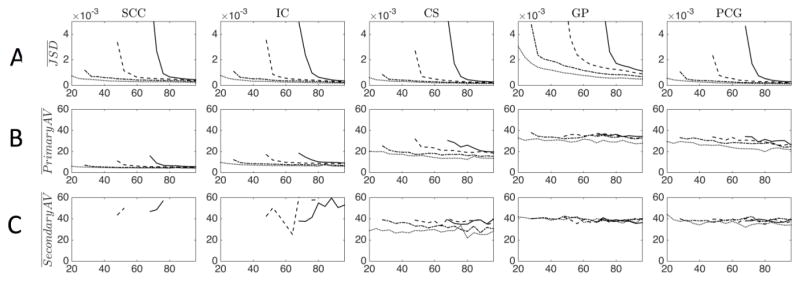
The ROI-based results across all directions are plotted in Figure 6 for b=2,000 s/mm2. The JSD (Figure 6a) decreases as the number of direction increases. Again, oversampling is required by approximately 4–14 directions (depending on SH order) for the ODF reproducibility to be on par with that from 4th order fit. In all cases, the JSD for a higher order fit is always larger than that of the lower order fits, and the JSD increases as b-value increases (see Supplementary Figure 4 for all b-value plots).
Figure 6.
The effect of number of gradient directions, and maximum SH order on JSD, AV1, and AV2 are presented for an ROI-based analysis for a b-value of 2,000 s/mm2. Maximum SH order fit is indicated by line style (dotted line=4th order; dash-dot=6th order; dashed=8th order; solid=10th order). For results at all b-values, see Supplementary Figure 4.
The WM ROIs (SCC, IC) showed greater precision (typically less than 10°) in estimation of the primary diffusion direction (Figure 6b), while the GM ROIs (GP, PCG) had a much larger angular variation (25–35°). The CS had an intermediate variation of 15–30°, depending on acquisition and analysis model. In all cases, lower SH order had a higher precision. In WM, significant reduction in variation was achieved after acquisition of 36, 52, and 76–80 acquired images for 6th, 8th, and 10th order fits. The angular precision of the secondary peak (Figure 6c) was always poor, with variation typically greater than 30° for all acquisition and processing schemes, all ROIs, and all b-values (note that data points are not shown if second peaks are not present in the ROI for a given scheme).
3.3 Effects of Diffusion Weighted Scheme
GFA and Crossing Angle
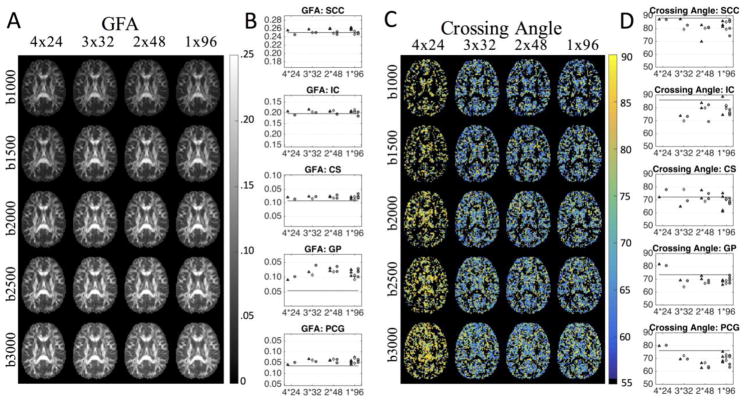
Representative maps of the GFA and crossing angle are shown in Figure 7 for the four different STE schemes. There is little noticeable difference in GFA across schemes (Figure 7a), however, there is an apparent increase in GFA in GM regions as b-value increases, reducing contrast between GM and WM. Figure 7b shows plots of the observed GFAs within ROIs by each diffusion scheme, with the gold standard contrast designated as a horizontal line (results are shown for a b-value of 2,000 s/mm2, for results at all b-values, see Supplementary Figure 5). With the exception of the GP, the gold standard GFA increases for all ROIs as b-value increases. In the simple WM regions of the SCC and IC, there appears to be no bias between any scheme and the gold standard. However, there is an upward bias of GFA for CS, GP, and PCG for all STE scans.
Figure 7.
The effects of diffusion-weighting scheme on GFA and crossing angle for four scan-time-equivalent sessions are shown as representative axial slices and as ROI-based analysis. The three separate scan sessions are represented as triangles, diamonds, and circles. Horizontal lines indicate the mean over three sessions of all data acquired per session, for the corresponding STE, ROI, and b-value. Note that axis limits are different across ROIs (while the scale remains the same). Results are shown for a b-value of 2,000 s/mm2, for results at all b-values, see Supplementary Figure 5. If no second fiber crossing is detected, no data point is shown.
Maps of fiber crossing angles (Figure 7c) highlight the near orthogonal crossings resolved in the 4X24 protocol, as opposed to the more acute angles resolved across the other STEs. Here, b-value appears to have little effect on intra-voxel crossings. Plots of the average crossing angle in each ROI (Figure 7d) confirm that the 4X24 acquisition tends to resolve fibers as crossing at more orthogonal angles, particularly in the GP, and PCG (for results at all b-values, see Supplementary Figure 5). Note that both the SCC and ICC are expected to be predominantly single fiber regions. However, a small number of voxels in the “gold standard” dataset still exhibited multiple fiber populations in these regions (average crossing angle is shown as a solid horizontal line), which are likely the result of false positive peaks. Finally, the results in the CS, where multiple fiber populations are expected, show general consistency across all schemes.
Trace of Hessian and Number of Peaks
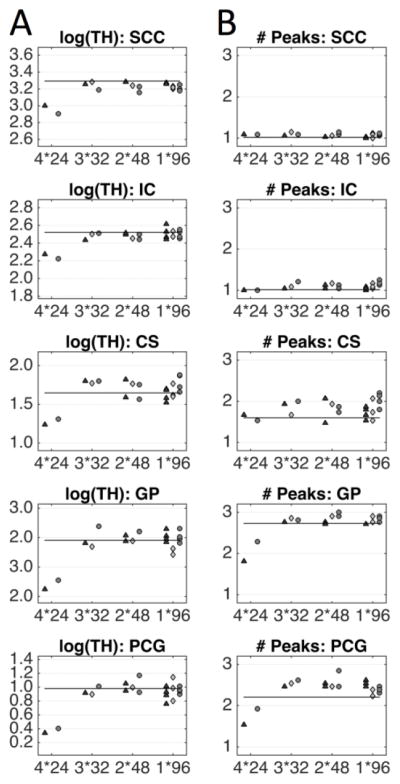
Maps of the log(TH) for varying STE sessions (Supplementary Figure 6a) appear to be of similar quality, and differences across schemes or b-values are not discernable by visual inspection alone. However, ROI analysis of the mean log(TH) shows that the 4X24 protocol has a downward bias in TH relative to the gold standard contrast, for both GM and WM. All other STEs show no apparent bias, and little to no difference across schemes (Figure 8a). ROI analysis for all b-values is shown in Supplementary Figure 6b.
Figure 8.
The effects of diffusion-weighting scheme on Number of Peaks and log(TH) for four scan-time-equivalent sessions are shown as ROI-based analysis. The three separate scan sessions are represented as triangles, diamonds, and circles. Horizontal lines indicate the mean over three sessions of all data acquired per session, for the corresponding STE, ROI, and b-value. Note that axis limits are different across ROIs (while the scale remains the same). Results are shown for a b-value of 2,000 s/mm2, for results at all b-values, see Supplementary Figure 6.
When looking at the number of identified peaks Supplementary Figure 6c), it is clear that the 4X24 analysis consistently resolves less fiber populations. This is clearly seen in plots of the CS, GP, and PCG (Figure 8b), where this scheme results in lower number of identified peaks. The higher angular resolution schemes tend to overestimate the number of peaks in these regions. It is interesting that the 4X24 protocol leads to the opposite effect at b-values of 2500 and 3000 in the SCC (Supplementary Figure 6d), resulting in overestimation of the numbers of peaks, where just a single fiber population is expected.
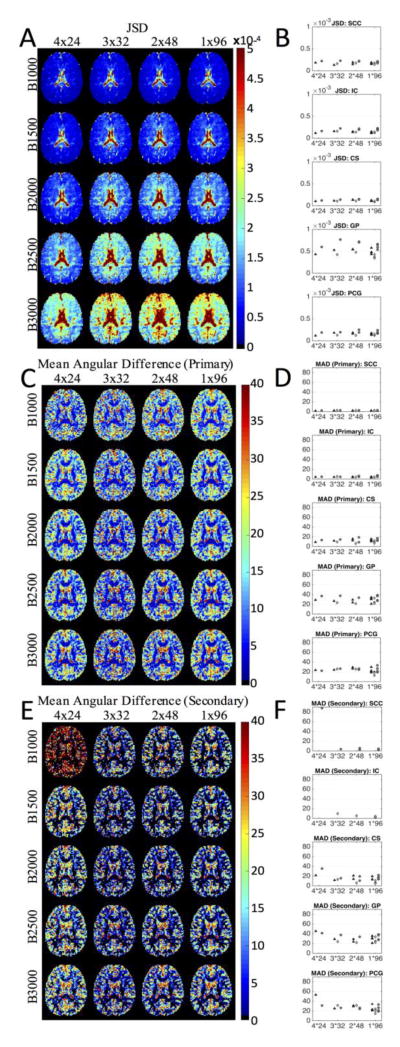
JSD, Mean Angular Difference 1, Mean Angular Difference 2
The STE results for JSD, MAD1, and MAD2 are shown qualitatively in Figure 9, along with plots of the mean quantity in the ROIs for all samples of the different schemes (for plots at all b-values, see Supplementary Figure 7). Visually, the JSD shows little difference across protocols (Figure 9a), although there is a slightly increased reproducibility with 4 averages of 24 directions. Similar to earlier results, JSD is higher in the central brain (particularly the ventricles), and increases as b-value increases. From ROI analysis (Figure 9b), we confirm little difference between the STEs, and notice a higher reproducibility in the WM relative to GM, with the highest JSD seen in the GP in the middle of the brain.
Figure 9.
The effects of diffusion-weighting scheme on JSD, MAD1, and MAD2 for four scan-time-equivalent sessions shown as representative axial slices and as ROI-based analysis at a b-value of 2,000 s/mm2. The three separate scan sessions are represented as triangles, diamonds, and circles. For ROI-based results at all b-values, see Supplementary Figure 7.
From the MAD1 images (Figure 9c), there is almost no difference detectable across differing STE acquisitions, and, in agreement with previous results, little to no difference across b-values. These results are confirmed in the ROI plots (Figure 9d). In addition, the SCC and IC have less than 10° average angular difference, the CS shows less than 20° MAD, while the GP and PCG show significant angular difference of 20–40°. The ROI analysis suggest that, for the primary direction detection, there is no benefit to increasing angular resolution, and overall low orientation accuracy in GM regions.
Visual inspection of the MAD2 images (Figure 9e) shows that the 4X24 protocol results in the largest angular differences, particularly in the b=1000 DWIs. There is little noticeable difference in the mean deviation from the second diffusion peak in all other schemes. This is confirmed in the ROI-based analysis (Figure 9f), where the 4X24 consistently results in larger angular deviation for both WM and GM regions. In CS, where multiple fiber populations are expected, the mean angular difference is typically less than 20° (for all schemes except 4X24), and there is slight reduction in MAD2 at larger b-values. For the GM ROIs (GP and PCG), the angular deviation is much larger, typically ranging from 20–40°. Thus, the second peak shows considerable deviation in the GM.
Discussion
The primary aim of this study was to assess the effects of the DW imaging acquisition and reconstruction method on QBI from a clinically feasible dataset. The question of the appropriate b-value and number of unique sampling directions needed to adequately describe the q-ball (and HARDI methods in general) is an open area of research. Tournier et al. (31) studies this by analyzing the angular structure of the diffusion-weighted signal in order to determine the highest angular frequency present in the DW-data. Given a certain angular frequency, the acquisition scheme needs only to contain as many coefficients necessary to recover the SH coefficients that contain that frequency. They show that at a b-value of 1000 s/mm2, spherical harmonics beyond order 4 are negligible in practice when characterizing the signal profile. Consequently, only 15 unique DWI’s are necessary to estimate these harmonics. Their results also suggest that the highest angular resolution achievable is obtained at a b-value of 3000 s/mm2, which can be fully characterized using order 8 SH (requiring only 45 DW-directions). However, with low-to-moderate SNR (SNR less than ~15 in the study), order <=6 terms capture the highest frequency, meaning that, in practice, only 28 DW-directions are needed to characterize any orientation information. However, they caution that these represent the bare minimum number of directions, and recommend acquiring more in order to meet SNR requirement of the intended reconstruction method.
Our findings clearly demonstrate the importance of oversampling the minimum number of directions for a given SH order. Without appropriate oversampling, the resulting SH coefficients fit the ODF (including noise errors) exactly, leading to low reproducibility of the ODF, increased measures of anisotropy, and spurious peaks. Specifically, to estimate SH coefficients of order 6, we recommend sampling 32–36 unique DW-directions (as opposed to the minimum 28 directions) in order to reduce variability of the ODF, reduce the number of peaks and angular variation of the peaks, and to regularize measures of anisotropy. We recommend even greater oversampling for higher orders, for example 52–56 directions for 8th order, and 76–80 for 10th order. However, we find no substantial advantages in fitting SH orders larger than 6. This means that in low-SNR empirical datasets, as few as 32–36 directions are required to reproducibly characterize the angular content of the ODF. In addition, we conclude that there is also no additional advantage to increasing the diffusion weighting beyond a b-value of 1,000 s/mm2. This is a surprising result, and is typically not the acquisition paradigm of choice when it comes to research studies, where higher b-values are used due to the theoretical increase in angular resolution at high b-values (10). Although there is a slightly increased ability to resolve crossing fibers at larger diffusion weightings, there is no significant improvement in accuracy or precision when estimating the primary or secondary fiber orientations. In addition, the higher b-values lead to less reproducible ODFs (larger JSD), likely due to the decreased SNR. It is interesting that the lowest reproducibility (lowest JSD) regions are located near the center of the brain where the lowest SNR is expected. It is also important to point out that all diffusion weightings in this study were acquired at the same 101 ms echo time (resulting in the same SNR for all b0 images). This is not optimal for the lower b-value shells, which can be acquired with a shorter TE and subsequent increase in SNR. Thus, the lower b-value shells in our study likely underestimate the reproducibility of these metrics that can be achieved with an optimized TE for a single low b-value shell, further emphasizing the importance of low b-value DWIs in low-SNR datasets. It is important to point out that these results regarding optimal b-value are not generalizable to high SNR datasets (often acquired in the research domain), where time is not as critical a factor as in the clinic. Overall, we find that QBI on our data sets performs quite poorly at resolving discrete measures of orientation information or discrete measures derived from these peaks. However, the overall shape of the ODF is fairly reproducible (as shown by the JSD), even with acquisition protocols similar to those recommended for DTI (low b-value, low number of directions).
Another question regarding QBI acquisition is whether it is more beneficial to increase angular sampling (more unique directions), or image SNR (more image averages) with a limited time available for scanning. Interestingly, we found very little difference in accuracy or precision of orientation or anisotropy metrics with 32 or more directions acquired. The only STE protocol that did not result in high accuracy was 4 averages of 24 directions. This suggests that, even with the increased SNR from 4 averages, 24 directions are not adequate to characterize that q-ball ODF. Again, we found little benefit to increasing b-value, even at the higher SNRs afforded by multiple averaging. It should be emphasized that our dataset represents the lower range of SNR expected in diffusion imaging, with SNR of approximately 10–12 for WM in the non-diffusion weighted image. However, this data was acquired quickly (>100 volumes in 4.5 minutes), and is in the range expected in a clinically relevant time-limited scan. It is expected that datasets with higher SNR will experience considerable improvement in reproducibly extracting discrete orientation measures from the ODF.
There are additional parameters that may exist for QBI that were not addressed in this study. For example, different approaches are proposed to use regularization methods while solving for the SH coefficients when large orders are estimated. For example, Tikhinov (28) and Laplace-Beltrami (29) regularization have been used to minimize the perturbing effects of high order fits, at the cost of a slightly reduced angular resolution (29). However, this was not introduced in our study for two reasons. First, several software and open-source implementations do not do this, and our goal was to reproduce what is arguably the most commonly implemented (and most simple) form of QBI, and second, it makes the parameter space to be investigated less tractable. Regularization will certainly reduce false peaks and reduce anisotropy, but it is likely that oversampling is still required for estimates of any SH order. One form of regularization that was applied is removing ODF peaks whose amplitude does not meet some threshold relative to the maximum ODF amplitude in each voxel (20% in this study). This is commonly done to reduce false positive peaks (37). Despite this threshold, false positive peaks were still prevalent in our data when fitting to high SH orders, even in regions known to contain a single dominant fiber population. In addition to regularization parameters, there are also multiple variations of the q-ball algorithm itself, some of which have been shown to result in sharper ODFs (45–47). Furthermore, QBI is just one of a large number of HARDI techniques (see (48) and (49) for validation and performance assessment of a comprehensive set of algorithms on phantom and simulated data). However, we have chosen to perform a thorough analysis on QBI, arguably the most commonly utilized HARDI technique.
A limitation of this study is the use of a single subject to compare different acquisition paradigms and analysis methods on QBI. This research follows on existing single-subject studies (41,42,44) that follow a single healthy subject in detail over repeated acquisitions in order to assess reproducibility and reliability of quantitative diffusion MRI measures. Thus, we expect these findings to be generalizable across multiple individuals. Decreased imaging time, which is a necessity for clinical relevance, is accompanied by an inevitable SNR loss, making the findings of this study particularly valuable for analyzing any set of DWIs that may have a low to moderate SNR. In addition, it is expect that advances resulting in higher SNR will be utilized to acquire higher resolution datasets(50), particularly if fiber tracking is the ultimate goal.
The ability of QBI to accurately and reliably describe the underlying fiber orientation in each voxel is critical for success of fiber tracking. The optimal HARDI acquisition in regards to accurate and reproducible characterization of the underlying fiber geometries is still an open question. In deterministic tracking, it is common to extract only the local maxima in the ODF (i.e., peaks) to use for propagating the track streamlines. Similarly, for probabilistic tractography, the peaks must be properly estimated, as well as some measure of uncertainty, such as the TH. Our results show that for DW-MRI scans achievable in a limited time (less than 5 minutes), the primary peaks in WM are reliably estimated; however, the secondary peak is often not extracted, and if detected, is not precise. In GM, neither primary nor secondary peaks are reproducible, or accurate, varying by as much as 30–50° across sessions. Fiber tracking from or into the cortex, or through deep brain structures, is unlikely to be anatomically accurate. Similarly, even in regions of crossing fibers, the resolved structure is heavily dependent on the SH fit used to model the ODF. Finally, the log(TH) was also heavily dependent on the SH order, and continually decreases as more directions were acquired. Thus, results of fiber tracking will be heavily dependent on the acquisition and analysis protocols. Great care must be taken when comparing clinical studies acquired at different b-values, using different acquisition schemes, and processed using different SH orders.
Conclusion
In this paper, the effects of acquisition and subsequent analysis models on the accuracy and precision of clinically feasible QBI were investigated. We find that the b-value and the number of DW-directions impact measures of the diffusion ODF orientation and anisotropy. In addition, we show the importance of oversampling DW directions to suppress noise when estimating the QBI ODF. Our results indicate that as few as 32 directions, and low b-values, can reproducibly capture a majority of the orientation information from the DW-measurements. These results provide guidance for determining optimal acquisition schemes and analysis protocols for QBI where scan time constraints exist, and low SNRs are expected.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health under award numbers R01EB017230 (Landman) and R01NS058639 (Anderson). This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. This project was supported in part by the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alexander AL, Lee JE, Lazar M, Field AS. Diffusion tensor imaging of the brain. Neurotherapeutics. 2007;4(3):316–329. doi: 10.1016/j.nurt.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annals of neurology. 1999;45(2):265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2000;44(4):625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 4.Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Raichle ME. Tracking neuronal fiber pathways in the living human brain. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(18):10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander AL, Hasan KM, Lazar M, Tsuruda JS, Parker DL. Analysis of partial volume effects in diffusion-tensor MRI. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2001;45(5):770–780. doi: 10.1002/mrm.1105. [DOI] [PubMed] [Google Scholar]
- 6.Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? NeuroImage. 2007;34(1):144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexander DC, Barker GJ, Arridge SR. Detection and modeling of non-Gaussian apparent diffusion coefficient profiles in human brain data. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2002;48(2):331–340. doi: 10.1002/mrm.10209. [DOI] [PubMed] [Google Scholar]
- 8.Anderson AW. Measurement of fiber orientation distributions using high angular resolution diffusion imaging. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2005;54(5):1194–1206. doi: 10.1002/mrm.20667. [DOI] [PubMed] [Google Scholar]
- 9.Ozarslan E, Shepherd TM, Vemuri BC, Blackband SJ, Mareci TH. Resolution of complex tissue microarchitecture using the diffusion orientation transform (DOT) NeuroImage. 2006;31(3):1086–1103. doi: 10.1016/j.neuroimage.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 10.Tuch DS. Q-ball imaging. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2004;52(6):1358–1372. doi: 10.1002/mrm.20279. [DOI] [PubMed] [Google Scholar]
- 11.Tournier JD, Calamante F, Gadian DG, Connelly A. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. NeuroImage. 2004;23(3):1176–1185. doi: 10.1016/j.neuroimage.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 12.Berman JI, Chung S, Mukherjee P, Hess CP, Han ET, Henry RG. Probabilistic streamline q-ball tractography using the residual bootstrap. NeuroImage. 2008;39(1):215–222. doi: 10.1016/j.neuroimage.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 13.Campbell JS, Siddiqi K, Rymar VV, Sadikot AF, Pike GB. Flow-based fiber tracking with diffusion tensor and q-ball data: validation and comparison to principal diffusion direction techniques. NeuroImage. 2005;27(4):725–736. doi: 10.1016/j.neuroimage.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Caverzasi E, Hervey-Jumper SL, Jordan KM, Lobach IV, Li J, Panara V, Racine CA, Sankaranarayanan V, Amirbekian B, Papinutto N, Berger MS, Henry RG. Identifying preoperative language tracts and predicting postoperative functional recovery using HARDI q-ball fiber tractography in patients with gliomas. J Neurosurg. 2016;125(1):33–45. doi: 10.3171/2015.6.JNS142203. [DOI] [PubMed] [Google Scholar]
- 15.Caiazzo G, Trojsi F, Cirillo M, Tedeschi G, Esposito F. Q-ball imaging models: comparison between high and low angular resolution diffusion-weighted MRI protocols for investigation of brain white matter integrity. Neuroradiology. 2016;58(2):209–215. doi: 10.1007/s00234-015-1616-3. [DOI] [PubMed] [Google Scholar]
- 16.Ding G, Chen J, Chopp M, Li L, Yan T, Davoodi-Bojd E, Li Q, Davarani SP, Jiang Q. White matter changes after stroke in type 2 diabetic rats measured by diffusion magnetic resonance imaging. J Cereb Blood Flow Metab. 2017;37(1):241–251. doi: 10.1177/0271678X15622464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southwell DG, Riva M, Jordan K, Caverzasi E, Li J, Perry DW, Henry RG, Berger MS. Language outcomes after resection of dominant inferior parietal lobule gliomas. J Neurosurg. 2017:1–9. doi: 10.3171/2016.8.JNS16443. [DOI] [PubMed] [Google Scholar]
- 18.Pujol S, Wells W, Pierpaoli C, Brun C, Gee J, Cheng G, Vemuri B, Commowick O, Prima S, Stamm A, Goubran M, Khan A, Peters T, Neher P, Maier-Hein KH, Shi Y, Tristan-Vega A, Veni G, Whitaker R, Styner M, Westin CF, Gouttard S, Norton I, Chauvin L, Mamata H, Gerig G, Nabavi A, Golby A, Kikinis R. The DTI Challenge: Toward Standardized Evaluation of Diffusion Tensor Imaging Tractography for Neurosurgery. J Neuroimaging. 2015;25(6):875–882. doi: 10.1111/jon.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calabrese E. Diffusion Tractography in Deep Brain Stimulation Surgery: A Review. Front Neuroanat. 2016;10:45. doi: 10.3389/fnana.2016.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bizzi A. Diffusion Imaging with MR Tractography for Brain Tumor Surgery. In: Stippich C, editor. Clinical Functional MRI: Presurgical Functional Neuroimaging. Berlin, Heidelberg: Springer Berlin Heidelberg; 2015. pp. 179–228. [Google Scholar]
- 21.Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40(5):885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 22.Kuo LW, Chen JH, Wedeen VJ, Tseng WY. Optimization of diffusion spectrum imaging and q-ball imaging on clinical MRI system. NeuroImage. 2008;41(1):7–18. doi: 10.1016/j.neuroimage.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Cho KH, Yeh CH, Tournier JD, Chao YP, Chen JH, Lin CP. Evaluation of the accuracy and angular resolution of q-ball imaging. NeuroImage. 2008;42(1):262–271. doi: 10.1016/j.neuroimage.2008.03.053. [DOI] [PubMed] [Google Scholar]
- 24.Tournier JD, Yeh CH, Calamante F, Cho KH, Connelly A, Lin CP. Resolving crossing fibres using constrained spherical deconvolution: validation using diffusion-weighted imaging phantom data. NeuroImage. 2008;42(2):617–625. doi: 10.1016/j.neuroimage.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Alexander DC, Barker GJ. Optimal imaging parameters for fiber-orientation estimation in diffusion MRI. NeuroImage. 2005;27(2):357–367. doi: 10.1016/j.neuroimage.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Prckovska V, Achterberg HC, Bastiani M, Pullens P, Balmashnova E, Ter Haar Romeny BM, Vilanova A, Roebroeck A. Optimal short-time acquisition schemes in high angular resolution diffusion-weighted imaging. Int J Biomed Imaging. 2013;2013:658583. doi: 10.1155/2013/658583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkins B, Lee N, Gajawelli N, Law M, Lepore N. Fiber estimation and tractography in diffusion MRI: development of simulated brain images and comparison of multi-fiber analysis methods at clinical b-values. NeuroImage. 2015;109:341–356. doi: 10.1016/j.neuroimage.2014.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess CP, Mukherjee P, Han ET, Xu D, Vigneron DB. Q-ball reconstruction of multimodal fiber orientations using the spherical harmonic basis. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2006;56(1):104–117. doi: 10.1002/mrm.20931. [DOI] [PubMed] [Google Scholar]
- 29.Descoteaux M, Angelino E, Fitzgibbons S, Deriche R. Regularized, fast, and robust analytical Q-ball imaging. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2007;58(3):497–510. doi: 10.1002/mrm.21277. [DOI] [PubMed] [Google Scholar]
- 30.Healy DM, Hendriks H, Kim PT. Spherical Deconvolution. Journal of Multivariate Analysis. 1998;67(1):1–22. [Google Scholar]
- 31.Tournier JD, Calamante F, Connelly A. Determination of the appropriate b value and number of gradient directions for high-angular-resolution diffusion-weighted imaging. NMR in biomedicine. 2013;26(12):1775–1786. doi: 10.1002/nbm.3017. [DOI] [PubMed] [Google Scholar]
- 32.Schilling KG, Nath V, Blaber J, Harrigan RL, Ding Z, Anderson AW, Landman BA. Effects of b-value and number of gradient directions on diffusion MRI measures obtained with Q-ball imaging. 2017 doi: 10.1117/12.2254545. 101330N-101330N-101337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20(2):870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 34.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 35.Cook PA, Symms M, Boulby PA, Alexander DC. Optimal acquisition orders of diffusion-weighted MRI measurements. Journal of magnetic resonance imaging: JMRI. 2007;25(5):1051–1058. doi: 10.1002/jmri.20905. [DOI] [PubMed] [Google Scholar]
- 36.Cook PA, YB, Nedjati-Gilani S, Seunarine KK, Hall MG, Parker GJ, Alexander DC. Camino: Open-Source Diffusion-MRI Reconstruction and Processing. Seattle, WA, USA: May, 2006. p. 2759. [Google Scholar]
- 37.Alexander DC. Multiple-Fiber Reconstruction Algorithms for Diffusion MRI. Annals of the New York Academy of Sciences. 2005;1064(1):113–133. doi: 10.1196/annals.1340.018. [DOI] [PubMed] [Google Scholar]
- 38.Jeurissen B, Leemans A, Tournier JD, Jones DK, Sijbers J. Investigating the prevalence of complex fiber configurations in white matter tissue with diffusion magnetic resonance imaging. Human brain mapping. 2013;34(11):2747–2766. doi: 10.1002/hbm.22099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dell’Acqua F, Simmons A, Williams SC, Catani M. Can spherical deconvolution provide more information than fiber orientations? Hindrance modulated orientational anisotropy, a true-tract specific index to characterize white matter diffusion Human brain mapping. 2013;34(10):2464–2483. doi: 10.1002/hbm.22080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker GJ, Alexander DC. Probabilistic anatomical connectivity derived from the microscopic persistent angular structure of cerebral tissue. Philos Trans R Soc Lond B Biol Sci. 2005;360(1457):893–902. doi: 10.1098/rstb.2005.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farrell JA, Landman BA, Jones CK, Smith SA, Prince JL, van Zijl PC, Mori S. Effects of signal-to-noise ratio on the accuracy and reproducibility of diffusion tensor imaging-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5 T. Journal of magnetic resonance imaging: JMRI. 2007;26(3):756–767. doi: 10.1002/jmri.21053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landman BA, Farrell JA, Jones CK, Smith SA, Prince JL, Mori S. Effects of diffusion weighting schemes on the reproducibility of DTI-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5T. NeuroImage. 2007;36(4):1123–1138. doi: 10.1016/j.neuroimage.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 43.Chiang MC, Barysheva M, Lee AD, Madsen S, Klunder AD, Toga AW, McMahon KL, de Zubicaray GI, Meredith M, Wright MJ, Srivastava A, Balov N, Thompson PM. Brain fiber architecture, genetics, and intelligence: a high angular resolution diffusion imaging (HARDI) study. Med Image Comput Comput Assist Interv. 2008;11(Pt 1):1060–1067. doi: 10.1007/978-3-540-85988-8_126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen-Adad J, Descoteaux M, Wald LL. Quality assessment of high angular resolution diffusion imaging data using bootstrap on Q-ball reconstruction. Journal of magnetic resonance imaging: JMRI. 2011;33(5):1194–1208. doi: 10.1002/jmri.22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canales-Rodriguez EJ, Melie-Garcia L, Iturria-Medina Y. Mathematical description of q-space in spherical coordinates: exact q-ball imaging. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2009;61(6):1350–1367. doi: 10.1002/mrm.21917. [DOI] [PubMed] [Google Scholar]
- 46.Aganj I, Lenglet C, Sapiro G, Yacoub E, Ugurbil K, Harel N. Reconstruction of the orientation distribution function in single- and multiple-shell q-ball imaging within constant solid angle. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2010;64(2):554–566. doi: 10.1002/mrm.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tristan-Vega A, Westin CF, Aja-Fernandez S. A new methodology for the estimation of fiber populations in the white matter of the brain with the Funk-Radon transform. NeuroImage. 2010;49(2):1301–1315. doi: 10.1016/j.neuroimage.2009.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daducci A, Canales-Rodriguez EJ, Descoteaux M, Garyfallidis E, Gur Y, Lin YC, Mani M, Merlet S, Paquette M, Ramirez-Manzanares A, Reisert M, Reis Rodrigues P, Sepehrband F, Caruyer E, Choupan J, Deriche R, Jacob M, Menegaz G, Prckovska V, Rivera M, Wiaux Y, Thiran JP. Quantitative comparison of reconstruction methods for intra-voxel fiber recovery from diffusion MRI. IEEE transactions on medical imaging. 2014;33(2):384–399. doi: 10.1109/TMI.2013.2285500. [DOI] [PubMed] [Google Scholar]
- 49.Ning L, Laun F, Gur Y, DiBella EV, Deslauriers-Gauthier S, Megherbi T, Ghosh A, Zucchelli M, Menegaz G, Fick R, St-Jean S, Paquette M, Aranda R, Descoteaux M, Deriche R, O’Donnell L, Rathi Y. Sparse Reconstruction Challenge for diffusion MRI: Validation on a physical phantom to determine which acquisition scheme and analysis method to use? Med Image Anal. 2015;26(1):316–331. doi: 10.1016/j.media.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heidemann RM, Anwander A, Feiweier T, Knosche TR, Turner R. k-space and q-space: combining ultra-high spatial and angular resolution in diffusion imaging using ZOOPPA at 7 T. NeuroImage. 2012;60(2):967–978. doi: 10.1016/j.neuroimage.2011.12.081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.