Abstract
Although generalized anxiety disorder (GAD) is heritable and aggregates in families, no genomic loci associated with GAD have been reported. We aimed to discover potential loci by conducting a genome-wide analysis of GAD symptoms in a large, population-based sample of Hispanic/Latino adults. Data came from 12,282 participants (aged 18–74) in the Hispanic Community Health Study/Study of Latinos. Using a shorted Spielberger Trait Anxiety measure, we analyzed: (1) a total trait anxiety score based on summing responses to all ten items; and (2) a GAD symptoms score restricted to the three items tapping diagnostic features of GAD as defined by DSM-V. We first calculated the heritability due to common variants (h2SNP) and then conducted a genome-wide association study (GWAS) of GAD symptoms. Replication was attempted in three independent Hispanic cohorts (Multi-Ethnic Study of Atherosclerosis, Women’s Health Initiative, Army STARRS). The GAD symptoms score showed evidence of modest heritability (7.2%; p=0.03), while the total trait anxiety score did not (4.97%; p=0.20). One genotyped SNP (rs78602344) intronic to Thrombospondin 2 (THBS2) was nominally associated (p=4.18×10−8) in the primary analysis adjusting for psychiatric medication use and significantly associated with the GAD symptoms score in the analysis excluding medication users (p=4.18×10−8). However, meta-analysis of the replication samples did not support this association. Although GWAS revealed a genome-wide significant locus in this sample, we were unable to replicate this finding. Evidence for heritability was also only detected for GAD symptoms, and not the trait anxiety measure, suggesting differential genetic influences within the domain of trait anxiety.
Keywords: genetic association study, anxiety, Hispanics/Latinos
Introduction
Generalized anxiety disorder (GAD) is a mental disorder characterized by persistent uncontrollable worry and symptoms of arousal (e.g., restlessness, insomnia, muscle tension, irritability) (Hoge, Ivkovic et al. 2012, American Psychiatric Association 2013, Stein and Sareen 2015). GAD is common in the United States and worldwide (Grant, Hasin et al. 2005, Kessler, Berglund et al. 2005, Kessler, Chiu et al. 2005, Wittchen and Jacobi 2005, Wittchen, Jacobi et al. 2011). Retrospective epidemiological studies suggest the past year prevalence of GAD is 3.1% and lifetime prevalence is 5.7% (Kessler, Berglund et al. 2005, Kessler, Chiu et al. 2005). Even higher estimates have been observed from prospective studies (14.2% lifetime; 4.2% past year) (Moffitt, Caspi et al. 2010). Though GAD is about half as common in Hispanics/Latinos compared to Whites (Grant, Hasin et al. 2005, Asnaani, Richey et al. 2010), Hispanics/Latinos represent one of the fastest growing populations in the US (Passel, Cohn et al. 2011, Brown 2014, June 26), making the population burden of GAD in the US therefore quite large. GAD is also a highly comorbid disorder, with about 90% of people with GAD experiencing at least one other DSM-IV Axis 1 or Axis 2 disorder (Grant, Hasin et al. 2005). Given its prevalence and profound social and economic costs (Hoffman, Dukes et al. 2008, Newman, Llera et al. 2013), it is of strong interest to identify factors associated with the development of GAD.
Exploration of the role of genetic factors in the etiology of GAD is warranted as GAD appears attributable, in part, to genetic variation (Shimada-Sugimoto, Otowa et al. 2015). Family studies have found people with GAD have six times the odds of those without the disorder to have a first degree relative who also has GAD (Hettema, Neale et al. 2001). Twin studies also suggest GAD is moderately heritable, with 32% of the variation in the population risk of GAD being attributable to genetic variation (Hettema, Neale et al. 2001). Despite evidence of family aggregation, there have not yet been any published genome-wide association studies (GWAS) of GAD or GAD symptoms. Given the recent success of GWAS for other anxiety disorders, notably post-traumatic stress disorder (Guffanti, Galea et al. 2013, Logue, Baldwin et al. 2013, Xie, Kranzler et al. 2013) and panic disorder (Otowa, Yoshida et al. 2009, Otowa, Tanii et al. 2010, Erhardt, Czibere et al. 2011), as well as efforts to examine domains related to GAD, including anxiety sensitivity (Davies, Verdi et al. 2015), or composite indicators of anxiety disorder (Otowa, Maher et al. 2014), we sought to identify genomic loci linked to GAD by conducting a genome-wide analysis of GAD symptoms. We used a dimensional measure of trait anxiety symptoms chosen to match DSM-5 criteria for GAD. Use of a dimensional measure enables an examination of the full range of quantitative variation, rather than extremes in this quantitative distribution (e.g., cases versus controls) and may be a statistically more powerful approach to identify variants associated with GAD (Plomin, Haworth et al. 2009).
In this report, we present results from the first GWAS of GAD symptoms, where we found a genome-wide significant association between a SNP intronic to Thrombospondin 2 (THBS2) and GAD symptoms in a large, diverse, and population-based sample of Hispanic/Latino adults. This finding did not replicate in a meta-analysis of three independent samples of Hispanic/Latino adults. We also present results from a SNP-chip heritability analysis, where we found evidence of modest heritability in GAD symptoms (7.2%), but no statistically significant heritability for a broader measure of trait anxiety symptoms.
Materials and Methods
Overview
The Hispanic Community Health Study/Study of Latinos (HCHS/SOL) is a community-based prospective cohort study following 16,415 self-identified Hispanic/Latino adults (aged 18–74 at screening) designed to examine the distribution and determinants of chronic health conditions, including diabetes, pulmonary disease, and cardiovascular disease. As described elsewhere (Lavange, Kalsbeek et al. 2010), participants were recruited via a stratified two-stage area probability sample of households across four cities in the United States (Chicago, IL; Miami, FL; Bronx, NY; San Diego, CA). The majority of the sample self-identified with the following background groups: Central American (n=1,730), Cuban (n=2,348), Dominican (n=1,460), Mexican (n=6,471), Puerto Rican (n=2,728), and South American (n=1,068). Baseline examinations were conducted between 2008 and 2011. Institutional Review Boards at each field center approved the study and all participants provided written informed consent. In the current study, we analyzed data from 12,254 respondents who consented to provide blood for the purpose of genotyping and had complete outcome and relevant covariates information (to be described later), as well as non-missing records of antianxiety and antidepressants medication use.
Phenotype Definition
Anxiety symptoms were assessed at baseline using a 10-item Spielberger State-Trait Anxiety Inventory (STAI-T) administered in the participant’s preferred language (Spanish or English) (Bromberger and Matthews 1996, Bergua, Meillon et al. 2015). This a short form version of the 20 item STAI-T (Spielberger 1989), which is a valid and commonly used measure of trait anxiety symptoms in population-based studies (see for example: (De Moor, Beem et al. 2006, Caravati-Jouvenceaux, Launoy et al. 2011)) that has been shown to correlate highly with other anxiety measures (Spielberger and Reheiser 2009). The abbreviated 10-item STAI-T short form has shown excellent internal consistency reliability in the full HCHS/SOL sample (alpha=0.93) and for both the English (alpha=0.92) and Spanish (alpha=0.94) versions of the instrument (Wassertheil-Smoller, Arredondo et al. 2014) and has been shown in other studies to correlate highly with the full version (α=0.96) (Bromberger and Matthews 1996). For each item, participants were asked to indicate how they generally feel (0=almost never; 1=sometimes; 2=often; 3=almost always). Using the STAI short form, we created a GAD symptoms score by summing the three items (i.e., feeling nervous or restless; worrying over things that don’t matter; getting in a state of tension or turmoil as you think about recent concerns and interests) that are diagnostic criteria for GAD as defined by the DSM-5 (American Psychiatric Association 2013). The GAD symptoms score demonstrated moderate internal consistency reliability (alpha=0.70) in the full HCHS/SOL sample. For comparison, we also examined a total trait anxiety score based on summing responses to all 10 items (i.e., the three GAD symptom score items noted above plus the following seven items: I feel satisfied with myself; I lack self confidence; I feel secure; I feel inadequate; I am a steady person; I wish I could be as happy as others seem to be; I feel like a failure). Both phenotypes were coded so that higher scores indicated higher levels of anxiety.
To account for the possibility that current use of antidepressant or anxiolytic medications might affect anxiety scores, we applied an imputation algorithm to increase the scores of medication users. This algorithm was used in a previous GWAS of depressive symptoms(Hek, Demirkan et al. 2013) and was similar to an algorithm used to adjust blood pressure for persons on antihypertensive medications (Levy, DeStefano et al. 2000). Antidepressant or anxiolytic medication use was determined by pill bottles brought by the participant to the baseline interview. Antidepressants were included, as this class of drugs are commonly prescribed to treat generalized anxiety symptoms (Kapczinski, Silva de Lima et al. 2003, Milea, Verpillat et al. 2010). This algorithm assumed that: (1) the anxiety score of a respondent taking these psychotropic medications is lower (i.e., indicating fewer symptoms) than would be expected if the respondent were not taking these medications (thus, we assume that the medications are effective in reducing symptoms); (2) respondents with high anxiety scores, on average, respond less to these medications than respondents with lower anxiety scores. It therefore replaced the anxiety score of respondents on medications (n=1,068) with the mean anxiety score of all respondents taking these medications that had the same or a higher anxiety score. For example, a medication user with an observed anxiety score of 10 would have a revised score of 21.07 (derived by taking the average anxiety score of medication users with an anxiety score value of 10 or greater). Anxiety scores for medication users were increased by 6.2 points on average above the raw score (raw scores ranged from 0–30).
SNP Genotyping, Quality Control and Imputation
Blood samples from consenting respondents were sent to Illumina Microarray Services for genotyping on the Illumina SOL HCHS Custom 15041502 B3 array. This array comprised the Illumina Omni 2.5M array (HumanOmni2.5-8v1-1) and additional custom content (e.g., ancestry-informative markers, variants characteristic of Amerindian populations, known GWAS hits, and other candidate gene markers) selected for HCHS/SOL.
Quality assurance/quality control (QA/QC) was performed by Illumina, LA Biomed, and the HCHS/SOL Genetic Analysis Center (GAC) according to established methods (Laurie, Doheny et al. 2010) to generate recommended SNP and sample-level quality filters. In brief, samples were checked for annotated versus genetic sex, gross chromosomal anomalies (Laurie, Laurie et al. 2012), call rates, batch effects, duplicate sample discordance, Mendelian errors, population structure, and relatedness (note: participants could have been genetically related due to being drawn from the same household or different households living in the same community). 12,803 unique study samples passed these criteria. SNPs that passed the Illumina/LA Biomed assay failure indicator were further checked for Hardy-Weinberg equilibrium, MAF, duplicate probe discordance, and missing call rate. A total of 2,232,944 SNPs passed both quality and informativeness filters (unduplicated on the array and polymorphic).
Genome-wide imputation was carried out on all 12,803 samples together using the 1000 Genomes Project phase 1 reference panel (Genomes Project, Abecasis et al. 2012) and IMPUTE2 software (Howie, Donnelly et al. 2009, Howie, Marchini et al. 2011). Genotypes were first pre-phased with SHAPEIT2 (v2.r644) and then imputed with IMPUTE2 (v2.3.0). Only variants with at least two copies of the minor allele present in any of the four 1000 Genomes continental panels were imputed, yielding a total of 25,568,744 imputed variants. Overall imputation quality was assessed both by looking at the distribution of imputed quality metrics by different MAF levels and by examining results from the IMPUTE2 internal masking experiments (as some genotyped variants were “masked”, meaning removed from the imputation basis).
Principal components (PCs) and kinship coefficients were computed in an iterative manner to estimate both population structure and relatedness between study individuals such that the PCs were not affected by relatedness, and kinship estimates are not affected by ancestry. The process began with estimating relatedness using KING-robust (Manichaikul, Mychaleckyj et al. 2010), followed by iterative estimation of PCs and kinship coefficients using PC-AiR (Conomos, Miller et al. 2015) and PC-Relate (https://www.bioconductor.org/packages/release/bioc/html/GENESIS.html), and is described comprehensively elsewhere (Conomos 2014). Consequently, 19 individuals who were identified to have primarily East Asian ancestry were excluded from analysis. For association analysis, the kinship matrix was based on an independent set of SNPs selected with LD pruning.
Statistical Analyses
All analyses used a linear mixed-effect model, which accounted for the correlations between individuals due to genetic relatedness (kinship), shared household, and the complex sampling design (Conomos, Laurie et al. 2016, Schick, Jain et al. 2016). The variance components were estimated using restricted maximum likelihood (REML). Fixed effects included the covariates: log(sampling weight), which reflect the differences in sampling probabilities of study individuals and is included to prevent potential selection bias; field center; age; sex; education (1=no high school diploma or GED – referent; 2=at most a High school diploma or GED, 3=greater than high school or GED; 4=bachelors degree, 5=masters, professional, or doctorate degree); and the top 5 PCs of ancestry. SNP annotation was performed using ANNOVAR (Wang, Li et al. 2010) (http://annovar.openbioinformatics.org/en/latest/).
Heritability Analysis
We estimated “SNP-chip heritability”, or the narrow-sense heritability due to the additive effect of common variants (genotyped and imputed), by first fitting a “null” linear mixed model that included all covariates, PCs, and random effects, but did not include genotypes, and then calculating the proportion of variance attributable to relatedness out of all phenotypic variance (Conomos, Laurie et al. 2016, Schick, Jain et al. 2016). For this analysis, the kinship matrix was calculated based on PC-relate using all autosomal SNPs, and the model was fit on a set of 10,414 unrelated individuals by removing participants so that the unrelated set did not have first-, second-, or third-degree relatives (Yang, Benyamin et al. 2010). We conducted this analysis examining the GAD symptoms score as well as the total trait anxiety score to evaluate and compare SNP-chip heritability estimates across these phenotypes.
GWAS Analysis
We performed a GWAS using the linear mixed-effect model approach. All SNPs were modeled additively and the standard 5×10−8 was used as the threshold for genome-wide statistical significance. In addition, we report the set of SNPs with p-value<1×10−6 according to the following selection criteria: out of SNPs that were less than 500,000 base pairs apart, and their correlation was higher than 0.5, we prioritized genotyped over imputed SNPs, we preferred imputed SNPs with higher quality score (info), lower p-values, and for SNPs with similar p-values and imputation quality score (or genotyped), we prioritized SNPs with higher MAF. Quantile-quantile (QQ) and Manhattan plots were generated using the R package GWASTools (Gogarten, Bhangale et al. 2012). Regional association plots were generated using Locus Zoom (Pruim, Welch et al. 2010).
Secondary Analysis
As a secondary analysis, we repeated our analyses in the subset of non-medication users (n=11,456; 91.5% of the sample) and using an untransformed score that did not consider medication use (i.e., the raw phenotype score).
Replication
We attempted replication of these results using data from three independent cohorts. Additional details about these cohorts are presented in Supplemental Materials. Briefly, the Women’s Health Initiative (1998, Wassertheil-Smoller, Shumaker et al. 2004) (WHI; www.whi.org) provided data on Hispanic/Latina women (n=3352; mean age 60.0; sd=6.57), where anxiety symptoms were measured using two items (i.e., Feeling nervous, anxious, on edge, or worrying a lot about different things; Have you been a very nervous person). The Multi-Ethnic Study of Atherosclerosis (Bild, Bluemke et al. 2002) (MESA; http://www.mesa-nhlbi.org) provided data from Hispanic/Latino adults (n=1449; mean age 61.38; sd=10.30) where anxiety symptoms were measured using a scale identical to the HCHS/SOL. Finally, the Army Study To Assess Risk and Resilience in Service members (Ursano, Colpe et al. 2014)(Army STARRS; http://www.armystarrs.org) provided data from Hispanic/Latino adults (n= 3394; mean age=25.98; sd=5.00), where anxiety symptoms were captured using a five-item scale designed to match DSM-IV criteria for GAD.
We meta-analyzed GWAS results across the three independent samples. As we were interested in testing whether the direction of effect was the same in the replication (as the discovery), one sided p-values were used (Heller, Bogomolov et al. 2015). Inverse variance weighted fixed-effect meta-analysis was conducted using METAL (http://www.sph.umich.edu/csg/abecasis/metal/;(Willer, Li et al. 2010)).
Results
A total of 12,282 Hispanic/Latino respondents were in the analysis. As expected, the GAD symptom score (skew=0.63; kurtosis=2.48) and total trait anxiety score (skew=0.87; kurtosis=3.21) were skewed towards lower values. No transformations of the outcome were performed as linear regression is robust to minor violations of normality (van Belle 2002).
Discovery Sample: SNP Heritability
As shown in Table I, the GAD symptom score showed evidence of modest heritability (h2SNP=7.2%; p=0.03), while the total trait anxiety score did not (h2SNP=4.97%; p=0.20). Building from these results, we conducted a GWAS only on the GAD symptom score.
Table I.
Results of genome-wide complex trait analysis
| Original scores | Accounting for medication use |
Medication users removed |
||||
|---|---|---|---|---|---|---|
|
|
||||||
| V(G)/Vp*100 | p | V(G)/Vp*100 | p | V(G)/Vp*100 | p | |
| Total trait anxiety score | 5.65 | 0.32 | 4.97 | 0.20 | 8.18 | 0.14 |
| GAD symptoms score | 7.57 | 0.12 | 7.20 | 0.03 | 8.15 | 0.06 |
V(G)/Vp*100 = SNP heritability estimate (h2SNP)*100. All phenotypes were treated as continuous. All models adjusted for sex, age, education (5-levels), principal components, study center, and sampling weights, and included random effects for the design variables kinship, household, and block unit, the three study design variables. P-values were calculated using the likelihood ratio test. The total trait anxiety score was derived by summing responses to all 10 items (see Supplemental Materials for listing of all items). The GAD symptoms score was based on summing three items (i.e., feeling nervous or restless; worrying over things that don’t matter; getting in a state of tension or turmoil as you think about recent concerns and interests) that are diagnostic criteria for GAD as defined by the DSM-V.
Discovery Sample: GWAS
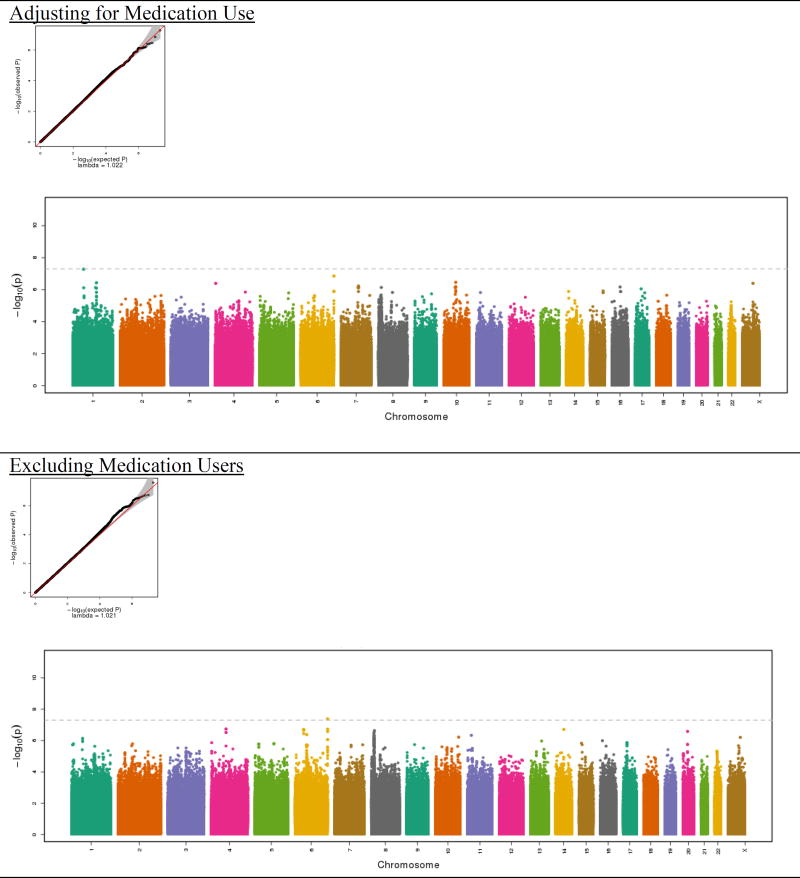
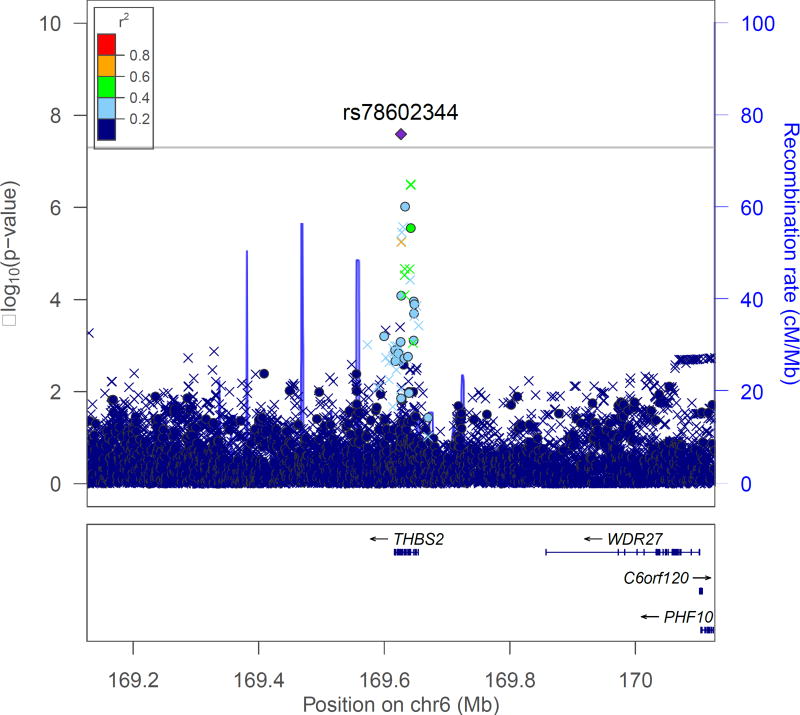
The Manhattan and QQ plots are shown in Figure 1. As shown in the QQ plots, there was no evidence of inflation in either the GWAS of the full sample or the analysis that excluded medication users (λ =1.02). No SNPs achieved genome-wide significance in the full sample, which included imputed scores for medication users (Table II). However, one genotyped SNP (rs78602344), located on chromosome 6 at position 169626581, emerged from both analyses. This SNP was the second most significant result in the full sample (p=1.41×10−7) and the most significant result (p=4.18×10−8) in the analysis excluding medication users (Table III). The SNP is intronic to Thrombospondin 2 (THBS2), a gene that mediates cell-to-cell and cell-to-matrix interactions. Several other SNPs in the region also showed support for association (Figure 2).
Figure 1.
Quantile-quantile (QQ) plots and Manhattan plots for GAD symptoms score from the Hispanic Community Health Study/Study of Latinos
The quantile–quantile plots (“QQ-plots”), which present the observed by expected P-values on the -log10 scale, indicate conformity of the observed results to what would be expected under the null. In the Manhattan plots, the x-axis is the chromosomal position and the y-axis is the -log10 p-value for the association between each SNP and core anxiety symptoms derived from the linear regression model. The dotted line shows the genome-wide significance level (5×10−8). The displayed p-value corresponds to SNPs with effective N > 30.
Table II.
Genome-wide association study (GWAS) results for the top loci (p<1×10−6) with the GAD symptoms score imputed for medication use
| SNP | CHR | position | alleleA | alleleB | MAF | minor allele |
geno. | n | Beta | SE | p-value | Location | Closest gene (<20kb) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10749727 | 1 | 62056653 | A | G | 0.002 | A | I | 12282 | 1.97 | 0.36 | 5.28E-08 | ||
| rs78602344 | 6 | 169626581 | T | C | 0.113 | B | G | 12282 | −0.26 | 0.05 | 1.41E-07 | intronic | THBS2 |
| rs10994985 | 10 | 63713219 | T | C | 0.046 | B | G | 12282 | −0.38 | 0.08 | 3.48E-07 | ||
| rs3753389 | 1 | 160807153 | T | C | 0.378 | A | I | 12282 | −0.16 | 0.03 | 3.70E-07 | intronic | CD244 |
| rs12353562 | X | 96354053 | G | A | 0.099 | B | I | 12269 | −0.23 | 0.05 | 4.04E-07 | intronic | DIAPH2 |
| rs144417828 | 4 | 3740444 | G | A | 0.010 | B | I | 12282 | −0.84 | 0.17 | 4.07E-07 | ||
| rs148349076 | 16 | 54271147 | C | T | 0.004 | B | I | 12282 | −1.29 | 0.26 | 6.78E-07 | ||
| rs7094998 | 10 | 61306319 | A | G | 0.210 | B | G | 12282 | 0.19 | 0.04 | 7.13E-07 | ||
| rs17729883 | 8 | 9256631 | T | C | 0.220 | B | G | 12281 | −0.19 | 0.04 | 7.29E-07 | ||
| rs11803917 | 1 | 156111578 | C | T | 0.071 | B | I | 12282 | 0.32 | 0.06 | 7.41E-07 | ||
| rs75192612 | 7 | 88641306 | A | G | 0.004 | B | I | 12282 | −1.19 | 0.24 | 7.49E-07 | ||
| rs3110650 | 17 | 36075905 | T | C | 0.002 | A | I | 12282 | 1.86 | 0.38 | 8.94E-07 |
CHR=chromosome. In the geno. (genotyping) column, G=genotyped and I=imputed. All imputed SNPs had info scores (indicating imputation quality) ≥ 0.70. AlleleA is the tested allele. Position is given in genome build GRCh37/hg19.
Table III.
Genome-wide association study (GWAS) results for the top loci (p<1×10−6) with the GAD symptoms score, after excluding medication users
| SNP | CHR | position | alleleA | alleleB | MAF | minor allele |
geno. | n | Beta | SE | p-value | Location | Closest gene (<20kb) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs78602344 | 6 | 169626581 | T | C | 0.115 | B | G | 11456 | −0.27 | 0.05 | 4.18E-08 | intronic | THBS2 |
| rs141474992 | 4 | 75958947 | A | C | 0.006 | B | I | 11456 | −1.04 | 0.20 | 1.83E-07 | ||
| rs79812562 | 14 | 64593580 | C | A | 0.008 | B | I | 11456 | −1.02 | 0.20 | 1.95E-07 | ||
| rs146964092 | 6 | 39754875 | A | G | 0.001 | B | I | 11456 | −2.16 | 0.42 | 1.96E-07 | ||
| rs7350124 | 8 | 10625045 | T | C | 0.182 | B | I | 11455 | −0.21 | 0.04 | 2.34E-07 | intronic | PINX1 |
| rs186222942 | 20 | 24548719 | G | A | 0.012 | B | I | 11456 | −0.77 | 0.15 | 2.61E-07 | ||
| rs11776020 | 8 | 8809696 | A | G | 0.455 | B | G | 11455 | −0.17 | 0.03 | 3.26E-07 | ||
| rs115013535 | 6 | 55628681 | C | A | 0.004 | B | I | 11456 | −1.30 | 0.26 | 4.16E-07 | ||
| rs186294317 | 11 | 20267060 | G | A | 0.002 | B | I | 11456 | −1.86 | 0.37 | 4.62E-07 | ||
| rs17729883 | 8 | 9256631 | T | C | 0.216 | B | G | 11455 | −0.19 | 0.04 | 5.09E-07 | ||
| rs189738814 | 10 | 126827710 | G | A | 0.004 | B | I | 11456 | −1.50 | 0.30 | 6.02E-07 | ||
| rs144369074 | X | 113716176 | A | T | 0.005 | B | I | 11445 | −1.03 | 0.21 | 6.24E-07 | ||
| rs115791358 | 1 | 64300191 | A | T | 0.004 | B | I | 11456 | −1.23 | 0.25 | 7.24E-07 | ||
| rs11756502 | 6 | 169633185 | C | T | 0.315 | B | G | 11456 | −0.16 | 0.03 | 8.66E-07 | intronic | THBS2 |
| rs62435218 | 6 | 169642301 | C | T | 0.179 | B | G | 11456 | −0.20 | 0.04 | 8.74E-07 | intronic | THBS2 |
| rs6601288 | 8 | 8943430 | A | T | 0.484 | A | G | 11455 | −0.16 | 0.03 | 9.59E-07 |
CHR=chromosome. In the geno. (genotyping) column, G=genotyped and I=imputed. All imputed SNPs had info scores (indicating imputation quality) ≥ 0.62. AlleleA is the tested allele. Position is given in genome build GRCh37/hg19.
Figure 2.
Regional association plot for the top SNP (rs78602344) identified in the analysis excluding medication users
The regional association plot was generated using LocusZoom (http://csg.sph.umich.edu/locuszoom/) The left-side y-axis refers to the -log of the p-value corresponding to the test of association between each SNP (denoted as a colored dot, if genotyped, or X, if imputed) and GAD symptoms. SNPs are colored based on the level of linkage disequilibrium (LD) between each SNP and the index, genotyped, SNP (purple diamond). r2 values are determined based on the HCHS/SOL data.
A second SNP with a low p-value in both analyses was rs17729883 (full sample p=7.29×10−7; excluding medication users p=5.09×10−7) located at chromosome 8 position 9256631. This genotyped SNP was located in an intron of an uncharacterized gene (LOC 106379231; Supplemental Figure 1).
All GWAS results at p<1×10−5 are shown in the Supplemental Materials for the GAD symptom score for the full sample (Supplemental Table I), excluding medication users (Supplemental Table II), and for the original, non-transformed score (Supplemental Table III).
To determine which SNPs to carry forward for replication, we estimated replication power for all SNPs with p-values <1×10−6 in at least one of the two analyses according to our selection criteria detailed above. Replication power estimates were based on the projected samples sizes of each replication dataset (Army STARRS=3000; WHI=3000; MESA=1500) and using MAF, outcome standard deviation, and estimated effect sizes from the discovery sample. Our power calculations incorporated a method (Zhong and Prentice 2008) to reduce bias due to “winner’s curse”, effectively attenuating the observed effect size. A prior study showed that attenuated effect size estimates tend to be closer than uncorrected estimates to effects seen in independent replication studies (Zhong and Prentice 2010).
Our power analysis suggested one SNP (rs78602344) would have excellent power in a meta analysis of the three replication cohorts after the winner’s curse bias correction (estimated power=0.96); all other SNPs had weak power (≤0.70). We therefore carried forward this single SNP for replication.
Replication Samples: GWAS Results
In the replication phase, one SNP (rs78602344) was evaluated in three independent samples. This SNP was not significantly associated with the GAD symptom score in a meta analysis of the replication sites (Table IV).
Table IV.
Replication results of rs78602344 for GAD symptoms
| A. Adjusting for medication use | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | position | alleleA | alleleB | MAF | minor allele | geno. | n | Beta | SE | pval | |
| discovery | rs78602344 | 6 | 169626581 | T | C | 0.113 | B | G | 12282 | −0.26 | 0.05 | 1.41E-07 |
|
| ||||||||||||
| Army NSS1 | rs78602344 | 6 | 169626581 | T | C | 0.108 | B | I | 1408 | 0.11 | 0.34 | 0.76 |
| Army NSS2 | rs78602344 | 6 | 169626581 | T | C | 0.111 | B | I | 453 | 1.38 | 0.68 | 0.04 |
| Army PPDS | rs78602344 | 6 | 169626581 | T | C | 0.108 | B | I | 1533 | 0.17 | 0.30 | 0.57 |
| MESA | rs78602344 | 6 | 169626581 | T | C | 0.133 | B | I | 1441 | 0.02 | 0.06 | 0.73 |
| WHI | rs9505953 | 6 | 108969803 | C | T | 0.206 | B | I | 2950 | −0.02 | 0.04 | 0.54 |
|
| ||||||||||||
| Meta Analysis | T | C | 7785 | 0.03 | 0.03 | 0.43 | ||||||
| B. Excluding medication users | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | position | alleleA | alleleB | MAF | minor allele | geno | n | Beta | SE | pval | |
| discovery | rs78602344 | 6 | 169626581 | T | C | 0.113 | B | G | 11456 | −0.27 | 0.05 | 4.18E-08 |
|
| ||||||||||||
| Army NSS1 | rs78602344 | 6 | 169626581 | T | C | 0.108 | B | I | 1372 | 0.11 | 0.34 | 0.74 |
| Army NSS2 | rs78602344 | 6 | 169626581 | T | C | 0.111 | B | I | 431 | 1.00 | 0.66 | 0.13 |
| Army PPDS | rs78602344 | 6 | 169626581 | T | C | 0.108 | B | I | 1430 | −0.04 | 0.27 | 0.88 |
| MESA | rs78602344 | 6 | 169626581 | T | C | 0.133 | B | I | 1369 | 0.01 | 0.07 | 0.92 |
| WHI | rs9505953 | 6 | 108969803 | C | T | 0.205 | B | I | 2513 | −0.01 | 0.04 | 0.77 |
|
| ||||||||||||
| Meta Analysis | T | C | 7115 | 0.01 | 0.03 | 0.71 | ||||||
CHR=chromosome. In the geno. (genotyping) column, G=genotyped and I=imputed. All imputed SNPs had info scores (indicating imputation quality) >0.83. AlleleA is the tested allele. The Army STARRs dataset was comprised of three different cohorts: the New Soldiers Study 1, the New Soldiers Study 2, and the Post Deployment Study. The SNP identified in the discovery analysis and carried forward to the replication (rs78602344) was neither genotyped nor imputed in WHI. We therefore used the best proxy SNP (rs9505953) in closest LD (r2=0.15).
Discussion
The current study involved three major innovations in efforts to identify the genetic basis of generalized anxiety. First, to our knowledge, this was the first GWAS of GAD symptoms. Prior genetic association studies of GAD have focused on candidate gene polymorphisms, most of which have showed inconsistent results (Smoller 2015). Among GWAS, extant studies have focused on other anxiety disorders, including post-traumatic stress disorder (Guffanti, Galea et al. 2013, Logue, Baldwin et al. 2013, Xie, Kranzler et al. 2013) and panic disorder (Otowa, Yoshida et al. 2009, Otowa, Tanii et al. 2010, Erhardt, Czibere et al. 2011), or have examined more global symptoms of trait anxiety in children (Trzaskowski, Eley et al. 2013) or composite indicators of anxiety disorder in adults (Otowa, Maher et al. 2014), but have not yet examined general symptoms of anxiety in adults. Second, our study was also the first to provide SNP-chip heritability estimates of GAD symptoms. Such analyses are important to provide upper- and lower-bound estimates of the additive genetic contribution to GAD. Finally, we conducted these genetic association analyses in Hispanics/Latinos, a large and growing US population group. Previous studies have largely focused on individuals of European ancestry.
Two findings emerged from the current study. First, results from the SNP-chip heritability analysis suggested that about 7.2% of the variance in GAD symptoms was explained by common genetic variants. This SNP heritability estimate is lower than those found for phobic anxiety (h2SNP=21%; p=0.01) (Walter, Glymour et al. 2013) and anxiety sensitivity (h2SNP=45%; 95% CI=32%, 56%) (Davies, Verdi et al. 2015) in adults, and also lower (though statistically significant) relative to estimates for a composite measure of anxiety traits in children, which was derived by summing measures of negative affect, negative cognition, fear, and social anxiety (h2SNP=16%; p=0.07) (Trzaskowski, Eley et al. 2013). The lower heritability estimates observed in this study relative to other studies conducted in adults may be due to the use of symptom scale, rather than a diagnostic measure of GAD. Interestingly, we also found that the total trait anxiety score, derived by summing all items on the scale (rather than just the three corresponding to GAD symptoms) carried no heritable signal. This result suggests that not all symptoms on existing anxiety scales may be equally influenced by additive genetic variation. Future studies using dimensional measures of anxiety symptoms may benefit from conducting similar analyses to determine whether an existing scale should be used in its entirety.
Second, we identified one genotyped SNP (rs78602344) located on chromosome 6 that was common to analyses accounting for psychiatric medication use or excluding medication users. Although not genome-wide significant in the former analysis, this SNP was genome-wide significant after excluding medication users (p=4.18×10−8). This SNP is intronic to Thrombospondin 2 (THBS2), a gene that mediates cell-to-cell and cell-to-matrix interactions. Several other SNPs in the region also showed support for association. However, the association of the lead SNP was not supported in a meta-analysis of the three independent Hispanic/Latino replication samples (n=7377). Although recent success from GWAS of other anxiety disorders suggest that genomic loci can be found, we suspect that GWAS of GAD symptoms will likely share a similar trajectory as depressive symptoms, where increasing larger sample sizes and refinement of the phenotype will lead to the identification of associated loci (CONVERGE Consortium 2015, Dunn, Brown et al. 2015).
We note several limitations of the current study. First, the outcomes were based on a brief inventory of trait anxiety symptoms. Although the widespread use of this anxiety measure in population-based studies allowed us to carry out the current analyses, future studies of diagnostic measures of GAD as well as more robust measures of GAD symptoms (from more detailed and specific measures or repeated phenotyping) are needed. Second, the replication samples were smaller and both more demographically and phenotypically heterogeneous than the HCHS/SOL discovery sample. Unfortunately, replication efforts are currently hampered by a lack of available data on anxiety symptoms in racial/ethnic minority populations. Third and relatedly, only one SNP was carried forward to the replication phase. This single SNP was the only one with high replication power. Had genetic and GAD symptoms data been available in more racial/ethnic minority samples, we could have had adequate power to attempt replication of other SNPs. Moreover, although no a limitation per se, greater insights are needed regarding the most optimal strategy to account for medication use in genetic association studies of quantitative traits. Future studies are needed to examine the suitability of different techniques and the extent to which different adjustment methods lead to different results (e.g., whether they substantially reduce variance if a substantial portion of the sample are assigned the same score; whether empirical data, such as medication efficacy, can be used to inform the adjustment strategy).
In conclusion, although the GWAS revealed a genome-wide significant locus in the discovery sample, we were unable to replicate this in independent samples. These findings underscore the need for even larger studies of GAD symptoms.
Supplementary Material
Acknowledgments
The Hispanic Community Health Study/Study of Latinos (HCHS/SOL) was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01- HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contribute to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. The Army Study of Risk and Resilience in Servicemembers (Army STARRS) was sponsored by the Department of the Army and funded under cooperative agreement number U01MH087981 with the U.S. Department of Health and Human Services, National Institutes of Health, and National Institute of Mental Health (NIH/NIMH).
The contents are solely the responsibility of the authors and do not necessarily represent the views of the Department of Health and Human Services, the National Institutes of Health, the Veterans Administration, Department of the Army, or the Department of Defense.
The current study is supported the National Institute Of Mental Health of the National Institutes of Health under Award Number K01MH102403 (Dunn) and by a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation (Dunn).
The authors thank Sandy Li and Jenna Kiely for their assistance in conducting the literature search for this paper.
The authors also thank the staff and participants of HCHS/SOL for their important contributions. A complete list of staff and investigators has been provided by Sorlie P., et al. in Annals of Epidemiology (2010), issue 20 (8), pages 629–641 and is also available on the study website http://www.cscc.unc.edu/hchs/.
Footnotes
Supplemental Information: see Supplemental Materials
Disclosure: Dr. Dunn takes responsibility for the integrity of the data and accuracy of the analyses. All authors have reviewed and approved the final manuscript. None of the authors had any financial or other conflicts of interest.
References
- Design of the Women's Health Initiative clinical trial and observational study. The Women's Health Initiative Study Group. Control and Clinical Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. Arlington, VA: American Psychiatric Publishing; 2013. [Google Scholar]
- Asnaani A, Richey JA, Dimaite R, Hinton DE, Hofmann SG. A cross-ethnic comparison of lifetime prevalence rates of anxiety disorders. Journal of Nervous and Mental Disease. 2010;198(8):551–555. doi: 10.1097/NMD.0b013e3181ea169f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergua V, Meillon C, Potvin O, Ritchie K, Tzourio C, Bouisson J, Dartigues JF, Amieva H. Short STAI-Y anxiety scales: validation and normative data for elderly subjects. Aging Ment Health. 2015:1–9. doi: 10.1080/13607863.2015.1051511. [DOI] [PubMed] [Google Scholar]
- Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, Nelson JC, O'Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- Bromberger JT, Matthews KA. A longitudinal study of the effects of pessimism, trait anxiety, and life stress on depressive symptoms in middle-aged women. Psychol Aging. 1996;11(2):207–213. doi: 10.1037//0882-7974.11.2.207. [DOI] [PubMed] [Google Scholar]
- Brown A. U.S. Hispanic and Asian populations growing, but for different reasons 2014 Jun 26; [Google Scholar]
- Caravati-Jouvenceaux A, Launoy G, Klein D, Henry-Amar M, Abeilard E, Danzon A, Pozet A, Velten M, Mercier M. Health-related quality of life among long-term survivors of colorectal cancer: A population-based study. The Oncologist. 2011;16:1626–1636. doi: 10.1634/theoncologist.2011-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos MP. Inferring, estimating and accounting for population and pedigree structure in genetic analyses. PhD. University of Washington; 2014. [Google Scholar]
- Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, Sofer T, Fernandez-Rhodes L, Justice AE, Graff M, Young KL, Seyerle AA, Avery CL, Taylor KD, Rotter JI, Talavera GA, Daviglus ML, Wassertheil-Smoller S, Schneiderman N, Heiss G, Kaplan RC, Franceschini N, Reiner AP, Shaffer JR, Barr RG, Kerr KF, Browning SR, Browning BL, Weir BS, Aviles-Santa ML, Papanicolaou GJ, Lumley T, Szpiro AA, North KE, Rice K, Thornton TA, Laurie CC. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet. 2016;98(1):165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos MP, Miller MB, Thorton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genetic Epidemiology. 2015 doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONVERGE Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523(7562):588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Verdi S, Burri A, Trzaskowski M, Lee M, Hettema JM, Jansen R, Boomsma DI, Spector TD. Generalised Anxiety Disorder--A Twin Study of Genetic Architecture, Genome-Wide Association and Differential Gene Expression. PLoS One. 2015;10(8):e0134865. doi: 10.1371/journal.pone.0134865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moor MH, Beem AL, Stubbe JH, Boomsma DI, De Geus EJ. Regular exercise, anxiety, depression and personality: a population-based study. Prev Med. 2006;42(4):273–279. doi: 10.1016/j.ypmed.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Dunn EC, Brown RC, Dai Y, Rosand J, Nugent NR, Amstadter AB, Smoller JW. Genetic determinants of depression: recent findings and future directions. Harv Rev Psychiatry. 2015;23(1):1–18. doi: 10.1097/HRP.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt A, Czibere L, Roeske D, Lucae S, Unschuld PG, Ripke S, Specht M, Kohli MA, Kloiber S, Ising M, et al. TMEM132D, a new candidate for anxiety phenotypes: evidence from human and mouse studies. Molecular Psychiatry. 2011;16(6):647–663. doi: 10.1038/mp.2010.41. [DOI] [PubMed] [Google Scholar]
- Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten SM, Bhangale T, Conomos MP, Laurie CA, McHugh CP, Painter I, Zheng X, Crosslin DR, Levine D, Lumley T, Nelson SC, Rice K, Shen J, Swarnkar R, Weir BS, Laurie CC. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28(24):3329–3331. doi: 10.1093/bioinformatics/bts610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF, Hasin DS, Stinson FS, Dawson DA, Ruan WJ, Goldstein RB, Smith SM, Saha TD, Huang B. Prevalence, correlates, co-morbidity, and comparative disability of DSM-IV generalized anxiety disorder in the USA: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Psychological Medicine. 2005;35:1747–1759. doi: 10.1017/S0033291705006069. [DOI] [PubMed] [Google Scholar]
- Guffanti G, Galea S, Yan L, Roberts AL, Solovieff N, Aiello AE, Smoller JW, De Vivo I, Ranu H, Uddin M, et al. Genome-wide association study implicates a novel RNA gene, the lincRNA AC068718.1, as a risk factor for post-traumatic stress disorder in women. Psychoneuroendocrinology. 2013;38(12):3029–3038. doi: 10.1016/j.psyneuen.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hek K, Demirkan A, Lahti J, Terracciano A, Teumer A, Cornelis MC, Amin N, Bakshis E, Baumert J, Ding J, Liu Y, Marciante K, Meirelles O, Nalls MA, Sun YV, Vogelzangs N, Yu L, Bandinelli S, Benjamin EJ, Bennett DA, Boomsma D, Cannas A, Coker LH, de Geus E, De Jager PL, Diez-Roux AV, Purcell S, Hu FB, Rimm EB, Hunter DJ, Jensen MK, Curhan G, Rice K, Penman AD, Rotter JI, Sotoodehnia N, Emeny R, Eriksson JG, Evans DA, Ferrucci L, Fornage M, Gudnason V, Hofman A, Illig T, Kardia S, Kelly-Hayes M, Koenen K, Kraft P, Kuningas M, Massaro JM, Melzer D, Mulas A, Mulder CL, Murray A, Oostra BA, Palotie A, Penninx B, Petersmann A, Pilling LC, Psaty B, Rawal R, Reiman EM, Schulz A, Shulman JM, Singleton AB, Smith AV, Sutin AR, Uitterlinden AG, Volzke H, Widen E, Yaffe K, Zonderman AB, Cucca F, Harris T, Ladwig KH, Llewellyn DJ, Raikkonen K, Tanaka T, van Duijn CM, Grabe HJ, Launer LJ, Lunetta KL, Mosley TH, Jr, Newman AB, Tiemeier H, Murabito J. A genome-wide association study of depressive symptoms. Biological Psychiatry. 2013;73(7):667–678. doi: 10.1016/j.biopsych.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller R, Bogomolov M, Benjamini Y, Sofer T. Testing for replicability in a follow-up study when the primary study hypotheses are two-sided 2015 [Google Scholar]
- Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. American Journal of Psychiatry. 2001;158:1568–1578. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- Hoffman DL, Dukes EM, Wittchen HU. Human and economic burden of generalized anxiety disorder. Depression and Anxiety. 2008;25(1):72–90. doi: 10.1002/da.20257. [DOI] [PubMed] [Google Scholar]
- Hoge EA, Ivkovic A, Frichione GL. Generalized anxiety disorder: Diagnosis and treatment. British Medical Journal. 2012;345:37500. doi: 10.1136/bmj.e7500. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3: Genes, Genomics, Genetics. 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapczinski FFK, Silva de Lima M, dos Santos Souza JJSS, Batista Miralha da Cunha AABC, Schmitt RRS. Antidepressants for generalized anxiety disorder. Cochrane Database of Systematic Reviews. 2003;2:CD003592. doi: 10.1002/14651858.CD003592. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Archives of General Psychiatry. 2005;62(6):593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Archives of General Psychiatry. 2005;62(6):617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, Gabriel SB, Harris EL, Hu FB, Jacobs KB, Kraft P, Landi MT, Lumley T, Manolio TA, McHugh C, Painter I, Paschall J, Rice JP, Rice KM, Zheng X, Weir BS, Investigators G. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34(6):591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, Ling H, Hetrick KN, Pugh EW, Amos C, Wei Q, Wang LE, Lee JE, Barnes KC, Hansel NN, Mathias R, Daley D, Beaty TH, Scott AF, Ruczinski I, Scharpf RB, Bierut LJ, Hartz SM, Landi MT, Freedman ND, Goldin LR, Ginsburg D, Li J, Desch KC, Strom SS, Blot WJ, Signorello LB, Ingles SA, Chanock SJ, Berndt SI, Le Marchand L, Henderson BE, Monroe KR, Heit JA, de Andrade M, Armasu SM, Regnier C, Lowe WL, Hayes MG, Marazita ML, Feingold E, Murray JC, Melbye M, Feenstra B, Kang JH, Wiggs JL, Jarvik GP, McDavid AN, Seshan VE, Mirel DB, Crenshaw A, Sharopova N, Wise A, Shen J, Crosslin DR, Levine DM, Zheng X, Udren JI, Bennett S, Nelson SC, Gogarten SM, Conomos MP, Heagerty P, Manolio T, Pasquale LR, Haiman CA, Caporaso N, Weir BS. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44(6):642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavange LM, Kalsbeek WD, Sorlie PD, Aviles-Santa LM, Kaplan RC, Barnhart J, Liu K, Giachello A, Lee DJ, Ryan J, Criqui MH, Elder JP. Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Annals of Epidemiology. 2010;20(8):642–649. doi: 10.1016/j.annepidem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, DeStefano AL, Larson MG, O'Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH. Evidence for a gene influencing blood pressure on chromosome 17: Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the Framingham Heart Study. Hypertension. 2000;36(4):477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- Logue MW, Baldwin C, Guffanti G, Melista E, Wolfe EJ, Reardon AF, Uddin M, Wildman D, Galea S, Koenen KC, Miller MW. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Molecular Psychiatry. 2013;18(8):937–942. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milea D, Verpillat P, Guelfucci F, Toumi M, Lamure M. Prescription patterns of antidepressants: findings from a US claims database. Curr Med Res Opin. 2010;26(6):1343–1353. doi: 10.1185/03007991003772096. [DOI] [PubMed] [Google Scholar]
- Moffitt TE, Caspi A, Taylor A, Kokaua J, Milne BJ, Polanczyk G, Poulton R. How common are common mental disorders? Evidence that lifetime prevalence rates are doubled by prospective versus retrospective ascertainment. Psychological medicine. 2010;40:899–909. doi: 10.1017/S0033291709991036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MG, Llera SJ, Erickson TM, Przeworski A, Castonguay LG. Worry and generalized anxiety disorder: A review and theoretical synthesis of evdience on nature, etiology, mechanisms, and treatment. Annual Review of Clinical Psychology. 2013;9:275–297. doi: 10.1146/annurev-clinpsy-050212-185544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otowa T, Maher BS, Aggen SH, McClay JL, van den Oord EJ, Hettema JM. Genome-wide and gene-based association studies of anxiety disorders in European and African American samples. PLoS One. 2014;9(11):e112559. doi: 10.1371/journal.pone.0112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otowa T, Tanii H, Sugaya N, Yoshida E, Inoue K, Yasuda S, Shimada T, Kawamura Y, Tochigi M, Minato T, et al. Replication of a genome-wide association study of panic disorder in a Japanese population. Journal of Human Genetics. 2010;55(2):91–96. doi: 10.1038/jhg.2009.127. [DOI] [PubMed] [Google Scholar]
- Otowa T, Yoshida E, Sugaya N, Yasuda S, Nishimura Y, Inoue K, Tochigi M, Umekage T, Miyagawa T, Nishida N, et al. Genome-wide association study of panic disorder in the Japanese population. J Hum Genet 54(2):122-6. Journal of Human Genetics. 2009;54(2):122–126. doi: 10.1038/jhg.2008.17. [DOI] [PubMed] [Google Scholar]
- Passel JS, Cohn DV, Lopez MH. Census 2010: 50 Million Latinos, Hispanics Account for More than Half of Nation's Growth in Past Decade, Pew Research Center, Pew Hispanic Center 2011 [Google Scholar]
- Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nature Reviews Genetics. 2009;10:872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick UM, Jain D, Hodonsky CJ, Morrison JV, Davis JP, Brown L, Sofer T, Conomos MP, Schurmann C, McHugh CP, Nelson SC, Vadlamudi S, Stilp A, Plantinga A, Baier L, Bien SA, Gogarten SM, Laurie CA, Taylor KD, Liu Y, Auer PL, Franceschini N, Szpiro A, Rice K, Kerr KF, Rotter JI, Hanson RL, Papanicolaou G, Rich SS, Loos RJ, Browning BL, Browning SR, Weir BS, Laurie CC, Mohlke KL, North KE, Thornton TA, Reiner AP. Genome-wide Association Study of Platelet Count Identifies Ancestry-Specific Loci in Hispanic/Latino Americans. Am J Hum Genet. 2016;98(2):229–242. doi: 10.1016/j.ajhg.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada-Sugimoto M, Otowa T, Hettema JM. Genetics of anxiety disorders: Genetic epidmeiological nd molecular studies in humans. Psychiatry and Clinical Neurosciences. 2015;69(7):388–401. doi: 10.1111/pcn.12291. [DOI] [PubMed] [Google Scholar]
- Smoller JW. The genetics of stress-related disorders: PTSD, depression, and anxiety disorders. Neuropsychopharmacology. 2015 doi: 10.1038/npp.2015.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielberger CD. State-Trait Anxiety Inventory: Bibliography. 2. Palo Alto, CA: Consulting Psychologists Press; 1989. Palo Alto, CA, Consulting Psychologists Press. [Google Scholar]
- Spielberger CD, Reheiser EC. Assessment of emotions: Anxiety, anger, depression, and curiosity. Applied Psychology: Health and Wellbeing. 2009;1(3):271–302. [Google Scholar]
- Stein MB, Sareen J. CLINICAL PRACTICE. Generalized Anxiety Disorder. N Engl J Med. 2015;373(21):2059–2068. doi: 10.1056/NEJMcp1502514. [DOI] [PubMed] [Google Scholar]
- Trzaskowski M, Eley TC, Davis OS, Doherty SJ, Hanscombe KB, Meaburn EL, Haworth CM, Price T, Plomin R. First genome-wide association study on anxiety-related behaviours in childhood. PLoS One. 2013;8(4):e58676. doi: 10.1371/journal.pone.0058676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursano RJ, Colpe LJ, Heeringa SG, Kessler RC, Schoenbaum M, Stein MB, Army Sc. The Army study to assess risk and resilience in servicemembers (Army STARRS) Psychiatry. 2014;77(2):107–119. doi: 10.1521/psyc.2014.77.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belle G. STRUTS: Statistical rules of thumb. New York, NY: John Wiley and Sons; 2002. [Google Scholar]
- Walter S, Glymour MM, Koenen K, Liang L, Tchetgen Tchetgen EJ, Cornelis M, Chang SC, Rimm E, Kawachi I, Kubzansky LD. Performance of polygenic scores for predicting phobic anxiety. PLoS One. 2013;8(11):e80326. doi: 10.1371/journal.pone.0080326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Research. 2010;(3):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassertheil-Smoller S, Arredondo E, Cai J, Castenada S, Choca JP, Gallo L, Jung M, LaVange LM, Lee-Rey ET, Mosley T, Jr, Penedo FJ, Santistaban D, Zee P. Depression, anxiety, antidepressant use, and cardiovascular disease among Hispanic men and women of different ethnic backgrounds: results from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) Annals of Epidemiology. 2014;11:822–830. doi: 10.1016/j.annepidem.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassertheil-Smoller S, Shumaker S, Ockene J, Talavera GA, Greenland P, Cochrane B, Robbins J, Aragaki A, Dunbar-Jacob J. Depression and cardiovascular sequela in postmenopausal women: The Women's Health Initiative (WHI) Archives of Internal Medicine. 2004;164:289–298. doi: 10.1001/archinte.164.3.289. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittchen HU, Jacobi F. Size and burden of mental disorders in Europe: A critical review and approaisal of 27 studies. European Neuropsychopharmacology. 2005:357–376. doi: 10.1016/j.euroneuro.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Wittchen HU, Jacobi F, Rehm J, Gustavsson A, Svensson M, Jonsson B, Olesen J, Allgulander C, Alonso J, Faravelli C, Fratiglioni L, Jennum P, Lieb R, Maercker A, van Os J, Preisig M, Salvador-Carulla L, Simon R, Steinhausen HC. The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur Neuropsychopharmacol. 2011;21(9):655–679. doi: 10.1016/j.euroneuro.2011.07.018. [DOI] [PubMed] [Google Scholar]
- Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biological Psychiatry. 2013;74(9):656–663. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Prentice RL. Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics. 2008;9(4):621–634. doi: 10.1093/biostatistics/kxn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Prentice RL. Correcting "winner's curse" in odds ratios from genomewide association findings for major complex human diseases. Genet Epidemiol. 2010;34(1):78–91. doi: 10.1002/gepi.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.