Abstract
Circadian clocks are endogenous oscillators that control 24-hour physiological and behavioral processes in organisms. These cell-autonomous clocks are composed of a transcription–translation-based autoregulatory feedback loop. With the development of next-generation sequencing approaches, biochemical and genomic insights into circadian function have recently come into focus. Genome-wide analyses of the clock transcriptional feedback loop have revealed a global circadian regulation of transcription factor occupancy, RNA polymerase II recruitment and initiation, nascent transcription and chromatin remodelling. The genomic targets of the circadian clock are pervasive and are intimately linked to the regulation of metabolism, cell growth and physiology.
Introduction
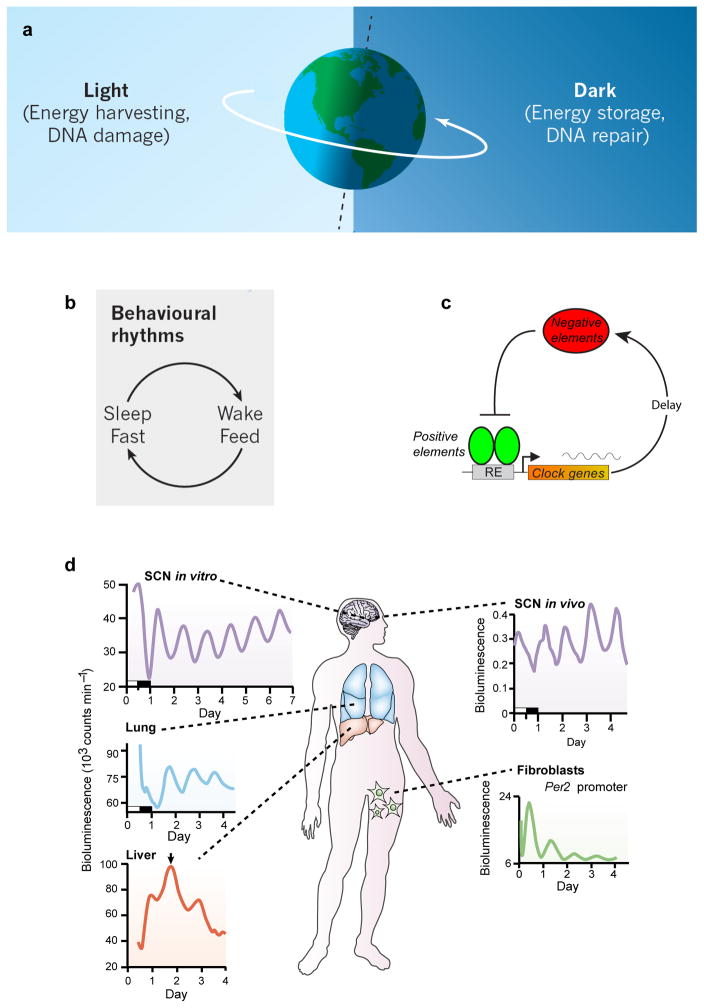
Life on earth evolved in the presence of a 24-hour energetic cycle driven by the solar day. To adapt and respond optimally to the daily environmental cycles, living systems utilize internal timing systems that resonate with the 24-hour day1 (Figure 1a,b). Across life forms, the internal timing system can be seen at the transcriptional level, giving rise to divergent gene networks in different organisms that oscillate on a 24-hour basis2. ‘Circadian clocks’, as these endogenous timers are known, seem to be the product of convergent evolution, as the design principles of circadian transcriptional circuits share a common network motif3 (that is, negative feedback with delay (Figure 1c)), although the specific components are divergent across the kingdoms of life2, 4–7. The link to the daily solar energetic cycle is most obvious in photosynthetic organisms8 but is also a conserved motif in the nutrient cycles of higher organisms9–11. Whether circadian clocks evolved to adapt to the energetic demands of the environment or to avoid the detrimental consequences of solar radiation remains a topic of debate1, 6, 11.
Figure 1.
Circadian rhythms are adaptations of organismal physiology to resonate with the 24-hour solar energetic cycle on earth. a) The earth’s 24-hour rotation leads to a diurnal cycle of light and darkness that drive an energy harvesting and energy storage daily rhythm. Solar irradiation also imposes a cycle of DNA damage and recovery. Reproduced with permission from REF.11. b) Animals have behavioral rhythms of sleep-wake and feeding and fasting occur on a 24-hour basis in synchrony with the solar day. Reproduced with permission from REF.11. c) A conserved network motif of circadian clocks involves a transcription-translation negative feedback loop with delay. d) In mammals, circadian clocks are cell autonomous and are found in all major organ systems and tissues of the body. A hierarchical organization exists in which the hypothalamic suprachiasmatic nucleus (SCN) acts as a master pacemaker to synchronize behavioral and physiological rhythms throughout the body. Modified with permission from REF.15.
Irrespective of why clocks evolved, it is evident today that clock genes and cell-autonomous clocks are ubiquitous. In mammals, for example, we no longer have a neuro-centric view of a master clock located only in the brain. Instead, the discovery of “clock genes”12 in the 1990s showed that virtually all cells express these genes and have the capacity to generate circadian oscillations13, 14. The circadian clock in mammals is now conceptualized as a hierarchical system in which a brain clock located in the hypothalamic suprachiasmatic nucleus (SCN) acts as a master pacemaker to synchronize or entrain peripheral clocks distributed throughout the body15–19 (Figure 1d). This system-wide architecture begs the questions: how is this hierarchy coupled and integrated, and what are the functions of clocks in tissue- and cell-specific contexts?
The molecular mechanism of circadian clocks in mammals is generated by a cell-autonomous transcriptional autoregulatory feedback loop. The ‘core’ clock genes include CLOCK and BMAL1, which encode activators, and PER1, PER2, CRY1 and CRY2, which encode repressors. Current topics of active investigation include the components of this core circadian clock gene network, how environmental and systemic inputs impinge upon the core clock mechanism20, 21, and how the core clock network regulates downstream output pathways. Recent work has provided insight into the genomic targets of the core clock pathway and has led to some surprises concerning the pervasiveness of circadian regulation in gene expression. A substantial fraction (~5–20%) of genes expressed in any particular cell or tissue have been found to undergo circadian oscillations at the mRNA level22; however, this level of regulation (that is, steady-state mRNA rhythms) is only one of a multitude of layers of circadian regulation in gene expression. Virtually every regulatory stage in gene expression that has been examined carefully — from transcription, splicing, termination, polyadenylation, nuclear export, microRNA regulation, translation to RNA degradation — has revealed additional layers of circadian control23, 24. Thus, both transcriptional and post-transcriptional regulation of circadian gene expression are key nodal points of control.
Our understanding of the mechanism of the circadian clock in mammals has been informed by progress on two fronts: first, the biochemical definition of components of the circadian activator and repressors complexes, and second, the determination of the structure of key domains of the clock proteins (Box 1). This Review will focus on molecular, biochemical and genomic advances in our understanding of circadian regulation in mammals. I will emphasize work on the genomic targets of the core circadian transcriptional pathway and will highlight studies revealing global circadian oscillations in RNA polymerase II (RNAPII)-mediated transcription and their relation to the dynamic circadian regulation of chromatin state and chromatin modifiers. For non-mammalian model systems and aspects of circadian biology focused on photic regulation, metabolism and immunity, there are other excellent reviews8, 25–37.
Box 1. Structural biology of clock proteins.
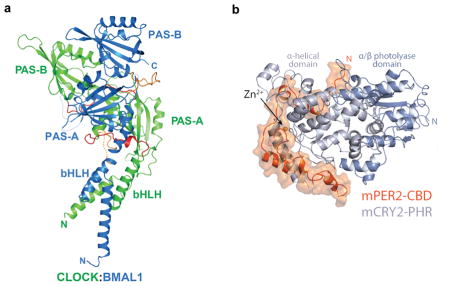
To achieve a mechanistic understanding of clock function, structural biology will be essential154, 155. The three-dimensional structures of a number of circadian proteins and their complexes have been solved using X-ray crystallography. These include the bHLH, PAS-A and PAS-B domains of the CLOCK:BMAL1 heterodimeric transcriptional activator complex (PDB code 4H10) (see figure a)156; the PAS-A/PAS-B domains of the mammalian PERIOD proteins157, 158, the photolyase homology region (PHR) of mammalian CRY1159, the PHR of CRY2 in complex with FBXL3160, and the PER2-CBD (CRY-binding domain) in complex with CRY1161 or CRY2 (PDB code 4U8H) (see figure)162. The structure of the CLOCK:BMAL1 heterodimeric complex is asymmetric with CLOCK wrapping around BMAL1. The PAS-A and PAS-B domains of the two subunits interact with their complementary domains but with two different interfaces: the PAS-A domains have symmetrical interactions involving the β-sheet surfaces; while the PAS-B domains have a head-to-tail interaction in which the β-sheet surface of BMAL1 interacts with the α-helical surface of CLOCK. The PAS-A/PAS-B domain configuration of BMAL1156 (but not CLOCK) is identical to that seen in the PAS-A/PAS-B domains of the mammalian PERIOD proteins157, 158 suggesting that PER could interact with CLOCK and replace BMAL1163. Interestingly, a crystal structure of a another member of the bHLH-PAS heterodimeric protein family, HIF-2α-ARNT, reveals a completely different overall domain arrangement when compared to CLOCK:BMAL1, but specific domain interface interactions are preserved between the respective bHLH, PAS-A and PAS-B domains of the two subunits164 (see REF.165 for a comparison of these bHLH-PAS structures). The recognition of the CACGTG E-box DNA binding motif of the CLOCK:BMAL1 bHLH region is similar to that seen for other bHLH proteins166.
The crystal structures of mammalian CRY2 have been determined in five functional states: 1) the photolyase homology region (PHR) by itself (apo); 2) the FAD-bound CRY2 PHR; 3) the CRY2-FBXL3-SKP1 complex; 4) the KL001-bound CRY2 PHR; and 5) CRY2 in complex with the PER2-CRY-binding domain (CBD)160, 162, 167. Both the CRY1 and CRY2 apo structures adopt the canonical photolyase fold and can be aligned most closely to the Drosophila (6–4) photolyase159, 160. However unlike (6–4) photolyases and cryptochromes from plants and Drosophila melanogaster, the mammalian CRY1 and CRY2 PHRs do not have the FAD cofactor bound. In CRY2, FAD can bind with low affinity, and the FAD-binding pocket has a more open conformation perhaps explaining the lower affinity of FAD160. CRY2 in complex with FBXL3 reveals an extensive interface with the FBXL3 leucine rich repeat (LRR) domain, but most interestingly is the C-terminal tail of FBXL3 inserting into the FAD-binding pocket revealing a novel substrate recognition motif by the SCFFbxl3 ubiquitin ligase160. The small molecule stabilizer of CRY, KL001168, binds the FAD pocket, but also extends beyond the pocket where the FBXL3 C-terminal tail enters, so as to be in a position to block the FBXL3-CRY2 interaction to stabilize CRY167. Finally, the known interaction between CRY and PER proteins, which mutually stabilize one another, has been illuminated by the recent crystal structures of the CRY1/PER2-CBD161 and CRY2/PER2-CBD162 complexes. The PER2-CBD forms an extended conformation on CRY2 that begins at a secondary pocket critical for CLOCK:BMAL1 binding and winds around the CRY2 C-terminal helix to sterically hinder the recognition of CRY2 by the SCFFbxl3 ubiquitin ligase (see figure b). Thus, the overlapping interfaces on CRY2 for PER2-CBD and FBXL3 provide a structural basis for the competition of PERs and FBXL3 for controlling the stability of CRY162
The existing crystal structures beg the question of how PER and CRY interact with CLOCK:BMAL1 as a quaternary complex. In different contexts, CRY1 can interact directly with CLOCK:BMAL1 without PER in vitro169 and while in other experiments PER has been shown to be essential for interaction with CLOCK:BMAL1 in vivo163. Likely PER is involved in reducing the occupancy of CLOCK:BMAL1 on DNA170 as seen in D. melanogaster171, while CRY1 may be able to repress CLOCK:BMAL1 on DNA170, 172. Because the native CLOCK:BMAL1/PER:CRY quaternary complexes are megadalton in size and involve other interacting proteins, and important domains of these proteins are intrinsically disordered, the solution of these complexes likely will require a combination of crystallography, NMR, and cryo electron microscopy methods in future work.
The core circadian clock gene network
Core components of the mammalian circadian clock
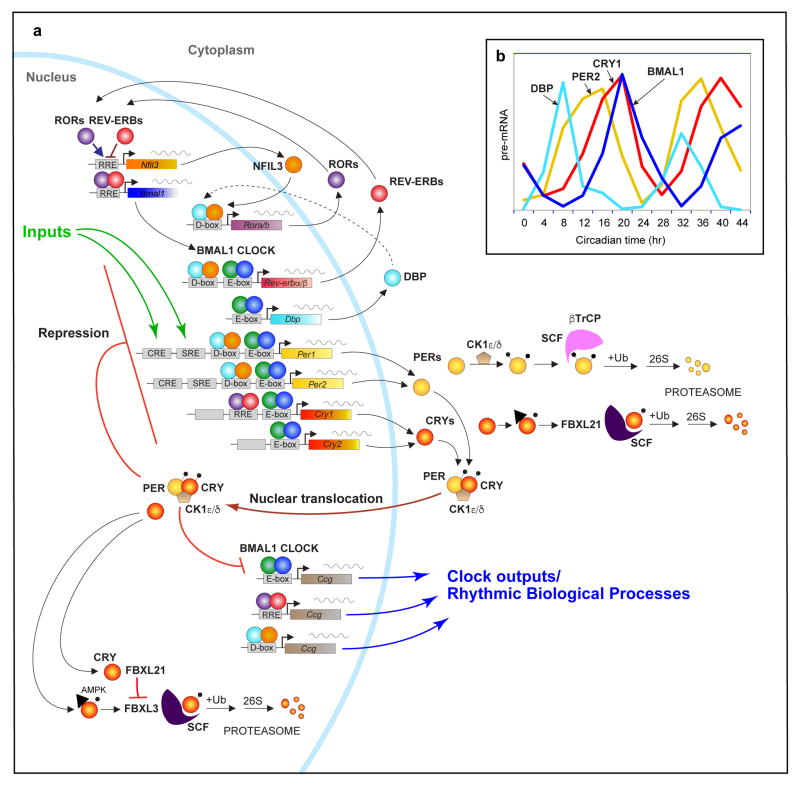
In mammals, the mechanism of the circadian clock consists of the activators, CLOCK (and its paralog NPAS2) and their heterodimeric partner BMAL1 (or ARNTL), which are basic-helix-loop-helix (bHLH) – PER-ARNT-SIM (PAS) transcription factors38, 39. The CLOCK:BMAL1 complex binds to regulatory elements containing E-boxes in a set of rhythmic genes encoding the repressor proteins PERIOD (Per1, Per2, Per3) and CRYPTOCHROME (Cry1, Cry2)39–41 (Figure 2a). In mouse, CLOCK:BMAL1 activation occurs in the daytime, leading to transcription of the Per and Cry genes in the afternoon and the accumulation of the PER and CRY proteins in the late afternoon or evening42. The PER and CRY proteins interact with each other as well as with the serine-threonine kinases Casein kinase 1δ (CK1δ) and CK1ε43, and translocate into the nucleus at night, where they interact with CLOCK:BMAL1 to repress their own transcription42, 44. As repression progresses, Per and Cry transcription decline, and the PER and CRY protein levels decrease since their half-lives are relatively short and specific E3 ligase complexes target the PER and CRY proteins for ubiquitination and subsequent degradation by the proteasome43, 45. Once negative feedback repression is relieved by turnover of the repressor complex, transcription by CLOCK:BMAL1 can begin anew to start a new cycle of transcription the next morning. Importantly, core genes such as PER2 and its regulatory kinase, CSNK1D, have been found to underlie human sleep timing disorders46, 47, demonstrating a clear role for clock genes in humans (Box 2).
Figure 2.
The circadian gene network in mammals. a) At the core, CLOCK and BMAL1 activate the Per1, Per2, Cry1 and Cry2 genes, whose protein products interact and repress their own transcription. The stability of the PER and CRY proteins are regulated by parallel E3 ubiquitin ligase pathways. CLOCK and BMAL1 also regulate the nuclear receptors, Rev-erbα/β, which rhythmically repress the transcription of Bmal1 and Nfil3 that is driven by the activators, RORa/b. NFIL3 in turn represses the PAR-bZip factor, DBP, to regulate a rhythm in the ROR nuclear receptors. These three interlocked transcriptional feedback loops represent the three major transcriptional regulators of the majority of cycling genes. Different combinations of these factors generate different phases of transcriptional rhythms as exemplified by the RNA profiles of Dbp, Per2, Cry1 and Bmal1 in the mouse liver (b). Additional rhythmic output genes (so call “Clock-controlled genes or Ccg’s) are transcriptionally regulated by the three loops acting on E-box, RRE and D-box elements in the regulatory regions of target genes.
Box 2. Human genetics of clock genes.
Validation of the core circadian transcriptional mechanism elucidated in rodents (Figure 2) has come from the Mendelian disorder, familial advanced sleep phase disorder (FASPD), in which the timing of sleep-wake cycles in affected individuals are shifted several hours earlier than normal173. Dominant mutations in both PER2 and CSNK1D have been shown to be linked to FASPD and to be causative in humans46, 47. In large-scale human genome-wide association studies (GWAS) for fasting glucose levels and increased risk of type 2 diabetes, common variants in the MTNR1B locus, which encodes melatonin receptor 1B, and in CRY2 have been documented174–177. Melatonin signalling is a major output pathway of the suprachiasmatic nucleus (SCN) in mammals, and levels of melatonin are circadian, with high levels at night and low levels in the day178 reflecting a physiological signal of darkness. The MTNR1B mutations have been proposed to act in pancreatic islets to impair insulin secretion but this remains subject to debate179–181. Recently, large-scale GWAS for ‘morningness’ and sleep timing preference (chronotype) have identified a number of genes known to be associated with circadian rhythms in animals182–184. These include RGS16, a regulator of G-protein signaling that is enriched in the SCN and has a long period phenotype with a loss-of-function mutation185; the core clock gene PER2; VIP (Vasoactive intestinal polypeptide), a neuropeptide in the SCN involved in oscillator coupling186; HCRTR2, the receptor for hypocretin or orexin that is linked to narcolepsy in humans and dogs187; and FBXL3, an F-box protein that is the recognition domain for an E3 ubiquitin ligase complex that degrades the CRY proteins85. GWAS using sleep duration as a phenotype have identified a few loci (PAX8, VRK2 and UFL1)183, 188, 189, but their relation to sleep mechanisms remains obscure. Overall, it is reassuring that a number of GWAS hits are associated with genes with known circadian and sleep functions, thus fortifying the notion that circadian transcriptional mechanisms will likely apply to human biology as well.
In addition to the Per and Cry target genes, CLOCK:BMAL1 also activate the nuclear receptors REV-ERBα and REV-ERBβ (encoded by Nr1d1 and Nr1d2, respectively), which compete at REV-ERB/retinoic acid-related orphan receptor (ROR) binding elements (ROREs) with RORα, RORβ and RORγ (encoded by Rora, Rorb, Rorc, respectively)48–50 (Figure 2a). The rhythmic expression of REV-ERBα/β leads to the repression of Bmal1 and Clock, which induces a rhythm in these genes that is in anti-phase with the Per gene expression rhythms48. These interlocked feedback loops of activators and repressors form a canonical positive- and negative-feedback gene network motif51. A third CLOCK:BMAL1-driven transcriptional loop involves the PAR-bZip (proline and acidic amino acid-rich basic leucine zipper) factors DBP (D-box binding protein), TEF (thyrotroph embryonic factor) and HLF (hepatic leukemia factor). These proteins interact at sites containing D-boxes with the repressor NFIL3 (Nuclear Factor, Interleukin 3 Regulated; or E4BP4), which is driven by the REV-ERB/ROR loop (Figure 2a) 52, 53. Together, these three interlocking transcriptional feedback loops can generate cycles of transcription with various phases of expression depending on the combination of cis-elements (E-box, RORE, D-box) in the promoters and enhancers of specific target genes (Figure 2b) 54.
Core clock protein interactions with chromatin and chromatin-modifying complexes
At the beginning of the transcription cycle, the activators, CLOCK and BMAL1, interact with the histone acetyltransferases (HAT) p300 and CREB binding protein (CBP), respectively, to acetylate histones and provide an accessible chromatin state for transcription55–58. CLOCK has been reported to have intrinsic HAT activity and to acetylate histone H3K9 and H3K1459, 60. The NAD+-dependent histone deacetylase (HDAC) sirtuin 1 (SIRT1) associates with CLOCK, BMAL1 and PER261, 62, and a circadian rhythm in NAD+ levels driven by the expression of the CLOCK:BMAL1 target gene Nampt leads to a rhythm in SIRT1 activity which feeds back to inhibit the CLOCK:BMAL1 complex63, 64.
CLOCK and BMAL1 have also been shown to be associated with the methyltransferase MLL1 (mixed lineage leukemia 1)65. Interestingly, MLL1 interacts with the δ19 activation domain of CLOCK and is thought to be important for the recruitment of CLOCK to DNA65. Moreover, MLL1 is associated with rhythmic H3K4 trimethylation (H3K4me3)65, a common histone marker associated with active gene promoters. Also, MLL1 is acetylated, and the metabolic regulator SIRT1 rhythmically deacetylates MLL1, thereby affecting its methyltransferase activity66. The related methyltransferase MLL3 exhibits circadian occupancy and is associated with promoters carrying rhythmic H3K4me3 marks; however, MLL3 recruitment to DNA does not depend on BMAL1 or CLOCK67.
CLOCK and BMAL1 also interact with the histone lysine demethylases JARID1a and LSD1, which are both thought to promote the recruitment of CLOCK:BMAL1 to Per gene promoters to enhance transcription68, 69. BMAL1 interacts with TRAP150 (thyroid hormone receptor-associated protein-150), a co-activator that promotes CLOCK:BMAL1 binding to circadian target genes and links CLOCK:BMAL1 to the Mediator complex subunit MED1 to recruit the general transcriptional machinery70. RNAPII is both recruited and initiated on a circadian basis as discussed in more detail below71, 72.
During the transition from activation to repression, the CLOCK:BMAL1 complex interacts with the PER-CRY repressor complex at the beginning of the night as the levels of PER and CRY accumulate and translocate into the nucleus42. The PER complex is large and contains at least 25 proteins73–75. Initially CLOCK:BMAL1 are associated with two components of the Mi-2/nucleosome remodelling and deacetylase (NuRD) transcriptional corepressor complex, CHD4 and MTA2. Interestingly, PER complexes without CLOCK:BMAL1 lack CHD4 and MTA2 but contain other members of the NuRD complex such as MBD2, GATAD2a, HDAC1 and RbAp4875, suggesting that during this transition phase there is a split NuRD complex which then comes together to form a reconstituted NuRD corepressor complex to inhibit CLOCK:BMAL1 transcription. The CLOCK:BMAL1 complex also recruits the polycomb repressor complex (PRC2), leading to the trimethylation of H3K27 at target genes and thereby inhibiting gene expression76, 77. In addition, CLOCK:BMAL1 recruits the Ddb1-Cullin-4 ubiquitin ligase complex to circadian target genes to promote H2B monoubiquitination at E-box sites to covalently mark genes for subsequent recruitment of the PER complex78. At the peak of repression in the middle of the night, the PER complex is also associated with the transcriptional repressor complexes, SIN3-HDAC79 and Hp1γ–Suv39h histone methyltransferase80, which contribute to CLOCK:BMAL1 repression by deacetylation of histone H3K9 and H4K5 as well as di- and trimethylation of H3K9. The PER repressor complex also includes NONO, a protein involved in RNA processing and WDR5, a component of a histone methyltransferase complex73. At later times during the repression phase, PER complexes include the RNA helicases DDX5 and DHX9, as well as the large subunit of RNAPII and SETX, a helicase that promotes transcriptional termination74. Thus, recruitment of a number of chromatin-modifying complexes as co-repressors is necessary for PER- and CRY-mediated repression of CLOCK:BMAL1. During the circadian cycle, transcriptional activation and repression are accompanied by dynamic epigenetic transitions in chromatin state that poise the genome for rhythmic gene expression.
Regulation of circadian period length by repressor stability
The stability of the core repressor proteins is an important determining factor in the dynamics of the transcriptional feedback loop, which in turn regulates the period length of the oscillation. The stability of PER and CRY is regulated by their phosphorylation state and by their ubiquitination by E3 ligase complexes (Figure 2a). For example, the tau mutation in CK1ε shortens circadian period length in vivo and destabilizes the PER proteins by phosphorylation of a regulatory site that marks the PER proteins for ubiquitination by β-TrCP and proteasomal degradation43, 81–83. By contrast, mutations in the F-box protein FBXL3, which is one of four subunits of the SCF (Skp1/Cullin/F-box protein) E3 ligase complex, lengthen circadian period and stabilize the CRY proteins by decreasing ubiquitination and subsequent proteosomal degradation84–86. In addition, CRY ubiquitination by SCFFbxl3 is triggered by AMPK-mediated phosphorylation, which provides a link to metabolism via nutrient-responsive AMPK activity87.
Stability and turnover of PER proteins is primarily regulated by CK1δ and CK1ε43, 83, 88, 89, by the balance between these two protein kinases and phosphatase 190, and by the stoichiometry of PER and the kinases91, 92. Recent work has shown that a dual-kinase, multisite phosphoswitch regulates PER stability and its ubiquitination by the β-TrCP E3 ubiquitin ligase complex93.
In the case of CRY, stability and turnover are regulated by the balance of the two paralogues FBXL3 and FBXL21 (Figure 2a) 94, 95. Contrary to expectation, FBXL21 antagonizes FBXL3 to reduce the E3 ligase activity of SCFFbxl3 on CRY1 and CRY2 to stabilize them95. Although SCFFbxl21 does possess E3 ligase activity, it is less efficient than SCFFbxl3 in the ubiquitination of CRY95. However, FBXL21 interacts more strongly with CRY than FBXL3, which protects CRY from SCFFbxl3 activity95. The subcellular localization of FBXL3 and FBXL21 also differ, with FBXL3 acting as the primary E3 ligase for CRY in the nucleus, whereas FBXL21 acts as the primary E3 ligase for CRY in the cytoplasm95. Finally, in genetic experiments exploring the interaction of the CK1εtau mutant with the Fbxl3Afh mutant, a broad range of periodicities were seen depending on the combination of alleles96, suggesting that the PER and CRY pathways independently affect the ultimate period length in vivo.
Genome-wide clock architecture
While previous work has examined the transcriptional regulation of CLOCK:BMAL1 target genes such as Dbp, Per1 and Nr1d1 in detail, identity of the circadian cistrome has only been recently defined. Using chromatin immunoprecipitation followed by sequencing (ChIP-seq), genome-wide DNA binding sites for BMAL1, CLOCK, NPAS2, PER1, PER2, CRY1 and CRY2 have been identified in mouse liver during the circadian cycle by a number of groups71, 97–100.
Genomic targets of the core circadian activators
Occupancy of CLOCK:BMAL1 at the Dbp locus, which encodes a transcription factor that modulates expression of clock output genes, is circadian, with peak binding occurring in the middle of the day101. At the single-cell level, CLOCK and BMAL1 recruitment to E-boxes in the Dbp gene are interdependent and circadian102. BMAL1 binding was observed to be dynamic and stochastic, and fluctuations in binding depend on the proteasome102, 103, which is consistent with a ‘kamikaze’ model of transcriptional activation99. On a genome-wide level, CLOCK and BMAL1 occupancy also peak within a narrow window of time during the middle of the day, and the phasing of CLOCK:BMAL1 occupancy is not tightly phase-locked with the RNA expression of their target genes71, 97, 98. This flexible phase relationship can be explained at least in part by the combinatorial action of other cis-acting elements (RORE and D-box) within the tripartite clock gene network54 (Figure 2a,b). For example, the Cry1 gene, which peaks much later in phase than the Per genes, is regulated by both E-box and RORE elements104.
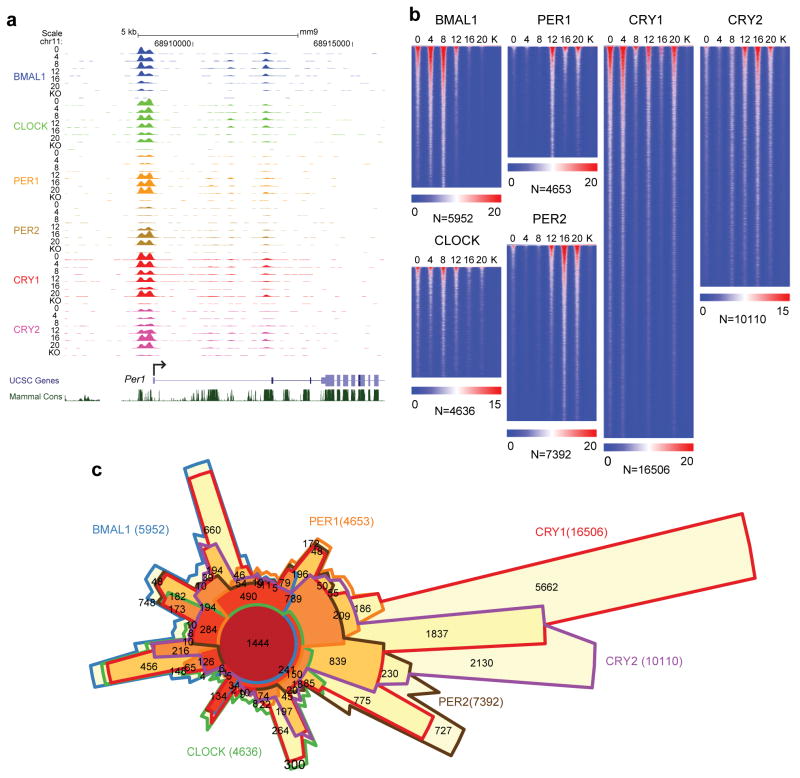
To view the dynamics of the occupancy of the core circadian activator complexes genomewide, all core clock proteins have been assessed together in the mouse liver71. At the Per1 gene, the activators BMAL1 and CLOCK bind in a cyclic manner at the promoter, between circadian time (CT) CT0 and CT12, with maximal binding observed at CT8. In genome-wide analyses, CLOCK and BMAL1 bind to over 4,600 and 5,900 sites, respectively, corresponding to ~3,000 unique genes in the liver71 (Figure 3b). At highly expressed target genes, CLOCK:BMAL1 DNA-binding motifs are enriched for a tandem E-box motif97. Native liver extracts show that CLOCK:BMAL1 bind tandem E-box sites cooperatively with higher affinity to more effectively compete with other E-box binding factors such as USF1105.
Figure 3.
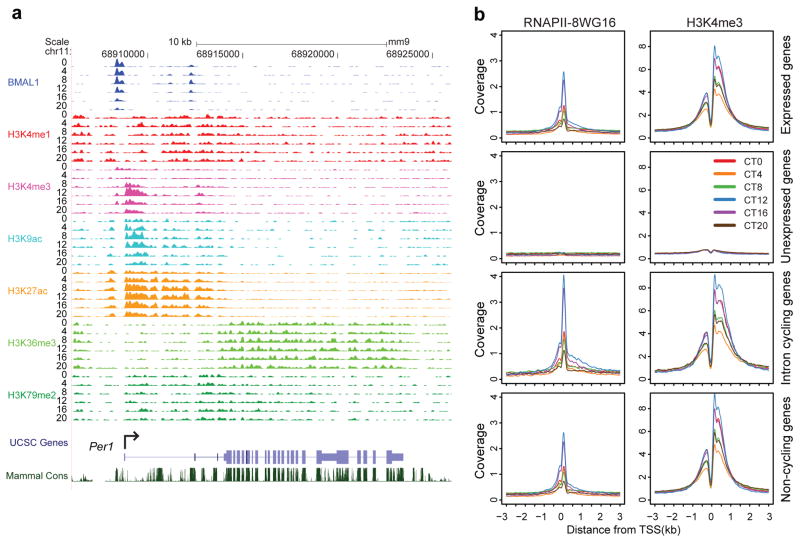
The circadian cistrome in the mouse liver. a) UCSC genome browser view of ChIP-seq profiles of circadian transcription factors at the Per1 gene at 6 circadian times of day [0, 4, 8, 12, 16, 20 CT (hr)]. The colors of the wiggle plots of ChIP-seq occupancy indicate the following transcriptional regulators: BMAL1 (blue), CLOCK (green), PER1 (orange), PER2 (gold), CRY1 (red), CRY2 (pink). KO indicates a ChIP-seq sample from a knockout mouse for each transcriptional regulator. b) Heat map views of genome-wide DNA binding for BMAL1, CLOCK, PER1, PER2, CRY1 and CRY2 measured over 500 bp fragments encompassing the binding sites. Each peak in the genome is represented as a horizontal line, ordered vertically by signal strength. Six time points are shown beginning at CT0 and ending at CT20 from left to right. Knockout (K) mouse control is shown on the far right of each panel. The number of peaks in the genome is indicated at the bottom of each panel. The blue-red gradient indicates the coverage for all binding sites in the genome. The number of binding sites (N) in the genome are indicated at the bottom of each heat map. c) Chow-Ruskey diagram showing the 6-way overlap of BMAL1, CLOCK, PER1, PER2, CRY1 and CRY2 peaks in the genome. The red circle in the middle represents the overlap of all six factors. Lighter shades of red, orange and yellow represent fewer overlaps of subsets. The boundaries for each protein are color coded: BMAL1 (blue), CLOCK (green), PER1 (orange), PER2 (brown), CRY1 (red) and CRY2 (purple), and the areas of each domain are proportional to the number of binding sites. Modified/Reproduced with permission from REF.71.
The hepatic steroid receptor coactivator 2 (SRC-2) is recruited to DNA in a diurnal cycle and overlaps extensively with CLOCK:BMAL1 sites106. SRC-2 acts as a potent transcriptional coactivator of CLOCK:BMAL1, and null mutations in Src-2 mice lead to an abnormally early phase of the activity rhythm106, suggesting that SRC-2 has a role within the circadian clock system.
In addition to the liver, the circadian cistrome has recently been described in mouse pancreatic islets107. Comparing the occupancy of BMAL1 between the liver and pancreas, about 40% of target genes overlapped, with disparate binding sites clustering at distal regulatory regions of tissue-specific genes. Interestingly, even within the overlapping set of target genes, BMAL1 was found to bind to distinct enhancers in liver and pancreas107, consistent with the existence of tissue-specific circadian cis-regulatory modules108.
Genomic targets of the core circadian repressors
In contrast to the activators, the repressors, PER1, PER2 and CRY2, bind genomewide at night between CT12 and CT20, peaking between CT15 and CT17)71 (Figure 3b). CRY1 has a bimodal pattern, with a major peak at CT0 (Figure 3b). The repressors CRY1 and CRY2 bind to significantly more sites in the genome than PER1 and PER2, with many thousands of these sites being independent of CLOCK:BMAL1 and containing DNA-binding motifs for nuclear receptors71 (Figure 3c). Consistent with this finding, CRY1 interacts with a number of nuclear receptors and is a corepressor for the glucocorticoid receptor109. The nuclear receptors REV-ERBα and REV-ERBβ also occupy DNA rhythmically, with a peak in the daytime110, 111. REV-ERBα competes with RORα for DNA binding at RORE and RevDR2 sites to regulate circadian gene target genes such as Bmal1 and Cry1, where it recruits N-CoR and HDAC3 to repress gene expression50, 111. Interestingly, REV-ERBα also interacts at other genomic sites that are enriched for tissue-specific transcription factors such as HNF6 in the liver50. At these sites, the action of REV-ERBα is independent of its DNA-binding domain and defines a different set of target genes that are directly involved in metabolism. Finally, SRC-2 has been proposed to modulate an oscillation in chromatin accessibility — via recruitment of PBAF (Polybromo-associated Brg/Brahma-associated factors) members of the SWI/SNF (SWItch/Sucrose Non-Fermentable) complex — to promote REV-ERB loading and enhance the amplitude of circadian gene expression112. Thus, the circadian cistrome is highly dynamic, and involves not only the core circadian E-box transcriptional regulators, but is interlinked via REV-ERBα/β as well as DBP and NFIL3 to secondary and tertiary gene targets with a wide range of phases of gene expression.
Genome-wide circadian RNA expression
To examine the functional consequences of the circadian cistrome, RNA sequencing (RNA-seq) has been used to profile cycling genes in the mouse liver71, 98, 99. For example at the Dbp and Per2 genes, strand-specific whole-transcriptome RNA-seq revealed a robust circadian oscillation using both the intron RNA signal as a proxy for pre-mRNA and the exon RNA signal as a proxy for mRNA71, 113, 114. As was previously observed in microarray experiments, thousands of genes cycle at the steady-state mRNA level in the liver as well as in many other tissues22, 115. As more regions in the mouse have been profiled, more than half of all genes have been found to cycle at the mRNA level in at least one tissue, and importantly, cycling genes are enriched for drug targets – more than half of the top selling 100 drugs on the market in the United States have a circadian target gene22. This emphasizes the importance of the optimizing the timing of drug administration for circadian drug targets, which can both enhance efficacy while at the same time reduce side effects.
A wide range of cycling RNA species has been found using RNA-seq. In addition to cycling protein-coding transcripts, oscillating long intergenic non-coding RNA (lincRNA), microRNA and antisense RNA transcripts have been found in mouse liver98, 99, 116. Using Global Run-On sequencing (GRO-seq)117 to identify enhancer RNA (eRNA) transcripts118, thousands of oscillating enhancers have been found, and the phase of these cycling eRNAs corresponds with the phases of transcription of nearby genes119. Motif analysis of cycling enhancers at different circadian phases revealed that there was phase-specific enrichment of transcription factor motifs and that four sets of motifs were enriched at different times119. E-box motifs were enriched between Zeitgeber time (ZT) ZT6 and ZT9, corresponding to the peak of CLOCK:BMAL1 occupancy. D-box motifs were enriched between ZT9 and ZT15, corresponding to the expression of DBP and the low point of NFIL3 repression. RORE and RevDR2 motifs were enriched between ZT18 and ZT24, corresponding with the low point of REV-ERB repression of the RORs. ETS (E26 transformation-specific) binding motifs were enriched between ZT0 and ZT3119, which may correspond to GM129 or CHRONO repression120–122. Overall, the circadian cistromic results agree well with their transcriptional readouts as assessed by eRNA and gene expression analysis: the tripartite core circadian transcriptional feedback loop (E-box, D-box, RORE) drives the majority of circadian gene expression on a genome-wide level.
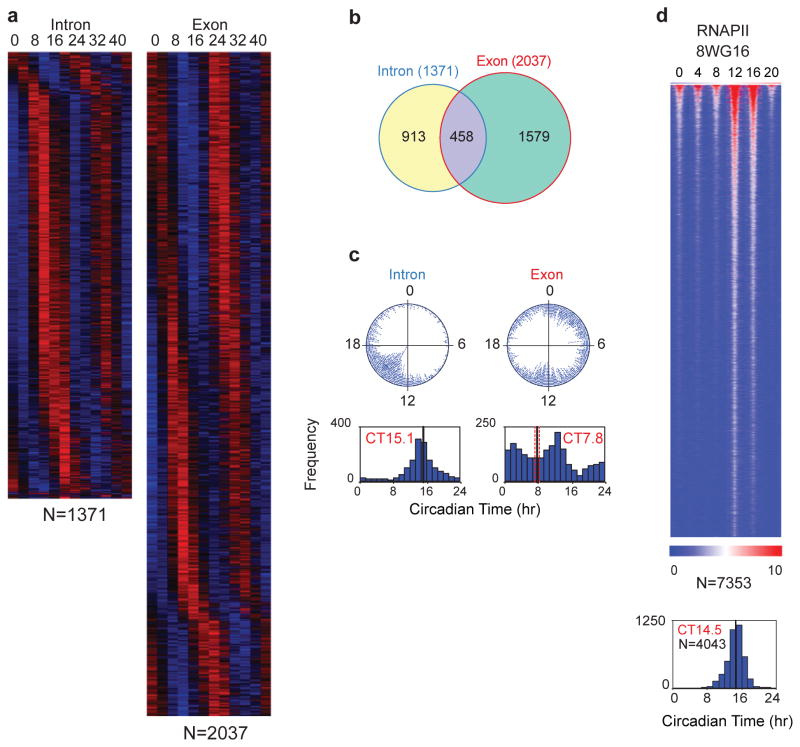
Using whole-transcriptome RNA-seq or nascent-seq combined with mRNA-seq, two divergent sets of cycling genes have been found (cycling nascent transcripts and cycling mRNA levels)71, 98 (Figure 4a). Only ~20% of cycling mRNAs also had a corresponding cycling nascent transcript71, 98, suggesting that other RNA processes likely contribute to steady-state circadian mRNA expression (Figure 4b). A number of different classes of cycling transcripts have been observed: 1) cycling transcription and cycling steady-state mRNA; 2) cycling transcription and no cycling mRNA, and 3) no detectable cycling transcription, but cycling mRNA levels. The first class is typical for the core circadian genes and their high-amplitude direct targets. The second class is typically seen in highly expressed genes in the liver that have mRNAs with long half-lives such as albumin. The third class represents transcripts that could become rhythmic via circadian variation in mRNA turnover, RNA splicing123, polyadenylation124, miRNA regulation116 and other post-transcriptional steps. Whether rhythmic translation or protein levels ultimately occur has also been revealing. For example, rhythmic diurnal polyadenylation occurs in 2.3% of expressed genes in the liver, and one-fifth of these genes (class III) lead to rhythmic protein expression correlated with polyA tail length in the absence of rhythmic mRNA levels124. Ribosomal profiling has also revealed substantial numbers of examples of circadian translation in U2OS cells and liver125, 126. Interestingly there are at least 150 examples of translationally driven oscillations in the liver for which no mRNA rhythms are present125. Thus, both transcriptional and post-transcriptional regulation are critical for circadian gene expression.
Figure 4.
Whole-transcriptome RNA-seq analysis of circadian gene expression in the mouse liver. a) Heat map view of intron (left) and exon (right) RNA cycling genes. Each gene is represented as a horizontal line, ordered vertically by phase in hours indicated at the top. Twelve time points are shown beginning at CT0 and ending at CT44 from left to right. “N” indicates the number of cycling genes in each category. b) Venn diagram of intron and exon cycling genes showing only 22% overlap. c) The phase distribution of cycling genes. The phase of each transcript rhythm is represented in a dot plot (top) and histogram plot (bottom). d) Heat map view of cycling RNAPII-8WG16 occupancy across the genome of the liver with a peak occurring at night. Modified/Reproduced with permission from REF.71.
Genome-wide circadian regulation of transcription, RNAPII and chromatin states
In the mouse liver, cycling pre-mRNA levels (using the intron RNA signal) were coordinately expressed in time with a peak at CT15 (Figure 4c). Thus, there is a peak of globally coordinated circadian transcription that occurs at the beginning of the night during the active or awake portion of the mouse circadian rhythm, which is not seen at the mRNA level (which has a bimodal pattern). To analyse the global rhythms in nascent transcription further, RNAPII occupancy has been measured during the circadian cycle71, 72. The large subunit of RNAPII (POLR2A) contains a C-terminal domain (CTD) that is post-translationally modified at different stages of transcription127–129. RNAPII is recruited into the pre-initiation complex in a hypophosphorylated state (recognized by the 8WG16 antibody)130. Surprisingly, RNAPII-8WG16 occupancy is highly circadian in the mouse liver, with a peak that is correlated with the intron RNA peak (Figure 4d). The first step in the initiation of RNAPII involves phosphorylation on serine 5 (Ser5P) of the CTD of RNAPII131. RNAPII-Ser5P occupancy is also circadian71, suggesting that both recruitment and initiation of RNAPII are under circadian control. Using an antibody against the RPB2 subunit of RNAPII (POLR2B), RNAPII occupancy for promoters and gene bodies is rhythmic and co-varies over time72. Rhythmic loading of RNAPII to promoters rather than a rhythmic transition from a paused state to productive elongation accounts for the majority of rhythmic transcription in the mouse liver72.
Given these circadian rhythms of RNAPII, chromatin states associated with RNAPII transcription have been examined during the circadian cycle71, 72, 99. Figure 5 illustrates genome views of histone modifications that are characteristic of promoter, enhancer and elongation states132–138. Histone H3K4me3, H3K9ac and H3K27ac are enriched at promoters and undergo circadian rhythms in histone modifications at the Per1 gene (Figure 5a). On a genome level, circadian rhythms in RNAPII occupancy as well as histone H3K4me3, H3K9ac and H3K27ac modifications are present in the majority of expressed genes irrespective of whether RNA cycling occurs or not71. Both RNAPII occupancy and H3K4me3 marks are highly dynamic and show circadian oscillations that peak in the early night (CT12) for all expressed genes, cycling intron RNA genes and non-cycling genes (Figure 5b).
Figure 5.
Circadian chromatin states in the mouse liver. a) UCSC genome browser view of histone methylation and acetylation at the Per1 gene at 6 circadian times of day [0, 4, 8, 12, 16, 20 CT (hr)]. The colors of the wiggle plots of ChIP-seq signal indicate the following: BMAL1 (blue), H3K4me1 (red), H3K4me3 (pink), H3K9ac (aqua), H3K27ac (orange), H3K36me3 (light green), H3K79me2 (dark green). b) Binding profiles of RNAPII-8WG16 and H3K4me3 at the transcription start site (TSS) +/− 3kb in 12,680 expressed genes (top row), 8,945 unexpressed genes (second row), 1371 intron cycling genes (third row) and 5,839 non-cycling genes (bottom row). Modified/Reproduced with permission from REF.71.
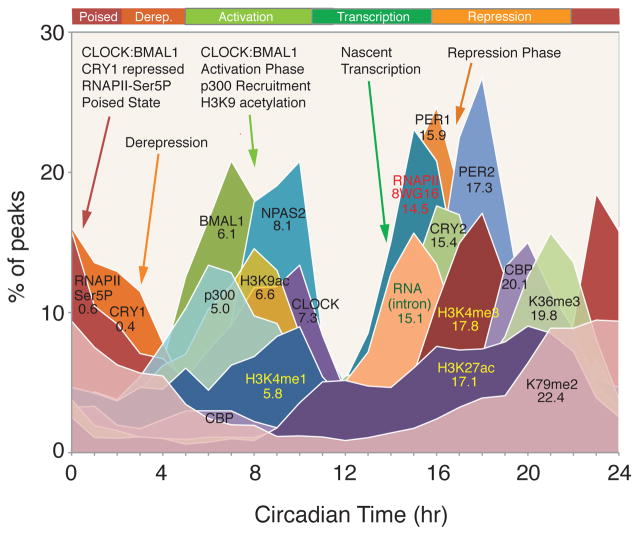
In summary, in the mouse liver there is a highly stereotyped, time-dependent pattern of core transcription factor binding, RNAPII occupancy, transcription and chromatin states (Figure 6). There are six distinctive phases of the circadian cycle: 1) a poised state at the beginning of the cycle (at CT0) in which CLOCK:BMAL1 and CRY1 bind to E-box sites in a transcriptionally silent state; 2) a derepression phase at CT4 as the levels of CRY1 decline; 3) a major activation phase at CT8 during which CLOCK:BMAL1 occupancy peak along with p300 and CBP recruitment and are accompanied by histone H3K4me1 and H3K9ac activation marks; 4) a temporally coordinated transcriptional activation phase at CT12–15 in which RNAPII recruitment and pre-mRNA transcript expression peak, followed by histone H3K4me3 and H3K27ac marks; 5) a classic repression phase at CT15–18 in which PER1, PER2 and CRY2 occupancy peaks and transcription declines; and 6) a transition between the end of repression and the beginning of the next cycle. Interestingly, the co-activator CBP interacts with the repressor PER2 at the end of the repression phase and CBP occupancy has a second peak at this time71, perhaps maintaining an open chromatin state during the transition from repression to activation. CRY1 occupancy also bridges this transition period and interacts with CLOCK:BMAL1 to begin the next cycle.
Figure 6.
Circadian transcriptional landscape in the mouse liver. Histograms show the phase distributions of each factor as a function of time of day. Reproduced with permission from REF.71
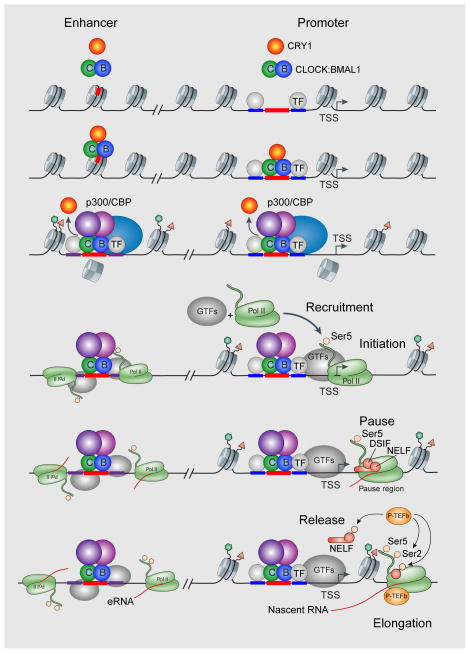
Unlike embryonic stem cells, where 30% of genes are paused and carry bivalent histone marks139, 140, genes that are unexpressed in mouse liver are not enriched for paused RNAPII (Figure 5b)71. Interestingly, the histone H3K4me3 promoter mark lags RNAPII occupancy by about 1 hour in cycling genes, and the H3K36me3 elongation mark lags about 3 hours72, consistent with the idea that these marks arise as a consequence of RNAPII transcription. Thus, circadian transcriptional regulators are clearly involved in RNAPII recruitment and initiation, as are the histone modifications associated with these events, but additional steps such as promoter proximal pausing, pause release and productive elongation probably have additional regulatory roles (Figure 7). At enhancer sites, CLOCK:BMAL1 is a pioneer transcription factor and can bind to nucleosomes to promote rhythmic chromatin remodelling and incorporation of the histone variant H2A.Z141 (Figure 7). Cycling enhancers also display circadian rhythms in eRNA expression in conjunction with tissue-specific transcription factors107, 119. In the case of cycling genes, RNAPII recruitment, initiation and productive elongation are all rhythmic and occur in phase, leading to associated rhythms in histone modifications72. In the case of expressed but non-cycling genes, RNAPII recruitment and initiation are also rhythmic, leading to rhythmic histone modifications at the promoter. However, subsequent steps in pausing, pause release and productive elongation may no longer be under circadian regulation71. Because RNAPII pausing at promoter-proximal regions and pause release are major regulatory steps in transcription, it will be important in the future to determine whether pausing factors such as negative elongation factor (NELF) and DRB-sensitivity-inducing factor (DSIF), as well as the positive transcription elongation factor-b (P-TEFb) complex, are under circadian control129, 142 (Figure 7).
Figure 7.
Circadian transcriptional regulation at enhancer and promoter sites during the CLOCK:BMAL1 activation phase in the daytime. CLOCK:BMAL1 act as pioneer transcription factors at enhancer sites. Circadian recruitment and initiation (as seen by RNAPII-Ser5 marks) of RNA polymerase II occurs at both distal enhancers and proximal promoter sites. However, the circadian regulation of pausing factors (NELF, DSIF) and pause release factors (P-TEFb) remain to be determined. Modified with permission from REF.129.
Conclusions
Circadian modulation of transcription occurs on a genome-wide scale far greater than that seen previously by gene expression profiling. The circadian clock in the liver regulates RNAPII occupancy on a genome-wide basis and drives genome-wide circadian modulation of chromatin states. These circadian rhythms in chromatin state poise the genome for transcription on a daily basis to coincide with the daily metabolic demands of the organism and to optimize the energy utilization of gene expression143. In addition to the circadian modulation of chromatin states, the three-dimensional architecture of the nucleus and the interaction of active and repressive chromosomal domains undergo circadian oscillations (Box 3) 144–148. Thus, the four-dimensional regulation of the genome and its nuclear architecture are ripe for study. In addition to the pervasive circadian regulation of metabolism11, 149, strong links of the circadian gene network to cell growth and cancer150–152, immune function29, 30 and signalling153 are apparent and will be important therapeutic targets in the future.
Box 3. Four-dimensional regulation of the genome.
It is well established that chromatin interactions occur in three dimensions (3D)190–192. However, temporal changes in 3D genomic interactions or in both space and time (the fourth dimension) are an emerging area (“the 4D nucleome”193). Circadian cycles in chromosome organization have been observed using circular chromosome conformation capture (4C) methods at the Dbp locus, a well-known CLOCK:BMAL1 target gene144. Although chromosome interactions surrounding the Dbp locus were not reported to change during the circadian cycle, the frequency of long-range inter-chromosomal interactions did vary. Inter-chromosomal interactions were highest at the peak of Dbp gene expression and were lowest at the opposite phase, when Dbp gene expression was low. The Dbp circadian interactome was dependent on Bmal1 since Bmal1 −/− knockout cells were similar to that seen at the trough of Dbp rhythms144. 4C analysis on a BMAL1-bound enhancer upstream of the circadian gene, Nr1d1 (which encodes REV-ERBα) also found that interactions were largely stable during the circadian cycle148. However, global analysis of cohesin and CTCF revealed that cohesin–CTCF co-binding sites insulated cycling genes with different phases of expression and that cohesin–non-CTCF sites were associated with high amplitude circadian cycling148. In addition, genome-wide chromosome conformation capture (Hi-C) combined with RNA-seq expression has been used in human fibroblasts to search for circadian interactions, and a Laplacian analysis framework has been implemented to quantify dynamical structure–function relationships in the genome145. Using multicolor 3D-FISH, CLOCK and PER2, which have rhythmic and antiphasic structure–function dynamics, were shown to also have a correlated rhythm in spatial distance between them145. Finally, using 4C to probe the H19 imprinting control region in human embryonic stem cells, a large inter-chromosomal interactome was identified146. The interactome was organized by PARP1 and CTCF, and interestingly active loci enriched for circadian genes were found to be recruited to repressed lamina-associated domains (LADs) in the nuclear periphery. In serum-synchronized HCT116 cells, there was a circadian rhythm of recruitment of circadian loci to the nuclear envelope that was dependent on PARP1 and CTCF. This was associated with the acquisition of H3K9me2 repressive marks and attenuation of transcription. Thus, circadian regulation of transcription also involves the recruitment and repression of circadian loci to LADs in the nuclear periphery146.
Key points.
The mammalian circadian clock mechanism is cell autonomous and composed of a transcription-translation negative feedback loop. These clocks are distributed throughout the body and regulate tissue-specific rhythmic functions.
The core circadian transcriptional regulators drive gene expression rhythms in thousands of genes. CLOCK:BMAL1 target genes in the mouse liver regulate genes in all fundamental metabolic pathways suggesting that the clock system in intimately imbedded in cellular metabolism.
Circadian activators and repressors recruit a wide array of chromatin modifiers leading to dynamic changes in the poise of the genome with time of day.
RNAPII is recruited and initiated genome-wide in a circadian manner in the mouse liver leading to genome-wide circadian changes in histone modifications.
Circadian CLOCK:BMAL1 gene targets are directly linked to metabolism, immune function, cell proliferation, cancer and signaling.
Acknowledgments
The author thanks Linda Koch and three anonymous reviewers for their constructive comments. Apologies to those whose work was not cited owing to content and length constraints. The author thanks Nobuya Koike and Tae-Kyung Kim for critical contributions to the research presented here. This work was supported by the Howard Hughes Medical Institute and NIH grants R01AG045795 (J.S.T.) and R21MH107672 (Genevieve Konopka and J.S.T.). J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Glossary
- convergent evolution
process in which unrelated organisms independently evolve similar traits as a result of having to adapt to similar environments or selective pressures
- network motif
As defined by Uri Alon, a small set of recurring regulatory patterns in a network
- oscillation
a repetitive variation of a variable in time with a stable frequency or period
- E-box
for CLOCK:BMAL1, the consensus cis-regulatory element is CACGTG
- REV-ERB/retinoic acid-related orphan receptor (ROR) binding elements (ROREs)
the cis-regulatory element for the nuclear receptors REV-ERB/ROR
- D-box
the cis-regulatory element for the PAR-zip transcription factors, DBP, HLF and TEF
- phases of expression
the time of the day when peak gene expression occurs
- period
the length of the circadian rhythm measured from specific phase points in each cycle such as the peak-to-peak interval
- phosphoswitch
in the case of the PER2 protein, a phosphoswitch model is proposed where two competing phosphorylation sites (one for the FASPD site (Box 2) and one for the β-TrCP binding site) determine whether PER2 has a fast or slow degradation rate.
- ‘kamikaze’ model of transcriptional activation
Ubiquitin-dependent proteolysis of transcriptional activators suggesting a role for activator degradation in RNAPII elongation and requirement for reloading of newly synthesized activators
- cistrome
the in vivo genome-wide location of transcription factor binding-sites
- Global Run-On sequencing (GRO-seq)
a nuclear run-on assay method that captures nascent transcripts from initiated RNAPII followed by next-gen sequencing
- Circadian time (CT)
A standard of time based on the free-running period of a rhythm (oscillation). By convention, CT0 is the beginning of the subjective day, and CT12 is the beginning of the subjective night.
- nascent-seq
next-gen sequencing of nascent RNA transcripts based on chromatin-associated RNA transcripts, nuclear run-on global transcripts such as GRO-seq or PRO-seq, or RNA sequencing of transcripts immunoprecipitated with RNA polymerase II
- Zeitgeber time (ZT)
A standard of time based on the period of an environmental synchronizer or zeitgeber, such as the 24-hour diurnal cycle of light and darkness. Under standard light-dark cycles, the time of ‘lights on’ usually defines zeitgeber time zero (ZT 0) for diurnal organisms and the time of lights off defines zeitgeber time twelve (ZT 12) for nocturnal animals.
- Laplacian analysis
a framework used in many disciplines to quantify topologies in which autonomous entities reach a consensus without a central direction. In the 4D nucleome, it is used to describe the underlying topology of Hi-C interactions in the genome.
- lamina-associated domains (LADs)
regions of condensed chromatin that are bound by the nuclear lamina. LADs are enriched for repressive histone marks such as H3K9me2.
Biography
Joseph S. Takahashi received his Ph.D. in Neuroscience from the University of Oregon, Eugene, in the laboratory of Michael Menaker. He is currently the Loyd B. Sands Distinguished Chair in Neuroscience, an Investigator in the Howard Hughes Medical Institute, and Chair of the Department of Neuroscience at the University of Texas Southwestern Medical Center in Dallas. His laboratory is known for the use of forward genetics and positional cloning in the mouse as a tool for discovery of genes underlying behavior and for the discovery of the mouse and human clock genes that led to a description of a conserved circadian clock mechanism in animals. He is a member of the US National Academy of Sciences, the American Academy of Arts and Sciences and the National Academy of Medicine.
Footnotes
Competing interests
The author declares competing interests. See the article online for details.
FURTHER INFORMATION
Link 1: http://www.genecards.org/Search/Keyword?queryString=circadian%20clock
Link 2: CircaDB, doi: 10.1093/nar/gks1161, http://circadb.hogeneschlab.org
Link 3: BioGPS Circadian Genomics Screen, doi: 10.1016/j.cell.2009.08.031, http://biogps.org/plugin/406/circadian-genomics-screen/
Competing interests
The author is a co-founder and scientific advisory board member of Reset Therapeutics, Inc.
Subject categories
Biological sciences / Cell biology / Circadian rhythms [URI /631/80/105]
Biological sciences / Genetics / Gene expression [URI /631/208/199]
References
- 1.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 2.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 3.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 4.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 5.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7:e62. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doherty CJ, Kay SA. Circadian control of global gene expression patterns. Annu Rev Genet. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenham K, McClung CR. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet. 2015;16:598–610. doi: 10.1038/nrg3976. [DOI] [PubMed] [Google Scholar]
- 9.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 10.Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- 11.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi JS. Finding new clock components: past and future. J Biol Rhythms. 2004;19:339–347. doi: 10.1177/0748730404269151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowrey P, Takahashi J. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Gen. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 16.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 18.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74:246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Gerber A, et al. Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell. 2013;152:492–503. doi: 10.1016/j.cell.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 21.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014;45:16219–16224. doi: 10.1073/pnas.1408886111. This paper shows that 43% of protein coding genes in the genome show circadian oscillation in at least one tissue from profiling circadian gene expression in 12 mouse tissues. They also estimate that the majority (56%) of best-selling drugs directly target the products of rhythmic genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim C, Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci. 2013;16:1544–1550. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- 24.Kojima S, Green CB. Circadian genomics reveal a role for post-transcriptional regulation in mammals. Biochemistry. 2015;54:124–133. doi: 10.1021/bi500707c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer A, Merrow M, editors. Circadian Clocks. Springer; 2013. [Google Scholar]
- 28.Eckel-Mahan K, Sassone-Corsi P. Metabolism and the Circadian Clock Converge. Physiological Reviews. 2013;93:107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. 2013;13:190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LAJ. Circadian Clock Proteins and Immunity. Immunity. 2014;40:178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Johnson CH, Egli M. Metabolic compensation and circadian resilience in prokaryotic cyanobacteria. Annu Rev Biochem. 2014;83:221–247. doi: 10.1146/annurev-biochem-060713-035632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeGates TA, Fernandez DC, Hattar S. Light as a central modulator of circadian rhythms, sleep and affect. Nat Rev Neurosci. 2014;15:443–454. doi: 10.1038/nrn3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy AB, Rey G. Metabolic and Nontranscriptional Circadian Clocks: Eukaryotes. Annu Rev Biochem. 2014;83:165–189. doi: 10.1146/annurev-biochem-060713-035623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 35.Cha J, Zhou M, Liu Y. Mechanism of the Neurospora circadian clock, a FREQUENCY-centric view. Biochemistry. 2015;54:150–156. doi: 10.1021/bi5005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurley J, Loros JJ, Dunlap JC. Dissecting the mechanisms of the clock in Neurospora. Meth Enzymol. 2015;551:29–52. doi: 10.1016/bs.mie.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shultzaberger RK, Boyd JS, Diamond S, Greenspan RJ, Golden SS. Giving Time Purpose: The Synechococcus elongatus Clock in a Broader Network Context. Annu Rev Genet. 2015;49:485–505. doi: 10.1146/annurev-genet-111212-133227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King DP, et al. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 40.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 41.Shearman LP, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 42.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 43.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 44.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preussner M, Heyd F. Post-transcriptional control of the mammalian circadian clock: implications for health and disease. Pflugers Arch. 2016;468:983–991. doi: 10.1007/s00424-016-1820-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toh K, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 47.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 48.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 49.Sato T, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, et al. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science. 2015;348:1488–1492. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novak B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol. 2008;9:981–991. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gachon F, et al. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nature Genetics. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 55.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 56.Curtis AM, et al. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- 57.Lee J, et al. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol. 2008;28:6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hosoda H, et al. CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol Brain. 2009;2:34. doi: 10.1186/1756-6606-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 60.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 61.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 63.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aguilar-Arnal L, Katada S, Orozco-Solis R, Sassone-Corsi P. NAD(+)-SIRT1 control of H3K4 trimethylation through circadian deacetylation of MLL1. Nat Struct Mol Biol. 2015 doi: 10.1038/nsmb.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valekunja UK, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci USA. 2013;110:1554–1559. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ditacchio L, et al. Histone Lysine Demethylase JARID1a Activates CLOCK-BMAL1 and Influences the Circadian Clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nam HJ, et al. Phosphorylation of LSD1 by PKCalpha is crucial for circadian rhythmicity and phase resetting. Mol Cell. 2014;53:791–805. doi: 10.1016/j.molcel.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 70.Lande-Diner L, Boyault C, Kim JY, Weitz CJ. A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc Natl Acad Sci USA. 2013;110:16021–16026. doi: 10.1073/pnas.1305980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. One of five papers (REFs 71, 72, 97, 98, 99) that define the circadian transcriptome, cistrome and epigenome in the mouse liver. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Martelot G, et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10:e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown SA, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 74.Padmanabhan K, Robles M, Westerling T, Weitz C. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337:599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 75.Kim JY, Kwak PB, Weitz CJ. Specificity in Circadian Clock Feedback from Targeted Reconstitution of the NuRD Corepressor. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.10.017. One of five papers (REFs 74,75, 78, 79, 80) on the PER repressor complex. [DOI] [PubMed] [Google Scholar]
- 76.Etchegaray JP, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 77.Nguyen KD, et al. Circadian Gene Bmal1 Regulates Diurnal Oscillations of Ly6Chi Inflammatory Monocytes. Science. 2013;341:1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tamayo AG, Duong HA, Robles MS, Mann M, Weitz CJ. Histone monoubiquitination by Clock–Bmal1 complex marks Per1 and Per2 genes for circadian feedback. Nat Struct Mol Biol. 2015;22:759–766. doi: 10.1038/nsmb.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duong H, Robles M, Knutti D, Weitz C. A molecular mechanism for circadian clock negative feedback. Science. 2011;332:1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duong HA, Weitz CJ. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol. 2014;21:126–132. doi: 10.1038/nsmb.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shirogane T, Jin J, Ang X, Harper J. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 82.Reischl S, et al. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 83.Meng QJ, et al. Setting clock speed in mammals: the CK1ε tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 85.Siepka S, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Godinho S, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 87.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lowrey PL, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Etchegaray JP, et al. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 2009;29:3853–3866. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee HM, et al. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci USA. 2011;108:16451–16456. doi: 10.1073/pnas.1107178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem. 2011;286:7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.D’Alessandro M, et al. A tunable artificial circadian clock in clock-defective mice. Nat Comm. 2015;6:8587. doi: 10.1038/ncomms9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou M, Kim JK, Eng GWL, Forger DB, Virshup DM. A Period2 Phosphoswitch Regulates and Temperature Compensates Circadian Period. Mol Cell. 2015;60:1–13. doi: 10.1016/j.molcel.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 94.Hirano A, et al. FBXL21 Regulates Oscillation of the Circadian Clock through Ubiquitination and Stabilization of Cryptochromes. Cell. 2013;152:1106–1118. doi: 10.1016/j.cell.2013.01.054. These two papers (REFs 94 and 95) identify FBXL21 as a component of a second E3 ubiquitin ligase complex for the CRY circadian repressor proteins. [DOI] [PubMed] [Google Scholar]
- 95.Yoo SH, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maywood ES, et al. Tuning the period of the mammalian circadian clock: additive and independent effects of CK1epsilonTau and Fbxl3Afh mutations on mouse circadian behavior and molecular pacemaking. J Neurosci. 2011;31:1539–1544. doi: 10.1523/JNEUROSCI.4107-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rey G, et al. Genome-Wide and Phase-Specific DNA-Binding Rhythms of BMAL1 Control Circadian Output Functions in Mouse Liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vollmers C, et al. Circadian Oscillations of Protein-Coding and Regulatory RNAs in a Highly Dynamic Mammalian Liver Epigenome. Cell Metab. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoshitane H, et al. CLOCK-controlled polyphonic regulation of circadian rhythms through canonical and noncanonical E-boxes. Mol Cell Biol. 2014;34:1776–1787. doi: 10.1128/MCB.01465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 102.Stratmann M, Suter DM, Molina N, Naef F, Schibler U. Circadian Dbp transcription relies on highly dynamic BMAL1-CLOCK interaction with E boxes and requires the proteasome. Mol Cell. 2012;48:277–287. doi: 10.1016/j.molcel.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 103.Thomas D, Tyers M. Transcriptional regulation: Kamikaze activators. Curr Biol. 2000;10:R341–343. doi: 10.1016/s0960-9822(00)00462-0. [DOI] [PubMed] [Google Scholar]
- 104.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 105.Shimomura K, et al. Usf1, a suppressor of the circadian Clock mutant, reveals the nature of the DNA-binding of the CLOCK:BMAL1 complex in mice. eLife. 2013;2:e00426. doi: 10.7554/eLife.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stashi E, et al. SRC-2 Is an Essential Coactivator for Orchestrating Metabolism and Circadian Rhythm. Cell Reports. 2014;6:633–645. doi: 10.1016/j.celrep.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perelis M, et al. Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science. 2015;350:aac4250–aac4250. doi: 10.1126/science.aac4250. This paper defines the circadian cistrome in pancreatic beta cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet. 2012;13:469–483. doi: 10.1038/nrg3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lamia K, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhu B, et al. Coactivator-Dependent Oscillation of Chromatin Accessibility Dictates Circadian Gene Amplitude via REV-ERB Loading. Mol Cell. 2015;60:769–783. doi: 10.1016/j.molcel.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ameur A, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 114.Gaidatzis D, Burger L, Florescu M, Stadler MB. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat Biotechnol. 2015;33:722–729. doi: 10.1038/nbt.3269. [DOI] [PubMed] [Google Scholar]
- 115.Hughes ME, et al. Harmonics of Circadian Gene Transcription in Mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Du NH, Arpat AB, De Matos M, Gatfield D. MicroRNAs shape circadian hepatic gene expression on a transcriptome-wide scale. eLife. 2014;3:e02510. doi: 10.7554/eLife.02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim TK, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015;162:948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fang B, et al. Circadian Enhancers Coordinate Multiple Phases of Rhythmic Gene Transcription In Vivo. Cell. 2014;159:1140–1152. doi: 10.1016/j.cell.2014.10.022. This paper identifies circadian enhancer RNAs using GRO-seq. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Anafi RC, et al. Machine Learning Helps Identify CHRONO as a Circadian Clock Component. PLoS Biol. 2014;12:e1001840. doi: 10.1371/journal.pbio.1001840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Annayev Y, et al. Gene Model 129 (Gm129) Encodes a Novel Transcriptional Repressor that Modulates Circadian Gene Expression. J Biol Chem. 2014;289:5013–5024. doi: 10.1074/jbc.M113.534651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Goriki A, et al. A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 2014;12:e1001839. doi: 10.1371/journal.pbio.1001839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Preuβner M, et al. Rhythmic U2af26 Alternative Splicing Controls PERIOD1 Stability and the Circadian Clock in Mice. Mol Cell. 2014;54:651–662. doi: 10.1016/j.molcel.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 124.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26:2724–2736. doi: 10.1101/gad.208306.112. This paper shows an important example of circadian post-transcriptional regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 2015;25:1848–1859. doi: 10.1101/gr.195404.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jang C, Lahens NF, Hogenesch JB, Sehgal A. Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res. 2015;25:1836–1847. doi: 10.1101/gr.191296.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 128.Fuda N, Ardehali M, Lis J. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jones JC, et al. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem. 2004;279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chapman RD, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 132.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 135.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 136.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 140.Rahl PB, et al. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Menet JS, Pescatore S, Rosbash M. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev. 2014;28:8–13. doi: 10.1101/gad.228536.113. This paper shows that CLOCK:BMAL1 act as pioneer transcription factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang GZ, et al. Cycling transcriptional networks reduce the synthetic cost of genomes. Cell Reports. 2015;13:1868–1880. doi: 10.1016/j.celrep.2015.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Aguilar-Arnal L, et al. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat Struct Mol Biol. 2013;20:1206–1213. doi: 10.1038/nsmb.2667. This paper is the first to provide evidence for circadian rhythms in chromosomal conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen H, et al. Functional organization of the human 4D Nucleome. Proc Natl Acad Sci USA. 2015;112:8002–8007. doi: 10.1073/pnas.1505822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao H, et al. PARP1- and CTCF-Mediated Interactions between Active and Repressed Chromatin at the Lamina Promote Oscillating Transcription. Mol Cell. 2015;59:984–997. doi: 10.1016/j.molcel.2015.07.019. This paper shows that there is a circadian rhythm of recruitment of circadian loci to the nuclear envelope that is dependent on PARP1 and CTCF. [DOI] [PubMed] [Google Scholar]
- 147.Aguilar-Arnal L, Sassone-Corsi P. Chromatin landscape and circadian dynamics: Spatial and temporal organization of clock transcription. Proc Natl Acad Sci USA. 2015;112:6863–6870. doi: 10.1073/pnas.1411264111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xu Y, et al. Long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression. PLoS Genet. 2016;12:e1005992. doi: 10.1371/journal.pgen.1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sancar A, et al. Circadian Clock, Cancer, and Chemotherapy. Biochemistry. 2014;54:110–123. doi: 10.1021/bi5007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Masri S, Kinouchi K, Sassone-Corsi P. Circadian clocks, epigenetics, and cancer. Curr Opin Oncol. 2015;27:50–56. doi: 10.1097/CCO.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Papagiannakopoulos T, et al. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016;24:324–331. doi: 10.1016/j.cmet.2016.07.001. Demonstration that circadian disruption promotes tumoregenesis in a well-validated genetically engineered mouse model of lung cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schibler U, et al. Clock-Talk: Interactions between Central and Peripheral Circadian Oscillators in Mammals. Cold Spring Harb Symp Quant Biol. 2015;80:223–232. doi: 10.1101/sqb.2015.80.027490. [DOI] [PubMed] [Google Scholar]
- 154.Crane BR, Young MW. Interactive features of proteins composing eukaryotic circadian clocks. Annu Rev Biochem. 2014;83:191–219. doi: 10.1146/annurev-biochem-060713-035644. [DOI] [PubMed] [Google Scholar]
- 155.Gustafson CL, Partch CL. Emerging models for the molecular basis of mammalian circadian timing. Biochemistry. 2015;54:134–149. doi: 10.1021/bi500731f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang N, et al. Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex. Science. 2012;337:189–194. doi: 10.1126/science.1222804. First crystal structure of a heterodmeric bHLH-PAS transcription factor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hennig S, et al. Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol. 2009;7:e1000094. doi: 10.1371/journal.pbio.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kucera N, et al. Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function. Proc Natl Acad Sci USA. 2012;109:3311–3316. doi: 10.1073/pnas.1113280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Czarna A, et al. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell. 2013;153:1394–1405. doi: 10.1016/j.cell.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 160.Xing W, et al. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496:64–68. doi: 10.1038/nature11964. Crystal structure for CRY2 in complex with FBXL3 revealing a novel mode of substrate recognition for an SCF E3 ubiquitin ligase complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schmalen I, et al. Interaction of Circadian Clock Proteins CRY1 and PER2 Is Modulated by Zinc Binding and Disulfide Bond Formation. Cell. 2014;157:1203–1215. doi: 10.1016/j.cell.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 162.Nangle SN, et al. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. eLife. 2014;3:e03674. doi: 10.7554/eLife.03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen R, et al. Rhythmic PER Abundance Defines a Critical Nodal Point for Negative Feedback within the Circadian Clock Mechanism. Mol Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015;524:303–308. doi: 10.1038/nature14883. [DOI] [PubMed] [Google Scholar]
- 165.Wu D, Rastinejad F. Structural characterization of mammalian bHLH-PAS transcription factors. Curr Opin Struct Biol. 2016;43:1–9. doi: 10.1016/j.sbi.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang Z, Wu Y, Li L, Su XD. Intermolecular recognition revealed by the complex structure of human CLOCK-BMAL1 basic helix-loop-helix domains with E-box DNA. Cell Res. 2012;23:213–224. doi: 10.1038/cr.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nangle S, Xing W, Zheng N. Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res. 2013;23:1417–1419. doi: 10.1038/cr.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hirota T, et al. Identification of small molecule activators of cryptochrome. Science. 2012;337:1094–1097. doi: 10.1126/science.1223710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ye R, Selby C, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem. 2011;286:25891–25902. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chiou YY, et al. Mammalian Period represses and de-represses transcription by displacing CLOCK-BMAL1 from promoters in a Cryptochrome-dependent manner. Proc Natl Acad Sci U S A. 2016;113:E6072–E6079. doi: 10.1073/pnas.1612917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ye R, et al. Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev. 2014;28:1989–1998. doi: 10.1101/gad.249417.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jones CR, Huang AL, Ptacek LJ, Fu YH. Genetic basis of human circadian rhythm disorders. Exp Neurol. 2013;243:28–33. doi: 10.1016/j.expneurol.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bouatia-Naji N, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 175.Lyssenko V, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pevet P, Challet E. Melatonin: both master clock output and internal time-giver in the circadian clocks network. J Physiol Paris. 2011;105:170–182. doi: 10.1016/j.jphysparis.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 179.Bonnefond A, et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet. 2012;44:297–301. doi: 10.1038/ng.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tuomi T, et al. Increased Melatonin Signaling Is a Risk Factor for Type 2 Diabetes. Cell Metab. 2016;23:1067–1077. doi: 10.1016/j.cmet.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 181.Bonnefond A, Karamitri A, Jockers R, Froguel P. The Difficult Journey from Genome-wide Association Studies to Pathophysiology: The Melatonin Receptor 1B (MT2) Paradigm. Cell Metab. 2016;24:345–347. doi: 10.1016/j.cmet.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 182.Hu Y, et al. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat Commun. 2016;7:10448. doi: 10.1038/ncomms10448. Three papers (REFS 182, 183, 184) report GWAS loci including known circadian genes for morningness or early chronotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jones SE, et al. Genome-Wide Association Analyses in 128,266 Individuals Identifies New Morningness and Sleep Duration Loci. PLoS Genet. 2016;12:e1006125. doi: 10.1371/journal.pgen.1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lane JM, et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat Commun. 2016;7:10889. doi: 10.1038/ncomms10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Doi M, et al. Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat Commun. 2011;2:327. doi: 10.1038/ncomms1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chabas D, Taheri S, Renier C, Mignot E. The genetics of narcolepsy. Annu Rev Genomics Hum Genet. 2003;4:459–483. doi: 10.1146/annurev.genom.4.070802.110432. [DOI] [PubMed] [Google Scholar]
- 188.Gottlieb DJ, et al. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol Psychiatry. 2015;20:1232–1239. doi: 10.1038/mp.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Spada J, et al. Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE Adult Study. J Sleep Res. 2016 doi: 10.1111/jsr.12421. [DOI] [PubMed] [Google Scholar]
- 190.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- 193.Cremer T, et al. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS letters. 2015;589:2931–2943. doi: 10.1016/j.febslet.2015.05.037. [DOI] [PubMed] [Google Scholar]