Introduction
GBF1 is ubiquitously expressed in eukaryotic cells and is essential for cellular and organismal life. Depletion of GBF1 from cultured cells induces apoptosis, while mouse GBF1 knockout and D. melanogaster knockout of GARZ (the fly GBF1 ortholog) cause embryonic lethality.1,2 Silencing Garz in only the salivary glands leads to severely stunted and disorganized glands, with disorganized lumens and dramatic disorganization of the actin cytoskeleton in the epithelial salivary cells.3
GBF1 belongs to a sub-family of 3 “large GEFs” in a 15 member family of ARF activators.4,5 Only the large GEFs that include GBF1, BIG1 and BIG2 are present in all eukaryotes, consistent with their fundamental roles in organellogenesis and membrane traffic. All large GEFs are inhibited by the fungal metabolite Brefeldin A (BFA), a feature that distinguishes them from the other ARF GEFs.
GBF1 regulates membrane traffic at the ER-Golgi interface and is the only GEF capable of sustaining ARF activation required for the recruitment of the COPI coat.6-11 Inactivation of GBF1 through mutation of the catalytic Sec 7-domain or BFA treatment leads to the dissociation of COPI from membranes, the collapse of the Golgi into the ER, and inhibition of secretory traffic. We and others have shown that in fibroblastic cells such as HeLa, GBF1 is mostly (∼90%) cytosolic, with the remaining ∼10% localized to the Golgi complex.9,12 The 2 pools exist in equilibrium and GBF1 cycles between cytosol and Golgi membranes with rapid turnover.12,13 Immunofluorescence and EM immunogold analyses of cells in culture and tissues indicate that GBF1 is concentrated at the Golgi complex, with a preferred localization to the cis- face of the Golgi stack.14
In addition to the well-documented localization and function of GBF1 at the Golgi, GBF1 was also detected within ∼100 nm proximity to the plasma membrane in D. melanogaster S2R+ cells, and was essential for constitutive fluid-phase endocytosis from cell surface in a process that is dependent on active actin remodeling.15 Furthermore, GBF1 has been detected at the leading edge in HL60 neutrophils stimulated with N-formyl-methionyl-leucyl-phenylalanine (fMLP).16 Specifically, fMLP binding to G-Protein Coupled Receptors (GPCRs) leads to the recruitment and stimulation of Phosphatidyl Inositol 3-Kinase γ (PI3Kγ). The PI3Kγ–mediated production of Phosphatidyl Inositol 3-Phosphates (PIP3s) at the leading edge then facilitates GBF1 recruitment. PI3Kγ activity is essential for GBF1 recruitment as inactivation of PI3Kγ either with the drug AS-604850 or by siRNA-mediated knockdown blocked fMLP-induced GBF1 localization to the leading edge. Importantly, the PM recruitment of GBF1 was observed only in HL60 cells stimulated with fMLP, while GBF1 was found exclusively at the Golgi in non-stimulated cells.
One of the key characteristics of stimulated HL60 cells is the establishment of directional polarity, suggesting that GBF1 recruitment to the PM may occur in other cells that exhibit directional motility or extrude directional processes. This model predicts that in cells that exhibit strong polarized architecture, GBF1 might localize to PM domains independent of acute chemotactic stimulation. Thus, we examined GBF1 localization in cell lines derived from human glioblastomas (GBM), focusing on the possible recruitment of GBF1 to PM regions specialized for adhesion and/or migration.
Results
Endogenous GBF1 localizes to the Golgi and cell protrusions in GBM cells
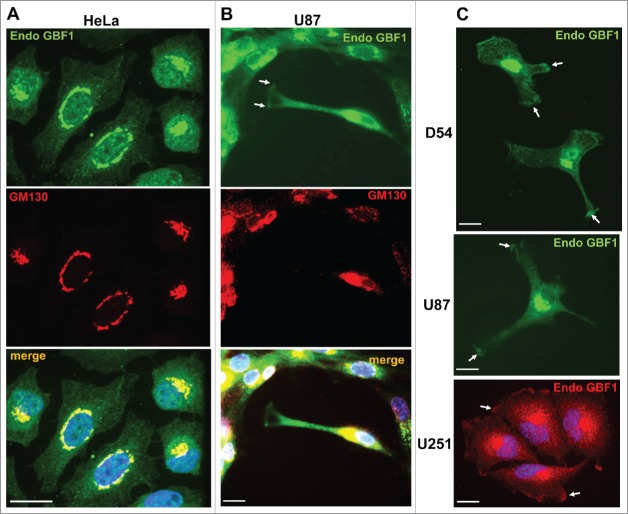
We used immunofluorescence to compare the localization of endogenous GBF1 in HeLa cells and in 3 cell lines derived from human glioblastomas (WHO grade IV astrocytoma), specifically D54, U87 and U251. Prior studies showed GBF1 localization at the Golgi in all cells examined to date.6-8,17 Consistent with these findings, we detected GBF1 in a peri-nuclear region where it co-localized with the cis-Golgi marker GM130 in HeLa (Fig. 1A) and U87 (Fig. 1B) cells. In U87 cells, GBF1 staining was also evident at the plasma membrane (PM), specifically at the tips of extensions (Fig. 1B, arrows). In contrast, GM130 was not detected at the PM, suggesting that GBF1 location is specific for this cellular component and does not represent a general relocation of Golgi proteins (these images were purposefully overexposed to show the lack of GM130 on the cell surface).
Figure 1.
Dual Golgi and PM localization of GBF1 in GBM cells. HeLa (A) and U87 (B) cells were probed by double label IF with polyclonal rabbit anti-GBF1 and monoclonal mouse anti-GM130 antibodies. GBF1 co-localizes with GM130 to the Golgi, but is also present at tips of protrusions in U87 cells (arrows). (C) D54, U87 and U251 cells were processed for IF with monoclonal mouse anti-GBF1 antibodies. GBF1 localizes to the peri-nuclear Golgi and at tips of protrusions and the leading edge (arrows). Representative images from more than 3 independent experiments. Bar is 10 μm.
To determine if GBF1 localization to the PM was specific to the U87 cell line or more broadly applicable to GBM cells, we examined GBF1 distribution in D54 and U251 cells. As shown in Fig. 1C, GBF1 was readily detected in a peri-nuclear region consistent with Golgi localization in all cells. However, GBF1 was also visible at tips of cellular extensions in cells with spindle-like architecture or containing multiple long extensions, and at the leading edge in cells with more compact architecture (arrows). All cells with clearly extended protrusions contained GBF1 at the tips of such extensions, confirming the generality of this observation. We stress that all cells were grown in normal culture medium and were not acutely treated with chemotactic stimuli, indicating that GBF1 recruitment to the PM occurs under standard culture conditions.
The same Golgi and peripheral staining was observed with polyclonal anti-GBF1 antibodies raised in rabbits (Fig. 1B), as with monoclonal affinity-purified anti-GBF1 antibodies raised in mice (Fig. 1C), reducing the possibility that the staining is spurious. To ensure that the secondary antibodies did not contribute to the staining, we omitted the primary anti-GBF1 antibodies and processed the coverslips with only secondary anti-rabbit IgG or anti-mouse IgG antibodies. No signal whatsoever was detected (data not shown), indicating that the staining does not result from non-specific binding of the secondary antibodies. To further ascertain the specificity of the GBF1 signal, we performed an antigen-inhibition study. His-tagged full-length GBF1 was expressed in HEK293 cells and purified by affinity adsorption on Ni-agarose beads. The isolation protocol generated an enriched fraction of GBF1, as judged by Coomassie Blue staining of an SDS-PAGE gel (Fig. 2A, lane 4). As a control, analogous purification protocol was performed from cells not transfected with His-GBF1, and the material eluted from the Ni-agarose beads represents the control eluate (lane 8). Purified GBF1 or the control eluate were preincubated with monoclonal anti-GBF1 antibodies, and such antibodies were subsequently used for IF on U87 cells. While a GBF1 signal was detected at the Golgi and at the tips of cellular extensions when IF was done with antibodies pre-incubated with the control eluate (Fig. 2B), no visible Golgi or peripheral signal was detected when IF was done with antibodies presorbed with purified GBF1 (Fig. 2C). We stress that the same settings of the microscope were used during the acquisition of all images in this figure. This antigen competition results strongly suggest that the IF signal observed with the anti-GBF1 antibodies is specific.
Figure 2.
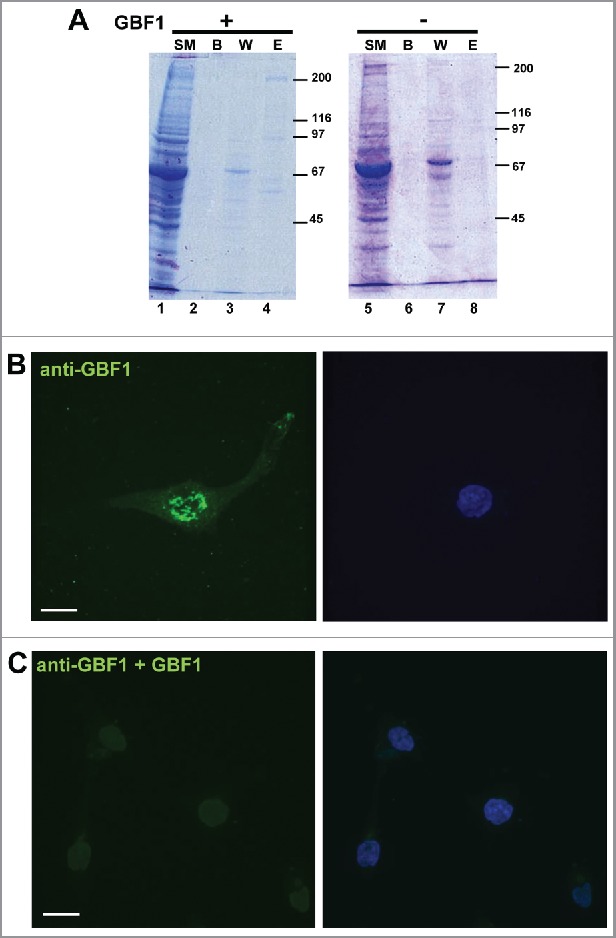
Specificity of GBF1 antibodies person (A) HEK293 cells were transfected with His-tagged GBF1 (lanes 1–4) or mock-transfected (lanes 5–8), lysed 48 h later, and the lysates subject to Ni-agarose purification. Equivalent volume of indicated sample was processed by SDS-PAGE. (SM: starting material; B: beads after elution; W: wash E: elution. An ∼200 kD GBF1 band is visible in lane 4, but not in lane 8. (B) D54 cells were probed with monoclonal mouse anti-GBF1 antibodies that were pre-incubated with eluate from mock-transfected cells. (C) D54 cells were probed with monoclonal mouse anti-GBF1 antibodies that were pre-incubated with purified GBF1. Peri-nuclear Golgi staining and PM staining is evident in cells processed with antibodies pre-incubated with control eluate, but both signals are abrogated when the antibodies are pre-incubated with purified GBF1. Representative images from 2 independent experiments. Bar is 10 μm.
Endogenous GBF1 co-localizes with cellular adhesion proteins
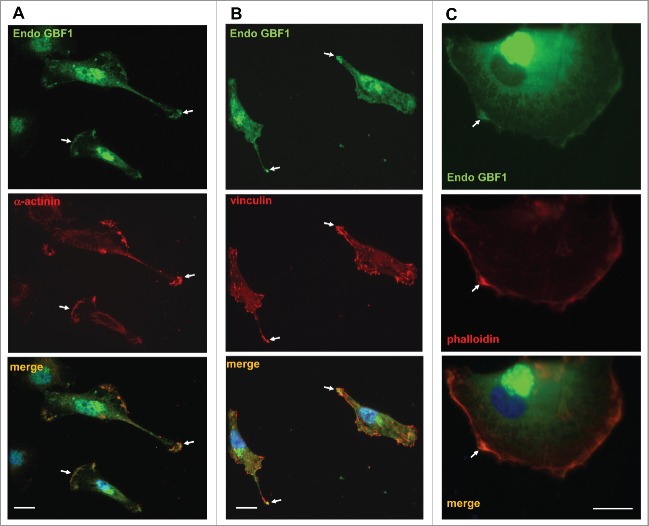
Tips of cellular protrusions and the leading edge are areas of substrate adhesion that allow cells to extend processes by tethering their PM to components of the extracellular matrix. Migration and extension of cellular processes occurs in response to extracellular signals and is mediated by localized actin polymerization. This propels the extension of lamellipodia and necessitates the formation of focal adhesions to anchor the newly-formed protrusions. Focal adhesions are foci of functional linkage between the extracellular matrix and the actin cytoskeleton. The bridging function is provided by integrins, heterodimeric transmembrane proteins that bind matrix components through their extracellular domains and interact with cytosolic focal adhesion proteins through their cytoplasmic tails (reviewed in.18-20) One of the key components of focal adhesions is α-actinin, which binds both integrins and actin filaments to directly link the cytoplasmic tails of integrins to the actin cytoskeleton.21 α-actinin is a homodimer that bundles actin filaments to increase the stiffness of stress fibers originating from focal adhesions. Focal adhesions are also enriched in paxillin that directly binds to integrins and recruits vinculin, which in turn, directly interacts with actin filaments to facilitate the stabilization of focal adhesions.
Thus, we examined whether the surface localized GBF1 co-localized with these adhesion proteins. As shown in Fig. 3, endogenous GBF1 was concentrated at regions enriched in α-actinin (A) and vinculin (B) and was largely absent from other domains of the cell surface (arrows). Focal adhesions and the leading edge of cells are enriched in actin filaments that can be selectively visualized with phalloidin, a bicyclic heptapeptide that binds and stabilizes filamentous actin (F-actin) and prevents the depolymerization of actin fibers.22,23 As shown in Fig. 3C, endogenous GBF1 was concentrated at the leading edge as determined by enriched phalloidin staining (arrows).
Figure 3.
Co-localization of GBF1 with adhesion proteins in GBM cells person D54 cells were probed by double label IF with polyclonal anti-GBF1 and either monoclonal anti-α-actinin, monoclonal anti-vincullin or phalloidin staining. In addition to peri-nuclear Golgi staining, GBF1 localizes to focal adhesions and the leading edge containing adhesion proteins or actin-rich foci (arrows). Representative images from more than 2 independent experiments. Bar is 10 μm.
Exogenously expressed GBF1 is recruited to adhesion sites
As GBF1 localization to adhesion sites has not been described previously to our knowledge, we sought to further ensure that the staining was not an artifact of antibody cross-reactivity. We therefore transfected GBM cells with GFP-tagged GBF1 and assessed the localization of the construct with anti-GFP antibodies. In agreement with Golgi localization of endogenous GBF1 in all cells, GFP-GBF1 co-localized with the Golgi marker GM130 in D54 cells (Fig. 4A). In addition, and confirming results evaluating the localization of endogenous GBF1, the exogenously expressed GFP-GBF1 was detected at the tips of cellular extensions (arrows). Echoing the localization of endogenous GBF1 to protrusions in all examined cells, GFP-GBF1 also was detected at tips of protrusions in all transfected cells. These GBF1-containing foci represent adhesion sites, as shown by the co-localization of GFP-GBF1 with paxillin, a protein found in focal adhesions and other actin-rich structures such as peripheral ruffles, dorsal ruffles and invadopodia/podosomes (Fig. 4B). Analogous results were obtained in U87 cells expressing GFP-GBF1: the construct was detected at the Golgi where it co-localized with GM130 and also at the tips of cellular projections (Fig. 4C). The cell surface centers of GFP-GBF1 represent adhesion sites as shown by their content of paxillin (Fig. 4D). We stress that we only observed staining at the Golgi and at the tips of cellular extensions in cells expressing GFP-GBF1, while untransfected cells had no visible signal. This indicates that the primary and secondary antibodies used in these IFs do not recognize any endogenous antigen that could generate a non-specific signal.
Figure 4.
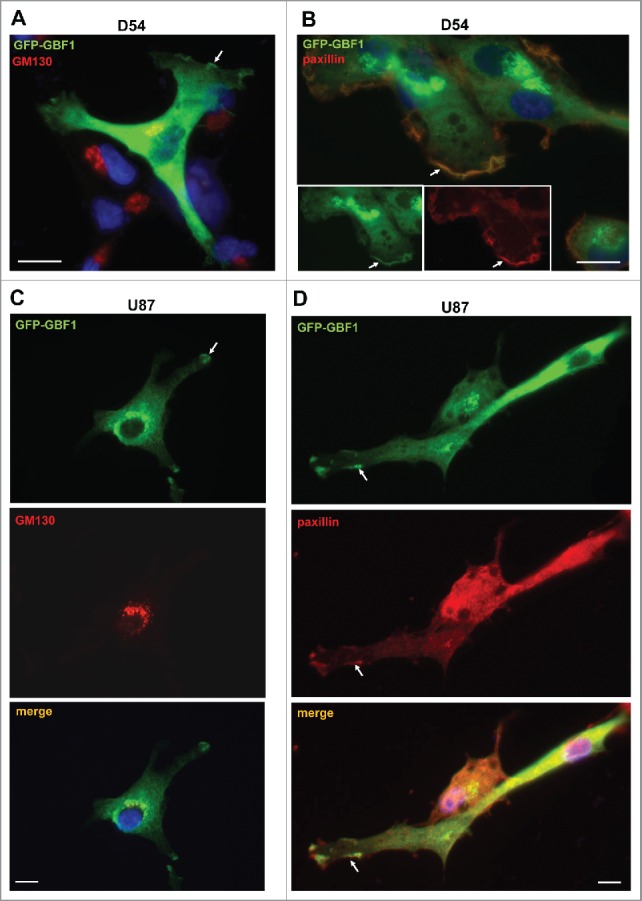
Golgi and PM localization of exogenously expressed GBF1 in GBM cells person D54 (A and B) and U87 (C and D) cells were transfected with GFP-tagged GBF1 and probed by double label IF with polyclonal anti-GFP and either monoclonal anti-GM130 (A and C) or monoclonal anti-paxillin antibodies (B and D). Exogenous GFP-GFP1 co-localizes with GM130 at the Golgi and co-localizes with paxillin at tips of cellular protrusions and the leading edge (arrows). Representative images from more than 3 independent experiments. Bar is 10 μm.
Taken together, our data show that both endogenous and exogenously expressed GBF1 show dual localization within GBM cells, with a pool of GBF1 concentrated at the Golgi complex and another pool localized to cellular adhesion sites at the tips of PM protrusions and the leading edge.
Discussion
GBF1 and its orthologues have been shown to localize to the Golgi complex in all examined cells ranging from mammalian to D. melanogaster and yeast (7, 8, 17, 24). The Golgi localization is consistent with the well characterized role of GBF1 in COPI recruitment to membranes, a process essential for the formation of COPI vesicles and ER-Golgi traffic. The essential Golgi function of GBF1 is underscored by the collapse and disassembly of the Golgi and inhibition in membrane traffic when GBF1 is inactivated or depleted.10,24,25
However, in HL60 neutrophil cell line acutely stimulated with fMLP, GBF1 was recruited to the leading edge oriented toward the chemotactic stimulus and was required for directional motility.16 Importantly, GBF1 was not detected at the PM in unstimulated HL60 cells, suggesting that only acute activation of surface GPCRs results in GBF1 relocation. These data also suggested that GBF1 recruitment to the PM may be specific to only neutrophils migrating toward a chemotactic stimulus.
Herein, we report that GBF1 is found at the leading edge and in adhesion sites in several human GBM cell lines grown under standard conditions and without acute stimulation. This represents a novel finding since the previous report has shown GBF1 localization to the cell surface only in stimulated human neutrophils, but not in unstimulated cells.16 Our results support and extend data in Drosophila, where Garz (the fly GBF1) has been detected close to the cell surface (within ∼100 nm) and facilitated fluid-phase pinocytosis in non-stimulated S2R+ cells.15
The potential functional relevance of GBF1 recruitment to adhesion sites in GBM cells remains to be determined, but it is possible that GBF1 directly or indirectly affects cell motility. This is consistent with our earlier finding that siRNA-mediated depletion of GBF1 from D54 cells inhibited the migratory potential of these cells.10 However, in those experiments, depletion of GBF1 would affect GBF1 functions throughout the cell (at the Golgi and at adhesions) making it impossible to assign the motility defect as inhibition of GBF1 function in the cell periphery. GBF1 is a large multi-domain protein and could impact motility directly by providing a scaffolding function to organize proteins that regulate actin dynamics. Alternatively, GBF1 could function indirectly, by activating ARF, which then could directly impact motility. Activated ARF1 has been shown to recruit the RhoGap domain-containing protein, ARHGAP10, which modulates Cdc42 dynamics at the cell surface.26 The exact role that GBF1 plays at the periphery of GBM cells and how GBF1 activity may contribute to GBM invasiveness remain to be determined.
GBF1 may affect actin dynamics and assembly, as suggested by our previous report that depletion of Garz in salivary glands of D. melanogaster causes marked disruption of the actin network.3 A putative function for GBF1 in actin dynamics is also supported by the described previously links between Gea1p and Gea2p (yeast orthologs of GBF1) and actin architecture in S. cerevisiae.27 First, GEA1 and GEA2 were identified as multicopy suppressors of profilin deletion that could rescue the disassembly of actin cables and the lack of polarized actin cortical patches in S. cerevisiae lacking profilin. Second, cells deleted of GEA2 and expressing inactive mutant alleles of GEA1 were defective in actin cytoskeleton and budding (an actin-dependent process). Third, overexpression of GEA1 or GEA2 in wild-type cells increased the appearance of actin cable-like structures. The mechanisms through which GEA1/2 influence actin dynamics remain to be determined.
Precise regulation of actin and focal adhesion dynamics is required for cell migration during normal physiologic processes such as developmental morphogenesis, but it also contributes to pathologies by increasing the invasive potential of cancer cells. GBM is the most common and aggressive primary brain cancer in adults. GBM remains a lethal disease due in part to the ability of these cells to infiltrate healthy brain tissue making surgical removal of all tumor cells impossible. GBM cells form PM protrusions and invadopodia to facilitate cell-cell interactions and motility. Our finding that GBF1 is concentrated at adhesion sites in GBM cells raises the possibility that it might be involved in processes regulating actin dynamics at those sites and, consequently, may be fundamentally involved in the invasive capacity of GBM cells.
GBF1 joins an ever-increasing number of proteins with multiple cellular localizations and functions. Such “moonlighting” proteins are exemplified by ARL2, which regulates both mitochondrial fusion and microtubule dynamics,28-30 Orc6 which participates in DNA replication and cytokinesis31-34 and the exocyst complex which facilitates polarized deliver of secretory cargoes as well as regulates actin dynamics.35-37 It is likely that such functional duality reflects the cellular need for coordination since the utilization of a single protein in multiple processes provides a mechanism for cross-talk and coordination of distinct cellular responses.
Materials and methods
Antibodies
The following antibodies were commercially obtained: rabbit polyclonal anti-GFP (Abcam, Ab290), mouse monoclonal affinity-purified anti-GBF1 (BD Bioscience, 612116; this antibody detected a single band on Western blots), mouse monoclonal affinity purified anti-GM130 (BD Transduction Laboratories, 610823), mouse monoclonal anti-paxillin (ThermoFisher, AHO0492), mouse monoclonal anti-vinculin (Abcam, Ab18058) and mouse monoclonal anti-α-actinin (Abcam, Ab18061). Polyclonal anti GM130 were raised in a rabbit and have been described previously.38 Secondary anti-rabbit or anti-mouse antibodies conjugated with Alexa 488 or Alexa 594 were obtained from Invitrogen (A11001, A11034, A11037, A11032). Phalloidin conjugated with Alexa 594 was purchased from Invitrogen (A12381). In some experiments, monoclonal anti-GBF1 antibodies (0.05μg) were incubated with 0.3μg of purified GBF1 or with equivalent volume of control eluate (material from a control purification; see below) for 2 hours at 4°C before being used in IF.
Plasmids
N-terminally GFP-tagged GBF1 (GFP-GBF1) was constructed by sub-cloning human GBF1 into the pEGFP vector using XhoI and Xmas restriction enzymes and has been described in.12 C-teminally his-tagged GBF1 was generated by subcloning human GBF1 into the pcDNA4-myc-His B vector using EcoRI and XhoI restriction enzymes.
Cell culture and transfection
D54 cells originated from a surgical resection from a GBM patient. D54 is a commonly studied cell line that has been extensively characterized. U-87 cell line is also derived from a grade 4 patient. Similarly, U-251 is derived from a human malignant glioblastoma multiforme isolated from a patient with grade III-IV malignant tumor by explant technique. D54, U87 and U251 cells were grown in cell culture dishes, some containing glass coverslips (ᴓ12mm), in Dulbecco's Modified Eagle's Medium, supplemented with L-glutamine (10–090-CV), 20% fetal bovine serum (35–010 CV), 100 units/mL penicillin/streptomycin (30–001-CI). All these reagents were purchased from Corning. Cells were grown at 37°C in 5% CO2 until ∼75% confluent and were transfected with Mirus TransIT-X2 Transfection Reagent (Mirus Bio Corporation, MIR6004), according to the manufacturer's instructions. After transfection, cells were incubated for 24 hours and processed for immunofluorescence.
HEK (GripTite™ 293 MSR, R79507) cells were purchased from ThermoFisher scientific, NY, USA. Cells were cultured in vitro in DMEM Eagle medium (Cellgro, Manassas, VA) supplemented with L- glutamine, 10% fetal bovine serum, 100/units/ml penicillin, 100 mg/ml streptomycin, and 1 mM sodium pyruvate (Cellgro, Manassas, VA) at 37°C in humidified atmosphere and transfected with Mirus TransIT-LT1 Transfection Reagent (Mirus Bio Corporation, Madison, WI), according to the manufacturer's instructions.
GBF1 purification
His-GBF1 was purified from HEK293 cells 48 hours after transfection. Cells were lysed in 50 mM HEPES (pH 7.4), 100 mM NaCl, protease inhibitor, scraped and lysed by passage 5 times through 21G needles (BD Bioscience, CA, USA) and twice through 27G needles (BD Bioscience, CA, USA). Cell debris was removed by centrifugation at 14 000 rpm for 15 min at 4°C. Supernatants were pre-cleaned using Pierce Glutatione Agarose (Thermo Scientific, IL, USA); at 4°C for 1 hour and centrifuged 1 000 rpm for 2 mins. Proteins were purified using Ni-NTA Agarose beads (Qiagen, CA, USA) for 3 hours at 4°C. Beads were recovered by centrifugation at 1 000 rpm for 1 min and washed with 20mM HEPES (pH 7.4), 100mM NaCl, 20mM imidazole buffer 5 times 5 min at 4°C, then centrifuged at 1 000 rpm for 2 min. Proteins were eluted from the beads with 25 mM HEPES (pH 7.4), 100 mM NaCl, 250 mM imidazole, 3 times 5 mins at 4°C and centrifuged 2 000rpm for 1 min. The same protocol was used to prepare “control eluate” sample from cells not transfected with GBF1. Purified proteins were stored at −80°C. For antigen competition analysis, 0.05 μg monoclonal anti-GBF1 antibodies were incubated with 0.3 μg of purified GBF1 (> 4-fold molar excess) or with equivalent volume of eluate from mock-transfectedcells. The mixtures were incubated at 4°C for 1 hour and then used for IF as described below.
SDS-PAGE
Proteins were resolved on 8% SDS-PAGE as described previously.39
Immunofluorescence and confocal microscopy
Cells were processed for IF as described in8: cells on coverslips were washed 3 times in PBS, fixed in 3% paraformaldehyde in PBS for 10 minutes and quenched with 10mM ammonium chloride in PBS for another 10 min. Subsequently cells were washed 3 times with PBS. Cells were then permeabilized in 0.1% Triton X-100 in PBS for 7 min. The coverslips were then washed in PBS, blocked in PBS containing 2.5% goat serum and 0.2% Tween 20 for 5 min, and in PBS, 0.4% fish skin gelatin, 0.2% Tween-20 for another 5 min. Cells were incubated with primary antibody diluted in 0.4% fish skin gelatin for 1 hour at room temperature, washed in PBS 0.2% Tween-20, and blocked like described above. Subsequently cells were incubated with secondary antibodies diluted in 2.5% goat serum for 45 minutes at room temperature. For coverslips processed with phalloidin, the phalloidin was diluted in 2.5% goat serum and cells were incubated with this phalloidin for 15 minutes at room temperature. Nuclei were stained using Hoechst; coverslips were washed with PBS-0.2% Tween-20 and mounted on slides in ProLong Gold antifade reagent (Invitrogen, P36930).
Fluorescence was visualized with a Leitz Orthoplan epifluorescence microscope (Wetzlar, Germany). Images were captured with a 12 bit CCD camera from Q imaging using iVision-Mac software. Confocal imaging studies were performed using a Perkin Elmer Ultraview ERS 6FE spinning disk confocal attached to a Nikon TE 2000-U microscope equipped with laser and filter sets for FITC, TRITC and DAPI fluorescence. Images were captured using a Hamamatsu C9100–50 EM-CCD camera (Hamamatsu Photonics K.K) and 60X or 100X Plan APO oil-immersion objectives. The imaging system was controlled by Velocity 6.2 software (Perkin Elmer).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Susan Nozell for the gift of D54, U87 and U251 cells and her invaluable advice on cell culture and transfection of these cells.
Funding
This work was supported by MCB0744471 to ES and 1R21NS096531 to ABH.
References
- [1].Wang S, Meyer H, Ochoa-Espinosa A, Buchwald U, Onel S, Altenhein B, Heinisch JJ, Affolter M, Paululat A. GBF1 (Gartenzwerg)-dependent secretion is required for Drosophila tubulogenesis. J Cell Sci 2012; 125:461-72; PMID:22302994; http://dx.doi.org/ 10.1242/jcs.092551 [DOI] [PubMed] [Google Scholar]
- [2].Armbruster K, Luschnig S. The Drosophila Sec 7 domain guanine nucleotide exchange factor protein Gartenzwerg localizes at the cis-Golgi and is essential for epithelial tube expansion. J Cell Sci 2012; 125:1318-28; PMID:22349697; http://dx.doi.org/ 10.1242/jcs.096263 [DOI] [PubMed] [Google Scholar]
- [3].Szul T, Burgess J, Jeon M, Zinn K, Marques G, Brill JA, Sztul E. The Garz Sec 7 domain guanine nucleotide exchange factor for Arf regulates salivary gland development in Drosophila. Cell Logist 2011; 1:69-76; PMID:21686256; http://dx.doi.org/ 10.4161/cl.1.2.15512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wright J, Kahn RA, Sztul E. Regulating the large Sec 7 ARF guanine nucleotide exchange factors: the when, where and how of activation. Cell Mol Life Sci 2014; 71:3419-38; PMID:24728583; http://dx.doi.org/ 10.1007/s00018-014-1602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bui QT, Golinelli-Cohen MP, Jackson CL. Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Mol Genet Genomics 2009; 282:329-50; PMID:19669794; http://dx.doi.org/ 10.1007/s00438-009-0473-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Claude A, Zhao BP, Kuziemsky CE, Dahan S, Berger SJ, Yan JP, Armold AD, Sullivan EM, Melancon P. GBF1: A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J Cell Biol 1999; 146:71-84; PMID:10402461 [PMC free article] [PubMed] [Google Scholar]
- [7].Kawamoto K, Yoshida Y, Tamaki H, Torii S, Shinotsuka C, Yamashina S, Nakayama K. GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic 2002; 3:483-95; PMID:12047556; http://dx.doi.org/ 10.1034/j.1600-0854.2002.30705.x [DOI] [PubMed] [Google Scholar]
- [8].Garcia-Mata R, Szul T, Alvarez C, Sztul E. ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol Biol Cell 2003; 14:2250-61; PMID:12808027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhao X, Claude A, Chun J, Shields DJ, Presley JF, Melancon P. GBF1, a cis-Golgi and VTCs-localized ARF-GEF, is implicated in ER-to-Golgi protein traffic. J Cell Sci 2006; 119:3743-53; PMID:16926190; http://dx.doi.org/ 10.1242/jcs.03173 [DOI] [PubMed] [Google Scholar]
- [10].Szul T, Grabski R, Lyons S, Morohashi Y, Shestopal S, Lowe M, Sztul E. Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J Cell Sci 2007; 120:3929-40; PMID:17956946; http://dx.doi.org/ 10.1242/jcs.010769 [DOI] [PubMed] [Google Scholar]
- [11].Saenz JB, Sun WJ, Chang JW, Li J, Bursulaya B, Gray NS, Haslam DB. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat Chem Biol 2009; 5:157-65; PMID:19182783; http://dx.doi.org/ 10.1038/nchembio.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Szul T, Garcia-Mata R, Brandon E, Shestopal S, Alvarez C, Sztul E. Dissection of Membrane Dynamics of the ARF-Guanine Nucleotide Exchange Factor GBF1. Traffic 2005; 6:374-85; PMID:15813748; http://dx.doi.org/ 10.1111/j.1600-0854.2005.00282.x [DOI] [PubMed] [Google Scholar]
- [13].Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. Dynamics of GBF1, a Brefeldin A-Sensitive Arf1 Exchange Factor at the Golgi. Mol Biol Cell 2005; 16:1213-22; PMID:15616190; http://dx.doi.org/ 10.1091/mbc.E04-07-0599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lowery J, Szul T, Styers M, Holloway Z, Oorschot V, Klumperman J, Sztul E. The Sec 7 guanine nucleotide exchange factor GBF1 regulates membrane recruitment of BIG1 and BIG2 to the trans-Golgi network (TGN). J Biol Chem 2013; 288(16):11532-45; PMID:23386609; http://dx.doi.org/ 10.1074/jbc.M112.438481 doi:M112.438481 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gupta GD, Swetha MG, Kumari S, Lakshminarayan R, Dey G, Mayor S. Analysis of endocytic pathways in Drosophila cells reveals a conserved role for GBF1 in internalization via GEECs. PLoS One 2009; 4:e6768; PMID:19707569; http://dx.doi.org/ 10.1371/journal.pone.0006768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mazaki Y, Nishimura Y, Sabe H. GBF1 bears a novel phosphatidylinositol-phosphate binding module, BP3K, to link PI3Kgamma activity with Arf1 activation involved in GPCR-mediated neutrophil chemotaxis and superoxide production. Mol Biol Cell 2012; 23:2457-67; PMID:22573891; http://dx.doi.org/ 10.1091/mbc.E12-01-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhao X, Lasell TK, Melancon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic. Mol Biol Cell 2002; 13:119-33; PMID:11809827; http://dx.doi.org/ 10.1091/mbc.01-08-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li Z, Lee H, Zhu C. Molecular mechanisms of mechanotransduction in integrin-mediated cell-matrix adhesion. Exp Cell Res 2016; 349:85-94; PMID:27720950; http://dx.doi.org/ 10.1016/j.yexcr.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Arnaout MA, Goodman SL, Xiong JP. Structure and mechanics of integrin-based cell adhesion. Curr Opin Cell Biol 2007; 19:495-507; PMID:17928215; http://dx.doi.org/ 10.1016/j.ceb.2007.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Critchley DR, Holt MR, Barry ST, Priddle H, Hemmings L, Norman J. Integrin-mediated cell adhesion: the cytoskeletal connection. Biochem Soc Symp 1999; 65:79-99; PMID:10320934 [PubMed] [Google Scholar]
- [21].Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev 2008; 88:489-513; PMID:18391171; http://dx.doi.org/ 10.1152/physrev.00021.2007 [DOI] [PubMed] [Google Scholar]
- [22].Chazotte B. Labeling cytoskeletal F-actin with rhodamine phalloidin or fluorescein phalloidin for imaging. Cold Spring Harb Protoc 2010; 2010:pdb prot4947 [DOI] [PubMed] [Google Scholar]
- [23].VanBuren P, Begin K, Warshaw DM. Fluorescent phalloidin enables visualization of actin without effects on myosin's actin filament sliding velocity and hydrolytic properties in vitro. J Mol Cell Cardiol 1998; 30:2777-83; PMID:9990547; http://dx.doi.org/ 10.1006/jmcc.1998.0856 [DOI] [PubMed] [Google Scholar]
- [24].Peyroche A, Courbeyrette R, Rambourg A, Jackson CL. The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J Cell Sci 2001; 114:2241-53; PMID:11493664 [DOI] [PubMed] [Google Scholar]
- [25].Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. Dynamics of GBF1, a Brefeldin A-sensitive Arf1 Exchange Factor at the Golgi. Mol Biol Cell 2004; 16(3):1213-22; PMID:15616190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kumari S, Mayor S. ARF1 is directly involved in dynamin-independent endocytosis. Nat Cell Biol 2008; 10:30-41; PMID:18084285; http://dx.doi.org/ 10.1038/ncb1666 [DOI] [PubMed] [Google Scholar]
- [27].Zakrzewska E, Perron M, Laroche A, Pallotta D. A role for GEA1 and GEA2 in the organization of the actin cytoskeleton in Saccharomyces cerevisiae. Genetics 2003; 165:985-95; PMID:14668359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell 2006; 17:2476-87; PMID:16525022; http://dx.doi.org/ 10.1091/mbc.E05-10-0929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Francis JW, Turn RE, Newman LE, Schiavon C, Kahn RA. Higher order signaling: ARL2 as regulator of both mitochondrial fusion and microtubule dynamics allows integration of 2 essential cell functions. Small GTPases 2016; 7(4):188-96; http://dx.doi.org/ 10.1080/21541248.2016.1211069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Newman LE, Zhou CJ, Mudigonda S, Mattheyses AL, Paradies E, Marobbio CM, Kahn RA. The ARL2 GTPase is required for mitochondrial morphology, motility, and maintenance of ATP levels. PLoS One 2014; 9:e99270; PMID:24911211; http://dx.doi.org/ 10.1371/journal.pone.0099270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Akhmetova K, Balasov M, Huijbregts RP, Chesnokov I. Functional insight into the role of Orc6 in septin complex filament formation in Drosophila. Mol Biol Cell 2015; 26:15-28; PMID:25355953; http://dx.doi.org/ 10.1091/mbc.E14-02-0734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huijbregts RP, Svitin A, Stinnett MW, Renfrow MB, Chesnokov I. Drosophila Orc6 facilitates GTPase activity and filament formation of the septin complex. Mol Biol Cell 2009; 20:270-81; PMID:18987337; http://dx.doi.org/ 10.1091/mbc.E08-07-0754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol Cell Biol 2007; 27:3143-53; PMID:17283052; http://dx.doi.org/ 10.1128/MCB.02382-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci U S A 2003; 100:9150-55; PMID:12878722; http://dx.doi.org/ 10.1073/pnas.1633580100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Biondini M, Sadou-Dubourgnoux A, Paul-Gilloteaux P, Zago G, Arslanhan MD, Waharte F, Formstecher E, Hertzog M, Yu J, Guerois R, Gautreau A, Scita G, Camonis J, Parrini MC. Direct interaction between exocyst and Wave complexes promotes cell protrusions and motility. J Cell Sci 2016; 129:3756-69; PMID:27591259; http://dx.doi.org/ 10.1242/jcs.187336 [DOI] [PubMed] [Google Scholar]
- [36].Lizunov VA, Lisinski I, Stenkula K, Zimmerberg J, Cushman SW. Insulin regulates fusion of GLUT4 vesicles independent of Exo70-mediated tethering. J Biol Chem 2009; 284:7914-19; PMID:19155211; http://dx.doi.org/ 10.1074/jbc.M806460200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec 4p, targeting secretory vesicles to sites of exocytosis. EMBO J 1999; 18:1071-80; PMID:10022848; http://dx.doi.org/ 10.1093/emboj/18.4.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Alvarez C, Garcia-Mata R, Hauri HP, Sztul E. The p115-interactive proteins GM130 and giantin participate in endoplasmic reticulum-Golgi traffic. J Biol Chem 2001; 276:2693-2700; PMID:11035033; http://dx.doi.org/ 10.1074/jbc.M007957200 [DOI] [PubMed] [Google Scholar]
- [39].Gao YS, Alvarez C, Nelson DS, Sztul E. Molecular cloning, characterization, and dynamics of rat formiminotransferase cyclodeaminase, a Golgi-associated 58-kDa protein. J Biol Chem 1998; 273:33825-34; PMID:9837973; http://dx.doi.org/ 10.1074/jbc.273.50.33825 [DOI] [PubMed] [Google Scholar]