ABSTRACT
Hyperpolarization-activated cyclic-nucleotide gated channel (HCN) proteins are important regulators of both neuronal and cardiac excitability. Among the 4 HCN isoforms, HCN4 is known as a pacemaker channel, because it helps control the periodicity of contractions in vertebrate hearts. Although the physiological role of HCN4 channel has been studied in adult mammalian hearts, an earlier role during embryogenesis has not been clearly established. Here, we probe the embryonic roles of HCN4 channels, providing the first characterization of the expression profile of any of the HCN isoforms during Xenopus laevis development and investigate the consequences of altering HCN4 function on embryonic pattern formation. We demonstrate that both overexpression of HCN4 and injection of dominant-negative HCN4 mRNA during early embryogenesis results in improper expression of key patterning genes and severely malformed hearts. Our results suggest that HCN4 serves to coordinate morphogenetic control factors that provide positional information during heart morphogenesis in Xenopus.
KEYWORDS: bioelectricity, cardiogenesis, HCN4, ion channel, membrane potential, morphogenesis, positional identity
Introduction
Hyperpolarization-activated cyclic nucleotide-gated channels (HCN) are a family of ion channels that have been extensively investigated for their roles in cardiac physiology.1-3 Expressed predominantly in the nervous system and heart, 4 genes encoding HCN isoforms (HCN1–4) have been identified in mammals, which have differential tissue-specific expression and functionality. Functional channels are composed of 4 subunits that are arranged around a centrally located pore. HCN channels are permeable to Na+, K+ and Ca2+, and produce currents (known as If [funny current] in the heart and Ih in the nervous system) that have a variety of physiological functions, such as regulating the spontaneous depolarization of adult heart and nerve cells.4,5 HCN4 and HCN2 are the most abundant isoforms in mammalian adult hearts and are distributed in all regions of the heart that undergo spontaneous firing (known as the cardiac conduction system).6
In addition to their well-characterized roles in adult tissues, several studies have reported that HCN4 and HCN2 are also expressed during early stages of both murine and chick embryogenesis.7-10 For example, in mouse embryos, HCN4 mRNA is first expressed in progenitors of the primary heart field during gastrulation, well before the heart begins to beat.11 At the beginning of cardiac looping, it is asymmetrically expressed on the left side of the sinus venosus, but this asymmetry is reversed to the right side by the end of looping. Although HCN2 becomes the dominant isoform in the hearts of mouse neonates, HCN4 continues to be expressed in the developing heart, with its expression becoming progressively restricted to the cardiac conductive system.11-13 Thus, in addition to its pacemaker function, HCN4 channels likely play an earlier role during the development of the embryonic heart. Because HCN4 knockout in mice leads to lethality before heart maturation, investigating novel embryonic roles of HCN4 has been challenging in animals with in utero development.14,15
To investigate a potential role for HCN4 channels during embryonic heart development, we took advantage of the externally developing vertebrate, Xenopus laevis. The underlying molecular mechanisms that direct heart development in Xenopus are highly conserved in all vertebrate organisms, making it an ideal translational system for mammalian models.16-20 Importantly, unlike other model organisms, Xenopus embryos develop into tadpoles in the absence of a working circulatory system. Thus, it is possible to study early cardiovascular defects in vivo without the complication of secondary effects arising from a lack of circulation.
To create a functional heart during development, precursor cells need to differentiate into cardiac cell types, and tissues must be properly positioned within the body. In Xenopus, cardiac progenitors arise during gastrulation, and subsequently migrate as 2 bilateral patches along the lateral plate mesoderm to the ventral side of the body after the completion of neurulation.21,22 As development proceeds, these 2 heart precursor fields merge at the ventral midline to create the primary heart tube. Before the bilateral precardiac fields merge, the expression of transcription factors such as Nkx2.5 and GATA-4 initiate the program for differentiation of the cardiac mesoderm. In addition to the signaling cascades that direct the differentiation of cardiac cells, patterning genes are simultaneously expressed that restrict, coordinate, and regulate the orientation and position of the heart in the body.23 Specifically, the left-sided expression of genes such as Xnr-1 and Lefty, establish an asymmetric field of gene expression between the left and right sides of an organism. Downstream effector genes like Pitx and BMP-4, reinforce this asymmetry and translate it to asymmetric morphogenesis during heart looping and chamber differentiation. HCN4's asymmetric expression the cardiac crescent in mice before heart tube formation11 suggests that it might also regulate organ position within the body.
Further evidence suggesting a role for HCN4 during development is the fact that bioelectric signaling through ion channels has been shown to play a critical role during tissue formation, patterning, and organogenesis.24-32 These signals establish membrane potentials and voltage gradients in cells through the unequal distribution of ions across the plasma membrane. Mutations in ion channels can lead to serious impairments during embryonic development. For example, mutating a Na+/K+-ATPase ion pump leads to abnormalities in heart tube extension, cardiomyocyte differentiation and cardiac function in zebrafish embryos.33,34 Mutations in numerous ion channels induce a variety of channelopathies which manifest in human patients and model species as birth defects affecting various organs' patterning.35-38 Although most ion channels have been studied in the contexts of neurobiology and physiology, many are present during embryogenesis and could therefore have instructive patterning roles.39,40
Here we investigate the roles of HCN4 during embryogenesis, providing the first characterization of the expression profile of any of the HCN isoforms during Xenopus development and functionally investigating the consequences of altering HCN4 function during heart development. Both overexpressing HCN4 and injection of dominant-negative HCN4 mRNA disrupt the localization of key morphogenetic patterning genes, Xnr-1, Lefty, Pitx2, and BMP-4, resulting in hearts with incorrect positional orientation and morphology. Together, our data reveal a novel role for HCN4 channels in coordinating the distribution of critical morphogenetic control factors during heart development and directing normal heart morphology.
Results
HCN4 is expressed asymmetrically in the developing heart field of Xenopus
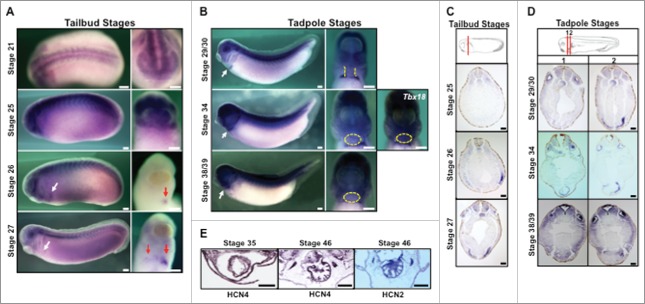
Because mice lacking functional HCN4 channels die during embryonic development between E9.5-E11.5, it has been postulated that this lethality results from atypical cardiac function.14,15 While HCN4 function is essential for normal pacemaker activity, in both mice and chickens HCN4 is expressed very early in development, well before heart beating is initiated.10,12,13 To investigate the possibility that HCN4 channels play an additional and earlier role during cardiogenesis, a spatio-temporal analysis via whole-mount in situ hybridization of HCN4 mRNA expression was performed in Xenopus laevis during embryogenesis. HCN4 transcripts were first robustly detected in the developing craniofacial region, in 2 bilateral stripes along the neural tube, and in the developing somites at the end of neurulation (NF stage 21, Fig. 1A). The craniofacial and somite expression pattern remained consistent throughout later tailbud and tadpole stages (NF stages 25–39, Fig. 1A and B). Expression in the notochord was discernible in tailbud stages (e,g., NF stages 25 and 27) and persisted throughout tadpole stages (Fig. 1A–D). Beginning at NF stage 26, HCN4 transcripts were present in the presumptive heart region, where they were restricted to the left side of the developing cardiac field. Shortly thereafter expression became bilateral in the heart field (NF stage 27, Fig. 1A) but remained higher on the left side throughout the later stages of heart development (Fig. 1B). HCN4 expression was diffusely present in the myocardium at the completion of heart tube formation (NF stage 29/30, Fig. 1B). This myocardial expression remained throughout cardiac looping and chamber septation (NF stage 34 and stage 38/39, Fig. 1B). Unlike what has been observed in mice,11 HCN4 expression never became more highly expressed on the right side of the developing heart. As the sections in Fig. 1C illustrate, when cardiac differentiation is beginning during late tailbud stages, HCN4 expression in the developing cardiac mesoderm was similar to what has been described for other primary heart field markers including: Nkx2.5, MLC2a, Tbx5, and Tbx20.18,41 However, as differentiation and looping progress, HCN4 expression was observed in both the myocardial tissue, as well as the septum transversum, the area where proepicardial precursor cells are located (Fig. 1D). The pattern of HCN4 expression observed at these stages is nearly identical to another marker of the septum transversurm, the epicardial-associated transcription factor Tbx18 (NF stage 34, Fig. 1B).42
Figure 1.
HCN4 expression during embryogenesis (A) In situ hybridization analysis reveals HCN4 is expressed in the developing head and along the dorsal midline at the completion of neurulation embryos (NF stage 21 lateral and frontal views). At early tailbud stages, HCN4 expression is observed primarily in the craniofacial region, in 2 stripes along the neural tube, and in the developing somites. This expression pattern is maintained as development progresses (NF stage 25, lateral and anterior/ventral views). At NF stage 26, HCN4 mRNA begins to be expressed on the left side of the developing heart field (arrow; lateral and ventral views). Expression becomes bilateral beginning at NF stage 27/28 (arrows; lateral and ventral views) but remains more highly expressed on the left side of the body. Scale bars = 200μm. (B) As cardiogenesis progresses during tadpole stages (NF stages 29–39, lateral and ventral views, arrows denote heart), HCN4 expression is diffusely present in the developing myocardium (outlined in yellow dashed lines/circles), but the highest expression is in the septum transversum where other proepicardial genes are expressed (Tbx18, NF stage 34). Scale bars = 200μm. (C,D) Histological sections confirm HCN4 expression in the cardiac mesoderm at tailbud stages, and in the myocardium and proepicardial region in tadpole stages. HCN4 expression can also be observed in the notochord and somites. Red lines in schematic diagrams denote the plane of section. Sections = 10μm, scale bars = 100μm. (E) Immunohistochemistry shows HCN4 protein is expressed at the completion of cardiac looping (NF stage 35) and is expressed throughout the entire ventricle of mature hearts (NF stage 46). Another important cardiac isoform, HCN2, is also found throughout the ventricle of mature hearts, similar to what has been described in mammalian systems. Transverse sections = 15 µm, scale bars = 100 µm. Schematic images are modified from Xenbase.20
We further confirmed HCN4s myocardial expression by examining the protein distribution in the heart via immunohistochemistry. HCN4 protein was expressed in the myocardium at NF stage 35, and later at NF stage 46 when heart maturation was complete (Fig. 1E). In both stages, HCN4 was robustly expressed throughout the myocardium. Interestingly, similar to what has been described in neonatal mouse and rat hearts,43 the HCN2 protein was also highly expressed in the myocardium in NF stage 46 tadpoles (Fig. 1E). Although HCN2 was robustly expressed in the developing nervous system and notochord at tailbud stages, HCN2 channel expression was not detected in the developing heart region during early stages of cardiogenesis in Xenopus (between NF stages 26–34, Supplemental Figure 1). Thus, in addition to its established role in patterning the cardiac conduction system, the expression of HCN4 and not HCN2, in the developing heart field before the formation of a beating heart tube is consistent with additional embryonic roles during Xenopus cardiogenesis.
Functional expression of HCN4-WT and HCN4-DN(AAA) mutant in HEK293 cells
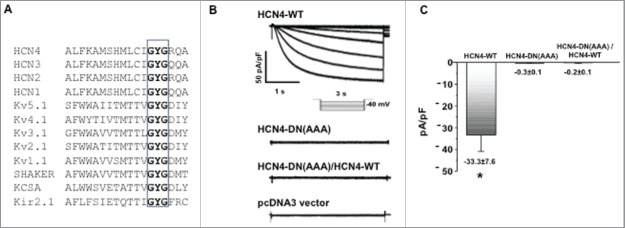
In humans, inherited mutations in the gene encoding HCN4 have been shown to cause sinus node dysfunction. For example, patients who carry a missense D553N mutation44 or a G480R point mutation45 develop cardiac arrhythmias. G480 is one of the conserved amino acid residues within the “GYG” signature cation selectivity motif of the pore region across species. This motif is common to an array of potassium-permeable channels, and has previously been used to generate dominant negative constructs for other HCN isoforms (Fig. 2A).46 We generated the triple amino acid mutant (“GYG” to “AAA”), and used voltage clamping in HEK293 cells, which do not have endogenous HCN expression, to characterize whether the HCN4-DN(AAA) mutant displayed a dominant-negative effect (Fig. 2B). Characteristic IHCN4 currents were generated in cells transfected with wildtype HCN4 in response to successive hyperpolarizing voltage steps, where larger voltage steps produced larger channel conductance. In contrast, in the HCN4-DN(AAA) expressing cells, the mutant channel showed little IHCN4 currents, with currents that were at the same magnitude as the negative vector control (pcDNA3, Fig. 2B). We then tested whether the HCN4-DN(AAA) could act as a true dominant-negative construct by co-transfecting it with the wildtype channel. If the HCN4-AAA mutant successfully integrated with wildtype subunits to form nonfunctional channels, then the currents observed with just HCN4-WT expression alone should be abolished. Indeed, when the mutant channel was co-expressed with the HCN4-WT channel (at 1:1 DNA ratio during transfection), the hybrid channel did not yield significant IHCN4 currents (Fig. 2B–C; n = 4, *p < 0.05). These data demonstrate that the HCN4-AAA mutant can result in a dominant negative effect. Thus, the HCN4-DN(AAA) mutation was subsequently used to manipulate channel function in Xenopus embryos.
Figure 2.
HCN4 dominant-negative effect test in HEK293 cells. (A) Sequence alignment of mouse HCN channels and other K+ channel examples that have a signature cation selectivity “GYG” motif at the channel pore region. (B) IHCN currents were elicited by a stepwise hyperpolarization to −120 mV from a holding potential of −40 mV; and then normalized to membrane capacitance. Representative IHCN4 current traces recorded in HEK293 cells expressing HCN4-WT, HCN4-DN(AAA) mutant, both HCN4-DN(AAA)/HCN4-WT, or a vector control are shown. Transfection with HCN4-WT produced robust IHCN4 currents, whereas transfection with HCN4-DN(AAA) did not produce any current greater than the vector control. When HCN4-WT and HCN4-DN(AAA) were co-transfected, IHCN4 was abolished. (C) Summary data for the peak IHCN4 currents recorded from HEK293 cells expressing WT-HCN4, HCN4-AAA mutant, HCN4-AAA/WT-HCN4 or vector control, confirming the ability of HCN4-DN(AAA) to act as a dominant-negative construct. n = 4–5, *p < 0.05.
Altering HCN4 leads to morphological and physiological defects in mature Xenopus hearts, but does not affect mesodermal or myocardial differentiation
To ascertain a possible role for HCN4 channels during the formation of the embryonic heart, we examined whether overexpressing wildtype HCN4 (HCN4-OE) or injecting the dominant-negative construct (HCN4-DN[AAA]) resulted in changes in heart morphology and physiology. For these experiments, we injected mRNAs encoding either the wild-type version of HCN4 (HCN4-[OE]) or a dominant-negative HCN4 mutant (HCN4-DN[AAA]) into one blastomere of 2-cell embryos. To determine if the sidedness of perturbation was important, membrane-bound RFP (mem-RFP) was co-injected as a lineage tracer to confirm location of expression in injected animals, and embryos were sorted for fluorescence on the left vs. the right side of the body. Heart morphology was examined after the completion of chamber septation (NF stage 46) using immunohistochemistry (IHC) for a Cardiac Troponin-T antibody (myocardial tissue) that allows visualization of mature heart structures. Both injection conditions induced abnormal cardiac phenotypes including twisted hearts, rotated hearts, unlooped hearts, and hearts with double ventricles (Fig. 3Aii-v respectively).
Figure 3.
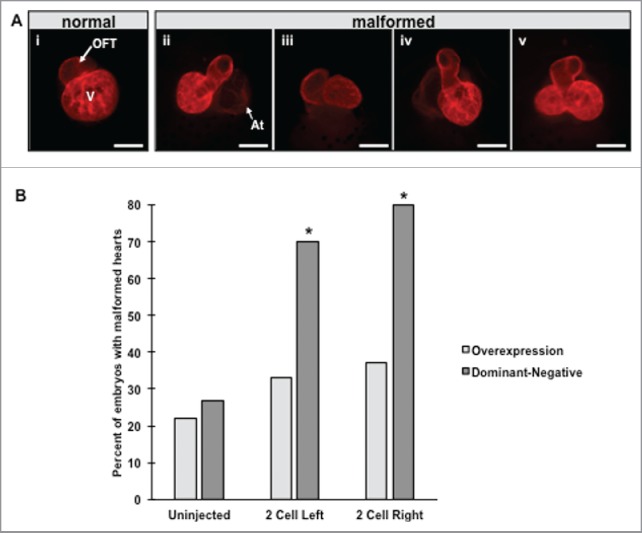
Overexpression and injection of dominant-negative HCN4 mRNA induces aberrant heart morphology. (A) Mature NF stage 46 tadpole hearts were visualized by immunohistochemistry using a cardiac troponin-T antibody to detect mature cardiac muscle. In control tadpoles, the ventricle (V) and outflow tract (OFT) are readily visible with the atria located directly behind the ventricle (i). Altering HCN4 expression induced a variety of morphological defects including hearts that were twisted into the dorso-ventral (D-V) body axis (ii), hearts rotated in the same plane of the D-V axis (iii), unlooped hearts (iv), and double ventricle phenotypes (v). In twisted hearts (ii) the atria are no longer positioned behind the ventricle and can now be seen (At = atria). (B) Both injection conditions of the HCN4-DN(AAA) construct induced significantly higher percentages of morphological defects compared with uninjected tadpoles, whereas overexpression of HCN4 did not cause significant occurrences of malformed hearts χ2 = 58.174, df = 4, * p < 0.05; n = 75 embryos per condition across 3 biological replicates.
Although tadpoles with malformed hearts were seen when HCN4 was overexpressed, abnormal heart phenotypes were observed less frequently in HCN-OE injected animals than in the HCN4-DN(AAA) expressing tadpoles (Fig. 3B). In HCN4-DN(AAA) expressing animals, a significant percentage of mispatterned hearts was observed compared with stage-matched, uninjected tadpoles (Fig. 3B, χ2 = 55.709, df = 2, * p < 0.01). There was no statistical difference between left-sided expressing and right-side expressing animals in both HCN4-OE or HCN4-DN(AAA) conditions. Remarkably, irrespective of where the HCN4-OE (data not shown) or HCN4-DN(AAA) (Fig. 4) mRNAs were targeted in the tadpole, severely malformed hearts were observed, including animals without any detectable injected HCN4 in the ventral-anterior region where the heart is forming. Thus, altering the endogenous area of HCN4 function during early stages of cardiogenesis results aberrant heart phenotypes, even when injected mRNAs are expressed outside of the heart region in mature tadpoles. We conclude that HCN4 function is an important endogenous input into heart patterning, and that HCN4-dependent signaling functions at considerable distance (non-cell-autonomously).
Figure 4.
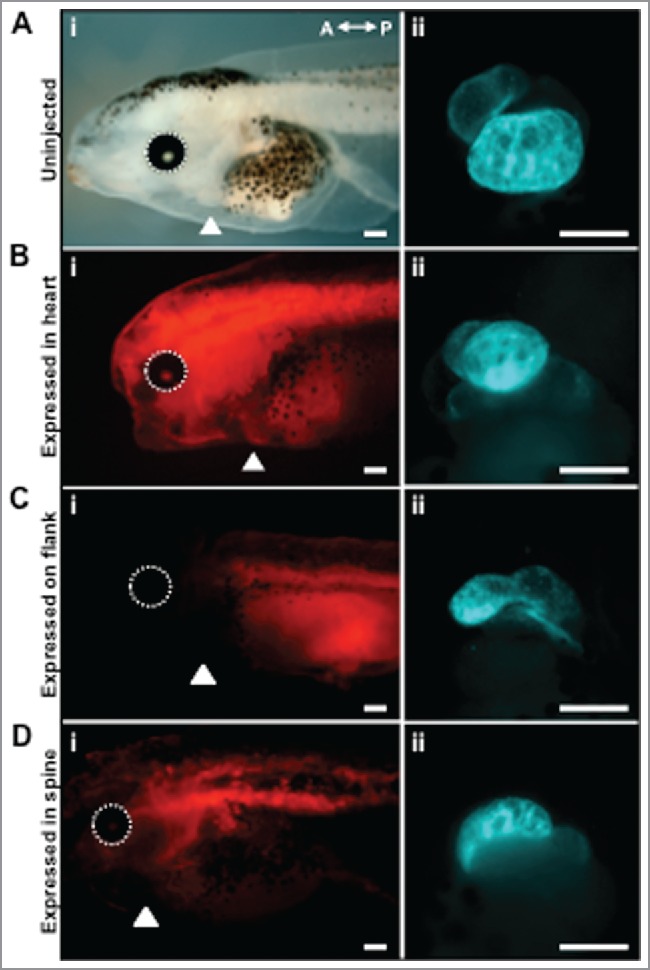
Inhibiting HCN4 function in different locations disrupts heart morphology. (A) Uninjected tadpoles have a clear chest cavity at NF stage 43/44, making the heart easily visible laterally (arrowhead, i). Hearts have normal ventricle and outflow tract morphology as seen via IHC for cardiac troponin-T antibody (ventral view, ii). The left eye is circled (white dashed line) in all images as a landmark. (B-D) HCN4-DN(AAA) injected embryos display a variety of heart defects, irrespective of the location of the lineage label membrane-bound RFP. Examples of tadpoles with mem-RFP located in the: heart (arrowhead, B, i), posterior flank (C, i), and dorsal region (D, i) of HCN4-DN(AAA) injected animals. White arrowheads denote position of heart. Ventral views of malformed heart phenotypes: symmetric midline hearts (B, ii), unlooped hearts (C, ii), and twisted hearts (D, ii). n = 18–31embryos per injection condition. Scale bars = 200 µm.
To examine whether the abnormal heart morphology resulted from the global mispatterning of embryonic tissues, the expression of several genes expressed during different stages of development was examined in HCN4-DN(AAA)-injected embryos (Supplemental Figure 2). The expression pattern of both Xbra, a panmesodermal gene activated during gastrulation,47 and Otx2, a broad anterior structure marker expressed during gastrulation and neurulation,48 was similar to uninjected control embryos (Supplemental Figure 2A). Similarly, Nkx2.5 expression, which is an important cardiac field specification gene, was not affected by injection of HCN4-DN(AAA) during early cardiogenesis (NF stage 27), or later cardiac looping (NF stage 32). In addition, the presence of a myocardial-specific protein (CT3) in HCN4-altered animals (Figs. 3 and 4) provides further support that myocardial differentiation proceeded normally.
Since perturbing HCN4 channel function resulted in altered heart morphology, heart rates were analyzed to determine if cardiac conduction was also compromised (Fig. 5). There was no statistical difference between embryos with HCN4 overexpressed and uninjected, sibling embryos (Fig. 5A). However, all tadpoles injected with the HCN4-DN(AAA) channel had significantly faster heart rates (tachycardia) than uninjected sibling tadpoles confirming the ability of our injected-DN construct to modulate HCN4-dependent bioelectric events in vivo (Welch's test, *p < 0.05) (Fig. 5B).
Figure 5.
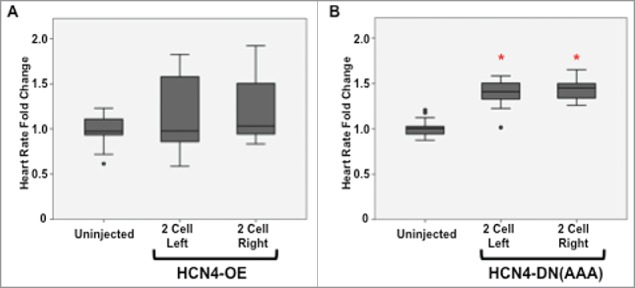
Injection of HCN4-DN(AAA) results in altered cardiac function. (A) Injecting wild-type HCN4 (HCN4-OE) into 2-cell embryos did not significantly alter heart rate. (B) Inhibiting HCN4 ion channel function (HCN4-DN[AAA]) resulted in significantly higher heart rates in all injection conditions (Welch's test, *p < 0.05). Dots represent outlier individuals who were greater or less than 1.5 times the upper and lower quartiles respectively. n = 30 embryos per condition across 3 biological replicates.
Altering HCN4 expression disrupts the distribution of left/right and cardiac morphogenetic patterning genes
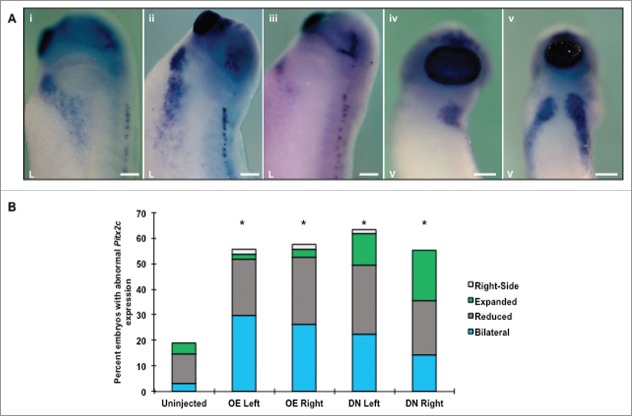
A critical step of creating functional organs during development is the final placement of organs inside the body cavity. All organs are dependent on surrounding tissues to provide the spatial cues needed to regulate growth, cell fate, and determine organ position. Previous research has demonstrated that altering the location of proteins that direct laterality decisions during development often results in the misplacement of organs. Because altering HCN4 channel function resulted in the formation of mispositioned, but properly differentiated myocardial tissue at swimming tadpole stages, and the endogenous channel is asymmetrically expressed in the heart field during early cardiogenesis, we investigated whether HCN4 interacts with canonical determinants of organ laterality during embryogenesis. Xnr-1 is one of the earliest regulators of laterality, and its left-sided expression is required for cardiogenesis to progress normally.49-51 Therefore, we examined the expression pattern of Xnr-1 at stages when it is asymmetrically expressed in the lateral plate mesoderm at the completion of neurulation (NF stage 21) in animals injected with either the HCN4-OE or HCN4-DN(AAA) (Fig. 6). Injection of HCN4-DN(AAA) induced significantly higher percentages of abnormal, ectopic patterns of Xnr-1 expression compared with uninjected sibling embryos, including: complete bilateral fields, right-sided expression, and punctate Xnr-1 expression in both the lateral plate mesoderm as well as in dorsal region of the larvae (χ2 = 31.0207, df = 4, *p < 0.05). Interestingly, injecting the dominant-negative mRNA on the right-side of the body induced the highest percentage of abnormal Xnr-1 expression phenotypes (Fig. 6B) with over 50% of HCN4-DN (right) injected embryos with abnormal ectopic Xnr-1 expression, although this was not statistically different than injection on the left side of the body. While Xnr-1 expression was occasionally altered in HCN4-OE injected animals, these were not statistically different from uninjected embryos (Fig. 6B).
Figure 6.
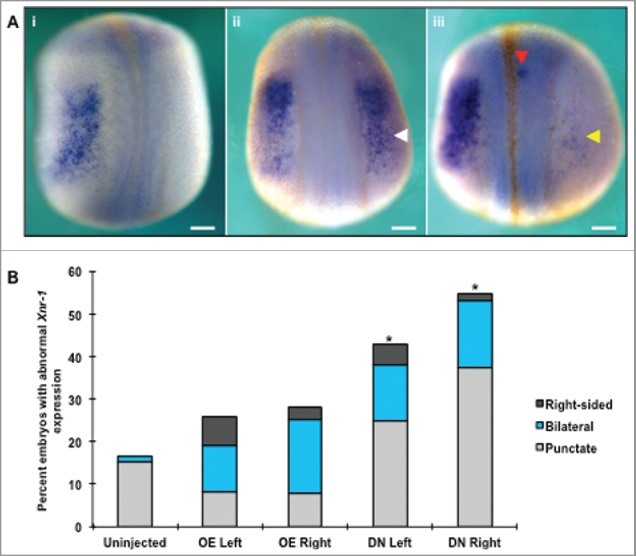
HCN4 helps coordinate the distribution of Xenopus nodal-related 1 (Xnr-1) expression. (A) During normal development, Xnr-1 is expressed exclusively in the left lateral plate mesoderm following the completion of neurulation as seen via in situ hybridization (NF stage 21, i). Both overexpression and injection of HCN4-DN(AAA) resulted in bilateral expression of Xnr-1, with the secondary Xnr-1 field ranging from a small partial field containing few cells (yellow arrow, iii) to an equivalent right-sided field (white arrow, ii). In addition to generating a second field of Xnr-1, punctate dots of expression were observed along the fused neural tube (iii, red arrow) and flanks of embryos. These ectopic dots were found on both sides of embryos regardless of the presence of endogenous Xnr-1 expression. Dorsal views, anterior at top. (B) While abnormal (bilateral, punctate, and right-sided reversed) expression was observed in all injection conditions, only HCN4-DN(AAA) injection caused a significant increase in abnormal phenotypes compared with uninjected embryos (χ2 = 31.0207, df = 4, *p < 0.05). n = 73–84 per condition across 3 biological replicates. OE = HCN4-OE, DN = HCN4-DN(AAA). Scale bars = 200 μm.
The ectopic, bilateral expression of Xnr-1 ranged from small partial fields with few Xnr-1 expressing cells (Fig. 6A iii, yellow arrowhead) to complete Xnr-1 fields that matched their left-sided counterparts (Fig. 6A ii, white arrowhead). We also observed punctate dots of Xnr-1 expression in injected embryos that were located outside of the normal Xnr-1 field including: along the midline (Fig. 6A, iii red arrowhead), in the posterior, and along the flanks of embryos. This ectopic, spotty Xnr-1 expression outside of the lateral plate mesoderm ranged from one or 2 punctate areas of expression to multiple spots. To our knowledge, this is the first report of ectopic, punctate Xnr-1 expression, as other previous studies report only the presence of bilateral expression or reversed, right-sided expression in response to genetic manipulations.49-51
Lefty is another patterning gene, which is activated by Xnr-1 and acts to limit the spatial distribution and duration of Xnr-1 signaling.52 Its left-sided expression acts as a barrier to Xnr-1 at the midline, preventing the transduction of “left” signals to the right half of the body. Lefty is expressed throughout organogenesis, propagating the polarity established by Xnr-1 through interactions with other downstream factors such as Pitx2 and BMP-4.53,54 To determine if HCN4 regulates the spatial organization of Lefty expression in a similar fashion to Xnr-1, we investigated Lefty expression at stages when both HCN4 and Lefty transcripts are present in the cardiac mesoderm (NF stage 27, Figs. 1 and 7, respectively). Both overexpression and injection of the dominant-negative mRNA resulted in bilateral Lefty expression. As observed with Xnr-1, there was a range in the severity of bilateral expression. Some embryos had a partial second Lefty field with a gap in expression along the midline (Fig. 7A, ii). However, in other embryos, Lefty expression appeared extremely disorganized and encompassed a large area of the anterior-ventral surface of the animal, including the midline (Fig. 7A, iii). All injection conditions generated significantly higher percentages of abnormal Lefty expression compared with uninjected sibling controls (Fig. 7B, χ2 = 19.0086, df = 4, p < 0.05). There was no statistical difference between overexpression, dominant-negative or sidedness of injection.
Figure 7.
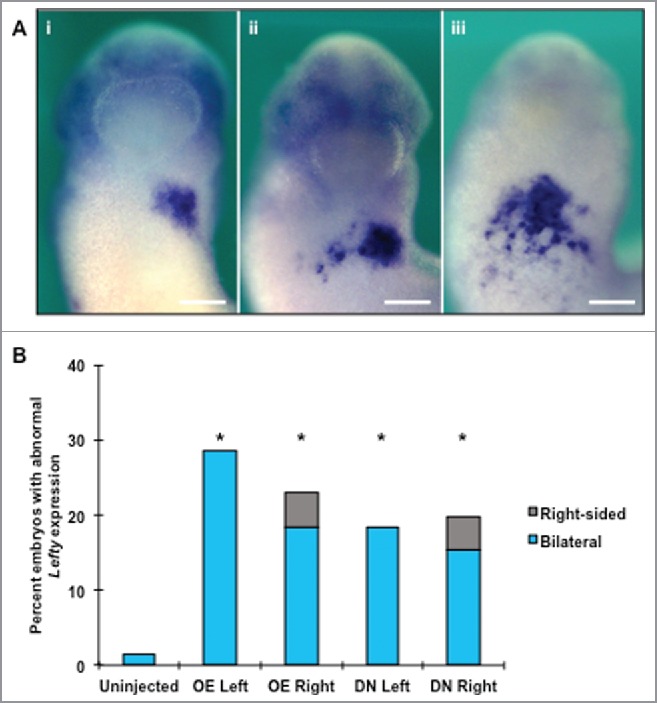
Altering normal HCN4 expression results in aberrant Lefty expression. (A) As visualized via in situ hybridization analysis, Lefty is normally expressed unilaterally in the left lateral plate mesoderm in NF stage 27 embryos (i). Altering HCN4 expression induced both full or partial bilateral and abnormal Lefty expression but did not result in a complete reversal of expression (solely right-sided). One of the most frequently observed bilateral phenotypes was a partial secondary field on the right side of embryos with the midline maintained by an absence of Lefty expression (ii). In the other commonly seen phenotype, Lefty transcripts were expressed across the anterior ventral surface of animals in an undefined shape with no visible midline gap (iii). Images are ventral views with anterior at top. (B) Overexpressing HCN4 and HCN4-DN(AAA) injection led to significant percentages of abnormal embryos compared with uninjected embryos (χ2 = 19.0086, df = 4, *p < 0.05). There was no statistical difference between overexpression and dominant-negative phenotypes. n = 56–71 embryos per condition across 3 biological replicatesOE = HCN4-OE, DN = HCN4-DN(AAA). Scale bars = 200 μm.
After axis-patterning genes such as Xnr-1 and Lefty help to establish sidedness in the body, organ morphogenesis must also be carefully regulated such that final organ asymmetry occurs correctly. Two critical mediators of these events, Pitx2 and BMP-4, directly translate asymmetric gene expression into asymmetric morphogenesis in multiple species through independent and coordinated interactions.53,55-62 Both Pitx2 and BMP-4 are regulated by Xnr-1 signaling, and work together to impart laterality to the developing heart tube.56,61 Both of these genes are asymmetrically expressed along the left side of the heart tube, providing laterality to the tube just before the onset of rightward looping. Because misexpression of either gene leads to severe positional defects in embryonic hearts, we examined the expression of Pitx2 and BMP-4 in HCN4-OE and HCN4-DN(AAA) injected embryos.
To characterize the relationship of HCN4 expression to Pitx2 expression, we examined NF stage 27 embryos in HCN4-OE and HCN4-DN(AAA) injected animals. Both overexpression and injection of the dominant-negative construct resulted in significantly higher percentages of abnormal Pitx2 expression patterns compared with uninjected sibling control animals (Fig. 8, χ2 = 64.71, df = 4, *p < 0.05). There was no statistical difference between the overexpression and dominant-negative conditions, or sidedness of injection, all of which resulted in altered Pitx2 expression. Phenotypes included an expansion or reduction in the size of the endogenous field and bilateral expression (Fig. 8A, ii, iii, v). Similar to the Xnr-1 and Lefty results, the bilateral fields were not always equivalent in size. Many embryos had only a small, partial field on the right side of the body in addition to the endogenous field. No animals were observed to have a complete ablation of Pitx2 expression.
Figure 8.
Altering HCN4 expression leads to improper distribution of the homeodomain transcription factor Pitx2. (A) In situ hybridization of representative embryos displaying normal and examples of the most commonly observed Pitx2 expression defects. Uninjected embryos have asymmetric, left-sided expression (i lateral view; iv ventral view). Overexpression and injection of the dominant-negative construct resulted in a variety of molecular phenotypes including expansion or reduction of the endogenous field (ii, iii) and bilateral Pitx2 expression (v). Anterior at top of images. (B) All manipulations caused significantly higher percentages of abnormal Pitx2 expression compared with uninjected embryos (χ2 = 64.71, df = 4, *p < 0.05). There was no statistical difference between overexpression and dominant-negative phenotypes. n = 54–86 embryos per condition across 3 biological replicates. OE = HCN4-OE, DN = HCN4-DN(AAA). Scale bars = 200 μm.
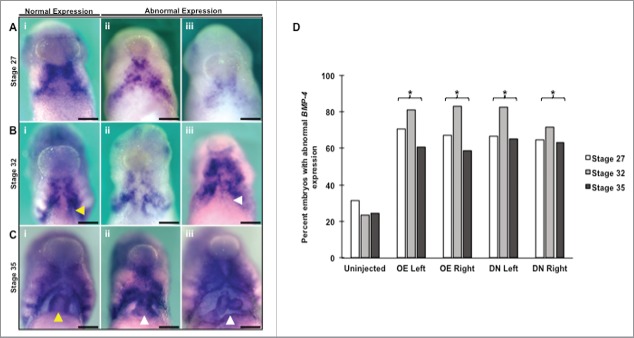
Because BMP-4 transcripts have a dynamic spatio-temporal pattern during critical stages of heart development and morphogenesis,63 we examined its expression profile in injected embryos at 3 different stages of heart development: before the onset of looping (NF stage 27), during looping while expression is asymmetric in the myocardium (NF stage 32), and after the completion of cardiac looping (NF stage 35). In uninjected embryos, BMP-4 has a bilateral, wing-shaped pattern beginning at NF stage 27 (Fig. 9A, i). This pattern becomes increasingly more distinct as looping progress with an area of asymmetric expression arising in the myocardium during these stages (Fig. 9B, i, yellow arrow). At the completion of looping (NF stage 35), the wing-pattern remains in the tissue surrounding the heart, and expression becomes more uniform around the outside layer of the myocardium (Fig. 9C, i). As Fig. 9D illustrates, all 3 stages investigated had significantly higher percentages of abnormal expression of BMP-4 in injected animals (HCN4-OE and HCN4-DN[AAA]) compared with uninjected sibling controls (NF stage 27: χ2 = 32.4768, df = 4; NF stage 32: χ2 = 82.2366, df = 4; NF stage 35: χ2 = 36.0791, df = 4, All: *p < 0.05). Similar abnormal BMP-4 expression phenotypes were observed in animals injected with the overexpression or the dominant-negative mRNA (no statistical difference between HCN4-OE and HCN4-DN[AAA] conditions, or left vs. right sided injections). Altering HCN4 function resulted in a global mispatterning of BMP-4 expression that included either a reduction in BMP-4 expression or loss of definition in the boundaries of the field (Fig. 9A ii-iii, 9B ii-iii, 9C ii-iii). There was no clear pattern in the location of reduction in embryos; BMP-4 was reduced on either side or both sides in individual embryos. When there was a loss in field definition the canonical wing-shaped BMP-4 pattern was no longer distinguishable; however, the area of BMP-4 expression that lost definition was not consistent across individuals. Frequently, BMP-4 expression was a disorganized smear across the ventral side, without wings or separation along the midline. We also observed a randomization or loss of asymmetric BMP-4 expression in the myocardium at stage 32, which may have important implications for final heart morphology. Finally, by stage 35, although the majority of embryos had uniform BMP-4 expression in the myocardium, heart looping was abnormal (Fig. 9C ii-iii).
Figure 9.
BMP-4 expression is reduced and incorrectly localized in response to altering HCN4 expression. (A) At NF stage 27 BMP-4 is expressed in 2 bilateral patches around the developing heart field (i). Altering HCN4 caused a loss of definition to the fields (ii) as well as an overall reduction in the size of the fields and intensity of expression (iii). (B) BMP-4 remains bilaterally expressed during heart looping (NF stage 32) with an additional asymmetric area of expression in the myocardium (yellow arrow, i). This left-sided myocardial expression helps to direct and orient heart looping. Overexpression of HCN4 and expression of HCN4-DN(AAA) not only disrupted the lateral BMP-4 fields of expression (ii), it also caused both a randomization and loss of myocardial expression (white arrow, iii). (C) By the end of looping at NF stage 35, BMP-4 is evenly expressed in the myocardium and retains its bilateral expression in the surrounding tissues (i). Similar to stage 27 and 32, disruption of HCN4 led to both a reduction in expression as well as a loss of field definition (ii, iii). Injected embryos frequently displayed malformed hearts who did not complete looping properly (yellow vs. white arrows). All abnormal images are from HCN4-DN(AAA) injected embryos but similar phenotypes were observed with HCN4-OE embryos. (A-C) Images are ventral views with anterior at top. (D) All stages and injection conditions induced significantly higher percentages of abnormal phenotypes compared with controls (Stage 27: χ2 = 32.4768, df = 4; Stage 32: χ2 = 82.2366, df = 4; Stage 35: χ2 = 36.0791, df = 4; All: *p < 0.05). There was no statistical difference between overexpression and dominant-negative phenotypes. n = 58–79 embryos per condition across 3 biological replicates. OE = HCN4-OE, DN = HCN4-DN(AAA). Scale bars = 200 μm.
Interestingly, the disruption of normal dorsal-anterior development, resulting in a reduction of anterior structures and loss of anterior notochord, results in abnormal heart left-right asymmetry.64 During embryogenesis, Shh signaling helps to establish the dorsoventral axis, and patterns the midline, including the central nervous system and notochord.65 Thus, to examine if the aberrant heart phenotypes observed in our experiments were due to a global mispatterning of the anterior, dorsal-midline, we examined the expression of Shh at several times during heart development. Embryos injected with HCN4-OE or HCN4-DN(AAA) were examined for Shh expression at 3 time points: during gastrulation, at the completion of neurulation, and during cardiac looping (Supplemental Figure 2B). In all stages examined, Shh expression was similar in uninjected control and injected animals suggesting the dorsal midline structures were not altered in these experiments.
Discussion
Taken together, these data illustrate that altering HCN4 channel function during embryogenesis results in atypical positional orientation and morphogenesis of the heart. Importantly, the phenotypes observed in HCN4-DN(AAA) injected animals have a unique heart morphology when compared with what has been previously observed in studies that disrupted canonical laterality genes. For example, ectopic right-sided Xnr-1 expression randomizes heart situs but the hearts are still properly looped.66 Similarly, overexpression of Lefty on either side of the body induces heart reversals (situs inversus) but proper looping and patterning is again maintained.53 In contrast, overexpression of Pitx2 can cause reversal in heart laterality, straightened outflow tracts, and leads to chamber malrotation.67 Likewise, inhibition of BMP-4 or its receptors results in a lack of cardiac looping and misplacement of the atria along the midline.56,57 Remarkably, the phenotypes we observed in embryos where HCN4 expression is disrupted appear to be a combination of inappropriate signals during both the patterning of early embryonic fields as well as later events during cardiac morphogenesis such as looping (Fig. 3). It is important to note that these data also illustrate that heart mispatterning observed is not a matter of incorrect myocardial differentiation or gross morphological alteration of all organs and tissues. Both HCN4-OE and HCN4-DN(AAA) injected embryos expressed the mature myocardial protein, cardiac troponin-T, as well as the cardiac field marker Nkx2.5, indicating that differentiation occurred. In addition, the indistinguishable expression patterns of Xbra, Otx2, and Shh between injected animals and sibling controls suggest that HCN4 isn't arbitrarily disrupting axis formation, or germ layer specification, but rather it is specifically interacting with the genes that determine organ positioning.
Physiologically, only injection of HCN4-DN(AAA) significantly affected heart rate, with injected embryos displaying tachycardia. Tachycardia has also been described in human HCN4 loss of function mutations: a c-linker interaction mutant and a trafficking-defective mutant.44,68 Importantly, both mutations occurred endogenously in patients, and the c-linker mutation was inherited across 4 generations of affected individuals. This suggests that the presence of a functioning HCN4 channel is required to properly pattern the cardiac conduction system, and this role is highly conserved across vertebrates; however, it is also possible that the abnormal heart rate is due to disrupted heart morphology or vice versa.
Our results demonstrating the altered expression of numerous morphogenetic control factors suggest a novel role for HCN4 during embryogenesis: coordinating the positional identity of cells needed to create functional organs via directing the distribution of important patterning molecules. The distribution of key signaling factors appears to be dependent on the correct function of HCN4 as both perturbations (overexpression and dominant-negative) induced similar phenotypes. Although Lefty, Pitx2, and BMP-4 expression was disrupted in embryos overexpressing HCN4, it seems that embryos can somewhat compensate for these molecular aberrations because many of the embryos in those populations were able to form normal hearts. This form of compensation has recently been described in Xenopus organ laterality, where disruptions in upstream gene signaling did not always induce downstream changes in organ situs.69 In sharp contrast, embryos with the dominant-negative construct ultimately have abnormal heart morphology, positioning and heart physiology. Combined, these data indicate that HCN4 channel function is required for the correct distribution of morphogenetic patterning genes during cardiogenesis, and contributes to directing final heart morphology. It should be noted that because Xnr-1 is upstream of Lefty, Pitx2, and BMP-4, it is possible that HCN4 interacts with Xnr-1 primarily, and subsequent changes in gene expression are a result of disrupted Xnr-1 signaling.
These findings serve to expand the known roles of this important channel family, and reveal a new set of bioelectric inputs into cardiac organogenesis. This is not the first time an ion channel has been implicated in the distribution of patterning genes. For examples, during Drosophila wing formation, the inward rectifying potassium channel Irk2 is necessary for the correct dispersal of Dpp, the Drosophila homolog of BMP.70 Knocking down the channel led to phenotypes that were similar to BMP mutant phenotypes, and reducing Dpp signaling exacerbated abnormal phenotypes. Similar developmental defects were also observed when the vertebrate homolog of the channel, Kir2.1, was disrupted during both Xenopus and mouse embryogenesis.70-72 Again, the morphological defects were similar to those seen in defective TGFβ signaling. Taken together, these results combined with our study suggest that ion channel activity can play a significant role impacting gene expression patterns that feed into crucial aspects of embryogenesis.
One question that remains is how changes in ion channel function might instruct changes in the distribution of critical developmental genes. HCN4 could potentially regulate either the generation or the propagation of morphogenetic factors as altering its expression frequently led to a reduction in gene expression. Previous studies have shown that ion channels have reciprocal interactions with integrins found in the membrane, and these interactions affect downstream signaling responses.73 In fact, in adult hearts HCN4 complexes with β2-adrenergic receptors to modulate the funny current and subsequent heart rate.74 Therefore, it is possible that during embryogenesis fluxes through ion channels help to coordinate the distribution of ligands and receptors in the membrane that interact with morphogenetic proteins. For example, the BMP antagonist Noggin binds to heparin sulfate proteoglycans in cell membranes to modify the distribution of BMP, and this interaction can only occur within a specific concentration of Na+ present.75 Potentially, channels such as HCN4 change the concentration of specific ionic species, altering not only membrane potential but also the localization, modification state, or interactions of critical transmembrane receptors. Likewise, modulation of resting potential by a variety of ion channels is known to regulate transcription of genes such as Notch76 and of many other targets conserved from amphibian to human.77 Many studies have also implicated bioelectrical control of the movement of small molecule signals (such as calcium, butyrate, and serotonin) that transduce voltage changes into second messenger cascades and alterations of transcription39,78 of numerous downstream targets that are well-conserved among human and amphibian bioelectrical signaling responses.79
One other important hypothesis to consider is the importance of temporal, as well as spatial, patterning of bioelectrical activity. Numerous studies have shown HCN channels regulate membrane potential in a cyclical manner.4 This type of clock-like signaling might be necessary in developing tissues to modulate patterning genes both spatially and temporally. This is consistent with a potential voltage threshold or cyclic pulsing of membrane potential that must occur to achieve correct positional identity; hypotheses that will be tested with further development of in vivo optogenetic tools that may allow a finer temporal dissection of ionic signaling in development.80 Finally, although we have presented evidence that altering HCN4 expression during embryogenesis causes incorrect positioning of hearts, future studies are needed to dissect out the molecular mechanism and timing of this phenotype. Overall, the data presented here highlight a novel role for the clinically important HCN4 channel, and suggest early embryonic physiology as a tractable target for modulation of key development gene expression programs.
Materials and methods
Animal husbandry and embryo culture
All animals were treated humanely and with regard for alleviation of suffering according to approved protocols (Institutional Animal Care and Use Committee Protocol Tufts University). Adult female Xenopus laevis were induced to ovulate via injection of chorionic gonadotropin hormone (Chorulon) and eggs were fertilized in vitro. After fertilization, eggs were dejellied in a 2% cysteine solution (pH 8) and reared in 0.1X Marc's Modified Ringer's solution (Stock solution: 10X MMR; 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 5 mM HEPES, 10 μM EDTA, pH 7.4) at 14–18°C. Embryos and tadpoles were staged according to Nieuwkoop and Faber.81
Molecular cloning of constructs
A mouse HCN4-WT cDNA clone was kindly provided by Dr. Richard Robinson (Columbia University). Using a standard strategy for abolishing channel function by altering the highly conserved cation selectivity sequence in the pore domain (changed from GYG349–351 to AAA349–351;,46,82,83 the HCN4-(AAA) mutant was generated in WT-HCN4 using primers: 5′ CACATGCTGTGCATTGAGGACGAACGTCAGGCA-3′ (forward) and 5′-TGCCTGA CGTTCGTCCTCAATGCACAGCATGTG-3′ (reverse). For recordings of IHCN4 currents in mammalian cell culture experiments, a WT or HCN4(AAA) mutant construct was sub-cloned into a bi-cistronic vector, pIRESGFP1 (a kind gift from Dr. David Johns, John Hopkins University), to create a CMVp-HCN4-IRES-eGFP fusion for subsequent expression studies. An empty vector pcDNA3 (Life Technologies, Grand Island, NY) was used as a negative control in electrophysiological recording experiments.
HCN4 constructs were used to transcribe HCN4-WT (overexpression [OE]) or HCN4-(AAA) (dominant negative) mRNAs for Xenopus laevis microinjection, both HCN constructs were subcloned into a pCS2 vector. The membrane-bound RFP that was microinjected as a lineage tracer was a gift from Dr. John Wallingford (University of Texas-Austin).
Cell culture, transfection and recording of IHCN4 currents
HEK293 cells were routinely maintained in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum, 100 ug/ml penicillin and 100 ug/ml streptomycin at 37°C as described previously.84 Transfection was performed using a Fugene6 kit following manufacturer's instructions (Roche, Indianapolis, IN). For voltage clamping analysis, approximately 1×105 cells were seeded in a 35-mm culture dish. 0.7 µg DNA was transiently transfected into HEK293 cells. In all experiments, the transfected cells were cultured for 24 hours to allow protein expression.
Whole cell-based HCN4 currents (IHCN4) were recorded from transfected HEK293 cells at room temperature as described previously.85,86 The extracellular recording solution contained: 110mM NaCl, 0.5 mM MgCl2, 30 mM KCl, 1.8 mM CaCl2, and 5mM HEPES. Recording pipettes contained: 10 mM NaCl, 0.5mM MgCl2, 130 mM KCl, 5 mM HEPES, and 1mM EGTA. Cells expressing eGFP were subjected to voltage clamping measurements. IHCN4 was evoked in response to hyperpolarizing voltage steps to potentials between −60 and −130 mV from a holding potential of −40 mV. Data were collected by an Axopatch 200B amplifier interfaced to a Digidata 1322A acquisition system, and analyzed using a pCLAMP10 software package (Molecular Devices, Sunnyvale, CA).
Microinjection of mRNA into Xenopus embryos
Capped mRNAs were synthesized in vitro using the Sp6 mMessage Machine kit (Invitrogen). Cleavage stage embryos were injected in either the left or right blastomere at the 2-cell stage with mRNAs encoding either 350pg wild-type mouse HCN4 (HCN4-WT) or 500pg dominant-negative mouse HCN4 pore mutant (HCN4-DN[AAA]). Embryos injected with HCN4 mRNAs were co-injected with membrane-bound RFP (300pg) as a lineage tracer.
After microinjection into cleavage stage Xenopus embryos, embryos healed in 1X MMR overnight. Before the onset of gastrulation, the MMR was gradually reduced to 0.1X MMR, and embryos were raised in 0.1X MMR until they reached the desired stage, at which point they were sorted for fluorescent expression to confirm the presence and location of injected constructs. Prior to molecular analysis, embryos were killed with 5% tricaine (pH 7.2, MS-222, Acros Organics) and fixed for 1 hour at room temperature in MEMFA (0.1 M MOPS pH 7.4, 2 mM EGTA, 1 mM MgSO4, 3.7% formaldehyde).
Heart rate analysis
To determine if heart function was affected, the heart rate of 10 individual tadpoles per injection group was measured for a total of 30 embryos across 3 replicates. N.F. stage 46/47 tadpoles were lightly anesthetized in a petri dish with 0.1X MMR. 10-second videos of the beating hearts were recorded using a Canon RebelT5i camera. MPEG Streamclip software was used to reduce video speed for heart-rate analysis. Heart rates were calculated by dividing the number of beats by the time difference between the last and first diastole (when the ventricle is the least contracted). To account for replicate variation and differences in external factors such as season and room temperature, all experimental rates were normalized to the mean of control rates in each replicate.
In situ hybridization (ISH)
The localization of mRNA transcripts was evaluated by whole-mount ISH as described previously.87 Briefly, tadpoles were hybridized with digoxigenin-labeled antisense riboprobes, and expression was visualized using an anti-digoxigenin antibody conjugated to alkaline-phosphatase (Roche) and a purple BCIP/NBT precipitate. After the chromogenic reaction finished, embryos were fixed in MEMFA for 1 hour at room temperature or overnight at 4°C and either kept in 1X PBS at 4°C or dehydrated in 100% methanol for long-term storage at −20°C. Antisense RNA probes included: BMP-4,88 Pitx2,61 Xnr-1,66 Lefty,89 Otx2,48 Xbra, Shh90 and HCN2 (X. laevis hcn2.L IMAGE clone 5514485, which was purchased from Dharmacon and a HindIII fragment was deleted, leaving exons 2–4 and part of exon 5 as probe). HCN4 transcripts were detected using an antisense HCN4 probe that was constructed as a synthetic DNA (Invitrogen) and cloned into vector PCRII Topo (Invitrogen). The synthetic sequence corresponded to parts of exon 1 and 3, and all of exon 2, of the RNA model Xelaev18020786m, of the gene annotated as hcn4.S, at position chr3S:59295253–59295679 of X. laevis genome build 9.1 (http://www.xenbase.org). The region synthesized was chosen because it had high similarity to homolog hcn4.L. Sense control probes were developed and tested to ensure the specificity of the probes.
Immunohistochemistry, immunofluorescence, and heart morphology scoring
Immunohistochemistry (IHC) and immunofluorescence was used to detect the localization of proteins in whole mount and on section. Euthanized tadpoles were permeabilized with PBTr (1X PBS, 0.1% Triton, 2 mg/ml bovine serum albumin [BSA]), blocked with 20% heat-inactivated goat serum in PBTr, and incubated with the primary antibody overnight at 4°C. After 5 hrs of PBTr washes, tadpoles were blocked and incubated overnight in goat anti-mouse IgG secondary antibodies conjugated to Alexa Fluor 555 (AF-555; 1:300; Invitrogen). Then, tadpoles were again washed in PBTr for 5 hours, rinsed in 1X PBS, and post fixed in MEMFA, for 1 hr. Protein expression was detected through fluorescence using a Zeiss Imager.M1 scope and Cy3 filter cube. For paraffin embedded sections, slides were first dewaxed in xylene, rehydrated through a series of ethanol washes, and then chromogenic IHC was performed as described previously.91 Expression was detected through the presence of a purple precipitate from the chromogenic reaction. The following markers were assessed through IHC: CT3 (Developmental Studies Hybridoma Bank), HCN4 (Abcam ab69054), and HCN2 (Abcam ab84817). Hearts were examined for any aberrant phenotypes using a stringent scoring rubric that classified deviations from normal heart situs (in any direction) as abnormal, even if they were mild. Hearts were characterized as twisted when the chambers were displaced along the dorso-ventral (DV) axis. Rotated hearts were displaced along the left-right (LR) axis but in the same DV plane. Hearts were classified as unlooped when the outflow tract was improperly folded with respect to the ventricle.
Histology
For sectioning, dehydrated embryos were gradually transferred to 100% ethanol. Next, embryos were cleared in 2 subsequent 45-min Xylene washes and tissues were infiltrated with paraffin wax in 2 20-min incubations at 60°C. Embryos were then wax-embedded in molds and allowed to solidify overnight. Using a Leica 2255 rotary microtome, 10 or 15 µm sections were generated and fixed on Superfrost Plus glass slides. The slides were dewaxed using Xylene and mounted with Permount (Fisher Scientific). Sections were examined using a Nikon SMZ1700 microscope and pictures were taken using SpotIn-light Color Camera software.
Statistical analysis
All statistical analyses were performed using IBM SPSS statistics version 22 or R version 3.1.2, Pumpkin helmet.92 For all categorical phenotype data, significance was assessed using a chi-square test (R, statistics package). In each chi-square analysis all abnormal categories were grouped into one to make it a binary comparison (normal vs. abnormal). The fifer package was used to analyze all pairwise comparisons in each chi-square test.93 For heart rate data, significance was assessed using a Welch's test due to unequal variance in our data sets (SPSS).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank members of the McLaughlin and Levin laboratories for discussions and support.
Funding
This research was conducted with Government support under and awarded by DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a (E. Pitcairn). This work was supported by awards from IOS-NSF (KM), AHA (ML/KM, NQS, BY), and by the Allen Discovery Center program through The Paul G. Allen Frontiers Group (ML/KM).
References
- [1].Biel M, Wahl-Schott C, Michalakis S, Zong X. Hyperpolarization-activated cation channels: from genes to function. Physiol Rev 2009; 89:847-85; PMID:19584315; https://doi.org/ 10.1152/physrev.00029.2008 [DOI] [PubMed] [Google Scholar]
- [2].Postea O, Biel M. Exploring HCN channels as novel drug targets. Nat Revi Drug Discov 2011; 10:903-14; PMID:22094868 [DOI] [PubMed] [Google Scholar]
- [3].Baruscotti M, Bucchi A, DiFrancesco D. Physiology and pharmacology of the cardiac pacemaker (“funny”) current. Pharmacol Ther 2005; 107:59-79; https://doi.org/ 10.1016/j.pharmthera.2005.01.005 [DOI] [PubMed] [Google Scholar]
- [4].Herrmann S, Schnorr S, Ludwig A. HCN Channels—modulators of cardiac and neuronal excitability. Int J Mol Sci 2015; 16:1429-47; PMID:25580535; https://doi.org/ 10.3390/ijms16011429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bucchi A, Barbuti A, DiFrancesco D. Funny current and cardiac rhythm: insights from HCN knockout and transgenic mouse models. Front Physiol 2012; 49-58; PMID:22457651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Accili E, Proenza C, Baruscotti M, DiFrancesco D. From funny current to HCN channels: 20 years of excitation. Physiology 2002; 17:32-7 [DOI] [PubMed] [Google Scholar]
- [7].Cerbai E, Pino R, Sartiani L, Mugelli A. Influence of postnatal-development on If occurrence and properties in neonatal rat ventricular myocytes. Cardiovasc Res 1999; 42:416-23; PMID:10533577; https://doi.org/ 10.1016/S0008-6363(99)00037-1 [DOI] [PubMed] [Google Scholar]
- [8].Robinson R, Yu H, Chang F, Cohen IS. Developmental change in the voltage-dependence of the pacemaker current, if, in rat ventricle cells. Pflügers Arch 1997; 433:533-5; PMID:9000433; https://doi.org/ 10.1007/s004240050309 [DOI] [PubMed] [Google Scholar]
- [9].Yasui K, Liu W, Opthof T, Kada K, Lee JK, Kamiya K, Kodama I. If current and spontaneous activity in mouse embryonic ventricular myocytes. Circ Res 2001; 88:536-42; PMID:11249878; https://doi.org/ 10.1161/01.RES.88.5.536 [DOI] [PubMed] [Google Scholar]
- [10].Vicente-Steijn R, Passier R, Wisse LJ, Schalij MJ, Poelmann RE, Gittenberger-de Groot AC, Jongbloed MR. Funny current channel HCN4 delineates the developing cardiac conduction system in chicken heart. Heart Rhythm 2011; 8:1254-63; PMID:21421080; https://doi.org/ 10.1016/j.hrthm.2011.03.043 [DOI] [PubMed] [Google Scholar]
- [11].Garcia-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expr Patterns 2003; 3:777-83; PMID:14643687; https://doi.org/ 10.1016/S1567-133X(03)00125-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liang X, Evans SM, Sun Y. Insights into cardiac conduction system formation provided by HCN4 expression. Trends Cardiovasc Med 2015; 25:1-9; PMID:25442735; https://doi.org/ 10.1016/j.tcm.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y, Evans SM. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res 2013; 113:399-407; PMID:23743334; https://doi.org/ 10.1161/CIRCRESAHA.113.301588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stieber J, Herrmann S, Feil S, Löster J. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci 2003; 100:15235-40; https://doi.org/ 10.1073/pnas.2434235100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Harzheim D, Pfeiffer KH, Fabritz L, Kremmer E, Buch T, Waisman A, Kirchhof P, Kaupp UB, Seifert R. Cardiac pacemaker function of HCN4 channels in mice is confined to embryonic development and requires cyclic AMP. EMBO J 2008; 27:692-703; PMID:18219271; https://doi.org/ 10.1038/emboj.2008.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Afouda BA, Hoppler S. Xenopus explants as an experimental model system for studying heart development. Trends Cardiovasc Med 2009; 19:220-6; PMID:20382345; https://doi.org/ 10.1016/j.tcm.2010.01.001 [DOI] [PubMed] [Google Scholar]
- [17].Duncan AR, Khokha MK Xenopus as a model organism for birth defects—Congenital heart disease and heterotaxy. Semin Cell Dev Biol 2016; 51:73-79; https://doi.org/ 10.1016/j.semcdb.2016.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mohun T, Leong L, Weninger W, Sparrow D. The morphology of heart development in Xenopus laevis. Dev Biol 2000; 218:74-88; PMID:10644412; https://doi.org/ 10.1006/dbio.1999.9559 [DOI] [PubMed] [Google Scholar]
- [19].Kolker SJ, Tajchman U, Weeks DL. Confocal imaging of early heart development in Xenopus laevis. Dev Biol 2000; 218:64-73; PMID:10644411; https://doi.org/ 10.1006/dbio.1999.9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Karpinka JB, Fortriede JD, Burns KA, James-Zorn C, Ponferrada VG, Lee J, Karimi K, Zorn AM, Vize PD. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res 2015; 43:D756-63; PMID:25313157; https://doi.org/ 10.1093/nar/gku956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mohun T, Orford R, Shang C. The origins of cardiac tissue in the amphibian, Xenopus laevis. Trends Cardiovasc Med 2003; 13:244-8; PMID:12922021; https://doi.org/ 10.1016/S1050-1738(03)00102-6 [DOI] [PubMed] [Google Scholar]
- [22].Warkman AS, Krieg PA. Xenopus as a model system for vertebrate heart development. Seminars Cell Dev Biol 2007; 18:46-53; https://doi.org/ 10.1016/j.semcdb.2006.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol 2003; 258:1-19; PMID:12781678; https://doi.org/ 10.1016/S0012-1606(03)00112-X [DOI] [PubMed] [Google Scholar]
- [24].Pitcairn E, McLaughlin KA. Bioelectric signaling coordinates patterning decisions during embryogenesis. Trends Dev Biol 2016; 9:1-9 [Google Scholar]
- [25].Levin M. Molecular bioelectricity in developmental biology: new tools and recent discoveries: control of cell behavior and pattern formation by transmembrane potential gradients. BioEssays 2012; 34:205-17; PMID:22237730; https://doi.org/ 10.1002/bies.201100136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Levin M, Stevenson C. Regulation of cell behavior and tissue patterning by bioelectrical signals: challenges and opportunities for biomedical engineering. Annu Rev Biomed Eng 2012; 14:295-323; PMID:22809139; https://doi.org/ 10.1146/annurev-bioeng-071811-150114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Levin M. Large-scale biophysics: ion flows and regeneration. Trends Cell Biol 2007; 17:261-70; PMID:17498955; https://doi.org/ 10.1016/j.tcb.2007.04.007 [DOI] [PubMed] [Google Scholar]
- [28].Funk RH, Thiede C. Ion gradients and electric fields-an intrinsic part of biological processes. J Clin Exp Oncol 2014; S1, 004; https://doi.org/ [DOI] [Google Scholar]
- [29].Levin M. Endogenous bioelectrical networks store non-genetic patterning information during development and regeneration. J Physiol 2014; 592:2295-305; PMID:24882814; https://doi.org/ 10.1113/jphysiol.2014.271940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Perathoner S, Daane JM, Henrion U, Seebohm G, Higdon CW, Johnson SL, Nüsslein-Volhard C, Harris MP. Bioelectric signaling regulates size in zebrafish fins. PLoS Genet 2014; 10:e1004080; PMID:24453984; https://doi.org/ 10.1371/journal.pgen.1004080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pai VP, Lemire JM, Paré JF, Lin G, Chen Y, Levin M. Endogenous gradients of resting potential instructively pattern embryonic neural tissue via notch signaling and regulation of proliferation. J Neurosci 2015; 35:4366-85; PMID:25762681; https://doi.org/ 10.1523/JNEUROSCI.1877-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pai VP, Aw S, Shomrat T, Lemire JM, Levin M. Transmembrane voltage potential controls embryonic eye patterning in Xenopus laevis. Development 2012; 139:313-323; PMID:22159581; https://doi.org/ 10.1242/dev.073759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shu X, Cheng K, Patel N, Chen F, Joseph E. Na, K-ATPase is essential for embryonic heart development in the zebrafish. Development 2003; 130:6165-73; PMID:14602677; https://doi.org/ 10.1242/dev.00844 [DOI] [PubMed] [Google Scholar]
- [34].Yuan S. The small heart mutation reveals novel roles of Na+/K+-ATPase in maintaining ventricular cardiomyocyte morphology and viability in zebrafish. Circ Res 2004; 95:595-603; PMID:15297381; https://doi.org/ 10.1161/01.RES.0000141529.48143.6e [DOI] [PubMed] [Google Scholar]
- [35].Chong JX, McMillin MJ, Shively KM, Beck AE, Marvin CT, Armenteros JR, Buckingham KJ, Nkinsi NT, Boyle EA, Berry MN, et al.. De novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia, and developmental delay. Am J Hum Genet 2015; 96:462-73; PMID:25683120; https://doi.org/ 10.1016/j.ajhg.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Barel O, Shalev SA, Ofir R, Cohen A, Zlotogora J, Shorer Z, Mazor G, Finer G, Khateeb S, Zilberberg N, et al.. Maternally inherited Birk Barel mental retardation dysmorphism syndrome caused by a mutation in the genomically imprinted potassium channel KCNK9. Am J Hum Genet 2008; 83:193-9; PMID:18678320; https://doi.org/ 10.1016/j.ajhg.2008.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Masotti A, Uva P, Davis-Keppen L. Keppen-lubinsky syndrome is caused by mutations in the inwardly rectifying K+ channel encoded by KCNJ6. Am J Hum Genet 2015; 96:295-300; PMID:25620207; https://doi.org/ 10.1016/j.ajhg.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tristani-Firouzi M, Etheridge SP. Kir 2.1 channelopathies: the Andersen–Tawil syndrome. Pflügers Archiv-Euro J Physiol 2010; 460:289-94; PMID:20306271; https://doi.org/ 10.1007/s00424-010-0820-6 [DOI] [PubMed] [Google Scholar]
- [39].Levin M. Molecular bioelectricity: how endogenous voltage potentials control cell behavior and instruct pattern regulation in vivo. Mol Biol Cell 2014; 25:3835-50; PMID:25425556; https://doi.org/ 10.1091/mbc.E13-12-0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bates E. Ion channels in development and cancer. Annu Rev Cell Dev Biol 2015; 31:231-47; PMID:26566112; https://doi.org/ 10.1146/annurev-cellbio-100814-125338 [DOI] [PubMed] [Google Scholar]
- [41].Lee YH, Saint-Jeannet JP. Cardiac neural crest is dispensable for outflow tract septation in Xenopus. Development 2011; 138:2025-34; PMID:21490068; https://doi.org/ 10.1242/dev.061614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tandon P, Miteva YV, Kuchenbrod LM, Cristea IM, Conlon FL. Tcf21 regulates the specification and maturation of proepicardial cells. Development 2013; 140:2409-21; PMID:23637334; https://doi.org/ 10.1242/dev.093385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Shi W, Wymore R, Yu H, Wu J, Wymore RT, Pan Z, Robinson RB, Dixon JE, McKinnon D, Cohen IS. Distribution and prevalence of hyperpolarization-activated cation channel (HCN) mRNA expression in cardiac tissues. Circ Res 1999; 85:e1-6; PMID:10400919; https://doi.org/ 10.1161/01.RES.85.1.e1 [DOI] [PubMed] [Google Scholar]
- [44].Ueda K, Nakamura K, Hayashi T, Inagaki N, Takahashi M, Arimura T, Morita H, Higashiuesato Y, Hirano Y, Yasunami M, et al.. Functional characterization of a trafficking-defective HCN4 mutation, D553N, associated with cardiac arrhythmia. J Biol Chem 2004; 279:27194-8; PMID:15123648; https://doi.org/ 10.1074/jbc.M311953200 [DOI] [PubMed] [Google Scholar]
- [45].Nof E, Luria D, Brass D, Marek D, Lahat H, Reznik-Wolf H, Pras E, Dascal N, Eldar M, Glikson M. Point mutation in the HCN4 cardiac ion channel pore affecting synthesis, trafficking, and functional expression is associated with familial asymptomatic sinus bradycardia. Circulation 2007; 116:463-70; PMID:17646576; https://doi.org/ 10.1161/CIRCULATIONAHA.107.706887 [DOI] [PubMed] [Google Scholar]
- [46].Xue T, Marbán E, Li RA. Dominant-negative suppression of HCN1- and HCN2-encoded pacemaker currents by an engineered HCN1 construct: insights into structure-function relationships and multimerization. Circ Res 2002; 90:1267-73; PMID:12089064; https://doi.org/ 10.1161/01.RES.0000024390.97889.C6 [DOI] [PubMed] [Google Scholar]
- [47].Smith J, Price B, Green J, Weigel D, Herrmann B. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell 1991; 67:79-87; PMID:1717160; https://doi.org/ 10.1016/0092-8674(91)90573-H [DOI] [PubMed] [Google Scholar]
- [48].Pannese M, Polo C, Andreazzoli M, Vignali R, Kablar B, Barsacchi G, Boncinelli E. The Xenopus homologue of Otx2 is a maternal homeobox gene that demarcates and specifies anterior body regions. Development 1995; 121:707-20; PMID:7720578 [DOI] [PubMed] [Google Scholar]
- [49].Foley AC, Korol O, Timmer AM, Mercola M. Multiple functions of Cerberus cooperate to induce heart downstream of Nodal. Dev Biol 2007; 303:57-65; PMID:17123501; https://doi.org/ 10.1016/j.ydbio.2006.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sampath K, Cheng A, Frisch A, Wright C. Functional differences among Xenopus nodal-related genes in left-right axis determination. Development 1997; 124:3293-302; PMID:9310324 [DOI] [PubMed] [Google Scholar]
- [51].Lohr JL, Danos MC, Yost HJ. Left-right asymmetry of a nodal-related gene is regulated by dorsoanterior midline structures during Xenopus development. Dev Suppl 1997; 124:1465-72 [DOI] [PubMed] [Google Scholar]
- [52].Wagner M, Siddiqui MA. Signal transduction in early heart development (I): cardiogenic induction and heart tube formation. Exp Biol Med 1997; 232:852-65 [PubMed] [Google Scholar]
- [53].Branford WW, Essner JJ, Yost HJ. Regulation of gut and heart left–right asymmetry by context-dependent interactions between Xenopus lefty and BMP4 signaling. Dev Biol 2000; 223:291-306; PMID:10882517; https://doi.org/ 10.1006/dbio.2000.9739 [DOI] [PubMed] [Google Scholar]
- [54].Cheng A, Thisse B, Thisse C, Wright C. The lefty-related factor Xatv acts as a feedback inhibitor of nodal signaling in mesoderm induction and LR axis development in xenopus. Development 2000; 127:1049-61; PMID:10662644 [DOI] [PubMed] [Google Scholar]
- [55].Chang H, Zwijsen A, Vogel H, Huylebroeck D, Matzuk MM. Smad5 is essential for left–right asymmetry in mice. Dev Biol 2000; 219:71-8; PMID:10677256; https://doi.org/ 10.1006/dbio.1999.9594 [DOI] [PubMed] [Google Scholar]
- [56].Breckenridge RA, Mohun TJ, Amaya E. A role for BMP signalling in heart looping morphogenesis in Xenopus. Dev Biol 2001; 232:191-203; PMID:11254357; https://doi.org/ 10.1006/dbio.2001.0164 [DOI] [PubMed] [Google Scholar]
- [57].Chocron S, Verhoeven MC, Rentzsch F, Hammerschmidt M, Bakkers J. Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev Biol 2007; 305:577-88; PMID:17395172; https://doi.org/ 10.1016/j.ydbio.2007.03.001 [DOI] [PubMed] [Google Scholar]
- [58].Chen JN, van Eeden FJ, Warren KS, Chin A, Nüsslein-Volhard C, Haffter P, Fishman MC. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 1997; 124:4373-82; PMID:9334285 [DOI] [PubMed] [Google Scholar]
- [59].Ai D, Liu W, Ma L, Dong F, Lu MF, Wang D, Verzi MP, Cai C, Gage PJ, Evans S, et al.. Pitx2 regulates cardiac left–right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol 2006; 296:437-49; PMID:16836994; https://doi.org/ 10.1016/j.ydbio.2006.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shiratori H, Yashiro K, Shen MM, Hamada H. Conserved regulation and role of Pitx2 in situs-specific morphogenesis of visceral organs. Development 2006; 133:3015-25; PMID:16835440; https://doi.org/ 10.1242/dev.02470 [DOI] [PubMed] [Google Scholar]
- [61].Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, et al.. The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development 1999; 126:1225-34; PMID:10021341 [DOI] [PubMed] [Google Scholar]
- [62].Dagle JM, Sabel JL, Littig JL, Sutherland LB, Kolker SJ, Weeks DL. Pitx2c attenuation results in cardiac defects and abnormalities of intestinal orientation in developing Xenopus laevis. Dev Biol 2003; 262:268-81; PMID:14550790; https://doi.org/ 10.1016/S0012-1606(03)00389-0 [DOI] [PubMed] [Google Scholar]
- [63].Shi Y, Katsev S, Cai C, Evans S. BMP signaling is required for heart formation in vertebrates. Dev Biol 2000; 224:226-37; PMID:10926762; https://doi.org/ 10.1006/dbio.2000.9802 [DOI] [PubMed] [Google Scholar]
- [64].Danos MC, Yost HJ. Linkage of Cardiac Left-Right Asymmetry and Dorsal-Anterior Development in Xenopus. Development 1995; 121:1467-74; PMID:7789276 [DOI] [PubMed] [Google Scholar]
- [65].Peyrot SM, Wallingford JB, Harland RM. A revised model of Xenopus dorsal midline development: differential and separable requirements for Notch and Shh signaling. Dev Biol 2011; 352:254-66; PMID:21276789; https://doi.org/ 10.1016/j.ydbio.2011.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sampath K, Cheng AM, Frisch A, Wright CV. Functional differences among Xenopus nodal-related genes in left-right axis determination. Development 1997; 124:3293-302; PMID:9310324 [DOI] [PubMed] [Google Scholar]
- [67].Essner JJ, Branford WW, Zhang J, Yost HJ. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development 2000; 127:1081-93; PMID:10662647 [DOI] [PubMed] [Google Scholar]
- [68].Duhme N, Schweizer PA, Thomas D, Becker R, Schröter J, Barends TR, Schlichting I, Draguhn A, Bruehl C, Katus HA, et al.. Altered HCN4 channel C-linker interaction is associated with familial tachycardia–bradycardia syndrome and atrial fibrillation. Euro Heart J 2013; 34:2768-75; PMID:23178648; https://doi.org/ 10.1093/eurheartj/ehs391 [DOI] [PubMed] [Google Scholar]
- [69].McDowell GS, Rajadurai S, Levin M. From cytoskeletal dynamics to organ asymmetry: a non-linear, regulative pathway underlies left-right patterning. bioRxiv 2016; 371(1710):pii: 20150409; 052191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dahal GJ, Rawson J, Gassaway B, Kwok B, Tong Y, Ptácek LJ, Bates E. An inwardly rectifying K+ channel is required for patterning. Development 2012; 139:3653-64; PMID:22949619; https://doi.org/ 10.1242/dev.078592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bates EA. A potential molecular target for morphological defects of fetal alcohol syndrome: Kir2. 1. Curr Opin Genet Dev 2013; 23:324-9; https://doi.org/ 10.1016/j.gde.2013.05.001 [DOI] [PubMed] [Google Scholar]
- [72].Adams DS, Uzel SG, Akagi J, Wlodkowic D, Andreeva V, Yelick PC, Devitt-Lee A, Pare JF, Levin M. Bioelectric signalling via potassium channels: a mechanism for craniofacial dysmorphogenesis in KCNJ2-associated Andersen-Tawil Syndrome. J Physiol 2016; 594(12):3245-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Arcangeli A, Becchetti A. Complex functional interaction between integrin receptors and ion channels. Trends Cell Biol 2006; 16:631-9; PMID:17064899; https://doi.org/ 10.1016/j.tcb.2006.10.003 [DOI] [PubMed] [Google Scholar]
- [74].Greene D, Kang S, Kosenko A, Hoshi N. Adrenergic regulation of HCN4 channel requires protein association with β2-adrenergic receptor. J Biol Chem 2012; 287:23690-7; PMID:22613709; https://doi.org/ 10.1074/jbc.M112.366955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Paine-Saunders S, Viviano BL, Economides AN, Saunders S. Heparan sulfate proteoglycans retain Noggin at the cell surface a potential mechanism for shaping bone morphogenetic protein gradients. J Biol Chem 2002; 277:2089-96; PMID:11706034; https://doi.org/ 10.1074/jbc.M109151200 [DOI] [PubMed] [Google Scholar]
- [76].Raya A, Kawakami Y, Rodríguez-Esteban C, Ibañes M, Rasskin-Gutman D, Rodríguez-León J, Büscher D, Feijó JA, Izpisúa Belmonte JC. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature 2004; 427:121-8; PMID:14712268; https://doi.org/ 10.1038/nature02190 [DOI] [PubMed] [Google Scholar]
- [77].Pai VP, Martyniuk CJ, Echeverri K, Sundelacruz S, Kaplan DL, Levin M. Genome-wide analysis reveals conserved transcriptional responses downstream of resting potential change in Xenopus embryos, axolotl regeneration, and human mesenchymal cell differentiation. Regeneration 2015; 3(1):3-25; PMID:27499876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tseng AS, Levin M. Transducing bioelectric signals into epigenetic pathways during tadpole tail regeneration. Anat Rec 2012; 295:1541-51; https://doi.org/ 10.1002/ar.22495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Pai VP, Martyniuk CJ, Echeverri K, Sundelacruz S, Kaplan DL, Levin M. Genome-wide analysis reveals conserved transcriptional responses downstream of resting potential change in Xenopus embryos, axolotl regeneration, and human mesenchymal cell differentiation. Regeneration (Oxf) 2016; 3:3-25; PMID:27499876; https://doi.org/ 10.1002/reg2.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Spencer AD, Lemire JM, Kramer RH, Levin M. Optogenetics in Developmental Biology: using light to control ion flux-dependent signals in Xenopus embryos. Int J Dev Biol 2014; 58:851-61; PMID:25896279; https://doi.org/ 10.1387/ijdb.140207ml [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Niewkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin: ). (Garland Publishing, Inc., 1994) [Google Scholar]
- [82].Kuzhikandathil EV, Oxford GS. Dominant-negative mutants identify a role for GIRK channels in D3 dopamine receptor-mediated regulation of spontaneous secretory activity. J Gen Physiol 2000; 115:697-706; PMID:10828244; https://doi.org/ 10.1085/jgp.115.6.697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Preisig-Müller R, Schlichthörl G, Goerge T, Heinen S, Brüggemann A, Rajan S, Derst C, Veh RW, Daut J. Heteromerization of Kir2.x potassium channels contributes to the phenotype of Andersen's syndrome. Proc Natl Acad Sci U S A 2002; 99:7774-9; PMID:12032359; https://doi.org/ 10.1073/pnas.102609499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ye B, Valdivia CR, Ackerman MJ, Makielski JC. A common human SCN5A polymorphism modifies expression of an arrhythmia causing mutation. Physiol Genomics 2003; 12:187-93; PMID:12454206; https://doi.org/ 10.1152/physiolgenomics.00117.2002 [DOI] [PubMed] [Google Scholar]
- [85].Ye B, Balijepalli RC, Foell JD, Kroboth S, Ye Q, Luo YH, Shi NQ. Caveolin-3 associates with and affects the function of hyperpolarization-activated cyclic nucleotide-gated channel 4. Biochemistry 2008; 47:12312-8; PMID:19238754; https://doi.org/ 10.1021/bi8009295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ye B, Nerbonne JM. Proteolytic processing of HCN2 and co-assembly with HCN4 in the generation of cardiac pacemaker channels. J Biol Chem 2009; 284:25553-9; PMID:19574228; https://doi.org/ 10.1074/jbc.M109.007583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Harland R. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol 1991; 36:685-95; PMID:1811161 [DOI] [PubMed] [Google Scholar]
- [88].Dale L, Howes G, Price BM, Smith JC. Bone morphogenetic protein 4: a ventralizing factor in early Xenopus development. Development 1992; 115:573-85; PMID:1425340 [DOI] [PubMed] [Google Scholar]
- [89].Meno C, Ito Y, Saijoh Y, Matsuda Y, Tashiro K, Kuhara S, Hamada H. Two closely-related left-right asymmetrically expressed genes, lefty-1 and lefty-2: their distinct expression domains, chromosomal linkage and direct neuralizing activity in Xenopus embryos. Genes Cells 1997; 2:513-24; https://doi.org/ 10.1046/j.1365-2443.1997.1400338.x [DOI] [PubMed] [Google Scholar]
- [90].Roelink H, Augsburger A, Heemskerk J, Korzh V, Norlin S, Ruiz i Altaba A, Tanabe Y, Placzek M, Edlund T, Jessell TM, et al.. Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell 1994; 76:761-75; PMID:8124714; https://doi.org/ 10.1016/0092-8674(94)90514-2 [DOI] [PubMed] [Google Scholar]
- [91].Kelly OG, Melton DA. Development of the pancreas in Xenopus laevis. Dev Dyn 2000; 218:615-27; PMID:10906780; https://doi.org/ 10.1002/1097-0177(2000)9999:9999%3c::AID-DVDY1027%3e3.0.CO;2-8 [DOI] [PubMed] [Google Scholar]
- [92].Team RC. R: A language and environment for statistical computing. 2013; https://CRAN.R-project.org/doc/FAQ/R-FAQ.html [Google Scholar]
- [93].Fife D. Fife package. R Package Version 1.0 2015; https://cran.r-project.org/web/packages/fifer/fifer.pdf [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.