ABSTRACT
In their natural environment, plants have to continuously face constraints such as biotic and abiotic stresses. To achieve their life cycle, plants have to perceive and interpret the nature, but also the strength of environmental stimuli to activate appropriate physiological responses. Nowadays, it is well established that signaling pathways are crucial steps in the implementation of rapid and efficient plant responses such as genetic reprogramming. It is also reported that rapid raises in calcium (Ca2+) levels within plant cells participate in these early signaling steps and are essential to coordinate adaptive responses. However, to be informative, calcium increases need to be decoded and relayed by calcium-binding proteins also referred as calcium sensors to carry-out the appropriate responses. In a recent study, we showed that CML8, an Arabidopsis calcium sensor belonging to the calmodulin-like (CML) protein family, promotes plant immunity against the phytopathogenic bacteria Pseudomonas syringae pv tomato (strain DC3000). Interestingly, other CML proteins such as CML9 were also reported to contribute to plant immunity using the same pathosystem. In this addendum, we propose to discuss about the specific contribution of these 2 CMLs in stress responses.
KEYWORDS: Arabidopsis thaliana, biotic and abiotic stress responses, calmodulin-like protein, calcium sensor, pseudomonas syringae
It is now well documented that most of external biotic and abiotic stimuli induce a rapid increase in free calcium levels within plant cells.1 Calcium is considered as an ubiquitous and versatile second messenger. Indeed, calcium variations are at the heart of sophisticated signaling networks, integrating information from a diverse range of developmental cues and environmental challenges that ultimately will impact gene expression and cell physiology.2 Interestingly, these variations can harbour stimulus-specific properties3 and according to the nature and strength of the stimuli, calcium variations display differences in frequencies, amplitudes, cellular localization and they were defined as «Ca2+ signature».4 These Ca2+ signatures have been proven to encode a first layer of response specificity through their spatio-temporal properties.3,5 A second layer of specificity is brought by the decoding processes requiring calcium-binding proteins also referred as calcium sensors and their downstream targets. Ca2+ sensors are characterized by the presence in their sequence of the canonical Ca2+-binding motif called EF-hand.6 The Arabidopsis genome encodes at least 250 EF-hand proteins, which is much higher than in mammals, arguing for higher importance of this class of proteins in plants.7 These Ca2+ sensors are classified into 4 major groups, the calcineurin-B like, the calcium-dependent kinases, the calmodulin group and a closely related group, the CalModulin-Like protein (CML) family, which is specific to plants.8 In 2017, we reported that the ectopic CML8 expression (P35S:: cds CML8) confers to transgenic plants a better resistance to a virulent phytopathogenic bacteria (P. syringae pv tomato, Pst) compared with wild type (Col8) or the cml8 knock-down and knockout lines.9 Collectively, our results support the idea that CML8 acts as a positive regulator of plant immunity. To better understand the molecular processes related to CML8, we used Pst strains unable to inject effectors into the plant host cells or deficient for some effectors known to target the salicylic acid (SA) defense pathway. Our results led us to propose a role for CML8 in SA-dependent processes probably by modulating the effect of bacterial effectors rather than through PAMP-Triggered Immunity (PTI). Interestingly, we demonstrated in a previous work, that CML9, another Arabidopsis CML that belongs to the same subgroup than CML8 (Fig. 1A) contributes, as CML8, to positively regulate plant immunity in response to Pst inoculation.10 Nevertheless, in this case, we show that CML9 contributes to PTI (via a flagellin dependent pathway) but also to SA-dependent processes.10 Interestingly, CML9 acts as a multifunctional Ca2+ sensor since this CML is also involved, as a negative regulator, in drought and salt stress responses in plant.11 The involvement of CML8 was also evaluated in response to abiotic stress treatments but in this case, we showed that CML8 does not act as a main regulator of plant responses to salinity.9 Thus, these functional analyses clearly indicate that 2 closely related CMLs (Fig. 1A) might participate to distinct signaling pathways as positive regulators of plant defense and act as a negative regulator (i.e. CML9) in abiotic stresses (Fig. 1B). To date, the roles of most of the CMLs remain mainly unknown although recent studies using the model plant Arabidopsis thaliana have pointed out the involvement for few of them in physiological processes associated with stress (biotic and abiotic) responses.12 Interestingly, CML24, classified into a more divergent subgroup than CML8 and 9, was reported to take part in abiotic stress responses13 as well as in plant immune responses following Pst inoculation. However, it is interesting to note that compared with CML8 and CML9, the contribution of CML24 in response to Pst is linked both to innate immunity and hypersensitive responses.14 The Arabidopsis CML42 and CML37 were also reported to antagonistically contribute to plant defense and drought response. While CML42 acts as a negative regulator of plant defense against herbivorous insects, CML37 acts as a positive regulator.15-17 In response to drought stress, CML37 and CML42 also act as positive and negative regulators respectively.15,16
Figure 1.
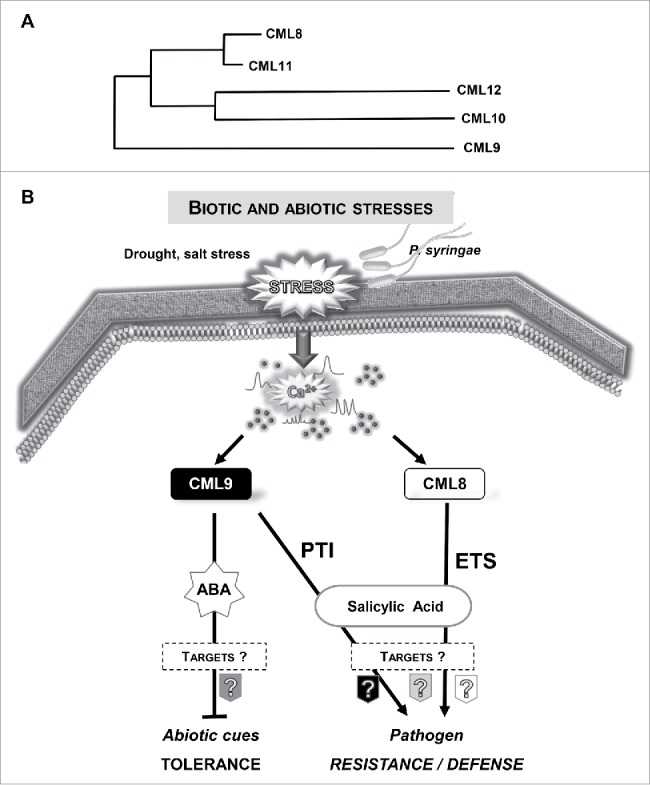
(A) Phylogenetic tree built with protein sequences from CML8, CML9, CML10, CML11 and CML12. The tree was obtained using Phylogeny program. (B) Model for CML8 and CML9 functions in response to biotic and abiotic stresses. Environmental signals can trigger Ca2+ increases that are relayed and decoded by calcium sensors such as CMLs. CML9 is involved as a negative regulator in drought and salt stresses by acting in an ABA dependent pathway11 and as a positive regulator in plant innate immunity (PTI) against P. syringae through a flagellin-dependent signaling pathway.10 CML8 positively regulates P. syringae defense responses but, contrary to CML9, CML8 could be involved in effector triggered susceptibility (ETS) through a SA dependent pathway.9 Moreover, CML8 do not act as a key actor in salt stress responses.9
Collectively, all these data highlight the complexity of calcium signaling in plant and support the idea that CMLs could be at the crossroads of plant biotic and abiotic stresses signaling pathways. CMLs can exhibit a dual function as either positive and/or negative regulators in plant stresses and thus, might be involved in the fine tuning of plant physiological responses to pathogens under fluctuating parameters of the environment (i.e., temperature, water availability). If these proteins can be considered as multifunctional proteins, this is certainly due to the diversity of their target repertoire. Indeed, CMLs do not have intrinsic activity by themselves but act by interacting with downstream targets to modulate their activities that embrace a broad spectrum of functions such as the regulation of transcription, transport, trafficking, enzyme activities and phosphorylation events. Only few targets have been identified and characterized so far for CMLs but to our knowledge, only a single study was performed to identify CML interacting proteins using a systematic approach.18 Current data obtained clearly show that the emergent CML's target repertoire is more extensive than previously predicted. Concerning CML9 and CML8, we identified PRR2 as a common target.19 PRR2 is a pseudo-response regulator containing several features that are typical of plant transcription factors but its function in biotic and/or abiotic stress responses is still under investigation.
CML8 and CML9 belong to the same CML subgroup in Arabidopsis as well as 3 others CMLs (CML10, CML11 and CML12) (Fig. 1A). Whereas the role of these 5 proteins is not fully characterized, our data indicate that there is probably not a complete functional redundancy between these proteins. Indeed, thanks to the global expression data and transgenic reporter lines, we observed that the spatio-temporal gene expression patterns of these CMLs are not similar. At the protein level these CMLs also exhibit some specificities. CML10 and CML12 encompass 4 and 6 EF-hand domains respectively and contains 191 and 324 amino acids whereas others exhibit an average of 150 amino acids with only 4 EF-hand motifs. These differences at protein level might be important both in the ability to sense different Ca2+ signatures, through EF-hands affinity but also to specifically interact with partners. Thus, CML10 was reported to interact with a phosphomannomutase, an enzyme able to modulate ascorbic acid levels20 but not with PRR2, a common target of CML8 and 9.19
Concluding remarks
In 2015, to further understand the function of CMLs in plant, we exploited plant genome sequencing data to analyze CaM and CML evolution among the green lineage.8 The results show that the abundance of CaMs and CMLs evolved during terrestrial colonization of plants and that the emergence of new CML classes appear throughout the green lineage that correlate with land colonization by plants and with the acquisition of novel biologic traits. We can then speculate that each CMLs may be associated to specific functions during plant development and stress response as it was the case for CML8 and CML9.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
Many thanks to C. Mazars for critical reading of the manuscript.
Funding
This work was supported by the Universite Paul Sabatier (Toulouse, France), the CNRS (France) and by the French Laboratory of Excellence project “TULIP” (ANR-10-LABX-41; ANR-11-IDEX-0002–02). Funding for this work was provided by the China Scholarship Council (CSC) and by the French Ministry of Education and Research to XZ and MP respectively.
References
- 1.Kudla J, Batistic O, Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell 2010; 22:541-63. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2861448; PMID:20354197; https://doi.org/ 10.1105/tpc.109.072686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodd AN, Kudla J, Sanders D. The language of calcium signaling. Annu Rev Plant Biol 2010; 61:593-620; PMID:20192754; https://doi.org/ 10.1146/annurev-arplant-070109-104628 [DOI] [PubMed] [Google Scholar]
- 3.McAinsh MR, Pittman J. Shaping the calcium signature. New Phytol 2009; 181:275-94. http://onlinelibrary.wiley.com/doi/10.1111/; PMID:19121028; https://doi.org/ 10.1111/j.1469-8137.2008.02682.x [DOI] [PubMed] [Google Scholar]
- 4.McAinsh MR, Hetherington A. Encoding specificity in Ca2+ signalling systems. Trends Plant Sci 1998; 3:32-6. http://www.sciencedirect.com/science/article/pii/S1360138597011503; https://doi.org/ 10.1016/S1360-1385(97)01150-3 [DOI] [Google Scholar]
- 5.Ng CK, McAinsh MR. Encoding specificity in plant calcium signalling: Hot-spotting the ups and downs and waves. Ann Bot 2003; 92:477-85. https://academic.oup.com/aob/article/92/4/477/223078/; PMID:12933365; https://doi.org/ 10.1093/aob/mcg173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakayama S, Kretsinger R. Evolution of the EF-Hand family of proteins. Annu Rev Biophys Biomol Struct 1994; 23:473-507. http://www.annualreviews.org/doi/abs/10.1146/; PMID:7919790; https://doi.org/ 10.1146/annurev.bb.23.060194.002353 [DOI] [PubMed] [Google Scholar]
- 7.Day IS, Reddy VS, Shad Ali G, Reddy AS. Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol 2002; 3:RESEARCH0056. https://genomebiology.biomedcentral.com/articles/10.1186/; PMID:12372144; https://doi.org/ 10.1186/gb-2002-3-10-research0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu X, Dunand C, Snedden W, Galaud JP. CaM and CML emergence in the green lineage. Trends Plant Sci 2015; 20:483-9. http://www.ncbi.nlm.nih.gov/pubmed/26115779; PMID:26115779; https://doi.org/ 10.1016/j.tplants.2015.05.010 [DOI] [PubMed] [Google Scholar]
- 9.Zhu X, Robe E, Jomat L, Aldon D, Mazars C, Galaud JP. CML8, an Arabidopsis calmodulin-like protein, plays a role in Pseudomonas syringae plant immunity. Plant Cell Physiol 2017; 58:307-19. http://www.ncbi.nlm.nih.gov/pubmed/27837097; PMID:27837097; https://doi.org/ 10.1093/pcp/pcw189 [DOI] [PubMed] [Google Scholar]
- 10.Leba LJ, Cheval C, Ortiz-Martín I, Ranty B, Beuzón CR, Galaud JP, Aldon D. CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J 2012; 71:976-89. http://www.ncbi.nlm.nih.gov/pubmed/22563930; PMID:22563930; https://doi.org/ 10.1111/j.1365-313X.2012.05045.x [DOI] [PubMed] [Google Scholar]
- 11.Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud JP, Aldon D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J 2008; 56:575-89. http://www.ncbi.nlm.nih.gov/pubmed/18643966; PMID:18643966; https://doi.org/ 10.1111/j.1365-313X.2008.03622.x [DOI] [PubMed] [Google Scholar]
- 12.Bender KW, Snedden WA. Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol 2013; 163:486-95. http://www.plantphysiol.org/cgi/doi/10.1104/pp.113.221069; PMID:23908390; https://doi.org/ 10.1104/pp.113.221069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delk NA, Johnson KA, Chowdhury NI, Braam J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol 2005; 139:240-53. http://www.plantphysiol.org/content/139/1/240; PMID:16113225; https://doi.org/ 10.1104/pp.105.062612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma W, Smigel A, Tsai YC, Braam J, Berkowitz GA. Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol 2008; 148:818-28. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid = 2556846; PMID:18689446; https://doi.org/ 10.1104/pp.108.125104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholz SS, Vadassery J, Heyer M, Reichelt M, Bender KW, Snedden WA, Boland W, Mithöfer A. Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Mol Plant 2014; 7:1712-26. https://doi.org/10.1093/mp/ssu102; PMID:25267731; https://doi.org/ 10.1093/mp/ssu102 [DOI] [PubMed] [Google Scholar]
- 16.Vadassery J, Reichelt M, Hause B, Gershenzon J, Boland W, Mithofer A. CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol 2012; 159:1159-75. http://www.plantphysiol.org/content/159/3/1159; PMID:22570470; https://doi.org/ 10.1104/pp.112.198150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vadassery J, Scholz SS, Mithöfer A. Multiple calmodulin-like proteins in Arabidopsis are induced by insect-derived (Spodoptera littoralis) oral secretion. Plant Signal Behav 2012; 7:1277-80. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3493413/; PMID:22902684; https://doi.org/ 10.4161/psb.21664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popescu SC, Popescu G V, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA 2007; 104:4730-5. http://www.pnas.org/content/104/11/4730; PMID:17360592; https://doi.org/ 10.1073/pnas.0611615104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perochon A, Dieterle S, Pouzet C, Aldon D, Galaud JP, Ranty B. Interaction of a plant pseudo-response regulator with a calmodulin-like protein. Biochem Biophys Res Commun 2010; 398:747-51. http://www.ncbi.nlm.nih.gov/pubmed/20627089; PMID:20627089; https://doi.org/ 10.1016/j.bbrc.2010.07.016 [DOI] [PubMed] [Google Scholar]
- 20.Cho KM, Nguyen HT, Kim SY, Shin JS, Cho DH, Hong SB, Shin JS, Ok SH. CML10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytol 2016; 209:664-78. http://onlinelibrary.wiley.com/doi/10.1111/; PMID:26315131; https://doi.org/ 10.1111/nph.13612 [DOI] [PubMed] [Google Scholar]


