ABSTRACT
Genetic pathways relevant to flowering of Arabidopsis are under the control of environmental cues such as day length and temperatures, and endogenous signals including phytohormones and developmental aging. However, genes and even regulatory pathways for flowering identified in crops show divergence from those of Arabidopsis and often do not have functional equivalents to Arabidopsis and/or existing species- or genus-specific regulators and show modified or novel pathways.
Orchids are the largest, most highly evolved flowering plants, and form an extremely peculiar group of plants. Here, we briefly summarize the flowering pathways of Arabidopsis, rice and wheat and present them alongside recent discoveries/progress in orchid flowering and flower developmental processes including our transgenic Phalaenopsis orchids for LEAFY overexpression. Potential biotechnological applications in flowering/flower development of orchids with potential target genes are also discussed from an interactional and/or comparative viewpoint.
KEYWORDS: Flower development, flowering, orchid, orchid biotechnology, orchid transformation
Importance of floral transition in orchid breeding
Orchids are a valuable floricultural crop and some wild species are under constant threat of extinction due to over-collection for commercial trade.1 Large and complex polyploid genomes, low transformation efficiency, slow growth and long life cycles mean that it is challenging to overcome the risk of extinction or generate new varieties containing desirable traits with commercial value by either traditional breeding or genetic engineering techniques.2,3 For example, it takes more than 2 y for popular orchid cultivars with high commercial value (e.g., Phalaenopsis orchids) to switch from the vegetative to the reproductive phase.3,4
The structure of orchid flowers has unique diversification among flowering plants. Moreover, most orchids have defined favorable seasons for floral induction as well as inflorescence and flower development. Although several genes affecting flower development have been identified in orchids, the function of the genes during orchid floral transition still remains to be studied through the establishment of reliable protocols to induce alterations in flowering time of orchids.5,6,7 Molecular and genetic studies of orchid flowering are invaluable not only for understanding the molecular mechanisms of flower development including evolutionary trends but also for assisting molecular breeding to produce orchids with desirable traits.
Floral transition in a long-day model plant, Arabidopsis
In Arabidopsis, photoperiodic information for floral transition is specified through the interaction of the circadian clock and light. Both of these factors converge to regulate the expression and the activity of the CONSTANS (CO) transcription factor.8 CO activates SUPPRESSOR OF CONSTANS OVEREXPRESSION1 (SOC1) and APETALA1 (AP1) through FLOWERING LOCUS T (FT) to promote flowering. FT encoding a small protein that belongs to the phosphatidyl-ethanolamine binding protein (PEBP) family is expressed in the leaves and the polypeptide moves to the shoot apical meristem and forms a complex with a bZIP transcription factor, FD.9,10 Subsequently, the floral meristem identity genes such as AP1, LEAFY (LFY), FRUITFULL (FUL) and CAULIFLOWER (CAL) are induced in the floral meristems emerging on the flanks of the shoot apex.10,11
A MADS-domain protein, FLOWERING LOCUS C (FLC) represses flowering by preventing the transcription of FT in leaves and SOC1 and FD in the shoot apex. FRIGIDA (FRI) is a positive regulator of FLC. However, the mechanisms of cold-mediated repression including epigenetic modifications and antisense transcriptions of FLC are not yet fully understood.12,13 Other MADS proteins, AGAMOUS-like19 (AGL19) and AGL24, closely related to SOC1 and SHORT VEGETATIVE PHASE (SVP), respectively, have positive effects on flowering. Of note, their transcripts are accumulated during vernalization and this process is likely to be FLC-independent.14,15 Summer annual Arabidopsis can flower without vernalization but an active repression of flowering takes place at lower ambient temperatures. FLOWERING LOCUS M (FLM) and SVP are central to repression of flowering under the low ambient temperature. In particular, FLM-β, an alternatively spliced form of the flowering repressor FLM, interacts with SVP to respond to ambient temperature changes and SVP delays flowering by repressing FT and SOC1 transcription.16 Therefore, the decrease of SVP-FLM-β repressor complexes at higher temperatures leads to flowering.17 The distribution of flowering time genes in Arabidopsis is shown in Fig. 1A.
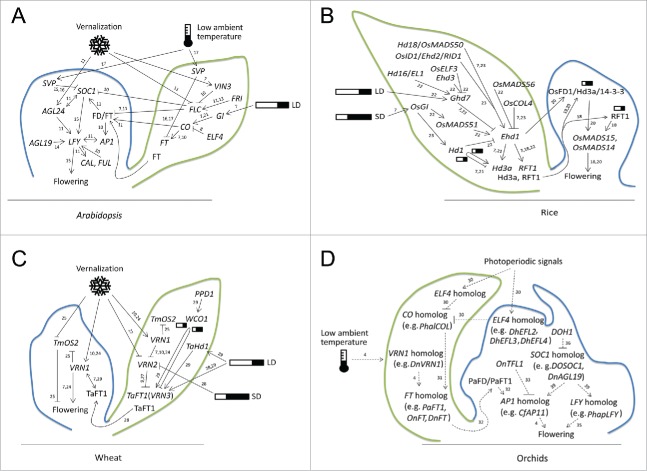
Figure 1.
Major flowering genes in the photoperiodic and temperature network in Arabidopsis, rice, wheat and orchids. (A) Flowering of Arabidopsis is promoted by long days (LDs). The photoperiod signaling cascade involves GI and CO with modulation by EARLY FLOWERING4 (ELF4). ELF4 negatively regulates CO expression by sequestering GI from the CO promoter. CO positively regulates expression of floral integrator FT that mediates SOC1activation. The vernalization response in Arabidopsis through VERNALIZATION INSENSITIVE3 (VIN3) family proteins represses FLC gene family members, which represses FT and SOC1. In addition, AGL24 participates in the FLC–independent vernalization response to regulate LFY expression. The ambient temperature affects flowering time in Arabidopsis through SVP-FLM-β repressor complexes that delay flowering by repressing FT and SOC1 transcription at low ambient temperature. (B) In rice, a short day plant, the OsGI -Hd1 (CO in Arabidopsis)-Hd3a (FT in Arabidopsis) cascade is well conserved. Hd1 and Ehd1 positively regulate Hd3a and promote flowering under inductive short days (SDs). However, under LDs, Ghd7 is a major flowering suppressor that represses Ehd1 and Hd1 also represses Hd3a expression. Instead, RFT1 is activated through OsMADS50 and Ehd1 for eventual flowering under LDs. OsMADS50 and OsMADS56 function antagonistically to affect flowering under LDs by controlling expression of Ehd1. Rice AP1 homologs such as OsMADS14 and OsMADS15 are activated by florigens (e.g., Hd3a and RFT1) in the SAM. (C) In wheat, the TaFT1/VRN3 gene integrates photoperiod (via PPD1–WCO) and vernalization (via VRN1–VRN2). The photoperiodic signals involving PPD1 are transmitted to negative regulator WCO1 and positive regulator TaHd1 to control TaFT1/VRN3 expression under SD. VRN2 prevents flowering before vernalization but later, vernalization induces VRN1, which is followed by the downregulation of VRN2, thereby releasing TaFT1/VRN3 which leads to flowering. ODDSOC2 (TmOS2/TaAGL33) functions as a vernalization-regulated flowering repressor. It is downregulated by cold independently of VRN1. (D) In orchids, Phalaenopsis PhalCOL and PaFT1 were regulated by photoperiod and low ambient temperature, respectively. Doritaenopsis DhEFL2, 3 and 4, as floral repressors, coordinate floral transition in the photoperiodic flowering pathway. DnVRN1 and DnFT from Dendrobium nobile are linked to low temperature (10°C)-induced floral transition. Dendrobium Chao Praya Smile DOSOC1 is expressed specifically in floral meristemsduring the transition from the vegetative to the reproductive phase and coupled with the upregulation of its LFY. DnAGL19 expression of D. nobile repressed by polycomb-group complexes is activated after vernalization reminiscent of Arabidopsis AGL19 pathway to activate LFY and AP1 for flowering. Arrows and T-shaped bars are regulatory links for the promotion and repression of gene transcription, respectively. The numbers next to the arrows and T-shaped bars are references to related studies and the dashed lines indicate predictions of the flowering regulation networks. Prediction of orchids flowering networks should be confirmed by more extensive studies.
Floral transition in a short-day model plant, rice
As shown in Fig. 1B, in rice, Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1) are responsible for floral induction under short-day (SD) and long-day (LD) conditions, respectively.18 The basic leucine-zipper (bZIP) domain transcription factor, OsFD1 interacts with Hd3a through 14–3–3 proteins to form a florigen activation complex (FAC) acting on the induction of OsMADS15, a rice AP1 ortholog in the shoot apex during floral induction.19,20 Early heading date 1 (Ehd1) acts as a flowering activator by upregulating Hd3a and RFT1 and also controls the inflorescence architecture irrespective of photoperiod.21 Interestingly, Heading date 1 (Hd1), a rice ortholog of Arabidopsis CO has a dual function: it acts as floral activator by activating Hd3a under inductive SD conditions but is converted to a repressor of Hd3a under long days (LDs).18,22 Grain number, Plant height and Heading date 7 (Ghd7) and Oryza sativa CONSTANS-LIKE4 (OsCOL4) are upstream suppressors of Ehd1 whereas Oryza sativa INDETERMINATE1 (OsID1)/Ehd2/Rice INDETERMINATE1 (RID1) and OsMADS50, a homolog of Arabidopsis SOC1 are activators of Ehd1 (Fig. 1B).23 Another MADS-box gene, OsMADS51 functioning downstream of Oryza sativa GIGANTEA (OsGI) under short days (SDs) promotes flowering by inducing Ehd1 expression. Moreover, Hd16/EARLY FLOWERING1 (EL1) acts as a flowering repressor by phosphorylating Ghd7 resulting in the reduction of Ehd1 expression. Rice Ehd3 and Early flowering3 (ELF3) were also identified as repressors of Ghd7.22
Floral transition in a seasonal flowering plant, wheat
In wheat, MADS-domain transcription factor VERNALIZATION1 (VRN1), a wheat ortholog of Arabidopsis AP1, is a central regulator of vernalization-induced flowering.24 The grass-specific MADS-box gene ODDSOC2 (TmOS2/TaAGL33) and its splice variant TaAGL22, the FLC orthologs, are downregulated by cold independently of VRN1, but VRN1represses ODDSOC2 during the developmental stage under LDs and normal temperature.25,26 The VRN2 containing a zinc-finger motif and a CCT (CONSTANS, CO-like, TOC1) - domain is a flowering repressor downregulated by both vernalization and SDs.27 Cold signals coordinately activate VRN1 and repress VRN2 expression during vernalization. Also, VRN1 down-regulates VRN2 to control the activity of the photoperiodic flowering pathway (Fig. 1C). VRN2 represses flowering under SDs, and confers the downregulation of VRN2 and the upregulation of VRN1 and VRN3 after the LDs of spring. VRN3, an ortholog of Arabidopsis FT, is a flowering activator and also encodes a mobile protein that is transported from the leaves to the shoot apical meristem (SAM).28 Pseudo-response regulator (PRR) family PHOTOPERIOD1 (PPD1) encodes a CCT-domain protein domain that influences day length sensitivity by altering expression of CO-like genes: Wheat CO (WCO1) negatively regulates and Triticum aestivum HEADING DATE 1 (TaHd1) positively regulates VRN3 expression associated with late flowering under SD conditions.29
Genes that control flowering in orchids
Not so many functional studies of orchid flowering/flower developmental genes have been reported. Recently, it was reported that expression of Doritaenopsis hybrid EARLY FLOWERING4-like4 (DhEFL4) is regulated by photoperiod and overexpression in Arabidopsis delays flowering.30 Ectopic expression of Phalaenopsis CO-like (PhalCOL) encoding a protein with 2 B-box zinc finger motifs and a CCT domain in Phalaenopsis hybrida (cv. Wedding Promenade) caused an early-flowering phenotype in tobacco31 and Phalaenopsis aphrodite FLOWERING LOCUS T1 (PaFT1) that is upregulated under inductive low ambient temperature but is not subject to photoperiodic control showed precocious flowering in Arabidopsis and rice when it was ectopically expressed.32 Moreover, phloem-specific expression of PaFT1 in Arabidopsis suppresses the late flowering effect by an active FRI allele and SVP overexpression. SVP forms a complex with FLM, a floral repressor of FT, by ambient temperature in Arabidopsis.17 A bZIP domain transcription factor PaFD has been isolated as a PaFT1-interacting protein and was able to partially complement the late flowering phenotype of Arabidopsis fd-3.32 Oncidium Gower Ramsey FLOWERING LOCUS T (OnFT) and TERMINAL FLOWER 1 (OnTFL1) encoding floral activator FT and repressor TERMINAL FLOWER1 (TFL1) homologs, respectively were shown to play opposite roles in Arabidopsis flowering.33 Recently, Dendrobium nobile FLOWERING LOCUS T (DnFT) and MOTHER OF FT (DnMFT) expressed preferentially in the auxiliary buds and leaves have been reported. Of note, the expression of the 2 genes in the leaves responded to temperature in an opposite manner. The low temperature (10°C) required for flowering of D. nobile Lindl: increased DnFT expression but decreased DnMFT expression.34
The transcript of floral meristem identity gene Phalaenopsis aphrodite LEAFY (PhapLFY) is accumulated in the floral meristem primordia to stimulate flower initiation. It is also believed that PhapLFY acts in the early stages of floral organ development.35 Expression of DENDROBIUM ORCHID HOMEOBOX1 (DOH1) from D. Madame Thong-In is detected in leaf primordia and downregulated in the shoot apex during floral transition as a possible upstream regulator of DOMADS1 expressed in the transitional shoot apical meristem, advancing the floral transition and flower development (Fig. 1D).36 No homologs of FLC have been reported but homologs of Arabidopsis AGL19 were identified in D. nobile. In addition, expression of OncidiumMADS1 (OMADS1) belonging to the AP1/AGL9 group of MADS-box genes is detected in the apical meristem and in the lip and carpel of flowers and OMADS1 also interacts with OMADS3 associated with floral initiation of Oncidium Gower Ramsey.37 Table 1 provides a summary of the current understanding of orchid flowering genes which are either functional orthologs /homologs of Arabidopsis, rice and wheat genes.
Table 1.
Key flowering genes in orchids.
| Orchids | Gene Name | Accession No. | Homologa | Refsb |
|---|---|---|---|---|
| Phalaenopsis | PaFT1 | KJ609179 | The PaFT1 protein showed 70%, 76% and 89% identity to Arabidopsis FT (BAA77838), rice Hd3a (BAB61030), and Oncidium orchid OnFT (ACC59806), respectively. | 32 |
| Oncidium | OnFT | KJ909968 | OnFT cDNA encodes a 176 amino acid protein that shows 70% and 79% identity to Arabidopsis FT (AT1G65480) and rice Hd3a (Os06g0157700), respectively. | 33 |
| Cymbidium | CgFT | HM120863 | The CgFT protein was 94% with OnFT (EU583502) from Oncidium Gower Ramsey, 79% with Hd3a (BAB61028.1) from Oryza sativa, and 74% with FT (BAA77838.1) from Arabidopsis thaliana. | 42 |
| Cypripedium | CfFT | CFTC014733 | The partial CfFT align 155 amino acids and showed 82.58% identity to wheat TaFT (ABK32205). | Orch |
| Phalaenopsis | PaFD | KJ609180 | PaFD protein shows 34.7% identity to Arabidopsis FD (AT4G35900). | 32 |
| Dendrobium | DnFD | DNTC011756 | The partial DnFD align 169 amino acids and showed 65.1% and 32.2% identity to Phalaenopsis FD (KJ609180) and Arabidopsis FD (AT4G35900), respectively. | Vect |
| Phalaenopsis | PhalCOL | FJ469986 | The PhalCOL protein showed 46% and 45% identity to Arabidopsis COL4 (Q940T9.2) and rice Hd1 (ABB17664), respectively. | 31 |
| Cymbidium | CeCOL | CETC010739 | The CeCOL protein showed 84.5%, 34.9%, and 36.3% identity to Phalaenopsis COL (FJ469986), Arabidopsis CO (AT5G15840) and rice Hd1 (Os06g0275000), respectively. | Vect |
| Cypripedium | CfFLC | CFTC002458 | The partial CfFLC align 145 amino acids and showed 45.5% identity to Arabidopsis FLC (AT5G10140) and 41.2% with wheat TaAGL33 (ABF57950). | Orch Vect |
| Oncidium | OgFLC | OGTC046040 | The partial OgFLC align 145 amino acids and showed 44.8% identity to Arabidopsis FLC (AT5G10140) and 38.6% with wheat TaAGL33 (ABF57950).. | Orch Vect |
| Cypripedium | CfVRN2 | CFTC009002 | The partial CfVRN2 align 106 amino acids and showed 50% identity to wheat VRN2 (AAS58481). | Orch |
| Phalaenopsis | PmVRN2 | PMTC002168 | The partial PmVRN2 align 142 amino acids and showed 41.55% identity to wheat VRN2 (AAS58481). | Orch |
| Dendrobium | DOSOC1 | KC121576 | DOSOC1 shared 52% sequence identity with Arabidopsis SOC1 (AT2G45660), and 57% identity to Oryza sativa OsSOC1 (Os03g0122600). | 39 |
| Phalaenopsis | PlSOC1 | PLTC039163 | PlSOC1 shared 87.9%, 48.4%, and 54.3% identity to Dendrobium (KC121576), Arabidopsis (AT2G45660) and rice (Os03g0122600), respectively. | Vect |
| Dendrobium | DnAGL19 | KU373056 | DnAGL19 protein shows 48% identity to Arabidopsis AGL19 (AT4G22950). | 4 |
| Oncidium | OnTFL1 | KM233713 | OnTFL1 encodes a 173 amino acid protein that shows 71% and 79% identity to Arabidopsis TFL1 (AT5G03840) and rice OsFDR1 (AF159883), respectively. | 33 |
| Vanilloideae | VpTFL1 | VPTC023176 | The partial VpTFL1 align 160 amino acids and showed 81.8%, 69.9%, and 83.6% identity to Oncidium TFL1 (KM233713), Arabidopsis TFL1 (AT5G03840) and rice FDR1 (AF159883), respectively. | Vect |
| Cymbidium | CfAPl1 | JQ031272.1 | CfAPl1 shared 45% sequence identity with Arabidopsis AP1 (AT1G69120), and 52% identity to Oryza sativa OsMADS14 (Os03g0752800). | 43 |
| Phalaenopsis | PsAP1 | PSTC034274 | The PsAP1 protein showed 55.8%, 47.6% and 58.3% identity to Cymbidium AP11 (JQ031272.1), Arabidopsis AP1 (AT1G69120) and rice MADS14 (Os03g0752800), respectively. | Vect |
| Doritaenopsis | DnVRN1 | DNTC013820 | The partial DnVRN1 align 251 amino acids and showed 61.75% identity to wheat VRN1 (AAZ76881). | Orch |
| Phalaenopsis | PaVRN1 | PATC201550 | The partial PaVRN1 align 250 amino acids and showed 60.4% identity to wheat VRN1 (AAZ76881). | Orch |
| Cymbidium | ChLFY | AGE45851 | The ChLFY protein showed 51% and 48% identity to Arabidopsis LFY (AT5G61850) and rice RFL (Os04g0598300), respectively. | Vect |
| Phalaenopsis | PhalLFY | FJ469985 | PhalLFY protein has 60% identity to rice RFL (BAA21547), 54% identity to Arabidopsis LFY (NP_200993), and a high identity with LFY (68% and 67%) from Anacamptis (BAC55081) and Orchis (BAC54955), respectively. | 44 |
| Phalaenopsis | PhapLFY | KP893636 | PhapLFY protein has 78.5% identity to ChLFY (AGE45851), a Cymbidium orchid LFY homolog, 53.2% identity to RFL (BAA21547), a rice LFY homolog, and 47.0% identity to Arabidopsis LFY (AAM27941). | 35 |
| Doritaenopsis | DhEFL2 | KP728997 | DhEFL homologs showed that DhEFL4 (KP010003) and DhEFL2 (KP728997) are similar with 72% identical amino acids, whereas DhEFL3 (KP728998) is divergent with 72% similarity with DhEFL2 and 68% similarity with DhEFL4. | 45 |
| Doritaenopsis | DhEFL3 | KP728998 | The DhEFL3 protein showed 31.4% identity to Arabidopsis ELF4 (AT2G40080). | Vect |
| Doritaenopsis | DhEFL4 | KP010003 | The DhEFL4 protein showed 37% identity to Arabidopsis ELF4(AT2G40080). | Vect |
| Phalaenopsis | PsEhd1 | PSTC018079 | The partial PsEhd1 align 266 amino acids and showed 40% identity to rice Ehd1 (Os10g0463400). | Orch |
| Cymbidium | CsEhd1 | CSTC008880 | The partial CsEhd1 align 263 amino acids and showed 49.5% identity to rice Ehd1 (Os10g0463400). | Orch |
The homology is based on each reference and Orchidstra 2.0, a transcriptome database of orchid species (http://orchidstra2.abrc.sinica.edu.tw/orchidstra2/orchid_blast.php). Amino acid sequences of flowering in Arabidopsis, rice and wheat were obtained from GenBank (http://www.ncbi.nlm.nih.gov/) and Orchidstra 2.0.
Orch, homology gained through Orchidstra 2.0; Vect, homology gained by analysis using Vector NTI program.
Orchid breeding strategies
Generally, orchids grow slowly and have long life cycles. Consequently, it requires a long period of time to generate new cultivars using the traditional breeding process. Transformation technology has been developed for orchids and recently, a few successful methods38,39 that use virus-induced gene-silencing (VIGS) approaches have been demonstrated as efficient strategies for functional studies of genes in orchids.40 Thus, we may now speed up the orchid breeding process by suppressing the expression of floral repressors resulting in early flowering. Also, virus-induced flowering (VIF) approaches to accelerate the transition to reproductive growth may facilitate research and/or breeding in orchids.41 For example, either gain of function of FT or loss of function of TFL1 using transient methods such as VIGS/VIF may contribute to a faster orchid breeding process. Using inducible gene expression systems, we may expect to control flowering of orchids with transgenic lines regardless of the ambient environment.32 Specific alleles can be selected for breeding and/or modified by induced mutation (genome editing) to acquire orchid cultivars containing desirable traits when current transformation procedures are stabilized.11 In our recent reports, we have shown that the VIGS of PaFT1 exhibits delayed spiking of P. aphrodite subsp. formosana and the VIGS-PhapLFY flowers displayed increased chlorophyll content with the aberrant epidermal cell shape.32,35 Furthermore, we successfully produced transgenic Phalaenopsis for PhapLFY overexpression (Fig. 2).
Figure 2.
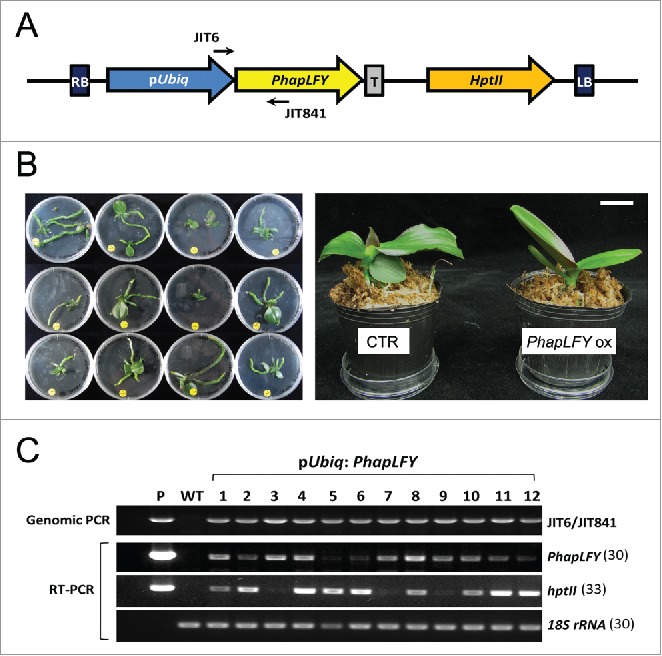
Overexpression of PhapLFY in Phalaenopsis. (A) Plasmid construction for PhapLFY overexpression in Phalaenopsis orchids. pUbiq means maize ubiquitin promoter and T indicates nos terminator. RB and LB indicate right and left T-DNA borders. The hygromycin phosphotransferase II (hptII) expressing cassette was used for hygromycin resistance in selection of transgenic orchids. (B) Transgenic Phalaenopsis orchids overexpressing PhapLFY gene. Each transgenic orchid seedling used for analyses in C (left) and young orchid plantlets. Bar = 2cm. (C) Verification of transgenic Phalaenopsis orchids by genomic PCR and RT-PCR. Expression of PhapLFY and hygromycin resistance gene (hptII) was examined by RT-PCR together with 18s rRNA in leaves of each plant. The location of primers used for genomic PCR is parked in (A). P indicates plasmid DNA used for transformation as a positive control for PCR and WT indicates a non-transgenic Phalaenopsis orchid in the same developmental stage with transgenic orchids. JIT6: 5′TTGTCGATGCTCACCCTG3′, JIT841: 5′CTAGCCGCTCCTCTCTGTCTCCGAC3′, PhapLFY-F: 5′GAGGAGGAGGTGGACGATATGATG3′, PhapLFY-R: 5′GCTTGTTTATGTAGCTTGCTCCTAC3′, hptII-F: 5′GATTCCGGAAGTGCTTGACATTG3′, hptII-R: 5′GCATCAGCTCATCGAGAGCCTG3′, 18S rRNA-F: 5′TTAGGCCACGGAAGTTTGAGG3′, 18S rRNA-R: 5′ACACTTCACCGGACCATTCAA3′.
Conclusion and future directions
The Orchidaceae is the largest and the most diverse family of flowering plants and most orchids have their own flowering seasons: D. nobile requires vernalization for flowering whereas D. phalaenopsis flowers at high temperatures. Spiking of P. aphrodite subsp formosana is significantly inhibited under warm conditions while the natural flowering period of P. sanderiana is through the summer in the Philippines. The molecular mechanisms underlying flowering among various orchid species largely remain elusive. Research on flowering to induce early flowering in orchids has been aimed not only at a better understanding molecular and genetic mechanisms of orchid flowering but also at assisting orchid breeding programs.1 In vitro flowering or floral transition in orchids is affected by various plant growth regulators (PGRs) although the molecular working mechanisms of those PGRs underlying orchid floral induction are unclear. Molecular genetic approaches are required to shed some light on the roles of putative key flowering genes of orchids and will also provide a platform for application of genetic resources to the orchid industry.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Ms. Miranda Loney for help with English editing.
Funding
This work was supported in part by a core grant from BCST of ABRC, Academia Sinica, Taiwan.
References
- 1.Hossain MM, Kant R, Van PT, Winarto B, Zeng S, Teixeira da Silva JA. The application of biotechnology to orchids. Crit Rev Plant Sci 2013; 32:69-139; https://doi.org/ 10.1080/07352689.2012.715984 [DOI] [Google Scholar]
- 2.Pan IC, Liao DC, Wu FH, Daniell H, Singh ND, Chang C, Shih MC, Chan MT, Lin CS. Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding. PLoS One 2012; 7:e34738; PMID:22496851; https://doi.org/ 10.1371/journal.pone.0034738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu HC, Chen HH, Tsai WC, Chen WH, Su HJ, Chang CN, Yeh HH. Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol 2007; 143:558-69; PMID:17189336; https://doi.org/ 10.1104/pp.106.092742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang S, Ye QS, Li RH, Leng JY, Li MR, Wang XJ, Li HQ. Transcriptional regulations on the low-temperature-induced floral transition in an Orchidaceae species, Dendrobium nobile: An expressed sequence tags analysis. Comp Funct Genomics 2012; 2012:757801; PMID:22550428; https://doi.org/ 10.1155/2012/757801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su CL, Chen WC, Lee AY, Chen CY, Chang YC, Chao YT, Shih MC. A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite. PLoS One 2013; 8:e80462; PMID:24265826; https://doi.org/ 10.1371/journal.pone.0080462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu HF, Hsu WH, Lee YI, Mao WT, Yang JY, Li JY, Yang CH.. Model for perianth formation in orchids. Nat Plant 2015; 1:15046; https://doi.org/ 10.1038/nplants.2015.46 [DOI] [Google Scholar]
- 7.Jarillo JA, Piñeiro M. Timing is everything in plant development. The central role of floral repressors. Plant Sci 2011; 181:364-78; PMID:21889042; https://doi.org/ 10.1016/j.plantsci.2011.06.011 [DOI] [PubMed] [Google Scholar]
- 8.Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002; 419:74-77; PMID:12214234; https://doi.org/ 10.1038/nature00954 [DOI] [PubMed] [Google Scholar]
- 9.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Ann Rev Plant Biol 2008; 59:573-94; PMID:18444908; https://doi.org/ 10.1146/annurev.arplant.59.032607.092755 [DOI] [PubMed] [Google Scholar]
- 10.Jung C, Müller AE “Flowering time control and applications in plant breeding”. Trends Plant Sci 2009; 14:563-73; PMID:19716745; https://doi.org/ 10.1016/j.tplants.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 11.Parcy F. Flowering: A time for integration. Int J Dev Biol 2005; 49:585-93; PMID:16096967; https://doi.org/ 10.1387/ijdb.041930fp [DOI] [PubMed] [Google Scholar]
- 12.Helliwell CA, Anderssen RS, Robertson M, Finnegan EJ. How is FLC repression initiated by cold? Trends Plant Sci 2015; 20:76-82; PMID:25600480; https://doi.org/ 10.1016/j.tplants.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 13.Bouché F, Woods D, Amasino RM. Winter memory throughout the plant kingdom: Different paths to flowering. Plant Physiol 2017; 173:27-35; PMID:27756819; https://doi.org/ 10.1104/pp.16.01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schönrock N, Bouveret R, Leroy O, Borghi L, Köhler C, Gruissem W, Hennig L. Polycomb-group proteins repressthe floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev 2006; 20:1667-78; PMID:16778081; https://doi.org/ 10.1101/gad.377206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bluemel M, Dally N, Jung C. Flowering time regulation in crops—what did we learn from Arabidopsis? Curr Opin Biotechnol 2015; 32:121-29; PMID:25553537; https://doi.org/ 10.1016/j.copbio.2014.11.023 [DOI] [PubMed] [Google Scholar]
- 16.Jang S, Torti S, Coupland G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J 2009; 60:614-25; PMID:19656342; https://doi.org/ 10.1111/j.1365-313X.2009.03986.x [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Ryu HS, Chung KS, Posé D, Kim S, Schmid M, Ahn JH. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 2013; 342:628-32; PMID:24030492; https://doi.org/ 10.1126/science.1241097 [DOI] [PubMed] [Google Scholar]
- 18.Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 2009; 136:3443-50; PMID:19762423; https://doi.org/ 10.1242/dev.040170 [DOI] [PubMed] [Google Scholar]
- 19.Taoka K, Ohki I, Tsuji H, Kojima C, Shimamoto K. Structure and function of florigen and the receptor complex. Trends Plant Sci 2013; 18:287-94; PMID:23477923; https://doi.org/ 10.1016/j.tplants.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 20.Tamaki S, Tsuji H, Matsumoto A, Fujita A, Shimatani Z, Terada R, Sakamoto T, Kurata T, Shimamoto K. FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice. Proc Natl Acad Sci USA 2015; 112:E901-10; PMID:25675495; https://doi.org/ 10.1073/pnas.1417623112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endo-Higashi N, Izawa T. Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol 2011; 52:1083-94; PMID:21565907; https://doi.org/ 10.1093/pcp/pcr059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun C, Chen D, Fang J, Wang P, Deng X, Chu C. Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell 2014; 5:889-98; PMID:25103896; https://doi.org/ 10.1007/s13238-014-0068-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi SC, Lee S, Kim SR, Lee YS, Liu C, Cao X, An G. Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol 2014; 164:1326-37; PMID:24420930; https://doi.org/ 10.1104/pp.113.228049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 2003; 100:6263-68; PMID:12730378; https://doi.org/ 10.1073/pnas.0937399100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenup AG, Sasani S, Oliver SN, Talbot MJ, Dennis ES, Hemming MN, Trevaskis B. ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol 2010; 153:1062-73; PMID:20431086; https://doi.org/ 10.1104/pp.109.152488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma N, Ruelens P, Dhauw M, Maggen T, Dochy N, Torfs S, Kaufmann K, Rohde A, Geuten K. A Flowering locus C homolog is a vernalization-regulated repressor in Brachypodium and is cold-regulated in wheat. Plant Physiology 2017; 173:1301-15; PMID:28034954; https://doi.org/ 10.1104/pp.16.01161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 2004; 303:1640-44; PMID:15016992; https://doi.org/ 10.1126/science.1094305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 2006; 103:19581-86; PMID:17158798; https://doi.org/ 10.1073/pnas.0607142103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitagawa S, Shimada S, Murai K. Effect of Ppd-1 on the expression of flowering-time genes in vegetative and reproductive growth stages of wheat. Genes Genet Syst 2012; 87:161-68; PMID:22976391; https://doi.org/ 10.1266/ggs.87.161 [DOI] [PubMed] [Google Scholar]
- 30.Chen W, Qin Q, Zheng Y, Wang C, Wang S, Zhou M, Zhang C, Cui Y.. Overexpression of Doritaenopsis hybrid EARLY FLOWERING 4-like4 gene, DhEFL4, postpones flowering in transgenic Arabidopsis. Plant Mol Biol Rep 2016; 34:103-17; https://doi.org/ 10.1007/s11105-015-0899-1 [DOI] [Google Scholar]
- 31.Zhang JX, Wu KL, Tian LN, Zeng SJ, Duan J. Cloning and characterization of a novel CONSTANS-like gene from Phalaenopsis hybrida. Acta Physiol Plant 2011; 33:409-17; https://doi.org/ 10.1007/s11738-010-0560-4 [DOI] [Google Scholar]
- 32.Jang S, Choi SC, Li HY, An G, Schmelzer E. Functional characterization of Phalaenopsis aphrodite flowering genes PaFT1 and PaFD. PLoS One 2015; 10:e0134987; PMID:26317412; https://doi.org/ 10.1371/journal.pone.0134987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou CJ, Yang CH. Functional analysis of FT and TFL1 orthologs from orchid (Oncidium Gower Ramsey) that regulate the vegetative to reproductive transition. Plant Cell Physiol 2009; 50:1544-57; PMID:19570813; https://doi.org/ 10.1093/pcp/pcp099 [DOI] [PubMed] [Google Scholar]
- 34.Li R, Wang A, Sun S, Liang S, Wang X, Ye Q, Li H. Functional characterization of FT and MFT ortholog genes in orchid (Dendrobium nobile Lindl) that regulate the vegetative to reproductive transition in Arabidopsis. Plant Cell Tissue Organ Cult 2012; 111:143-51; https://doi.org/ 10.1007/s11240-012-0178-x [DOI] [Google Scholar]
- 35.Jang S. Functional characterization of PhapLEAFY, a FLORICAULA/LEAFY ortholog in Phalaenopsis aphrodite. Plant Cell Physiol 2015; 56:2234-47; PMID:26493518; https://doi.org/ 10.1093/pcp/pcv130 [DOI] [PubMed] [Google Scholar]
- 36.Yu H, Yang SH, Goh CJ. DOH1, a class 1 knox gene, is required for maintenance of the basic plant architecture and floral transition in orchid. Plant Cell 2000; 12:2143-59; PMID:11090215; https://doi.org/ 10.1105/tpc.12.11.2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu HF, Huang CH, Chou LT, Yang CH. Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol 2003; 44:783-94; PMID:12941870; https://doi.org/ 10.1093/pcp/pcg099 [DOI] [PubMed] [Google Scholar]
- 38.Thiruvengadam M, Chung IM, Yang CH. Overexpression of Oncidium MADS box (OMADS1) gene promotes early flowering in transgenic orchid (Oncidium Gower Ramsey). Acta Physiol Plant 2012; 34:1295-02; https://doi.org/ 10.1007/s11738-012-0926-x [DOI] [Google Scholar]
- 39.Ding L, Wang Y, Yu H. Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid Dendrobium Chao Parya Smile. Plant Cell Physiol 2013; 54:595-608; PMID:23396600; https://doi.org/ 10.1093/pcp/pct026 [DOI] [PubMed] [Google Scholar]
- 40.Lu HC, Chen HH, Tsai WC, Chen WH, Su HJ, Chang DCN, Yeh HH. Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol 2007; 143:558-69; PMID:17189336; https://doi.org/ 10.1104/pp.106.092742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGarry RC, Klocko AL, Pang M, Strauss SH, Ayre BG. Virus-induced flowering: An application of reproductive biology to benefit plant research and breeding. Plant Physiol 2017; 173:47-55; PMID:27856915; https://doi.org/ 10.1104/pp.16.01336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang W, Fang Z, Zeng S, Zhang J, Wu K, Chen Z, Teixeira da Silva JA, Duan J. Molecular cloning and functional analysis of three FLOWERING LOCUS T (FT) homologous genes from Chinese Cymbidium. Int J Mol Sci 2012; 13:11385-98; PMID:23109860; https://doi.org/ 10.3390/ijms130911385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian Y, Yuan X, Jiang S, Cui B, Su J. Molecular cloning and spatiotemporal expression of an APETALA1/FRUITFULL-like MADS-box gene from the orchid (Cymbidium faberi). Sheng Wu Gong Cheng Xue Bao 2013; 29:203-13; PMID:23697165. [PubMed] [Google Scholar]
- 44.Zhang JX, Wu KL, Zeng SJ, Duan J, Tian LN. Characterization and expression analysis of PhalLFY, a homologue in Phalaenopsis of FLORICAULA/LEAFY genes. Sci Horti 2010; 124:482-89; https://doi.org/ 10.1016/j.scienta.2010.02.004 [DOI] [Google Scholar]
- 45.Chen W, Qin Q, Zhang C, Zheng Y, Wang C, Zhou M, Cui Y. DhEFL2, 3 and 4, the three EARLY FLOWERING4-like genes in a Doritaenopsis hybrid regulate floral transition. Plant Cell Rep 2015; 34:2027-41; PMID:26205509; https://doi.org/ 10.1007/s00299-015-1848-z [DOI] [PubMed] [Google Scholar]