ABSTRACT
This addendum discusses the compartmentation of γ-aminobutyrate (GABA) metabolism, highlighting recent progress with Arabidopsis thaliana and raising new questions about the roles of mitochondria, plastids and peroxisomes in abiotic stress tolerance.
KEYWORDS: Aldehyde dehydrogenase family 10, Four-Aminobutyrate, GABA biosynthesis, subcellular localization
Four-Aminobutyrate (GABA) is a ubiquitous, 4-carbon, non-proteinogenic amino acid that has been linked to stress, signaling and storage in plants.1-3 Previously, we proposed a model for the compartmentation of GABA metabolism, based on the known distribution and biochemical function of specific enzymes and transporters.4 Here, we briefly review new evidence for multiple pathways involved in GABA metabolism, and propose an updated model for its compartmentation in Arabidopsis thaliana.
Subcellular fractionation of protoplasts from developing soybean cotyledons has demonstrated that GABA is synthesized in the cytosol via the glutamate decarboxylase reaction (GAD),5 and to our knowledge, there is no evidence for the presence of an organelle-targeting domain in Arabidopsis GADs. While GAD activity is greatest at a slightly acidic pH of pH 5.8, Arabidopsis GAD1/2/4 possess a C-terminal calmodulin (CaM)-binding domain, which allows in vitro GAD activity to be activated at neutral pH by Ca2+/CaM.1 In silico analysis suggests that GAD3 and GAD5 proteins are CaM independent. The CaM-binding domain in GAD is responsible for at least part of GABA accumulation which occurs in response to elevated cytosolic Ca2+ during stress.1,6 There is some preliminary evidence of a plasma membrane location for GAD in Arabidopsis leaves and petioles and tobacco pollen tubes,7,8 but further research is required to substantiate these findings. Recent evidence indicates that a atgad1/2 double mutant accumulates much less GABA, is oversensitive to prolonged drought, and displays salinity-induced increases in GAD4 expression, suggesting that the pathway from glutamate to GABA plays an important role in the stress response.9,10
While the biosynthesis of GABA from glutamate is generally considered to be the primary source of GABA, there is also evidence for its synthesis from the polyamine putrescine (Put). In the October issue of Scientific Reports we reported that peroxisomal aldehyde dehydrogenase 10A9 (ALDH10A9) catalyzes the final step of Put oxidation in Arabidopsis by converting 4-aminobutanal (ABAL) to GABA, and suggested that a protein with similar biochemical function, ALDH10A8, is translocated from the cytosol to the plastid, perhaps in response to stress-induced post-translational modification (Fig. 1).11 The conversion of Put to ABAL is catalyzed by copper-containing amine oxidase (CuAO). Arabidopsis possesses 10 putative CuAOs, 4 of which (AO1, CuAO1, CuAO2 and CuAO3) have been characterized to date.12 Two of these, CuAO2 and CuAO3, are peroxisomal.13 Five Arabidopsis FAD-dependent polyamine oxidases (PAO1–5) have been reported to be involved in the back conversion of spermidine and spermine to Put; however, only PAO2–4 are peroxisomal.14 In silico analysis predicts that 3 putative CuAOs (At1g31670, At1g31710 and At4g12290) could be plastidial (Fig. 1).11 We have proposed that arginine would be an appropriate source of Put in plastids, but localization of agmatine imidohydrolase and N-carbamoylputrescine amidohydrolase needs to be established.11 Notably, the carbon flux through the CuAOs and ALDH10As could be regulated by a combination of O2 availability and redox balance.15 Recently, we demonstrated that GABA accumulates in shoots of single ataldh10A8 and ataldh10A9 mutants and root growth is oversensitive to salinity, suggesting that the pathway from polyamines to GABA plays a role in the stress response.11
Figure 1.
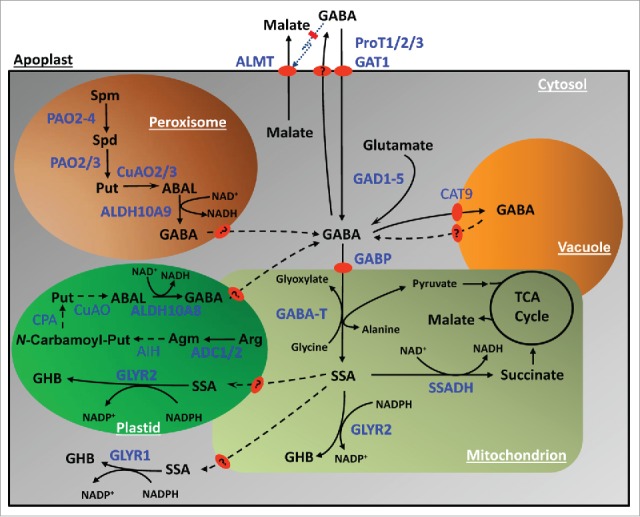
Subcellular compartmentation of GABA metabolism.Enzymes are shown in blue letters, with biochemically characterized enzymes indicated by bold lettering. Dashed arrows and question marks indicate enzymes and transporters which have not been studied. The dotted line with a red bar represents negative regulation. Abbreviations: ABAL, 4-aminobutanal; ADC, arginine decarboxylase; ALDH, aldehyde dehydrogenase; Agm, agmatine; AIH, agmatine iminohydrolase; ALMT, aluminum-acticated malate transporter; Arg, arginine; CT, cationic amino acid transporter; N-Carbamoyl-Put, N-Carbamoylputrescine; CPAH, N-Carbamoylputrescine amidohydrolase; CuAO, copper amine oxidase; GABA, 4-aminobutyrate; GAD, glutamate decarboxylase; GABP, GABA permease; GABA-T, GABA transaminase; GAT, GABA transporter; GHB, 4-hydroxybutyrate; GLYR, glyoxylate/succinic semialdehyde reductase; PAO, polyamine oxidase; ProT, proline transporter; Put, putrescine; SSA, succinate semialdehyde; SSADH, succinate semialdehyde dehydrogenase; Spd, spermidine; Spm, spermine; TCA, tricarboxylic acid (adapted from refs. 4, 11 and 20).
Regardless of the source of GABA, it appears to be catabolized to succinic semialdehyde (SSA) in Arabidopsis via a single mitochondrial pyruvate/glyoxylate-dependent GABA transaminase (GABA-T) (Fig. 1).16 Notably, the atgaba-t mutant has elevated GABA, impaired central carbon metabolism, and increased sensitivity to salinity.17,18 These findings are consistent with the oxidation of SSA to succinate via a mitochondrial NAD-dependent SSA dehydrogenase in mitochondria.19 Alternatively, SSA can be metabolized to 4-hydroxybutyrate via 2 NADPH-dependent glyoxylate/succinate semialdehyde reductases (GLYR1 and GLYR2). Recently, we confirmed that GLYR1 is localized to the cytosol and demonstrated that GLYR2 is localized to both plastids and mitochondria of Arabidopsis cells (Fig. 1).20 The operation of this path for SSA metabolism would be facilitated by elevated NADH/NAD+ and NADPH/NADP+ ratios that accompany many abiotic stress conditions.1 For example, the growth of plantlets of various Arabidopsis lines with altered GLYR activity respond differentially to succinic semialdehyde and glyoxylate under chilling conditions (Zarei A and Shelp BJ, unpublished), highlighting their potential regulation by NADPH/NADP+ ratios in planta, and their roles in the reduction of toxic aldehydes within distinct subcellular compartments.20 Interestingly, glycolate, which is also formed by the SSA metabolizing enzyme when it utilizes glyoxylate, accumulates in response to hypoxia.21
Together, these findings suggest that cytosolic GABA is at least in part, derived from polyamine catabolism in the peroxisome, polyamine anabolism in the plastid, and glutamate in the cytosol, although the relative importance of these different routes could depend on the specific stress and developmental stage. If so, they suggest the existence of outward-flowing GABA transporters in peroxisomal and plastid membranes (Fig. 1). High-affinity proton-coupled GABA transporters have been localized to plasma and mitochondrial membranes of Arabidopsis, and while these are very important for carbon-nitrogen interactions, the potential involvement of other GABA transporters such as AtProT2 has not been excluded.22-26 There is evidence for GABA uptake via SlCAT9 (cationic amino acid transporter) into tomato vacuoles in exchange for glutamate or aspartate, but further research is required to establish which, if any, of the tonoplast-localized members of the CAT family transport GABA in Arabidopsis.27 Notably, AtCAT9 is 68% identical to SlCAT9 at the amino acid level, whereas AtCAT2/4/8 are 27–32% identical, suggesting that AtCAT9 is the most promising candidate, thereby contributing to the regulation of GABA storage in the vacuole. Cytosolic GABA also crosses the plasma membrane,28 but the transporter has not yet been identified. All these transport activities would contribute to the status of GABA both in the cytosol and the apoplast, and contribute to control of the aluminum-activated malate transporter and export of carbon from the cell.3,29,30 Finally, multiple locations for GLYR activity suggest the existence of outward- and inward-flowing transporters for SSA in mitochondria and plastids, respectively.
In summary, recent research, much of it involving the characterization of recombinant proteins and Arabidopsis knockout mutants, has demonstrated the existence of multiple pathways which could influence the status of GABA in plant cells. It raises new issues regarding the roles of mitochondria, plastids and peroxisomes in abiotic stress tolerance.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Fundamental research on GABA metabolism and function in the Shelp laboratory has been supported by the Natural Science and Engineering Research Council (NSERC) of Canada.
References
- 1.Shelp BJ, Bozzo GG, Zarei A, Simpson JP, Trobacher CP, Allan WL. Strategies and tools for studying the metabolism and function of γ-aminobutyrate in plants. II. Integrated analysis. Botany 2012; 90:781-93; https://doi.org/ 10.1139/B2012-041 [DOI] [Google Scholar]
- 2.Shelp BJ, Bozzo GG, Trobacher CP, Chiu G, Bajwa VS. Strategies and tools for studying the metabolism and function of γ-aminobutyrate in plants. I. Pathway structure. Botany 2012; 90:651-68; https://doi.org/ 10.1139/B2012-030 [DOI] [Google Scholar]
- 3.Gilliham M, Tyerman SD. Linking metabolism to membrane signaling: The GABA-malate connection. Trends Plant Sci 2016; 21:295-301; PMID:26723562; https://doi.org/ 10.1016/j.tplants.2015.11.011 [DOI] [PubMed] [Google Scholar]
- 4.Shelp BJ, Mullen RT, Waller JC. Compartmentation of GABA metabolism raises intriguing questions. Trends Plant Sci 2012; 17:57-9; PMID:22226724; https://doi.org/ 10.1016/j.tplants.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 5.Breitkreuz KE, Shelp BJ. Subcellular compartmentation of the 4-aminobutyrate shunt in protoplasts from developing soybean cotyledons. Plant Physiol 1995; 108:99-103; PMID:12228455; https://doi.org/ 10.1104/pp.108.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy ASN, Ali GS, Celesnik H, Day IS. Coping with stresses: Roles of calcium- and calcium-calmodulin-regulated gene expression. Plant Cell 2011; 23:2010-32; PMID:21642548; https://doi.org/ 10.1105/tpc.111.084988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandersson E, Saalbach G, Larsson C, Kjellbom P. Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol 2004; 45:1543-56; PMID:15574830; https://doi.org/ 10.1093/pcp/pch209 [DOI] [PubMed] [Google Scholar]
- 8.Yu G-H, Zou J, Feng J, Peng X-B, Wu J-Y, Wu Y-L, Palanivelu R, Sun M-X. Exogenous γ-aminobutyric acid (GABA) affects pollen tube growth via modulating putative Ca2+-permeable membrane channels and is coupled to negative regulation on glutamate decarboxylase. J Exp Bot 2014; 65:3235-48; PMID:24799560; https://doi.org/ 10.1093/jxb/eru171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mekonnen DW, Flügge UI, Ludewig F. Gamma-aminobutyric acid depletion affects stomata closure and drought tolerance of arabidopsis thaliana. Plant Sci 2016; 245:25-34; PMID:26940489; https://doi.org/ 10.1016/j.plantsci.2016.01.005 [DOI] [PubMed] [Google Scholar]
- 10.Zarei A, Chiu GZ, Yu GH, Trobacher CP, Shelp BJ. Salinity-regulated expression of genes involved in GABA metabolism and signaling. Botany 2017; https://doi.org/ 10.1139/cjb-2016-0304 [DOI] [Google Scholar]
- 11.Zarei A, Trobacher CP, Shelp BJ. Arabidopsis aldehyde dehydrogenase 10 family members confer salt tolerance through putrescine-derived 4-aminobutyrate (GABA). Sci Rep 2016; 6:35115; PMID:27725774; https://doi.org/ 10.1038/srep35115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiburcio AF, Altabella T, Bitrián M, Alcázar R. The roles of polyamines during the lifespan of plants: From development to stress. Planta 2014; 240:1-18; PMID:24659098; https://doi.org/ 10.1007/s00425-014-2055-9 [DOI] [PubMed] [Google Scholar]
- 13.Planas-Portell J, Gallart M, Tiburcio AF, Altabella T. Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana. BMC Plant Biol 2013; 13:109; PMID:23915037; https://doi.org/ 10.1186/1471-2229-13-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fincato P. Moschou PN, Spedaletti V, Tavazza R, Angelini R, Federico R, Roubelakis-Angelakis KA, Tavladoraki P. Functional diversity inside the arabidopsis polyamine oxidase gene family. J Exp Bot 2011; 62:1155-68; PMID:21081665; https://doi.org/ 10.1093/jxb/erq341 [DOI] [PubMed] [Google Scholar]
- 15.Shelp BJ, Bozzo GG, Trobacher CP, Zarei A, Deyman KL, Brikis CJ. Hypothesis/review: Contribution of putrescine to 4-aminobutyrate (GABA) production in response to abiotic stress. Plant Sci 2012; 193-194:130-5; PMID:22794926; https://doi.org/ 10.1016/j.plantsci.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 16.Clark SM, Di Leo R, Dhanoa PK, Van Cauwenberghe OR, Mullen RT, Shelp BJ. Biochemical characterization, mitochondrial localization, expression, and potential functions for an Arabidopsis gamma-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. J Exp Bot 2009; 60:1743-57; PMID:19264755; https://doi.org/ 10.1093/jxb/erp044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renault H, Roussel V, El Amrani A, Arzel M, Renault D, Bouchereau A, Delu C. The Arbidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol 2010; 10:20; PMID:20122158; https://doi.org/ 10.1186/1471-2229-10-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renault H, El Amrani A, Berger A, Mouille G, Soubigout-Taconnat L, Bouchereau A, Delu C. γ-Aminobutyric acid transaminase deficiency impairs central carbon metabolism and leads to cell wall defects during salt stress in arabidopsis roots. Plant Cell Environ 2013; 36:1009-18; PMID:23148892; https://doi.org/ 10.1111/pce.12033 [DOI] [PubMed] [Google Scholar]
- 19.Bouché N, Fait A, Bouchez D, Møller SG, Fromm H. Mitochondrial succinic-semialdehyde dehydrogenase of the γ-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc Natl Acad Sci USA 2003; 100:6843-8; PMID:12740438; https://doi.org/ 10.1073/pnas.1037532100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brikis CJ, Zarei A, Trobacher CP, DeEll JR, Akama K, Mullen RT, Bozz GG, Shelp BJ. Ancient plant glyoxylate/succinic semialdehyde reductases: GLYR1s are cytosolic, whereas GLYR2s are localized to both mitochondria and plastids. Front Plant Sci 2017; 8:601; https://doi.org/ 10.3389/fpls.2017.00601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narsai R, Howell KA, Carroll A, Ivanova A, Millar AH, Whelan J. Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol 2009; 151:306-22; PMID:19571305; https://doi.org/ 10.1104/pp.109.142026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breitkreuz KE, Shelp BJ, Fischer WN, Schwack R, Rentsch D. Identification and characterization of GABA, proline and quaternary ammonium compound transporters from Arabidopsis thaliana. FEBS Lett 1999; 450:280-84; PMID:10359089; https://doi.org/ 10.1016/S0014-5793(99)00516-5 [DOI] [PubMed] [Google Scholar]
- 23.Grallath S, Weimar T, Meyer A, Gumy C, Suter-Grotemeyer M, Neuhaus J-M, Rentsch D. The AtProT family. Compatible solute transporters with similar substrate specificity but differential expression patterns. Plant Physiol 2005; 137:117-26; PMID:15618414; https://doi.org/ 10.1104/pp.104.055079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer A, Eskandari S, Grallath S, Rentsch D. AtGAT1, a high affinity transporter for γ-aminobutyric acid in Arabidopsis thaliana. J Biol Chem 2006; 281:7197-204; PMID:16407306; https://doi.org/ 10.1074/jbc.M510766200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaeli S, Fait A, Lagor K, Nunes-Nesi A, Grillich N, Yellin A, Bar D, Khan M, Fernie AR, Turano FJ, et al.. A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism. Plant J 2011; 67:485-98; PMID:21501262; https://doi.org/ 10.1111/j.1365-313X.2011.04612.x [DOI] [PubMed] [Google Scholar]
- 26.Batushansky A, Kirma M, Grillich N, Pham PA, Rentsch D, Galili G, Fernie AR, Fait A. The transporter GAT1 plays an important role in GABA-meditated carbon-nitrogen interactions in arabidopsis. Front Plant Sci 2015; 6:785; PMID:26483804; https://doi.org/ 10.3389/fpls.2015.00785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snowden CJ, Thomas B, Baxter CJ, Smith JAC, Sweetlove LJ. A tonoplast Glu/Asp/GABA exchanger that affects tomato fruit amino acid composition. Plant J 2015; 81:651-60; PMID:25602029; https://doi.org/ 10.1111/tpj.12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung I, Bown AW, Shelp BJ. The production and efflux of 4-aminobutyrate in isolated mesophyll cells. Plant Physiol 1992; 99:659-64; PMID:16668937; https://doi.org/ 10.1104/pp.99.2.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramesh SA, Tyerman SD, Xu B, Bose J, Kaur S, Conn V, Domingos P, Ullah S, Wege S, Shabala S, et al.. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat Commun 2015; 6:7879; PMID:26219411; https://doi.org/ 10.1038/ncomms8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bown AW, Shelp BJ. Plant GABA: Not just a metabolite. Trends Plant Sci 2016; 21:811-3; PMID:27542324; https://doi.org/ 10.1016/j.tplants.2016.08.001 [DOI] [PubMed] [Google Scholar]


