Abstract
We have previously found that Epigallocatechin-3-gallate (EGCg), an abundant catechin in green tea, reduced apoptotic signaling and improved muscle recovery in response to reloading after hindlimb suspension (HS). In this study, we investigated if EGCg altered autophagy signaling in skeletal muscle of old rats in response to HS or reloading after HS. Fischer 344 x Brown Norway inbred rats (age 34 mo.) were given 1 ml/day of purified EGCg (50 mg/kg body weight), or the same sample volume of the vehicle by gavage. One group of animals received HS for 14 days and the second group of rats received 14 days of HS, then the HS was removed and they were allowed to recover by ambulating normally around the cage for two weeks. EGCg decreased a small number of autophagy genes in control muscles, but it increased the expression of other autophagy genes (e.g., ATG16L2, SNCA, TM9SF1, Pink1, PIM-2) and HS did not attenuate these increases. HS increased Beclin1, ATG7 and LC3-II/I protein abundance in hindlimb muscles. Relative to vehicle treatment, EGCg treatment had greater ATG12 protein abundance (35.8%, P<0.05), but decreased Beclin1 protein levels (−101.1%, P<0.05) after HS. However, in reloaded muscles, EGCg suppressed Beclin1 and LC3-II/I protein abundance as compared to vehicle treated muscles. EGCg appeared to “prime” autophagy signaling before and enhance autophagy gene expression and protein levels during unloading in muscles of aged rats, perhaps to improve the clearance of damaged organelles. However, EGCg suppressed autophagy signaling after reloading, potentially to increase the recovery of hindlimb muscles mass and function after loading is restored.
Keywords: Autophagy, muscle wasting, aging, muscle reloading, nutrition
1. Introduction
Sarcopenia independently lowers mobility (Murphy et al., 2014a; Murphy et al., 2014b; Roshanravan et al., 2016) and increases mortality in elderly persons (Metter et al., 2002; Murphy et al., 2014a). An abrupt decrease in physical activity leading to forced bedrest or hospitalization exacerbates muscle wasting in both middle aged (Arentson-Lantz et al., 2016) and elderly persons (Pisot et al., 2016), and significantly exacerbates their health problems (Inouye et al., 1993; Pisani et al., 2016). Furthermore, aging attenuates or delays recovery of muscle mass and function after disuse in elderly humans and aged animal models (Alway et al., 2014a; Hao et al., 2011; Pisot et al., 2016; Tanner et al., 2015; White et al., 2015; Zarzhevsky et al., 2001).
The mechanisms that regulate the poor recovery of muscle mass and function after inactivity in aging are not fully known. However, there is evidence that apoptotic signaling in myonuclei and satellite cells may have a role in regulating muscle loss in response to forced disuse (Alway et al., 2014a; Alway et al., 2015; Alway et al., 2011; Alway et al., 2014b; Hao et al., 2011; Marzetti et al., 2013a; Marzetti et al., 2012; Wang et al., 2011). Loss of autophagy signaling in satellite cells/muscle stem cells in aging muscle (Garcia-Prat et al., 2016; Sousa-Victor et al., 2014) might increase the susceptibility of satellite cells for apoptosis and reduce the regenerative capacity of muscle. A general suppression of autophagy in aging is also associated with muscle atrophy and reduced function in thigh muscles of older women (77–83 yrs.) (Drummond et al., 2014) and older mice (12 mo. male imprinting control region mice) (Kim et al., 2013) and this appears to permit increased apoptosis in aging muscles.
Autophagic removal of dysfunctional organelles such as mitochondria (mitophagy) appears to be an important suppressor of the apoptotic death-signaling program (Dutta et al., 2014; Lee 2016). Mitophagy is the selective removal of dysfunctional mitochondria by autophagic processes (Pryde et al. 2016; Wang et al. 2011) that requires tagging of dysfunctional mitochondria so that they can be recognized for disassembly. The selectivity of mitophagy is controlled by the loss of the mitochondrial membrane potential (Δψm) and by several important proteins such as BCL2 interaction myosin/moesin like coiled-coil protein 1 (Beclin1), in conjugation with autophagy-related gene (ATG) family members which support lipidation of microtubule associated protein light chain 3 (LC3)-I to LC3-II. Dysfunctional mitochondria are engulfed in the double membrane autophagosome (Amaya et al., 2015; Levine et al., 2015). The autophagosome fuses with a lysosome, and the proteolytic contents of the lysosome are emptied into the autophagosome, which then digests the dysfunctional mitochondria (Pryde et al. 2016; Wang et al. 2011).
Modulation of autophagy protein abundance in aging and muscle wasting is a complex process. For example, elevated autophagy signaling has been shown to occur in response to muscle disuse (Kang et al., 2016a; Kang et al., 2016b; White et al., 2015) and severe muscle wasting (Lokireddy et al., 2012; Wang et al., 2015). Autophagy has also been reported to increase in aging muscle (Pagano et al., 2015). However, autophagy also has been reported to be unchanged in skeletal muscles from elderly persons (Fry et al., 2013), and reduced in some pathological conditions of muscle loss including dystrophy (De et al., 2012; Spitali et al., 2013). Furthermore, reduced autophagy has been proposed to contribute to reduced muscle stem cell function (Garcia-Prat et al., 2016; Tang et al., 2014) and aging-associated muscle loss and muscle dysfunction (Carnio et al., 2014; Drummond et al., 2014; Rubinsztein et al., 2011; Sebastian et al., 2016; Wohlgemuth et al., 2010).
Lower autophagy/mitophagy would be expected to lead to an accumulation of dysfunctional muscle mitochondria, which in turn would activate mitochondrial-associated apoptotic signaling pathways. We hypothesized that part of the depressed recovery of muscle after forced inactivity such as that imposed by hindlimb suspension (HS) in aged rats (Alway et al., 2014a; Hao et al., 2011; Pisot et al., 2016; Tanner et al., 2015; White et al., 2015; Zarzhevsky et al., 2001) could be due to high levels of apoptosis and that increasing autophagy/mitophagy during reloading after disuse could remove dysfunctional mitochondria as the apoptotic initiators, thereby improving muscle recovery in aged animals or humans. However, the role of muscle autophagy/mitophagy during recovery from disuse-atrophy in aging is not clear because similar autophagy protein levels have been reported in hindlimb muscles of aged rats as compared to younger rats during recovery from disuse, despite a lower restoration of muscle mass in aging (White et al., 2015). Nevertheless, because removing unhealthy mitochondria and leaving healthy mitochondria in muscle (Vainshtein et al., 2015b) potentially could reduce aging-induced sarcopenia (Marzetti et al., 2013b; Wohlgemuth et al., 2010), it is still possible that the regulation mitophagy may have a role in determining the extent of muscle recovery from atrophied conditions in aging (Alway et al., 2017).
Clinically, it is important to identify strategies to reduce muscle loss during disuse and improve muscle recovery during reloading from disuse in aging. Green tea catechins were reported to reduce the loss of soleus muscle force during a period of HS in mice (Ota et al., 2011) and in aged rats (Alway et al., 2015). Epigallocatechin-3-gallate (EGCg) is the primary catechin found in green tea, and EGCg has been shown to improve muscle restoration after HS-induced inactivity in aged rats (Alway et al., 2014a). Both disuse and reloading greatly increase the oxidative stress in muscles (Andrianjafiniony et al., 2010; Jackson et al., 2010; Pellegrino et al., 2011) and oxidant stress is a powerful initiator of mitochondrial damage leading to apoptosis and muscle wasting (Herbst et al., 2016; Lee 2016; Wenz et al., 2009). EGCg has strong antioxidant and anti-inflammatory properties and oxidative stress is reduced in muscles of mice after eccentric exercise (Haramizu et al., 2011) that were given green tea catechins. Thus, EGCg is a potential therapeutic candidate for improving muscle recovery after inactivity by suppressing apoptosis, potentially by mediating oxidative stress. Nevertheless, we did not know if the EGCg mediated attenuation of apoptotic signaling (Alway et al., 2014a), was accomplished in concert with altered autophagy signaling or if if EGCg had any effect in regulating autophagy during HS or recovery from disuse in aged muscles.
In this study, we tested the hypothesis that EGCg would increase autophagy signaling during unloading but suppress autophagy signaling in hindlimb muscles of aged rats during recovery from unloading. The data generally support our hypothesis and suggest that EGCg increased autophagy during unloading and reduced autophagy in rodent hindlimb muscles during recovery after disuse. Taken together with our previous observations that EGCg catechins suppressed apoptosis signaling during reloading after disuse (Alway et al., 2014a; Alway et al., 2015), these findings provide a rationale to suggest that muscle recovery may be improved by pre-treating elderly subjects with EGCg before prolonged bedrest, or immobilization such as planned surgery, to potentially suppress both apoptosis and autophagy during muscle reloading.
2. Material and methods
2.1. Animal care
The National Institute on Aging has recommended the Fischer 344 Brown Norway (FBN) rat for aging research because it is a long-lived strain that has a low incidence of pathologies, a mean life span of approximately 32 months and a maximum life span of 41 months (Lipman et al., 1996). Male 34 months of age were obtained from the National Institute on Aging colony at Harlan. This age of rat is at the beginning of the senescent period. While older rats (~36 mo.) are more sarcopenic (Lushaj et al., 2008), our experience is that rats that are 34–35 months old handle the stress of hindlimb suspension quite well, but older rats become more stressed, do not eat well, and lose excessive body weight during the hindlimb unloading period. The Institutional Animal Care and Use Committee from the West Virginia University approved the experiments described in this paper. The standards for the animal care of laboratory animals as advocated by the American Association for Accreditation of Laboratory Animal Care (AAALAC) were followed in this study and the animals were housed in the institutional AAALAC accredited facility.
2.2. Hindlimb suspension (HS) and reloading (recovery)
Senescent rats were randomly divided into three groups: cage control (C, n=20), hindlimb suspended (HS, n=20) for 14 days, and 14 days of recovery (R, n=20) after HS. HS was conducted as described previously (Alway et al., 2014a; Alway et al., 2015; Hao et al., 2011; Siu et al., 2008). The animals had free access to water and food and they were housed individually. The suspension height was adjusted so that the torso angle did not exceed 30° with respect to the cage floor. The animals were euthanized either after 14 days of HS or after 14 days of normal cage ambulation exercise (recovery, R) following HS. The cage control animals moved freely around their cages until they were euthanized. The plantaris muscles were removed, frozen in liquid nitrogen, and stored until used for analyses.
2.3. Nutritional treatment
Ten animals in each experimental group received 1 ml of EGCg (50 mg/kg-Teavigo®, DSM/day) dissolved in distilled water, or 1 ml/day of the vehicle (distilled water) by gavage feeding once a day (Alway et al., 2014a; Alway et al., 2015). The animals in the HS and recovery groups were pretreated EGCg, or the vehicle for seven days before HS.
2.4. Autophagy microarrays
The Rat Autophagy RT2 Profiler™ PCR Array (Qiagen/SAbiosciences, Valencia, CA) was used to measure mRNA signals from 84 autophagy specific genes in control and treated muscles according to the manufacturer’s recommendation. Cage control and hindlimb suspended muscles were compared. The signals were detected by an Applied Biosystems 7300 Real-Time PCR System. RNA was purified from rat muscles using an RNeasy Plus Mini Kit (Qiagen). The A260:A280 ratio of the RNA was 1.8 to 2.0. One μg total RNA was used for cDNA synthesis. The ΔCT was calculated for each gene and its’ housekeeping gene and the fold change between cage control and treatment groups. The 2(−ΔΔCT) value was calculated with Qiagen software and the data were reported for each gene and reported as a fold change in gene expression relative to the appropriate control sample.
2.5. Immunoblots
Approximately 50 μg of muscle was homogenized in RIPA buffer (1% Triton x-100, 150 mM NaCl, 5 mM EDTA, 10 mM Tris; pH 7.4), containing a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). The protein content of the homogenate was measured using the BioRad DC protein assay (BioRad, Hercules, CA). Forty micrograms of protein were loaded into each well of a 4–12% gradient polyacrylamide gel (Invitrogen, Carlsbad, CA) and separated by routine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for 1 hour at 120V. The proteins were transferred to a nitrocellulose membrane for 1.5 hours at 25V. Non-specific protein binding was blocked by incubating the membranes in 5% nonfat milk in Tris-buffered saline containing 0.05% Tween 20 (TBST). The membranes were incubated with antibodies against Beclin1 (1:1000; Cell Signaling Technology, MA), LC3A/B (1:1000; Cell Signaling Technology, MA), ATG12 (1:1500; Cell Signaling Technology, MA), ATG7 (1:1000; Cell Signaling Technology, MA), MURF1 (1:250, Santa Cruz), peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α) (1:1000, Santa Cruz, CA), copper-zinc superoxide dismutase (CuZnSOD) (1:200, Santa Cruz), MURF1 (1:400, Santa Cruz, CA), manganese superoxide dismutase (MnSOD) (1:1000; Cell Signaling Technology, MA), and Sirt1 (1:1000; EMD Millipore, MA).
The membranes were incubated with appropriate dilutions of secondary antibodies (diluted in 5% non-fat milk) conjugated to horseradish peroxidase (Sigma-Aldrich, MO). The signals were developed using a chemiluminescent substrate (Lumigen TMA-6; Lumigen, MI) and visualized on a U:Genius photo-documentation system (Syngene, MD). The digital images were quantified using Image J, and each band was normalized to the GAPDH signal. The data were expressed as optical density (OD) relative to the OD of the GAPDH signal and were expressed as arbitrary units (AU).
2.6 Statistical analysis
The data are presented as means ± SE. Differences in means between groups were determined using Sigma Stat 3.5 (Point Richmond, CA) by a Two-way analysis of variance (ANOVA) with Hotelling’s T-Square test with groups (Vehicle, EGCg) and conditions (Control, HS or Recovery). Bonferroni post hoc analyses were conducted between significant means. Significance was established as P≤0.05.
3. Results
The animals received 14 days of HS or 14 days of HS followed by 14 days of reloading (Alway et al., 2014a; Alway et al., 2015; Alway et al., 2013). The animal characteristics and changes in muscle morphology have been reported previously for the animals that were given EGCg (Alway et al., 2014a). The amount of food that was consumed by the rats was reduced from 22.1 ± 2 g/day to 14.2 ± 1.6 g/day during HS and this is consistent with our observations in other studies (Alway et al., 2015; Alway et al., 2013). During recovery, the animals eat 16.4 ± 1.4 g/day of food but this was still significantly lower than the amount that they eat before HS. There was no difference in the amount of food that was consumed between animals that were given EGCg or the vehicle for any condition.
3.1. Gene array analysis of autophagy signaling proteins
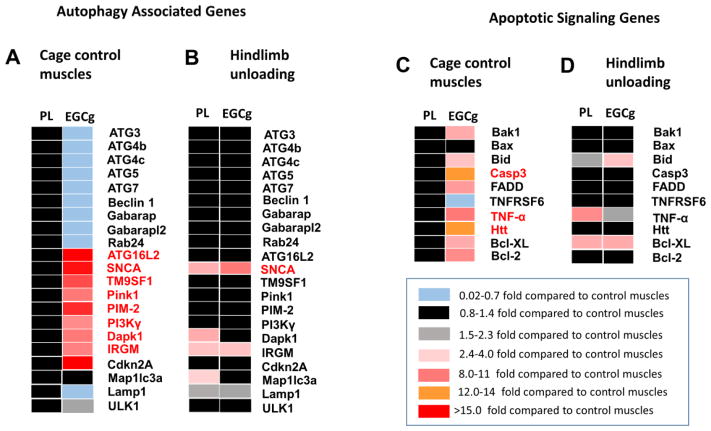
Gene array data were obtained from cage control, vehicle, and EGCg treated plantaris muscles after HS. The data showed changes in both autophagy and apoptosis gene expression by EGCg treatment alone in cage control animals (Figure 1A) and differential EGCg interactions of several autophagy associated genes in HS animals (Figure 1B). Only HS conditions were examined as a result of a freezer failure, which led to an insufficient amount of tissue to conduct analysis for muscles from the 14-day recovery group.
Figure 1. Autophagy genes that were regulated by EGCg.
The relative expression levels of autophagy-associated genes (A, B) and apoptosis associate genes (C,D) were determined in plantaris muscles of old rats by PCR array analysis. Data are shown for muscles from cage control animals (A, C), in plantaris muscles that were exposed to 14 days of HS (B,D). Rats were fed EGCg or a vehicle one week before and throughout 14 days of HS. mRNA was extracted from plantaris muscles and analyzed by PCR array the global data set were evaluated from 84 genes. The data are represented as a heat map, with significant increases indicated by shade of red, no change in black and small decreases indicated by blue as indicated by the scale. Cage control treatments are normalized to vehicle treatments. Hindlimb unloading data are normalized to the respective cage control treated gene of the same treatment.
3.1.1 EGCg down regulation of autophagy genes
EGCg tended to downregulate several autophagy-associated genes in plantaris muscles of cage control rats as compared to vehicle treated control animals (Figure 1A). These included Akt1, and protein transport associated with autophagy (ATG 4b, ATG4c, ATG7, Gabarap, Gabarapl2, Rab24), although these were all less than a one-fold, difference from vehicle treated cage control muscles.
3.1.2. EGCg upregulation of autophagy related genes
EGCg also increased the number of important autophagy signaling genes. This included an 89 fold increase in Autophagy related 16-like 2 (ATG16L2), which is an isoform of ATG16L. Autophagy related 16-like 1 (ATG16L1), is important for regulating lipidation of LC3 for autophagosome formation (Fujita et al., 2008) and ATG16L2 is part of a complex consisting of ATG5, ATG12 and ATG16L1 that is required for elongation of phagophores during autophagy (Ishibashi et al., 2011). The EGCg-induced increase in ATG16L2 in muscles of control rats as compared to vehicle treated control muscles, was sustained throughout HS, so that there was no difference between muscle ATG16L2 gene levels from cage control and HS animals treated with EGCg. The gene expression of alpha synuclein (SNCA) was increased by 32.6 fold by EGCg in cage control muscles. SNCA mRNA was further increased by 4.4 fold greater than cage control muscles by HS in EGCg treated animals. In addition, SNCA was increased by HS alone in muscles of vehicle treated animals, as SNCA gene expression was 3.6 fold greater in muscles from HS than cage control animals that were vehicle treated. The ubiquitously expressed TM9SF1 (MP70) may participate in autophagosome induction (He et al., 2009; Wang et al., 2015a). TM9SF1 gene expression was increased by 7 fold in EGCg treated cage control muscles, and while HS did not further upregulate this gene, it maintained gene expression at this high level because there was no difference in HS and vehicle treated EGCg muscles (Figure 1A, B). Furthermore, PTEN-induced putative kinase 1 (Pink1), a mitophagy signaling ubiquination related protein (Liu et al., 2016; Wu et al., 2015) was upregulated by 7.8 fold in cage control muscles treated with EGCg but not further changed by HS. Proviral Integrations of Moloney virus 2 (PIM-2) is another positive regulatory gene of autophagy was upregulated by 24.7 fold by EGCg in cage control animals. PIM-2 gene expression was not increased further by HS but rather, PIM-2 was maintained at this high level throughout HS. PIM-2 is thought to promote expression and organization of LC3 and Beclin1 and enhance lysosomal acidification (Bohensky et al., 2007).
Inhibition of phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma (PI3Kγ) has been suggested to induce autophagy in muscle (Hou et al., 2014; Zhao et al., 2007). PI3Kγ was increased by 4.1 fold in control EGCg treated muscles, but this was not altered by HS in either vehicle or EGCg treated muscles as compared to the control muscles, respectively, on these same treatments.
Death associated protein kinase 1 (Dapk1) gene expression was increased by 5.6 fold by EGCg in cage control muscles relative to muscles from vehicle treated control muscles. Dapk1 gene levels were increased by 2.4 fold after HS in vehicle-treated muscles, but Dapk1 gene expression was not different after HS in muscles from EGCg treated rats as compared to Dapk1 levels in control EGCg treated muscles.
Interferon gamma (IFNγ) mRNA was elevated by 54.5 fold by EGCg in cage control muscles as compared to muscles from vehicle-treated control rats. IFNγ tended to be depressed after HS in muscles of EGCg treated animals as compared to control muscles of EGCg treated animals. Although the role of Immunity-related GTPase family M protein (IRGM) in skeletal muscle is not clear, it is thought to play a role in the immune response to IFNγ by regulating autophagy based caspase 3 that is involved in cell removal after DNA damage (Lin et al., 2016). IRGM gene expression was elevated by 5.5 fold after EGCg treatment in cage control animals as compared to vehicle treated cage control rats. IRGM mRNA was further increased by 2.8 fold by HS in EGCg treated animals vs. the EGCg treated cage control muscles. HS also increased IRGM gene expression to a similar level in vehicle treated muscles, as there was a 2.6 fold greater gene expression level of IRGM in HS vs. cage control vehicle treated muscles. A similar pattern was seen for SNCA gene expression, where EGCg alone elevated SNCA mRNA levels by 32.5 fold relative to vehicle treated control muscles, and HS increased the gene expression level of SNCA mRNA by 3.6 and 4.4 fold in HS muscles after vehicle or EGCg treatment, respectively. Tumor necrosis factor (TNF) gene expression was increased by 32 fold after EGCg treatment alone. TNF gene expression was maintained by HS because there was no difference between EGCg treated cage control and HS EGCg-treated muscles. However, HS increased TNF gene expression by 3.1 fold in vehicle treated muscles of old rats as compared to vehicle treated cage control muscles.
Cyclin-dependent kinase inhibitor 2A (Cdkn2A) has a cell regulatory function, but when the levels of this gene are suppressed in aging, autophagy is inhibited in muscle stem cells (Garcia-Prat et al., 2016; Sousa-Victor et al., 2014). EGCg increased Cdkn2A gene expression by 61 fold in cage control muscles, but Cdkn2A was not significantly elevated by HS in either vehicle or EGCg treated muscles as compared to their control muscles.
3.1.3. EGCg upregulation of apoptotic genes
Although we have previously found that EGCg decreased apoptotic signaling in recovery after disuse, in this study we observed that EGCg upregulated a number of pro-apoptotic proteins in cage control muscles (Figure 1C,D). This included BCL2-antagonist/killer 1, (Bak1) which was increased by 2.2 fold compared to cage control vehicle treated muscles. Bak1 is thought to induce the opening of the mitochondrial voltage-dependent anion channel, leading to the release of cytochrome c from the mitochondria in apoptosis (Buytaert et al., 2006).
The gene expression of BH3 interacting domain death agonist (Bid) interacts with another Bax resulting in an insertion of Bax into the outer mitochondrial membrane. Bid is also involved in p53 regulated apoptosis (Sax et al., 2002). Bid mRNA was increased by 2.9 fold in EGCg treated cage control muscles as compared to vehicle treated control muscles. While EGCg increased the gene expression of Bid in HS muscles by 3.2 fold, Bid gene expression was not different in HS and control muscles of vehicle treated animals.
The effector pro-apoptotic cysteine-aspartic acid protease-3 (Casp3) is a well-studied regulator of apoptosis in muscle wasting (Siu et al., 2005; Siu et al., 2006; Siu et al., 2009). Casp3 mRNA increased by 14 fold in EGCg treated control muscles as compared to vehicle treated control muscles but the gene expression of Casp3 was maintained at this high level during HS-induced disuse in EGCg treated muscles as it was unchanged by HS vs. the cage control muscles.
Fas associated via death domain (FADD) was upregulated by 6.7 fold after EGCg treatment. FADD is an adaptor protein that bridges members of the TNF receptor superfamily, and the Fas-receptor, to procaspases 8 and 10 to form the death-inducing signaling complex (DISC) during apoptosis. Another closely associated pro-apoptotic protein, Fas, also called tumor necrosis factor receptor superfamily member 6 (TNFRSF6) assists FADD in forming the DISC upon ligand binding and allows FADD to bind to the death domain of Fas. Tumor necrosis factor-α (TNF-α) is a pro-apoptotic cytokine that was increased by 32 fold by EGCg in cage control animals as compared to control muscles of vehicle treated animals. HS increased TNF-α in vehicle treated muscles by three-fold but TNF-α was not different between control and HS muscles of EGCg treated rats.
The ubiquitously expressed Huntingtin gene (Htt) may be another important regulator of mitochondrial associated apoptosis (Costa et al., 2010; Gil et al., 2008). The Htt mRNA gene level was increased by 11.5 fold in EGCg-treated cage control muscles as compared to vehicle treated cage control muscles and remained at this gene level in HS EGCg-treated muscles from old animals. However, Htt was not changed from control levels in vehicle-treated HS muscles. The only autophagy-associated gene that was identified to increase solely from HS was microtubule-associated protein 1 light chain 3 alpha (Map1lc3a), which increased by two-fold in vehicle treated muscles as compared to control vehicle treated muscles. In contrast, Map1lc3a gene expression decreased slightly in EGCg treated muscles from cage control animals, as compared to vehicle treated control muscles and this was not changed by HS (Figure 1 A,B)
The mRNA level for anti-apoptotic proteins B-cell lymphoma 2 (Bcl-2) and Bcl2-like 1 (Bcl-XL) were increased by 3.8 fold and 2.5 fold in cage control muscles after EGCg treatment. While HS did not increase Bcl-2 further, after either vehicle or EGCg treatment. HS increased Bcl-XL to similar levels by HS in both vehicle and EGCg treated muscles.
3.2. Autophagy signaling proteins in unloaded and reloaded muscles
3.2.1. Beclin1
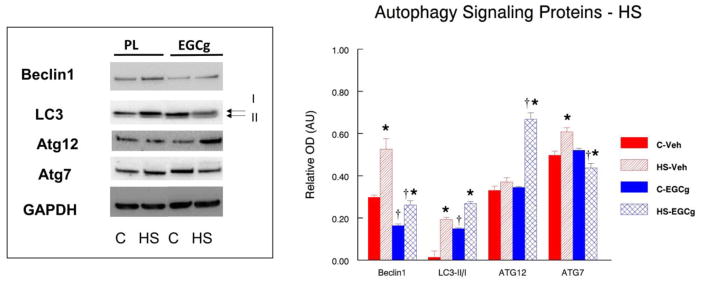
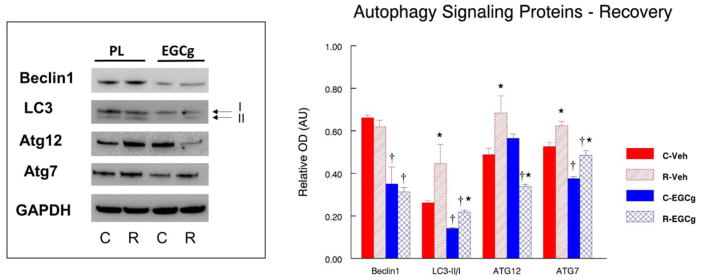
We conducted Western blots on some of the key proteins that are known to be important in autophagy signaling and other proteins that had genes differentially expressed by the array analysis. Beclin1 (ATG6) is a well-known key regulator of autophagy that acts in part to recruit additional ATG proteins to form autophagosomes although it may have other roles than just autophagy (Wirawan et al., 2012). Beclin1 protein abundance was 45% lower (P<0.05) in muscles of EGCg fed cage control animals as compared to placebo fed cage control animals (Figure 2). HS increased Beclin1 by 76.5% (P<0.05) and 59.8% (P<0.05) in muscles of vehicle and EGCg-treated animals, respectively. However, Beclin1 protein abundance was 50.2% lower (P<0.05) in muscles of HS animals fed EGCg as compared to muscle from HS animals given the vehicle. Unlike HS, Beclin1 was not different in the reloaded muscles as compared to muscles of cage control animals in vehicle or EGCg fed animals (Figure 3). However, Beclin1 was suppressed by EGCg because Beclin1 remained lower in the muscles of cage control EGCg fed animals as compared to cage control vehicle treated animals (−47.1%). Beclin1 was also 49.5% lower (P<0.05) in recovery muscles of EGCg fed animals as compared to reloaded muscles from vehicle treated animals.
Figure 2. Autophagy signaling proteins after hindlimb unloading.
The abundance of autophagy signaling proteins was determined by Western blot analysis in plantaris muscles of rats that had received 14 days of hindlimb suspension induced muscle disuse. The animals received epigallocatechin gallate (EGCg) or the vehicle (Veh) daily by gavage. GAPDH was used as a loading control. The density and area from the respective apoptotic signaling protein bands were quantified and the signals were normalized to GAPDH. Representative blots and summarized group data are presented. The data are displayed as mean ± SE for plantaris. *p<0.05, Control vs. treatment group; †p<0.05, Vehicle vs. treatment group of the same condition.
Figure 3. Autophagy signaling after 14 days of recovery that followed 14 days of hindlimb suspension.
The abundance of autophagy signaling proteins was determined by Western blot analysis in plantaris muscles of rats that had received 14 days of hindlimb suspension induced muscle disuse. The animals received epigallocatechin gallate (EGCg) or the placebo-vehicle (Veh) daily by gavage. GAPDH was used as a loading control. The density and area from the respective apoptotic signaling protein bands were quantified and the signals were normalized to GAPDH. Representative blots and summarized group data are presented. The data are displayed as mean ± SE for plantaris. * p<0.05, Control vs. treatment group; †p<0.05, Vehicle vs. treatment group of the same condition.
3.2.2. LC3
The ratio of LC3-II/I is a marker for an increased propensity for autophagic signaling. EGCg appeared to increase autophagy signaling even without other interventions because the ratio of LC3-II/I was significantly greater (193.3%, P<0.05) in muscles of cage control EGCg treated animals vs. muscles of vehicle-treated cage control animals. While HS increased the LC3-II/I ratio to a greater extent in unloaded muscles of placebo (278.9%, P<0.05) as compared to EGCg (80.1%, P<0.05) treated animals, there was no difference between the LC3-II/1 ratios of unloaded muscles of vehicle and EGCg treated muscles after HS (Figure 2). After 14 days of reloading, the ratio of LC3-II/I was 237.9% (P<0.05) greater in recovery muscles as compared to cage control muscles of vehicle-treated animals. The LC3-II/I ratio in animals that were treated with EGCg was 54.6% greater (P<0.05) in muscles from reloaded rats as compared to muscles from cage control animals. Interestingly, the LC3-II/I ratio was lower in reloaded muscles of animals that were fed EGCG as compared to animals that were vehicle treated (−51.1%, P<0.05) (Figure 3) which suggests that EGCg suppressed autophagy during reloading.
3.2.3. ATG12
EGCg did not alter ATG12 protein abundance in muscles from cage control animals. However, ATG12 protein abundance was significantly greater after unloading in EGCg treated muscles (93.6%, P<0.05) as compared to control muscles in EGCg treated animals, yet there was no HS-induced increase in ATG12 protein levels in vehicle treated muscles. Furthermore, ATG12 protein abundance was 80.1% (P<0.05) greater in HS-unloaded muscles of EGCg as compared to HS-unloaded muscles from vehicle treated animals. This suggested a potential for a heightened autophagy response of ATG12 to unloading in EGCg treated muscles.
ATG12 protein abundance was very different in reloaded muscles as compared to muscles after HS. After reloading, the ATG12 protein abundance was 40.2% greater (P<0.05) in the muscles from recovery animals fed the vehicle as compared to the control animals fed the vehicle treatment. In contrast, ATG12 protein abundance was 40.0% (P<0.05) lower in reloaded muscles of EGCg treated animals as compared to control muscles from EGCg treated animals. Furthermore, after HS, ATG12 protein abundance was 28% lower in muscles from EGCg treated as compared to vehicle treated animals (Figure 3). This indicates that ATG12 was reduced in reloaded muscles of EGCg treated animals as compared to vehicle treated animals.
3.2.4. ATG7
EGCg treatment did not alter the level of ATG7 proteins under control conditions because the protein abundance of ATG7 was similar in muscles from cage control animals that received the vehicle or EGCg treatment (Figure 2). ATG7 protein abundance in vehicle treated animals was 22.3% (P<0.05) greater in HS muscles than muscles from cage control animals. In contrast, after HS, the muscles from EGCg treated animals had a 15.9% lower (P<0.05) ATG7 protein abundance than muscles from cage control animals (Figure 2). Following 14 days of recovery after HS, ATG7 protein abundance was 18.6% (P<0.05) greater in the reloaded muscles of vehicle treated animals as compared to muscles from the vehicle treated cage control animals. ATG7 protein abundance was 29.3% greater (P<0.05) in reloaded muscles of animals that received the EGCg treatment as compared to muscles of cage control animals that received EGCg but did not undergo the initial HS or the reloading. The protein abundance of ATG7 was 28.0% (P<0.05) lower in HS muscles of EGCg treated animals as compared to vehicle treated HS animals. Similarly, the protein abundance of ATG7 was 22.1% (P<0.05) lower in reloaded muscles of EGCg treated animals as compared to reloaded vehicle treated animals (Figure 3). Thus, relative to vehicle treatment, EGCg suppressed ATG7 protein abundance in muscles after both HS and reloading.
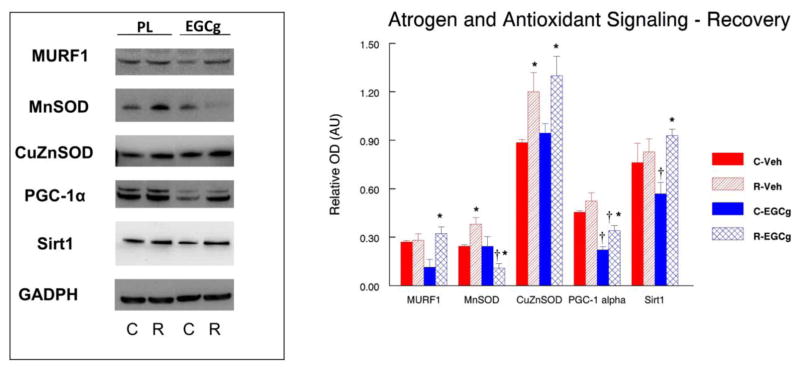
3.3. Atrogen and antioxidant signaling after hindlimb unloading and recovery from unloading
Basic levels of reactive oxygen species (ROS) may promote cell survival and adaptation to stress, but an excessive level of ROS can induce autophagy and apoptosis (Andrianjafiniony et al., 2010; Calvani et al., 2013; Kocturk et al., 2008; Zhang et al., 2015). Basal ROS levels are already high in aging muscle and are increased even further with disuse and overload (Alway et al., 2015; Jackson et al., 2011; Jackson et al., 2010; Ryan et al., 2011; Ryan et al., 2010), and therefore ROS levels might be a mitigating factor in determining autophagy levels in aging muscle. Therefore, we examined muscle RING finger 1 (MURF1), an E3 ubiquitin ligase that is an autophagic sensitive atrogen, and markers of antioxidant enzymes, and expected that if ROS levels were elevated, MnSOD and CuZnSOD would be increased as a countermeasure to the increased oxidative stress. Furthermore, as PGC1-α has been linked to autophagy, and Sirt1 is a known activator of PGC1-α we wanted to determine if either of these proteins increased with unloading or reloading.
3.3.1. MURF1
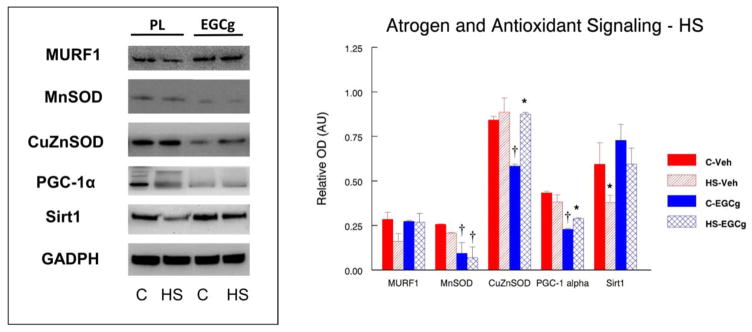
Although surprising, our data (Figure 4) suggest that the protein abundance of MURF1 was not increased by hindlimb suspension in any of the experimental conditions in old animals, and further, EGCg treatment did not change MURF1 protein abundance in unloaded muscles of the old rats as compared to vehicle treated HS muscles. However, MURF1 was significantly greater (64.9%, P<0.05) in reloaded muscles from animals that received the EGCg treatment as compared to the EGCg treated control muscles.
Figure 4. Atrogen and stress signaling after 14 days of hindlimb suspension.
The abundance of the atrogen RING-finger protein-1 (MURF1), mitochondrial antioxidant manganese superoxide dismutase (MnSOD), cytosolic antioxidant copper/zinc superoxide dismutase (CuZnSOD), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and silent mating type information regulation 2 homolog 1 (Sirt1) proteins was determined by western blot analysis in plantaris muscles of rats that had received 14 days of hindlimb suspension induced muscle disuse. The animals received epigallocatechin gallate (EGCg) or the placebo-vehicle (Veh) daily by gavage. GAPDH was used as a loading control. The density and area from the respective apoptotic signaling protein bands were quantified and the signals were normalized to GAPDH. Representative blots and summarized group data are presented. The data are displayed as mean ± SE for plantaris. * p<0.05, Control vs. treatment group; †p<0.05, Placebo vs. treatment group of the same condition.
3.3.2. MnSOD
The mitochondrial housed antioxidant MnSOD was not different in control and unloaded muscles from vehicle treated rat (Figure 4). However, MnSOD was suppressed in EGCg treated muscles in both control (−196.4%, P<0.05) and hindlimb suspended (−686.6%) muscles of EGCg treated rats as compared to muscles in placebo treated animals. MnSOD was 35.9% (P<0.05) greater in control muscles from reloaded placebo treated animals as compared to the control muscles, of vehicle treated animals (Figure 5). MnSOD protein abundance was lower in reloaded muscles of EGCg treated animals as compared to control muscles of the EGCg treated animals.
Figure 5. Atrogen and stress signaling after 14 days of recovery, which followed 14 days of hindlimb suspension.
The abundance of the atrogen RING-finger protein-1 (MURF1), mitochondrial antioxidant manganese superoxide dismutase (MnSOD), cytosolic antioxidant copper/zinc superoxide dismutase (CuZnSOD), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and silent mating type information regulation 2 homolog 1 (Sirt1) proteins was determined by Western blot analysis in plantaris muscles of rats that had received 14 days of hindlimb suspension induced muscle disuse. The animals received epigallocatechin gallate (EGCg) or the placebo-vehicle (Veh) daily by gavage. GAPDH was used as a loading control. The density and area from the respective apoptotic signaling protein bands were quantified and the signals were normalized to GAPDH. Representative blots and summarized group data are presented. The data are displayed as mean ± SE for plantaris. * p<0.05, Control vs. treatment group; †p<0.05, Vehicle vs. treatment group of the same condition.
3.3.3. CuZnSOD
The protein abundance of CuZnSOD was 44.2% lower (P<0.05) in EGCg treated cage control muscles as compared to placebo control muscles (Figure 4). CuZnSOD protein abundance was 33.3%, (P<0.05) greater in HS muscles of EGCg treated rats as compared to the control muscles of EGCg treated animals, but CuZnSOD did was not different between muscles from control and suspended animals that were vehicle treated. CuZnSOD protein abundance was significantly greater in reloaded plantaris muscles than control muscles after both vehicle (26.2%, P<0.05) and EGCg (27.4%, P<0.05) treatments as compared to their respective control muscles (Figure 5).
3.3.4. PGC-1α
Relative to control muscles from vehicle treated rats, PGC1α protein abundance was suppressed by EGCg treatment in cage control muscles. However, HS increased the PGC1α protein abundance by 15.3% (P<0.05) in HS muscles of EGCg treated muscles as compared to the EGCg treated control muscles (Figure 4). PGC-1α protein abundance was greater in reloaded muscles of EGCg treated animals (38.7%, P<0.05) as compared to EGCg treated control muscles. However, PGC-1α protein levels remained suppressed in cage control, EGCg treated muscles (−105.5%, P<0.05) as compared to control placebo treated muscles (Figure 4, 5) throughout the 28 days of the reloading study.
3.3.5. Sirt1
EGCg did not improve Sirt1 protein abundance in muscles of cage control animals as compared to placebo treated animals (Figure 4). In vehicle treated animals, Sirt1 protein abundance was significantly lower (−56.6%, P<0.05) in muscles after HS as compared to the control muscles. Sirt1 protein abundance was unchanged by HS in EGCg treated animals as compared to EGCg treated control muscles. Reloading in EGCg treated muscles increased Sirt1 protein (38.7%, P<0.05) as compared to the control muscles of EGCg treated animals but this was similar to vehicle treated Sirt1 muscle levels after reloading (Figure 5).
4. Discussion
The results of our current study suggest that when compared to vehicle treatment, EGCg increased the expression of several autophagy-associated genes (e.g., ATG16L2, SNCA, TM9SF1, Pink1, PIM-2). Additionally, the abundance of autophagy signaling proteins generally increased during HS in both vehicle and EGCg treatment. However, EGCg treatment suppressed autophagy signaling during reloading after disuse as compared to vehicle treatment and this was associated with improved recovery of muscle mass in aging. These results suggest that the modulation of autophagy by EGCg and similar catechins may provide a therapeutic intervention that would potentially improve recovery after disuse (e.g., HS, forced bedrest).
4.1. Regulation (“priming”) of autophagy signaling by EGCg in control muscles
Under basal (cage control) conditions in the current study, EGCg increased muscle gene levels of several autophagy genes as compared to vehicle treated animals. For example, Pink1 was ~8 fold greater in control muscles of animals treated with EGCg as compared to control muscles from vehicle treated animals. The greater Pink1 levels in EGCg treated muscles may represent an elevation in a targeting control pathway to eliminate dysfunctional mitochondria by mitophagy (Kang et al., 2016b). Other autophagy genes like PIM-2 were ~25 fold greater in control muscles of EGCg treated animals as compared to control muscles from vehicle treated animals. This is important because PIM-2 promotes the expression and organization of autophagic proteins LC3, and Beclin1 (Bohensky et al., 2007). Although Beclin1 protein and gene expression levels were lower in EGCg treated as compared to vehicle treated control muscles, LC3-II/I was higher in control muscles of EGCg treated animals as compared to vehicle treated animals. The higher Pink1 and PIM-2 levels might indicate that EGCg increased autophagy/mitophagy signaling, in basal muscles, essentially “priming” these control muscles for mitophagy even without additional external stimuli. This priming of autophagy signaling in control muscles before disuse may be an important strategy for reversing the suppression of autophagy signaling that has been reported in aged muscles under control conditions (Carnio et al., 2014; Drummond et al., 2014; Garcia-Prat et al., 2016; Rubinsztein et al., 2011; Sebastian et al., 2016; Tang et al., 2014; Wohlgemuth et al., 2010). Further support for this idea comes from observations that overexpression of PGC-1α reduces Pink1 levels in response to immobilization (reducing mitophagy) followed by remobilization (exercise) (Kang et al., 2016a), and in our study, we found that PGC1-α protein abundance was lower in EGCg treated control and HS muscles (increasing autophagy signaling) and indeed, most autophagy proteins were elevated in EGCg as compared to vehicle treated muscles after HS. This observation is consistent with the idea that EGCg promotes autophagy signaling in control and HS muscles. Nevertheless, not all autophagy genes were upregulated by EGCg in resting muscle (Figure 1).
4.2. Modulation of autophagy signaling by EGCg after HS
Autophagy genes that were greater in control EGCg treated muscles as compared to vehicle treated control muscles, largely maintained this higher level during HS. It is important to note that the HS data for EGCg treated animals were normalized to the cage control samples that were treated with EGCg, so the general lack of change in the gene expression after HS (Figure 1) indicates that the genes that had an EGCg-mediated increase in autophagy gene expression were not reduced by HS. Rather HS conditions maintained high autophagy genes at the same levels that were achieved by EGCg in cage control muscles. Thus, the initial “priming” of at least part of the autophagy pathway by EGCg in the pretreatment before HS (e.g., in control muscles) was maintained or increased during HS presumably for removing dysfunctional proteins and damaged organelles including mitochondria that are damaged during disuse and muscle wasting.
While different from the gene array data, Western blot data showed that HS increased Beclin1 protein abundance in both EGCg and vehicle treated muscles. However, Beclin1 protein abundance remained lower in both control and HS muscles of EGCg treated animals relative to muscles from vehicle treated control or HS animals, respectively. Nevertheless, EGCg appeared to prime the autophagy pathway prior to HS, because both control muscles and muscles from HS animals had higher LC3-II/I ratios as compared to vehicle treated control or HS muscles respectively.
ATG12 protein abundance was greater in muscles from EGCg treated animals after HS as compared to vehicle treated animals, and this is consistent with the idea that EGCg increased autophagic signaling during HS. In contrast, ATG7 increased with HS in vehicle treated muscles compared to their control muscles, but it was reduced by HS in EGCg treated muscles as compared to muscles from EGCg treated animals. Thus, while there was a general pattern for an increase in autophagy signaling in EGCg treated muscles during HS, this was not universal for all autophagy proteins.
4.3. Autophagy protein signaling by EGCg during recovery following HS
The protein abundances of Beclin1, LC3-II/I, ATG12, and ATG7 were all significantly lower in reloaded muscles of EGCg treated as compared to vehicle treated animals. While reloading increased LC3-II/I and ATG7 as compared to control muscles in EGCg treated animals, the abundance of these autophagy proteins in recovery muscles was still lower than found in vehicle treated muscles for these conditions. While our data support the idea that reduced autophagy signaling may have promoted improved muscle recovery in EGCg treated animals, we cannot rule out the possibility that autophagy protein abundance may not be linearly related to muscle adaptation in our model. For example, data from White et al. show that ATG7 protein levels were similar in young and old muscles during a period of recovery after atrophy, despite suppressed muscle protein accumulation with aging (Ballak et al., 2015; White et al., 2015) and this suggested that protein levels and muscle outcomes were not linear. We do not know if the lower autophagy protein levels in EGCg treated muscles directly affected muscle recovery or occurred in response to muscle recovery, and this will take additional studies to clarify this point.
4.4 Apoptotic regulation
An elevation of apoptotic gene expression levels has been reported previously in conditions of aging and muscle disuse (Alway et al., 2014a; Bennett et al., 2013; Hao et al., 2011; Jackson et al., 2010; Leeuwenburgh et al., 2005; Marzetti et al., 2012; Ogata et al., 2009; Pistilli et al., 2006). The PCR array data in the current study identified an EGCg-induced elevation in a number of apoptotic genes in muscles of cage control animals. However, there was also an increase in Bcl-2 and Bcl-XL, two anti-apoptotic genes, which would presumably have the potential to temper the increase in apoptotic signaling in EGCg treated muscles. Similar to autophagy signaling, the apoptotic “priming” that occurred with EGCg treatment appeared to be sustained during HS, as gene levels in muscles after HS were similar in unloaded and control EGCg treated muscles (Figure 1 C, D). Thus, EGCg appeared to elevate both basal apoptotic- and autophagy-signaling genes but this did not exacerbate muscle loss with HS.
4.5 Atrogene and mitochondrial associated stress signaling
MURF1 is ROS sensitive autophagy related ubiquitin ligase that is greatly increased in atrophying muscles (Bedard et al., 2015; Gomes et al., 2012; Smith et al., 2014; Tanner et al., 2015). ROS, and in sarcopenic muscles from old rats (Alway et al., 2014a; Alway et al., 2015; Hao et al., 2011; Ziaaldini et al., 2015). However, in the current study, MURF1 showed little changes after HS or reloading in vehicle treated muscles as compared to control muscles from rats whereas, EGCg appeared to suppress MURF1 in control muscles of recovery rats. As muscles from young animals were not studied, it is not possible to rule out that the longer treatment with EGCg reduced the catabolic signaling that was already at very high levels in control muscles from old animals, and that unloading reached a celling effect for MURF1.
MURF1 levels are increased with ROS (Doyle et al., 2011; Li et al., 2003), but MURF1 also has a role in regulating mitochondrial induced ROS production (Mattox et al., 2014). As ROS is increased during HS and reloading, we examined whether antioxidant protein levels that are associated primarily with the mitochondria (MnSOD) or cytosol (CuZnSOD) regions of the muscle would change along with MURF1 protein levels. Interestingly, when compared to vehicle treated muscles, the protein abundance of MnSOD and CuZnSOD were lower in control muscles of EGCg treated animals as compared to vehicle treated animals, but HS failed to increase the muscle levels of MnSOD in EGCg treated animals. This suggests that EGCg was acting as a direct or indirect buffer against the additional mitochondrial associated ROS imposed by HS. In contrast, the greater abundance of the muscle cytosolic CuZnSOD protein from EGCg treated animals after HS (Figure 4), suggests that EGCg did not buffer cytosolic ROS and this required an elevation in CuZnSOD. Furthermore, these antioxidant gene responses occurred without any change in MURF1 protein abundance in muscles from control, HS- or recovery-treated animals as compared to vehicle treatment (Figure 4). These data suggest that EGCg regulation of antioxidant proteins did not affect MURF1 atrogen protein abundance during HS or recovery.
While PGC1α increases mitochondrial biogenesis, it may also affect transcriptional regulation of autophagy genes (Fedorova et al., 2013; Scott et al., 2014). Furthermore, overexpression of PGC1-α can reduce muscle wasting (Cannavino et al., 2014), and increase autophagy (Vainshtein et al., 2015b), but the loss of PGC1-α can reduce autophagy (Vainshtein et al., 2015a). Contrary to our expectation, PGC1-α was lower in EGCg treated control muscles as compared to vehicle treated control muscles. Although PGC1-α increased with HS and reloading in muscles of EGCg treated rats, it remained lower than vehicle treated muscles. As Sirt1 is a known activator of PGC1-α, we examined if EGCg altered Sirt1 levels in muscles of the old rats in this study. Indeed Sirt1 protein abundance did not increase after HS as compared to control muscles of EGCg treated animals. Both PGC1-α and Sirt1 were lower in control muscles of recovery animals fed EGCg as compared to vehicle treated animals, but these two proteins increased in response to reloading in muscles from the EGCg but not the vehicle treated animals. Together, these findings suggest that there may be a link between autophagy, PGC1-α, Sirt1 and improved muscle recovery in EGCg-treated muscles and this implicates PGC1-α, and Sirt1 in autophagy signaling pathways, potentially for removing dysfunctional mitochondria via autophagic (mitophagy) pathways (Lo et al., 2014).
5. Conclusions
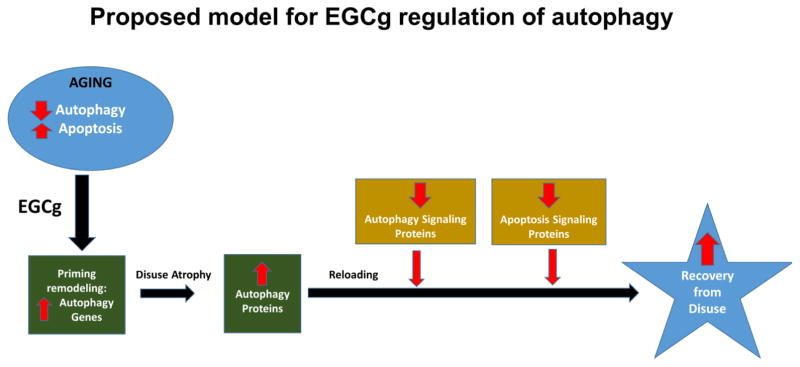
This study showed that EGCg modulates autophagic signals in unloaded and reloaded muscles of old rats. We propose that pre-treatment with these compounds (perhaps for several weeks prior to planned surgery) could provide a strategy to prime the signaling pathways that will be needed to clear dysfunctional proteins and organelles to improve recovery after a period of muscle disuse (Figure 6). Strategies that reduce the extent of muscle wasting or muscle recovery during rehabilitative efforts is clinically relevant to aging populations, especially after periods of prolonged bed rest or immobilization. However, additional studies are needed to clarify whether autophagy is essential to derive benefits from EGCg treatment during muscle recovery after forced disuse. Moreover, additional work is warranted to determine if EGCg treatment prior to scheduled bed rest (e.g., after a planned surgery) will “prime” autophagy signaling in skeletal muscle thereby improving recovery during reloading.
Figure 6. Proposed model of EGCg regulation of autophagy in aged muscles.
Muscles from aged animals have greater levels of apoptosis signaling and many studies suggest a lower overall autophagy-signaling program. We propose that EGCg treatment primes muscle by increasing several autophagy genes. The upregulation of autophagy genes and proteins by EGCg persists during muscle disuse. This EGCg induced elevation in autophagy proteins likely improves the clearance of damaged mitochondrial and other proteins during muscle disuse. This clearance provides a better cellular environment so that when muscle reloading occurs, muscle recovery from reloading is improved. The suppression of autophagy and apoptotic death signaling during reloading permits a greater accumulation of muscle protein and restoration during reloading.
Highlights.
Epigallocatechin-3-gallate (EGCg) supplementation increased autophagy expression in aged muscles of control rodents.
EGCg treatment maintained muscle autophagy signaling at high levels in during muscle disuse in aging.
EGCg suppressed autophagy protein abundance when muscles were reloaded after disuse in aging.
EGCg treatment primed autophagy before unloading and suppressed muscle autophagy signaling after reloading, potentially to increase the recovery of muscle mass and function after loading is restored.
Acknowledgments
Funding.
This project has been funded in grants from the Genomics Core Facility and WV-INBRE (NIH grant P20 GM103434), WV Program to Stimulate Competitive Research (PSCoR) and Abbott Nutrition R&D to SEA. The salaries for HT and YS were funded by the program of Strategic Fosterage of Young Researchers who Innovate System of Livestock Production (S2305, Young Researcher Overseas Visits Program for Vitalizing Brain Circulation) which was awarded to TG. Abbott Nutrition R&D provided salary support for SLP. We would like to acknowledge the facilities and staff of the West Virginia University Microscope Imaging Facility, which is supported by the Mary Babb Randolph Cancer Center and NIH grants P20 RR016440 and P30 RR032138/GM103488. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations
- ATG
Autophagy-related gene
- Bak1
BCL2-antagonist/killer 1
- Bcl-2
B-cell lymphoma 2
- Bcl-XL
Bcl2-like 1
- Beclin1
BCL2 interaction myosin/moesin like coiled-coil protein 1
- Cdkn2A
Cyclin-dependent kinase inhibitor 2A
- Dapk1
Death associated protein kinase 1
- EGCg
epigallocatechin gallate
- FADD
Fas associated via death domain
- Htt
Huntingtin gene
- IFNγ
Interferon gamma
- IRGM
Immunity-related GTPase family M protein
- LC3
Microtubule-associated protein light chain 3
- Map1lc3a
Microtubule-associated protein 1 light chain 3 alpha
- MURF1
Atrogen/RING-finger protein-1
- MnSOD
Mitochondrial antioxidant manganese superoxide dismutase
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PI3Kγ
Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma
- PIM-2
Proviral Integrations of Moloney virus 2
- ROS
reactive oxygen species
- Sirt1
silent mating type information regulation 2 homolog 1
- TNF
Tumor necrosis factor
- TNF-α
Tumor necrosis factor-α
- TNFRSF6
Tumor necrosis factor receptor superfamily member 6
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alway SE, Bennett BT, Wilson JC, Edens NK, Pereira SL. Epigallocatechin-3-gallate improves plantaris muscle recovery after disuse in aged rats. Exp Gerontol. 2014a;50:82–94. doi: 10.1016/j.exger.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alway SE, Bennett BT, Wilson JC, Sperringer J, Mohamed JS, Edens NK, Pereira SL. Green tea extract attenuates muscle loss and improves muscle function during disuse, but fails to improve muscle recovery following unloading in aged rats. J Appl Physiol (1985) 2015;118:319–330. doi: 10.1152/japplphysiol.00674.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alway SE, Mohamed JS, Myers MJ. Mitochondria hold the keys for driving sarcopenia. Exerc Sport Sci Rev. 2017 doi: 10.1249/JES.0000000000000101. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alway SE, Morissette MR, Siu PM. Aging and apoptosis in muscle. Handbook of the Biology of Aging. 2011;7:63–118. [Google Scholar]
- Alway SE, Myers MJ, Mohamed JS. Regulation of satellite cell function in sarcopenia. Frontiers of Aging Neuroscience. 2014b;6:1–15. doi: 10.3389/fnagi.2014.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alway SE, Pereira SL, Edens NK, Hao Y, Bennett BT. β–Hydroxy-β-methylbutyrate (HMB) enhances the proliferation of satellite cells in fast muscles of aged rats during recovery from disuse atrophy. Exp Gerontol. 2013;48:973–984. doi: 10.1016/j.exger.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Andrianjafiniony T, Dupre-Aucouturier S, Letexier D, Couchoux H, Desplanches D. Oxidative stress, apoptosis, and proteolysis in skeletal muscle repair after unloading. Am J Physiol Cell Physiol. 2010;299:C307–C315. doi: 10.1152/ajpcell.00069.2010. [DOI] [PubMed] [Google Scholar]
- Arentson-Lantz EJ, English KL, Paddon-Jones D, Fry CS. Fourteen days of bed rest induces a decline in satellite cell content and robust atrophy of skeletal muscle fibers in middle-aged adults. J Appl Physiol (1985) 2016;120:965–975. doi: 10.1152/japplphysiol.00799.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballak SB, Jaspers RT, Deldicque L, Chalil S, Peters EL, de HA, Degens H. Blunted hypertrophic response in old mouse muscle is associated with a lower satellite cell density and is not alleviated by resveratrol. Exp Gerontol. 2015;62:23–31. doi: 10.1016/j.exger.2014.12.020. [DOI] [PubMed] [Google Scholar]
- Bedard N, Jammoul S, Moore T, Wykes L, Hallauer PL, Hastings KE, Stretch C, Baracos V, Chevalier S, Plourde M, Coyne E, Wing SS. Inactivation of the ubiquitin-specific protease 19 deubiquitinating enzyme protects against muscle wasting. FASEB J. 2015;29:3889–3898. doi: 10.1096/fj.15-270579. [DOI] [PubMed] [Google Scholar]
- Bennett BT, Mohamed JS, Alway SE. Effects of resveratrol on the recovery of muscle mass following disuse in the plantaris muscle of aged rats. PLoS One. 2013;8:e83518. doi: 10.1371/journal.pone.0083518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohensky J, Shapiro IM, Leshinsky S, Watanabe H, Srinivas V. PIM-2 is an independent regulator of chondrocyte survival and autophagy in the epiphyseal growth plate. J Cell Physiol. 2007;213:246–251. doi: 10.1002/jcp.21117. [DOI] [PubMed] [Google Scholar]
- Buytaert E, Callewaert G, Vandenheede JR, Agostinis P. Deficiency in apoptotic effectors Bax and Bak reveals an autophagic cell death pathway initiated by photodamage to the endoplasmic reticulum. Autophagy. 2006;2:238–240. doi: 10.4161/auto.2730. [DOI] [PubMed] [Google Scholar]
- Calvani R, Joseph AM, Adhihetty PJ, Miccheli A, Bossola M, Leeuwenburgh C, Bernabei R, Marzetti E. Mitochondrial pathways in sarcopenia of aging and disuse muscle atrophy. Biol Chem. 2013;394:393–414. doi: 10.1515/hsz-2012-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavino J, Brocca L, Sandri M, Bottinelli R, Pellegrino MA. PGC1-alpha over-expression prevents metabolic alterations and soleus muscle atrophy in hindlimb unloaded mice. J Physiol. 2014;592:4575–4589. doi: 10.1113/jphysiol.2014.275545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnio S, LoVerso F, Baraibar MA, Longa E, Khan MM, Maffei M, Reischl M, Canepari M, Loefler S, Kern H, Blaauw B, Friguet B, Bottinelli R, Rudolf R, Sandri M. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep. 2014;8:1509–1521. doi: 10.1016/j.celrep.2014.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa V, Giacomello M, Hudec R, Lopreiato R, Ermak G, Lim D, Malorni W, Davies KJ, Carafoli E, Scorrano L. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol Med. 2010;2:490–503. doi: 10.1002/emmm.201000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De PC, Morisi F, Cheli S, Pambianco S, Cappello V, Vezzoli M, Rovere-Querini P, Moggio M, Ripolone M, Francolini M, Sandri M, Clementi E. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis. 2012;3:e418. doi: 10.1038/cddis.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A, Zhang G, Abdel Fattah EA, Eissa NT, Li YP. Toll-like receptor 4 mediates lipopolysaccharide-induced muscle catabolism via coordinate activation of ubiquitin-proteasome and autophagy-lysosome pathways. FASEB J. 2011;25:99–110. doi: 10.1096/fj.10-164152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MJ, Addison O, Brunker L, Hopkins PN, McClain DA, Lastayo PC, Marcus RL. Downregulation of E3 Ubiquitin Ligases and Mitophagy-Related Genes in Skeletal Muscle of Physically Inactive, Frail Older Women: A Cross-Sectional Comparison. J Gerontol A Biol Sci Med Sci. 2014 doi: 10.1093/gerona/glu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Xu J, Dirain ML, Leeuwenburgh C. Calorie restriction combined with resveratrol induces autophagy and protects 26-month-old rat hearts from doxorubicin-induced toxicity. Free Radic Biol Med. 2014;74:252–262. doi: 10.1016/j.freeradbiomed.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova LV, Sodhi K, Gatto-Weis C, Puri N, Hinds TD, Jr, Shapiro JI, Malhotra D. Peroxisome proliferator-activated receptor delta agonist, HPP593, prevents renal necrosis under chronic ischemia. PLoS One. 2013;8:e64436. doi: 10.1371/journal.pone.0064436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CS, Drummond MJ, Glynn EL, Dickinson JM, Gundermann DM, Timmerman KL, Walker DK, Volpi E, Rasmussen BB. Skeletal muscle autophagy and protein breakdown following resistance exercise are similar in younger and older adults. J Gerontol A Biol Sci Med Sci. 2013;68:599–607. doi: 10.1093/gerona/gls209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, Sandri M, Munoz-Canoves P. Autophagy maintains stemness by preventing senescence. Nature. 2016;529:37–42. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- Gil JM, Rego AC. Mechanisms of neurodegeneration in Huntington’s disease. Eur J Neurosci. 2008;27:2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- Gomes AV, Waddell DS, Siu R, Stein M, Dewey S, Furlow JD, Bodine SC. Upregulation of proteasome activity in muscle RING finger 1-null mice following denervation. FASEB J. 2012;26:2986–2999. doi: 10.1096/fj.12-204495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Jackson JR, Wang Y, Edens N, Pereira SL, Alway SE. β-Hydroxy-β-methylbutyrate reduces myonuclear apoptosis during recovery from hind limb suspension-induced muscle fiber atrophy in aged rats. Am J Physiol Regul Integr Comp Physiol. 2011;301:R701–R715. doi: 10.1152/ajpregu.00840.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramizu S, Ota N, Hase T, Murase T. Catechins attenuate eccentric exercise-induced inflammation and loss of force production in muscle in senescence-accelerated mice. J Appl Physiol. 2011;111:1654–1663. doi: 10.1152/japplphysiol.01434.2010. [DOI] [PubMed] [Google Scholar]
- Herbst A, Wanagat J, Cheema N, Widjaja K, McKenzie D, Aiken JM. Latent mitochondrial DNA deletion mutations drive muscle fiber loss at old age. Aging Cell. 2016 doi: 10.1111/acel.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye SK, Wagner DR, Acampora D, Horwitz RI, Cooney LM, Jr, Hurst LD, Tinetti ME. A predictive index for functional decline in hospitalized elderly medical patients. J Gen Intern Med. 1993;8:645–652. doi: 10.1007/BF02598279. [DOI] [PubMed] [Google Scholar]
- Jackson JR, Ryan MJ, Alway SE. Long-term supplementation with resveratrol alleviates oxidative stress but does not attenuate sarcopenia in aged mice. J Gerontol A Biol Sci Med Sci. 2011;66:751–764. doi: 10.1093/gerona/glr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JR, Ryan MJ, Hao Y, Alway SE. Mediation of endogenous antioxidant enzymes and apoptotic signaling by resveratrol following muscle disuse in the gastrocnemius muscles of young and old rats. Am J Physiol Regul Integr Comp Physiol. 2010;299:R1572–R1581. doi: 10.1152/ajpregu.00489.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Ji LL. PGC-1alpha overexpression via local transfection attenuates mitophagy pathway in muscle disuse atrophy. Free Radic Biol Med. 2016a;93:32–40. doi: 10.1016/j.freeradbiomed.2015.12.032. [DOI] [PubMed] [Google Scholar]
- Kang C, Yeo D, Ji LL. Muscle Immobilization Activates Mitophagy and Disrupts Mitochondrial Dynamics in Mice. Acta Physiol (Oxf) 2016b doi: 10.1111/apha.12690. [DOI] [PubMed] [Google Scholar]
- Kim YA, Kim YS, Oh SL, Kim HJ, Song W. Autophagic response to exercise training in skeletal muscle with age. J Physiol Biochem. 2013;69:697–705. doi: 10.1007/s13105-013-0246-7. [DOI] [PubMed] [Google Scholar]
- Kocturk S, Kayatekin BM, Resmi H, Acikgoz O, Kaynak C, Ozer E. The apoptotic response to strenuous exercise of the gastrocnemius and solues muscle fibers in rats. Eur J Appl Physiol. 2008;102:515–524. doi: 10.1007/s00421-007-0612-7. [DOI] [PubMed] [Google Scholar]
- Lee MS. Role of mitochondrial function in cell death and body metabolism. Front Biosci(Landmark Ed) 2016;21:1233–1244. doi: 10.2741/4453. [DOI] [PubMed] [Google Scholar]
- Leeuwenburgh C, Gurley CM, Strotman BA, Dupont-Versteegden EE. Age-related differences in apoptosis with disuse atrophy in soleus muscle. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1288–R1296. doi: 10.1152/ajpregu.00576.2004. [DOI] [PubMed] [Google Scholar]
- Li YP, Chen Y, Li AS, Reid MB. Hydrogen peroxide stimulates ubiquitin-conjugating activity and expression of genes for specific E2 and E3 proteins in skeletal muscle myotubes. Am J Physiol Cell Physiol. 2003;285:C806–C812. doi: 10.1152/ajpcell.00129.2003. [DOI] [PubMed] [Google Scholar]
- Lin CF, Chien SY, Chen CL, Hsieh CY, Tseng PC, Wang YC. IFN-gamma Induces Mimic Extracellular Trap Cell Death in Lung Epithelial Cells Through Autophagy-Regulated DNA Damage. J Interferon Cytokine Res. 2016;36:100–112. doi: 10.1089/jir.2015.0011. [DOI] [PubMed] [Google Scholar]
- Lipman RD, Chrisp CE, Hazzard DG, Bronson RT. Pathologic characterization of brown Norway, brown Norway x Fischer 344, and Fischer 344 x brown Norway rats with relation to age. J Gerontol A Biol Sci Med Sci. 1996;51:B54–B59. doi: 10.1093/gerona/51A.1.B54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo VF, Carnio S, Vainshtein A, Sandri M. Autophagy is not required to sustain exercise and PRKAA1/AMPK activity but is important to prevent mitochondrial damage during physical activity. Autophagy. 2014;10:1883–1894. doi: 10.4161/auto.32154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokireddy S, Wijesoma IW, Teng S, Bonala S, Gluckman PD, McFarlane C, Sharma M, Kambadur R. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 2012;16:613–624. doi: 10.1016/j.cmet.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Lushaj EB, Johnson JK, McKenzie D, Aiken JM. Sarcopenia accelerates at advanced ages in Fisher 344xBrown Norway rats. J Gerontol A Biol Sci Med Sci. 2008;63:921–927. doi: 10.1093/gerona/63.9.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti E, Calvani R, Cesari M, Buford TW, Lorenzi M, Behnke BJ, Leeuwenburgh C. Mitochondrial dysfunction and sarcopenia of aging: from signaling pathways to clinical trials. Int J Biochem Cell Biol. 2013a;45:2288–2301. doi: 10.1016/j.biocel.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti E, Csiszar A, Dutta D, Balagopal G, Calvani R, Leeuwenburgh C. Role of mitochondrial dysfunction and altered autophagy in cardiovascular aging and disease: from mechanisms to therapeutics. Am J Physiol Heart Circ Physiol. 2013b;305:H459–H476. doi: 10.1152/ajpheart.00936.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti E, Lees HA, Manini TM, Buford TW, Aranda JM, Jr, Calvani R, Capuani G, Marsiske M, Lott DJ, Vandenborne K, Bernabei R, Pahor M, Leeuwenburgh C, Wohlgemuth SE. Skeletal muscle apoptotic signaling predicts thigh muscle volume and gait speed in community-dwelling older persons: an exploratory study. PLoS ONE. 2012;7:e32829. doi: 10.1371/journal.pone.0032829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattox TA, Young ME, Rubel CE, Spaniel C, Rodriguez JE, Grevengoed TJ, Gautel M, Xu Z, Anderson EJ, Willis MS. MuRF1 activity is present in cardiac mitochondria and regulates reactive oxygen species production in vivo. J Bioenerg Biomembr. 2014;46:173–187. doi: 10.1007/s10863-014-9549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metter EJ, Talbot LA, Schrager M, Conwit R. Skeletal muscle strength as a predictor of all-cause mortality in healthy men. J Gerontol A Biol Sci Med Sci. 2002;57:B359–B365. doi: 10.1093/gerona/57.10.b359. [DOI] [PubMed] [Google Scholar]
- Murphy RA, Register TC, Shively CA, Carr JJ, Ge Y, Heilbrun ME, Cummings SR, Koster A, Nevitt MC, Satterfield S, Tylvasky FA, Strotmeyer ES, Newman AB, Simonsick EM, Scherzinger A, Goodpaster BH, Launer LJ, Eiriksdottir G, Sigurdsson S, Sigurdsson G, Gudnason V, Lang TF, Kritchevsky SB, Harris TB. Adipose tissue density, a novel biomarker predicting mortality risk in older adults. J Gerontol A Biol Sci Med Sci. 2014a;69:109–117. doi: 10.1093/gerona/glt070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RA, Reinders I, Register TC, Ayonayon HN, Newman AB, Satterfield S, Goodpaster BH, Simonsick EM, Kritchevsky SB, Harris TB. Associations of BMI and adipose tissue area and density with incident mobility limitation and poor performance in older adults. Am J Clin Nutr. 2014b;99:1059–1065. doi: 10.3945/ajcn.113.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Machida S, Oishi Y, Higuchi M, Muraoka I. Differential cell death regulation between adult-unloaded and aged rat soleus muscle. Mech Ageing Dev. 2009;130:328–336. doi: 10.1016/j.mad.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Pagano TB, Wojcik S, Costagliola A, De BD, Iovino S, Iovane V, Russo V, Papparella S, Paciello O. Age related skeletal muscle atrophy and upregulation of autophagy in dogs. Vet J. 2015;206:54–60. doi: 10.1016/j.tvjl.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Pellegrino MA, Desaphy JF, Brocca L, Pierno S, Camerino DC, Bottinelli R. Redox homeostasis, oxidative stress and disuse muscle atrophy. J Physiol. 2011;589:2147–2160. doi: 10.1113/jphysiol.2010.203232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani MA, Albuquerque A, Marcantonio ER, Jones RN, Gou RY, Fong TG, Schmitt EM, Tommet D, Aizpurua Isaza, II, Alsop DC, Inouye SK, Travison TG. Association Between Hospital Readmission and Acute and Sustained Delays in Functional Recovery During 18 Months After Elective Surgery: The Successful Aging after Elective Surgery Study. J Am Geriatr Soc. 2016 doi: 10.1111/jgs.14549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisot R, Marusic U, Biolo G, Mazzucco S, Lazzer S, Grassi B, Reggiani C, Toniolo L, di Prampero PE, Passaro A, Narici M, Mohammed S, Rittweger J, Gasparini M, Gabrijelcic BM, Simunic B. Greater loss in muscle mass and function but smaller metabolic alterations in older compared with younger men following 2 wk of bed rest and recovery. J Appl Physiol (1985) 2016;120:922–929. doi: 10.1152/japplphysiol.00858.2015. [DOI] [PubMed] [Google Scholar]
- Pistilli EE, Siu PM, Alway SE. Molecular regulation of apoptosis in fast plantaris muscles of aged rats. J Gerontol A Biol Sci Med Sci. 2006;61:245–255. doi: 10.1093/gerona/61.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshanravan B, Patel KV, Fried LF, Robinson-Cohen C, de Boer IH, Harris T, Murphy RA, Satterfield S, Goodpaster BH, Shlipak M, Newman AB, Kestenbaum B. Association of Muscle Endurance, Fatigability, and Strength With Functional Limitation and Mortality in the Health Aging and Body Composition Study. J Gerontol A Biol Sci Med Sci. 2016 doi: 10.1093/gerona/glw210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Ryan MJ, Jackson JR, Hao Y, Leonard SS, Alway SE. Inhibition of xanthine oxidase reduces oxidative stress and improves skeletal muscle function in response to electrically stimulated isometric contractions in aged mice. Free Radic Biol Med. 2011;51:38–52. doi: 10.1016/j.freeradbiomed.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MJ, Jackson JR, Hao Y, Williamson CL, Dabkowski ER, Hollander JM, Alway SE. Suppression of oxidative stress by resveratrol after isometric contractions in gastrocnemius muscles of aged mice. J Gerontol A Biol Sci Med Sci. 2010;65:815–831. doi: 10.1093/gerona/glq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol. 2002;4:842–849. doi: 10.1038/ncb866. [DOI] [PubMed] [Google Scholar]
- Scott I, Webster BR, Chan CK, Okonkwo JU, Han K, Sack MN. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289:2864–2872. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian D, Sorianello E, Segales J, Irazoki A, Ruiz-Bonilla V, Sala D, Planet E, Berenguer-Llergo A, Munoz JP, Sanchez-Feutrie M, Plana N, Hernandez-Alvarez MI, Serrano AL, Palacin M, Zorzano A. Mfn2 deficiency links age-related sarcopenia and impaired autophagy to activation of an adaptive mitophagy pathway. EMBO J. 2016;35:1677–1693. doi: 10.15252/embj.201593084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu PM, Alway SE. Mitochondria-associated apoptotic signalling in denervated rat skeletal muscle. J Physiol. 2005;565:309–323. doi: 10.1113/jphysiol.2004.081083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu PM, Alway SE. Deficiency of the Bax gene attenuates denervation-induced apoptosis. Apoptosis. 2006;11:967–981. doi: 10.1007/s10495-006-6315-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu PM, Pistilli EE, Alway SE. Age-dependent increase in oxidative stress in gastrocnemius muscle with unloading. J Appl Physiol. 2008;105:1695–1705. doi: 10.1152/japplphysiol.90800.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu PM, Wang Y, Alway SE. Apoptotic signaling induced by H2O2-mediated oxidative stress in differentiated C2C12 myotubes. Life Sci. 2009;84:468–481. doi: 10.1016/j.lfs.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HK, Matthews KG, Oldham JM, Jeanplong F, Falconer SJ, Bass JJ, Senna-Salerno M, Bracegirdle JW, McMahon CD. Translational signalling, atrogenic and myogenic gene expression during unloading and reloading of skeletal muscle in myostatin-deficient mice. PLoS One. 2014;9:e94356. doi: 10.1371/journal.pone.0094356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitali P, Grumati P, Hiller M, Chrisam M, Aartsma-Rus A, Bonaldo P. Autophagy is Impaired in the Tibialis Anterior of Dystrophin Null Mice. PLoS Curr. 2013:5. doi: 10.1371/currents.md.e1226cefa851a2f079bbc406c0a21e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AH, Rando TA. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 2014;33:2782–2797. doi: 10.15252/embj.201488278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner RE, Brunker LB, Agergaard J, Barrows KM, Briggs RA, Kwon OS, Young LM, Hopkins PN, Volpi E, Marcus RL, LaStayo PC, Drummond MJ. Age-related differences in lean mass, protein synthesis and skeletal muscle markers of proteolysis after bed rest and exercise rehabilitation. J Physiol. 2015;593:4259–4273. doi: 10.1113/JP270699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainshtein A, Desjardins EM, Armani A, Sandri M, Hood DA. PGC-1alpha modulates denervation-induced mitophagy in skeletal muscle. Skelet Muscle. 2015a;5:9. doi: 10.1186/s13395-015-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainshtein A, Tryon LD, Pauly M, Hood DA. Role of PGC-1alpha during acute exercise-induced autophagy and mitophagy in skeletal muscle. Am J Physiol Cell Physiol. 2015b;308:C710–C719. doi: 10.1152/ajpcell.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DT, Yang YJ, Huang RH, Zhang ZH, Lin X. Myostatin Activates the Ubiquitin-Proteasome and Autophagy-Lysosome Systems Contributing to Muscle Wasting in Chronic Kidney Disease. Oxid Med Cell Longev. 2015;2015:684965. doi: 10.1155/2015/684965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hao Y, Alway SE. Suppression of GSK-3beta activation by M-cadherin protects myoblasts against mitochondria-associated apoptosis during myogenic differentiation. J Cell Sci. 2011;124:3835–3847. doi: 10.1242/jcs.086686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci USA. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- White JR, Confides AL, Moore-Reed S, Hoch JM, Dupont-Versteegden EE. Regrowth after skeletal muscle atrophy is impaired in aged rats, despite similar responses in signaling pathways. Exp Gerontol. 2015;64:17–32. doi: 10.1016/j.exger.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan E, Lippens S, Vanden Berghe T, Romagnoli A, Fimia GM, Piacentini M, Vandenabeele P. Beclin1: a role in membrane dynamics and beyond. Autophagy. 2012;8:6–17. doi: 10.4161/auto.8.1.16645. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Seo AY, Marzetti E, Lees HA, Leeuwenburgh C. Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise. Exp Gerontol. 2010;45:138–148. doi: 10.1016/j.exger.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzhevsky N, Menashe O, Carmeli E, Stein H, Reznick AZ. Capacity for recovery and possible mechanisms in immobilization atrophy of young and old animals. Ann NY Acad Sci. 2001;928:212–225. doi: 10.1111/j.1749-6632.2001.tb05651.x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang K, Lei Y, Li Q, Nice EC, Huang C. Redox signaling: Potential arbitrator of autophagy and apoptosis in therapeutic response. Free Radic Biol Med. 2015;89:452–465. doi: 10.1016/j.freeradbiomed.2015.08.030. [DOI] [PubMed] [Google Scholar]
- Ziaaldini MM, Koltai E, Csende Z, Goto S, Boldogh I, Taylor AW, Radak Z. Exercise training increases anabolic and attenuates catabolic and apoptotic processes in aged skeletal muscle of male rats. Exp Gerontol. 2015;67:9–14. doi: 10.1016/j.exger.2015.04.008. [DOI] [PubMed] [Google Scholar]