Abstract
Recent advances in microfluidic approaches have enabled the efficient isolation and detailed molecular characterization of circulating tumor cells (CTCs) in the peripheral blood of patients with cancer. Single-cell molecular analyses of CTCs reveal a tremendous degree of intracellular heterogeneity in CTC populations, reflective of heterogeneity across different patients as well as the underlying heterogeneity of tumors within each individual patient. These studies have enabled the identification of heterogeneous drug resistance mechanisms that can coexist in treatment refractory tumors. CTC analyses also enable serial noninvasive monitoring in patients and can capture the emergence of tumor heterogeneity over time, whether due to tumor evolution through genetic instability or through cellular plasticity. The presence and extent of intratumoral heterogeneity as revealed through the study of CTCs have important clinical implications for understanding and predicting the development of treatment resistance in a variety of solid tumors and for formulating appropriate therapeutic strategies in the effective treatment of cancer.
Major advances in our molecular understanding of cancer have led to the development of effective new therapies that target tumor-specific genetic alterations, often resulting in profound responses in subsets of patients with specific oncogenic drivers in their tumors (Haber et al. 2011; Hyman et al. 2017). However, acquired resistance to therapy remains a pressing problem, and the emergence of resistant subclones within heterogeneous tumors represents a significant barrier to the effective treatment of cancer. Detailed next-generation sequencing studies have showed dramatic spatial and temporal heterogeneity in metastatic and primary tumors from individual patients (Gerlinger et al. 2012; Wu et al. 2012), but such analyses present technical and logistical challenges to routine clinical implementation, particularly for cancers that frequently metastasize to the bone, brain, or lungs. Tumor heterogeneity is especially difficult to study in patients with multiple metastases, because capturing the full range of relevant tumor clones would require invasive biopsies of multiple different individual sites of metastatic disease. Furthermore, traditional tissue biopsies sample only a small portion of a tumor, and thus the range of clones within a heterogeneous tumor may not be fully represented.
The sampling of tumor cells that circulate in the peripheral blood, or “circulating tumor cells” (CTCs), provides an elegant solution to the study of tumor heterogeneity, because CTCs may be a more representative sample of invasive tumor cell populations derived from heterogeneous tumors in multiple sites within an individual patient. In addition, the ability to sample cells in the blood noninvasively at multiple time points allows for the longitudinal study of tumor evolution over time. Recent advances in microfluidic engineering have made possible the development of novel platform technologies that efficiently isolate rare CTCs at the single-cell level, thus enabling the study of single CTCs to gain insights into tumor heterogeneity and treatment resistance in a variety of cancers.
Microfluidic Isolation of Circulating Tumor Cells
Although the existence of circulating populations of tumor cells in the peripheral blood was first posited in 1869 (Ashworth 1869), the isolation and study of CTCs has been a challenging endeavor because of their rarity and fragility. In the past several years, a variety of methods and technologies have been developed to isolate and enumerate CTCs. In this concise review, we focus on microfluidic CTC isolation technologies that were developed in our laboratories; other CTC isolation technologies have been reviewed in detail elsewhere (Alix-Panabieres and Pantel 2014, 2016; Haber and Velculescu 2014; Miyamoto et al. 2014; Li et al. 2015). In collaboration with our bioengineering colleagues at the Massachusetts General Hospital, we developed a series of microfluidic devices aimed at the gentle and efficient isolation of CTCs from blood. The first- and second-generation microfluidic devices, the μpCTC-Chip and the HBCTC-Chip, process blood through microfluidic channels and rely on the physical capture and immobilization of CTCs onto microfluidic surfaces coated with antibodies directed against tumor-specific cell surface antigens (Nagrath et al. 2007; Stott et al. 2010). The third-generation device, the CTC-iChip, is a tumor antigen-independent CTC isolation technology based on the concept of negative depletion, in which leukocytes, erythrocytes, platelets, and noncellular objects are removed from the blood, resulting in the enrichment of an untagged population of CTCs (Ozkumur et al. 2013; Karabacak et al. 2014). Because the CTC-iChip enables the efficient, gentle isolation of intact, untagged CTCs in suspension, it has allowed for detailed single-cell molecular analyses of CTCs from cancer patients and mouse models, thus revealing new insights into tumor heterogeneity at the single-cell level.
The microfluidic CTC-iChip uses three integrated sequential steps, consisting of continuous deterministic lateral displacement for size-based separation of CTCs and leukocytes from whole blood, inertial focusing for precise alignment of cells in a microchannel, and microfluidic magnetophoresis for removal of leukocytes prebound to magnetic beads labeled with anti-CD45, anti-CD16, and anti-CD66b antibodies (Karabacak et al. 2014). This tumor antigen-independent purification method enables the isolation of CTCs without assumptions regarding the nature of cell surface epitopes present on the tumor cells. Using the integrated microfluidic CTC-iChip, up to 107 cells per second can be sorted, enabling the sensitive, efficient, and high-throughput isolation of CTCs. The purity of CTCs within the enriched product ranges between 0.01% and 10%, depending on the burden of CTCs present in the patient's circulation. Importantly, CTCs isolated using this method are intact and untagged and amenable to a variety of downstream biological and molecular analyses including immunofluorescence, RNA-in situ hybridization, single-cell RNA-seq, and in vitro culture (Ozkumur et al. 2013; Aceto et al. 2014; Ting et al. 2014; Yu et al. 2014; Miyamoto et al. 2015). These sensitive analyses are possible because the gentle microfluidic enrichment processes of the CTC-iChip preserves the integrity and viability of isolated cells, in contrast to other methods that require cellular fixation and antibody-mediated binding of magnetic beads to cell membrane epitopes. Of note, our single CTC RNA-seq studies suggest that approximately one-half of CTCs have RNA at various stages of degradation even when isolated using this gentle microfluidic method, indicating that many cells in the circulation are nonviable (Ting et al. 2014; Miyamoto et al. 2015). Nevertheless, the quality and integrity of RNA in CTCs isolated using the CTC-iChip are well-preserved compared with prior methods, thus enabling single-cell RNA expression analyses, including the study of intracellular heterogeneity.
Insights Into Intratumoural Heterogeneity Through CTCs
Prostate Cancer
Prostate cancer is a heterogeneous entity, with primary tumors that are frequently multifocal and arise from divergent cancer clones (Andreoiu and Cheng 2010; Cooper et al. 2015). Metastases in prostate cancer are likely established through polyclonal seeding of divergent clones (Gundem et al. 2015), although some studies point to the conservation of driver lesions in metastatic lesions within individuals (Kumar et al. 2016). Consistent with substantial intratumoral heterogeneity, we observed through single-cell immunofluorescence analysis heterogeneous and varying degrees of androgen receptor (AR) signaling in CTCs from patients with castration-resistant prostate cancer (CRPC), in striking contrast to relatively homogeneous CTC populations with activated AR signaling in patients with untreated prostate cancer (Miyamoto et al. 2012). Similar levels of heterogeneity have been observed in prostate CTCs with respect to cellular morphologic criteria, related to disease status and cell differentiation state (Chen et al. 2015; Beltran et al. 2016). Single-cell RNA-seq of CTCs isolated from CRPC patients revealed a tremendous degree of heterogeneity in expression profiles, both in CTC populations from individual patients and across different patients (Miyamoto et al. 2015). Indeed, upon unsupervised hierarchical clustering of transcriptional profiles, the mean correlation coefficient for single CTCs from individual patients was significantly lower than the mean correlation coefficient of single cells from prostate cancer cell lines, and it was similar to that of single cells across multiple different cell lines, suggesting a much higher level of heterogeneity in CTCs. Nevertheless, these single CTCs clustered based on their patient of origin, indicative of shared transcriptional programs among CTCs derived from any given individual patient.
Heterogeneity at the single CTC level was also observed in the acquisition of varied mechanisms of resistance to AR-targeted therapies (Miyamoto et al. 2015). In an analysis of specific molecular aberrations in CTCs, including previously reported AR splice variants and AR mutations (Antonarakis et al. 2014; Robinson et al. 2015), we noted that single CTCs from individual patients expressed remarkable heterogeneity in their expression of different AR splice variants. More than half of patients had multiple CTCs expressing different AR splice variants, and about one of six single CTCs had expression of multiple different AR splice variants simultaneously. Through the analysis of transcriptional differences between CTCs from patients who were resistant to the anti-androgen therapy enzalutamide compared with patients who were enzalutamide naïve, we identified two AR-independent pathways associated with resistance to enzalutamide: we confirmed the previously reported activation of glucocorticoid receptor (GR) (Arora et al. 2013) and discovered elevated noncanonical Wnt (ncWnt) signaling, finding that both of these resistance pathways were activated in different subsets of cells. Thus, we found that complex and heterogeneous drug resistance mechanisms exist in advanced prostate cancer, as revealed by the study of heterogeneous populations of CTCs. The degree of heterogeneity observed in CTCs in CRPC patients is consistent with intratumoral heterogeneity observed in patients with advanced metastatic prostate cancer, suggestive of the polyclonal seeding of divergent clones (Lohr et al. 2014; Gundem et al. 2015; Jiang et al. 2015), and is reflective of the variety of molecular alterations that may occur in parallel in tumor cells during disease progression and the development of resistance to therapy.
Pancreatic Cancer
We observed similar intracellular heterogeneity in single CTCs isolated from the genetically engineered LSL-KrasG12D, Trp53flox/flox or +, Pdx1-Cre (KPC) mouse model of pancreatic cancer (Bardeesy et al. 2006; Ting et al. 2014). Although the genetic driver mutations were identical in CTCs from different mice because of their shared genetic background, there was nevertheless a significant level of heterogeneity observed within CTCs, with specific clustering of CTCs derived from different animals (Ting et al. 2014). These findings suggest the potential importance of somatically acquired genetic and epigenetic changes in defining the heterogeneity of CTC populations, despite a shared genetic background.
In the KPC pancreatic cancer mouse model, we noted three subsets of CTCs: a major “classical CTC” group with strong expression of epithelial markers, a group with enrichment of platelet markers, and a group associated with proliferation signatures (Ting et al. 2014). Notably, although the classical CTCs showed clear loss of the epithelial marker E-cadherin (Cdh1) consistent with epithelial-to-mesenchymal transition (EMT), they did not lose expression of other epithelial markers such as cytokeratins, and they showed heterogeneous expression of mesenchymal genes across single cells. Thus, many classical CTCs appear to be arrested in a bipheno-typic EMT state. Nevertheless, they also showed great diversity in their transcriptional programs, including expression of cancer stem cell markers Aldh1a1 and Aldh1a2 in some cells, and surprising enrichment of extracellular matrix (ECM) transcripts in many CTCs. Indeed, this finding of ECM gene expression in CTCs was recapitulated in human pancreatic, breast, and prostate cancer patients, with enrichment of the core matrisome protein SPARC in 100% of pancreatic CTCs. Together, these studies of CTCs in the KPC genetic mouse model showed substantial intracellular heterogeneity in CTCs from mice despite a conserved genetic background and strong expression of ECM transcripts in a majority of pancreatic CTCs, suggestive of a remarkable ability of CTCs to make their own contributions to tumor stromal remodeling and establishment of a hospitable microenvironment at metastatic sites.
Breast Cancer
Considerable heterogeneity has also observed in single CTCs from breast cancer patients using a variety of methods, including microfluidic transcriptional profiling, targeted mutation detection, and next-generation sequencing (Powell et al. 2012; Deng et al. 2014; De Luca et al. 2016). We used an RNA-in situ hybridization (ISH) assay to observe a spectrum of expression of epithelial and mesenchymal markers in single breast CTCs (Yu et al. 2013), ranging from exclusively epithelial to exclusively mesenchymal CTCs, as well as CTCs in an intermediate state with dual expression of epithelial and mesenchymal markers, similar to the biphenotypic EMT state identified in the pancreatic cancer mouse model (Ting et al. 2014). Interestingly, the EMT features of CTCs varied according to histological subtype of breast cancer, where CTCs from ER+/PR+ and HER2+ cancers were predominantly epithelial, and CTCs from triple-negative (ER−/PR−/HER2−) breast cancer were predominantly mesenchymal. Examination of changes in EMT states before and after systemic therapy suggested that patients who responded to therapy showed a decrease in CTC numbers and/or a proportional decrease in mesenchymal compared with epithelial markers, and that those with progressive disease despite therapy showed an increase in mesenchymal markers in CTCs post-treatment. Thus, the EMT status of CTCs may serve as a potential biomarker of therapeutic response, with the degree of heterogeneity of cells reflective of their susceptibility to treatment.
Progressive disease in cancer patients is often accompanied by the appearance of multicellular clusters of CTCs (Molnar et al. 2001; Cho et al. 2012), and the presence of these CTC clusters in breast cancer patients is associated with a worse overall survival (Aceto et al. 2014). A detailed examination of CTC clusters in mouse models revealed that they have a critical role as mediators of cancer metastasis, because they have a higher propensity for seeding metastatic disease compared with single CTCs in circulation (Fidler 1973; Liotta et al. 1976; Aceto et al. 2014). Of note, cellular tagging and mixing studies in mice showed that nearly all CTC clusters were derived from oligoclonal precursor cells, indicative of their heterogeneous composition and origin (Aceto et al. 2014). Nevertheless, RNA-seq expression studies of breast cancer patient-derived single CTCs and CTC clusters showed a strong clustering pattern by patient of origin, and a high level of concordance in expression patterns between matched CTC clusters and single CTCs from individual breast cancer patients, with the exception of a few candidate cluster-related genes including XBP1, AGR2, HER3, and plakoglobin (Aceto et al. 2014). Indeed, knockdown of plakoglobin in mouse models suppressed CTC cluster formation and development of metastases, pointing to an important role of this protein in tumor dissemination. Thus, two distinct populations of CTCs appear to coexist in the circulation, including single CTCs and CTC clusters with much higher metastatic potential, derived from oligoclonal precursor cells representative of the inherent heterogeneity of the primary tumor.
Monitoring Tumor Evolution and Heterogeneity with CTCs
Although the evolution of tumors in response to initially effective targeted therapies has become increasingly appreciated, either through the selection of rare preexisting subclonal populations or through molecular tumor evolution (Hata et al. 2016), the routine sampling of tumors at multiple time points remains a major technical challenge. CTCs provide a uniquely accessible mechanism to perform noninvasive serial sampling of tumors over time, and hence they could prove revolutionary in monitoring tumor evolution in patients undergoing treatments. Although circulating tumor DNA (ctDNA) is another type of “liquid biopsy” that may be used to assess for the emergence of specific tumor mutations (Bardelli and Pantel 2017), tumor evolution occurs not just through genetic changes, but also from phenotypic changes through epigenetic cellular plasticity (Fig. 1). Such functional changes cannot be easily ascertained using ctDNA but rather require the use of functional assays in live tumor cells. Thus, the analysis of CTCs, CTC-derived cell lines, and patient-derived xenograft models are invaluable tools in the study of tumor evolution.
Figure 1.
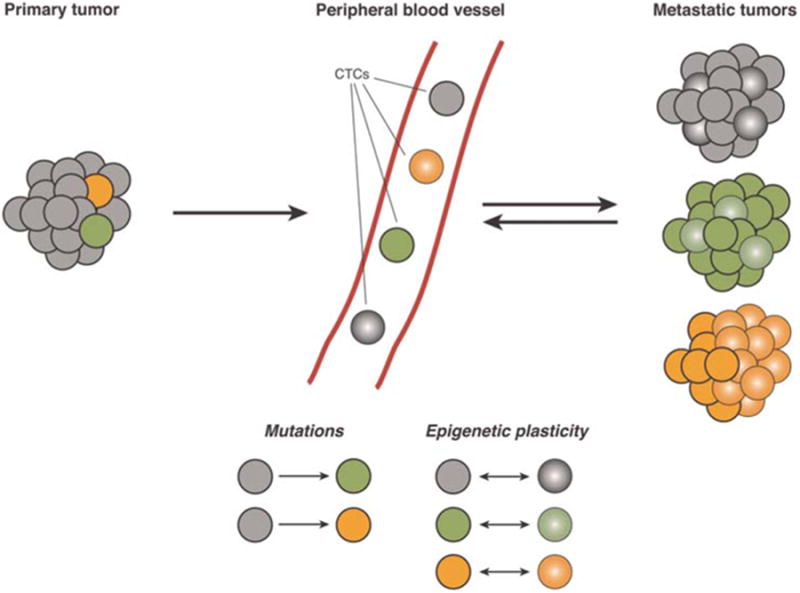
Schematic of heterogeneous tumors giving rise to circulating tumor cells (CTCs). CTCs arise from intravasation of cancer cells into peripheral blood vessels from primary or metastatic tumors. Single-cell heterogeneity can arise from genetic mutations (represented by gray circles becoming green or orange circles) or from epigenetic plasticity (represented by circles converting into spheres, and the reverse conversion of spheres to circles). Mutations and epigenetic changes can also occur simultaneously in the same cancer cell (represented by green and orange circles converting into green and orange spheres). Metastatic tumors may consist of heterogeneous groups of cancer cells that have undergone changes because of any combination of mutations, epigenetic plasticity, or both.
In breast cancer, the emergence of HER2 expressing subpopulations has been observed in CTCs from patients with initially HER2− tumors after exposure to multiple courses of therapy (Fehm et al. 2010; Lindstrom et al. 2012). Analysis of CTCs in such patients revealed discrete HER2+ and HER2− subpopulations, which had the ability to interconvert spontaneously when maintained in culture (Jordan et al. 2016). Thus, a dynamic equilibrium of HER2+ and HER2− cell populations exist within a heterogeneous tumor cell population, driven by spontaneous interconversion between these phenotypes. HER2+ CTCs were more rapidly proliferative with activation of multiple receptor tyrosine kinase pathways, whereas HER2− CTCs showed resistance to cytotoxic chemotherapy and activation of Notch and DNA damage pathways. These two cell populations had comparable tumor initiating potential and similar expression of the stem cell marker ALDH1, suggesting that an underlying cellular plasticity leads to disease progression and drug resistance in breast cancer, rather than preexisting drug-resistant sub-clones in a hierarchical cancer stem-cell model.
Whether resistant subclones arise within heterogeneous tumors through genetic instability or through cellular plasticity, their emergence portends the development of therapeutic resistance. The early detection of resistant subclonal populations may allow the early implementation of combination therapies that simultaneously target multiple different oncogenic pathways before a dominant resistant clone emerges. This strategy requires careful repeated monitoring of patients while on treatment, which may be accomplished using serial CTC analyses. In addition, the deployment of aggressive treatments earlier in the disease course before tumor evolution, such as adjuvant therapy following local therapy, may become an increasingly important approach guided by the monitoring of patients through CTC analyses.
Conclusion
In the new world of precision oncology, the development of novel therapeutics and diagnostics often merge with one another and are codependent. The emergence of heterogeneity in evolving tumors suggests the inadequacy of a “one-size-fits-all” approach to cancer therapy and necessitates the development of sophisticated molecular tests to guide the precision selection of appropriate therapeutic strategies. The driving force for the effective use of new targeted therapies is novel diagnostic strategies to identify the presence of specific molecular lesions in tumors, but at the same time there is no impetus to develop such diagnostic tests until clear therapeutic options have been developed that make these molecular lesions worth uncovering. This coevolution model in oncology is unprecedented and requires the integrated development of therapeutics and diagnostics at the levels of discovery, validation, and practical implementation in the clinic. Ultimately, the careful application of CTC-based diagnostic tools may help overcome treatment resistance mediated by tumor heterogeneity by guiding the rational selection and application of effective new targeted therapies.
Acknowledgments
This work was supported by grants from the Prostate Cancer Foundation (D.T.M., D.A.H., S.M., M.T.), Charles Evans Foundation (D.A.H.), Department of Defense (D.T.M., D.T.T.), Howard Hughes Medical Institute (D.A.H.), Burroughs Wellcome Fund (D.T.T.), National Institute of Biomedical Imaging and Bioengineering (NIBIB) EB008047 (M.T.), and the MGH-Johnson & Johnson Center for Excellence in CTC Technologies (M.T., S.M.).
References
- Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- Alix-Panabieres C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- Andreoiu M, Cheng L. Multifocal prostate cancer: Biologic, prognostic, and therapeutic implications. Hum Pathol. 2010;41:781–793. doi: 10.1016/j.humpath.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, Shah N, Cai L, Efstathiou E, Logothetis C, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155:1309–1322. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aust Med J. 1869;14:146–149. [Google Scholar]
- Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardelli A, Pantel K. Liquid biopsies, what we do not know (yet) Cancer Cell. 2017;31:172–179. doi: 10.1016/j.ccell.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Beltran H, Jendrisak A, Landers M, Mosquera JM, Kossai M, Louw J, Krupa R, Graf RP, Schreiber NA, Nanus DM, et al. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res. 2016;22:1510–1519. doi: 10.1158/1078-0432.CCR-15-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Ho H, Lichterman J, Lu YT, Zhang Y, Garcia MA, Chen SF, Liang AJ, Hodara E, Zhau HE, et al. Sub-classification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer. 2015;121:3240–3251. doi: 10.1002/cncr.29455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EH, Wendel M, Luttgen M, Yoshioka C, Marrinucci D, Lazar D, Schram E, Nieva J, Bazhenova L, Morgan A, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys Biol. 2012;9:016001. doi: 10.1088/1478-3975/9/1/016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alex-androv LB, Kremeyer B, Butler A, Lynch AG, Camacho N, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca F, Rotunno G, Salvianti F, Galardi F, Pestrin M, Gabel-lini S, Simi L, Mancini I, Vannucchi AM, Pazzagli M, et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget. 2016;7:26107–26119. doi: 10.18632/oncotarget.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G, Krishnakumar S, Powell AA, Zhang H, Mindrinos MN, Telli ML, Davis RW, Jeffrey SS. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC Cancer. 2014;14:456. doi: 10.1186/1471-2407-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehm T, Muller V, Aktas B, Janni W, Schneeweiss A, Stickeler E, Lattrich C, Lohberg CR, Solomayer E, Rack B, et al. HER2 status of circulating tumor cells in patients with meta-static breast cancer: A prospective, multicenter trial. Breast Cancer Res Treat. 2010;124:403–412. doi: 10.1007/s10549-010-1163-x. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis. Eur J Cancer. 1973;9:223–227. doi: 10.1016/s0014-2964(73)80022-2. [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Hognas G, Annala M, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber DA, Velculescu VE. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4:650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang HE, Krishnamurthy Radhakrishna V, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell. 2017;168:584–599. doi: 10.1016/j.cell.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Lu YT, Ho H, Li B, Chen JF, Lin M, Li F, Wu K, Wu H, Lichterman J, et al. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget. 2015;6:44781–44793. doi: 10.18632/oncotarget.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y, Yu M, Sundaresan TK, Licausi JA, Desai R, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102–106. doi: 10.1038/nature19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen PI, et al. Micro-fluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, Etzioni R, Bolouri H, Montgomery B, White T, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gregory SG, Garcia-Blanco MA, Armstrong AJ. Using circulating tumor cells to inform on prostate cancer biology and clinical utility. Crit Rev Clin Lab Sci. 2015;52:191–210. doi: 10.3109/10408363.2015.1023430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom LS, Karlsson E, Wilking UM, Johansson U, Hartman J, Lidbrink EK, Hatschek T, Skoog L, Bergh J. Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol. 2012;30:2601–2608. doi: 10.1200/JCO.2011.37.2482. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Saidel MG, Kleinerman J. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res. 1976;36:889–894. [PubMed] [Google Scholar]
- Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, Smas ME, Lord JB, Brannigan BW, Trautwein J, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Sequist LV, Lee RJ. Circulating tumour cells-monitoring treatment response in prostate cancer. Nat Rev Clin Oncol. 2014;11:401–412. doi: 10.1038/nrclinonc.2014.82. [DOI] [PubMed] [Google Scholar]
- Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar B, Ladanyi A, Tanko L, Sreter L, Tulassay Z. Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients. Clin Cancer Res. 2001;7:4080–4085. [PubMed] [Google Scholar]
- Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, et al. Single cell profiling of circulating tumor cells: Transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K, et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–1918. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, Croul S, Bouffet E, Fults DW, Eberhart CG, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]


