Abstract
Chromatin-based mechanisms offer therapeutic targets in acute myeloid leukemia (AML) that are of great current interest. In this study, we conducted an RNAi-based screen to identify druggable chromatin regulator-based targets in leukemias marked by oncogenic rearrangements of the MLL gene. In this manner, we discovered the H4K16 histone acetyltransferase (HAT) MOF to be important for leukemia cell growth. Conditional deletion of Mof in a mouse model of MLL-AF9-driven leukemogenesis reduced tumor burden and prolonged host survival. RNA sequencing showed an expected downregulation of genes within DNA damage repair pathways that are controlled by MOF, as correlated with a significant increase in yH2AX nuclear foci in Mof-deficient MLL-AF9 tumor cells. In parallel, Mof loss also impaired global H4K16 acetylation in the tumor cell genome. Rescue experiments with catalytically inactive mutants of MOF showed that its enzymatic activity was required to maintain cancer pathogenicity. In support of the role of MOF in sustaining H4K16 acetylation, a small molecule inhibitor of the HAT component MYST blocked the growth of both murine and human MLL-AF9 leukemia cell lines. Furthermore Mof inactivation suppressed leukemia development in a NUP98-HOXA9 driven AML model. Taken together, our results establish that the HAT activity of MOF is required to sustain MLL-AF9 leukemia and may be important for multiple AML subtypes. Blocking this activity is sufficient to stimulate DNA damage, offering a rationale to pursue MOF inhibitors as a targeted approach to treat MLL-rearranged leukemias.
Keywords: MOF, H4K16ac, MLL-AF9, leukemia, acetyltransferase
INTRODUCTION
Chromosomal rearrangements at 11q23 are associated with the development of acute leukemia and led to the discovery of lysine (K)-specific methyltransferase 2A (KMT2A also known as MLL1).1,2 MLL translocations are found in about 10% of all patients with acute leukemia and generally associated with an unfavorable prognosis.3 MLL translocations are more frequently present in infant and pediatric patients with the highest frequency (80%) in infant acute lymphoblastic leukemia (ALL).4
It is evident that chromatin modifications, including DNA methylation and histone modifications, enforce oncogenic gene expression programs and substantially contribute to the initiation and maintenance of leukemia cells.5,6 Epigenomic studies utilizing in vivo and in vitro models of MLL-rearranged leukemia have revealed that direct targets of MLL fusion proteins such as HOXA cluster genes are associated with aberrantly high levels of histone 3 lysine 79 dimethylation (H3K79me2).7,8 DOT1L was found to be the key regulator of H3K79me2 and MLL-rearranged cells were shown to be highly dependent on Dot1l for leukemia initiation and maintenance.9–12 This discovery led to the development of small molecule inhibitors targeting DOT1L, one of which is currently undergoing early phase trials.13,14 Similarly, MLL-AF9 leukemia maintenance was shown to be dependent upon expression of other chromatin regulators such as Lysine (K) Demethylase 1A (Kdm1a)15 and Bromodomain Containing 4 (Brd4)16. KDM1A is an H3K4 and H3K9 demethylase and BRD4 is a well-known member of the bromodomain family. The bromodomain family consists of epigenetic “readers”, important for recognizing posttranslational chromatin modifications and recruiting downstream effector proteins to specific loci to activate gene expression programs.17 Multiple bromodomain inhibitors are currently under investigation in early phase clinical trials.17
Findings such as these indicate the importance of chromatin regulation in leukemia. In order to identify novel druggable epigenetic targets in MLL-rearranged leukemia, we conducted a chromatin regulator-focused RNAi screen in murine MLL-AF9 leukemia cells and found hairpins targeting (K) Lysine Acetyltransferase 8 (Kat8, also known as Mof) and the previously identified target Brd416, to be the most potent suppressors of cell growth. MOF is a histone 4 lysine 16 (H4K16) acetyltransferase and member of the MYST family of lysine acetyltransferases. The MYST family is named for its founding members MOZ, YBF2, SAS2 and TIP60, proteins that all contain a MYST region with a canonical acetyl coenzyme A (CoA) binding site and C2HC-type zinc finger motif.18 MOF is one of the best-characterized MYST-family proteins and was shown to be crucial for murine embryogenesis. MOF functions as a cell-type dependent regulator of chromatin state and controls various cellular processes such as T-cell differentiation19, DNA damage response20–24, cell cycle progression20,25 and embryonic stem cell self-renewal and pluripotency26.
The role of MOF in tumorigenesis seems complex. Studies in breast carcinoma27, medulloblastoma27 and ovarian cancer28 suggest that tumor progression is associated with downregulation of MOF and H4K16 acetylation (H4K16ac). On the other hand, studies in lung29,30 and oral31 carcinoma have associated high expression of MOF with carcinogenesis and suppression of MOF with cancer cell death. This suggests that MOF may regulate tumorigenesis in a cell- and tissue- dependent manner.
While MOF’s enzymatic activity is druggable by small molecules32 and our RNAi screen suggests a crucial role for MOF in MLL-AF9 leukemogenesis, we studied the role of Mof in detail using a conditional murine Mof knockout system33. Our current findings indicate a strong dependency of MLL-AF9 leukemic cells on Mof. Gene expression and immunofluorescence data suggest that the importance of MOF in MLL-AF9 leukemogenesis may be through DNA damage repair. Mof knockout in MLL-AF9 transformed cells led to loss of global H4K16ac and in line with this finding, rescue experiments with histone acetyltransferase (HAT) domain mutated MOF illustrated that the HAT activity of MOF is indispensable for MLL-AF9 leukemia maintenance. Finally, experiments with the selective MYST protein HAT inhibitor MG149, showed a strong anti-proliferative effect on murine as well as human MLL-AF9 leukemia cell lines. MOF HAT activity may be a good target for novel small molecule inhibitor development to improve the treatment of patients with MLL-rearranged leukemia.
MATERIALS AND METHODS
RNAi screen
A customized shRNA library (TRMPV-Neo system) focused on mouse chromatin-regulating genes, together with control shRNAs, was designed as previously described.15,34 After sequence verification the virus library containing 2,252 shRNAs targeting 468 genes (four to six shRNAs per gene), was pooled. The virus pool was transduced into monoclonal mouse MLL-AF9 (MA9) leukemic cells (two replicates), stably expressing rtTA3 (Tet-on system), at a viral titer that on average causes a single viral transduction per cell and at which each shRNA is represented in at least 2,000 cells. The infected cells were selected for two days in 1 mg/mL Neomycin (G418 sulfate, Corning, NY) and subsequently shRNA expression was induced by adding 1 ug/mL Doxycycline (Sigma-Aldrich, St. Louis, MO). The shRNA-expressing cells (dsRed and Venus double positive) were sorted (T0) using a FACS ARIA (BD, San Jose, CA), and cultured for 12 days (T12).35 The integrated shRNA sequences in T0 and T12 cell samples were assessed by high-throughput sequencing (HiSeq) using the Illumina Next-Gen Sequencing HiSeq platform (Illumina, San Diego, CA), as previously described.15,34 shRNA screen output data can be found in Table S1.
Mice
The generation of Mof conditional knockout mouse in a C57Bl/6 background has been described.33 To generate Mx1-Cre; Moff/f mice, Moff/f mice were crossed to Mx1-Cre mice (B6.Cg-Tg(Mx1-Cre)1Cgn/J strain from Jackson Laboratory, Bar Harbor, ME) and Cre was maintained as a heterozygous allele. Genotyping strategies were previously described.33 All transplant experiments were performed with 6–8 week old female wild-type C57Bl/6 mice, purchased from Taconic, Hudson, NY. All animal experiments in this study were approved by, and adhered to guidelines of the Memorial Sloan-Kettering Cancer Center Animal Care and Use Committee.
RNA extraction and RNA sequencing
Trizol (Invitrogen, Carlsbad, CA) was used to extract RNA from viable cells. RNA was QCed on the Agilent Bioanalyzer 2100 platform (Agilent, Santa Clara, CA) and Poly-A tail selection was performed. Sequencing (RNAseq) was done using the Illumina Next-Gen Sequencing HiSeq platform (Illumina, San Diego, CA) with 30–45 million 50bp, paired-end reads.
Data Analysis and Statistical Methods
GraphPad Prism software was used for statistical analysis. Statistical significance between 2 groups was determined by unpaired 2-tailed Student’s t-test. The Kaplan-Meier method was applied to plot survival curves for murine leukemic transplant data, and the log-rank test to determine statistical significance.
RNAseq raw reads were aligned to NCBI37/mm9 and normalized using STAR. Differential expression data were obtained using the DEseq algorithm. These analyses were done through the platform from Basepair (New York City, NY). DEseq output tables are included in Table S2 and raw data were deposited in GEO (GSE80671). Gene ontology analyses were performed using the PANTHER gene analysis tool.
RESULTS
Chromatin regulator focused RNAi screen in MLL-AF9 leukemia identifies Mof as a key regulator
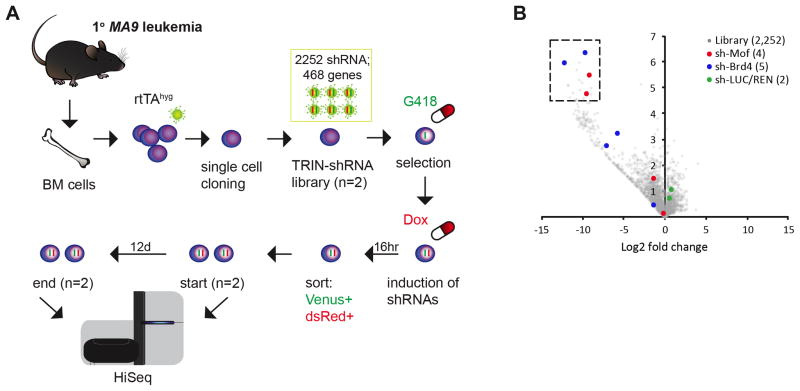
To identify novel druggable targets within epigenetic pathways required for MLL-AF9 (MA9) leukemia maintenance, we conducted a customized, chromatin regulator focused RNAi screen in murine, MA9 leukemia cells (Figure 1A). An shRNA library containing 2,252 shRNAs targeting 468 known chromatin regulators was constructed in TRMPV-Neo and transduced as one pool into Tet-on-competent, monoclonal mouse MA9 leukemic cells. After Neomycin (G418) selection, shRNA expression was induced by Doxycycline treatment. shRNA expressing cells were then sorted and changes in shRNA library representation after 12 days of culture were assessed by high-throughput sequencing (HiSeq) of shRNA guide strands as previously described.15,34 Using the scoring criterion of more than 256-fold depletion in each of two independent replicates, 20 shRNAs targeting 18 genes were strongly depleted (Figure 1B). Only two out of these 18 had two independent shRNAs that showed strong depletion, namely Mof and the previously identified target Brd415 (Figure 1B, S1A, S1B). This RNAi screen strongly suggests that the lysine acetyltransferase MOF is crucial for MA9 leukemic cell growth.
Figure 1. Chromatin regulator focused RNAi screen in MLL-AF9 leukemia identifies Mof as a key regulator.
(A) Schematic outline of chromatin regulator focused shRNA library screen coupled with high-throughput sequencing (HiSeq) in murine primary bone marrow (BM) MA9 monoclonal leukemia cells.
(B) Volcano plot depicting the changes in representation (x-axis) and significance (y-axis) of each shRNA construct in the screen before versus 12 days after hairpin-induced knockdown. One dot represents the mean for two independent experiments. The dotted area contains the 20 most significantly depleted hairpins in the screen (more than 256-fold depletion, significance (defined as −log10 of the p-value) ≥ 4.5). Total library (gray; 2,252 shRNAs, 468 genes), shMof (red; four shRNAs) and shBRD4 (blue; five shRNAs) are highlighted as the only two genes with two hits.
G418: Neomycin; Dox: Doxycycline.
Mof loss in a murine MLL-AF9 leukemia model leads to impaired colony-forming capacity, a phenotype rescued by exogenous full-length Mof
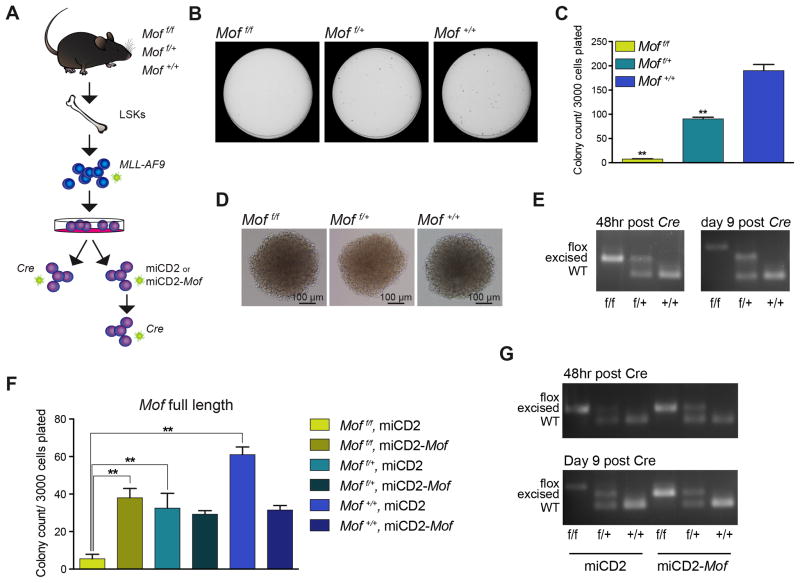
To study the role of MOF in MA9 leukemogenesis and validate the results of our RNAi screen we used a well-described conditional Mof knockout (KO) mouse in a C57Bl/6 background.33 For the initial in vitro experiments, Moff/f, Moff/+ and wild type (Mof+/+) adult mice were euthanized and Lin−, SCA1+, cKIT+ cells (LSKs) were purified from the bone marrow (BM) (Figure 2A). These fresh LSKs were infected with MA9 in an MSCV-Migr1 retroviral vector containing the GFP selection marker. Following a few days of liquid culture, cells were sorted for GFP positivity. These stable MA9 transformed cells grow indefinitely and were capable of forming dense, round colonies (blast colonies, data not shown).
Figure 2. Mof loss in murine MLL-AF9 leukemia model leads to impaired colony-forming capacity, a phenotype rescued by exogenous full-length Mof.
(A) Schematic for in vitro Mof knockout and full-length Mof rescue experiments.
(B) Day seven of methylcellulose colony-forming assay of MA9 in vitro transformed Moff/f, Moff/+ or wild type (Mof+/+) LSKs plated immediately upon sorting Cre positive cells. Representative 35 mm petri dishes are shown.
(C) Bar graph indicating mean number of colonies per 35mm dish after seven days. 3000 cells were plated per dish. Data are representative of four individual experiments.
(D) Colonies at day seven of CFU assay. Representative images are shown.
(E) PCR analysis illustrating excision at indicated time points of the 7-day colony-forming experiment. Representative gel images are shown.
(F) Day seven of CFU assay of MA9 in vitro transformed Moff/f, Moff/+ or Mof+/+ LSKs that were infected with full-length Mof (miCD2-Mof) or empty vector control (miCD2) and selected by sorting hCD2 positive cells. Bar graph indicates mean number of colonies per 35mm dish after seven days. 3000 cells were plated per dish. Data are representative of three individual experiments.
(G) PCR analysis illustrates excision throughout the duration of the 7-day colony-forming experiment. Representative gel images are shown.
** p < 0.01. Error bars represent SD of mean.
For in vitro excision of Mof, MA9 cells were infected with an MSCV-dTomato retrovirus containing Cre. 48 hours after infection, Cre positive Moff/f, Moff/+ and Mof+/+ cells were sorted, counted and immediately plated in myeloid cytokine supplemented methylcellulose for colony-forming unit (CFU) assays. Homozygous Mof loss significantly reduced colony-forming capacity of MA9 cells (Figure 2B and C). No difference in colony morphology was observed (Figure 2D). Heterozygous loss of Mof (Moff/+) also led to a reduction of total colony number (Figure 2C), albeit less dramatic compared to full Mof deletion, suggesting a potential gene dosage effect of Mof on the clonogenic capacity and cell growth of MA9 cells. However, Mof excision PCR data consistently illustrated that although at time of plating, all cells in Moff/f and Moff/+ were completely excised (Figure 2E), all Moff/f colonies that had formed at day 7 of the CFU assay were in fact unexcised, whereas the floxed-allele in the Moff/+ colonies remained fully excised (Figure 2E). These PCR data indicate strong selective pressure against homozygous, but not heterozygous Mof loss in an MA9 leukemic setting.
To assess specificity of the observed phenotype, a Mof full-length rescue experiment was performed. Moff/f, Moff/+ and Mof+/+ MA9 cells were transduced with either an MSCV-miCD2 retrovirus containing full-length, human influenza hemagglutinin (HA)-tagged Mof or the empty vector (Figure 2A). hCD2 positive cells were sorted and subsequent Western blot analysis for the HA-tagged MOF indicated that exogenous MOF was expressed (Figure S2A). These cells were then transduced with Cre and Cre positive cells were used for CFU assays as before. Figure 2F shows full rescue of the phenotype observed in Moff/f, miCD2 cells by exogenous expression of full length Mof (Moff/f, miCD2-Mof). Mof excision PCR analysis confirms this rescue. While Moff/f, miCD2 cells have fully lost excision at day 9 post Cre infection, Moff/f, miCD2-Mof cells are still largely excised (Figure 2G). Taken together, these findings demonstrate that Mof is required for the colony-forming capacity of MA9 leukemic cells.
Mof loss leads to reduced tumor burden and prolonged survival in an in vivo MLL-AF9 leukemia model
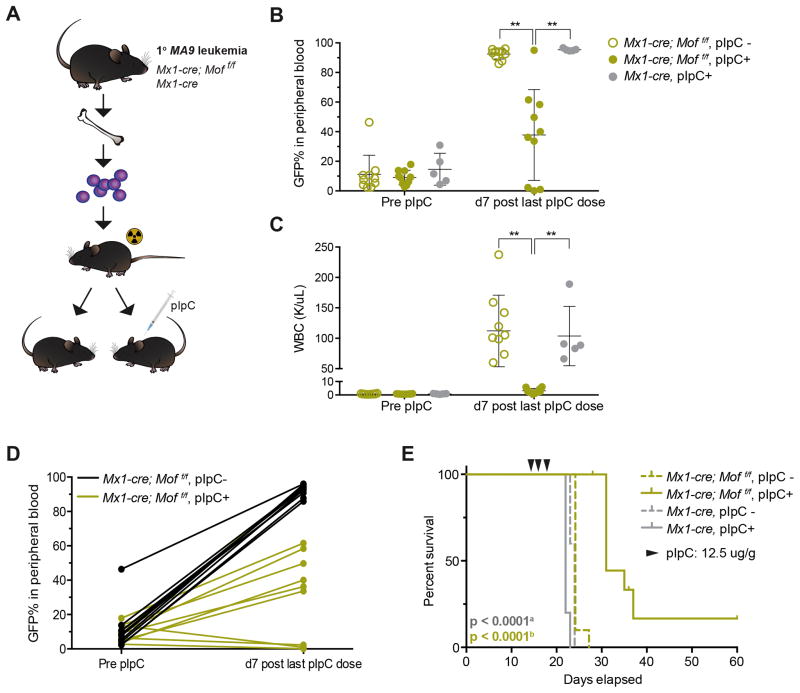
Mof dependency in MA9 leukemia was further assessed using an in vivo secondary, murine leukemia model. Mx1-Cre; Moff/f and Mx1-Cre mouse BM LSKs were transduced with GFP-tagged MA9 and 48 hours after infection, GFP+ cells were sorted and injected into sub-lethally irradiated (600 cGy) C57Bl/6 mice. These mice developed acute myeloid leukemia (AML) within four to eight weeks post transplant (data not shown). Leukemic mice were sacrificed and BM cells obtained. 45,000 GFP+ primary leukemic Mx1-Cre; Moff/f or Mx1-Cre BM cells were then injected into sub-lethally irradiated C57Bl/6 mice (Figure 3A). At day 14 post transplant, mice were bled to check engraftment (pre pIpC, Figure 3B). That same day, half of the mice per group (n = 10 for Mx1-Cre, n = 20 for Mx1-Cre; Moff/f) were injected with poly(I:C) (pIpC) to induce Cre expression. pIpC treated mice received a total of three dosages, one every other day and were bled again, seven days after the last dose. The white blood cell (WBC) count (Figure 3C) and GFP% in peripheral blood (PB) (Figure 3B) illustrate the lethality of Mof loss to MA9 leukemic cells. Seven days after pIpC treatment Mx1-Cre; Moff/f, pIpC+ mice had a mean WBC count of 3 K/uL versus 112 K/uL (Mx1-Cre; Moff/f, pIpC−) and 104 K/uL (Mx1-Cre, pIpC+), and a significantly lower GFP% of 38% versus 92% (Mx1-Cre; Moff/f, pIpC−) and 95% (Mx1-Cre, pIpC+). Several mice within the Mx1-Cre; Moff/f, pIpC+ group had an actual reduction of GFP% in the PB after receiving pIpC injections (Figure 3D) indicating a reduction of tumor burden upon Mof loss.
Figure 3. Mof loss leads to reduced tumor burden and prolonged survival in an in vivo MLL-AF9 leukemia model.
(A) Schematic for in vivo Mof excision experiment. Primary, GFP+ MA9 leukemia BM cells in an Mx1-Cre; Moff/f or Mx1-Cre background were injected into sub-lethally irradiated C57Bl/6 mice (n=10 for Mx1-Cre and n=20 for Mx1-Cre; Moff/f). Half of the mice per group were treated with poly (I:C) pIpC at day 14 post transplant to induce Mof excision.
(B) GFP% of live cells in peripheral blood (PB) of mice before and after pIpC treatment. A dot represents a single mouse in the experiment.
(C) White blood cell (WBC) counts before and after pIpC treatment.
(D) GFP% of live cells in PB of mice before and after pIpC treatment. A line connects the two values for a single mouse in the experiment.
(E) Survival curve of mice in secondary leukemia experiment. Data are representative of two individual experiments. Arrows indicate pIpC treatment.
a.Log-rank test p-value comparing survival of Mx1-Cre; Moff/f mice treated with pIpC to either of the Mx1-Cre control groups.
b.Log-rank test p-value comparing survival of Mx1-Cre; Moff/f mice treated with pIpC to the untreated group.
All mice eventually succumbed to AML. At time of death, mice had elevated WBC counts (data not shown) and splenomegaly (Figure S3A). FACS analysis showed more than 90% GFP+ cells in BM (Figure S3B), spleen and PB (data not shown). The majority of GFP+ cells in BM, spleen and PB expressed myeloid cell markers MAC1, GR1 and cKIT, indicating a myeloid leukemic phenotype (Figure S3C and D). Animals in all three control groups died with a median survival of 22 (Mx1-Cre, pIpC+), or 24 days (Mx1-Cre; Moff/f and Mx1-Cre, pIpC−). Mx1-Cre; Moff/f, pIpC+ mice lived significantly longer with a median survival of 31 days (p < 0.0001; Figure 3E). At time of death, GFP+ BM cells of Mx1-Cre; Moff/f, pIpC+ mice showed incomplete excision (Figure S3E), illustrating that Mof-deficient MA9 cells have a strong disadvantage of forming leukemia in vivo. When we ended the experiment (day 60), the last living mouse in the Mx1-Cre; Moff/f, pIpC+ cohort did not have any GFP+ cells in the PB or BM. Together these in vivo data demonstrate that Mof is important for maintenance of MA9 driven AML.
Homozygous Mof loss in MLL-AF9 transformed mouse LSKs leads to cell death and DNA damage
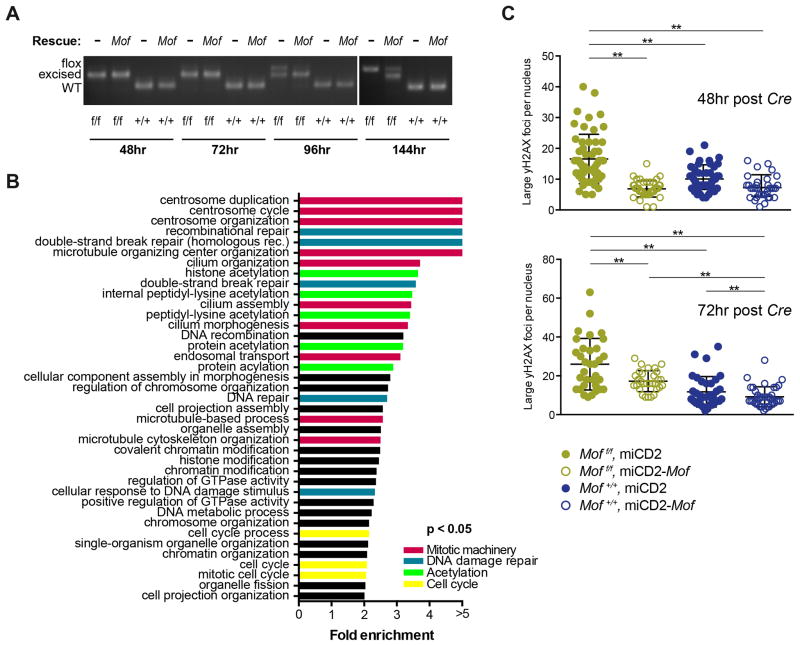
Our CFU assay data show a strong dependence for Mof in MA9 cells. To study the potential underlying mechanism, we performed in vitro excision of Mof in MA9 transformed murine LSKs as described above. While the strong dependency caused a rapid loss of Mof excision (Figure 2E and G), we first had to define the latest time point after Cre infection at which MA9 cells are still fully excised. PCR analysis indicated complete Mof excision up to 72 hours after Cre infection (Figure 4A). We next performed RNA sequencing (RNAseq) on Moff/f and Mof+/+ MA9 cells, harvested 72 hours after Cre infection (Table S2). Gene ontology (GO) analysis comparing MA9 cells with homozygous Mof loss to the wild type control, showed a significant enrichment of cell division and DNA damage repair pathways in genes that are downregulated in MA9 Mof KO cells. To verify this finding, we stained MA9 transformed LSKs at 48 or 72 hours after Cre infection with an immunofluorescent-labeled yH2AX antibody (Figure S4A and S4B). Confocal microscopy revealed significantly more yH2AX foci per cell nucleus in Mof-deficient MA9 cells compared to controls (p<0.01; Figure 4C), indicative of more DNA damage. This increase in DNA damage was largely rescued by overexpression of full-length Mof (Figure 4C). These experiments demonstrate that there is evidence of cell death and DNA damage in MA9 cells upon Mof loss.
Figure 4. Homozygous Mof loss in MLL-AF9 transformed mouse LSKs leads to cell death and DNA damage.
(A) MA9 in vitro transformed Moff/f or Mof+/+ LSKs were infected with full-length Mof or empty vector control (miCD2) and selected by sorting hCD2 positive cells. Cells were then plated in liquid culture immediately upon sorting dTomato positive cells, 48 hours after infection with dTomato-Cre. PCR analysis illustrates excision at 48, 72, 96 and 144 hours post Cre infection.
(B) Gene ontology analysis with a differential expression analysis list of significantly (p < 0.05) downregulated genes comparing Moff/f cells to Mof+/+ cells at 72 hours post Cre infection.
(C) Number of large yH2AX foci per cell nucleus at 48 or 72 hours post Cre infection. A dot represents a single cell.
** p < 0.01. Error bars represent SD of mean.
Mof histone acetyltransferase activity is required for colony formation of MLL-AF9 transformed mouse LSKs
MOF has been identified as the major H4K16 acetyltransferase in humans, mice and Drosophila.25,33,36–38 MOF contains a HAT domain with a CoA binding site that was found to be crucial for its acetyltransferase activity.18,36 While MOF possesses acetyltransferase activity on various histones and nucleosomes, depletion of MOF in HeLa cells was shown to lead to a dramatic decrease in H4K16ac whereas other acetylation sites appeared to be unaffected.25
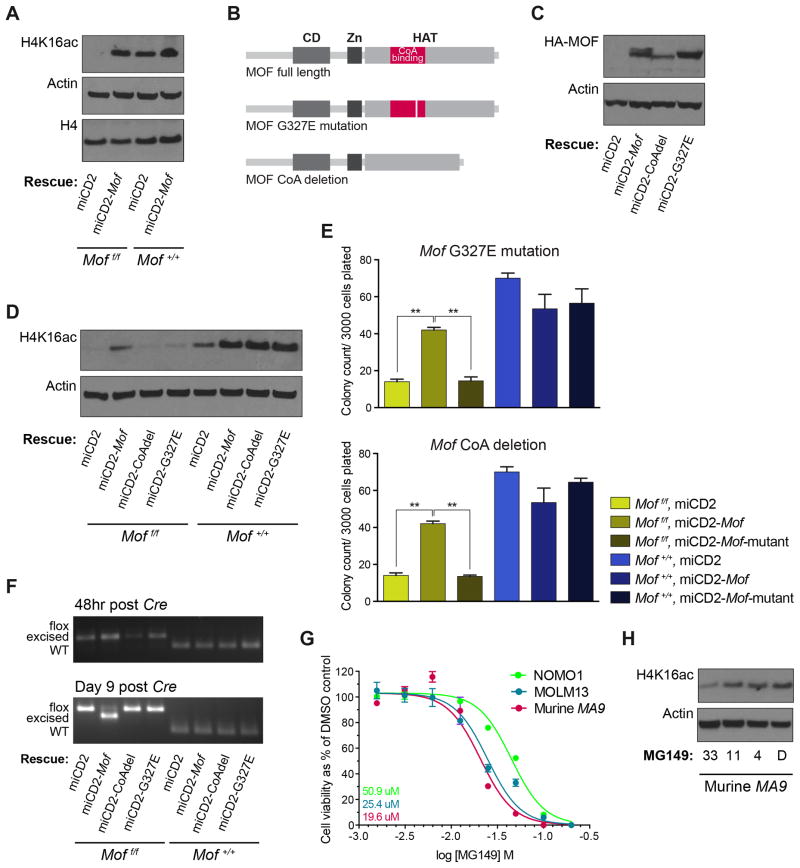
Since our data indicate a strong Mof dependence in MA9 leukemic cells, we set out to establish whether this dependence is through MOF HAT activity. When assessing changes in global H4K16ac upon Mof loss in MA9 cells we found a significant decrease of H4K16ac (Figure 5A), a decrease that was averted by expression of exogenous Mof (Moff/f, miCD2-Mof). Next, we designed two HA-tagged, HAT inactivated Mof retroviral constructs (Figure 5B) in which either the CoA binding site was deleted (Mof-CoAdel) or a HAT inactivating point mutation (G327E) was introduced (Mof-G327E). The HAT inactivating point mutation in the MOF CoA binding domain was first described in Drosophila36 and later used in a human MOF construct where it was also found to diminish H4K16ac39. Alignment of Drosophila, murine and human MOF illustrates that the point mutation involves replacement of glycine by glutamic acid on position 327 (G327E) in murine, as well as human MOF. The constructs were packaged in an miCD2-MSCV retroviral vector with the hCD2 selection marker (miCD2-Mof-CoAdel and miCD2-Mof-G327E).
Figure 5. Mof histone acetyltransferase activity is required for colony formation of MLL-AF9 transformed mouse LSKs.
(A) MA9 in vitro transformed Moff/f or Mof+/+ LSKs were infected with full-length Mof or empty vector control (miCD2) and selected by sorting hCD2 positive cells. Cells were then infected with dTomato-Cre, 48 hours later sorted and another 24 hours later harvested. Western blot was performed on whole protein lysates.
(B) Schematic illustrating full length Mof and two Mof mutants with either a G327E point mutation or a CoA binding-site deletion.
(C) MA9 in vitro transformed Moff/f or Mof+/+ LSKs were infected with HA-tagged full-length Mof, CoA deleted Mof (CoAdel), Mof with a G327E mutation or empty vector control, and selected by sorting hCD2 positive cells. Western blot for HA confirms presence of these exogenous constructs.
(D) Cells were infected with dTomato-Cre and 48 hours later sorted and harvested. Western blot illustrates global H4K16ac and Actin in protein lysates.
(E) Day seven of CFU assay. Cells were plated immediately upon sorting dTomato positive cells, 48 hours after infection with dTomato-Cre. Bar graph indicates mean number of colonies per 35mm dish after seven days. Data are representative of three individual experiments.
(F) PCR analysis illustrating excision throughout the duration of the 7-day colony-forming experiment. Representative gel images are shown.
(G) MOLM13, NOMO1 and murine polyclonal MA9 cells were plated in liquid culture and treated with various concentrations of MG149. Plotted is the IC50 curve for cell viability as a percentage of the vehicle (DMSO) control at day three of treatment. Numbers indicate the IC50 per cell type (by color).
(H) Western blot showing global H4K16ac and Actin in murine MA9 cells after three days of MG149 treatment at various concentrations.
** p < 0.01. Error bars represent SD of mean.
We infected unexcised Moff/f and Mof+/+ MA9 cells with either miCD2-Mof, miCD2-CoAdel, miCD2-G327E or miCD2 and sorted hCD2 positive cells. Western blot analysis for HA-tagged MOF indicated that exogenous MOF was expressed at similar levels in miCD2-Mof and miCD2-G327E, though MOF expression seemed a little lower with the miCD2-CoAdel construct (Figure 5C). Upon in vitro Mof KO by transduction with retroviral Cre, Western blot analysis confirmed that exogenous full-length MOF was capable of restoring H4K16ac levels upon genetic Mof loss, while both HAT domain mutant Mof constructs were not (Figure 5D). When using these Cre transduced MA9 cells for a CFU assay, full-length MOF indeed rescued colony formation of Moff/f cells while exogenous, HAT-deficient MOF could not (Figure 5E). Mof excision PCR analysis reaffirms these findings where Moff/f miCD2, miCD2-CoAdel and miCD2-G327E cells have fully lost excision at day 9 post Cre infection, but miCD2-Mof cells are still largely excised (Figure 5F). In summary, we found a loss of global H4K16ac upon Mof KO and in line with this finding of H4K16ac loss, rescue experiments with HAT domain mutated MOF illustrated that the HAT activity of MOF is indispensable for MA9 colony-forming capacity.
Our finding that MOF HAT activity is required for MA9 leukemia suggests that targeting MOF HAT activity could provide us with a new approach in treating MA9 leukemia patients. In 2011 a selective small molecule inhibitor of MYST protein HAT activity, MG149, was designed.32 While this small molecule is not particularly potent, we decided to use this inhibitor to test its effect on cell viability in the human MA9 AML cell lines NOMO1 and MOLM13 and in murine, polyclonal MA9 primary leukemia cells. A dose response curve showed a strong anti-proliferative effect on all three MA9 cells with an IC50 of 19.6 uM (murine MA9), 25.4 uM (MOLM13) and 50.9 uM (NOMO1). We further repeated the MG149 in vitro assay on the human non-MLL-rearranged leukemia cell lines Kasumi-1 (AML1-ETO fusion), U937 (CALM-AF10 fusion) and K562 (BCR-ABL fusion). All three cell lines showed an apparent sensitivity to the small molecule MYST protein HAT inhibitor (Figure S5G). In addition, MG149 inhibition induced global H4K16ac loss in murine MA9 cells (Figure 5H). Together, these results indicate that MG149 effectively targets MOF HAT activity with an anti-proliferative effect on human as well as murine leukemia cells, suggesting that MOF HAT activity may be a viable target in not only MLL-AF9 driven, but also various acute leukemias.
Mof is required for NUP98-HOXA9 driven leukemogenesis in vitro and in vivo
Given that human leukemia cell lines with various translocations are sensitive to MOF HAT inhibitor MG149, we then test whether MOF is required for non-MLL-rearranged leukemogenesis. To assess this question we performed Mof knockout experiments in a NUP98-HOXA9 leukemia model. Similar to what we found for MA9 cells, loss of Mof in NUP98-HOXA9 transformed cells lead to impaired colony formation (Figure S5A–E). An in vivo secondary NUP98-HOXA9/Meis1a leukemia experiment, in set-up similar to the MA9 in vivo experiment (Figure 3), illustrated significantly prolonged survival in the Mof-deficient group (Mx1-Cre; Moff/f, pIpC+, median survival 117 days, p < 0.001) compared to all five control groups that died with a median survival of 28 to 39 days (Figure S5F). These results indicate that Mof is required for maintenance of NUP98-HOXA9-transformed cells in vitro and NUP98-HOXA9 driven AML in vivo. The indispensable role of Mof in NUP98-HOXA9 driven AML demonstrated here is in line with our recent findings that the NUP98 fusions physically interact with MLL1 and the non-specific lethal (NSL) histone-modifying complexes 50. It will be interesting to further examine whether and how MOF affects the oncogenic program in NUP98-fusion driven leukemia.
DISCUSSION
To identify novel druggable targets in MLL-rearranged leukemia, we performed an RNAi screen focused on chromatin regulators. This resulted in the identification of Mof as a critical gene for cell growth in a murine MA9 leukemia model. We found that homozygous Mof loss led to a significant decrease in colony-forming ability, reduced tumor burden and prolonged survival in mice. RNAseq of Mof-deficient MA9 cells showed significant downregulation of genes associated with DNA damage repair pathways, and upon validation by immunofluorescence we indeed found a significant increase of yH2AX foci in Mof-deficient MA9 cells. In addition, we uncovered a loss of global H4K16ac upon Mof KO. Rescue experiments with HAT domain mutated MOF illustrated that MOF HAT activity is indispensable for MA9 leukemia maintenance. This new insight could be utilized in the development of a small molecule inhibitor for the treatment of leukemia patients carrying an MLL translocation.
Gene expression analysis in Mof-deficient MA9 cells indicated that MOF loss does not lead to clear downregulation of MA9 target genes such as Meis1 and Hoxa cluster genes. Our RNAseq data suggested that Mof loss leads to impairment of more general biological processes required for cellular integrity implying that Mof may not only be required for MA9 leukemogenesis, but for a broad range of acute leukemias, This is further supported by the facts that Mof is required for NUP98-HOXA9 driven leukemia, and non-MLL-rearranged human leukemia cells are also sensitive to HAT inhibitor MG149. We identified DNA damage repair as a possible mechanism of cell death in Mof-deficient leukemia cells (Figure 4C), though our findings are not conclusive with regard to the mechanism of Mof-dependence. Mof-null murine embryonic fibroblasts (MEFs) were previously shown to be deficient in DNA damage repair after ionizing radiation.22 In wild type MEFs, radiation induced DNA damage led to an increase of global H4K16ac22, suggesting that a MOF-mediated increase of H4K16ac may be essential for an appropriate DNA damage response. Unacetylated H4K16 is required to achieve the maximum tendency of in vitro nucleosome arrays to fold into secondary or tertiary chromatin structures.40 In contrast, 30% H4K16 acetylation alleviates compaction of the chromatin fiber.41 Therefore it may be that the MOF-loss induced global H4K16ac depletion influences the DNA damage response and/or chromatin integrity by increasing chromatin compaction.
GO analysis on our RNAseq data suggested that loss of Mof in MA9 leukemic cells may not only lead to DNA damage, but also general chromosomal instability. This could contribute to the rapid cell death we observed in Mof-deficient MA9 cells. Genetic and biochemical data underscore the importance of unacetylated acidic histone tails in gene silencing.42 Interestingly, the yeast ortholog of MOF, SAS2, was previously shown to lead to telomeric silencing43 and it is well established that telomeric dysfunction can lead to chromosomal instability44. Future experiments will be required to assess how Mof-loss induced H4K16ac depletion may contribute to chromosomal instability in Mof-deficient MA9 cells, and whether telomeric silencing is involved.
MOF was shown to functionally and physically interact with the histone methyltransferase MLL1. The interaction between MOF and MLL1 is important for the chromatin regulatory function of both enzymes.45 In normal hematopoiesis, Mll1 is essential for development and maintenance of both embryonic and adult progenitors and hematopoietic stem cells (HSCs).46 In addition, it has been suggested that wild type MLL1 may play a role in MLL-rearranged leukemogenesis.47 However, in both normal and malignant hematopoiesis, the H3K4 methyltransferase activity of MLL1 is dispensable.48 Given the identified MA9 leukemia dependence on the enzymatic activity of MOF, it may be that, in the setting of MLL-rearranged leukemia, it is in fact the HAT activity of MOF that is required for leukemogenesis, and MLL1 merely functions as a scaffolding protein, recruiting MOF to the targets of the oncogenic fusion.
Over the last decade, many advances have been made in the field of cancer epigenomics. Our vastly expanding knowledge on the role of chromatin regulation in cancer has led to the development of various drug compounds that target the cancer epigenome, several of which are currently in clinical trials.5,49 Here we have established that MOF HAT activity is required for MA9 leukemia maintenance and loss of MOF HAT activity leads to elevated DNA damage. In addition, we have successfully inhibited cell growth of human and murine MA9 leukemia cells, but also other fusion-driven leukemias by using a first generation small molecule MYST protein HAT inhibitor. Based on our findings, we believe that inhibiting MOF HAT activity by small molecules may prove to be an effective, novel approach for the treatment of patients with MLL-rearranged and perhaps other leukemias.
Supplementary Material
Acknowledgments
FINANCIAL SUPPORT:
This work was supported by a CURE Childhood Cancer Research Grant (D.G.V.); by a DoD Bone Marrow Failure Postdoctoral Fellowship Award (W81XWH-12-1-0568) (H.X.); NIH grants PO1 CA66996 and R01 CA140575 (S.A.A.); the Leukemia and Lymphoma Society (S.A.A.); Gabrielle’s Angel Research Foundation (S.A.A.); NIH RO1 CA129537 and RO1 GM109768 (TKP); and an NIH Memorial Sloan Kettering Cancer Center Support Grant (P30 CA008748).
We would like to thank C.M. Woolthuis for critically reading the manuscript and Z. Feng for administrative assistance.
Footnotes
AUTHORSHIP CONTRIBUTIONS
D.G.V., H.X. and S.A.A. conceived the study and designed the experiments; D.G.V., H.X., T.H., M.E.E. and C.D. performed experiments; C.C., C.H., A.L. and S.W.L. designed and analyzed the RNAi screen; D.G.V. performed all other data analysis; H.X., C.C., M.C. and A.J.D. gave conceptual advice; G.Z. provided MG149 compound; T.K.P. generated the conditional Mof knockout mouse model; D.G.V. and S.A.A. wrote the manuscript; H.X. edited the manuscript.
DISCLOSURE OF CONFLICTS OF INTEREST:
S.A.A. is a consultant for Epizyme Inc. and Imago Biosciences. Other authors declare no conflict of interest.
References
- 1.Djabali M, Selleri L, Parry P, Bower M, Young BD, Evans GA. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat Genet. 1992;2(2):113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 2.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71(4):691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 3.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greaves MF. Infant leukaemia biology, aetiology and treatment. Leukemia. 1996;10(2):372–377. [PubMed] [Google Scholar]
- 5.Brien Gerard L, Valerio Daria G, Armstrong Scott A. Exploiting the Epigenome to Control Cancer-Promoting Gene-Expression Programs. Cancer Cell. 2016;29(4):464–476. doi: 10.1016/j.ccell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi P, Allis CD, Wang GG. Covalent histone modifications-miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guenther MG, Lawton LN, Rozovskaia T, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22(24):3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14(5):355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernt KM, Zhu N, Sinha AU, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshpande AJ, Deshpande A, Sinha AU, et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell. 2014;26(6):896–908. doi: 10.1016/j.ccell.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang MJ, Wu H, Achille NJ, et al. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010;70(24):10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117(18):4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daigle SR, Olhava EJ, Therkelsen CA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122(6):1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daigle SR, Olhava EJ, Therkelsen CA, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris WJ, Huang X, Lynch JT, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21(4):473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13(5):337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 18.Yang XJ, Ullah M. MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells. Oncogene. 2007;26(37):5408–5419. doi: 10.1038/sj.onc.1210609. [DOI] [PubMed] [Google Scholar]
- 19.Gupta A, Hunt CR, Pandita RK, et al. T-cell-specific deletion of Mof blocks their differentiation and results in genomic instability in mice. Mutagenesis. 2013;28(3):263–270. doi: 10.1093/mutage/ges080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A, Hunt CR, Hegde ML, et al. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell Reports. 2014;8(1):177–189. doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta A, Sharma GG, Young CS, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25(12):5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Corsa CA, Pan PW, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30(22):5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma GG, So S, Gupta A, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30(14):3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhadra MP, Horikoshi N, Pushpavallipvalli SN, et al. The role of MOF in the ionizing radiation response is conserved in Drosophila melanogaster. Chromosoma. 2012;121(1):79–90. doi: 10.1007/s00412-011-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taipale M, Rea S, Richter K, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25(15):6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Li L, Pandey R, et al. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11(2):163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfister S, Rea S, Taipale M, et al. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. International Journal of Cancer. 2008;122(6):1207–1213. doi: 10.1002/ijc.23283. [DOI] [PubMed] [Google Scholar]
- 28.Cai M, Hu Z, Liu J, et al. Expression of hMOF in different ovarian tissues and its effects on ovarian cancer prognosis. Oncology reports. 2015;33(2):685–692. doi: 10.3892/or.2014.3649. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Liu X, Zhang Y, Cheng Y, Li Y. RNAi screening identifies KAT8 as a key molecule important for cancer cell survival. Int J Cancer. 2013;6(5):870–877. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao L, Wang DL, Liu Y, Chen S, Sun FL. Histone acetyltransferase hMOF promotes S phase entry and tumorigenesis in lung cancer. Cell Signal. 2013;25(8):1689–1698. doi: 10.1016/j.cellsig.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, Sun H, Shu Y, Zou X, Zhao Y, Ge C. hMOF (human males absent on the first), an oncogenic protein of human oral tongue squamous cell carcinoma, targeting EZH2 (enhancer of zeste homolog 2) Cell Prolif. 2015;48(4):436–442. doi: 10.1111/cpr.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghizzoni M, Wu J, Gao T, Haisma HJ, Dekker FJ, George Zheng Y. 6-alkylsalicylates are selective Tip60 inhibitors and target the acetyl-CoA binding site. Eur J Med Chem. 2012;47(1):337–44. doi: 10.1016/j.ejmech.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta A, Guerin-Peyrou TG, Sharma GG, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol. 2008;28(1):397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang CH, Lujambio A, Zuber J, et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28(16):1800–1814. doi: 10.1101/gad.244368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CW, Koche RP, Sinha AU, et al. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat Med. 2015;21(4):335–343. doi: 10.1038/nm.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5(2):367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 37.Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25(21):9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol. 2008;28(16):5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Su J, Wang F, et al. Crosstalk between NSL histone acetyltransferase and MLL/SET complexes: NSL complex functions in promoting histone H3K4 di-methylation activity by MLL/SET complexes. PLoS Genet. 2013;9(11):e1003940. doi: 10.1371/journal.pgen.1003940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 41.Robinson PJ, An W, Routh A, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381(4):816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pillus LaMG. Chromatin Structure & Gene Expression. Oxford: Oxford University Press; 1995. Chromatin structure and epigenetic regulation in yeast; pp. 123–146. [Google Scholar]
- 43.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14(1):42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 44.Greenberg RA. Telomeres, crisis and cancer. Curr Mol Med. 2005;5(2):213–218. doi: 10.2174/1566524053586590. [DOI] [PubMed] [Google Scholar]
- 45.Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121(6):873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 46.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiel AT, Blessington P, Zou T, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild type Mll allele. Cancer Cell. 2010;17(2):148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra BP, Zaffuto KM, Artinger EL, et al. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Reports. 2014;7(4):1239–1247. doi: 10.1016/j.celrep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai SF, Chen CW, Armstrong SA. Drugging Chromatin in Cancer: Recent Advances and Novel Approaches. Mol Cell. 2015;60(4):561–570. doi: 10.1016/j.molcel.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, Chen CW, Chu SH, Brien GL, Park CY, Hsieh JJ, Ernst P, Armstrong SA. NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis. Cancer Cell. 2016;30(6):863–878. doi: 10.1016/j.ccell.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.