Abstract
Cell-based assays are finding wider use in evaluating compounds in primary screens for drug development, yet it is still challenging to measure enzymatic activities as an end point in a cell-based assay. This paper reports a strategy that combines state-of-the-art cantilever free polymer pen lithography (PPL) with self-assembled monolayer laser desorption–ionization (SAMDI) mass spectrometry to guide cell localization and measure cellular enzymatic activities. Experiments are conducted with a 384 spot array, in which each spot is composed of ∼400 nanoarrays and each array has a 10 × 10 arrangement of 750 nm features that present extracellular matrix (ECM) proteins surrounded by an immobilized phosphopeptide. Cells attach to the individual nanoarrays, where they can be cultured and treated with small molecules, after which the media is removed and the cells are lysed. Phosphatase enzymes in the proximal lysate can then act on the immobilized phosphopeptide substrate to convert it to the dephosphorylated form. After the lysate is removed, the array is analyzed by SAMDI mass spectrometry to identify the extent of dephosphorylation and, therefore, the amount of enzyme activity in the cell. This novel approach of using nanopatterning to mediate cell adhesion and SAMDI to record enzyme activities in the proximal lysate will enable a broad range of cellular assays for applications in drug discovery and research not possible with conventional strategies.
Keywords: Self-assembled monolayer, SAMDI mass spectrometry, phosphatase inhibitor, polymer pen nanolithography
Assays that evaluate the biological effects of small molecules in cell cultures are important in many applications, including studying the mechanisms of action of natural products, elucidating signal transduction pathways, and screening small-molecule libraries in drug-discovery programs.1,2 However, it is still difficult to measure many biochemical activities in cell-based assays, and therefore, these assays cannot be applied to many targets of interest. Indeed, most assays report on a phenotypic behavior, including cell differentiation,3 cell death,4 and migration5 and in those cases, they do not measure the inhibition or activation of specific enzymes. Detection methods based on optical sensors6 or fluorescent proteins7 have allowed the real-time observation of metabolite secretion and specific protein and enzyme activities as a result of chemical or mechanical stimuli,8−10 but it remains challenging to develop these reagents and many biochemical activities have not yet been targeted with these approaches. In this paper, we describe a strategy wherein adherent cells can be treated with small molecules, cultured, lysed, and then analyzed by mass spectrometry to measure the activities of endogenous enzymes. The implementation of this method relies on the use of surfaces that are nanopatterned with extracellular matrix (ECM) proteins to mediate cell attachment and with a peptide that is a substrate for the desired enzyme activity in the lysate.
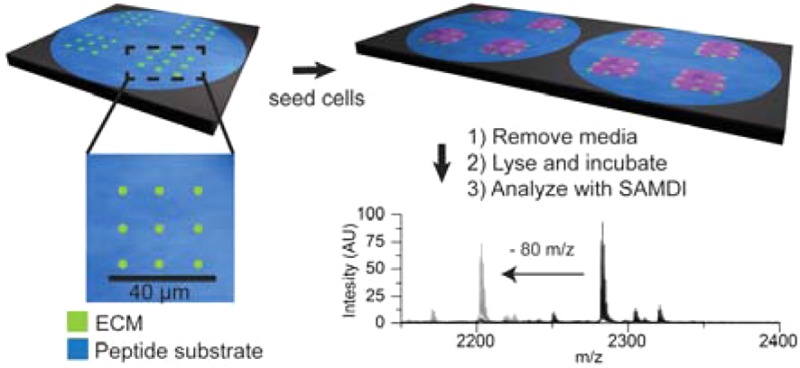
Our approach is based on monolayers having two distinct properties; they must present proteins that mediate cell adhesion, and they must also present peptides that are substrates for enzymes whose activities will be measured. Because these two functions are not compatible since the adhesion proteins would obstruct access of the enzyme to the immobilized peptide, it is necessary to pattern the monolayer into two regions. By the use of the emerging state-of-the-art cantilever-free polymer pen lithography (PPL)11−15 technique to create nanopatterns of the adhesive protein in 750 nm features, cells can still attach and spread, but the majority of the monolayer still presents the phosphopeptide substrate that is measured by self-assembled monolayer laser desorption–ionization (SAMDI) mass spectrometry (Figures 1 and S1).16 In this way, cells adhere to the surface by way of interactions with the matrix proteins,17 while the other regions of the surface remain available for recording the enzyme activity (Figure 2a,b). A further benefit of this approach is that it can be used to define sites for adsorption of virtually any matrix protein, and therefore, it allows the tandem culture and lysis self-assembled monolayer laser desorption–ionization (TCAL-SAMDI) method to be applied to assays using any adherent cell line.18
Figure 1.
This work reports the use of surfaces that are nanopatterned with extracellular matrix proteins that support cell adhesion, and where the intervening regions present a peptide substrate for an enzyme, to enable cell-based assays using SAMDI mass spectrometry. Note that this work used nanoarrays that have 100 fibronectin features. Cells that are adherent to the nanoarrays are cultured and treated with small molecules. The media is then removed, and a lysis buffer is applied to each region of cells, where enzymes in the lysate can modify the peptide in the intervening regions. The surface is then rinsed and analyzed with SAMDI mass spectrometry to determine the extent of conversion of the peptide substrate and, therefore, the amount of enzyme activity in the lysate.
Figure 2.
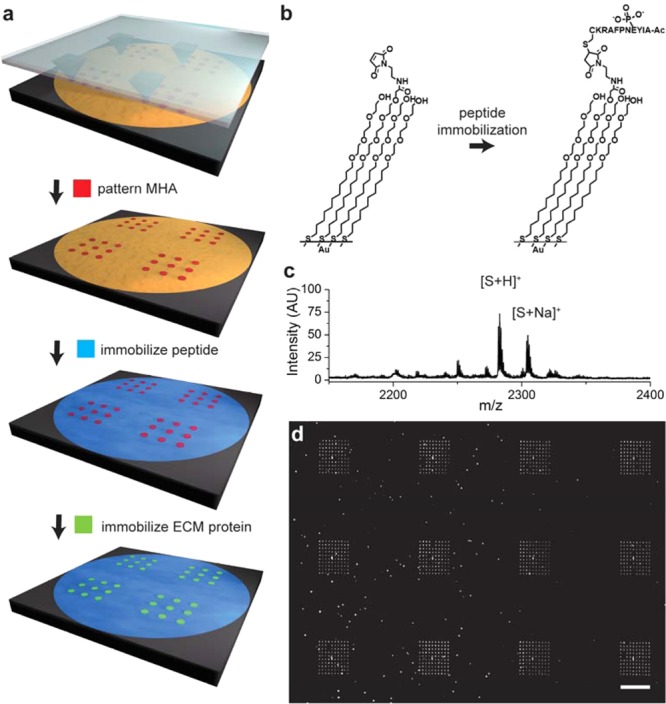
Nanoarrays were prepared by using PPL to pattern mercaptohexadecanoic acid (MHA) on a gold-coated surface in many 10 × 10 arrays where each spot was 750 nm in diameter and where neighboring spots had a center-to-center spacing of 4.4 μm (a). The remaining areas of gold were then modified with a monolayer presenting maleimide groups against a background of tri(ethylene glycol) groups and used to immobilize a cysteine terminated phosphopeptide (b). The surface was then treated with a solution of fibronectin to allow the adsorption of the extracellular matrix protein to the MHA nanoarray. A SAMDI spectrum of the monolayer confirms immobilization of the peptide (c). The fluorescence micrograph shows fibronectin patterned nanorrays stained with mouse antifibronectin antibody and AlexaFluor568-conjugated goat anti-mouse IgG (d). The scale bar is 40 μm.
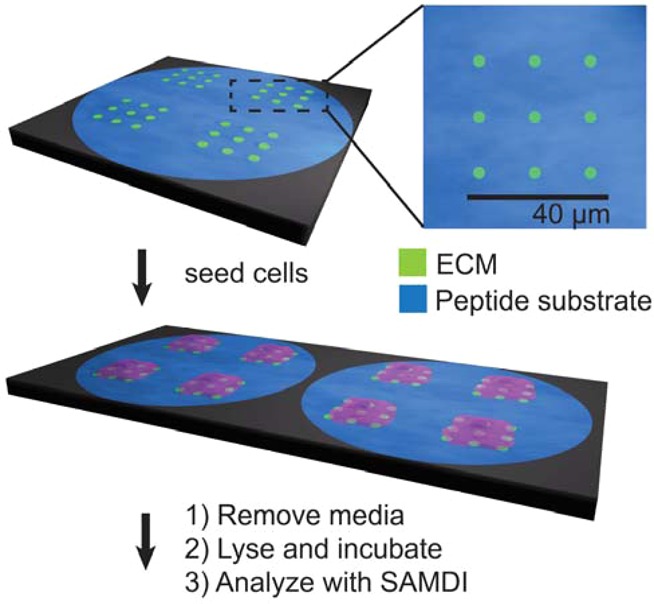
To prepare the array plates, we first evaporated titanium onto a glass slide and then deposited gold through a mask having an array of holes arranged in the standard 384 well format.19 The slide was then immersed in a solution of hexadecylphosphonic acid (10 mM in ethanol) for 10 min to form a hydrophobic monolayer on the titanium dioxide areas surrounding the gold circular regions. This monolayer serves to confine aqueous solutions to the circular regions of gold and to isolate each reaction. Next, we used PPL to create patterns of a mercaptohexadecanoic acid (MHA) monolayer on the gold-coated regions of the glass plate. This technique has proven useful for patterning proteins, peptides, oligonucleotides, and small molecules for a wide variety of biological applications.20−25 In PPL, an elastomeric pen array is coated with a molecular “ink” and subsequently mounted to a scanning probe instrument and pressed onto a gold-coated slide to create an array of circular MHA monolayer features. This step can be repeated with translational movement of the array to create arbitrary patterns.11,15 The feature size can be easily controlled and customized by adjusting the amount of force applied to the pen array and the time the pen array remains in contact with the surface.13 Here, we used a SAMDI array that has a portion of a microtiter plate with 384 gold islands, wherein each island is 2.8 mm in diameter.26 PPL was then used to pattern MHA features within each island. In a typical experiment, a poly(dimethylsiloxane) (PDMS) pen array (1.2 × 1.2 cm2) having 10 000 pens, corresponding to a pen-to-pen distance of 120 μm and each coated with a solution of MHA (10 mM in ethanol), was used to generate 428 regions containing 10 × 10 square arrays of MHA features, each measuring 750 nm in diameter and spaced by a center-to-center distance of 4.4 μm within each gold island (Figure S1). These MHA features, when later modified with the appropriate ECM protein, mediate the attachment of an individual HeLa cell to each square array.27 We verified the fidelity of the patterning step by chemically etching a portion of the substrate with a mixed aqueous solution of iron nitrate (13.3 mM) and thiourea (20 mM) to remove the nonpatterned gold film (Figure S2). The nonpatterned gold areas were functionalized with a mixed monolayer that presents maleimide groups at a density of 10% against a background of tri(ethylene glycol) groups.
Finally, a peptide substrate for phosphotyrosine phosphatases (AIpYENPFARKC, where p denotes phosphorylation of the tyrosine residue)28−30 was covalently immobilized by a conjugate addition of the terminal cysteine residue to the maleimide groups present on the monolayer.31 SAMDI mass spectrometry confirmed that peptide immobilization was complete, and X-ray photoelectron spectroscopy (XPS) characterization showed the presence of sulfur and nitrogen peaks in the resulting monolayer consistent with the presence of thiols and amide bonds, respectively (Figures 2c and S3). Finally, the patterned substrates were immersed in a solution of fibronectin (30 μg/mL in PBS) to allow the nonspecific adsorption of protein to the patterned MHA features. Immunofluorescent labeling of fibronectin confirmed the adsorption only to the regions of MHA (Figure 2d). In general, this approach is applicable to other ECM attachment proteins, such as collagen and laminin, which can also adsorb to self-assembled monolayers by way of nonspecific interactions.
We seeded HeLa cells on the fibronectin nanopatterned substrates and cultured the cells for 2 h under standard media conditions (Figure 3a). The cells spread fully within the 10 × 10 nanoarrays of fibronectin, and they remained adherent during the culture (Figure 3b). We only observed cells on the patterned regions presenting the fibronectin, and cells remained confined to those regions of the substrate, showing that the tri(ethylene glycol)-terminated monolayers were effective at preventing cell adhesion and spreading beyond the patterned matrix. After 2 h in culture, the monolayers were rinsed with PBS to remove the media, and then a lysis buffer containing a protease inhibitor cocktail was applied to each patterned region. The solutions were kept at 37 °C for 1 h to allow enzymes in the lysate to interact with the phosphopeptides on the monolayer. The mixed monolayer was rinsed with PBS buffer and then treated with 2,4,6-trihydroxyacetophenone (THAP) matrix (30 mg/mL in acetone) and analyzed with SAMDI mass spectrometry.
Figure 3.
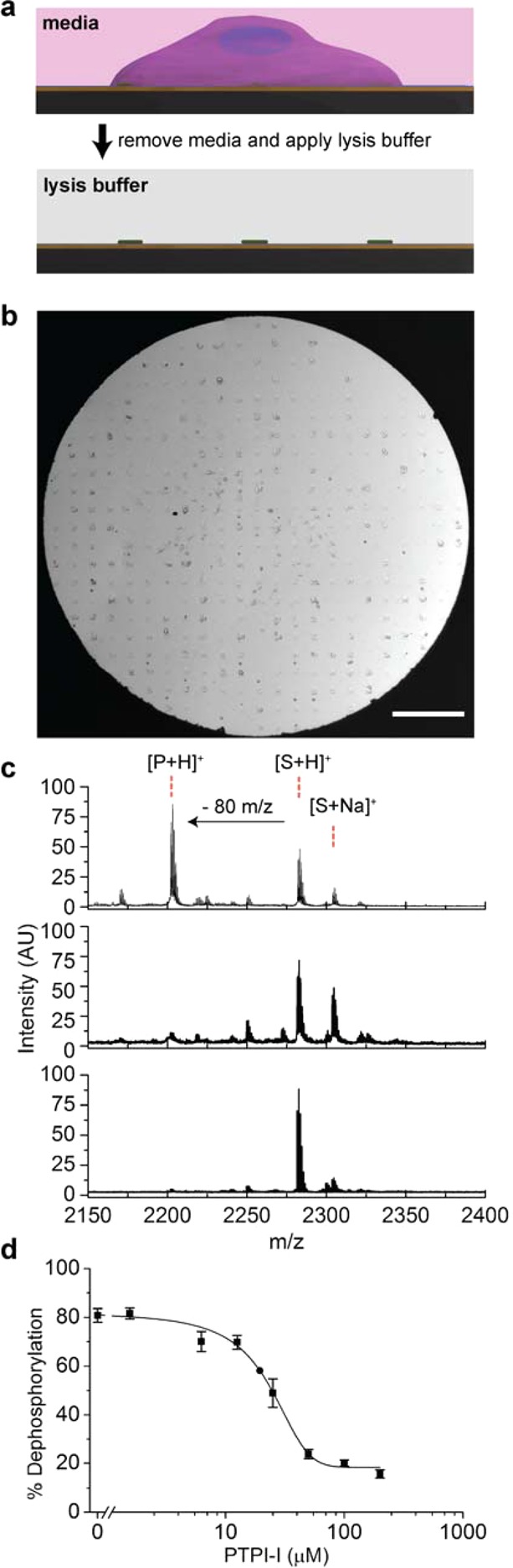
Cell culture and lysis on mixed monolayers. Cells were cultured on patterned monolayers as described in Figure 2a. Individual cells attached to each 10 × 10 fibronectin nanoarray and remained confined to these regions of the substrate (b). The media was then removed from the entire plate and a lysis buffer was added to each spot of the 384 spot array to allow phosphatase enzymes in the lysate to act on peptides immobilized on the monolayer. The scale bar is 500 μm. SAMDI spectra of the surface after removal of the lysate showed a peak corresponding to generation of the dephosphorylated product (c, top). Addition of the phosphatase inhibitor PTPI-I to the lysis buffer resulted in a loss of phosphatase activity (middle) as did proteolytic removal of the cells without lysis (bottom). Separately, populations of HeLa cells were treated with PTPI-I in concentrations ranging from 0 to 200 μM and then lysed and analyzed with SAMDI-MS. A dose–response curve shows half-maximum inhibition at concentration of approximately 22 μM. Standard errors were determined from three independent experiments with at least five spots per condition.
We first analyzed a control array that was not seeded with cells and we observed peaks in the SAMDI spectrum that corresponded to asymmetric disulfides terminated in one phosphopeptide and one tri(ethylene glycol) group (m/z = 2282) as well as the Na+ (m/z = 2304) and K+ (m/z = 2320) adducts of this molecule (Figure 2c). For arrays that were treated with cells that had been lysed, the SAMDI spectra revealed corresponding peaks appearing at 80 Da lower mass, which is consistent with dephosphorylation of the peptide (Figure 3c, top). The spectra were similar to those acquired from a monolayer that only presented the phosphopeptide against the tri(ethylene glycol) background and that was treated with a lysate isolated in the conventional manner. Hence, the nanopatterned fibronectin features did not interfere with the enzyme action on the peptide or with the SAMDI mass spectrometry analysis of the intervening monolayer. This was expected because the protein was present on approximately 1% of the patterned surface, leaving most of the monolayer available for analysis by SAMDI, and also because the protein would be observed at a much higher mass range in the spectrum.
We confirmed that the phosphatase activity we observed was due to enzymes present in the cell lysate. For example, when cells were cultured for 2 h and then removed by treatment with the protease TrypLE, a selective protease that reduces the digestion of cell surface proteins, the resulting surfaces had essentially no dephosphorylated peptide, showing that potential secretion of phosphatases by the cell did not significantly contribute to our measurements (Figure 3c, middle). Similarly, we assayed conditioned media obtained from cell cultures and did not observe phosphatase activity. We also introduced a known phosphotyrosine phosphatase inhibitor during cell culture to confirm that the activity was due to cellular phosphatases. PTP Inhibitor I (PTPI-I), a covalent inhibitor,32 was added to cell cultures (300 μM) during the 2 h culture period. Following lysis and analysis as described above, we observed a 92% decrease in phosphatase activity (Figure 3c, bottom). To assess the use of the assay to quantitatively characterize the effect an inhibitor has in cultured cells, we cultured several populations of HeLa cells on the nanopatterned monolayers and treated each with a distinct concentration of the PTPI-I inhibitor. We then lysed the cells and used SAMDI mass spectrometry to determine the extent of the reaction. The degree of inhibition showed the expected sigmoidal dependence on the concentration of the inhibitor, with an IC50 of 22 μM (Figure 3d). Furthermore, the experiment was performed three independent times, and measurement of the standard error revealed good reproducibility in the measurements. Together, these experiments demonstrate that the TCAL assay quantitatively measures enzyme activities present in the cell lysate.
The nanopatterned substrates reported here are significant because they expand the use of the TCAL assay to a broad range of cell types.18 Whereas the TCAL assay had previously been limited to the use of cells that could be cultured on monolayers presenting short peptides that mediate cell adhesion (for example, the RGD motif),33 we now show that monolayers that are patterned with nanoarrays of ECM proteins can support the adhesion and culture of cells and still be analyzed with SAMDI mass spectrometry. Hence, established cultures that use glass or plastic substrates that are uniformly modified with a layer of ECM can be readily translated to the TCAL assay with these nanopatterned substrates. This approach is also significant because it can measure activities in lysates prepared from as few as ten cells and because there is no processing or delay between generation and assay of the lysate, which often leads to loss of protein activity.18 The use of SAMDI-MS provides a label-free assay of a broad range of enzyme activities, making this format quite general for applications in different drug development targets.34,35 The tri(ethylene glycol)-terminated monolayers have been shown to remain inert for up to 1 week in culture, making this approach compatible with most cell-based assay protocols.36 Finally, the TCAL-SAMDI method is not limited to the use of peptides as substrates for the relevant enzyme but can also use carbohydrates,37 small molecules, and protein substrates38 because each of these molecules can be immobilized to a monolayer and characterized with SAMDI mass spectrometry.
Traditionally, cell-based assays have been employed when the phenotype of interest could not be translated to an enzyme activity; for example, a validated target for blocking metastasis is still lacking. They have not been used when a validated target is available because molecular assays are faster, less expensive, and far less limited as to the molecular activities that can be assayed. The novel strategy we report here narrows this gap between cell-based and molecular assays and promises to increase the use of cell-based assays in the first phase of drug discovery programs. The ability to assay compounds in cells, which reveals aspects of entry, trafficking, and effects owing to interaction with other cellular proteins but with a molecular readout, combines the advantages of molecular and cellular assays and represents a significant advance in both drug discovery and for fundamental studies of signal transduction.
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award no. U54CA199091. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This material is also based upon work supported by the AFOSR awards nos. FA9550-12-1-0141 and FA9550-16-1-0150. This work also made use of the Keck-II facility of the NUANCE Center at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205); the MRSEC program (NSF DMR-1121262) at the Materials Research Center; the International Institute for Nanotechnology (IIN); the Keck Foundation; and the State of Illinois through the IIN. We thank A. Ivankin for useful discussions.
Glossary
ABBREVIATIONS
- SAMDI-MS
self-assembled monolayer laser desorption–ionization mass spectrometry
- TCAL
tandem culture and lysis
- PPL
polymer pen lithography
- ECM
extracellular matrix
- PTP
phosphotyrosine phosphatase
- PBS
phosphate-buffered saline
- THAP
2,4,6-trihydroxyacetophenone
- MHA
mercaptohexadecanoic acid
- PTPI-I
phosphatase inhibitor I
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.6b04176.
Author Contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Inglese J.; Johnson R. L.; Simeonov A.; Xia M. H.; Zheng W.; Austin C. P.; Auld D. S. Nat. Chem. Biol. 2007, 3 (8), 466–479. 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- Mahmoud L.; Al-Saif M.; Amer H. M.; Sheikh M.; Almajhdi F. N.; Khabar K. S. A. J. Virol. 2011, 85 (18), 9268–9275. 10.1128/JVI.00772-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meli L.; Barbosa H. S.; Hickey A. M.; Gasimli L.; Nierode G.; Diogo M. M.; Linhardt R. J.; Cabral J. M.; Dordick J. S. Stem Cell Res. 2014, 13 (1), 36–47. 10.1016/j.scr.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepp O.; Galluzzi L.; Lipinski M.; Yuan J.; Kroemer G. Nat. Rev. Drug Discovery 2011, 10 (3), 221–37. 10.1038/nrd3373. [DOI] [PubMed] [Google Scholar]
- Liang C. C.; Park A. Y.; Guan J. L. Nat. Protoc. 2007, 2 (2), 329–33. 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- Lussier F.; Brule T.; Vishwakarma M.; Das T.; Spatz J. P.; Masson J. F. Nano Lett. 2016, 16 (6), 3866–71. 10.1021/acs.nanolett.6b01371. [DOI] [PubMed] [Google Scholar]
- Tsien R. Y. Annu. Rev. Biochem. 1998, 67, 509–544. 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Hannoush R. N. PLoS One 2008, 3 (10), e3498. 10.1371/journal.pone.0003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman G. R.; Moerke N. J.; Hsia M.; Shamu C. E.; Blenis J. Assay Drug Dev. Technol. 2010, 8 (2), 186–99. 10.1089/adt.2009.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma V. P.; Liu Y.; Yehl K.; Galior K.; Zhang Y.; Salaita K. Angew. Chem., Int. Ed. 2016, 55 (18), 5488–92. 10.1002/anie.201600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo F.; Zheng Z.; Zheng G.; Giam L. R.; Zhang H.; Mirkin C. A. Science 2008, 321 (5896), 1658–60. 10.1126/science.1162193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig A. B.; Huo F.; Mirkin C. A. Nat. Chem. 2009, 1 (5), 353–8. 10.1038/nchem.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X.; Braunschweig A. B.; Zheng Z.; Mirkin C. A. Small 2010, 6 (10), 1082–6. 10.1002/smll.200901538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giam L. R.; Mirkin C. A. Angew. Chem., Int. Ed. 2011, 50 (33), 7482–5. 10.1002/anie.201100839. [DOI] [PubMed] [Google Scholar]
- Eichelsdoerfer D. J.; Liao X.; Cabezas M. D.; Morris W.; Radha B.; Brown K. A.; Giam L. R.; Braunschweig A. B.; Mirkin C. A. Nat. Protoc. 2013, 8 (12), 2548–60. 10.1038/nprot.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrksich M. ACS Nano 2008, 2 (1), 7–18. 10.1021/nn7004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C.; Stewart K. M.; Weaver V. M. J. Cell Sci. 2010, 123 (24), 4195–4200. 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns E. J.; Cabezas M. D.; Mrksich M. Small 2016, 12 (28), 3811–8. 10.1002/smll.201502940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurard-Levin Z. A.; Scholle M. D.; Eisenberg A. H.; Mrksich M. ACS Comb. Sci. 2011, 13 (4), 347–50. 10.1021/co2000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piner R. D.; Zhu J.; Xu F.; Hong S.; Mirkin C. A. Science 1999, 283 (5402), 661–3. 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- Lee K. B.; Lim J. H.; Mirkin C. A. J. Am. Chem. Soc. 2003, 125 (19), 5588–9. 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- Lim J. H.; Ginger D. S.; Lee K. B.; Heo J.; Nam J. M.; Mirkin C. A. Angew. Chem., Int. Ed. 2003, 42 (20), 2309–12. 10.1002/anie.200351256. [DOI] [PubMed] [Google Scholar]
- Ginger D. S.; Zhang H.; Mirkin C. A. Angew. Chem., Int. Ed. 2004, 43 (1), 30–45. 10.1002/anie.200300608. [DOI] [PubMed] [Google Scholar]
- Salaita K.; Wang Y.; Mirkin C. A. Nat. Nanotechnol. 2007, 2 (3), 145–55. 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- Vega R. A.; Shen C. K.; Maspoch D.; Robach J. G.; Lamb R. A.; Mirkin C. A. Small 2007, 3 (9), 1482–5. 10.1002/smll.200700244. [DOI] [PubMed] [Google Scholar]
- Gurard-Levin Z. A.; Mrksich M. Annu. Rev. Anal. Chem. 2008, 1, 767–800. 10.1146/annurev.anchem.1.031207.112903. [DOI] [PubMed] [Google Scholar]
- Giam L. R.; Massich M. D.; Hao L.; Shin Wong L.; Mader C. C.; Mirkin C. A. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (12), 4377–82. 10.1073/pnas.1201086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z.; Shoelson S. E.; Chaudhuri M.; Gish G.; Pawson T.; Haser W. G.; King F.; Roberts T.; Ratnofsky S.; Lechleider R. J.; et al. Cell 1993, 72 (5), 767–78. [DOI] [PubMed] [Google Scholar]
- Songyang Z.; Carraway K. L.; Eck M. J.; Harrison S. C.; Feldman R. A.; Mohammadi M.; Schlessinger J.; Hubbard S. R.; Smith D. P.; Eng C.; Lorenzo M. J.; Ponder B. A. J.; Mayer B. J.; Cantley L. C. Nature 1995, 373 (6514), 536–539. 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- Li S.; Liao X.; Mrksich M. Langmuir 2013, 29 (1), 294–8. 10.1021/la3034066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman B. T.; Gawalt E. S.; Mrksich M. Langmuir 2003, 19 (5), 1522–1531. 10.1021/la0262304. [DOI] [Google Scholar]
- Arabaci G.; Guo X. C.; Beebe K. D.; Coggeshall K. M.; Pei D. J. Am. Chem. Soc. 1999, 121 (21), 5085–5086. 10.1021/ja9906756. [DOI] [Google Scholar]
- Ruoslahti E. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- Cabrera-Pardo J. R.; Chai D. I.; Liu S.; Mrksich M.; Kozmin S. A. Nat. Chem. 2013, 5 (5), 423–7. 10.1038/nchem.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.; Sherrill J.; Mrksich M.; Scholle M. D. J. Biomol. Screening 2015, 20 (7), 842–8. 10.1177/1087057115588512. [DOI] [PubMed] [Google Scholar]
- Mrksich M. Acta Biomater. 2009, 5 (3), 832–41. 10.1016/j.actbio.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban L.; Pettit N.; Li L.; Stuparu A. D.; Cai L.; Chen W.; Guan W.; Han W.; Wang P. G.; Mrksich M. Nat. Chem. Biol. 2012, 8 (9), 769–73. 10.1038/nchembio.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.; Mrksich M. Biochemistry 2004, 43 (50), 15811–21. 10.1021/bi049174+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


