SYNOPSIS
Clostridium difficile infections (CDI) have emerged as one of the principal threats to the health of hospitalized and immunocompromised patients. Nucleic acid testing for C. difficile toxin genes has eclipsed traditional clinical diagnostics for CDI in sensitivity and is now widespread in clinical use, but preliminary evidence suggests that this may have come at a cost of substantially reduced positive predictive value. The importance of C. difficile colonization is increasingly recognized not only as a source for false positive clinical testing but also as a source of new infections within hospitals and other healthcare environments. In the last five years, several new treatment strategies that capitalize on the increasing understanding of the altered microbiome and host defenses in CDI patients have completed clinical trials, including fecal microbiota transplantation (FMT). This article highlights the changing epidemiology, laboratory diagnostics, pathogenesis, and treatment of CDI.
Keywords: Clostridium difficile, CDI, diagnosis, epidemiology, fecal microbiota transplantation, FMT
Overview
Clostridium difficile was identified as the leading cause of antibiotic-associated diarrhea and colitis in 1978, but since 2001 C. difficile infections (CDIs) have evolved from sporadic complications of antimicrobial therapy to severe, sometimes fatal, events that have become an endemic threat to the health of hospitalized and immunosuppressed patients worldwide. This article discusses the changing epidemiology, clinical and laboratory diagnosis, pathogenesis and treatment of CDI.
Microbiology
Clostridium difficile is an obligate anaerobic, spore-forming, Gram-positive rod first described in 1935 as Bacillus difficilis in the fecal flora of healthy infants.1 The organism remained unrecognized as a cause of human infection until 1977 when it was identified as the cause of what had previously been referred to as antibiotic-associated colitis.2, 3 Hall and O’Toole’s species name reflected difficulty of isolating C. difficile from other anaerobic and facultative stool flora, which was mostly attributable to its relatively slow growth (40–70 minutes doubling time).1 C. difficile is exquisitely aero-intolerant during logarithmic growth phases when vegetative cells predominate,4 making it difficult for laboratories not equipped with anaerobic chambers to passage the organism before sporulation occurs at approximately 48–72 hours. Laboratories equipped with anaerobic jar systems are delayed in the ability to isolate the organism because of the need for a minimum 48 hours between passages in order to avoid fatal oxygen intoxication of fresh cultures.
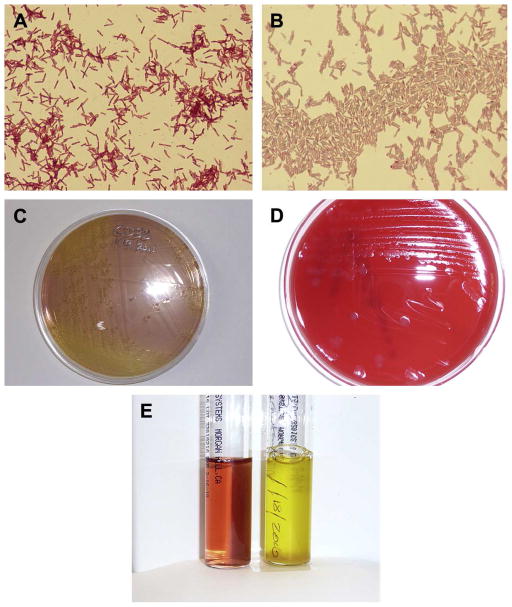
Culture of C. difficile has been unfairly perceived as difficult, however, as selective media have been refined to isolate the organism from fecal and environmental specimens (Fig. 1).5–7 These media capitalize on the organism’s intrinsic resistance to cefoxitin and cycloserine and the ability of C. difficile to utilize fructose and mannitol as carbohydrate sources; Stickland reactions play a central role in the organism’s biosynthetic pathways and account for the organism’s alkalization of undefined peptone-based media which are widely used with neutral red to indicate C. difficile grows in both liquid and agar media (Fig. 1), usually within 48 hours of incubation.8 Sodium taurocholate, a bile salt, has been shown to be vital to the germination of C. difficile endospores and is an essential component in selective media.9, 10
Fig. 1.
(A) Gram stain of C. difficile from 24-hour growth on trypticase soy agar with 5% sheep blood. The vegetative cell bodies are often gram negative during early growth; note the abundant subterminal endospores that do not swell the parent cell. ( B ) Malachite green stain of 48-hour growth of C. difficile. The s afranin counterstain renders vegetative cells pink, whereas the endospores stain green, revealing their ovoid shape. (C) A 48-hour growth of C. difficile on typical selective agar medium, C. difficile basal agar with moxalactam and norfloxacin, CDMN, that uses norfloxacin and moxalactam as selective antibiotics to allow primary isolation from stool specimens. Other media use combinations of cycloserine and cefoxitin as selective antibiotics (cycloserine cefoxitin fructose agar). Neutral red is turned yellow by C. difficile growth. (D) Typical appearance of C. difficile colonies on trypticase soy agar with 5% sheep blood at 48 hours. Colonies are nonhemolytic. (E) A selective broth medium for isolation of C. difficile, cycloserine cefoxitin mannitol broth with taurocholate and lysozyme. The tube at right shows turbidity and growth of C. difficile at 24 hours, with obvious alkalinization of the neutral red indicator. The tube at left is a negative control.
Once isolated, C. difficile can be readily sub-cultured to non-selective media such as 5% sheep blood agar on which it assumes its characteristic irregular ground-glass colonial morphology without hemolysis (Fig. 2). While C. difficile colonies vary greatly in size (more motile strains have maximum colony widths of 12 to 15 mm), they fluoresce under UV illumination and exhibit a characteristic odor often referred to as “horse dung” or “barn” odor. This phenotypic feature results from the ability of the organism to ferment tyrosine into p-cresol, a phenolic compound for which the species has considerable tolerance and which can be used to distinguish the organism from other clostridia using high performance liquid chromatography.11, 12 Smaller colonies of C. difficile can be difficult to distinguish morphologically from those of other non-hemolytic Clostridium spp, but rapid biochemical confirmation with L-proline aminopeptidase spot disk activity can provide confirmation of suspected C. difficile.13
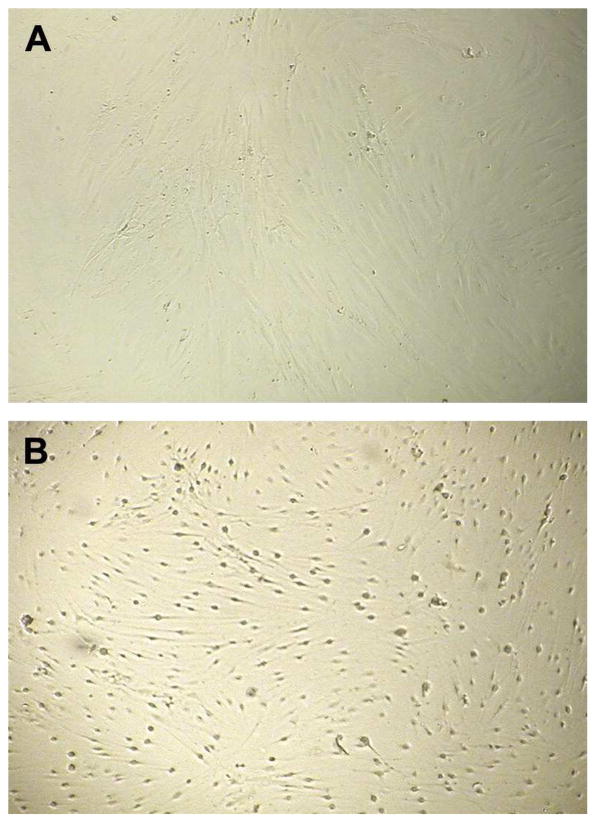
Fig. 2.
(A) Negative cell culture cytotoxicity assay. Human fibroblasts remain spindle-shaped and in contact with each other. (B) Positive cell culture cytotoxicity assay revealing cytopathic effects of C. difficile toxin B causing cell rounding and separation. (Courtesy of Ray Hariri, PhD.)
Chromogenic commercial media have also been developed which allow for more rapid isolation of C. difficile (usually as colorless or grey-black colonies on a blue background within 24 hours), but these media often allow for growth of non-difficile clostridia.14 Matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF) platforms are readily able to distinguish C. difficile from other clostridia in these circumstances.14–16
Culture of C. difficile is primarily a research tool used in the evaluation of diagnostic tests as well as to recover isolates for outbreak investigation and molecular epidemiology. C. difficile isolates can be maintained nearly indefinitely at room temperature as spore stocks or as vegetative cells in chopped meat medium, although many laboratories are in the habit of storing the organism in frozen glycerol stocks.17, 18 Because non-toxigenic strains of C. difficile are commonplace, culture of the organism for clinical purposes must be followed by confirmation of toxin production. This can be done either using molecular tests for the presence of toxin genes or by performing cytotoxicity assays or enzyme immunoassays on cell-free culture supernatants(see Pathogenesis and Diagnosis below). For phenotypic toxin testing, use of complex media without glucose, fructose, and mannitol (present in most of the primarily selective media for the organism) may be important to minimize the effect of rapidly metabolized carbon sources in repression of toxin synthesis.19 The glucosyltransferase activities of toxins A and B in C. difficile have been used to devise selective media that can identify toxigenic strains in a single step using the presence of an insoluble blue product around colonies of toxigenic strains at 48 hours of incubation, but to date this methodology has not undergone a wide-scale validation and may miss some strains expressing toxin at low levels.20
Molecular epidemiology and strain typing
For the purposes of tracking global and local molecular epidemiology of C. difficile, several strain typing systems have been devised (Table 1). For tracking global linages, pulsed field gel electrophoresis (PFGE) is the method generally preferred by the US Centers for Disease Control and Prevention,21, 22 while European centers and others have preferred PCR ribotyping.23 Both methods are band-based and require sets of reference isolates and suffer from a degree of subjectivity, but both methods are able to separate C. difficile lineages that correspond to a large degree with the 380 lineages of C. difficile identified using a more objective and technically demanding method based on Sanger sequencing of C. difficile housekeeping genes, multilocus sequence typing (MLST).24 A single locus typing scheme based on sequencing of the tcdC gene has been devised and can serve to distinguish the lineages of toxigenic strains, but this scheme is not suited for typing of non-toxigenic strains.25, 26 MLST and tcdC typing are both entirely portable, with a public database of sequence types maintained by the University of Oxford (http://pubmlst.org/cdifficile). PFGE, ribotyping, MLST, and tcdC genotyping are insufficiently discriminatory for source attributions or tracking hospital outbreaks, for which restriction endonuclease analysis (REA), multilocus variable number of tandem repeats analysis (MLVA), and whole genome sequencing (WGS) are more suited, each representing progressively increased discriminatory capacity.27–30 There has been considerable enthusiasm for adoption of WGS-based methods, but the high cost and lack of a standardized analysis scheme for WGS data are barriers to the wide-scale adoption of this method.
Table 1.
Laboratory typing methods for CDI
| Method | High-throughput | Portable | Determines Lineage | Discriminatory capacity | Reference |
|---|---|---|---|---|---|
| Toxinotyping | No | Yes | No | Low | 188 |
| Pulsed field gel electrophoresis (PFGE) | No | No | Yes | Low-moderate | 22 |
| Restriction endonuclease analysis (REA) | No | No | Yes | Moderate-high | 27 |
| PCR ribotyping | Yes | No | Yes | Low-moderate | 23 |
| Multilocus sequence typing (MLST) | Yes | Yes | Yes | Low | 24, 189 |
| tcdC genotyping | Yes | Yes | Yes | Low | 25, 26 |
| Multiple locus variable number of tandem repeats analysis (MLVA) | Yes | Yes | No | High | 29, 30 |
| Whole genome sequencing | No | Yes | Yes | High | 28, 190 |
Ecology, host range, and distribution
C. difficile is ubiquitous and widely distributed in nature (Table 2).31 It can be found easily in soil and sewage and has been found in the feces of most mammals.31 C. difficile has occasionally been described as a veterinary pathogen, particularly in piglets, calves, and some avian species,32, 33 but its epidemiology in animals is not nearly as well-described as it is for C. perfringens in animal husbandry. The original animal model for CDI was the Syrian hamster, which suffers a lethal colitis after antibiotic pre-exposure. 34 More recently, mouse models of CDI have been developed which more closely parallel non-lethal CDI in humans, including the development of relapsed infection after recovery.35 To date, however, there is scant evidence that CDI occurs in nature in the absence of antimicrobial use. Despite initial concern that C. difficile is widespread in the food supply; most prevalence studies suggest that the prevalence of C. difficile is low and occurs at very low colony counts (Table 3).36, 37 C. difficile strain typing has suggested that some high initial prevalence estimates of C. difficile in meat products may have arisen from laboratory contamination events, and thus far there is no direct evidence that C. difficile is a foodborne illness.38, 39 In contrast, environmental studies have consistently identified healthcare environments, including outpatient clinics, as heavily contaminated with C. difficile.6, 40, 41
Table 2.
Prevalence of toxigenic C. difficile from sampling of various sources in South Wales.
| Source | N | toxigenic C. difficile (%) |
|---|---|---|
| Domestic animals | 200 | 3 (1.5) |
| Farm animals | 524 | 4 (0.8) |
| Fish | 107 | 0 |
| Soil | 104 | 9 (8.6) |
| Hospitals | 380 | 72 (18.9) |
| Nursing homes | 275 | 4 (1.5) |
| Houses | 350 | 3 (0.9) |
| Dorms | 200 | 3 (1.5) |
| Water* | 110 | 36 (32.7) |
| Vegetables | 300 | 5 (1.7) |
| Total | 2580 | 140 (5.4) |
Adapted from al Saif N, Brazier JS. The distribution of Clostridium difficile in the environment of South Wales. J Med Microbiol 1996;45(2):133–137; with permission.
Table 3.
Prevalence of C. difficile in the human food supply.
| Prevalence (%) | 95 % CI | Region | Product sampled | Reference: |
|---|---|---|---|---|
| 37/88 (42.0) | 31.6 to 53.1 | Tucson, AZ | ground meat | 191 |
| 8/500 (1.6) | 0.69 to 3.1 | Netherlands | retail meats | 192 |
| 5/111 (4.5) | 1.5 to 10.2 | Ontario, CA | Vegetables | 193 |
| 0/46 (0) | 0 to 7.7 | Switzerland | ground beef | 194 |
| 26/203 (12.8) | 8.5 to 18.2 | Ontario, CA | retail chicken | 195 |
| 3/100 (3) | 0.6 to 8.5 | Switzerland | ground meat | 196 |
| 3/50 (6) | 1.3 to 16.6 | Pennsylvania | ground veal | 197 |
| 2/82 (2.4) | 0.3 to 8.5 | Sweden | ground meat | 198 |
| 4/32 (12.5) | 3.5 to 29.0 | Texas | poultry meat | 199 |
| 3/40 (7.5) | 1.6 to 20.4 | Texas | ground meat | 200 |
| 2/102 (2.0) | 2.4 to 6.9 | Pennsylvania | ground meat | 36 |
Disinfection, Survival, and Laboratory Infection
The endospores of C. difficile are resistant to heat, 70% ethanol used in hand sanitizers, and the quaternary ammonium detergents used as hospital and laboratory disinfectants, but sodium hypochlorite-based solutions are capable of inactivating spores.42 C. difficile spores are known to have nearly indefinite viability, demonstrating only 0.5log10 reduction in viability after 14 months of storage on steel disks at room temperature.18 The viability of C. difficile spores contaminating hospital environments also declines over time, but not at a rate that is below the infectious inoculum (ID50 =5 spores/cm2) in the mouse model of CDI.41, 43 Sodium hypochlorite (chlorine bleach) at a concentration of 5000 ppm (achieved with 10% aqueous solution of standard household bleach in distilled, deionized water) at 10 minutes of contact time results in a 6 log10 (≥ 99.9999%) reduction in the viability of C. difficile spores, although lesser concentrations of bleach take up to 30 minutes for the same level of activity.44
Occupancy of a room previously occupied by both known CDI patients and by patients previously receiving antimicrobials have both been identified as significant risks for development of CDI among subsequent hospital room occupants, highlighting the need to provide adequate terminal disinfection to prevent transmission to new patients.45, 46 Previous studies have reported adoption of sodium hypocholorite cleaning of hospital environments as key elements in the control of CDI epidemics in hospitals,47 but more recently less caustic alternative agents such as combinations of hydrogen peroxide and peracetic acid have been developed as terminal disinfectants.48 The development of fully automated ultraviolet irradiation devices and hydrogen peroxide vapor systems for disinfection of hospital rooms holds promise as a means to enhance the effectiveness of hospital disinfection, but these devices do not eliminate the need to perform routine cleaning of the organic soil burden on hospital surfaces and add 30–60 minutes to the process of cleaning a hospital room, adding considerably to the cost of this approach.49, 50 Nonetheless, centers that have adopted these systems have observed significant decreases in healthcare-associated CDI rates.51–53
Laboratory-acquired CDI has been anecdotally reported. Both reported cases involved young females who were working with C. difficile on open benches and using alcohol (which is inactive against C. difficile spores) to disinfect laboratory work surfaces. 54 Only one of the two cases had known antibiotic exposures, although the case in which no antibiotic exposure was noted was self-limited and resolved without treatment. 54 Nonetheless, Bouza et al. recommended that all laboratory work with C. difficile take place in biological safety cabinets using appropriate barrier precautions and appropriate terminal disinfectants active against C. difficile spores.54
Asymptomatic carriage of C. difficile
Healthy, non-hospitalized ambulatory adults are commonly colonized with C. difficile, and the colonization of 5–15% of adults is often transient.55–57 A prospective survey found that 26% of 428 hospitalized patients in a medical ward acquired C. difficile. In this study, only 38% developed symptoms consistent with CDI by 11 months, indicating that 62% of patients who acquired C. difficile were asymptomatically colonized. 58 In a later study by Clabots and colleagues,59 21% of all admissions to a single medical ward at a Veterans Affairs hospital were positive for C. difficile carriage during a 9-month period. Of these, 86% were asymptomatic carriers. The predominant mode of acquisition in this study was hospital-associated in 41% of admissions. Molecular typing of recovered isolates revealed 19 instances of within-hospital transmission among asymptomatic patients. Acquisition of the same strains was documented within the same hospital room separated in time by up to 24 weeks, suggesting that environmental contamination contributes to C. difficile transmission. The study also documented transmission between pairs of patients who were in rooms far separated from each other, suggesting transmission of C. difficile spores on the hands of health care workers to new ward admissions.59 Subsequent mathematical models of C. difficile infection have suggested that transmission within hospitals solely from symptomatic C. difficile patients is inadequate to account for sustained endemic transmission within hospitals; the contribution of asymptomatic carriers was estimated to be particularly significant if transmission from these carriers was common.60
The contribution of asymptomatic carriers to incident C. difficile infections has been further estimated in recent studies. Eyre and colleagues performed a population-based study of 1250 CDI cases occurring 2007–2011 in Oxfordshire, UK. Using whole genome sequencing of C. difficile isolates, they showed that 45% of all incident CDI cases were genetically distinct from all previous cases diagnosed in the region, suggesting that sources beyond symptomatic patients were important in CDI transmission.28 Using MLVA genotyping of incident CDI and a concurrent cohort of individuals colonized with C. difficile at a single medical center in Pittsburgh, Pennsylvania over a 119-day period, Curry and colleagues determined that 17/56 (30%) and 16/56 (29%) of incident CDI cases could be traced to other symptomatic CDI patients and C. difficile carriers, respectively.61 In a 15-month prospective study in six Canadian hospitals, 117/4143 (2.8%) and 123/4143 (3.0%) had healthcare-associated C. difficile infection and colonization, respectively, although direct links between these cohorts was not established by strain typing.62 Longtin and colleagues performed a controlled intervention study to examine the effect of screening incoming admissions to a single Canadian hospital for C. difficile colonization 2013–2015, demonstrating that simple contact isolation measures for the 4.8% of admissions identified as carriers of C. difficile resulted in an observed rate of hospital-acquired CDI of 3.0 infections/10,000 patient-days compared to 6.9 infections/10,000 patient-days in the pre-intervention period.63 Vertical controls based on active surveillance for C. difficile within healthcare facilities hold promise as a means to control CDI.
Clinical presentation and prognosis
As above, asymptomatic carriers are by far the most prevalent group among those individuals who are culture-positive for toxin-producing C. difficile, representing 62% to 86% of hospitalized individuals with C. difficile -positive stools in previous prospective studies.58, 59 For the minority who develop CDI, the infection exists on a continuum from mild diarrhea to fulminant colitis. Few descriptive series of CDI have been published, but in the pre-2001 era, development of semi-formed diarrhea more than 7 days after antimicrobial exposure was the most common presentation among a series of 43 inpatients.64 Leukocytosis is a common feature of CDI, which has been found to be the most common cause for unexplained leukocytosis among inpatients and the fourth most common cause for leukocytosis overall. 65, 66 Fever is seen in only 50% of cases, and bloody diarrhea is seen in a distinct minority of cases.64 Rapid increases in leukocytosis, abdominal distension or pain, and sudden cessation of diarrhea are poor prognostic signs that have been anecdotally reported in cases of fulminant colitis.67
Several severity score indices for CDI have been developed to best predict CDI patients at risk for death, colectomy, or intensive care unit admission attributable to CDI; fever, leukocytosis, hypoalbuminemia, and abdominal distension were independent predictors of severe CDI in one early study.68 Subsequent multicenter cohort studies have suggested that a 3-point scale based on age≥65 years, peak serum creatinine ≥2 mg/dL, and peak peripheral blood leukocyte count≥20,000 cells/μL can be used to predict ~75% of patients at risk for severe CDI.69 A subsequent prospective cohort has resulted in a bedside scoring system based on age, treatment with systemic antibiotics, leukocyte count, albumin, and serum creatinine (ATLAS score 0 to 10) to predict cure among CDI patients,70 and similar prediction rules for recurrence have also been developed.71 The clinical utility of these scores in management of CDI, an infection characterized by occasionally abrupt changes in clinical severity, is not yet established. The incubation period for CDI remains unknown, particularly since the incubation period can be defined either from the time of first exposure to the organism, from the first exposure to antibiotics, or from the first appearance of organism in stools until the onset of symptoms. In studies of healthy volunteers without diarrhea, oral administration of non-toxigenic C. difficile spores results in fecal shedding within 2–4 days,72 but this may not reflect the dynamics of fecal shedding in hospitalized patients. Early studies of hospitalized patients suggested that CDI probably occurred within 7 days of the first appearance of C. difficile in stool,73 and beyond this several inpatient cohort studies suggested that patients who did not manifest symptoms of CDI by 7 days of appearance in stool were at reduced risk of subsequent CDI compared to non-colonized patients in the same cohorts.74 Observational evidence indicates that patients remain at risk for CDI for up to 12 weeks after exposure to antimicrobials, however,75, 76 with the onset of CDI occurring up to 60 days (median 20.3 days) from the time of hospital discharge.77 The discrepancies in these observations regarding the potential incubation period for CDI may reflect that follow-up in the initial hospital cohort studies was limited to hospital discharge, and the cohorts were relatively small and may not have included large numbers of severely immunosuppressed patients at high risk for CDI.
Epidemiology of CDI
Age ≥ 65, exposure to antimicrobials, and length of stay in a health care facility have all been independently associated with risk for CDI.78 In addition, age greater than 65 years has been repeatedly associated with an increased risk of symptomatic infection.79, 80 CDI epidemiology changed dramatically after 2000, with an increase in disease incidence and severity, including fulminant colitis, colectomy, and death described at several large hospitals worldwide.21, 81, 82 In the United States, C. difficile incidence doubled from 1996 to 2003.79 A newly emergent epidemic strain of C. difficile, which was rarely encountered before 2000 which became the most prevalent strain causing 30% to 50% of CDI cases, was associated with this phenomenon.21, 81 The epidemic strain has been named NAP1 by PFGE, 027 by PCR ribotyping, and BI by REA typing, and ST1 by MLST/tcdC genotyping and is usually referred to as NAP1/BI/027.83
The causal association of the emergence of this epidemic lineage of C. difficile and severe CDI is uncertain. In a large population-based surveillance study of 2057 CDI cases with available stain typing data, severe outcomes and death occurred in 4.9 and 2.7% of cases, respectively, and NAP1 strains (28.4% of cases) were associated with severe outcomes and death after adjusting for other confounding risks with adjusted odds ratios of 1.66 and 2.12, respectively.84 A European study noted that ribotype 078 CDI had higher observed mortality than other lineages, including ribotype 027, however.85 Several studies have failed to observe any relationship between epidemic lineage CDI and severe clinical outcome.86–88 Enhanced toxin production originally observed for epidemic lineage stains actually varies widely across strains of ribotype 027, and the significance of lineage with respect to clinical outcome has been cast into considerable question.88–90 An association between epidemic lineage CDI and recurrence, however, has been consistently observed in many study settings.91, 92
In 2005, severe CDI was reported in patients previously assumed to be at a low risk for CDI, including pregnant women and outpatients without known exposures to antimicrobials.93 Population-based studies were subsequently undertaken which showed rates of community disease ranging between 6.9 and 23.4 cases per 100,000 person-years, but these were all based on laboratory surveillance.94–97 These estimates of the incidence of community-acquired C. difficile are far exceeded by incidence rates of hospital-acquired C. difficile, which are approximately 5000 times higher. A subsequent study of the prevalence of CDI among community-dwellers testing positive for CDI in the evaluation of acute diarrheal syndromes showed that 43 of 1091 (3.9 %) such patients tested positive for C. difficile, but only 3 patients (0.3%) remained as possible CDI patients when more recognized causes of outpatient gastroenteritis such as norovirus were excluded.98 CDI remains extremely unlikely as a cause of outpatient gastroenteritis based on available evidence.
Among patients with traditional risk factors for CDI (Table 4), several sub-groups who experience exceptional rates of CDI have emerged, including solid organ transplant patients, bone marrow transplant patients, and patients with inflammatory bowel disease (Crohn’s disease and ulcerative colitis) (Table 5). The rates of CDI observed in these populations can be readily expressed in denominators of 100, reflecting disease incidences 100 to 1000 times those observed in general inpatient populations and long-term care facilities. The unifying clinical feature of these groups is widespread use of broad-spectrum antimicrobials needed to manage conditions such as febrile neutropenia and opportunistic infections, although all of these groups also share significant immunosuppression and exposure to healthcare facilities. Unfortunately, most clinical trials for CDI have traditionally excluded patients with underlying diarrheal disorders such as inflammatory bowel disease, creating an evidence gap in the management of these patients. The impact of CDI on these populations can be significant. In one study of kidney recipients, the attributable mortality of CDI was 0.7%, higher than the observed loss of transplant grafts due to thrombosis and rejection.99 In a study of lung transplant recipients, 54% of CDI occurred within the first 6 months of transplant; and these early CDI patients had a higher risk of death.100 In this cohort, 70% of CDI occurred after discharge from the initial hospitalization, such as in the outpatient setting or during readmissions for complications, rather than immediately post-operatively.100
Table 4.
Clinical risk factors for Clostridium difficile infection (CDI) and recurrent CDI
| Any episode of CDI | Recurrent CDI |
|---|---|
| Age ≥65 | Age 65 |
| Length of inpatient stay | Number of prior CDI episodes |
Antibiotic exposures:
|
Use of non CDI antibiotics after CDI diagnosis |
| Proton pump inhibitors | Fluoroquinolone use |
| Renal insufficiency | |
| Female gender | |
| NAP1/ribotype 027/BI/MLST1/tcdC1 lineage |
Table 5.
Populations at exceptional risk for Clostridium difficile infection.
The public health impact of CDI is substantial. The burden of CDI has been estimated at $2,454 to $6,326 in excess healthcare costs per case, resulting in additional US healthcare expenditures of $4.8 billion per year for acute care facilities alone.101–103 Beyond the economic impact, CD results in an estimated 453,000 cases and 29,000 deaths annually in the United States and has replaced methicillin-resistant Staphylococcus aureus as the most common cause of healthcare-associated in U.S. hospitals.104–106 In 2013, CDI was classified by Centers for Disease Control and Prevention as one of three urgent antibiotic resistance threats in the U.S. that “require urgent public health attention to identify infections and to limit transmission.”107
Pathogenesis
The secretory diarrhea and colonic inflammation seen in CDI is attributable to the effects of 2 large clostridial toxins, toxin A and toxin B, both encoded by tcdA and tcdB respectively as part of the 19.6-kb pathogenicity locus (PaLoc) on the C. difficile chromosome.108–110 Non-toxigenic strains of C. difficile in which the entire PaLoc is replaced with a 115-bp sequence are commonly encountered both in colonized humans and the environment.31, 111 TcdA and TcdB, similar to other large clostridial toxins, glycosylate small guanosine triphosphatases in the Rho and Ras families when endocytosed into epithelial cells, leading to actin filament disassembly, disruption of tight junctions, and ultimately cell death.112 While toxin B is observed to intoxicate a wide variety of mammalian cell lines, candidate receptors responsible for cell surface binding and endocytosis in intestinal epithelial cells have only recently been characterized. Using an insertional mutagenesis technique in Caco-2 cells, LaFrance and colleagues demonstrated that poliovirus receptor-like 3 (PVRL3) is involved in binding of TcdB to its target, disruption of or pre-treatment by antibodies to which creates resistance to toxin B in this cell line.113 Tao and colleagues, however, failed to confirm this finding in HeLa or Caco-2 cells.114 Instead, their CRISPR– Cas9-mediated genome-wide screens identified members of the Wnt-binding frizzled family, particularly FZD1, 2 and 7, as the predominant receptors for TcdB. The finding that TcdB may compete directly with Wnt for FZD binding suggests that colonic epithelial disruption may be a direct consequence of TcdB, as Wnt signaling is particularly important for maintaining colonic stem cells.114
Previous studies using pure TcdB and TcdA pointed to a synergistic role for the 2 toxins in CDI, but the recent ability to create gene knockouts in C. difficile has led to the demonstration that TcdB is necessary and sufficient to cause disease in both the hamster and mouse models.115, 116 This finding is concordant with the clinical observation that TcdA−TcdB+ C. difficile strains are fully capable of causing human disease. 117–119
In wild-type strains of C. difficile, tcdA and tcdB are expressed only in late-logarithmic and stationary growth phases.109, 120 Toxin production is under the immediate control of 2 other genes on the PaLoc, tcdR and tcdC, which serve as positive and negative regulators, respectively. 109, 120–122 BI/NAP1/027 strains have been associated with nonsense mutations in tcdC that lead to a truncated, dysfunctional TcdC and thus TcdB production at all phases of growth, but these tcdC mutations are found in many other non-epidemic lineages.25, 123 There is also in vitro evidence that BI/NAP1/027 strains produce more toxins than wild-type strains, although many other lineages including the type stain VPI 10463 make substantially more toxin than epidemic lineages.121, 122, 124
C. difficile also has potential virulence factors outside the PaLoc, including a binary toxin, encoded by two genes cdtA and cdtB, which is also prevalent among BI/NAP1/027 strains.125–127 Evidence for a role of binary toxin in CDI, however, is limited by the observation that CDT+TcdA−TcdB− strains are incapable of reproducing CDI in the hamster model and that these strains have not yet been associated with human disease.128 Preliminary evidence also points to increased adherence to intestinal mucosa mediated by mutations in slpA, a surface layer protein, in BI/NAP1/027 strains of C. difficile.129
BI/NAP1/027 strains may have an enhanced sporulation capacity compared with wild-type strains, but this finding has not been reproduced in later studies which have noted substantial variation within the epidemic lineage.130, 131 Antibiotic resistance, particularly to clindamycin, macrolides, fluoroquinolones, and rifampin, is also more common in the BI/NAP1/027 strains and leads to potentiation of the spread of the organism within hospitals where such antibiotics are in common use. 21, 81, 132
The clear association of antibiotic use with CDI has focused attention on the relationship of the human microbiome to CDI risk. Using culture-independent 16S rRNA phylogenetic analysis of microbial components of stool flora, it was shown that the stool flora of patients with recurrent episodes of CDI were significantly less diverse than that of patients with initial episodes of CDI whose stool flora was, in turn less diverse than control subjects.133 In a subsequent study, pure culture of a single species of the Lachnospiraceae cultivated from mice that survived C. difficile challenge were able to confer partial colonization resistance in a germfree mouse model, although the mechanism for this colonization resistance was not determined.134
The role of conjugated primary bile salts (taurocholate and glycocholate) in recovering C. difficile in the clinical microbiology laboratory has been known for some time;135 early investigators hypothesized that the effect of antibiotics on the transit of these bile salts to the human large intestine after antibiotic use might be significant in the pathogenesis of CDI.9, 10 The significance of bile salts in both the germination (by primary bile salts) and inhibition of C. difficile outgrowth by chenodeoxycholate has recently been confirmed,136, 137 and subsequent studies in the mouse model of CDI correlated antibiotic challenge in mice with a shift from a predominance of deoxycholate, which inhibits C. difficile outgrowth, to taurocholate, which is essential to germination, in the murine cecum.138 Microbiome analysis of patients colonized with C. difficile has suggested that Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium, is associated with resistance to CDI through its production of secondary bile acids,139 a finding that awaits confirmation in formal trials of this candidate probiotic organism.
Laboratory Diagnosis
The laboratory diagnosis of CDI has undergone considerable upheaval since the first characterization of CDI. Cell culture cytotoxicity neutralization assays (CCCNAs) (see Fig. 2) were the only available clinical tests for CDI following the discovery of CDI; these tests recapitulate the process used to discover the disease. For CCCNAs, patient stool is filtered and incubated with human fibroblast cells with and without C. difficile (originally C. sordellii) antitoxin for up to 72 hours, and if the antitoxin-free well shows a cytopathic effect and the antitoxin well does not, the presence of C. difficile toxin in stool is confirmed (Fig. 2). CCCNA is technically demanding, takes 2–4 days to turn around a negative result, and is generally only performed by large clinical laboratories with the capacity to maintain cell cultures. Compared with identification of toxin-producing C. difficile using anaerobic culture (termed toxigenic culture, see Microbiology), CCCNA was observed to be 67% to 78% sensitive,140, 141 but the extent to which either CCCNA or toxigenic culture could be considered a reference standard was never established. Both CCCNA and toxigenic culture identify patients with colonization and are therefore susceptible to false positives for the endpoint of CDI. Eventually, CCCNA came to be regarded as a clinical gold standard in its own right to which more rapid immunoassays (EIA) were compared as they were introduced into clinical use in the 1980s.
Cell culture cytotoxicity assays were replaced by most clinical microbiology laboratories in North America and Europe by EIAs for TcdA or TcdA+TcdB because of their ease of use and rapid turnaround time of approximately 2 hours. Compared to CCCNA as a reference standard, EIA has a comparatively low sensitivity of 80% to 90%,141–143 and it is even less sensitive when compared with toxigenic culture. Clinicians frequently sought to overcome the perceived sensitivity issue of immunoassays for CDI through use of repeated testing. Submission of multiple samples from the same patient within days became a common strategy for both CCCNA and immunoassays, but these were observed to have a marginal return.144–147 Various 2-step testing algorithms incorporating a rapid antigen test for C. difficile glutamate dehydrogenase (GDH) activity followed by reflex testing of positives to confirm toxin production by EIA or cell culture cytotoxicity were also devised,148–150 but sensitivity of GDH screening varied by C. difficile lineage, casting some doubt on this strategy.151, 152
The introduction of nucleic acid testing for C. difficile into clinical practice in 2008 (Table 6) initially held significant promise for improving the sensitivity of testing relative to EIA platforms and substantially reducing the turn-around time and technical complexity of CCCNA. The latest generation of molecular tests has emphasized the detection of tcdB, although platforms that are designed to detect only tcdA are still able to detect toxinA-/B+ strains because of a conserved fragment of tcdA present in these lineages. Almost all currently approved platforms incorporate sample lysis, DNA extraction, target amplification, and interpretation of internal controls into a single sample vessel or cartridge, substantially reducing the technical complexity of the tests compared to earlier generations of real-time PCR instruments. Assays based on toxin gene detection exclude false-positive results from detection of nontoxigenic strains of C. difficile. The sensitivity of GDH antigen, PCR, and toxigenic culture were all recently shown to be decreased substantially by empiric therapy, with 45% of patients converting initially positive tests to negative within three days of therapy in one cohort of CDI patients.153 The impact of empiric therapy has been poorly accounted for in most prior diagnostic test evaluations for CDI.
Table 6.
Selected FDA-approved clinical diagnostic nucleic acid testing platforms for diagnosis of CDI
| Device | Manufacturer | Approval | Platform | Gene target(s) |
|---|---|---|---|---|
| Cobas | Roche | 5/2015 | RT-PCR | tcdB |
| BD MAX | BD Diagnostics | 4/2013 | RT-PCR | tcdB |
| Verigene | Nanosphere | 12/2012 | PCR + nanoparticle hybridization | tcdA/tcdB/tcdC/cdt |
| Simplexa | Focus | 4/2012 | RT-PCR | tcdB |
| Illumigene | Meridian | 7/2010 | LAMP | tcdA |
| Xpert | Cepheid | 7/2009 | RT-PCR | tcdB/tcdC |
| Progastro CD | Prodesse | 4/2009 | RT-PCR | tcdB |
| BD Geneohm | BD Diagnostics | 12/2008 | RT-PCR | tcdB |
Recent studies of C. difficile testing performance have focused on strategies to incorporate clinical outcome or symptoms into test performance given increasing concerns about the lack of specificity of laboratory diagnostics for CDI. Dubberke and colleagues performed a prospective study of 150 patients being tested for CDI, including prospective evaluation of tested patients’ symptoms using blinded physician interviews as part of a reference standard for evaluating 2 different EIAs, 3 nucleic acid amplificiation tests, CCCNA, and toxigenic culture.154 Among the 150 patients, 44 were positive by toxigenic culture, but only 35 of these had clinically significant diarrhea.154 Using a composite reference standard of ≥4 assays positive, 50/150 patients were positive, but only 40 of these patients had clinically significant diarrhea. As expected from prior studies, CCCNA had the lowest observed sensitivity regardless of reference standard used (55.0–62.9%) while the nucleic acid tests had very high sensitivity (100%). Somewhat unexpectedly, the sensitivity of varying EIAs was actually higher than CCCNA, and the inclusion of clinical symptoms into the reference standard increased the specificity and positive predictive value of all tested assays. The authors observed that 15 patients whose diagnosis of CDI was rejected by EIA but positive by the composite reference standard (mostly by nucleic acid tests) had no diagnosis of CDI within 60 days despite 10/15 patients having clinically significant diarrhea and 4/15 patients receiving empiric therapy for CDI, suggesting that less sensitive tests like EIA may actually predict typical CDI features more reliably.154 Planche and colleagues conducted a much larger multicenter prospective observational study of 12,420 fecal samples from 6,522 inpatient episodes of diarrhea.155 Mortality was observed in 72/435 (16.6%) patients with positive CCCNAs compared to 20/207 (9.7%) positive by toxigenic culture with negative CCCNAs and 503/5880 (8.6%) patients negative by both methods.155 The authors also observed that patients with negative CCCNA but positive nucleic acid tests did not differ in their observed mortality, length of stay, or peak leukocyte count from patients negative on both assays; given that CDI is not 100% fatal this observation is not clear-cut evidence that CCCNA remains the most relevant clinical standard for diagnosis, nor could algorithm combining EIA, GDH antigen, and nucleic acid testing fully replicate the sensitivity and specificity of CCCNA itself in their cohort.155 Finally, Polage and colleagues conducted a prospective cohort study at a single center of 1416 hospitalized adult patients tested for CDI using EIA and nucleic acid testing (PCR) during which the PCR results were withheld from ordering physicians.156 In this study 293/1416 (21%) were positive by PCR while 131/1416 (9.3%) were EIA-positive.156 The authors observed that the EIA-/PCR+ patients had shorter duration of diarrhea than in EIA+/PCR+ patients despite minimal empiric treatment in the former group, and no CDI complications (attributable death, colectomy, ICU stay) were observed vs 10 complications in patients with concordant tests.156 Recurrent positive C. difficile testing was observed in only 5 (3.1%) EIA-/PCR+ patients vs 14 (10.7%) of EIA+/PCR+ patients; deaths due to recurrent CDI occurred in both groups, but at significantly lower rates in the EIA-/PCR+ group.156 The study did not include prospective use of CCCNA or toxigenic culture as a reference standard, however, which may have misclassified patients in the EIA-/PCR− group with mild CDI as control group patients, but overall this study adds to a body of evidence that nucleic acid tests may substantially over-diagnose CDI. The available evidence does not yet support that a return to EIA as the principal diagnostic test for CDI would not result in significant loss of sensitivity for diagnosis of patients with mild CDI, however. The need for laboratory correlates of colonization versus infection remains acute, and CDI will remain partly a clinical diagnosis until additional diagnostics are developed (see Differential Diagnosis).
Treatment, prognosis, and long-term outcome
Prior to recent shifts in observed incidence and severity after 2001, CDI was often regarded as a self-limited disease so long as the inciting antimicrobial agents could be stopped. Early therapeutic trials sometimes contained placebo arms, and head-to-head comparisons of the 2 main C. difficile antimicrobials, enteral vancomycin and metronidazole, failed to show superiority for either.157 Metronidazole was generally recommended by expert guidelines as first-line therapy for reasons of cost and out of concern that widespread use of vancomycin would promote acquisition of vancomycin-resistant enterococci.158 A randomized, double-blind trial of metronidazole versus vancomycin showed superior response rates for vancomycin in a subset of patients with severe disease, however.159( Metronidazole was also found to be inferior to metronidazole?) for the endpoint of clinical success (defined as cessation of diarrhea for ≥ 2 days) in all CDI patients in a multisite blinded, randomized trial with 72.7 vs 81.1% success rates observed, respectively.160 Fidaxomicin, a new macrocyclic antimicrobial, was observed to be non-inferior to vancomycin in three randomized trials, but it did not demonstrate superiority for the endpoint of clinical success; while much has been made of the observation that fidaxomicin has a ~10% reduced risk for recurrence compared to vancomycin, this was not consistent across studies and not tested in patients with multiple CDI recurrences, who were excluded from these trials.161–163 Current clinical guidelines stress the use of vancomycin monotherapy for patients observed to be at risk for severe disease based on varying clinical criteria, although metronidazole use in patients with milder CDI is still recommended.164, 165
The debates regarding which anti– C. difficile antimicrobial is preferred in CDI management have been largely eclipsed by the observation of high rates of success for management of CDI, particularly multiply recurrent cases, with fecal microbiota transplantation (FMT).166–171 In contrast to defined probiotic formulations, high-quality evidence has emerged showing that FMT is superior to vancomycin in management of recurrent CDI (Table 7). The regulatory status of FMT with the Food and Drug Administration remains uncertain, however, and the risk of short and long-term donor-derived infection, particularly in immunosuppressed CDI patients, remains a concern for this treatment modality. The optimal dose, route of delivery, donor source for FMT have yet to be determined in well-controlled randomized trials, as is the use of FMT for first episodes of CDI. The risks of non-disclosure of sexual risk-taking and self-reported diarrheal illness in paid FMT donors as well as the use of small numbers of FMT donors for large numbers of recipients for the sake of improved patient access to this treatment modality may prove to be short-sighted in assessing the long-term risks of FMT in transmission of undiscovered pathogens. Nonetheless, FMT has emerged as the most effective single therapy for management of patients with recurrent CDI.
Table 7.
Key trials of fecal microbiota transplantation (FMT) for treatment of recurrent CDI.
| N= | Design | Comparator | Dose (estimated stool mass) | Route | Followup | Observed successes (per protocol) | Comments | Reference |
|---|---|---|---|---|---|---|---|---|
| 42 | Randomized open label | Vancomycin +/− laxative | 500 ml (~50g) | Nasoduodenal tube | 10 weeks | 13/16 (81%) single dose 15/16 (94%) overall 7/26 (27%) patients in comparator arms |
3 patients required 2nd dose | 169 |
| 20 | Open label | None | 48g | Oral capsules (n=30) | 8 weeks | 14/20 single dosing 19/20 2nd dose |
15 capsules × 2 days=“1 dose” | 170 |
| 20 | Randomized nastogastric tube vs. colonoscopy | None | 90 ml (41g) | Nasogastric tube Colonoscopy |
8 weeks | 8/10 colonoscopic 6/10 nasogastric tube |
All unrelated donors Non-significant difference in success |
171 |
| 80 | Retrospective multicenter case series | None | varied | Colonoscopy | 12 weeks | 62/80 (78%) single dose 70/80 (89%) multiple doses |
First series to report inclusion of immunocompromised (n=19 organ transplants, 3 HIV+) | 167 |
| 43 | Case series, directed fresh donors vs. standardized frozen donors | None | 250 ml (50g) | Colonoscopy | 8 weeks | 7/10 directed 30/33 stoolbank p=0.12 |
30% inflammatory bowel disease Unrelated donors from stool bank | 166 |
| 46 | Randomized controlled double-blind | Autologous stool (recipient’s) | 300 ml (64g) | Colonoscopy | 8 weeks | 22/22 (90.9%) donor FMT 15/24 (62.5%) autologous FMT |
IBD and age ≥75 patients excluded 90% success for autologous stool reported at a single center | 168 |
| 219 | Randomized, double-blind frozen vs fresh FMT dose | None | 50 ml (12.5g) | Retention enema | 13 weeks | 76/91 (83.5%) frozen FMT 74/87 (85.1%) fresh FMT |
Single dose response rates much lower (62–63%) | 204 |
The prognosis for most patients with CDI remains favorable, but adverse event rates for CDI (colectomy/death) reached as high as 44 of 253 (17.3%) for inpatients with hospital-acquired CDI at the University of Pittsburgh during a 2-year period.82 A 30-day attributable mortality of 6.9% was observed in 20 hospitals in Quebec subsequently.81 For patients who require colectomy, all-cause mortality is 50%.67 Recently surgical interventions have been devised that allow for delivery of vancomycin to the cecum of patients with severe, fulminant CDI using a loop ileostomy technique; this has a substantially improved observed success rates compared to historical controls who underwent colectomy. 172
Differential diagnosis
The high prevalence of C. difficile colonization among inpatients and more modest colonization among outpatients often obscures a number of infectious and non-infectious causes of diarrhea and gastroenteritis when a positive C. difficile diagnostic test is generated, resulting in substantial diagnostic test bias. Among outpatients presenting with acute gastroenteritis, particularly with vomiting, norovirus remains the most prevalent etiology. Most new molecular diagnostic platforms that are replacing stool cultures for routine enteric pathogens in clinical microbiology laboratories are incorporating tests for not only C. difficile but also norovirus, astrovirus, sapovirus, and others. The vast majority of patients without traditional risk factors for CDI who test positive for both a viral etiology and C. difficile are probably colonized with the latter, and treatment for C. difficile in these patients should be avoided. Treatment with vancomycin has been observed to prolong carriage with toxigenic C. difficile and, in one instance, to actually provoke CDI in a patient with previously asymptomatic carriage.173
Among inpatients who present with diarrhea >3 calendar days after admission, CDI is widely cited as the most prevalent infectious cause. With the increasing focus on early and aggressive diagnosis of CDI, however, it is often overlooked that non-infectious causes of diarrhea predominate, particularly post-antibiotic diarrhea not associated with C. difficile. Osmotic diarrhea from enteral feeding formulations, malabsorption related to ischemic colitis, and inflammatory diarrhea from ulcerative colitis and Crohn’s disease should always be considered in the differential diagnosis for diarrheal illnesses presenting in hospitalized patients. Even laxative use has been observed in 20% of hospitalized patients being tested for CDI, however,154 and the rate of diagnoses other than CDI was >90% in the largest diagnostic testing cohort yet reported,155 indicating that CDI remains an uncommon finding among hospitalized patients with diarrhea. Patients with any of these causes of diarrhea are frequently colonized with C. difficile and are found on diagnostic testing, and to date there is no laboratory assay that can distinguish colonization from CDI. Fecal leukocytes, fecal calprotectin levels, and C-reactive protein have all been proposed as biomarkers to make this distinction, for example, but none of these can distinguish CDI from inflammatory bowel disease flares.174, 175 Thus, the diagnosis of CDI remains partly a clinical one. The absence of known risk factors, intact serum albumin levels, normal peripheral leukocyte counts, and a failure to exhibit any improvement after 10 days of antimicrobial therapy for CDI should raise suspicion that a patient with a positive C. difficile stool test does not have CDI.
In the era of FMT, making the distinction between colonization and CDI is particularly important; patients with alternate causes of diarrhea are highly unlikely to benefit from FMT based on available data. In one outpatient study of 117 patients referred 2013–2014 to an FMT clinic, 29 patients (25%) were determined to have an alternative diagnosis, most frequently irritable bowel syndrome(IBS).176 Because many patients with a history of multiple positive C. difficile tests may have had inappropriate tests of cure while experiencing IBS after recovery from CDI, careful history-taking must distinguish patients who exhibit a pattern of worsening symptoms days to weeks after cessation of CDI antibiotics from those who have “refractory CDI” regardless of their treatment status. While common in clinical parlance, “chronic,” “treatment-refractory,” and “treatment-resistant” CDI is unlikely to exist based on available evidence, and the preponderance of the of patients in CDI clinical trials who “fail” vancomycin and fidaxomicin (~10% in most recent trials) are probably among the patients who did not have CDI given the low observed mortality among these patients.
Immunity and reinfection
The most troubling aspect of CDI is the high relapse rate of 10–35% after a first episode, ~40% after a first recurrence, and 60–100% following 2 or more recurrences.177 Patients with multiple CDI have been strongly associated with a failure to demonstrate an anamnestic IgG response against TcdA.178–182 Besides age and immune senescence, continued exposure to antibiotics is a powerful predictor of CDI relapse.183 Efforts to devise a C. difficile toxoid vaccine have met with initial success in individuals with multiple relapses,184 but to date there are no published trials using vaccines for primary prevention of CDI. The debilitating effects of multiple CDI relapses were the principal drivers of the development of FMT, which has emerged as the only reliable treatment modality to date. A randomized, placebo-controlled trial of passive immunotherapy for recurrent CDI using monoclonal antibodies to TcdA and TcdB showed an 18% reduction in the absolute risk of recurrence.185 Two phase 3 evaluations of these antibodies revealed that the antibody to TcdB (bezlotoxumab) was the active component, reducing recurrence rates by 10% compared to placebo.186, 187 Bezlotoxumab is available only parenterally, must be used with standard antimicrobial therapies for CDI, and is likely to be substantially more costly than (and likely less effective than) FMT in managing recurrent CDI, however, and its role in clinical care of CDI patients has yet to be established.
Summary
CDIs have emerged as one of the principal threats to the health of hospitalized and immunocompromised patients. Nucleic acid testing for C. difficile toxin genes has eclipsed traditional clinical diagnostics for CDI in sensitivity and is now widespread in clinical use, but preliminary evidence suggests that this may have come at a cost of substantially reduced positive predictive value. The importance of C. difficile colonization is increasingly recognized not only as a source for false positive clinical testing but also as a source of new infections within hospitals and other healthcare environments. In the last five years, fecal microbiota transplantation has emerged as the most effective treatment for patients with multiply recurrent CDI. The increasing understanding of the microbiome and colonization resistance as one of the host defenses against CDI will likely result in improved therapeutics for CDI in the next decade.
KEY POINTS.
Clostridium difficile is widely distributed in nature but concentrated within healthcare environments
Asymptomatic carriage of humans with C. difficile is common, posing a significant challenge for the laboratory diagnosis of C. difficile infection (CDI)
CDI poses a substantial threat to the health of hospitalized patients and other sub-groups of immunocompromised patients.
The collateral protective effect of an individual’s microbiome may be as important as host immune defenses for CDI.
Fecal microbiota transplantation (FMT) is emerging as the most effective treatment for patients with recurrent CDI.
Footnotes
DISCLOSURE STATEMENT
The Author has nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hall I, O’Toole E. Intestinal flora in newborn infants with a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Dis Child. 1935;49:390–402. [Google Scholar]
- 2.Bartlett JG, Chang TW, Gurwith M, et al. Antibiotic-associated pseudo-membranous colitis due to toxin-producing clostridia. N Engl J Med. 1978;298(10):531–534. doi: 10.1056/NEJM197803092981003. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett JG, Onderdonk AB, Cisneros RL, et al. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J Infect Dis. 1977;136(5):701–705. doi: 10.1093/infdis/136.5.701. [DOI] [PubMed] [Google Scholar]
- 4.Jump RL, Pultz MJ, Donskey CJ. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrob Agents Chemother. 2007;51(8):2883–2887. doi: 10.1128/AAC.01443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arroyo LG, Rousseau J, Willey BM, et al. Use of a selective enrichment broth to recover Clostridium difficile from stool swabs stored under different conditions. J Clin Microbiol. 2005;43(10):5341–5343. doi: 10.1128/JCM.43.10.5341-5343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riggs MM, Sethi AK, Zabarsky TF, et al. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis. 2007;45(8):992–998. doi: 10.1086/521854. [DOI] [PubMed] [Google Scholar]
- 7.Hink T, Burnham CA, Dubberke ER. A systematic evaluation of methods to optimize culture-based recovery of Clostridium difficile from stool specimens. Anaerobe. 2013;19:39–43. doi: 10.1016/j.anaerobe.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson S, Calos M, Myers A, et al. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J Bacteriol. 2006;188(24):8487–8495. doi: 10.1128/JB.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson KH. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol. 1983;18(4):1017–1019. doi: 10.1128/jcm.18.4.1017-1019.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol. 1982;15(3):443–446. doi: 10.1128/jcm.15.3.443-446.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson LF, Stabler RA, Wren BW. Assessing the role of p-cresol tolerance in Clostridium difficile. J Med Microbiol. 2008;57(Pt 6):745–749. doi: 10.1099/jmm.0.47744-0. [DOI] [PubMed] [Google Scholar]
- 12.Sivsammye G, Sims HV. Presumptive identification of Clostridium difficile by detection of p-cresol in prepared peptone yeast glucose broth supplemented with p-hydroxyphenylacetic acid. J Clin Microbiol. 1990;28(8):1851–1853. doi: 10.1128/jcm.28.8.1851-1853.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedorko DP, Williams EC. Use of cycloserine-cefoxitin-fructose agar and L-proline-aminopeptidase (PRO Discs) in the rapid identification of Clostridium difficile. J Clin Microbiol. 1997;35(5):1258–1259. doi: 10.1128/jcm.35.5.1258-1259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JH, Cheng VC, Wong OY, et al. The importance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for correct identification of Clostridium difficile isolated from chromID C. difficile chromogenic agar. J Microbiol Immunol Infect. 2016 doi: 10.1016/j.jmii.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Coltella L, Mancinelli L, Onori M, et al. Advancement in the routine identification of anaerobic bacteria by MALDI-TOF mass spectrometry. Eur J Clin Microbiol Infect Dis. 2013;32(9):1183–1192. doi: 10.1007/s10096-013-1865-1. [DOI] [PubMed] [Google Scholar]
- 16.Kim YJ, Kim SH, Park HJ, et al. MALDI-TOF MS is more accurate than VITEK II ANC card and API Rapid ID 32 A system for the identification of Clostridium species. Anaerobe. 2016;40:73–75. doi: 10.1016/j.anaerobe.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Edwards AN, McBride SM. Isolating and Purifying Clostridium difficile Spores. Methods Mol Biol. 2016;1476:117–128. doi: 10.1007/978-1-4939-6361-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez J, Springthorpe VS, Sattar SA. Clospore: a liquid medium for producing high titers of semi-purified spores of Clostridium difficile. J AOAC Int. 2011;94(2):618–626. [PubMed] [Google Scholar]
- 19.Bouillaut L, Dubois T, Sonenshein AL, et al. Integration of metabolism and virulence in Clostridium difficile. Res Microbiol. 2015;166(4):375–383. doi: 10.1016/j.resmic.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darkoh C, Dupont HL, Kaplan HB. Novel one-step method for detection and isolation of active-toxin- producing Clostridium difficile strains directly from stool samples. J Clin Microbiol. 2011;49(12):4219–4224. doi: 10.1128/JCM.01033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353(23):2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 22.Corkill JE, Graham R, Hart CA, et al. Pulsed-field gel electrophoresis of degradation-sensitive DNAs from Clostridium difficile PCR ribotype 1 strains. J Clin Microbiol. 2000;38(7):2791–2792. doi: 10.1128/jcm.38.7.2791-2792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stubbs SL, Brazier JS, O’Neill GL, et al. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol. 1999;37(2):461–463. doi: 10.1128/jcm.37.2.461-463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths D, Fawley W, Kachrimanidou M, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48(3):770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curry SR, Marsh JW, Muto CA, et al. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol. 2007;45(1):215–221. doi: 10.1128/JCM.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dingle KE, Griffiths D, Didelot X, et al. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One. 2011;6(5):e19993. doi: 10.1371/journal.pone.0019993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clabots CR, Johnson S, Bettin KM, et al. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J Clin Microbiol. 1993;31(7):1870–1875. doi: 10.1128/jcm.31.7.1870-1875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eyre DW, Cule ML, Wilson DJ, et al. Diverse sources of C difficile infection identified on whole-genome sequencing. N Engl J Med. 2013;369(13):1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsh JW, O’Leary MM, Shutt KA, et al. Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in Hospitals. J Clin Microbiol. 2006;44(7):2558–2566. doi: 10.1128/JCM.02364-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Berg RJ, Schaap I, Templeton KE, et al. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J Clin Microbiol. 2007;45(3):1024–1028. doi: 10.1128/JCM.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.al Saif N, Brazier JS. The distribution of Clostridium difficile in the environment of South Wales. J Med Microbiol. 1996;45(2):133–137. doi: 10.1099/00222615-45-2-133. [DOI] [PubMed] [Google Scholar]
- 32.Moono P, Foster NF, Hampson DJ, et al. Clostridium difficile Infection in Production Animals and Avian Species: A Review. Foodborne Pathog Dis. 2016 doi: 10.1089/fpd.2016.2181. [DOI] [PubMed] [Google Scholar]
- 33.Hammitt MC, Bueschel DM, Keel MK, et al. A possible role for Clostridium difficile in the etiology of calf enteritis. Vet Microbiol. 2008;127(3–4):343–352. doi: 10.1016/j.vetmic.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larson HE, Price AB, Honour P, et al. Clostridium difficile and the aetiology of pseudomembranous colitis. Lancet. 1978;1(8073):1063–1066. doi: 10.1016/s0140-6736(78)90912-1. [DOI] [PubMed] [Google Scholar]
- 35.Sun X, Wang H, Zhang Y, et al. Mouse relapse model of Clostridium difficile infection. Infect Immun. 2011;79(7):2856–2864. doi: 10.1128/IAI.01336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curry SR, Marsh JW, Schlackman JL, et al. Prevalence of Clostridium difficile in uncooked ground meat products from Pittsburgh, Pennsylvania. Appl Environ Microbiol. 2012;78(12):4183–4186. doi: 10.1128/AEM.00842-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weese JS, Avery BP, Rousseau J, et al. Detection and enumeration of Clostridium difficile spores in retail beef and pork. Appl Environ Microbiol. 2009;75(15):5009–5011. doi: 10.1128/AEM.00480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marsh JW. Counterpoint: Is Clostridium difficile a food-borne disease? Anaerobe. 2013;21:62–63. doi: 10.1016/j.anaerobe.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Marsh JW, Tulenko MM, Shutt KA, et al. Multi-locus variable number tandem repeat analysis for investigation of the genetic association of Clostridium difficile isolates from food, food animals and humans. Anaerobe. 2011;17(4):156–160. doi: 10.1016/j.anaerobe.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Jury LA, Sitzlar B, Kundrapu S, et al. Outpatient healthcare settings and transmission of Clostridium difficile. PLoS One. 2013;8(7):e70175. doi: 10.1371/journal.pone.0070175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim KH, Fekety R, Batts DH, et al. Isolation of Clostridium difficile from the environment and contacts of patients with antibiotic-associated colitis. J Infect Dis. 1981;143(1):42–50. doi: 10.1093/infdis/143.1.42. [DOI] [PubMed] [Google Scholar]
- 42.Fawley WN, Underwood S, Freeman J, et al. Efficacy of hospital cleaning agents and germicides against epidemic Clostridium difficile strains. Infect Control Hosp Epidemiol. 2007;28(8):920–925. doi: 10.1086/519201. [DOI] [PubMed] [Google Scholar]
- 43.Lawley TD, Clare S, Deakin LJ, et al. Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl Environ Microbiol. 2010;76(20):6895–6900. doi: 10.1128/AEM.00718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez J, Springthorpe VS, Sattar SA. Activity of selected oxidizing microbicides against the spores of Clostridium difficile: relevance to environmental control. Am J Infect Control. 2005;33(6):320–325. doi: 10.1016/j.ajic.2005.04.240. [DOI] [PubMed] [Google Scholar]
- 45.Shaughnessy MK, Micielli RL, DePestel DD, et al. Evaluation of hospital room assignment and acquisition of Clostridium difficile infection. Infect Control Hosp Epidemiol. 2011;32(3):201–206. doi: 10.1086/658669. [DOI] [PubMed] [Google Scholar]
- 46.Freedberg DE, Salmasian H, Cohen B, et al. Receipt of Antibiotics in Hospitalized Patients and Risk for Clostridium difficile Infection in Subsequent Patients Who Occupy the Same Bed. JAMA Intern Med. 2016;176(12):1801–1808. doi: 10.1001/jamainternmed.2016.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muto CA, Blank MK, Marsh JW, et al. Control of an outbreak of infection with the hypervirulent Clostridium difficile BI strain in a university hospital using a comprehensive “bundle” approach. Clin Infect Dis. 2007;45(10):1266–1273. doi: 10.1086/522654. [DOI] [PubMed] [Google Scholar]
- 48.Deshpande A, Mana TS, Cadnum JL, et al. Evaluation of a sporicidal peracetic acid/hydrogen peroxide-based daily disinfectant cleaner. Infect Control Hosp Epidemiol. 2014;35(11):1414–1416. doi: 10.1086/678416. [DOI] [PubMed] [Google Scholar]
- 49.Nerandzic MM, Cadnum JL, Pultz MJ, et al. Evaluation of an automated ultraviolet radiation device for decontamination of Clostridium difficile and other healthcare-associated pathogens in hospital rooms. BMC Infect Dis. 2010;10:197. doi: 10.1186/1471-2334-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davies A, Pottage T, Bennett A, et al. Gaseous and air decontamination technologies for Clostridium difficile in the healthcare environment. J Hosp Infect. 2011;77(3):199–203. doi: 10.1016/j.jhin.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 51.Levin J, Riley LS, Parrish C, et al. The effect of portable pulsed xenon ultraviolet light after terminal cleaning on hospital-associated Clostridium difficile infection in a community hospital. Am J Infect Control. 2013;41(8):746–748. doi: 10.1016/j.ajic.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 52.Miller R, Simmons S, Dale C, et al. Utilization and impact of a pulsed-xenon ultraviolet room disinfection system and multidisciplinary care team on Clostridium difficile in a long-term acute care facility. Am J Infect Control. 2015;43(12):1350–1353. doi: 10.1016/j.ajic.2015.07.029. [DOI] [PubMed] [Google Scholar]
- 53.Boyce JM, Havill NL, Otter JA, et al. Impact of hydrogen peroxide vapor room decontamination on Clostridium difficile environmental contamination and transmission in a healthcare setting. Infect Control Hosp Epidemiol. 2008;29(8):723–729. doi: 10.1086/589906. [DOI] [PubMed] [Google Scholar]
- 54.Bouza E, Martin A, Van den Berg RJ, et al. Laboratory-acquired Clostridium difficile polymerase chain reaction ribotype 027: a new risk for laboratory workers? Clin Infect Dis. 2008;47(11):1493–1494. doi: 10.1086/593109. [DOI] [PubMed] [Google Scholar]
- 55.Nakamura S, Mikawa M, Nakashio S, et al. Isolation of Clostridium difficile from the feces and the antibody in sera of young and elderly adults. Microbiol Immunol. 1981;25(4):345–351. doi: 10.1111/j.1348-0421.1981.tb00036.x. [DOI] [PubMed] [Google Scholar]
- 56.Ozaki E, Kato H, Kita H, et al. Clostridium difficile colonization in healthy adults: transient colonization and correlation with enterococcal colonization. J Med Microbiol. 2004;53(Pt 2):167–172. doi: 10.1099/jmm.0.05376-0. [DOI] [PubMed] [Google Scholar]
- 57.Galdys AL, Nelson JS, Shutt KA, et al. Prevalence and duration of asymptomatic Clostridium difficile carriage among healthy subjects in Pittsburgh, Pennsylvania. J Clin Microbiol. 2014;52(7):2406–2409. doi: 10.1128/JCM.00222-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McFarland LV, Mulligan ME, Kwok RY, et al. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989;320(4):204–210. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 59.Clabots CR, Johnson S, Olson MM, et al. Acquisition of Clostridium difficile by hospitalized patients: evidence for colonized new admissions as a source of infection. J Infect Dis. 1992;166(3):561–567. doi: 10.1093/infdis/166.3.561. [DOI] [PubMed] [Google Scholar]
- 60.Lanzas C, Dubberke ER, Lu Z, et al. Epidemiological model for Clostridium difficile transmission in healthcare settings. Infect Control Hosp Epidemiol. 2011;32(6):553–561. doi: 10.1086/660013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Curry SR, Muto CA, Schlackman JL, et al. Use of multilocus variable number of tandem repeats analysis genotyping to determine the role of asymptomatic carriers in Clostridium difficile transmission. Clin Infect Dis. 2013;57(8):1094–1102. doi: 10.1093/cid/cit475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loo VG, Bourgault AM, Poirier L, et al. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med. 2011;365(18):1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 63.Longtin Y, Paquet-Bolduc B, Gilca R, et al. Effect of Detecting and Isolating Clostridium difficile Carriers at Hospital Admission on the Incidence of C difficile Infections: A Quasi-Experimental Controlled Study. JAMA Intern Med. 2016;176(6):796–804. doi: 10.1001/jamainternmed.2016.0177. [DOI] [PubMed] [Google Scholar]
- 64.Manabe YC, Vinetz JM, Moore RD, et al. Clostridium difficile colitis: an efficient clinical approach to diagnosis. Ann Intern Med. 1995;123(11):835–840. doi: 10.7326/0003-4819-123-11-199512010-00004. [DOI] [PubMed] [Google Scholar]
- 65.Wanahita A, Goldsmith EA, Marino BJ, et al. Clostridium difficile infection in patients with unexplained leukocytosis. Am J Med. 2003;115(7):543–546. doi: 10.1016/s0002-9343(03)00420-0. [DOI] [PubMed] [Google Scholar]
- 66.Wanahita A, Goldsmith EA, Musher DM. Conditions associated with leukocytosis in a tertiary care hospital, with particular attention to the role of infection caused by Clostridium difficile. Clin Infect Dis. 2002;34(12):1585–1592. doi: 10.1086/340536. [DOI] [PubMed] [Google Scholar]
- 67.Dallal RM, Harbrecht BG, Boujoukas AJ, et al. Fulminant Clostridium difficile: an underappreciated and increasing cause of death and complications. Ann Surg. 2002;235(3):363–372. doi: 10.1097/00000658-200203000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fujitani S, George WL, Murthy AR. Comparison of clinical severity score indices for Clostridium difficile infection. Infect Control Hosp Epidemiol. 2011;32(3):220–228. doi: 10.1086/658336. [DOI] [PubMed] [Google Scholar]
- 69.Na X, Martin AJ, Sethi S, et al. A Multi-Center Prospective Derivation and Validation of a Clinical Prediction Tool for Severe Clostridium difficile Infection. PLoS One. 2015;10(4):e0123405. doi: 10.1371/journal.pone.0123405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller MA, Louie T, Mullane K, et al. Derivation and validation of a simple clinical bedside score (ATLAS) for Clostridium difficile infection which predicts response to therapy. BMC Infect Dis. 2013;13:148. doi: 10.1186/1471-2334-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu MY, Katchar K, Kyne L, et al. Prospective derivation and validation of a clinical prediction rule for recurrent Clostridium difficile infection. Gastroenterology. 2009;136(4):1206–1214. doi: 10.1053/j.gastro.2008.12.038. [DOI] [PubMed] [Google Scholar]
- 72.Villano SA, Seiberling M, Tatarowicz W, et al. Evaluation of an oral suspension of VP20621, spores of nontoxigenic Clostridium difficile strain M3, in healthy subjects. Antimicrob Agents Chemother. 2012;56(10):5224–5229. doi: 10.1128/AAC.00913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samore MH, DeGirolami PC, Tlucko A, et al. Clostridium difficile colonization and diarrhea at a tertiary care hospital. Clin Infect Dis. 1994;18(2):181–187. doi: 10.1093/clinids/18.2.181. [DOI] [PubMed] [Google Scholar]
- 74.Shim JK, Johnson S, Samore MH, et al. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet. 1998;351(9103):633–636. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 75.Kutty PK, Benoit SR, Woods CW, et al. Assessment of Clostridium difficile-associated disease surveillance definitions, North Carolina, 2005. Infect Control Hosp Epidemiol. 2008;29(3):197–202. doi: 10.1086/528813. [DOI] [PubMed] [Google Scholar]
- 76.McDonald LC, Coignard B, Dubberke E, et al. Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol. 2007;28(2):140–145. doi: 10.1086/511798. [DOI] [PubMed] [Google Scholar]
- 77.Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med. 1994;330(4):257–262. doi: 10.1056/NEJM199401273300406. [DOI] [PubMed] [Google Scholar]
- 78.Brown E, Talbot GH, Axelrod P, et al. Risk factors for Clostridium difficile toxin-associated diarrhea. Infect Control Hosp Epidemiol. 1990;11(6):283–290. doi: 10.1086/646173. [DOI] [PubMed] [Google Scholar]
- 79.McDonald LC, Owings M, Jernigan DB. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerg Infect Dis. 2006;12(3):409–415. doi: 10.3201/eid1203.051064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zilberberg MD, Shorr AF, Kollef MH. Increase in adult Clostridium difficile-related hospitalizations and case-fatality rate, United States, 2000–2005. Emerg Infect Dis. 2008;14(6):929–931. doi: 10.3201/eid1406.071447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loo VG, Poirier L, Miller MA, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353(23):2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 82.Muto CA, Pokrywka M, Shutt K, et al. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect Control Hosp Epidemiol. 2005;26(3):273–280. doi: 10.1086/502539. [DOI] [PubMed] [Google Scholar]
- 83.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7(7):526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 84.See I, Mu Y, Cohen J, et al. NAP1 strain type predicts outcomes from Clostridium difficile infection. Clin Infect Dis. 2014;58(10):1394–1400. doi: 10.1093/cid/ciu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walker AS, Eyre DW, Wyllie DH, et al. Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin Infect Dis. 2013;56(11):1589–1600. doi: 10.1093/cid/cit127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sirard S, Valiquette L, Fortier LC. Lack of association between clinical outcome of Clostridium difficile infections, strain type, and virulence-associated phenotypes. J Clin Microbiol. 2011;49(12):4040–4046. doi: 10.1128/JCM.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cloud J, Noddin L, Pressman A, et al. Clostridium difficile strain NAP-1 is not associated with severe disease in a nonepidemic setting. Clin Gastroenterol Hepatol. 2009;7(8):868–873. e862. doi: 10.1016/j.cgh.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 88.Walk ST, Micic D, Jain R, et al. Clostridium difficile ribotype does not predict severe infection. Clin Infect Dis. 2012;55(12):1661–1668. doi: 10.1093/cid/cis786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carlson PE, Jr, Walk ST, Bourgis AE, et al. The relationship between phenotype, ribotype, and clinical disease in human Clostridium difficile isolates. Anaerobe. 2013;24:109–116. doi: 10.1016/j.anaerobe.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aitken SL, Alam MJ, Khaleduzzaman M, et al. In the Endemic Setting, Clostridium difficile Ribotype 027 Is Virulent But Not Hypervirulent. Infect Control Hosp Epidemiol. 2015;36(11):1318–1323. doi: 10.1017/ice.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marsh JW, Arora R, Schlackman JL, et al. Association of relapse of Clostridium difficile disease with BI/NAP1/027. J Clin Microbiol. 2012;50(12):4078–4082. doi: 10.1128/JCM.02291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Petrella LA, Sambol SP, Cheknis A, et al. Decreased cure and increased recurrence rates for Clostridium difficile infection caused by the epidemic C difficile BI strain. Clin Infect Dis. 2012;55(3):351–357. doi: 10.1093/cid/cis430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Severe Clostridium difficile-associated disease in populations previously at low risk--four states, 2005. MMWR Morb Mortal Wkly Rep. 2005;54(47):1201–1205. [PubMed] [Google Scholar]
- 94.Surveillance for community-associated Clostridium difficile--Connecticut, 2006. MMWR Morb Mortal Wkly Rep. 2008;57(13):340–343. [PubMed] [Google Scholar]
- 95.Lambert PJ, Dyck M, Thompson LH, et al. Population-based surveillance of Clostridium difficile infection in Manitoba, Canada, by using interim surveillance definitions. Infect Control Hosp Epidemiol. 2009;30(10):945–951. doi: 10.1086/605719. [DOI] [PubMed] [Google Scholar]
- 96.Kutty PK, Woods CW, Sena AC, et al. Risk factors for and estimated incidence of community-associated Clostridium difficile infection, North Carolina, USA. Emerg Infect Dis. 2010;16(2):197–204. doi: 10.3201/eid1602.090953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fellmeth G, Yarlagadda S, Iyer S. Epidemiology of community-onset Clostridium difficile infection in a community in the South of England. J Infect Public Health. 2010;3(3):118–123. doi: 10.1016/j.jiph.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 98.Hirshon JM, Thompson AD, Limbago B, et al. Clostridium difficile infection in outpatients, Maryland and Connecticut, USA, 2002–2007. Emerg Infect Dis. 2011;17(10):1946–1949. doi: 10.3201/eid1710.110069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mitu-Pretorian OM, Forgacs B, Qumruddin A, et al. Outcomes of patients who develop symptomatic Clostridium difficile infection after solid organ transplantation. Transplant Proc. 2010;42(7):2631–2633. doi: 10.1016/j.transproceed.2010.04.057. [DOI] [PubMed] [Google Scholar]
- 100.Lee JT, Kelly RF, Hertz MI, et al. Clostridium difficile infection increases mortality risk in lung transplant recipients. J Heart Lung Transplant. 2013;32(10):1020–1026. doi: 10.1016/j.healun.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 101.Dubberke ER, Olsen MA. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis. 2012;55(Suppl 2):S88–92. doi: 10.1093/cid/cis335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dubberke ER, Reske KA, Olsen MA, et al. Short- and long-term attributable costs of Clostridium difficile-associated disease in nonsurgical inpatients. Clin Infect Dis. 2008;46(4):497–504. doi: 10.1086/526530. [DOI] [PubMed] [Google Scholar]
- 103.Kyne L, Hamel MB, Polavaram R, et al. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin Infect Dis. 2002;34(3):346–353. doi: 10.1086/338260. [DOI] [PubMed] [Google Scholar]
- 104.Lessa FC, Winston LG, McDonald LC, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372(24):2369–2370. doi: 10.1056/NEJMc1505190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Magill SS, Edwards JR, Bamberg W, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370(13):1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller BA, Chen LF, Sexton DJ, et al. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile Infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol. 2011;32(4):387–390. doi: 10.1086/659156. [DOI] [PubMed] [Google Scholar]
- 107.CDC. [Accessed 11 November 2016];Antibiotic resistance threats in the United States. 2013 Jan 31; http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf.
- 108.Braun V, Hundsberger T, Leukel P, et al. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene. 1996;181(1–2):29–38. doi: 10.1016/s0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- 109.Hundsberger T, Braun V, Weidmann M, et al. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur J Biochem. 1997;244(3):735–742. doi: 10.1111/j.1432-1033.1997.t01-1-00735.x. [DOI] [PubMed] [Google Scholar]
- 110.Soehn F, Wagenknecht-Wiesner A, Leukel P, et al. Genetic rearrangements in the pathogenicity locus of Clostridium difficile strain 8864--implications for transcription, expression and enzymatic activity of toxins A and B. Mol Gen Genet. 1998;258(3):222–232. doi: 10.1007/s004380050726. [DOI] [PubMed] [Google Scholar]
- 111.Cohen SH, Tang YJ, Silva J., Jr Analysis of the pathogenicity locus in Clostridium difficile strains. J Infect Dis. 2000;181(2):659–663. doi: 10.1086/315248. [DOI] [PubMed] [Google Scholar]
- 112.Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18(2):247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.LaFrance ME, Farrow MA, Chandrasekaran R, et al. Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc Natl Acad Sci U S A. 2015;112(22):7073–7078. doi: 10.1073/pnas.1500791112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tao L, Zhang J, Meraner P, et al. Frizzled proteins are colonic epithelial receptors for C difficile toxin B. Nature. 2016;538(7625):350–355. doi: 10.1038/nature19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lyras D, O’Connor JR, Howarth PM, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458(7242):1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carter GP, Chakravorty A, Pham Nguyen TA, et al. Defining the Roles of TcdA and TcdB in Localized Gastrointestinal Disease, Systemic Organ Damage, and the Host Response during Clostridium difficile Infections. MBio. 2015;6(3):e00551. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson S, Sambol SP, Brazier JS, et al. International typing study of toxin A-negative, toxin B-positive Clostridium difficile variants. J Clin Microbiol. 2003;41(4):1543–1547. doi: 10.1128/JCM.41.4.1543-1547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moncrief JS, Zheng L, Neville LM, et al. Genetic characterization of toxin A-negative, toxin B-positive Clostridium difficile isolates by PCR. J Clin Microbiol. 2000;38(8):3072–3075. doi: 10.1128/jcm.38.8.3072-3075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Samra Z, Talmor S, Bahar J. High prevalence of toxin A-negative toxin B-positive Clostridium difficile in hospitalized patients with gastrointestinal disease. Diagn Microbiol Infect Dis. 2002;43(3):189–192. doi: 10.1016/s0732-8893(02)00400-5. [DOI] [PubMed] [Google Scholar]
- 120.Hammond GA, Lyerly DM, Johnson JL. Transcriptional analysis of the toxigenic element of Clostridium difficile. Microb Pathog. 1997;22(3):143–154. doi: 10.1006/mpat.1996.0100. [DOI] [PubMed] [Google Scholar]
- 121.Dupuy B, Govind R, Antunes A, et al. Clostridium difficile toxin synthesis is negatively regulated by TcdC. J Med Microbiol. 2008;57(Pt 6):685–689. doi: 10.1099/jmm.0.47775-0. [DOI] [PubMed] [Google Scholar]
- 122.Matamouros S, England P, Dupuy B. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol. 2007;64(5):1274–1288. doi: 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 123.MacCannell DR, Louie TJ, Gregson DB, et al. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada. J Clin Microbiol. 2006;44(6):2147–2152. doi: 10.1128/JCM.02563-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Warny M, Pepin J, Fang A, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366(9491):1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 125.Barbut F, Decre D, Lalande V, et al. Clinical features of Clostridium difficile-associated diarrhoea due to binary toxin (actin-specific ADP-ribosyltransferase)-producing strains. J Med Microbiol. 2005;54(Pt 2):181–185. doi: 10.1099/jmm.0.45804-0. [DOI] [PubMed] [Google Scholar]
- 126.McEllistrem MC, Carman RJ, Gerding DN, et al. A hospital outbreak of Clostridium difficile disease associated with isolates carrying binary toxin genes. Clin Infect Dis. 2005;40(2):265–272. doi: 10.1086/427113. [DOI] [PubMed] [Google Scholar]
- 127.Rupnik M, Grabnar M, Geric B. Binary toxin producing Clostridium difficile strains. Anaerobe. 2003;9(6):289–294. doi: 10.1016/j.anaerobe.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 128.Geric B, Carman RJ, Rupnik M, et al. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J Infect Dis. 2006;193(8):1143–1150. doi: 10.1086/501368. [DOI] [PubMed] [Google Scholar]
- 129.Joost I, Speck K, Herrmann M, et al. Characterisation of Clostridium difficile isolates by slpA and tcdC gene sequencing. Int J Antimicrob Agents. 2009;33(Suppl 1):S13–18. doi: 10.1016/S0924-8579(09)70010-X. [DOI] [PubMed] [Google Scholar]
- 130.Akerlund T, Persson I, Unemo M, et al. Increased sporulation rate of epidemic Clostridium difficile Type 027/NAP1. J Clin Microbiol. 2008;46(4):1530–1533. doi: 10.1128/JCM.01964-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Burns DA, Heeg D, Cartman ST, et al. Reconsidering the sporulation characteristics of hypervirulent Clostridium difficile BI/NAP1/027. PLoS One. 2011;6(9):e24894. doi: 10.1371/journal.pone.0024894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Curry SR, Marsh JW, Shutt KA, et al. High frequency of rifampin resistance identified in an epidemic Clostridium difficile clone from a large teaching hospital. Clin Infect Dis. 2009;48(4):425–429. doi: 10.1086/596315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197(3):435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 134.Reeves AE, Koenigsknecht MJ, Bergin IL, et al. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun. 2012;80(11):3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kamiya S, Yamakawa K, Ogura H, et al. Effect of various sodium taurocholate preparations on the recovery of Clostridium difficile spores. Microbiol Immunol. 1987;31(11):1117–1120. doi: 10.1111/j.1348-0421.1987.tb01344.x. [DOI] [PubMed] [Google Scholar]
- 136.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190(7):2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol. 2009;191(3):1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Koenigsknecht MJ, Theriot CM, Bergin IL, et al. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun. 2015;83(3):934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peterson LR, Olson MM, Shanholtzer CJ, et al. Results of a prospective, 18-month clinical evaluation of culture, cytotoxin testing, and culturette brand (CDT) latex testing in the diagnosis of Clostridium difficile-associated diarrhea. Diagn Microbiol Infect Dis. 1988;10(2):85–91. doi: 10.1016/0732-8893(88)90045-4. [DOI] [PubMed] [Google Scholar]
- 141.Walker RC, Ruane PJ, Rosenblatt JE, et al. Comparison of culture, cytotoxicity assays, and enzyme-linked immunosorbent assay for toxin A and toxin B in the diagnosis of Clostridium difficile-related enteric disease. Diagn Microbiol Infect Dis. 1986;5(1):61–69. doi: 10.1016/0732-8893(86)90092-1. [DOI] [PubMed] [Google Scholar]
- 142.Shanholtzer CJ, Willard KE, Holter JJ, et al. Comparison of the VIDAS Clostridium difficile toxin A immunoassay with C difficile culture and cytotoxin and latex tests. J Clin Microbiol. 1992;30(7):1837–1840. doi: 10.1128/jcm.30.7.1837-1840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sloan LM, Duresko BJ, Gustafson DR, et al. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J Clin Microbiol. 2008;46(6):1996–2001. doi: 10.1128/JCM.00032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Aichinger E, Schleck CD, Harmsen WS, et al. Nonutility of repeat laboratory testing for detection of Clostridium difficile by use of PCR or enzyme immunoassay. J Clin Microbiol. 2008;46(11):3795–3797. doi: 10.1128/JCM.00684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Drees M, Snydman DR, O’Sullivan CE. Repeated enzyme immunoassays have limited utility in diagnosing Clostridium difficile. Eur J Clin Microbiol Infect Dis. 2008;27(5):397–399. doi: 10.1007/s10096-007-0452-8. [DOI] [PubMed] [Google Scholar]
- 146.Nemat H, Khan R, Ashraf MS, et al. Diagnostic value of repeated enzyme immunoassays in Clostridium difficile infection. Am J Gastroenterol. 2009;104(8):2035–2041. doi: 10.1038/ajg.2009.174. [DOI] [PubMed] [Google Scholar]
- 147.Renshaw AA, Stelling JM, Doolittle MH. The lack of value of repeated Clostridium difficile cytotoxicity assays. Arch Pathol Lab Med. 1996;120(1):49–52. [PubMed] [Google Scholar]
- 148.Goldenberg SD, Cliff PR, Smith S, et al. Two-step glutamate dehydrogenase antigen real-time polymerase chain reaction assay for detection of toxigenic Clostridium difficile. J Hosp Infect. 2010;74(1):48–54. doi: 10.1016/j.jhin.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 149.Novak-Weekley SM, Marlowe EM, Miller JM, et al. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol. 2010;48(3):889–893. doi: 10.1128/JCM.01801-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wren MW, Kinson R, Sivapalan M, et al. Detection of Clostridium difficile infection: a suggested laboratory diagnostic algorithm. Br J Biomed Sci. 2009;66(4):175–179. doi: 10.1080/09674845.2009.11730269. [DOI] [PubMed] [Google Scholar]
- 151.Crobach MJ, Dekkers OM, Wilcox MH, et al. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI) Clin Microbiol Infect. 2009;15(12):1053–1066. doi: 10.1111/j.1469-0691.2009.03098.x. [DOI] [PubMed] [Google Scholar]
- 152.Snell H, Ramos M, Longo S, et al. Performance of the TechLab C. DIFF CHEK-60 enzyme immunoassay (EIA) in combination with the C. difficile Tox A/B II EIA kit, the Triage C. difficile panel immunoassay, and a cytotoxin assay for diagnosis of Clostridium difficile-associated diarrhea. J Clin Microbiol. 2004;42(10):4863–4865. doi: 10.1128/JCM.42.10.4863-4865.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sunkesula VC, Kundrapu S, Muganda C, et al. Does empirical Clostridium difficile infection (CDI) therapy result in false-negative CDI diagnostic test results? Clin Infect Dis. 2013;57(4):494–500. doi: 10.1093/cid/cit286. [DOI] [PubMed] [Google Scholar]
- 154.Dubberke ER, Han Z, Bobo L, et al. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J Clin Microbiol. 2011;49(8):2887–2893. doi: 10.1128/JCM.00891-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Planche TD, Davies KA, Coen PG, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis. 2013;13(11):936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Polage CR, Gyorke CE, Kennedy MA, et al. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern Med. 2015;175(11):1792–1801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Teasley DG, Gerding DN, Olson MM, et al. Prospective randomised trial of metronidazole versus vancomycin for Clostridium-difficile-associated diarrhoea and colitis. Lancet. 1983;2(8358):1043–1046. doi: 10.1016/s0140-6736(83)91036-x. [DOI] [PubMed] [Google Scholar]
- 158.Cohen SH, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31(5):431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 159.Zar FA, Bakkanagari SR, Moorthi KM, et al. A comparison of vancomycin and metronidazole for the treatment of Clostridium difficile-associated diarrhea, stratified by disease severity. Clin Infect Dis. 2007;45(3):302–307. doi: 10.1086/519265. [DOI] [PubMed] [Google Scholar]
- 160.Johnson S, Louie TJ, Gerding DN, et al. Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: results from two multinational, randomized, controlled trials. Clin Infect Dis. 2014;59(3):345–354. doi: 10.1093/cid/ciu313. [DOI] [PubMed] [Google Scholar]
- 161.Cornely OA, Crook DW, Esposito R, et al. Fidaxomicin versus vancomycin for infection with Clostridium difficile in Europe, Canada, and the USA: a double-blind, non-inferiority, randomised controlled trial. Lancet Infect Dis. 2012;12(4):281–289. doi: 10.1016/S1473-3099(11)70374-7. [DOI] [PubMed] [Google Scholar]
- 162.Cornely OA, Miller MA, Louie TJ, et al. Treatment of first recurrence of Clostridium difficile infection: fidaxomicin versus vancomycin. Clin Infect Dis. 2012;55(Suppl 2):S154–161. doi: 10.1093/cid/cis462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Louie TJ, Miller MA, Mullane KM, et al. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med. 2011;364(5):422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 164.Bauer MP, Kuijper EJ, van Dissel JT, et al. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): treatment guidance document for Clostridium difficile infection (CDI) Clin Microbiol Infect. 2009;15(12):1067–1079. doi: 10.1111/j.1469-0691.2009.03099.x. [DOI] [PubMed] [Google Scholar]
- 165.Surawicz CM, Brandt LJ, Binion DG, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108(4):478–498. doi: 10.1038/ajg.2013.4. quiz 499. [DOI] [PubMed] [Google Scholar]
- 166.Hamilton MJ, Weingarden AR, Sadowsky MJ, et al. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012;107(5):761–767. doi: 10.1038/ajg.2011.482. [DOI] [PubMed] [Google Scholar]
- 167.Kelly CR, Ihunnah C, Fischer M, et al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol. 2014;109(7):1065–1071. doi: 10.1038/ajg.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kelly CR, Khoruts A, Staley C, et al. Effect of Fecal Microbiota Transplantation on Recurrence in Multiply Recurrent Clostridium difficile Infection: A Randomized Trial. Ann Intern Med. 2016;165(9):609–616. doi: 10.7326/M16-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 170.Youngster I, Russell GH, Pindar C, et al. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312(17):1772–1778. doi: 10.1001/jama.2014.13875. [DOI] [PubMed] [Google Scholar]
- 171.Youngster I, Sauk J, Pindar C, et al. Fecal microbiota transplant for relapsing Clostridium difficile infection using a frozen inoculum from unrelated donors: a randomized, open-label, controlled pilot study. Clin Infect Dis. 2014;58(11):1515–1522. doi: 10.1093/cid/ciu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Neal MD, Alverdy JC, Hall DE, et al. Diverting loop ileostomy and colonic lavage: an alternative to total abdominal colectomy for the treatment of severe, complicated Clostridium difficile associated disease. Ann Surg. 2011;254(3):423–427. doi: 10.1097/SLA.0b013e31822ade48. discussion 427–429. [DOI] [PubMed] [Google Scholar]
- 173.Johnson S, Homann SR, Bettin KM, et al. Treatment of asymptomatic Clostridium difficile carriers (fecal excretors) with vancomycin or metronidazole. A randomized, placebo-controlled trial. Ann Intern Med. 1992;117(4):297–302. doi: 10.7326/0003-4819-117-4-297. [DOI] [PubMed] [Google Scholar]
- 174.Swale A, Miyajima F, Roberts P, et al. Calprotectin and lactoferrin faecal levels in patients with Clostridium difficile infection (CDI): a prospective cohort study. PLoS One. 2014;9(8):e106118. doi: 10.1371/journal.pone.0106118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.D’Haens G, Ferrante M, Vermeire S, et al. Fecal calprotectin is a surrogate marker for endoscopic lesions in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18(12):2218–2224. doi: 10.1002/ibd.22917. [DOI] [PubMed] [Google Scholar]
- 176.Jackson M, Olefson S, Machan JT, et al. A High Rate of Alternative Diagnoses in Patients Referred for Presumed Clostridium difficile Infection. J Clin Gastroenterol. 2016;50(9):742–746. doi: 10.1097/MCG.0000000000000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McFarland LV, Surawicz CM, Rubin M, et al. Recurrent Clostridium difficile disease: epidemiology and clinical characteristics. Infect Control Hosp Epidemiol. 1999;20(1):43–50. doi: 10.1086/501553. [DOI] [PubMed] [Google Scholar]
- 178.Aboudola S, Kotloff KL, Kyne L, et al. Clostridium difficile vaccine and serum immunoglobulin G antibody response to toxin A. Infect Immun. 2003;71(3):1608–1610. doi: 10.1128/IAI.71.3.1608-1610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kelly CP. Immune response to Clostridium difficile infection. Eur J Gastroenterol Hepatol. 1996;8(11):1048–1053. doi: 10.1097/00042737-199611000-00004. [DOI] [PubMed] [Google Scholar]
- 180.Kyne L, Warny M, Qamar A, et al. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med. 2000;342(6):390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 181.Kyne L, Warny M, Qamar A, et al. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet. 2001;357(9251):189–193. doi: 10.1016/S0140-6736(00)03592-3. [DOI] [PubMed] [Google Scholar]
- 182.Salcedo J, Keates S, Pothoulakis C, et al. Intravenous immunoglobulin therapy for severe Clostridium difficile colitis. Gut. 1997;41(3):366–370. doi: 10.1136/gut.41.3.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Garey KW, Sethi S, Yadav Y, et al. Meta-analysis to assess risk factors for recurrent Clostridium difficile infection. J Hosp Infect. 2008;70(4):298–304. doi: 10.1016/j.jhin.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 184.Sougioultzis S, Kyne L, Drudy D, et al. Clostridium difficile toxoid vaccine in recurrent C difficile- associated diarrhea. Gastroenterology. 2005;128(3):764–770. doi: 10.1053/j.gastro.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 185.Lowy I, Molrine DC, Leav BA, et al. Treatment with monoclonal antibodies against Clostridium difficile toxins. N Engl J Med. 362(3):197–205. doi: 10.1056/NEJMoa0907635. [DOI] [PubMed] [Google Scholar]
- 186.Wilcox MH, et al. Phase 3 Double-blind Study of Actoxumab (ACT) & Bezlotoxumab (BEZ) for Prevention of Recurrent C. difficile Infection (rCDI) in Patients on Standard of Care (SoC) Antibiotics (MODIFY I). Presented at: Interscience Conference on Antimicrobial Agents and Chemotherapy; Sept. 17–21, 2015; San Diego. [Google Scholar]
- 187.Gerding DN, et al. Phase 3 Double-Blind Study of Bezlotoxumab (BEZ) Alone & with Actoxumab (ACT) for Prevention of Recurrent C. difficile Infection (rCDI) in Patients on Standard of Care (SoC) Antibiotics (MODIFY II). Presented at: Interscience Conference on Antimicrobial Agents and Chemotherapy; Sept. 17–21, 2015; San Diego. [Google Scholar]
- 188.Rupnik M, Avesani V, Janc M, et al. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol. 1998;36(8):2240–2247. doi: 10.1128/jcm.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lemee L, Dhalluin A, Pestel-Caron M, et al. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J Clin Microbiol. 2004;42(6):2609–2617. doi: 10.1128/JCM.42.6.2609-2617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Didelot X, Eyre DW, Cule M, et al. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol. 2012;13(12):R118. doi: 10.1186/gb-2012-13-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Songer JG, Trinh HT, Killgore GE, et al. Clostridium difficile in retail meat products, USA, 2007. Emerg Infect Dis. 2009;15(5):819–821. doi: 10.3201/eid1505.081071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.de Boer E, Zwartkruis-Nahuis A, Heuvelink AE, et al. Prevalence of Clostridium difficile in retailed meat in the Netherlands. Int J Food Microbiol. 2011;144(3):561–564. doi: 10.1016/j.ijfoodmicro.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 193.Metcalf DS, Costa MC, Dew WM, et al. Clostridium difficile in vegetables, Canada. Lett Appl Microbiol. 2010;51(5):600–602. doi: 10.1111/j.1472-765x.2010.02933.x. [DOI] [PubMed] [Google Scholar]
- 194.Hoffer E, Haechler H, Frei R, et al. Low occurrence of Clostridium difficile in fecal samples of healthy calves and pigs at slaughter and in minced meat in Switzerland. J Food Prot. 2010;73(5):973–975. doi: 10.4315/0362-028x-73.5.973. [DOI] [PubMed] [Google Scholar]
- 195.Weese JS, Reid-Smith RJ, Avery BP, et al. Detection and characterization of Clostridium difficile in retail chicken. Lett Appl Microbiol. 2010 doi: 10.1111/j.1472-765X.2010.02802.x. [DOI] [PubMed] [Google Scholar]
- 196.Jobstl M, Heuberger S, Indra A, et al. Clostridium difficile in raw products of animal origin. Int J Food Microbiol. 2010;138(1–2):172–175. doi: 10.1016/j.ijfoodmicro.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 197.Houser BA, Soehnlen MK, Wolfgang DR, et al. Prevalence of Clostridium difficile toxin genes in the feces of veal calves and incidence of ground veal contamination. Foodborne Pathog Dis. 2012;9(1):32–36. doi: 10.1089/fpd.2011.0955. [DOI] [PubMed] [Google Scholar]
- 198.Von Abercron SM, Karlsson F, Wigh GT, et al. Low occurrence of Clostridium difficile in retail ground meat in Sweden. J Food Prot. 2009;72(8):1732–1734. doi: 10.4315/0362-028x-72.8.1732. [DOI] [PubMed] [Google Scholar]
- 199.Harvey RB, Norman KN, Andrews K, et al. Clostridium difficile in poultry and poultry meat. Foodborne Pathog Dis. 2011;8(12):1321–1323. doi: 10.1089/fpd.2011.0936. [DOI] [PubMed] [Google Scholar]
- 200.Harvey RB, Norman KN, Andrews K, et al. Clostridium difficile in retail meat and processing plants in Texas. J Vet Diagn Invest. 2011;23(4):807–811. doi: 10.1177/1040638711407893. [DOI] [PubMed] [Google Scholar]
- 201.Nguyen GC, Kaplan GG, Harris ML, et al. A national survey of the prevalence and impact of Clostridium difficile infection among hospitalized inflammatory bowel disease patients. Am J Gastroenterol. 2008;103(6):1443–1450. doi: 10.1111/j.1572-0241.2007.01780.x. [DOI] [PubMed] [Google Scholar]
- 202.Paudel S, Zacharioudakis IM, Zervou FN, et al. Prevalence of Clostridium difficile infection among solid organ transplant recipients: a meta-analysis of published studies. PLoS One. 2015;10(4):e0124483. doi: 10.1371/journal.pone.0124483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Guddati AK, Kumar G, Ahmed S, et al. Incidence and outcomes of Clostridium difficile-associated disease in hematopoietic cell transplant recipients. Int J Hematol. 2014;99(6):758–765. doi: 10.1007/s12185-014-1577-z. [DOI] [PubMed] [Google Scholar]
- 204.Lee CH, Steiner T, Petrof EO, et al. Frozen vs Fresh Fecal Microbiota Transplantation and Clinical Resolution of Diarrhea in Patients With Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. JAMA. 2016;315(2):142–149. doi: 10.1001/jama.2015.18098. [DOI] [PubMed] [Google Scholar]