Abstract
Human noroviruses are the primary cause of epidemic and sporadic acute gastroenteritis. The worldwide high morbidity and mortality associated with norovirus infections, particularly among the elderly, immunocompromised patients and children, constitute a serious public health concern. There are currently no approved human vaccines or norovirus-specific small-molecule therapeutics or prophylactics. Norovirus 3CL protease has recently emerged as a potential therapeutic target for the development of anti-norovirus agents. We hypothesized that the S4 subsite of the enzyme may provide an effective means of designing potent and cell permeable inhibitors of the enzyme. We report herein the structure-guided exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design and synthesis of effective inhibitors of the protease.
1. Introduction
Human noroviruses belong to the family Caliciviridae and are the leading cause of acute gastroenteritis worldwide [1–2]. They are associated with high morbidity and a heavy economic burden [3–5]. In developing countries the mortality rate among children <5 years old due to diarrheal disease caused by noroviruses is estimated to account for 71,000 deaths annually [6–8]. Combating norovirus infections presents a challenge because of the facile foodborne and waterborne transmission of noroviruses, as well as their high genetic diversity and environmental stability [9–10]. The problem is further exacerbated by the current lack of diagnostics and norovirus-specific therapeutics and prophylactics, or vaccines. Thus, there is an urgent and unmet medical need for the discovery and development of anti-norovirus small-molecule therapeutics and prophylactics [11–14], as well as effective vaccines [15–16].
Both viral and host factors can in principle serve as a launching pad for the discovery of anti-norovirus therapeutics [11–14,17–20]. Prominent viral targets that are particularly well-suited as targets for anti-noroviral drug development include the viral-encoded 3CL protease (3CLpro) and RNA dependent RNA polymerase (RdRp) because of the essential roles they play in viral replication [18]. Following translation of the viral genome, the viral polyprotein is cleaved by 3CLpro, a cysteine protease with a prototypical catalytic triad (Cys 139, His30, Glu54) and a strong preference for a P1 Gln (or Glu) residue [21], to generate structural and nonstructural proteins [22–24] Norovirus 3CLpro has been the focus of intense investigations and an array of inhibitors of 3CLpro that display anti-norovirus activity have been reported [11–14,19], including peptidyl [25–26] and macrocyclic [27–28] transition state inhibitors and transition state mimics [29]. Importantly, in vivo proof of concept in a mouse model using a dipeptidyl transition state inhibitor of norovirus 3CLpro has also been demonstrated [25]. We describe herein the structure-guided exploration of the S4 subsite of 3CLpro and the subsequent deployment of structure-activity relationship and biochemical studies in the design and discovery of potent and permeable inhibitors of the enzyme.
2. Results and Discussion
2.1. Inhibitor design rationale
The design and optimization of the inhibitors was informed by the utilization of available crystal structures and the parallel acquisition of additional high resolution X-ray crystal structures. Thus, inspection of previously-determined high resolution X-ray crystal structures of dipeptidyl inhibitors bound to norovirus 3CL protease (PDB accession codes 4XBB, 4XBC and 4XBD) [25] revealed that the “cap” R group in inhibitor (I) projects toward the S4 subsite of the enzyme and suggested that its close proximity to a string of hydrophobic amino acids (Ala158, Ala160, Val168 and Ile109) can be exploited through appropriate cap modifications, including the use of sulfonamide functionalities. Additional design considerations included the use of key recognition elements, such as a Gln surrogate [30] and a cyclohexyl methyl residue at the P1 and P2 positions, respectively, capable of engaging in H-bonding and hydrophobic interactions. Finally, based on the recently-demonstrated in vivo efficacy of aldehyde bisulfite adducts [31], this moiety was incorporated into inhibitor (I) and, for comparative purposes, the corresponding aldehyde inhibitors were also included.
2.2. Chemistry
The synthesis of inhibitors 6a-7m was accomplished using the reaction sequence shown in Scheme 1. Briefly, N-Boc-L-cyclohexyl methyl alanine was coupled to glutamine surrogate 8 [30] to obtain 2, followed by removal of the Boc protective group to yield intermediate 3 which was subsequently reacted with a series of sulfonyl chlorides to yield sulfonamide esters 4a–m. Reduction with lithium borohydride generated the corresponding alcohols 5a–m. Dess-Martin periodinane oxidation of 5a–m proceeded smoothly to yield aldehydes 6a–m which were transformed to the corresponding aldehyde bisulfite adducts 7a–m. The synthesized sulfonamide derivatives are listed in Table 1. Carbamates 12a–c were synthesized according to Scheme 2 via reaction of intermediate 3 with an appropriate chloroformate and further elaboration to yield the corresponding aldehydes and aldehyde bisulfite adducts, which are listed in Table 2.
Scheme 1.
Synthesis of Inhibitors 6a–m and 7a–m
aEDCI/HOBt/DIEA/DMF then 8; b4M HCl in dioxane/2h; cRSO2Cl/TEA/DCM/35°C; d2M LiBH4/THF/CH3OH;eDess-Martin periodinane/DCM; fC2H5OH/EtOAc/NaHSO3
Table 1.
Activity of compounds 6a–m and 7a–m against NV 3CL protease and replicon harboring cells.
| Compound | R | IC50 (μM) | EC50 (μM) | CC50 (μM) |
|---|---|---|---|---|
| 6a | C6H5 | 8.5 | 9.5 | > 100 |
| 7a | 13.5 | 12.3 | > 100 | |
|
| ||||
| 6b | (C6H5) CH2 | 4.5 | 4.1 | > 100 |
| 7b | 4.3 | 4.3 | > 100 | |
|
| ||||
| 6c | (C6H5) CH2CH2 | 1.8 | 0.7 | > 100 |
| 7c | 2.1 | 0.8 | > 100 | |
|
| ||||
| 6d | p-F (C6H4) | 4.8 | 2 | > 100 |
| 7d | 6.2 | 2.1 | > 100 | |
|
| ||||
| 6e | m-Cl (C6H4) | 5.5 | 2.3 | > 100 |
| 7e | 5.7 | 1.9 | > 100 | |
|
| ||||
| 6f | p-Cl (C6H4) | 5.8 | 3.5 | > 100 |
| 7f | 5.1 | 7.2 | > 100 | |
|
| ||||
| 6g | p-Cl (C6H4) CH2 | 1.8 | 2.4 | > 100 |
| 7g | 1.2 | 2.2 | > 100 | |
|
| ||||
| 6h | m-Cl (C6H4) CH2CH2 | 2.3 | 1.1 | > 100 |
| 7h | 1.9 | 0.8 | > 100 | |
|
| ||||
| 6i | 2-Thiophene | 10.4 | 8.5 | > 100 |
| 7i | 8.5 | 8.1 | > 100 | |
|
| ||||
| 6j | Biphenyl-4- | 1.3 | 1.3 | > 100 |
| 7j | 1.1 | 1.2 | > 100 | |
|
| ||||
| 6k | 2-Phthalimidoethane | 2.5 | 2.3 | > 100 |
| 7k | 2.6 | 2.1 | > 100 | |
|
| ||||
| 6l | p-CH3O(C6H4)O(CH2)3 | 1.8 | 0.2 | > 100 |
| 7l | 1.6 | 0.2 | > 100 | |
|
| ||||
| 6m | CH3(CH2)7 | 1.2 | 0.3 | 33.8 |
| 7m | 1.1 | 0.2 | 35.4 | |
Scheme 2.
Synthesis of Inhibitors 11a–c and 12a–c
aROCOCl/TEA/DCM/ reflux 3 hrs; b2M LiBH4/THF/CH3OH;cDess-Martin periodinane/DCM; dC2H5OH/EtOAc/NaHSO3
Table 2.
Activity of compounds 11a–c and 12a–c against NV 3CL protease and replicon harboring cells
| Compound | R | IC50 (μM) | EC50 (μM) | CC50 (μM) |
|---|---|---|---|---|
| 11a | m-Cl (C6H4)CH2CH2 | 0.6 | 0.1 | 43.2 |
| 12a | 0.5 | 0.1 | 46.7 | |
|
| ||||
| 11b | (C6H5)CH2O(CH2)2 | 2.1 | 0.2 | > 100 |
| 12b | 1.6 | 0.3 | > 100 | |
|
| ||||
| 11c | CH3 (CH2)9 | 0.3 | 0.1 | 32.1 |
| 12c | 0.4 | 0.2 | 34.5 | |
2.3. Biochemical Studies
The inhibitory activity of the synthesized compounds against NV 3CLpro and their anti-norovirus activity in a cell-based replicon system, were evaluated as described in the experimental section [32]. The determined IC50, EC50 and CC50 values are listed in Tables 1 and 2 and they are the average of at least two determinations.
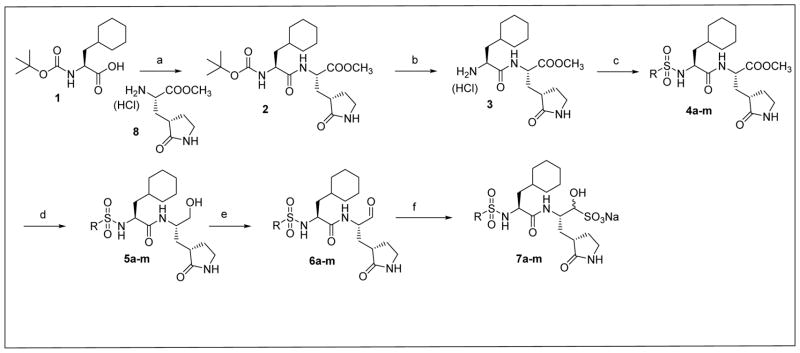
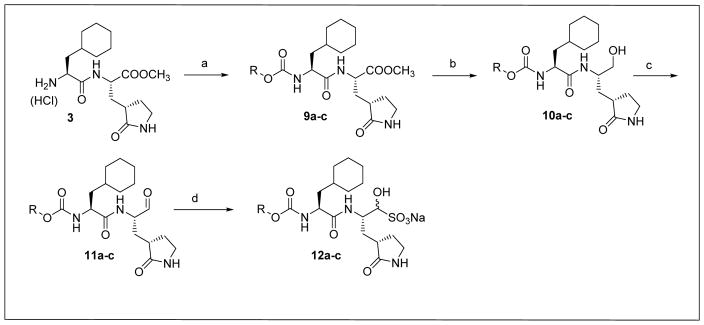
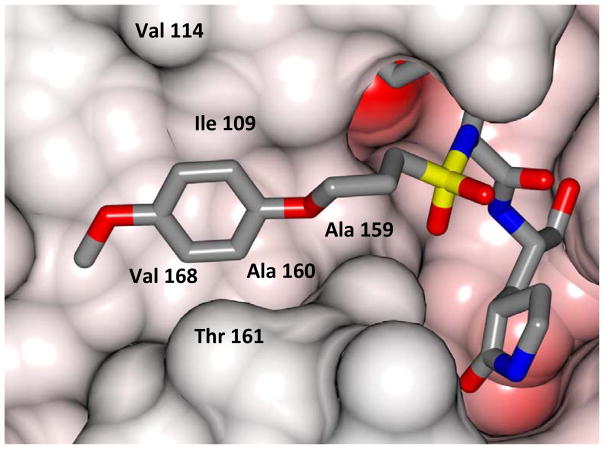
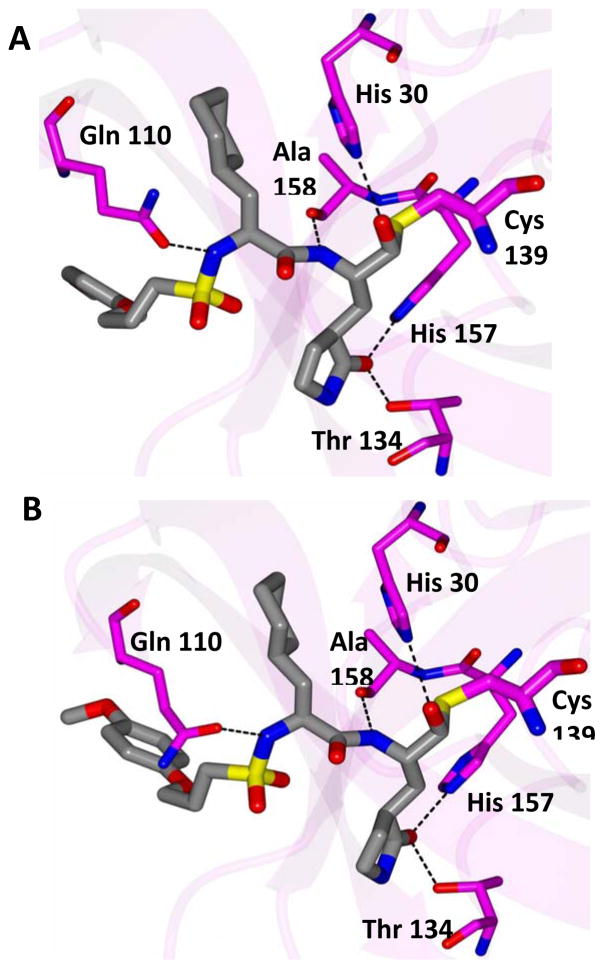
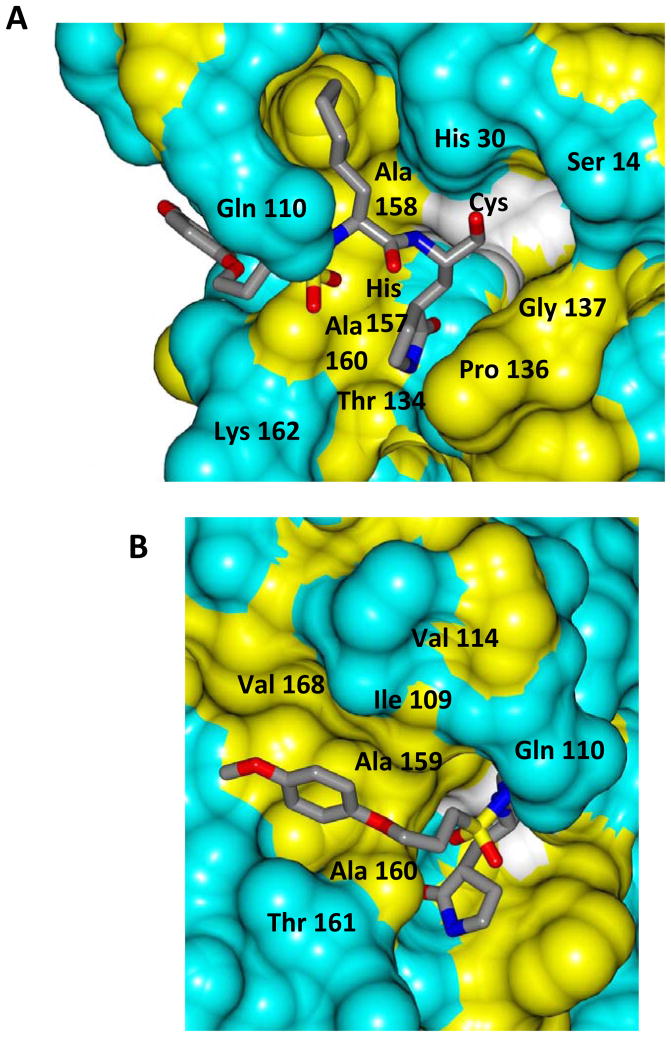
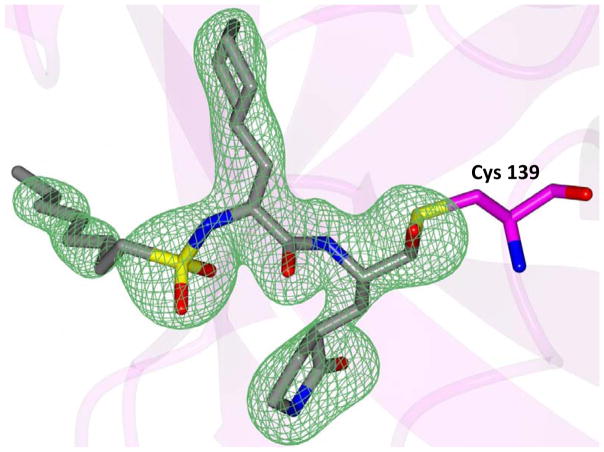
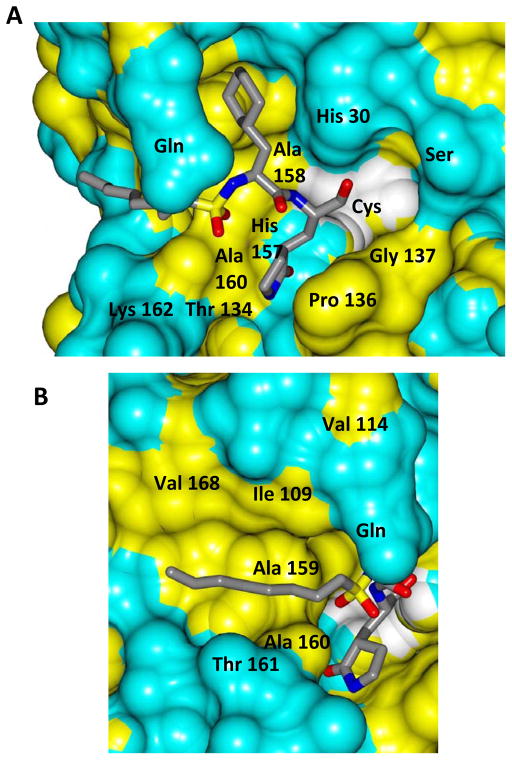
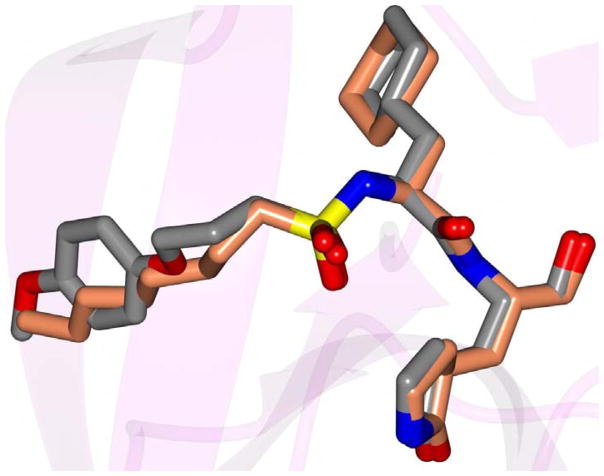
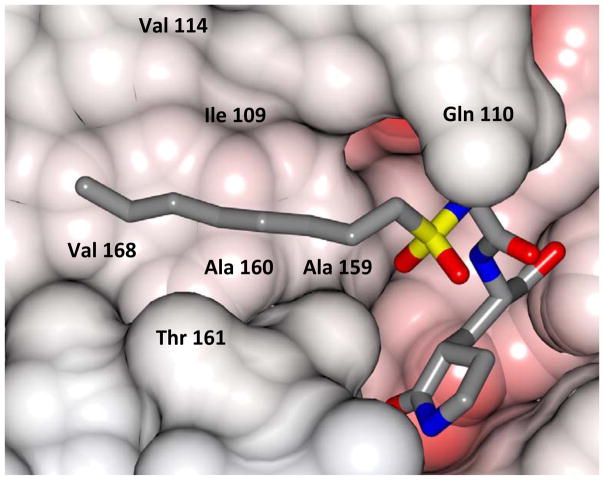
Inspection of the results in Table 1 permit the following inferences to be drawn: (a) the potency of the aldehyde bisulfite adducts is comparable to those of the precursor aldehydes, an observation noted previously [25,33]. Advantages accrued through the use of aldehyde bisulfite adducts include higher solubility and metabolic stability, as well as low cytotoxicity [31]. Whether the bisulfite adducts exert their action by functioning as transition state mimics or latent precursors of the aldehyde functionality, or both, has not been established as yet and is currently under investigation; (b) a significant increase in potency (15-fold) was observed when the phenyl ring is moved further into the S4 pocket (Table 1, compare the EC50 values of compounds 7a, 7b and 7c) and an additional 4-fold gain in potency was realized by extending the linker further (Table 1, compound 7l). These observations are congruent with the results of X-ray crystallographic studies with inhibitor 7l. Crystallization of NV 3CLpro with inhibitor 7l yielded hexagonal and orthorhombic crystals and the electron density clearly shows the sulfur atom of the active site Cys139 residue bound covalently to the carbonyl carbon of the P1 residue of inhibitor 7l, thereby establishing the mechanism of action of this class of inhibitors (Figure 2). The electrostatic surface representation shows the aryl ring to be optimally nestled in the hydrophobic S4 pocket of the enzyme and engaged in hydrophobic interactions with Ile109, Val168, Ala160 and Ala159 (Figure 3). Inhibitor 7l is bound to the active site of the enzyme via a network of hydrogen bonds in the manner of an antiparallel β-sheet and includes inhibitor backbone H-bonds with Gln110 and Ala158 which serve to position and orient the inhibitor correctly with respect to the catalytic residues (Figures 4 and 5). Additionally, a H-bond with His30 serves to stabilize the tetrahedral adduct and two critical H-bonds involving the P1 Gln surrogate ring oxygen with His157 and Thr134 are clearly evident; (c) halogen substitutions in the aromatic ring were generally beneficial (Table 1, compounds 6d/7d, 6e/7e and 6g/7g); (d) potency also improved when a 4-biphenyl or 2-pthalimidoethane moiety was used (Table 1, compounds 6j/7j and 6k/7k); (e) the significance of hydrophobic interactions involving R and the S4 subsite was further probed by using an n-octyl chain cap, resulting in inhibitors displaying submicromolar ED50 values (Table 1, compounds 6m/7m). The structural determinants accounting for the higher potency of compounds 6m/7m were identified by obtaining a high resolution X-ray crystal structure of the NV 3CLpro:7m complex (Figure 6). Inhibitor 7m adopts a binding mode in the active site similar to 7l (Figure 7) which is clearly evident from the superposition of the two inhibitors in the active site of the enzyme (Figure 8). Furthermore, inhibitor 7m engages in identical H-bond interactions as inhibitor 7l (Figure 9), hence the comparable potencies. Importantly, the n-octyl chain encompasses the S4 subsite and participates in hydrophobic interactions with the string of hydrophobic amino acids that make up the S4 pocket (Figure 10). The importance of hydrophobic interactions is similarly evident in carbamate derivatives 11c/12c (Table 2); and, (f) an eight to ten-fold increase in potency was realized when the sulfonamide linkage was replaced by a carbamate moiety (Table 2, compounds 11a/12a versus compounds 6h/7h in Table 1).
Fig. 2.
Fo-Fc omit map of inhibitor 7l (green mesh) contoured at 3σ for (A) NV 3CLpro:7l (hexagonal) and B) NV 3CLpro:7l (orthorhombic).
Fig. 3.
Electrostatic surface representation of NV 3CLpro showing the aryl ring region of inhibitor 7l positioned in the hydrophobic S4 pocket.
Fig. 4.
Hydrogen bond interactions (dashed lines) for (A) NV 3CLpro:7l (hexagonal) and B) NV 3CLpro:7l (orthorhombic).
Fig. 5.
Surface representation of NV 3CLpro:7l (hexagonal) with neighboring residues colored yellow (nonpolar), cyan (polar), and white (weakly polar). A) View of the inhibitor in the S1/S2 pocket and B) the S4 pocket.
Fig. 6.
Fo-Fc omit map of inhibitor 7m (green mesh) contoured at 3σ.
Fig. 7.
Surface representation of NV 3CLpro:7m with neighboring residues colored yellow (nonpolar), cyan (polar), and white (weakly polar). A) View of the inhibitor in the S1/S2 pocket and B) the S4 pocket.
Fig. 8.
Superposition of inhibitor 7l (gray) and 7m (coral) which adopt similar binding modes in the NV 3CLpro active site
Fig. 9.
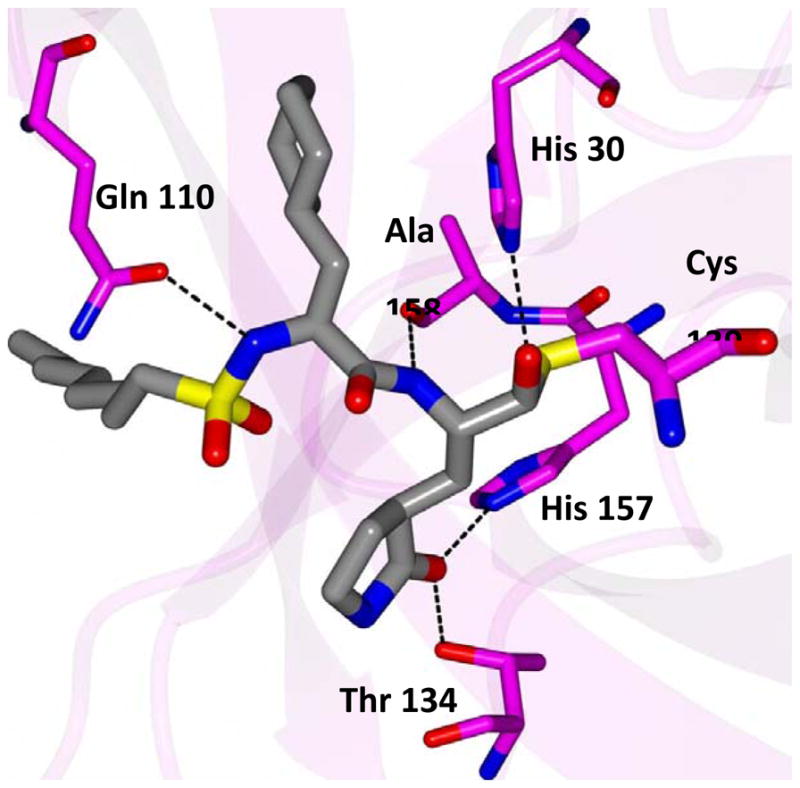
Hydrogen bond interactions (dashed lines) for NV 3CLpro:7m.
Fig. 10.
Electrostatic surface representation of NV 3CLpro showing the aliphatic chain of inhibitor 7m positioned in a hydrophobic S4 pocket.
3. Conclusion
Structural, synthetic, biochemical, and cell-based studies have been employed to illuminate the S4 subsite of NV 3CLpro and to gain insight and understanding into the significance and impact of hydrophobic interactions on pharmacological activity. In principle, further gains in potency can be realized through the use of constrained R moieties that mitigate the entropic penalty incurred in using long aliphatic chains. The results reported herein provide a sound framework and a solid foundation for conducting further studies aimed at identifying anti-norovirus drug candidates.
4. Experimental section
4.1. General
Reagents and dry solvents were purchased from various chemical suppliers (Chem-Impex, Acros Organics, TCI America, Aldrich, and Bachem) and were used as obtained. Silica gel (230–450 mesh) used for flash chromatography was purchased from Sorbent Technologies (Atlanta, GA). Thin layer chromatography was performed using Analtech silica gel plates (Newark, DE). Visualization was accomplished using UV light and/or iodine. NMR spectra were recorded in CDCl3 or DMSO-d6 using a Varian XL-400 spectrometer and are reported relative to TMS (δH= 0.00 ppm). Chemical shifts are reported in ppm and spin multiplicities are represented by the following signals: s (singlet), d (doublet), dd (doublets of doublet), t (triplet), q (quartet) and m (multiplet). Melting points were recorded on a Mel-Temp apparatus and are uncorrected. High resolution mass spectrometry (HRMS) was performed at the University of Kansas Mass Spectrometry lab using an LCT Premier mass spectrometer (Waters, Milford, MA) equipped with a time of flight mass analyzer and an electrospray ion source or in-house using a G6230B TOF MS (Agilent Technologies, Santa Clara, CA). The purity of the synthesized compounds was established using HPLC and was >95%.
4.1.1. Synthesis of compound 2
To a solution of compound 1 (10 mmol) in dry DMF (20 mL) was added EDCI (2.40 g, 12.5 mmol, 1.25 eq), HOBt (1.92 g, 12.5 mmol, 1.25 eq) and the mixture was stirred for 30 minutes at room temperature. In a separate flask, a solution of deprotected glutamine surrogate 8 (2.23 g, 10 mmol) in DMF (15 mL) cooled to 0–5 °C was treated with N,N-diisopropylethylamine (DIEA) (9.5 g, 40 mmol, 4 eq), stirred for 30 minutes, and then added to the reaction mixture containing (L) N-Boc cyclohexylalanine. The reaction mixture was stirred for 12 h while monitoring the reaction by TLC. The solvent was removed and the residue was dissolved in ethyl acetate (200 mL) and washed with 10% aqueous citric acid (2 × 40 mL). The ethyl acetate layer was further washed with saturated NaHCO3 (40 mL), followed by saturated NaCl (50 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated to yield a yellow-colored oily product. Purification by flash chromatography yielded ester 2 as a white solid.
4.1.1.1. Methyl (S)-2-((S)-2-((tert-butoxycarbonyl)amino)-3-cyclohexylpropanamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 2
White solid (yield 77%); M.p 72–74 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.84 – 1.04 (m, 2 H), 1.11 – 1.31 (m, 4 H), 1.44 (s, 9 H), 1.60 – 1.75 (m, 6 H), 1.78 – 1.94 (m, 3 H), 2.15 – 2.28 (m, 1 H), 2.35 – 2.48 (m, 2 H), 3.26 – 3.40 (m, 2 H), 3.73 (s, 3 H), 4.21 – 4.34 (m, 1 H), 4.53 (ddd, J=11.13, 7.37, 3.91 Hz, 1 H), 5.03 (d, J=7.23 Hz, 1 H), 6.34 (s, 1 H), 7.55 (d, J=5.66 Hz, 1 H). HRMS (ESI) calcd for C22H37N3O6: [M-]: 439.2682. Found: 439.2866.
4.1.2. Synthesis of compound 3
To a solution of compound 2 (10 mmol) in dry DCM (5 mL) was added 4M HCl in dioxane (10 mL) and the mixture was stirred for 2 h at room temperature. The solvent was removed and the residue was dried under high vacuum to yield the hydrochloride salt as a pale white solid.
4.1.2.1. Methyl (S)-2-((S)-2-amino-3-cyclohexylpropanamido)-3-((S)-2-oxopyrroli din-3-yl)propanoate hydrochloride 3
Pale white solid (yield 100%); M.p 60–62 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.87 (quin, J=10.76 Hz, 2 H), 1.09 – 1.29 (m, 3 H), 1.34 – 1.45 (m, 2 H), 1.52 – 1.60 (m, 2 H), 1.65 (d, J=8.25 Hz, 4 H), 1.80 (d, J=12.03 Hz, 1 H), 2.01 – 2.13 (m, 1 H), 2.12 – 2.24 (m, 2 H), 2.38 – 2.48 (m, 2 H), 3.07 – 3.23 (m, 2 H), 3.58 (s, 3 H), 3.79 (d, J=5.30 Hz, 1 H), 4.25 (br. s, 1 H), 4.36 – 4.46 (m, 1 H), 7.69 (s, 1 H), 8.33 (s, 1 H), 9.00 (d, J=7.74 Hz, 1 H). HRMS (ESI) calcd for C17H30ClN3O4: [M-]: 375.1925. Found: 375.1920.
4.1.3. Synthesis of compounds 4a–m
To a solution of compound 3 (5 mmol) in dry DCM (20 mL) kept at 0 °C was added triethylamine (TEA) (1.3 mL,11 mmol, 2.2 eq) slowly with stirring. The appropriate sulfonyl chloride derivative (5.5 mmol, 1.1 eq) was added and reaction mixture was stirred for 12 h at 35 °C. The solvent was removed and the residue was dissolved in ethyl acetate (50 mL) and washed with water (2 × 25 mL). The ethyl acetate layer was further washed with saturated NaCl (50 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated to yield a crude product. Purification by flash chromatography yielded esters 4a–m.
4.1.3.1. Methyl (S)-2-((S)-3-cyclohexyl-2-(phenylsulfonamido)propanamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4a
White solid (yield 73%); M.p 45–47 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.67 – 0.79 (m, 1 H), 0.82 – 0.94 (m, 1 H), 0.98 – 1.07 (m, 2 H), 1.10 – 1.22 (m, 1 H), 1.31 – 1.41 (m, 1 H), 1.43 – 1.65 (m, 6 H), 1.73 – 1.88 (m, 2 H), 2.02 – 2.13 (m, 2 H), 2.19 – 2.29 (m, 1 H), 2.34 (ddd, J=12.40, 8.40, 4.00 Hz, 1 H), 3.38 (dd, J=8.93, 4.74 Hz, 2 H), 3.70 (s, 3 H), 3.84 (td, J=9.30, 4.93 Hz, 1 H), 4.27 (ddd, J=11.23, 6.98, 3.86 Hz, 1 H), 5.30 (s, 1 H), 5.97 (d, J=9.18 Hz, 1 H), 6.44 (s, 1 H), 7.44 – 7.50 (m, 2 H), 7.51 – 7.57 (m, 1 H), 7.87 (d, J=7.23 Hz, 2 H). HRMS (ESI) calcd for C23H33N3O6S: [M+H]: 480.2168 Found: 480.2147.
4.1.3.2. Methyl (S)-2-((S)-3-cyclohexyl-2-((phenylmethyl)sulfonamido)propan amido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4b
White solid (yield 77%); M.p 41–43 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.82 – 0.98 (m, 2 H), 1.07 – 1.30 (m, 3 H), 1.42 – 1.51 (m, 2 H), 1.59 – 1.71 (m, 5 H), 1.77 (d, J=12.45 Hz, 1 H), 1.82 – 1.96 (m, 2 H), 2.17 (ddd, J=14.13, 11.74, 5.66 Hz, 1 H), 2.34 – 2.44 (m, 1 H), 2.45 – 2.54 (m, 1 H), 3.31 – 3.39 (m, 2 H), 3.73 (s, 3 H), 3.94 – 4.04 (m, 1 H), 4.22 – 4.33 (m, 2 H), 4.44 – 4.53 (m, 1 H), 5.51 (d, J=8.98 Hz, 1 H), 6.15 (s, 1 H), 7.33 – 7.39 (m, 3 H), 7.41 – 7.47 (m, 2 H), 7.99 (d, J=6.74 Hz, 1 H). HRMS (ESI) calcd for C24H35N3O6S: [M+H]: 494.2325 Found: 494.2302.
4.1.3.3. Methyl (S)-2-((S)-3-cyclohexyl-2-((2-phenylethyl)sulfonamido)propan amido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4c
White solid (yield 82%); M.p 45–47 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.84 – 1.06 (m, 2 H), 1.09 – 1.30 (m, 4 H), 1.47 – 1.59 (m, 2 H), 1.61 – 1.74 (m, 4 H), 1.75 – 1.94 (m, 3 H), 2.13 (ddd, J=14.08, 11.96, 5.69 Hz, 1 H), 2.19 – 2.29 (m, 1 H), 2.30 – 2.42 (m, 1 H), 3.06 – 3.16 (m, 2 H), 3.17 – 3.34 (m, 4 H), 3.64 (s, 3 H), 4.10 (td, J=9.02, 4.86 Hz, 1 H), 4.39 – 4.48 (m, 1 H), 5.68 (d, J=9.47 Hz, 1 H), 6.24 (s, 1 H), 7.17 – 7.25 (m, 2 H), 7.25 – 7.35 (m, 3 H), 8.15 (d, J=6.79 Hz, 1 H). HRMS (ESI) calcd for C25H37N3O6S: [M+H]: 508.2481 Found: 508.2460.
4.1.3.4. Methyl (S)-2-((S)-3-cyclohexyl-2-((4-fluorophenyl)sulfonamido)propan amido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4d
White solid (yield 78%); M.p 63–65 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.76 (d, J=10.40 Hz, 1 H), 0.85 – 0.96 (m, 1 H), 1.08 (d, J=9.52 Hz, 2 H), 1.13 – 1.22 (m, 1 H), 1.35 – 1.55 (m, 2 H), 1.55 – 1.66 (m, 5 H), 1.71 (br. s., 1 H), 1.79 – 1.92 (m, 2 H), 1.99 – 2.11 (m, 1 H), 2.19 – 2.28 (m, 1 H), 2.36 (ddd, J=12.55, 8.47, 4.37 Hz, 1 H), 3.39 (dd, J=8.93, 4.64 Hz, 2 H), 3.70 (s, 3 H), 3.81 (td, J=9.28, 4.93 Hz, 1 H), 4.21 – 4.29 (m, 1 H), 6.00 (d, J=9.23 Hz, 1 H), 6.31 (s, 1 H), 7.15 (t, J=8.54 Hz, 2 H), 7.86 – 7.92 (m, 2 H), 7.95 (d, J=6.59 Hz, 1 H). HRMS (ESI) calcd for C23H32FN3O6S: [M+H]: 498.2074 Found: 498.2050.
4.1.3.5. Methyl (S)-2-((S)-2-((3-chlorophenyl)sulfonamido)-3-cyclohexylpropan amido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4e
White solid (yield 81%); M.p 58–59 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.73 – 0.82 (m, 1 H), 0.84 – 0.94 (m, 1 H), 1.06 (t, J=8.98 Hz, 2 H), 1.13 – 1.20 (m, 1 H), 1.40 (d, J=16.50 Hz, 1 H), 1.44 – 1.50 (m, 1 H), 1.51 – 1.65 (m, 4 H), 1.72 (br. s., 2 H), 1.79 – 1.89 (m, 2 H), 1.97 – 2.08 (m, 1 H), 2.23 – 2.30 (m, 1 H), 2.35 (d, J=11.52 Hz, 1 H), 3.35 – 3.42 (m, 2 H), 3.68 (s, 3 H), 3.85 (d, J=4.20 Hz, 1 H), 4.15 – 4.23 (m, 1 H), 5.98 (d, J=9.08 Hz, 1 H), 6.27 (s, 1 H), 7.40 – 7.45 (m, 1 H), 7.47 – 7.53 (m, 1 H), 7.75 (d, J=7.62 Hz, 1 H), 7.83 (s, 1 H), 8.16 (d, J=5.96 Hz, 1 H). HRMS (ESI) calcd for C23H32ClN3O6S: [M+H]: 514.1779 Found: 514.1738.
4.1.3.6. Methyl (S)-2-((S)-2-((4-chlorophenyl)sulfonamido)-3-cyclohexylpropan amido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4f
White solid (yield 76%); M.p 52–54 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.70 – 0.83 (m, 1 H), 0.89 (q, J=11.00 Hz, 1 H), 1.03 – 1.12 (m, 1 H), 1.13 – 1.22 (m, 1 H), 1.37 – 1.54 (m, 2 H), 1.57 – 1.68 (m, 5 H), 1.71 (br. s., 1 H), 1.80 – 1.91 (m, 2 H), 1.97 – 2.09 (m, 2 H), 2.17 – 2.26 (m, 1 H), 2.32 – 2.45 (m, 1 H), 3.36 – 3.46 (m, 2 H), 3.70 (s, 3 H), 3.80 (td, J=9.25, 4.98 Hz, 1 H), 4.16 – 4.25 (m, 1 H), 6.00 (d, J=9.28 Hz, 1 H), 6.30 (s, 1 H), 7.44 (d, J=8.59 Hz, 2 H), 7.81 (d, J=8.59 Hz, 2 H), 8.04 (d, J=6.35 Hz, 1 H). HRMS (ESI) calcd for C23H32ClN3O6S: [M+H]: 514.1779 Found: 514.1770.
4.1.3.7. Methyl (S)-2-((S)-2-(((4-chlorophenyl)methyl)sulfonamido)-3-cyclohexyl propanamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4g
White solid (yield 71%); M.p 38–39 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.83 – 0.98 (m, 2 H), 1.08 – 1.29 (m, 3 H), 1.40 – 1.52 (m, 2 H), 1.60 – 1.71 (m, 5 H), 1.77 (d, J=12.30 Hz, 1 H), 1.84 – 1.98 (m, 2 H), 2.09 – 2.20 (m, 1 H), 2.34 – 2.45 (m, 1 H), 2.46 – 2.53 (m, 1 H), 3.37 (dd, J=8.81, 4.66 Hz, 2 H), 3.73 (s, 3 H), 3.95 – 4.03 (m, 1 H), 4.18 – 4.31 (m, 2 H), 4.40 – 4.48 (m, 1 H), 5.67 (d, J=8.93 Hz, 1 H), 6.21 (s, 1 H), 7.32 – 7.35 (m, 2 H), 7.38 – 7.42 (m, 2 H), 8.10 (d, J=6.49 Hz, 1 H). HRMS (ESI) calcd for C24H34ClN3O6S: [M+H]: 528.1835 Found: 528.1879.
4.1.3.8. Methyl (S)-2-((S)-2-((2-(3-chlorophenyl)ethyl)sulfonamido)-3-cyclohexyl propanamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4h
White solid (yield 71%); M.p 63–65 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.85 – 1.05 (m, 2 H), 1.12 – 1.31 (m, 3 H), 1.49 – 1.58 (m, 2 H), 1.62 – 1.75 (m, 6 H), 1.79 – 1.96 (m, 2 H), 2.13 (ddd, J=14.10, 12.00, 5.79 Hz, 1 H), 2.25 – 2.34 (m, 1 H), 2.36 – 2.45 (m, 1 H), 3.03 – 3.16 (m, 2 H), 3.21 – 3.29 (m, 2 H), 3.30 – 3.39 (m, 2 H), 3.68 (s, 3 H), 4.05 – 4.16 (m, 1 H), 4.39 – 4.47 (m, 1 H), 5.80 (d, J=9.52 Hz, 1 H), 6.28 (s, 1 H), 7.10 (d, J=6.83 Hz, 1 H), 7.19 – 7.26 (m, 3 H), 8.24 (d, J=6.69 Hz, 1 H). HRMS (ESI) calcd for C25H36ClN3O6S: [M+H]: 542.2092 Found: 542.2033.
4.1.3.9. Methyl (S)-2-((S)-3-cyclohexyl-2-(thiophene-2-sulfonamido)propanamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4i
White solid (yield 62%); M.p 45–47 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.72 – 0.85 (m, 1 H), 0.86 – 0.95 (m, 1 H), 1.04 – 1.15 (m, 2 H), 1.16 – 1.27 (m, 1 H), 1.35 – 1.46 (m, 1 H), 1.50 (dd, J=9.45, 4.76 Hz, 1 H), 1.54 – 1.67 (m, 5 H), 1.79 – 1.92 (m, 3 H), 2.04 – 2.18 (m, 1 H), 2.28 – 2.42 (m, 2 H), 3.35 – 3.41 (m, 2 H), 3.71 (s, 3 H), 3.95 (td, J=9.19, 4.91 Hz, 1 H), 4.34 (ddd, J=11.28, 7.08, 3.81 Hz, 1 H), 6.17 (d, J=8.93 Hz, 1 H), 6.53 (br. s., 1 H), 7.04 (dd, J=4.83, 3.91 Hz, 1 H), 7.56 (dd, J=4.96, 1.00 Hz, 1 H), 7.61 (dd, J=3.64, 1.05 Hz, 1 H), 7.89 (d, J=6.88 Hz, 1 H). HRMS (ESI) calcd for C21H31N3O6S2: [M+H]: 486.1733 Found: 486.1785.
4.1.3.10. Methyl (S)-2-((S)-2-([1,1′-biphenyl]-4-sulfonamido)-3-cyclohexylpropan amido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4j
White solid (yield 62%); M.p 89–92 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.73 (d, J=10.50 Hz, 1 H), 0.81 – 0.93 (m, 1 H), 0.98 – 1.07 (m, 2 H), 1.09 – 1.18 (m, 1 H), 1.31 – 1.43 (m, 1 H), 1.45 – 1.64 (m, 6 H), 1.67 – 1.84 (m, 3 H), 1.96 – 2.05 (m, 1 H), 2.08 – 2.19 (m, 2 H), 3.01 – 3.10 (m, 1 H), 3.17 – 3.25 (m, 1 H), 3.66 (s, 3 H), 3.81 (td, J=9.21, 4.86 Hz, 1 H), 4.16 – 4.24 (m, 1 H), 5.92 (d, J=9.13 Hz, 1 H), 6.18 (s, 1 H), 7.37 – 7.42 (m, 1 H), 7.43 – 7.48 (m, 2 H), 7.57 (d, J=7.32 Hz, 2 H), 7.66 (d, J=8.40 Hz, 2 H), 7.91 (d, J=8.40 Hz, 2 H), 8.00 (d, J=6.49 Hz, 1 H). HRMS (ESI) calcd for C29H37N3O6S: [M+H]: 556.2481 Found: 556.2430.
4.1.3.11. methyl (S)-2-((S)-3-cyclohexyl-2-((2-(1,3-dioxoisoindolin-2-yl)ethyl)sulfon amido)propanamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4k
White solid (yield 72%); M.p 59–60 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.85 – 1.03 (m, 2 H), 1.09 – 1.24 (m, 3 H), 1.58 (td, J=9.35, 5.74 Hz, 2 H), 1.63 – 1.77 (m, 5 H), 1.79 – 1.87 (m, 2 H), 1.94 (ddd, J=14.26, 7.49, 3.69 Hz, 1 H), 2.20 (ddd, J=14.04, 11.80, 5.77 Hz, 1 H), 2.35 – 2.45 (m, 1 H), 2.48 – 2.58 (m, 1 H), 3.30 – 3.47 (m, 4 H), 3.67 (s, 3 H), 4.03 – 4.19 (m, 2 H), 4.32 (dt, J=14.40, 7.03 Hz, 1 H), 4.43 – 4.54 (m, 1 H), 5.91 (d, J=9.11 Hz, 1 H), 6.37 (s, 1 H), 7.67 – 7.76 (m, 2 H), 7.81 – 7.89 (m, 2 H), 8.23 (d, J=6.71 Hz, 1 H). HRMS (ESI) calcd for C27H36N4O8S: [M+H]: 577.2332 Found: 577.2340.
4.1.3.12. Methyl (S)-2-((S)-3-cyclohexyl-2-((3-(4-methoxyphenoxy)propyl)sulfon amido)propanamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4l
White solid (yield 67%); M.p 49–51 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.86 – 1.02 (m, 2 H), 1.10 – 1.30 (m, 3 H), 1.47 – 1.59 (m, 2 H), 1.62 – 1.74 (m, 5 H), 1.79 – 1.90 (m, 2 H), 1.93 (dt, J=7.03, 3.56 Hz, 1 H), 2.13 (ddd, J=14.13, 11.79, 5.96 Hz, 1 H), 2.26 (dd, J=6.13, 3.25 Hz, 2 H), 2.31 – 2.39 (m, 1 H), 2.42 – 2.52 (m, 1 H), 3.17 – 3.26 (m, 2 H), 3.26 – 3.36 (m, 2 H), 3.70 (s, 3 H), 3.76 (s, 3 H), 4.00 (t, J=5.86 Hz, 2 H), 4.05 – 4.15 (m, 1 H), 4.39 – 4.50 (m, 1 H), 5.63 (d, J=9.42 Hz, 1 H), 6.16 (s, 1 H), 6.81 (s, 4 H), 8.23 (d, J=6.59 Hz, 1 H). HRMS (ESI) calcd for C27H41N3O8S: [M+H]: 568.2693 Found: 568.2633.
4.1.3.13. Methyl (S)-2-((S)-3-cyclohexyl-2-(octylsulfonamido)propanamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 4m
White solid (yield 75%); M.p 56–58 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.88 (t, J=6.79 Hz, 4 H) 0.91 – 1.04 (m, 2 H), 1.09 – 1.34 (m, 9 H), 1.36 – 1.44 (m, 1 H), 1.46 – 1.56 (m, 1 H), 1.58 – 1.65 (m, 6 H), 1.69 (d, J=14.06 Hz, 4 H), 1.77 – 1.89 (m, 2 H), 1.90 – 1.99 (m, 1 H), 2.09 – 2.21 (m, 1 H), 2.38 – 2.54 (m, 2 H), 3.00 (t, J=7.96 Hz, 2 H), 3.35 – 3.43 (m, 2 H), 3.73 (s, 3 H), 3.99 – 4.09 (m, 1 H), 4.42 – 4.50 (m, 1 H), 5.34 (d, J=9.32 Hz, 1 H), 6.04 (s, 1 H), 8.13 (d, J=6.54 Hz, 1 H). HRMS (ESI) calcd for C25H46N3O6S: [M+H]: 516.3107 Found: 516.3112.
4.1.4. Synthesis of alcohols 5a–m
To a solution of ester 4 (5 mmol) in anhydrous THF (30 mL) was added dropwise lithium borohydride (2M in THF, 7.5 mL, 15 mmol), followed by absolute ethyl alcohol (15 mL), and the reaction mixture was stirred at room temperature overnight. The reaction mixture was then acidified using 5% HCl and the pH was adjusted to ~2. Removal of the solvent left a residue which was taken up in ethyl acetate (100 mL) and the organic layer was washed with brine (25 mL), dried over anhydrous sodium sulfate, filtered, and concentrated to yield compounds 5a–m.
4.1.4.1. (S)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-(phenylsulfonamido)propanamide 5a
White solid (yield 95%); M.p 48–51 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.59 – 0.73 (m, 2 H), 0.76 – 0.94 (m, 2 H), 0.96 – 1.18 (m, 3 H), 1.31 (br. s, 1 H), 1.39 – 1.64 (m, 6 H), 1.75 – 1.87 (m, 1 H), 1.96 – 2.04 (m, 2 H), 2.30 – 2.44 (m, 2 H), 3.32 – 3.40 (m, 2 H), 3.42 – 3.49 (m, 1 H), 3.57 (dd, J=11.25, 3.25 Hz, 1 H), 3.69 – 3.78 (m, 1 H), 3.84 – 3.93 (m, 1 H), 6.28 (d, J=8.10 Hz, 1 H), 6.37 (s, 1 H), 7.48 – 7.53 (m, 2 H), 7.55 – 7.60 (m, 1 H), 7.71 (d, J=7.32 Hz, 1 H), 7.89 (d, J=7.47 Hz, 2 H). HRMS (ESI) calcd for C22H33N3O5S: [M+H]: 452.2219 Found: 452.2210.
4.1.4.2. (S)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-((phenylmethyl)sulfonamido)propanamide 5b
White solid (yield 94%); M.p 45–47 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.81 – 0.96 (m, 2 H), 1.07 – 1.23 (m, 4 H), 1.37 (br. s., 1 H), 1.43 – 1.52 (m, 1 H), 1.56 (dd, J=13.72, 6.74 Hz, 2 H), 1.62 – 1.76 (m, 6 H), 1.77 – 1.86 (m, 1 H), 2.38 (td, J=5.93, 3.47 Hz, 1 H), 2.46 (d, J=6.40 Hz, 1 H), 3.31 (d, J=7.62 Hz, 2 H), 3.54 – 3.60 (m, 1 H), 3.65 (d, J=8.49 Hz, 1 H), 3.83 – 3.93 (m, 1 H), 3.97 – 4.06 (m, 1 H), 4.26 (s, 2 H), 5.85 (d, J=7.08 Hz, 1 H), 6.26 (s, 1 H), 7.32 – 7.38 (m, 3 H), 7.38 – 7.43 (m, 2 H), 7.66 (d, J=7.18 Hz, 1 H). HRMS (ESI) calcd for C23H35N3O5S: [M+H]: 466.2376 Found: 466.2360.
4.1.4.3. (S)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-((2-phenylethyl)sulfonamido)propanamide 5c
White solid (yield 94%); M.p 49–50 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.84 – 1.01 (m, 2 H), 1.10 – 1.23 (m, 4 H), 1.55 (d, J=7.23 Hz, 2 H), 1.60 – 1.74 (m, 6 H), 1.77 – 1.87 (m, 2 H), 2.20 – 2.30 (m, 1 H), 2.33 – 2.42 (m, 1 H), 3.05 – 3.15 (m, 2 H), 3.18 – 3.33 (m, 2 H), 3.59 (d, J=18.50 Hz, 2 H), 3.83 (br. s., 1 H), 4.01 (d, J=4.39 Hz, 2 H), 4.14 – 4.22 (m, 2 H), 6.11 (d, J=5.42 Hz, 1 H), 6.45 (s, 1 H), 7.16 – 7.23 (m, 2 H), 7.23 – 7.33 (m, 3 H), 7.79 (d, J=6.59 Hz, 1 H). HRMS (ESI) calcd for C24H37N3O5S: [M+H]: 480.2532 Found: 480.2508.
4.1.4.4. (S)-3-cyclohexyl-2-((4-fluorophenyl)sulfonamido)-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 5d
White solid (yield 96%); M.p 66–68 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.63 – 0.77 (m, 1 H), 0.79 – 0.90 (m, 1 H), 0.91 – 1.18 (m, 3 H), 1.34 (d, J=11.81 Hz, 1 H), 1.42 – 1.52 (m, 2 H), 1.52 – 1.67 (m, 6 H), 1.79 – 1.89 (m, 1 H), 2.01 (d, J=6.40 Hz, 1 H), 2.27 – 2.42 (m, 2 H), 3.33 – 3.41 (m, 2 H), 3.46 – 3.55 (m, 2 H), 3.58 (br. s., 1 H), 3.68 – 3.78 (m, 1 H), 3.85 – 4.01 (m, 1 H), 6.33 (s, 1 H), 7.18 (t, J=8.28 Hz, 2 H), 7.80 (d, J=7.20 Hz, 1 H), 7.91 (dd, J=8.47, 5.00 Hz, 2 H), 8.25 (d, J=5.82 Hz, 1 H). HRMS (ESI) calcd for C22H32FN3O5S: [M+H]: 470.2125 Found: 470.2115.
4.1.4.5. (S)-2-((3-chlorophenyl)sulfonamido)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 5e
White solid (yield 94%); M.p 61–63 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.67 – 0.80 (m, 2 H), 0.86 (q, J=11.23 Hz, 1 H), 0.94 – 1.19 (m, 3 H), 1.38 – 1.51 (m, 3 H), 1.52 – 1.67 (m, 4 H), 1.84 (t, J=8.01 Hz, 2 H), 1.93 – 2.01 (m, 1 H), 2.32 – 2.43 (m, 2 H), 3.33 – 3.42 (m, 2 H), 3.44 – 3.50 (m, 1 H), 3.54 – 3.60 (m, 1 H), 3.77 (dt, J=8.86, 4.60 Hz, 1 H), 3.81 – 3.88 (m, 1 H), 4.21 (br. s, 1 H), 6.28 (s, 1 H), 7.45 (t, J=7.79 Hz, 1 H), 7.49 – 7.56 (m, 1 H), 7.78 (d, J=7.57 Hz, 1 H), 7.84 – 7.89 (m, 1 H), 7.97 (d, J=5.57 Hz, 1 H), 8.17 (d, J=7.20 Hz, 1 H). HRMS (ESI) calcd for C22H32ClN3O5S: [M+H]: 486.1829 Found: 486.1809.
4.1.4.6. (S)-2-((4-chlorophenyl)sulfonamido)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 5f
White solid (yield 93%); M.p 56–57 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.65 – 0.77 (m, 1 H), 0.78 – 0.93 (m, 1 H), 0.94 – 1.19 (m, 3 H), 1.34 – 1.42 (m, 1 H), 1.43 – 1.52 (m, 2 H), 1.53 – 1.64 (m, 5 H), 1.76 – 1.90 (m, 2 H), 1.91 – 2.02 (m, 1 H), 2.24 – 2.33 (m, 1 H), 2.38 (dd, J=8.30, 3.32 Hz, 1 H), 3.34 – 3.44 (m, 2 H), 3.45 – 3.52 (m, 1 H), 3.58 (d, J=8.15 Hz, 1 H), 3.82 (br. s, 1 H), 3.95 (d, J=3.91 Hz, 1 H), 4.17 – 4.25 (m, 1 H), 5.97 (d, J=5.20 Hz, 1 H), 6.33 (s, 1 H), 7.41 – 7.50 (m, 2 H), 7.79 – 7.89 (m, 2 H), 8.05 (d, J=7.20 Hz, 1 H). HRMS (ESI) calcd for C22H32ClN3O5S: [M+H]: 486.1829 Found: 486.1811.
4.1.4.7. (S)-2-(((4-chlorophenyl)methyl)sulfonamido)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 5g
White solid (yield 95%); M.p 47–48 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.82 – 0.95 (m, 2 H), 1.11 – 1.21 (m, 2 H), 1.37 (d, J=4.42 Hz, 1 H), 1.43 – 1.50 (m, 1 H), 1.53 – 1.61 (m, 3 H), 1.62 – 1.75 (m, 6 H), 1.83 (dd, J=11.42, 9.67 Hz, 1 H), 2.00 (br. s., 1 H), 2.35 – 2.52 (m, 2 H), 3.30 – 3.36 (m, 2 H), 3.55 – 3.61 (m, 1 H), 3.64 – 3.71 (m, 1 H), 3.84 – 3.93 (m, 1 H), 4.00 (d, J=3.56 Hz, 1 H), 4.23 (d, J=4.96 Hz, 2 H), 5.81 (d, J=8.40 Hz, 1 H), 6.11 (s, 1 H), 7.30 – 7.33 (m, 1 H), 7.34 – 7.36 (m, 3 H), 7.80 (d, J=7.03 Hz, 1 H). HRMS (ESI) calcd for C23H34ClN3O5S: [M+H]: 500.1986 Found: 500.1978.
4.1.4.8. (S)-2-((2-(3-chlorophenyl)ethyl)sulfonamido)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 5h
White solid (yield 96%); M.p 67–69 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.86 – 1.02 (m, 2 H), 1.09 – 1.21 (m, 2 H), 1.45 – 1.65 (m, 4 H), 1.70 (d, J=10.25 Hz, 5 H), 1.76 – 1.86 (m, 2 H), 1.99 (d, J=5.81 Hz, 1 H), 2.25 – 2.35 (m, 1 H), 2.35 – 2.45 (m, 1 H), 3.04 – 3.16 (m, 2 H), 3.19 – 3.28 (m, 2 H), 3.29 – 3.37 (m, 2 H), 3.49 (br. s, 1 H), 3.54 – 3.60 (m, 1 H), 3.62 – 3.69 (m, 1 H), 3.95 – 4.05 (m, 2 H), 5.87 (d, J=9.03 Hz, 1 H), 6.17 (s, 1 H), 7.10 (d, J=6.74 Hz, 1 H), 7.20 – 7.23 (m, 2 H), 7.24 (br. s., 1 H), 7.94 (d, J=7.03 Hz, 1 H). HRMS (ESI) calcd for C24H36ClN3O5S: [M+H]: 514.2142 Found: 514.2148.
4.1.4.9. (S)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-(thiophene-2-sulfonamido)propanamide 5i
White solid (yield 96%); M.p 48–50 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.65 – 0.84 (m, 2 H), 0.95 – 1.14 (m, 3 H), 1.29 – 1.38 (m, 4 H), 1.47 – 1.64 (m, 6 H), 1.67 – 1.78 (m, 1 H), 2.01 – 2.12 (m, 2 H), 3.04 – 3.19 (m, 2 H), 3.21 – 3.29 (m, 2 H), 3.63 (dd, J=6.96, 4.17 Hz, 1 H), 3.77 (q, J=7.53 Hz, 1 H), 4.65 (t, J=5.49 Hz, 1 H), 7.09 – 7.14 (m, 1 H), 7.53 – 7.54 (m, 1 H), 7.55 (s, 1 H), 7.73 (d, J=8.64 Hz, 1 H), 7.87 (dd, J=4.88, 1.03 Hz, 1 H), 8.08 (d, J=8.35 Hz, 1 H). HRMS (ESI) calcd for C20H31N3O5S2: [M+H]: 458.1783 Found: 458.1739.
4.1.4.10. (S)-2-([1,1′-biphenyl]-4-sulfonamido)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 5j
White solid (yield 92%); M.p 93–95 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.61 – 0.71 (m, 1 H), 0.72 – 0.82 (m, 1 H), 0.84 – 0.95 (m, 1 H), 0.98 – 1.12 (m, 2 H), 1.25 – 1.38 (m, 3 H), 1.40 – 1.53 (m, 4 H), 1.56 (d, J=9.28 Hz, 2 H), 1.66 – 1.79 (m, 2 H), 2.02 – 2.12 (m, 2 H), 2.86 – 2.97 (m, 1 H), 3.00 – 3.08 (m, 1 H), 3.09 – 3.16 (m, 1 H), 3.27 (dd, J=10.04, 4.70 Hz, 1 H), 3.56 – 3.66 (m, 1 H), 3.70 – 3.80 (m, 1 H), 4.64 (t, J=5.00 Hz, 1 H), 6.53 (s, 1 H), 7.41 – 7.46 (m, 1 H), 7.49 (d, J=5.30 Hz, 1 H), 7.51 – 7.55 (m, 2 H), 7.68 – 7.75 (m, 2 H), 7.82 – 7.89 (m, 4 H), 7.95 (d, J=8.37 Hz, 1 H). HRMS (ESI) calcd for C28H37N3O5S: [M+H]: 528.2532 Found: 528.2568.
4.1.4.11. (S)-3-cyclohexyl-2-((2-(1,3-dioxoisoindolin-2-yl)ethyl)sulfonamido)-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 5k
White solid (yield 94%); M.p 65–66 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.83 – 1.01 (m, 2 H), 1.06 – 1.21 (m, 4 H), 1.41 – 1.74 (m, 6 H), 1.80 (d, J=11.74 Hz, 2 H), 1.95 – 2.02 (m, 2 H), 2.30 – 2.51 (m, 2 H), 3.24 – 3.38 (m, 2 H), 3.50 – 3.59 (m, 3 H), 3.64 (d, J=4.15 Hz, 1 H), 3.84 – 3.97 (m, 1 H), 4.03 (br. s., 1 H), 4.21 – 4.34 (m, 1 H), 4.64 (s, 2 H), 5.88 (s, 1 H), 7.33 – 7.41 (m, 2 H), 7.48 – 7.56 (m, 1 H), 7.59 (d, J=6.18 Hz, 1 H), 7.71 – 7.76 (m, 1 H), 8.22 (d, J=7.30 Hz, 1 H). HRMS (ESI) calcd for C26H36N4O7S: [M+H]: 549.2383 Found: 549.2363.
4.1.4.12. (S)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-((3-(4-methoxyphenoxy)propyl)sulfonamido)propanamide 5l
White solid (yield 94%); M.p 58–61 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.87 – 1.02 (m, 2 H), 1.10 – 1.23 (m, 2 H), 1.42 – 1.58 (m, 3 H), 1.59 – 1.74 (m, 6 H), 1.78 – 1.87 (m, 2 H), 2.00 – 2.09 (m, 1 H), 2.20 – 2.30 (m, 2 H), 2.36 (ddd, J=9.18, 6.18, 3.34 Hz, 1 H), 2.41 – 2.51 (m, 1 H), 3.16 – 3.26 (m, 2 H), 3.27 – 3.33 (m, 2 H), 3.36 (d, J=6.05 Hz, 1 H), 3.54 – 3.61 (m, 1 H), 3.63 – 3.71 (m, 1 H), 3.76 (s, 3 H), 3.94 – 4.05 (m, 4 H), 5.75 (d, J=8.84 Hz, 1 H), 6.01 (s, 1 H), 6.78 – 6.86 (m, 4 H), 7.96 (d, J=6.98 Hz, 1 H). HRMS (ESI) calcd for C26H41N3O7S: [M+H]: 540.2743 Found: 540.2773.
4.1.4.13. (S)-3-cyclohexyl-N-((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-(octylsulfonamido)propanamide 5m
White solid (yield 91%); M.p 65–66 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.88 (t, J=6.71 Hz, 3 H) 0.93 – 1.03 (m, 2 H), 1.10 – 1.21 (m, 2 H), 1.22 – 1.33 (m, 10 H), 1.36 – 1.44 (m, 2 H), 1.44 – 1.51 (m, 1 H), 1.52 – 1.58 (m, 1 H), 1.59 – 1.75 (m, 5 H), 1.76 – 1.90 (m, 4 H), 2.01 – 2.11 (m, 2 H), 2.37 – 2.53 (m, 2 H), 2.99 (t, J=7.96 Hz, 2 H), 3.33 – 3.40 (m, 2 H), 3.54 – 3.61 (m, 1 H), 3.64 – 3.70 (m, 1 H), 3.93 (td, J=9.08, 5.22 Hz, 1 H), 3.97 – 4.06 (m, 1 H), 5.66 (d, J=8.98 Hz, 1 H), 6.12 (s, 1 H), 7.82 (d, J=7.27 Hz, 1 H). HRMS (ESI) calcd for C24H45N3O5S: [M+H]: 488.3158 Found: 488.3143.
4.1.5. Synthesis of aldehydes 6a–m
Compound 5 (5 mmol) was dissolved in anhydrous dichloromethane (50 mL) under a nitrogen atmosphere and cooled to 0 °C. Dess-Martin periodinane reagent (3.18 g, 7.5 mmol, 1.5 eq) was added to the reaction mixture with stirring. The ice bath was removed and the reaction mixture was stirred at room temperature for 3 h (monitoring by TLC indicated complete disappearance of the starting material). A solution of 10% aqueous sodium thiosulfate (20 mL) was added and the solution was stirred for 15 minutes. The aqueous layer was removed and the organic layer was washed with 10% aqueous sodium thiosulfate (20 mL), followed by saturated aqueous sodium bicarbonate (2 × 20 mL), water (2 × 20 mL) and brine (20 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated. The yellow residue was purified by flash chromatography using silica gel (methylene chloride/ethyl acetate/methanol) to yield a white solid 6a–m.
4.1.5.1. (S)-3-cyclohexyl-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-(phenylsulfonamido)propanamide 6a
White solid (yield 74%); M.p 69–71 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.73 (dd, J=11.52, 2.93 Hz, 1 H), 0.86 (td, J=11.91, 9.37 Hz, 1 H), 0.93 – 1.21 (m, 3 H), 1.24 – 1.36 (m, 1 H), 1.47 (ddd, J=14.16, 9.47, 4.88 Hz, 2 H), 1.51 – 1.66 (m, 4 H), 1.71 – 1.78 (m, 2 H), 1.81 – 1.92 (m, 2 H), 2.31 – 2.42 (m, 2 H), 3.34 – 3.43 (m, 2 H), 3.88 (td, J=9.28, 4.88 Hz, 1 H), 4.11 (dt, J=8.89, 6.10 Hz, 1 H), 6.14 (d, J=8.59 Hz, 1 H), 6.41 (s, 1 H), 7.45 – 7.58 (m, 3 H), 7.86 – 7.91 (m, 2 H), 8.36 (d, J=5.86 Hz, 1 H), 9.23 (d, J=0.78 Hz, 1 H). HRMS (ESI) calcd for C22H31N3O5S: [M-]: 449.1984. Found: 449.1981.
4.1.5.2. (S)-3-cyclohexyl-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-((phenylmethyl)sulfonamido)propanamide 6b
White solid (yield 76%); M.p 62–64 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.80 – 0.97 (m, 2 H), 1.09 – 1.30 (m, 3 H), 1.36 – 1.55 (m, 2 H), 1.57 – 1.79 (m, 6 H), 1.81 – 1.90 (m, 1 H), 1.92 – 2.02 (m, 2 H), 2.34 – 2.43 (m, 1 H), 2.47 – 2.57 (m, 1 H), 3.31 – 3.39 (m, 2 H), 3.98 (td, J=8.50, 6.05 Hz, 1 H), 4.22 – 4.32 (m, 2 H), 4.33 – 4.41 (m, 1 H), 5.61 (d, J=8.59 Hz, 1 H), 6.25 (s, 1 H), 7.33 – 7.38 (m, 3 H), 7.40 – 7.45 (m, 2 H), 8.26 (d, J=6.25 Hz, 1 H), 9.49 (s, 1 H). HRMS (ESI) calcd for C23H33N3O5S: [M-]: 463.2141 Found: 463.2143.
4.1.5.3. (S)-3-cyclohexyl-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-((2-phenylethyl)sulfonamido)propanamide 6c
White solid (yield 74%); M.p 60–62 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.84 – 1.04 (m, 2 H), 1.09 – 1.30 (m, 2 H), 1.43 – 1.61 (m, 2 H), 1.62 – 1.76 (m, 6 H), 1.78 – 1.89 (m, 2 H), 1.89 – 2.07 (m, 2 H), 2.20 – 2.33 (m, 1 H), 2.34 – 2.46 (m, 1 H), 3.02 – 3.17 (m, 2 H), 3.19 – 3.43 (m, 4 H), 4.12 (td, J=8.96, 5.42 Hz, 1 H), 4.33 (dt, J=10.23, 5.28 Hz, 1 H), 5.72 (d, J=9.08 Hz, 1 H), 6.30 (br. s., 1 H), 7.16 – 7.25 (m, 3 H), 7.26 – 7.34 (m, 2 H), 8.45 (d, J=5.96 Hz, 1 H), 9.48 (s, 1 H). HRMS (ESI) calcd for C24H35N3O5S: [M-]: 477.2297. Found: 477.2296.
4.1.5.4. (S)-3-cyclohexyl-2-((4-fluorophenyl)sulfonamido)-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 6d
White solid (yield 68%); M.p 68–70 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.64 – 0.91 (m, 2 H), 0.95 – 1.20 (m, 3 H), 1.29 – 1.38 (m, 1 H), 1.42 – 1.66 (m, 6 H), 1.71 – 1.77 (m, 1 H), 1.81 – 1.92 (m, 2 H), 2.28 – 2.47 (m, 2 H), 3.34 – 3.44 (m, 2 H), 3.79 – 3.89 (m, 1 H), 4.12 (q, J=7.08 Hz, 1 H), 6.15 (d, J=8.69 Hz, 1 H), 6.35 (br. s., 1 H), 7.11 – 7.23 (m, 2 H), 7.87 – 7.95 (m, 3 H), 8.44 (d, J=5.52 Hz, 1 H), 9.33 (s, 1 H). HRMS (ESI) calcd for C22H30N3O5FS: [M-]: 467.1890. Found: 467.1889.
4.1.5.5. (S)-2-((3-chlorophenyl)sulfonamido)-3-cyclohexyl-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 6e
White solid (yield 76%); M.p 52–53 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.70 – 0.96 (m, 3 H), 1.00 – 1.23 (m, 4 H), 1.33 – 1.43 (m, 1 H), 1.45 – 1.71 (m, 6 H), 1.80 – 1.91 (m, 2 H), 2.33 – 2.48 (m, 2 H), 3.35 – 3.45 (m, 2 H), 3.85 – 3.95 (m, 1 H), 3.99 – 4.09 (m, 1 H), 6.11 (d, J=8.84 Hz, 1 H), 6.27 (br. s., 1 H), 7.41 – 7.49 (m, 1 H), 7.49 – 7.57 (m, 1 H), 7.75 – 7.81 (m, 1 H), 7.85 – 7.90 (m, 1 H), 8.67 (d, J=4.98 Hz, 1 H), 9.27 (s, 1 H). HRMS (ESI) calcd for C22H30N3O5ClS: [M-]: 483.1595. Found: 483.1593.
4.1.5.6. (S)-2-((4-chlorophenyl)sulfonamido)-3-cyclohexyl-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 6f
White solid (yield 76%); M.p 72–74 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.69 – 0.93 (m, 2 H), 0.97 – 1.22 (m, 4 H), 1.25 – 1.38 (m, 1 H), 1.43 – 1.72 (m, 6 H), 1.81 – 1.91 (m, 2 H), 2.02 – 2.15 (m, 1 H), 2.24 – 2.34 (m, 1 H), 2.35 – 2.44 (m, 1 H), 3.36 – 3.47 (m, 2 H), 3.83 (td, J=9.01, 5.18 Hz, 1 H), 4.03 – 4.15 (m, 1 H), 6.26 (d, J=8.69 Hz, 1 H), 6.38 (s, 1 H), 7.41 – 7.53 (m, 2 H), 7.82 (d, J=8.54 Hz, 2 H), 8.51 (d, J=5.37 Hz, 1 H), 9.33 (s, 1 H). HRMS (ESI) calcd for C22H30N3O5ClS: [M-]: 483.1595. Found: 483.1591.
4.1.5.7. (S)-2-(((4-chlorophenyl)methyl)sulfonamido)-3-cyclohexyl-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 6g
White solid (yield 75%); M.p 52–53 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.80 – 0.97 (m, 2 H), 1.10 – 1.31 (m, 4 H), 1.36 – 1.54 (m, 1 H), 1.56 – 1.78 (m, 6 H), 1.80 – 1.90 (m, 1 H), 1.92 – 1.98 (m, 2 H), 2.36 – 2.46 (m, 1 H), 2.47 – 2.57 (m, 1 H), 3.32 – 3.42 (m, 2 H), 3.92 – 4.00 (m, 1 H), 4.12 (q, J=7.14 Hz, 2 H), 4.28 – 4.37 (m, 1 H), 5.69 (d, J=8.59 Hz, 1 H), 6.23 (s, 1 H), 7.30 – 7.43 (m, 4 H), 8.39 (d, J=5.66 Hz, 1 H), 9.49 (s, 1 H). HRMS (ESI) calcd for C23H33N3O5ClS: [M-]: 497.1751. Found: 497.1754.
4.1.5.8. (S)-2-((2-(3-chlorophenyl)ethyl)sulfonamido)-3-cyclohexyl-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 6h
White solid (yield 73%); M.p 53–55 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.84 – 1.03 (m, 2 H), 1.08 – 1.31 (m, 4 H), 1.43 – 1.57 (m, 2 H), 1.60 – 1.86 (m, 6 H), 1.89 – 1.97 (m, 2 H), 2.24 – 2.36 (m, 1 H), 2.36 – 2.48 (m, 1 H), 3.00 – 3.13 (m, 2 H), 3.16 – 3.41 (m, 4 H), 4.06 – 4.14 (m, 1 H), 4.27 – 4.36 (m, 1 H), 5.71 (d, J=9.28 Hz, 1 H), 6.22 (s, 1 H), 7.08 (d, J=6.83 Hz, 1 H), 7.16 – 7.24 (m, 3 H), 8.56 (d, J=5.76 Hz, 1 H), 9.47 (s, 1 H). HRMS (ESI) calcd for C24H34N3O5ClS: [M-]: 511.1908. Found: 511.1907.
4.1.5.9. (S)-3-cyclohexyl-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-2-(thiophene-2-sulfonamido)propanamide 6i
White solid (yield 75%); M.p 68–69 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.70 – 0.95 (m, 2 H), 1.01 – 1.25 (m, 3 H), 1.30 – 1.41 (m, 1 H), 1.46 – 1.70 (m, 6 H), 1.74 – 2.00 (m, 4 H), 2.32 – 2.51 (m, 2 H), 3.40 (d, J=9.08 Hz, 2 H), 3.98 (td, J=9.01, 4.64 Hz, 1 H), 4.15 – 4.25 (m, 1 H), 6.32 (d, J=8.49 Hz, 1 H), 6.48 (br. s., 1 H), 7.03 – 7.10 (m, 1 H), 7.55 – 7.60 (m, 1 H), 7.63 (dd, J=3.52, 0.98 Hz, 1 H), 8.35 (d, J=5.86 Hz, 1 H), 9.31 (s, 1 H). HRMS (ESI) calcd for C20H29N3O5S2: [M-]: 455.1549. Found: 455.1546.
4.1.5.10. (S)-2-([1,1′-biphenyl]-4-sulfonamido)-3-cyclohexyl-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 6j
White solid (yield 75%); M.p 56–58 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.68 – 0.80 (m, 2 H), 0.84 – 0.90 (m, 1 H), 0.91 – 0.97 (m, 1 H), 0.98 – 1.08 (m, 1 H), 1.09 – 1.20 (m, 2 H), 1.31 – 1.44 (m, 1 H), 1.45 – 1.68 (m, 6 H), 1.72 – 1.81 (m, 1 H), 1.81 – 1.89 (m, 1 H), 1.90 – 2.00 (m, 1 H), 2.17 – 2.37 (m, 2 H), 3.15 – 3.25 (m, 1 H), 3.27 – 3.38 (m, 1 H), 3.87 (td, J=9.00, 4.91 Hz, 1 H), 4.04 – 4.16 (m, 1 H), 6.13 (d, J=8.45 Hz, 1 H), 6.37 (br. s., 1 H), 7.39 – 7.44 (m, 1 H), 7.45 – 7.51 (m, 1 H), 7.58 (d, J=7.13 Hz, 2 H), 7.68 (d, J=8.35 Hz, 2 H), 7.94 (d, J=8.35 Hz, 2 H), 8.44 (d, J=5.61 Hz, 1 H), 9.30 (s, 1 H). HRMS (ESI) calcd for C28H35N3O5S: [M-]: 525.2297. Found: 525.2297.
4.1.5.11. (S)-3-cyclohexyl-2-((2-(1,3-dioxoisoindolin-2-yl)ethyl)sulfonamido)-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 6k
White solid (yield 73%); M.p 59–61 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.80 – 1.04 (m, 2 H), 1.09 – 1.28 (m, 3 H), 1.43 – 1.73 (m, 6 H), 1.80 (br. s., 2 H), 1.93 – 2.02 (m, 1 H), 2.16 – 2.23 (m, 1 H), 2.31 – 2.46 (m, 2 H), 2.48 – 2.62 (m, 1 H), 3.24 – 3.55 (m, 4 H), 3.63 – 3.76 (m, 1 H), 4.04 – 4.19 (m, 2 H), 4.25 – 4.40 (m, 1 H), 5.81 (d, J=8.69 Hz, 1 H), 6.23 (t, J=7.64 Hz, 1 H), 7.45 – 7.61 (m, 1 H), 7.67 – 7.79 (m, 1 H), 7.84 (dd, J=5.37, 3.03 Hz, 1 H), 7.94 (d, J=7.62 Hz, 1 H), 8.57 (d, J=5.86 Hz, 1 H), 9.48 (s, 1 H). HRMS (ESI) calcd for C26H34N4O7S: [M-]: 546.2148. Found: 546.2144.
4.1.5.12. (S)-3-cyclohexyl-2-((3-(4-methoxyphenoxy)propyl)sulfonamido)-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 6l
White solid (yield 73%); M.p 47–49 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.85 – 1.03 (m, 2 H), 1.06 – 1.31 (m, 2 H), 1.44 – 1.60 (m, 2 H), 1.62 – 1.76 (m, 6 H), 1.79 – 1.88 (m, 2 H), 1.91 – 2.00 (m, 2 H), 2.22 – 2.31 (m, 2 H), 2.31 – 2.41 (m, 1 H), 2.44 – 2.55 (m, 1 H), 3.15 – 3.28 (m, 2 H), 3.28 – 3.39 (m, 2 H), 3.76 (s, 3 H), 3.96 – 4.03 (m, 2 H), 4.09 (td, J=9.01, 5.42 Hz, 1 H), 4.29 – 4.37 (m, 1 H), 5.58 (d, J=9.18 Hz, 1 H), 6.11 (s, 1 H), 6.77 – 6.87 (m, 4 H), 8.53 (d, J=5.66 Hz, 1 H), 9.49 (s, 1 H). HRMS (ESI) calcd for C26H39N3O7S: [M-]: 537.2509. Found: 537.2508.
4.1.5.13. (S)-3-cyclohexyl-2-(octylsulfonamido)-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)propanamide 6m
sticky solid (yield 74%); 1H NMR (400 MHz, CDCl3-d): δ ppm 0.84 – 1.03 (m, 4 H), 1.11 – 1.35 (m, 8 H), 1.38 (d, J=6.44 Hz, 2 H), 1.48 – 1.61 (m, 3 H), 1.62 – 1.77 (m, 8 H), 1.77 – 1.89 (m, 4 H), 1.94 – 2.02 (m, 2 H), 2.38 – 2.48 (m, 1 H), 2.48 – 2.59 (m, 1 H), 3.00 (t, J=7.96 Hz, 2 H), 3.34 – 3.43 (m, 2 H), 4.05 (td, J=9.01, 5.22 Hz, 1 H), 4.33 – 4.41 (m, 1 H), 5.52 (d, J=9.18 Hz, 1 H), 6.21 (s, 1 H), 8.38 (d, J=5.96 Hz, 1 H), 9.51 (s, 1 H). HRMS (ESI) calcd for C24H43N3O5S: [M-]: 485.2923. Found: 485.2922.
4.1.6. Synthesis of bisulfite adducts 7a–m
To a solution of aldehyde 6 (5 mmol) in dry ethyl acetate (20 mL) was added absolute ethanol (12 mL) with stirring, followed by a solution of sodium bisulfite (540 mg; 5 mmol) in water (5 mL). The reaction mixture was stirred for 3 h at 50 °C and then allowed to cool to room temperature. The precipitate was vacuum filtered and the solid was thoroughly washed with absolute ethanol. The filtrate was dried over anhydrous sodium sulfate, filtered, and concentrated to yield a yellowish oil which was treated with ethyl ether (2 × 50 mL) to form a white solid. The white solid was stirred with ethyl ether (30 mL) and ethyl acetate (15 mL) for 5 minutes. Careful removal of the solvent using a pipette yielded compounds 7a–m.
4.1.6.1. Sodium (2S)-2-((S)-3-cyclohexyl-2-(phenylsulfonamido)propanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7a
White solid (yield 68%); M.p 154–156 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.57 – 0.82 (m, 2 H), 0.91 (d, J=9.18 Hz, 1 H), 1.00 – 1.14 (m, 2 H), 1.16 – 1.25 (m, 1 H), 1.29 (d, J=8.40 Hz, 1 H), 1.51 (d, J=11.33 Hz, 6 H), 1.68 – 1.87 (m, 1 H), 1.89 – 2.10 (m, 2 H), 2.95 – 3.15 (m, 2 H), 3.71 – 3.81 (m, 2 H), 3.87 (d, J=3.91 Hz, 1 H), 4.07 (t, J=9.23 Hz, 1 H), 5.35 (d, J=5.57 Hz, 1 H), 5.41 (d, J=5.86 Hz, 1 H), 7.45 (br. s., 1 H), 7.49 – 7.65 (m, 4 H), 7.71 – 7.93 (m, 3 H). HRMS (ESI) calcd for C22H32N3O8S2Na: [M-]: 553.1529. Found: 553.1531.
4.1.6.2. Sodium (2S)-2-((S)-3-cyclohexyl-2-((phenylmethyl)sulfonamido)propan amido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7b
White solid (yield 76%); M.p 137–139 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.85 (br. s., 2 H), 1.01 – 1.28 (m, 3 H), 1.31 – 1.48 (m, 2 H), 1.51 – 1.83 (m, 6 H), 1.86 – 2.26 (m, 4 H), 2.90 – 3.03 (m, 1 H), 3.11 (d, J=5.86 Hz, 1 H), 3.84 – 4.03 (m, 2 H), 4.19 – 4.30 (m, 1 H), 4.33 (s, 1 H), 5.46 (d, J=5.47 Hz, 1 H), 5.57 (d, J=6.25 Hz, 1 H), 7.35 (br. s., 5 H), 7.46 (d, J=8.20 Hz, 1 H), 7.51 (br. s., 1 H), 7.73 (d, J=8.98 Hz, 1 H), 7.97 (d, J=8.98 Hz, 1 H). HRMS (ESI) calcd for C23H34N3O8S2Na: [M-]: 567.1685. Found: 567.1682.
4.1.6.3. Sodium (2S)-2-((S)-3-cyclohexyl-2-((2-phenylethyl)sulfonamido)propan amido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7c
White solid (yield 65%); M.p 74–76 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.78 – 0.98 (m, 2 H), 1.04 – 1.29 (m, 4 H), 1.44 (br. s., 3 H), 1.65 (br. s., 4 H), 1.76 – 1.93 (m, 2 H), 2.00 (br. s., 1 H), 2.16 (br. s., 1 H), 2.69 (d, J=8.40 Hz, 1 H), 2.90 (d, J=9.96 Hz, 2 H), 3.05 (br. s., 4 H), 3.84 (br. s., 1 H), 3.96 (br. s., 1 H), 4.17 – 4.30 (m, 1 H), 7.26 (d, J=4.59 Hz, 5 H), 7.47 (br. s., 1 H), 7.59 – 7.64 (m, 1 H), 7.94 (br. s., 1 H), 8.64 (d, J=7.03 Hz, 1 H). HRMS (ESI) calcd for C24H36N3O8S2Na: [M-]: 581.1842. Found: 581.1841.
4.1.6.4. Sodium (2S)-2-((S)-3-cyclohexyl-2-((4-fluorophenyl)sulfonamido)propan amido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7d
White solid (yield 78%); M.p 75–77 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.64 – 0.84 (m, 2 H), 0.88 – 0.99 (m, 1 H), 1.04 – 1.13 (m, 3 H), 1.16 – 1.26 (m, 2 H), 1.28 – 1.39 (m, 2 H), 1.55 (d, J=9.57 Hz, 4 H), 1.69 – 1.85 (m, 1 H), 1.89 – 1.96 (m, 1 H), 2.05 – 2.14 (m, 1 H), 3.00 – 3.16 (m, 2 H), 3.54 – 3.63 (m, 1 H), 3.70 – 3.81 (m, 1 H), 3.98 – 4.08 (m, 1 H), 5.31 (dd, J=16.23, 5.93 Hz, 1 H), 7.32 – 7.46 (m, 2 H), 7.53 (d, J=4.74 Hz, 1 H), 7.63 – 7.70 (m, 1 H), 7.74 (s, 1 H), 7.79 – 7.90 (m, 2 H), 7.95 (br. s., 1 H). HRMS (ESI) calcd for C22H31N3O8FS2Na: [M-]: 571.1434. Found: 571.1435.
4.1.6.5. Sodium (2S)-2-((S)-2-((3-chlorophenyl)sulfonamido)-3-cyclohexylpropan amido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (7e)
White solid (yield 78%); M.p 74–75 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.64 – 0.84 (m, 3 H), 0.91 (d, J=6.40 Hz, 1 H), 1.01 – 1.21 (m, 4 H), 1.28 – 1.37 (m, 1 H), 1.42 – 1.63 (m, 4 H), 1.67 – 1.87 (m, 2 H), 1.88 – 2.14 (m, 2 H), 3.01 – 3.21 (m, 2 H), 3.75 – 3.90 (m, 1 H), 4.01 – 4.15 (m, 1 H), 4.26 – 4.42 (m, 1 H), 5.27 (d, J=5.27 Hz, 1 H), 6.02 – 6.10 (m, 1 H), 7.44 (br. s., 1 H), 7.50 – 7.60 (m, 1 H), 7.61 – 7.69 (m, 1 H), 7.70 – 7.75 (m, 1 H), 7.76 – 7.83 (m, 1 H), 8.05 – 8.17 (m, 1 H), 8.48 – 8.60 (m, 1 H). HRMS (ESI) calcd for C22H31N3O8ClS2Na: [M-]: 587.1139. Found: 587.1134.
4.1.6.6. Sodium (2S)-2-((S)-2-((4-chlorophenyl)sulfonamido)-3-cyclohexylpropan amido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7f
White soild (yield 61%); M.p 152–153 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.64 – 0.86 (m, 2 H), 1.01 – 1.12 (m, 4 H), 1.15 – 1.27 (m, 1 H), 1.29 – 1.37 (m, 2 H), 1.39 – 1.62 (m, 6 H), 1.71 – 1.93 (m, 2 H), 1.97 – 2.09 (m, 1 H), 3.01 – 3.13 (m, 2 H), 3.73 – 3.81 (m, 1 H), 3.87 (dd, J=5.53, 2.18 Hz, 1 H), 4.00 – 4.12 (m, 1 H), 5.28 – 5.39 (m, 1 H), 7.45 (d, J=1.88 Hz, 1 H), 7.56 – 7.65 (m, 2 H), 7.73 – 7.87 (m, 3 H), 8.03 (d, J=13.23 Hz, 1 H). HRMS (ESI) calcd for C22H31N3O8ClS2Na: [M-]: 587.1139. Found: 587.1137.
4.1.6.7. Sodium (2S)-2-((S)-2-(((4-chlorophenyl)methyl)sulfonamido)-3-cyclohexyl propanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7g
White solid (yield 61%); M.p 76–78 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.84 (br. s., 2 H), 1.04 – 1.24 (m, 6 H), 1.37 (d, J=8.59 Hz, 2 H), 1.65 (d, J=8.98 Hz, 6 H), 1.84 – 1.97 (m, 1 H), 2.08 – 2.35 (m, 3 H), 2.95 (d, J=7.42 Hz, 1 H), 3.04 – 3.18 (m, 1 H), 3.57 (dd, J=13.18, 6.83 Hz, 1 H), 3.74 (q, J=6.77 Hz, 1 H), 3.84 – 3.99 (m, 1 H), 4.17 – 4.38 (m, 1 H), 7.41 (br. s., 3 H), 7.50 – 7.55 (m, 1 H), 7.65 (br. s., 1 H), 7.92 (d, J=8.69 Hz, 1 H), 8.64 (d, J=6.83 Hz, 1 H). HRMS (ESI) calcd for C23H33N3O8ClS2Na: [M-]: 601.1295. Found: 601.1295.
4.1.6.8. Sodium (2S)-2-((S)-2-((2-(3-chlorophenyl)ethyl)sulfonamido)-3-cyclohexyl propanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7h
White solid (yield 71%); M.p 78–80 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.87 (br. s., 2 H), 1.02 – 1.23 (m, 5 H), 1.30 – 1.53 (m, 4 H), 1.54 – 1.71 (m, 4 H), 1.74 – 1.87 (m, 2 H), 1.92 – 2.10 (m, 1 H), 2.65 – 2.79 (m, 1 H), 2.86 – 3.00 (m, 2 H), 3.02 – 3.17 (m, 2 H), 3.73 (q, J=7.08 Hz, 1 H), 3.85 – 4.02 (m, 2 H), 4.20 (d, J=7.57 Hz, 1 H), 4.68 – 4.78 (m, 1 H), 7.21 – 7.37 (m, 3 H), 7.42 – 7.51 (m, 1 H), 7.55 – 7.62 (m, 1 H), 7.69 (d, J=8.79 Hz, 1 H), 7.90 – 7.98 (m, 1 H). HRMS (ESI) calcd for C24H35N3O8ClS2Na: [M-]: 615.1452. Found: 615.1459.
4.1.6.9. Sodium (2S)-2-((S)-3-cyclohexyl-2-(thiophene-2-sulfonamido)propan amido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7i
White solid (yield 67%); M.p 156–158 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.64 – 0.83 (m, 2 H), 0.92 – 1.13 (m, 4 H), 1.16 – 1.25 (m, 1 H), 1.28 – 1.38 (m, 1 H), 1.55 (d, J=11.38 Hz, 6 H), 1.70 – 1.95 (m, 2 H), 1.97 – 2.15 (m, 2 H), 2.97 – 3.19 (m, 2 H), 3.74 – 3.84 (m, 1 H), 3.84 – 3.94 (m, 1 H), 4.05 – 4.17 (m, 1 H), 5.31 (t, J=6.93 Hz, 1 H), 7.09 (t, J=4.00 Hz, 1 H), 7.45 (s, 1 H), 7.54 – 7.60 (m, 1 H), 7.73 (d, J=8.93 Hz, 1 H), 7.82 – 7.87 (m, 1 H), 8.05 (br. s., 1 H). HRMS (ESI) calcd for C20H30N3O8S3Na: [M-]: 599.1093. Found: 599.1094.
4.1.6.10. Sodium (2S)-2-((S)-2-([1,1′-biphenyl]-4-sulfonamido)-3-cyclohexylpropan amido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7j
White solid (yield 59%); M.p 154–156 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.66 (d, J=4.98 Hz, 1 H), 0.76 (d, J=10.64 Hz, 1 H), 0.87 (br. s., 1 H), 1.00 – 1.13 (m, 4 H), 1.16 – 1.24 (m, 1 H), 1.29 – 1.39 (m, 3 H), 1.41 – 1.62 (m, 6 H), 1.73 – 1.84 (m, 1 H), 1.89 – 2.05 (m, 1 H), 2.85 – 2.93 (m, 1 H), 2.97 – 3.09 (m, 1 H), 3.61 (dd, J=10.20, 3.37 Hz, 1 H), 3.71 – 3.83 (m, 1 H), 3.87 – 3.94 (m, 1 H), 5.31 (t, J=5.08 Hz, 1 H), 7.38 (d, J=3.81 Hz, 1 H), 7.42 – 7.47 (m, 2 H), 7.51 (d, J=2.05 Hz, 2 H), 7.58 – 7.66 (m, 1 H), 7.69 – 7.78 (m, 2 H), 7.80 – 7.90 (m, 2 H), 7.94 (d, J=7.42 Hz, 1 H). HRMS (ESI) calcd for C28H36N3O8S2Na: [M-]: 629.1842. Found: 629.1844.
4.1.6.11 Sdium (2S)-2-((S)-3-cyclohexyl-2-((2-(1,3-dioxoisoindolin-2-yl)ethyl)sulfon amido)propanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7k
White solid (yield 76%); M.p 74–76 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.85 (br. s., 2 H), 0.99 – 1.27 (m, 5 H), 1.41 (br. s., 2 H), 1.53 – 1.74 (m, 6 H), 1.80 – 1.92 (m, 1 H), 2.05 – 2.31 (m, 2 H), 2.96 – 3.14 (m, 2 H), 3.54 – 3.65 (m, 1 H), 3.74 (q, J=7.13 Hz, 2 H), 3.94 (d, J=6.05 Hz, 3 H), 4.31 – 4.41 (m, 1 H), 6.03 – 6.15 (m, 1 H), 6.63 (dd, J=9.03, 2.10 Hz, 1 H), 7.43 – 7.55 (m, 2 H), 7.59 – 7.74 (m, 1 H), 7.83 – 7.94 (m, 2 H), 8.62 – 8.71 (m, 1 H). HRMS (ESI) calcd for C26H35N4O10S2Na: [M-]: 650.1692. Found: 650.1693.
4.1.6.12. Sodium (2S)-2-((S)-3-cyclohexyl-2-((3-(4-methoxyphenoxy)propyl)sulfon amido)propanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7l
White solid (yield 63%); M.p 65–67 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.85 (d, J=10.74 Hz, 2 H), 1.02 – 1.30 (m, 3 H), 1.33 – 1.50 (m, 2 H), 1.52 – 1.69 (m, 6 H), 1.71 – 1.84 (m, 2 H), 1.88 – 2.22 (m, 5 H), 2.93 – 3.04 (m, 2 H), 3.05 – 3.17 (m, 2 H), 3.69 (s, 3 H), 3.80 (br. s., 1 H), 3.85 – 3.92 (m, 1 H), 3.95 – 4.07 (m, 2 H), 4.22 (br. s., 1 H), 5.47 (d, J=5.86 Hz, 1 H), 6.85 (s, 4 H), 7.37 – 7.51 (m, 1 H), 7.65 (d, J=8.98 Hz, 1 H), 7.94 (d, J=8.89 Hz, 1 H). HRMS (ESI) calcd for C26H40N3O10S2Na: [M-]: 641.2053. Found: 641.2051.
4.1.6.13. Sodium (2S)-2-((S)-3-cyclohexyl-2-(octylsulfonamido)propanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 7m
White solid (yield 65%); M.p 58–60 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.82 – 0.92 (m, 4 H), 1.04 – 1.19 (m, 2 H), 1.24 (br. s., 6 H), 1.36 – 1.50 (m, 2 H), 1.63 (d, J=11.42 Hz, 6 H), 1.77 (d, J=10.84 Hz, 2 H), 1.86 – 2.04 (m, 2 H), 2.05 – 2.14 (m, 3 H), 2.15 – 2.25 (m, 1 H), 2.83 – 2.96 (m, 2 H), 3.00 – 3.08 (m, 2 H), 3.14 (t, J=8.35 Hz, 2 H), 3.76 (d, J=8.30 Hz, 1 H), 3.85 (t, J=4.88 Hz, 2 H), 3.92 – 4.06 (m, 1 H), 4.22 (t, J=9.62 Hz, 1 H), 5.41 (d, J=5.86 Hz, 1 H), 7.21 – 7.29 (m, 1 H), 7.47 (d, J=5.47 Hz, 1 H), 7.62 (d, J=9.08 Hz, 1 H), 7.88 (d, J=8.98 Hz, 1 H). HRMS (ESI) calcd for C24H44N3O8S2Na: [M-]: 589.2468. Found: 589.2466.
4.1.7. Synthesis of esters 9a–c
To a solution of the hydrochloride salt of ester 3 (10 mmol) in anhydrous THF (40 mL) kept at 0 °C was added slowly N,N-diisopropylethylamine (DIEA) (2.6 mL,20 mmol, 2 eq) with stirring. The appropriate chloroformate (11 mmol, 1.1 eq) was added to the solution and the reaction mixture was stirred for 16 h at room temperature. The solvent was removed and the residue was dissolved in ethyl acetate (100 mL) and washed with water (2 × 50 mL). The ethyl acetate layer was further washed with 5% HCl (2 × 50 mL) and saturated NaCl (50 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated to yield a crude product which was purification by flash chromatography to yield esters 9a–c.
4.1.7.1. Methyl (S)-2-((S)-2-(((3-chlorophenethoxy)carbonyl)amino)-3-cyclohexylpropan amido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 9a
White solid (yield 73%); M.p 43–45 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.84 – 1.02 (m, 2 H), 1.09 – 1.23 (m, 3 H), 1.38 (d, J=2.64 Hz, 1 H), 1.45 – 1.53 (m, 1 H), 1.69 (d, J=10.20 Hz, 6 H), 1.79 – 1.95 (m, 2 H), 2.12 – 2.22 (m, 1 H), 2.36 – 2.46 (m, 2 H), 2.87 – 2.94 (m, 2 H), 3.34 (d, J=6.88 Hz, 2 H), 3.73 (s, 3 H), 4.20 – 4.35 (m, 3 H), 4.48 (ddd, J=11.12, 7.07, 3.88 Hz, 1 H), 5.29 (d, J=8.25 Hz, 1 H), 6.18 (s, 1 H), 7.08 – 7.13 (m, 1 H), 7.19 – 7.26 (m, 3 H), 7.84 (d, J=5.52 Hz, 1 H). HRMS (ESI) calcd for C26H37ClN3O6: [M+H]: 522.2371 Found: 522.2372.
4.1.7.2. Methyl (8S,11S)-8-(cyclohexylmethyl)-6,9-dioxo-11-(((S)-2-oxopyrrolidin-3-yl)methyl)-1-phenyl-2,5-dioxa-7,10-diazadodecan-12-oate9b
White solid (yield 67%); M.p 59–61 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.86 – 1.03 (m, 2 H), 1.09 – 1.29 (m, 4 H), 1.34 – 1.44 (m, 2 H), 1.45 – 1.55 (m, 1 H), 1.60 – 1.73 (m, 5 H), 1.75 – 1.95 (m, 2 H), 2.10 – 2.22 (m, 1 H), 2.35 – 2.46 (m, 2 H), 3.30 (d, J=8.40 Hz, 2 H), 3.67 (q, J=4.65 Hz, 2 H), 3.72 (s, 3 H), 4.18 – 4.28 (m, 2 H), 4.32 (d, J=5.66 Hz, 1 H), 4.49 (ddd, J=11.05, 7.02, 3.95 Hz, 1 H), 4.56 (s, 2 H), 5.40 (d, J=8.30 Hz, 1 H), 6.11 (s, 1 H), 7.28 – 7.31 (m, 1 H), 7.34 (s, 4 H), 7.76 (d, J=6.83 Hz, 1 H). HRMS (ESI) calcd for C27H39N3O7: [M+H]: 518.2866 Found: 518.2874.
4.1.7.3. Methyl (S)-2-((S)-3-cyclohexyl-2-undecanamidopropanamido)-3-((S)-2-oxop yrrolidin-3-yl)propanoate 9c
White solid (yield 60%); M.p 67–68 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.88 (t, J=6.44 Hz, 4 H), 0.91 – 1.03 (m, 2 H), 1.10 – 1.21 (m, 2 H), 1.23 – 1.36 (m, 14 H), 1.37 – 1.44 (m, 1 H), 1.45 – 1.54 (m, 1 H), 1.57 – 1.66 (m, 2 H), 1.70 (d, J=8.59 Hz, 2 H), 1.78 – 1.98 (m, 5 H), 2.11 – 2.26 (m, 2 H), 2.37 – 2.48 (m, 2 H), 3.35 (d, J=7.81 Hz, 2 H), 3.73 (s, 3 H), 3.98 – 4.12 (m, 2 H), 4.33 (d, J=4.78 Hz, 1 H), 4.46 – 4.56 (m, 1 H), 5.21 (d, J=7.91 Hz, 1 H), 6.23 (s, 2 H), 7.77 (d, J=5.66 Hz, 1 H). HRMS (ESI) calcd for C28H49N3O6: [M+H]: 524.3700 Found: 524.3712.
4.1.8. Synthesis of alcohols 10a–c
To a solution of ester 9 (5 mmol) in anhydrous THF (30 mL) was added dropwise lithium borohydride (2M in THF, 7.5 mL, 15 mmol), followed by absolute ethyl alcohol (15 mL), and the reaction mixture was stirred at room temperature overnight. The reaction mixture was acidified with 5% HCl and the pH adjusted to ~2. Removal of the solvent left a residue which was taken up in ethyl acetate (100 mL). The organic layer was washed with brine (25 mL), dried over anhydrous sodium sulfate, filtered, and concentrated to yield compounds 10a–c.
4.1.8.1. 3-chlorophenethyl ((S)-3-cyclohexyl-1-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)-1-oxopropan-2-yl)carbamate 10a
White solid (yield 93%); M.p 55–56 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.86 – 0.96 (m, 2 H), 1.18 (d, J=17.43 Hz, 3 H), 1.29 – 1.37 (m, 2 H), 1.39 – 1.52 (m, 1 H), 1.66 (br. s., 6 H), 1.76 – 1.88 (m, 2 H), 2.00 (br. s., 1 H), 2.34 – 2.49 (m, 2 H), 2.84 – 2.94 (m, 2 H), 3.33 (d, J=8.18 Hz, 2 H), 3.61 (d, J=15.55 Hz, 2 H), 3.94 – 4.02 (m, 1 H), 4.19 – 4.31 (m, 3 H), 5.36 (d, J=7.23 Hz, 1 H), 6.20 (s, 1 H), 7.10 (d, J=6.47 Hz, 1 H), 7.17 – 7.25 (m, 3 H), 7.77 (d, J=7.80 Hz, 1 H). HRMS (ESI) calcd for C25H36ClN3O5: [M+H]: 494.2422 Found: 494.2412.
4.1.8.2. 2-(benzyloxy)ethyl ((S)-3-cyclohexyl-1-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)-1-oxopropan-2-yl)carbamate 10b
White solid (yield 97%); M.p 68–69 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.83 – 1.04 (m, 2 H), 1.11 – 1.24 (m, 3 H), 1.31 – 1.41 (m, 1 H), 1.51 (dd, J=8.59, 5.27 Hz, 2 H), 1.68 (d, J=9.28 Hz, 6 H), 1.80 (d, J=8.79 Hz, 2 H), 2.02 (br. s., 1 H), 2.33 – 2.46 (m, 2 H), 3.23 – 3.34 (m, 2 H), 3.55 – 3.61 (m, 3 H), 3.66 (s, 2 H), 3.94 – 4.05 (m, 1 H), 4.24 (s, 2 H), 4.55 (s, 2 H), 5.59 (d, J=5.66 Hz, 1 H), 6.33 (s, 1 H), 7.28 – 7.41 (m, 5 H), 7.64 (d, J=6.44 Hz, 1 H). HRMS (ESI) calcd for C26H39N3O6: [M+H]: 490.2917 Found: 490.2907.
4.1.8.3. N-((S)-3-cyclohexyl-1-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)-1-oxopropan-2-yl)undecanamide 10c
White solid (yield 97%); M.p 78–80 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.88 (t, J=6.20 Hz, 3 H), 0.96 (d, J=10.94 Hz, 2 H), 1.09 – 1.19 (m, 3 H), 1.20 – 1.34 (m, 14 H), 1.51 (d, J=7.42 Hz, 1 H), 1.58 – 1.73 (m, 10 H), 1.80 (d, J=10.15 Hz, 2 H), 2.03 (br. s., 1 H), 2.36 – 2.55 (m, 2 H), 3.30 – 3.41 (m, 2 H), 3.63 (s, 3 H), 4.04 (t, J=7.00 Hz, 2 H), 4.19 – 4.28 (m, 1 H), 5.34 (d, J=7.80 Hz, 1 H), 6.45 (s, 1 H), 7.67 (d, J=5.60 Hz, 1 H). HRMS (ESI) calcd for C27H49N3O5: [M+H]: 496.3750 Found: 496.3745.
4.1.9. Synthesis of aldehydes 11a–c
Compound 10 (5 mmol) was dissolved in anhydrous dichloromethane (50 mL) under a nitrogen atmosphere and cooled to 0°C. Dess-Martin periodinane reagent (3.18 g, 7.5 mmol, 1.5 eq) was added to the reaction mixture with stirring. The ice bath was removed and the reaction mixture was stirred at room temperature for 3 h (monitoring by TLC indicated complete disappearance of the starting material). A solution of 10% aqueous sodium thiosulfate (20 mL) was added and the solution was stirred for another 15 minutes. The aqueous layer was removed and the organic layer was washed with 10% aqueous sodium thiosulfate (20 mL), followed by saturated aqueous sodium bicarbonate (2 × 20 mL), water (2 × 20 mL) and brine (20 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated. The yellow residue was purified by flash chromatography (silica gel/methylene chloride/ethyl acetate/methanol) to yield aldehydes 11a–c.
4.1.9.1. 3-chlorophenethyl ((S)-3-cyclohexyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)propan-2-yl)carbamate 11a
White solid (yield 76%); M.p 45–47 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.86 – 1.01 (m, 2 H), 1.10 – 1.27 (m, 2 H), 1.30 – 1.45 (m, 2 H), 1.47 – 1.58 (m, 1 H), 1.60 – 1.89 (m, 6 H), 1.92 – 2.00 (m, 2 H), 2.01 – 2.10 (m, 1 H), 2.36 – 2.51 (m, 2 H), 2.91 (t, J=6.69 Hz, 2 H), 3.31 – 3.43 (m, 2 H), 4.16 – 4.37 (m, 4 H), 5.25 (d, J=8.20 Hz, 1 H), 5.96 (br. s., 1 H), 7.10 (d, J=6.35 Hz, 1 H), 7.17 – 7.25 (m, 3 H), 8.36 (d, J=5.57 Hz, 1 H), 9.49 (s, 1 H). HRMS (ESI) calcd for C25H34ClN3O5: [M-]: 491.2187. Found: 491.2198.
4.1.9.2. 2-(benzyloxy)ethyl ((S)-3-cyclohexyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)propan-2-yl)carbamate 11b
White solid (yield 78%); M.p 38–40 °C; 1H NMR (400 MHz, CDCl3-d): δ ppm 0.84 – 1.04 (m, 2 H), 1.11 – 1.30 (m, 2 H), 1.39 (d, J=3.22 Hz, 1 H), 1.48 – 1.57 (m, 1 H), 1.71 (dd, J=13.57, 8.69 Hz, 6 H), 1.78 – 1.88 (m, 2 H), 1.93 – 1.98 (m, 1 H), 2.01 – 2.09 (m, 1 H), 2.32 – 2.52 (m, 2 H), 3.25 – 3.36 (m, 2 H), 3.67 (t, J=4.54 Hz, 4 H), 4.25 (d, J=2.93 Hz, 1 H), 4.30 – 4.39 (m, 1 H), 4.51 – 4.62 (m, 2 H), 5.37 (d, J=8.01 Hz, 1 H), 5.99 (br s, 1 H), 7.34 (s, 5 H), 8.24 (d, J=4.88 Hz, 1 H), 9.49 (s, 1 H). HRMS (ESI) calcd for C26H37N3O6: [M-]: 487.2682. Found: 487.2683.
4.1.9.3. N-((S)-3-cyclohexyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino) propan-2-yl)undecanamide 11c
Sticky solid (yield 71%); 1H NMR (400 MHz, CDCl3-d): δ ppm 0.88 (t, J=6.71 Hz, 2 H), 0.91 – 1.01 (m, 2 H), 1.07 – 1.36 (m, 16 H), 1.49 – 1.64 (m, 4 H), 1.69 (d, J=5.71 Hz, 6 H), 1.77 – 1.90 (m, 2 H), 1.91 – 1.99 (m, 1 H), 2.02 – 2.14 (m, 1 H), 2.27 – 2.58 (m, 3 H), 3.31 – 3.42 (m, 2 H), 3.94 – 4.12 (m, 2 H), 4.28 – 4.39 (m, 1 H), 4.41 – 4.53 (m, 1 H), 5.20 – 5.27 (m, 1 H), 6.16 – 6.24 (m, 1 H), 8.21 – 8.28 (m, 1 H), 9.57 (s, 1 H). HRMS (ESI) calcd for C27H47N3O5: [M-]: 493.3516. Found: 493.3518.
4.1.10. Synthesis of bisulfite adducts 12a–c
To a solution of aldehyde 11 (5 mmol) in dry ethyl acetate (20 mL) was added absolute ethanol (12 mL) with stirring, followed by a solution of sodium bisulfite (540 mg; 5 mmol) in water (5 mL). The reaction mixture was stirred for 3 h at 50 °C. The reaction mixture was allowed to cool to room temperature and then vacuum filtered. The solid was thoroughly washed with absolute ethanol and the filtrate was dried over anhydrous sodium sulfate, filtered, and concentrated to yield a yellowish oil. The oily product was treated with ethyl ether (2 × 50 mL) to form white solid. The white solid was stirred with ethyl ether (30 mL) and ethyl acetate (15 mL) for 5 minutes. Careful removal of the solvent using a pipette yielded compounds 12a–c.
4.1.10.1. Sodium (2S)-2-((S)-2-(((3-chlorophenethoxy)carbonyl)amino)-3-cyclohexylprop anamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 12a
White solid (yield 68%); M.p 76–78 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.76 – 0.92 (m, 2 H), 1.02 – 1.17 (m, 3 H), 1.25 (d, J=13.38 Hz, 1 H), 1.40 (d, J=4.69 Hz, 2 H), 1.51 – 1.75 (m, 6 H), 1.80 – 2.02 (m, 1 H), 2.03 – 2.23 (m, 2 H), 2.87 (d, J=3.81 Hz, 2 H), 2.99 – 3.08 (m, 1 H), 3.08 – 3.20 (m, 1 H), 3.69 – 3.87 (m, 2 H), 3.88 – 4.04 (m, 1 H), 4.14 (d, J=6.54 Hz, 3 H), 5.43 – 5.48 (m, 1 H), 7.19 – 7.38 (m, 4 H), 7.47 (br s, 1 H), 7.55 (dd, J=14.94, 9.08 Hz, 2 H). HRMS (ESI) calcd for C25H35ClN3O8SNa: [M-]: 595.1731. Found: 595.1733.
4.1.10.2. Sodium (8S,11S)-8-(cyclohexylmethyl)-12-hydroxy-6,9-dioxo-11-(((S)-2-oxo pyrrolidin-3-yl)methyl)-1-phenyl-2,5-dioxa-7,10-diazadodecane-12-sulfonate 12b
White solid (yield 73%); M.p 47–50 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.78 – 0.94 (m, 2 H), 1.01 – 1.22 (m, 4 H), 1.32 (br s, 1 H), 1.39 – 1.51 (m, 2 H), 1.53 – 1.72 (m, 6 H), 1.86 – 1.94 (m, 1 H), 2.00 – 2.34 (m, 2 H), 2.99 – 3.18 (m, 2 H), 3.60 (d, J=3.71 Hz, 4 H), 3.75 (q, J=7.13 Hz, 1 H), 4.00 – 4.22 (m, 2 H), 4.50 (s, 2 H), 7.25 – 7.38 (m, 5 H), 7.43 (d, J=7.13 Hz, 1 H), 7.54 (br s, 1 H), 7.64 (br s, 1 H), 8.47 (d, J=7.03 Hz, 1 H). HRMS (ESI) calcd for C26H38N3O9SNa: [M-]: 591.2226. Found: 591.2227.
4.1.10.3. Sodium (2S)-2-((S)-3-cyclohexyl-2-undecanamidopropanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate 12c
White solid (yield 67%); M.p 53–55 °C; 1H NMR (400 MHz, DMSO-d6): δ ppm 0.86 (t, J=6.13 Hz, 2 H), 1.07 – 1.14 (m, 2 H), 1.25 (br s, 16 H), 1.39 (d, J=19.72 Hz, 2 H), 1.52 (br s, 5 H), 1.62 (br. s., 6 H), 1.87 – 2.02 (m, 2 H), 2.12 (d, J=7.23 Hz, 2 H), 3.01 – 3.20 (m, 2 H), 3.82 – 4.00 (m, 2 H), 4.05 (br. s., 1 H), 4.17 – 4.29 (m, 1 H), 5.29 – 5.36 (m, 1 H), 5.73 – 5.78 (m, 1 H), 7.14 – 7.19 (m, 1 H), 7.47 (d, J=9.15 Hz, 1 H), 7.58 (s, 1 H). HRMS (ESI) calcd for C27H48N3O7SNa: [M-]: 597.3060. Found: 597.3063.
4.2. X-ray crystallographic studies. Crystallization and data collection
Purified norovirus 3CL Protease (NV 3CLpro) in 100 mM NaCl, 20 mM Tris pH 8.0 (11 mg/mL) was used for preparation of the inhibitor complexes. Stock solutions (100 mM) of each inhibitor were prepared in DMSO and added to NV 3CLpro by mixing 7 μL of the inhibitor (3 mM) with 243 μL (0.5 mM) of NV 3CLpro and incubating on ice for 1 hour. All crystallization experiments were conducted Compact Jr. (Rigaku Reagents) sitting drop vapor diffusion plates at 20 °C using equal volumes of protein and crystallization solution equilibrated against 75 μL of the latter. Hexagonal NV 3CLpro:7l (h) and orthorhombic crystals NV 3CLpro:7l (o) were obtained for the NV 3CLpro:7l complex in 2–3 days from the Index HT screen (Hampton Research) condition G5 (25% (w/v) PEG 3350, 100 mM Tris pH 8.5, 200 mM lithium sulfate) and Wizard 3–4 screen (Rigaku Reagents) condition A10 (20% (w/v) PEG 3350, 200 mM sodium thiocyanate) respectively. Crystals of the complex with inhibitor 7m (NV 3CLpro:7m) were also obtained from the Index HT G5 condition. Samples were transferred to a fresh drop composed of 80% crystallization solution and 20% (v/v) PEG 200 and stored in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source beamline 17-ID using a Dectris Pilatus 6M pixel array detector.
4.3. Structure Solution and Refinement
Intensities were integrated using XDS [34,35] via Autoproc [36] and the Laue class analysis and data scaling were performed with Aimless [37]. Structure solution was conducted by molecular replacement with Phaser [38] using a previously determined structure of inhibitor bound to NV 3CLpro (PDB: 3UR9) [32] as the search model. Each crystal form contained two molecules in the asymmetric unit. Structure refinement and manual model building were conducted with Phenix [39] and Coot [40], respectively. Disordered side chains were truncated to the point for which electron density could be observed. Structure validation was conducted with Molprobity [41] and figures were prepared using the CCP4MG package [42]. Structure superposition was conducted with GESAMT [43].
4.4. FRET protease assays
The FRET protease assay was performed by preparing stock solutions of the substrate (Edans-DFHLQ/GP-Dabcyl) and inhibitor in DMSO and diluting into assay buffer which was comprised of 20mM HEPES buffer, pH 8, containing NaCl (200 mM), 0.4 mM EDTA, glycerol (60%), and 6 mM dithiothreitol (DTT). The protease was mixed with serial dilutions of each compound or with DMSO in 25 μL of assay buffer and incubated at 37°C for 30 min, followed by the addition of 25 μL of assay buffer containing substrate. Fluorescence readings were obtained using an excitation wavelength of 360 nm and an emission wavelength of 460 nm on a fluorescence microplate reader (FLx800; Biotec, Winoosk, VT) 1 h following the addition of substrate. Relative fluorescence units (RFU) were determined by subtracting background values (substrate-containing well without protease) from the raw fluorescence values, as described previously [25,31–32]. The dose-dependent FRET inhibition curves were fitted with a variable slope by using GraphPad Prism software (GraphPad, La Jolla, CA) in order to determine the IC50 values of the inhibitors.
4.5. Cell-based inhibition assays
The effects of each inhibitor on virus replication were examined in the NV replicon harboring cells (HG23 cells) [32]. Briefly, confluent and semi-confluent cells were incubated with medium containing DMSO (<0.1%) or each compound (up to 100 μM) for 48 h. After the incubation, total RNA was extracted and viral genome was quantitated with real-time quantitative RT-PCR (qRT-PCR). The EC50 values were determined by GraphPadPrism software [32].
4.6. Nonspecific cytotoxic effects
The cytotoxic dose for 50% cell death (CC50) for each compound was determined for HG23 cells used in this study. Confluent cells grown in 96-well plates were treated with various concentrations (1 to 100 μM) of each compound for 72 h. Cell cytotoxicity was measured by a CytoTox 96 nonradioactive cytotoxicity assay kit (Promega, Madison, WI) and crystal violet staining. The in vitro therapeutic index was calculated by dividing the CC50 by the EC50.
4.7. Accession Codes
Coordinates and structure factors were deposited to the Worldwide Protein Data Bank (wwPDB) with the accession codes: NV 3CLpro-7l (h) (5T6D), NV 3CLpro-7l (o) (5T6F), and NV 3CLpro-7m (5T6G).
Supplementary Material
Fig. 1.
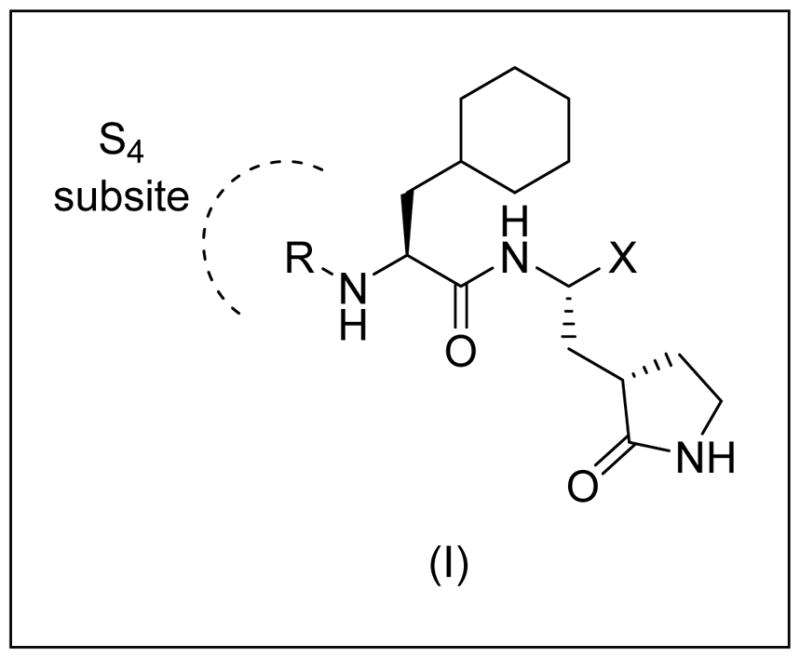
General structure of inhibitor (I)
Table 3.
Crystallographic Data for NV 3CL Protease-Inhibitor Structures
| NV 3CLpro:7l (h) | NV 3CLpro:7l (o) | NV 3CLPro:7m | |
|---|---|---|---|
| Data Collection | |||
| Unit-cell parameters (Å, °) | a=b=59.53, c=356.80 | a=37.51, b=64.80, c=125.37 | a=b=59.58, c=357.70 |
| Space group | P6122 | P212121 | P6122 |
| Resolution (Å)1 | 49.53–2.10 (2.16–2.10) | 45.05–1.90 (1.94–1.90) | 49.57–2.45 (2.55–2.45) |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 |
| Temperature (K) | 100 | 100 | 100 |
| Observed reflections | 434,656 | 158,261 | 276,664 |
| Unique reflections | 23,382 | 24,911 | 15,076 |
| <I/σ(I)>1 | 13.4 (1.9) | 13.7 (2.1) | 13.2 (1.9) |
| Completeness (%)1 | 100 (99.9) | 99.9 (100) | 100 (100) |
| Multiplicity1 | 18.6 (18.9) | 6.4 (5.8) | 18.4 (19.5) |
| Rmerge (%)1, 2 | 20.7 (186.3) | 9.9 (92.6) | 25.1 (197.3) |
| Rmeas (%)1, 4 | 21.3 (191.5) | 10.7 (101.8) | 25.8 (202.6) |
| Rpim (%)1, 4 | 4.9 (43.8) | 4.2 (41.6) | 6.0 (45.5) |
| CC1/2 1, 5 | 0.998 (0.897) | 0.999 (0.834) | 0.997 (0.714) |
| Refinement | |||
| Resolution (Å) 1 | 47.30–2.10 | 35.94–1.90 | 49.57–2.45 |
| Reflections (working/test)1 | 22,094/1,115 | 23,626/1,210 | 14,228/727 |
| Rfactor / Rfree (%)1,3 | 19.3/24.3 | 19.4/24.9 | 18.8/26.1 |
| No. of atoms (Protein/Ligand/Water) | 2,440/74/108 | 2,400/63/104 | 2,473/63/74 |
| Model Quality | |||
| R.m.s deviations | |||
| Bond lengths (Å) | 0.010 | 0.010 | 0.008 |
| Bond angles (°) | 1.040 | 1.031 | 0.924 |
| Average B-factor (Å2) | |||
| All Atoms | 32.6 | 35.4 | 38.1 |
| Protein | 32.3 | 35.3 | 38.1 |
| Ligand | 35.5 | 38.3 | 36.7 |
| Water | 38.2 | 37.2 | 39.7 |
| Coordinate error(maximum likelihood) (Å) | 0.23 | 0.22 | 0.28 |
| Ramachandran Plot | |||
| Most favored (%) | 96.9 | 96.0 | 94.9 |
| Additionally allowed (%) | 3.1 | 3.7 | 4.8 |
Values in parenthesis are for the highest resolution shell.
Rmerge = ΣhklΣi |Ii(hkl) - <I(hkl)>| / ΣhklΣi Ii(hkl), where Ii(hkl) is the intensity measured for the ith reflection and <I(hkl)> is the average intensity of all reflections with indices hkl.
Rfactor = Σhkl ||Fobs (hkl) | - |Fcalc (hkl) || / Σhkl |Fobs (hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
Acknowledgments
The generous financial support of this work by the National Institutes of Health (AI109039) is gratefully acknowledged. Use of the University of Kansas Protein Structure Laboratory was supported by a grant from the National Institute of General Medical Sciences (P30GM110761) from the National Institutes of Health. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-06CH11357.
References
- 1.Koo HL, Ajami N, Atmar RL, DuPont HL. Noroviruses: the leading cause of gastroenteritis worldwide. Discov Med. 2010;10:61–70. [PMC free article] [PubMed] [Google Scholar]
- 2.Hall AJ, Lopman BA, Payne DC, Patel MM, Gastanaduy PA, Vinje J, Parashar UD. Norovirus disease in the United States. Emerg Infect Dis. 2013;19:1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. Global economic burden of norovirus gastroenteritis. PLoS One. 2016;11(4):e0151219. doi: 10.1371/journal.pone.0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belliott G, Lopman BA, Ambert-Balay K, Pothier P. The burden of norovirus gastroenteritis: an important foodborne and healthcare-related infection. Clin Microbiol Infect. 2014;20:724–730. doi: 10.1111/1469-0691.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Institute of Health/U.S. National Library of Health/Medline Plus. Norovirus a Costly Bug. https://www.nih.gov/medlineplus/news/fullstory_158512.html.
- 6.Patel MM, Widdowson MA, Glass RL, Akazawa K, Vinje J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robilotti E, Deresinski S, Pinsky BA. Noroviruses. Clin Microbiol Rev. 2015;28:134–164. doi: 10.1128/CMR.00075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Aryee MKJ, Black RE. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLoS One. 2013;8:e72788. doi: 10.1371/journal.pone.0072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall AJ. Noroviruses: The perfect human pathogen? J Infect Dis. 2012;205(11):1622–1624. doi: 10.1093/infdis/jis251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore MD, Goulter RM, Jaykus LA. Human norovirus as a foodborne pathogen: challenges and developments. Ann Rev Food Sci Technol. 2015;6:411–433. doi: 10.1146/annurev-food-022814-015643. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y, Galasiti Kankanamalage AC, Chang K-O, Groutas WC. Recent advances in the discovery of norovirus therapeutics. J Med Chem. 2015;58:9438–9450. doi: 10.1021/acs.jmedchem.5b00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galasiti Kankanamalage AC, Weerawarna PM, Kim Y, Chang K-O, Groutas WC. Anti-norovirus therapeutics: a patent review (2010–2015) Expert Opin Ther Pat. 2016;26:297–308. doi: 10.1517/13543776.2016.1153065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkataraman Prasad BV, Shanker S, Muhaxhiri Z, Deng L, Choi J-M, Estes MK, Song Y, Palzkill T, Atmar RL. Antiviral targets of human noroviruses. Curr Opin Virol. 2016;18:117–125. doi: 10.1016/j.coviro.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weerasekara S, Prior AM, Hua DH. Current tools for norovirus drug discovery. Expert Opin Drug Discov. 2016;11:529–541. doi: 10.1080/17460441.2016.1178231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kocher J, Yuan L. Norovirus vaccines and potential anti-norovirus drugs: recent advances and future perspectives. Future Virol. 2015;10:899–913. doi: 10.2217/fvl.15.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atmar RL, Baehner F, Cramer JP, Song E, Borkowski A, Mendelman PM. Rapid responses to 2 virus-like particle norovirus vaccine candidate formulations in healthy adults: a randomized controlled trial. J Infect Dis. 2016;214(6):845–853. doi: 10.1093/infdis/jiw259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorne LG, Arias A, Goodfellow IG. Advances toward a norovirus antiviral: from classical inhibitors to lethal mutagenesis. J Infect Dis. 2016;213(Suppl 1):S27–31. doi: 10.1093/infdis/jiv280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorne LG, Goodfellow IG. Norovirus gene expression and replication. J Gen Virol. 2014;95:278–291. doi: 10.1099/vir.0.059634-0. [DOI] [PubMed] [Google Scholar]
- 19.Rocha-Pereira J, Neyts J, Jochmans D. Norovirus: targets and tools in antiviral drug discovery. Biochem Pharmacol. 2014;91:1–11. doi: 10.1016/j.bcp.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali ES, Rajapaksha H, Lundborg M, Carr JM, Petrovsky N. Norovirus drug candidates that inhibit viral capsid attachment to human histo-blood group antigens. Antiviral Res. 2016;133:14–22. doi: 10.1016/j.antiviral.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomenclature used is that of Schechter L, Berger A. Biochem Biophys Res Comm. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x.where S1, S2, S3, …. Sn and S1′, S2′, S3′, …. Sn′ correspond to the enzyme subsites on the N-terminus and C-terminus side, respectively, of the scissile bond. Each subsite accommodates a corresponding amino acid residue side chain designated P1, P2, P3,…..Pn and P1′, P2′, P3′,…..Pn′ of a substrate or inhibitor. P1 is the primary substrate specificity residue and P1-P1′ is the scissile bond.
- 22.Hussey RJ, Coates L, Gill RS, Erskine PT, Coker SF, Mitchell E, Cooper JB, Broadbridge R, Clarke IN, Lambden PR, Shoolingin-Jordan PM. Structural study of norovirus 3C specificity: binding of a designed active site-directed peptide inhibitor. Biochemistry. 2011;50:240–249. doi: 10.1021/bi1008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy ME, Crone TJ, Brower JE, Ettayebi K. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 2002;89:29–39. doi: 10.1016/s0168-1702(02)00114-4. [DOI] [PubMed] [Google Scholar]
- 24.Muhaxhiri Z, Deng L, Shanker S, Sankaran B, Estes MK, Palzkill T, Song Y, Venkataram Prasad BV. Structural basis of substrate specificity and protease inhibition in Norwalk virus. J Virol. 2013;87:4281–4292. doi: 10.1128/JVI.02869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galasiti Kankanamalage AC, Kim Y, Weerawarna PM, Uy RA, Damalanka VC, Mandadapu SR, Alliston KR, Mehzabeen N, Battaille KP, Lovell S, Chang K-O, Groutas WC. Structure-guided design and optimization of norovirus 3CL protease. Structure-activity relationships and biochemical, X-ray crystallographic, cell-based and in vivo studies. J Med Chem. 2015;58:3144–3155. doi: 10.1021/jm5019934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng L, Muhaxhiri Z, Estes MK, Palzkill T, Venkataram Prasad BV. Synthesis, activity, and structure-activity relationship of noroviral protease inhibitors. MedChemComm. 2013;4:1354–1359. doi: 10.1039/C3MD00219E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damalanka VC, Kim Y, Alliston KR, Weerawarna PM, Galasiti Kankanamalage AC, Lushington GH, Mehzabeen N, Battaille KP, Lovell S, Chang K-O, Groutas WC. Oxadiazole-based cell permeable macrocyclic transition state inhibitors of norovirus 3CL protease. J Med Chem. 2016;59:1899–1913. doi: 10.1021/acs.jmedchem.5b01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weerawarna PM, Kim Y, Galasiti Kankanamalage AC, Damalanka VC, Lushington GH, Alliston KR, Mehzabeen N, Battaille KP, Lovell S, Chang K-O, Groutas WC. Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies. Eur J Med Chem. 2016;119:300–318. doi: 10.1016/j.ejmech.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandadapu SR, Gunnam MR, Galasiti Kankanamalage AC, Uy RA, Alliston KR, Lushington GH, Kim Y, Chang K-O, Groutas WC. Potent inhibition of norovirus by dipeptidyl α-hydroxyphosphonate transiton state mimics. Bioorg Med Chem Lett. 2013;23:5941–5944. doi: 10.1016/j.bmcl.2013.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webster SE, Okano K, Little TL, Reich SH, Xin Y, Fuhrman SA, Matthews DA, Love RA, Hendrickson TF, Patick AK, Meador JW, Ferre RA, Brown EL, Ford CE, Binford SL, Worland ST. Tripeptide aldehyde inhibitors of human rhinovirus 3C protease: design, synthesis, biological evaluation, and cocrystal structure solution of P1 glutamine isosteric replacements. J Med Chem. 1998;41:2786–2805. doi: 10.1021/jm980071x. [DOI] [PubMed] [Google Scholar]
- 31.Kim Y, Liu H, Galasiti Kankanamalage AC, Weerasekara S, Hua DH, Groutas WC, Chang K-O, Pedersen NC. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12(3):e1005531. doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y, Lovell S, Tiew KC, Mandadapu SR, Alliston KR, Battaille KP, Groutas WC, Chang KO. Broad spectrum antivirals against 3Cl or 3C-like proteases of picornaviruses, noroviruses and coronaviruses. J Virol. 2012;6:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandadapu SR, Gunnam MR, Tiew KC, Uy RAZ, Prior AM, Alliston KR, Hua DH, Kim Y, Chang KO, Groutas WC. Inhibition of norovirus of norovirus 3CL protease by bisulfite adducts of transition state inhibitors. Bioorg Med Chem Lett. 2013;23:62–65. doi: 10.1016/j.bmcl.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabsch W. Automatic Indexing of Rotation Diffraction Patterns. J Appl Crystallogr. 1988;21:67–72. [Google Scholar]
- 35.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt4):293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn M, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, Afonine PV, Bunkoczi G, Chen VB, David IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowlan K, Emlsey P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular graphics project. Acta Crystallogr Sect D: Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 43.Krissinel E. Enhanced fold recognition using efficient short fragment clustering. J Mol Biochem. 2012;1:76–85. [PMC free article] [PubMed] [Google Scholar]
- 44.Evans P. Scaling and assessment of data quality. Acta Crystallogr Sect D: Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 45.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 46.Weiss MS. Global indicators of X-ray data quality. J Appl Crystallogr. 2001;34:130–135. [Google Scholar]
- 47.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans P. Biochemistry. Resolving some old problems in protein crystallography. Science. 2012;336:986–987. doi: 10.1126/science.1222162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.