Abstract
Purpose
Disease relapse remains a major challenge to successful outcomes in patients who undergo allogeneic hematopoietic cell transplantation (HCT). Donor natural killer (NK) cell alloreactivity in HCT can control leukemic relapse, but capturing alloreactivity in HLA-matched HCT has been elusive. HLA expression on leukemia cells—upregulated in the post-HCT environment—signals for NK cell inhibition via inhibitory killer immunoglobulin-like (KIR) receptors and interrupts their antitumor activity. We hypothesized that varied strengths of inhibition among subtypes of the ubiquitous KIR3DL1 and its cognate ligand, HLA-B, would titrate NK reactivity against acute myelogenous leukemia (AML).
Patients and Methods
By using an algorithm that was based on polymorphism-driven expression levels and specificities, we predicted and tested inhibitory and cytotoxic NK potential on the basis of KIR3DL1/HLA-B subtype combinations in vitro and evaluated their impact in 1,328 patients with AML who underwent HCT from 9/10 or 10/10 HLA-matched unrelated donors.
Results
Segregated by KIR3DL1 subtype, NK cells demonstrated reproducible patterns of strong, weak, or noninhibition by target cells with defined HLA-B subtypes, which translated into discrete cytotoxic hierarchies against AML. In patients, KIR3DL1 and HLA-B subtype combinations that were predictive of weak inhibition or noninhibition were associated with significantly lower relapse (hazard ratio [HR], 0.72; P = .004) and overall mortality (HR, 0.84; P = .030) compared with strong inhibition combinations. The greatest effects were evident in the high-risk group of patients with all KIR ligands (relapse: HR, 0.54; P < .001; and mortality: HR, 0.74; P < .008). Beneficial effects of weak and noninhibiting KIR3DL1 and HLA-B subtype combinations were separate from and additive to the benefit of donor activating KIR2DS1.
Conclusion
Consideration of KIR3DL1-mediated inhibition in donor selection for HLA-matched HCT may achieve superior graft versus leukemia effects, lower risk for relapse, and an increase in survival among patients with AML.
INTRODUCTION
As the only known curative therapy for most persons with acute myelogenous leukemia (AML), allogeneic hematopoietic cell transplantation (HCT) enlists an immune-mediated graft-versus-leukemia alloreactivity that is distinct from graft-versus-host disease (GVHD).1 Although GVHD can be significantly reduced with greater HLA matching, relapse remains responsible for 46% of deaths beyond 100 days post-transplant, which suggests that immune mediated AML control differs between donors.2 Understanding variations that govern graft-versus-leukemia reactivity may inform donor selection to improve HCT outcomes.
Natural killer (NK) cells are innate lymphocytes capable of recognizing transformed cells. Killer immunoglobulin-like receptors (KIRs) control NK function and are encoded by the highly polymorphic, multimembered KIR gene family.3 Interaction between self-specific inhibitory KIR and cognate HLA ligands is fundamental to NK education,4 where cells that express inhibitory KIR for self-HLA are licensed and more responsive than their unlicensed counterparts.4,5 Inflammatory cytokines induced after HCT6,7 activate unlicensed NK cells, but concurrently prompt HLA upregulation on the tumor, which puts the educated NK population at risk for inhibition.
In patients with AML who undergo HCT, lack of HLA ligand for donor KIR is associated with superior NK reactivity and lower relapse as a result of lack of NK inhibition.8-11 The missing ligand classification is defined by the patients’ unchangeable HLA; therefore, enlisting a similar effect in the 40% of patients who express all KIR ligands has been an elusive goal. We hypothesized that KIR allele variation between donors titrates the strength with which donor NK cells are inhibited by the HLA-laden tumor, which leads to differences in leukemotoxicity. We examined one common KIR (KIR3DL1) and its ligand (HLA-Bw4) for which allele subtype variation influences receptor and ligand expression, binding affinity, education, and inhibition.12-17 Compared with other KIR ligands, the lack of HLA-Bw4 conveys notably higher protection from leukemic relapse and solid tumor progression, which makes it likely that diversity in KIR3DL1/HLA-B interaction will have a clinical effect.18-21
The KIR3DL1/S1 gene is one of the most polymorphic KIRs22-24; subtypes are displayed at high (KIR3DL1-h) or low (KIR3DL1-l) cell-surface densities or retained within the cell (KIR3DL1-n).25,26 KIR3DS1 receptors are displayed on the cell surface but do not bind HLA-Bw4.14,27 Dimorphism between isoleucine and threonine at position 80 in HLA-Bw4 (Bw4-80I v Bw4-80T) is similarly associated with surface expression on healthy cells.13 Receptor density is broadly associated with affinity to HLA-Bw4 allomorphs. KIR3DL1-h receptors preferentially bind Bw4-80I in favor of Bw4-80T allotypes, but KIR3DL1-l receptors bind both HLA-Bw4 allomorphs similarly.13,28 Clinical data, however, suggest that both KIR3DL1-l and -h subtypes are impacted by coinherited HLA-Bw4 subtypes; therefore, affinity alone is unlikely to control receptor-ligand avidity and NK responses. Receptor density, receptor availability, ligand density, and affinity combine to influence NK education and effector function, with impacts on HIV control.13,29 These findings suggest a complex receptor-ligand interaction that may also impact inhibition and leukemia control.
Allelic combinations of KIR3DL1-h and Bw4-80I are enriched among patients with AML, which suggests that this is a strongly inhibiting combination that may predispose individuals to developing cancer.20 Furthermore, in patients with neuroblastoma, KIR3DL1 and HLA-B subtype combinations with predicted weak or no engagement are associated with increased disease-free survival compared with combinations with strong interaction.19
We now demonstrate that HLA-Bw4 subtypes differentially inhibit primary NK cells on the basis of the KIR3DL1 subtypes they express. In 1,328 patients with AML who received HLA-compatible allografts, donor-recipient KIR3DL1/HLA-B subtype combinations that demonstrate weak or no inhibition in vitro are associated with significantly lower relapse and higher survival compared with strong inhibition combinations. The benefit of weak or no KIR3DL1 inhibition is not driven by other known KIR-mediated benefits, including the activating KIR2DS1+HLA-C130,31; when combined, these configurations elicit superior outcomes compared with either one alone. In sum, we identify an influential axis that calibrates NK function against AML, elucidating a novel immunogenetic criterion with which stem cell donors can be chosen for maximum anti-AML activity and improved transplant outcome.
PATIENTS AND METHODS
Clinical Samples and Healthy Donor Peripheral Blood Mononuclear Cells
We evaluated 1,328 patients with AML who received an allograft from a 9/10 or 10/10 HLA-matched unrelated donor. The National Marrow Donor Program facilitated all transplants, and all donor-patient pairs for whom HLA typing and donor DNA were available were included in this study (Appendix Table A1, online only). Clinical data, HLA genotyping, sequence-based typing for KIR3DL1 alleles, and genomic DNA were provided by the Center for International Blood and Marrow Transplant Research. Studies were performed in compliance with federal regulations that pertained to the protection of human research participants and were approved by the National Marrow Donor Program institutional review board.
Patients and donors provided informed written consent for research. Healthy anonymous donor peripheral blood mononuclear cells (PBMCs) were collected from buffy coats obtained from the New York Blood Center (New York, NY), as described.13 Studies were approved by the Memorial Sloan Kettering Cancer Center institutional review board.
Donor KIR and KIR3DL1 Typing
KIR genotyping was performed by using sequence-specific PCR32,33 or KIR sequence-specific oligonucleotide probes (SSOP) (Thermo Fisher Scientific Life Sciences, Waltham, MA; and One Lambda, Canoga Park, CA). Sequence-based KIR3DL1 allele typing was available for 299 donors.34-36 By using multiplex PCR,37 1,029 donors were assessed for KIR3DL1 subtypes. Allele frequencies were similar to previous findings.38
KIR3DL1 alleles were classified as KIR3DS1, KIR3DL1-high (h), KIR3DL1-low (l), or KIR3DL1-null (n) subtypes on the basis of known polymorphisms and expression (Appendix Tables A1–A3, online only)37; Bw6, Bw4-80T and Bw4-80I epitopes were assigned by using the Immuno Polymorphism Database.39 KIR3DL1 and HLA-B were grouped on the basis of their compound subtypes, as described19,29 (Appendix Tables A2 and A3).
AML Cell Lines and Primary Blasts
AML blasts were collected from patient peripheral blood and bone marrow. Cell lines were confirmed to be mycoplasma negative, and HLA was determined by sequencing (Histogenetics, Ossining, NY) or KIR ligand (Olerup, West Chester, PA) typing. Cells were maintained in RPMI-1640 that was supplemented with 10% fetal bovine serum. To upregulate HLA expression, cells were cultured for 3 days with 1,000 IU/mL human interferon-γ (Peprotech, Rocky Hill, NJ).
Fluorescence-Activated Cell Sorting and In Vitro Cytotoxicity
NK and target cells were cocultured (1:1) with anti-CD107a to quantify degranulation. Where specified, AML cell lines were pretreated with 10 μg/mL anti-HLA-B and -C antibody (4E; Memorial Sloan Kettering Cancer Center Monoclonal Antibody and Bioresource core facility) to inhibit KIR engagement. After 6 hour coculture, PBMCs were stained for fluorescence-activated cell sorting by using live/dead fixable stain (Thermo Fisher Scientific Life Sciences) and fluorochrome-tagged antibodies (Appendix Table A4, online only).
To quantify cytotoxicity, target cells were stained by using CFSE (Sigma-Aldrich, St Louis, MO), cocultured with PBMCs (3:1 effector:target) for 48 hours at 37°C, 5% CO2, and counterstained with 4′,6-diamidino-2-phenylindole (Sigma-Aldrich). The anti-KIR3DL1/S1 antibody, Z27, was included to block KIR3DL1/HLA-Bw4 interaction.
Statistical Analysis
All models used Cox proportional hazards regression for time-to-event post-HCT outcomes for relapse and death. Probabilities of overall survival and relapse were obtained by using Kaplan-Meier and cumulative incidence estimates, respectively, where death without relapse was regarded as a competing risk for relapse. Multivariate analyses were adjusted for patient age, conditioning regimen, T-cell depletion, graft type, disease status, cytomegalovirus, gender match, and HLA-match. For functional studies, one-way ANOVA using Tukey’s post hoc test or Kruskal-Wallis nonparametric assessments with Dunn’s correction were used. To assess KIR3DL1-n–positive versus receptor-negative populations, paired Student’s t tests compared NK cells derived from the same donor. Clinical and functional analyses were completed in R and Prism 6 software, respectively, and P < 0.05 was considered statistically significant.
RESULTS
HLA-Bw4 Subtypes Hierarchically Inhibit Primary NK Cells
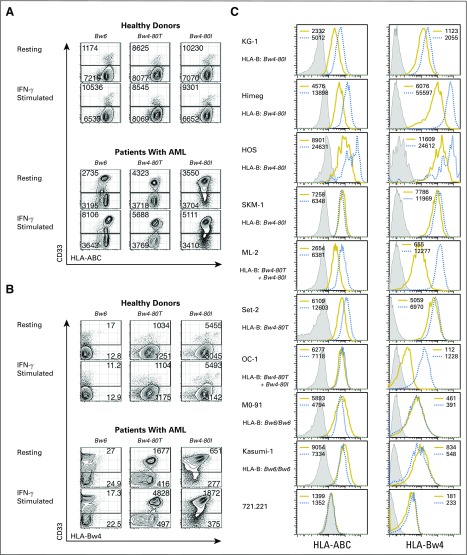
Patients with AML who lack HLA ligands for donor inhibitory KIR have lower relapse and higher survival after HCT compared with patients who exhibit all KIR ligands,10,11,40 which suggests that HLA expression on the tumor inhibits NK function in vivo. Indeed, we find that total HLA, specifically HLA-Bw4, is expressed on CD33+ AML cell blasts and cell lines (Appendix Fig. A1, online only). Treatment with interferon-γ to mimic inflammation in HCT6,7,41,42 further upregulates HLA.
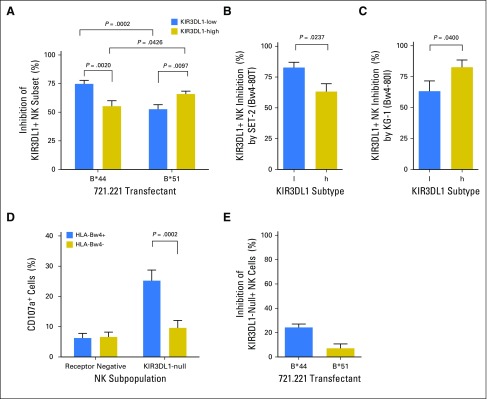
In HLA-matched HCT, educated NK cells are at risk of inhibition by HLA expressed on the tumor. To test the hypothesis that NK cells with specific KIR3DL1 subtypes are variably inhibited by HLA-Bw4 subtypes, we evaluated the inhibition of NK cells that were single positive (spNK) for KIR3DL1 by HLA-Bw4–positive target cells. To simulate the HLA-matched HCT setting, we challenged primary NK cells with high or low KIR3DL1 expression from Bw4-80T–positive or Bw4-80I–positive individuals with HLA-Bw4–matched targets. Among Bw4-80T donors, KIR3DL1-l–positive spNK cells were more inhibited than KIR3DL1-h–positive spNK cells by the 721.221 transfectant that expressed the Bw4-80T allele HLA-B*44:02 and by the Bw4-80T–positive AML cell line SET-2 (Figs 1A and 1B). The opposite was observed among Bw4-80I donors: KIR3DL1-h–positive spNK cells were more inhibited than KIR3DL1-l–positive spNK cells by 721.221 target cells that expressed the Bw4-80I allele HLA-B*51:01 and by the Bw4-80I–positive AML cell line KG-1 (Figs 1A and 1C). Given the higher affinity of KIR3DL1-h for Bw4-80I versus 80T,13,28 higher inhibition of KIR3DL1-h–positive NK cells by the former was expected and consistent with clinical correlations.19,21 Higher inhibition of KIR3DL1-l–positive NK cells by Bw4-80T relative to -80I, however, was not expected on the basis of binding affinity,28 but had been supported by previous clinical observations.19,29
Fig 1.
High and low subtypes of KIR3DL1 are differently sensitive to inhibition by Bw4-80I and Bw4-80T. (A) Primary KIR3DL1-low or -high natural killer (NK) cells from healthy Bw4-80T+ or Bw4-80I+ donors were challenged with 721.221 cells transfected with HLA-B*44 or HLA-B*51, respectively. Percentage of inhibition of KIR3DL1+ NK cells was calculated by comparing degranulation of the same NK cells toward parental 721.221 or 721.221-Bw4+ cells. Each bar represents 10 to 15 healthy donors. (B) Degranulation of KIR3DL1-low (l) or KIR3DL1-high (h)-positive NK cells derived from Bw4-80T+ donors in response to challenge with the Bw4-80T+ acute myelogenous leukemia (AML) cell line, SET-2. Percentage of inhibition was calculated by comparing NK degranulation in the presence and absence of the KIR3DL1 blocking antibody, DX9. (C) Degranulation of KIR3DL1-low (l) or KIR3DL1-high (h) NK cells derived from Bw4-80I+ donors in response to challenge with the Bw4-80I+ AML cell line, KG-1. (D) Response of KIR3DL1-null or inhibitory killer immunoglobulin-like receptor–negative cells to HLA-negative 721.221 target cells. NK cells are segregated on the basis of the presence or absence of HLA-Bw4 in the donor. (E) KIR3DL1-null-expressing NK cells from Bw4-80T+ or Bw4-80I+ donors were challenged using 721.221 cells transfected with HLA-B*44 or HLA-B*51, respectively. Percentage of inhibition is calculated by comparing responsiveness against parental 721.221 cells and Bw4-transfected cells. Each bar represents four to six donors and mean ± standard error of the mean.
KIR3DL1-Null–Positive NK Cells Are Cytotoxic, Yet Insensitive to Inhibition
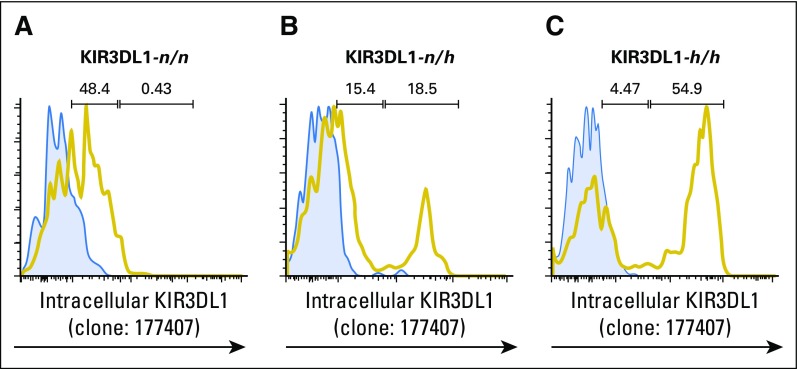
KIR3DL1-n receptor is retained intracellularly and does not signal for inhibition.22 We optimized staining for intracellular KIR3DL1 to investigate whether KIR3DL1-n would educate NK cells, following a recent finding that NK cells are educated by cell-intrinsic HLA43,44 (Appendix Fig A2, online only). KIR3DL1-n–positive spNK cells from HLA-Bw4–positive donors, but not HLA-Bw4–negative donors, were highly responsive to 721.221 targets (Fig 1D), but insensitive to inhibition by HLA-Bw4–positive 721.221 target cells (Fig 1E), which indicated that they are educated for effector response, though refractory to inhibition.
KIR3DL1 and HLA-Bw4 Subtype Combinations Predict Differential Leukemotoxicity
We next investigated how differences in the inhibition of KIR3DL1-expressing NK cells impacted AML killing. We assigned diploid haplotypes to KIR3DL1 subgroups KIR3DL1-L or KIR3DL1-H (Appendix Table A2).45 A third subgroup represented donors who exclusively exhibited KIR3DL1-n and/or KIR3DS1 subtypes (KIR3DL1-N). PBMCs from individuals who represented each subgroup were coincubated with HLA-Bw4 subtype-matched AML target cells.
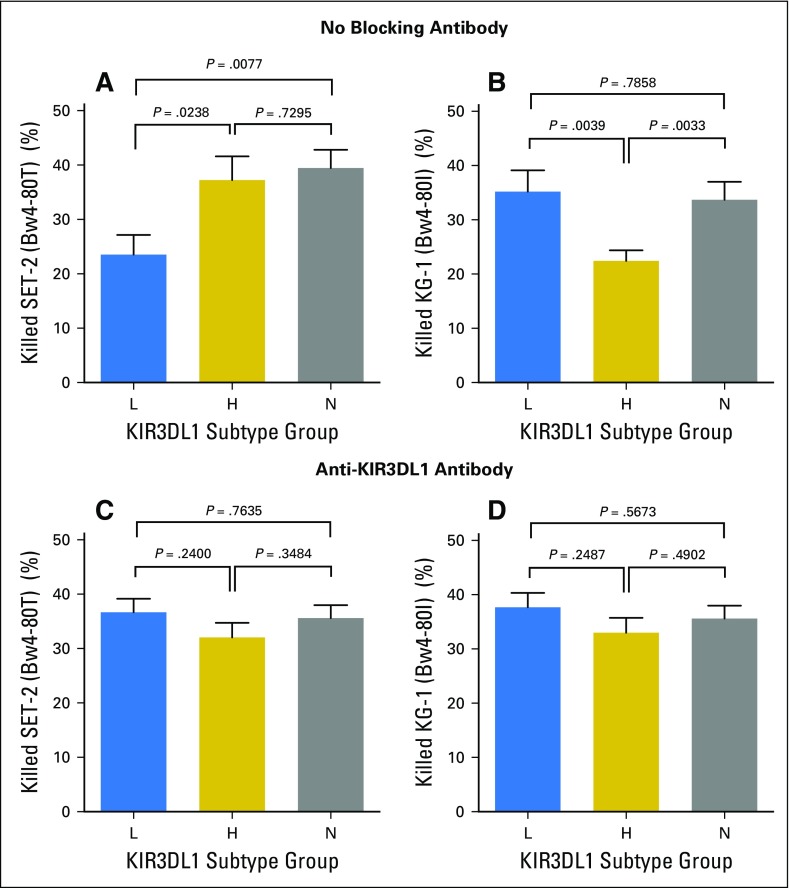
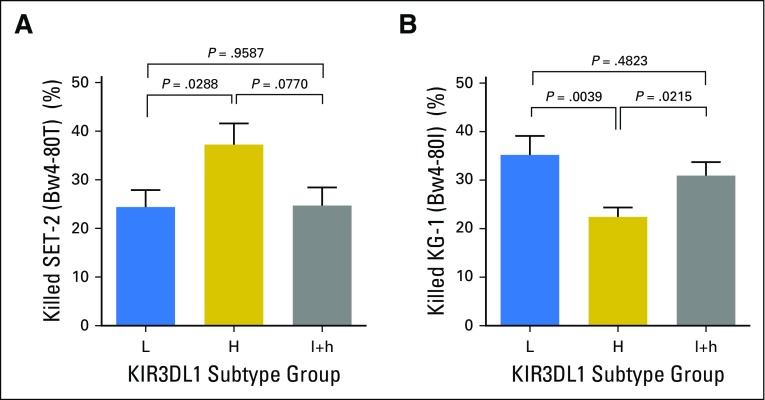
Among Bw4-80T individuals, KIR3DL1-H–positive PBMCs killed the Bw4-80T–positive AML target more efficiently than did KIR3DL1-L–positive PBMCs (Fig 2A). In contrast, among Bw4-80I individuals, KIR3DL1-L–positive PBMCs killed Bw4-80I–positive AML targets more efficiently than did KIR3DL1-H–positive PBMCs (Fig 2B). Antibody blockade of KIR3DL1 equalized target cell lysis between groups, which indicated that the differences in cytotoxicity could be explicitly attributed to differential inhibition of the KIR3DL1–positive cell population (Figs 2C and 2D). KIR3DL1-N PBMCs exhibited high cytotoxicity against both cell lines, unchanged by the addition of anti-KIR3DL1/S1, which reflected their simultaneous education and insensitivity to inhibition. NK cells that were heterozygous for KIR3DL1-l+h exhibited greater killing of Bw4-80I–positive targets than Bw4-80T–positive targets, which supported their inclusion in the KIR3DL1-L group19,29 (Appendix Fig A3, online only).
Fig 2.
Subtype combinations of KIR3DL1 and HLA-Bw4 predict differential inhibition and killing of leukemia target cells. Peripheral blood mononuclear cells (PBMCs) were cultured with Bw4 subtype-matched target cells for 48 hours and the viability of leukemia cells was measured thereafter. (A) Cytotoxicity of the Bw4-80T+ acute myelogenous leukemia (AML) cell line SET-2 by PBMCs from Bw4-80T+ donors. (B) Cytotoxicity of the Bw4-80I+ AML cell line KG-1 by PBMCs from Bw4-80I+ donors. (C) Cytotoxicity of the Bw4-80T+ AML cell line SET-2 by PBMCs from Bw4-80T+ donors in the presence of Z27 antibody. (D) Cytotoxicity of the Bw4-80I+ AML cell line KG-1 by PBMCs from Bw4-80I+ donors in the presence of Z27 antibody. All bars represent means ± standard error of the mean. Each bar represents a minimum of six independent healthy donors and HLA-C subtype groups are stratified equivalently between groups.
Strong Inhibitory Subtypes of KIR3DL1 and HLA-B Are Associated With Increased AML Relapse and Mortality
To determine whether the hierarchy of inhibitory sensitivities established by KIR3DL1 and HLA-Bw4 subtypes affect AML control, we retrospectively evaluated donor KIR3DL1 and HLA-B subtypes for 1,328 patients with AML who received an unrelated HLA-compatible HCT. Neither donor-recipient HLA-B epitope, Bw4 subtype, nor KIR3DL1 subtype alone was associated with overall mortality or relapse (data not shown). In contrast, any impact of KIR3DL1 on outcomes, particularly relapse, was dependent on the HLA-Bw4 subtype (test of interaction: P = .06).
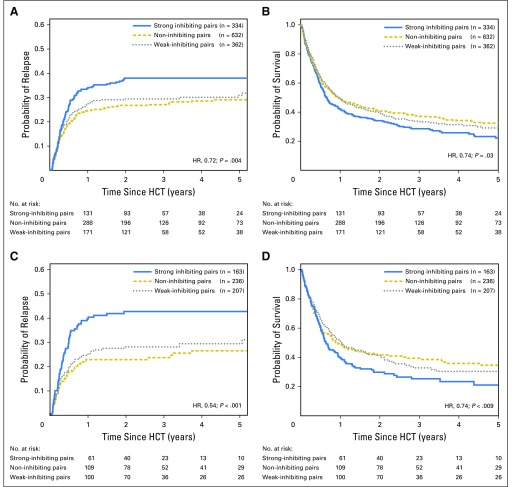
On the basis of their relative inhibitory and cytotoxic strengths against targets in vitro, KIR3DL1-H+Bw4-80I and KIR3DL1-L+Bw4-80T were considered collectively to be strong inhibiting pairs; reciprocal combinations were considered to be weak inhibiting pairs. A third noninhibiting classification was composed of donors with Bw6 and/or KIR3DL1-N. In a multivariable analysis, weak inhibiting pairs demonstrated significantly lower relapse (hazard ratio [HR], 0.73; P = .019) and mortality (HR, 0.83; P = .041) compared with strong inhibiting pairs; noninhibiting pairs were similarly beneficial (relapse: HR, 0.72; P = .008; and mortality: HR, 0.87; P = .064; Table 1). Combined, donors with weak or noninhibiting pairs were associated with superior outcomes compared with strong inhibiting pairs (relapse: HR, 0.72; P = .004; Fig 3A; mortality: HR, 0.84; P = .03; Fig 3B).
Table 1.
Impact of KIR3DL1/HLA-B Subtype Combinations
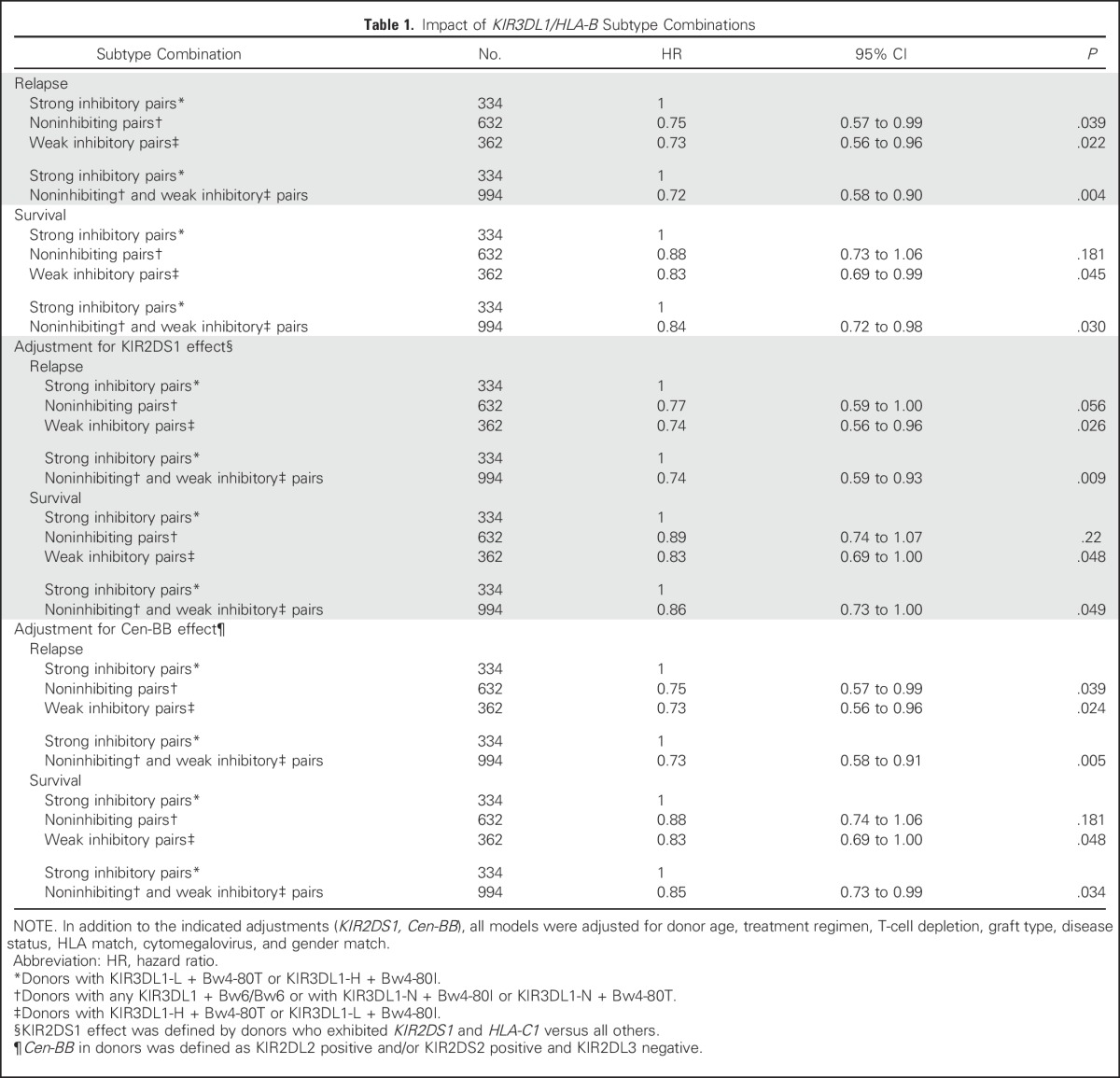
Fig 3.
KIR3DL1 and HLA-B subtype combinations predict outcomes post–hematopoietic cell transplantation (HCT). (A and B) Patients with acute myelogenous leukemia (AML; N = 1,328) who underwent HCT were segregated according to donor KIR3DL1 and HLA-B subtypes into strong inhibiting (blue lines), noninhibiting (dashed yellow lines), or weak inhibiting (dotted gray lines) subtype combinations. The number of donor-patient pairs per group is shown. (A) Cumulative incidence curves for relapse and (B) Kaplan-Meier plot for survival among all donor-patient pairs. (C and D) Patients with AML (n = 606) that exhibited HLA-C1 and -C2 who underwent HLA-matched HCT were segregated according to donor KIR3DL1I and HLA-B subtypes into strongly-interacting (blue lines), non-inhibiting (dashed yellow lines), or weakly interacting (dotted gray lines) subtype combinations. The number of donor-patient pairs per group is shown. (C) Cumulative incidence curves for relapse and (D) Kaplan-Meier plot for survival among donor-patient pairs exhibiting HLA-C1 and -C2. The indicated hazard ratios (HRs) and P values compare strong inhibiting pairs with weak and non-inhibiting pairs combined and reflect adjustment for patient’s age, conditioning regimen, T-cell depletion, graft type, disease status, cytomegalovirus match, and gender match. All curve comparisons were completed using Cox proportional hazards regression analysis for the time-to-event post-HCT outcomes.
Removal of the 55 KIR3DS1 homozygous donors did not alter our conclusions; therefore, the benefit of noninhibiting pairs was not a result of enrichment for KIR3DS1 or other activating receptors in positive linkage disequilibrium (Table 1). There was no impact of Bw4 epitopes encoded by HLA-A alleles. Previous studies have demonstrated an association between lower relapse and donors with the partial KIR haplotype that contains the KIR2DS2 and KIR2DL2 genes, cenB, where homozygosity (cenBB) confers particular protection.30 Correcting for cenBB did not alter KIR3DL1/Bw4 effects (Table 1).
Weak or Noninhibiting KIR3DL1/HLA-B Subtype Combinations Are Most Protective in HLA-C1/C2 HCT
Among HCT recipients, 40% exhibit HLA-Bw4/C1/C2, or all KIR ligands, a configuration that is associated with higher relapse and mortality compared with patients who lack at least one KIR ligand.30 Segregating HCT pairs according to HLA-C KIR ligands, we found that the protective effects of weak or noninhibiting versus strong inhibiting combinations for relapse (HR, 0.54; P < .001) and mortality (HR, 0.74; P = .009) were most evident in the high-risk HLA-C1/C2 transplant pairs (Figs 3C and 3D).
Benefits of KIR2DS1 and KIR3DL1/HLA-B Are Distinct
In our present cohort of 1,328 donor-patient pairs, 1,220 were assessed in our previous study of KIR2DS1, where we described a benefit of donor KIR2DS1+HLA-C1.31 This finding was unchanged within the larger cohort (relapse: HR, 0.79; P = .04; and mortality: HR, 0.89; P = .12). The protection associated with weak or noninhibiting KIR3DL1/HLA-B subtype combinations was not altered by correcting for KIR2DS1/HLA-C1 (Table 1).
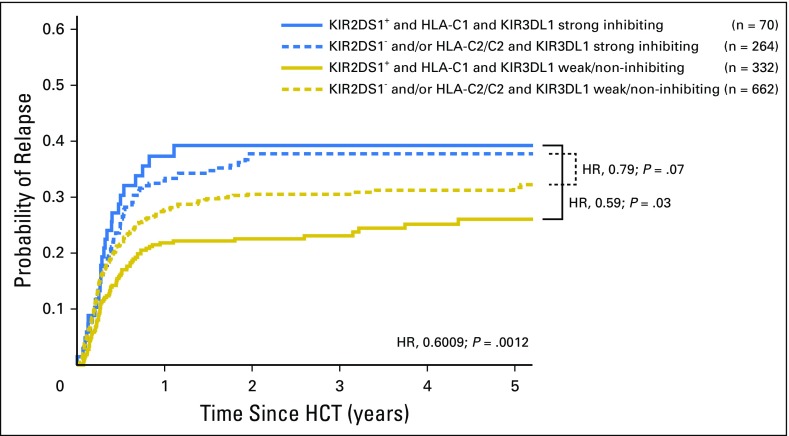
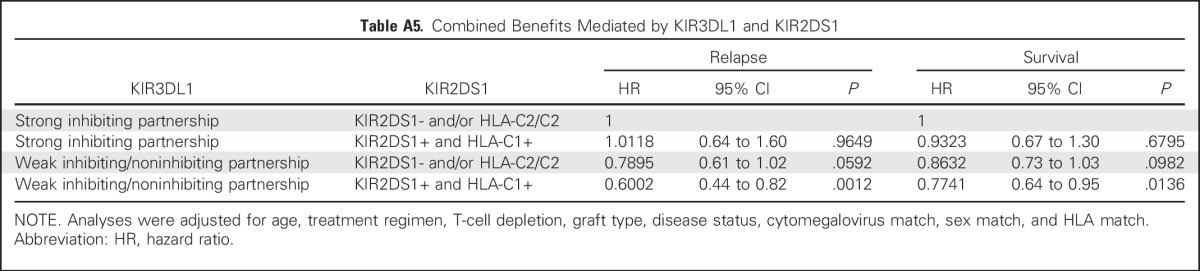
Donors who exhibited the combined benefits of weak or noninhibiting KIR3DL1/HLA-B with KIR2DS1/HLA-C1 conveyed the lowest relapse and highest survival to patients. Strong inhibiting KIR3DL1/HLA-Bw4 partnerships exhibited the highest relapse and mortality, which could not be improved by combination with KIR2DS1+HLA-C1 (Appendix Table A5, online only, and Fig 4). Therefore, although the benefits of KIR3DL1/HLA-B and KIR2DS1/HLA-C1 are separate, the former exhibits primacy in HCT outcomes.
Fig 4.
Independent and additive benefits of weak or noninhibiting KIR3DL1 and HLA-B pairs or KIR2DS1 and HLA-C1. Patients with acute myelogenous leukemia (N = 1,328) who underwent hematopoietic cell transplantation (HCT) from an unrelated donor were segregated on the basis of the presence/absence of KIR3DL1 and KIR2DS1-mediated benefits. A cumulative incidence curve for relapse is shown. Patients with beneficial KIR2DS1 + HLA-C1 are shown as solid lines; patients who lacked KIR2DS1 and/or HLA-C1 are shown in dashed lines. Patients with strong inhibiting partnerships of KIR3DL1 and HLA-B are shown as blue lines; patients with weak inhibiting or noninhibiting partnerships of KIR3DL1 and HLA-B are shown as yellow lines. Hazard ratios (HRs) compare the indicated groups and patients with neither KIR2DS1/KIR3DL1 benefit to those with both.
KIR3DL1 Subtypes: A Novel Donor Selection Criterion
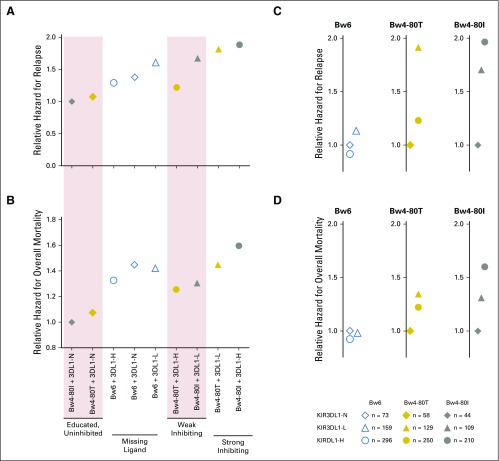
To examine the association of increased NK inhibition with risk of failure, we estimated relative hazards for AML relapse and survival among all KIR3DL1/HLA-B subtype combinations in HLA-matched HCT, clustering groups on the basis of in vitro assessments of education and inhibitory sensitivity (Figs 5A and 5B). Bw6 donor-patient pairs experienced intermediate protection, which reflected the known missing ligand benefit, but the lowest relapse and mortality was observed among KIR3DL1-N+HLA-Bw4, where cells were educated but refractory to inhibition. KIR3DL1-Bw4 partnerships that were predictive of weak inhibition (Bw4-80T+KIR3DL1-H or Bw4-80I+KIR3DL1-L) were associated with intermediate relapse and mortality, which implied a benefit of education and a weak sensitivity to inhibition. Strong inhibiting partnerships (Bw4-80I+KIR3DL1-H and Bw4-80T+KIR3DL1-L) were associated with the highest relapse and mortality, which implied that a strong inhibitory signal overrides the benefit of NK education.
Fig 5.
KIR3DL1 and HLA-B subtype combinations predict a spectrum of post-transplant outcomes. Relative hazards were calculated by Cox proportional hazards regression analysis to compare the impacts of donor and recipient HLA-B. (A) Overall relapse and (B) mortality among hematopoietic cell transplantation (HCT) pairs with specific donor KIR3DL1 and HLA-B subtype combinations are shown. (C) Relapse and (D) mortality segregated by recipient HLA-B subtype (Bw6, Bw4-80T, or Bw4-80I) stratified by donor KIR3DL1 subtypes (KIR3DL1-N, -L, or -H) are shown. Diamonds (◇) represent KIR3DL1-N donors, triangles (△) represent KIR3DL1-L donors, and circles (○) represent KIR3DL1-H donors. Open blue, yellow, and gray symbols represent donor-recipients who encode Bw6, Bw4-80T, or Bw4-80I, respectively, and the numbers of donor-patient pairs in each compound subgroup are shown. Relative hazards reflect adjustment for patient’s age, conditioning regimen, T-cell depletion, graft type, disease status, and cytomegalovirus and gender match. The legend indicates the number of patients present in each subgroup assessed.
We compared outcomes among HLA-B patient groups to understand whether donor selection on the basis of KIR3DL1 subtypes may be an effective intervention to improve AML control (Figs 5C and 5D). Predictably, there were no distinct advantages among donor KIR3DL1 subtypes in Bw6+ donor-recipient pairs. For Bw4-80T+ recipients, the greatest protection from relapse occurred if donors exhibited KIR3DL1-H (HR, 0.65; P = .031) or KIR3DL1-N (HR, 0.52; P = .058) compared with donors with KIR3DL1-L; overall mortality followed the same trend. For Bw4-80I+ recipients, KIR3DL1-N donors were most protective for relapse (HR, 0.52; P = .055) and mortality (HR, 0.64; P = .054) compared with KIR3DL1-H donors.
DISCUSSION
Disease relapse remains the leading cause of death after allogeneic HCT.2 It is increasingly evident that NK cells—educated by HLA and KIR—impact HCT outcomes for AML.46 We now demonstrate that primary KIR3DL1-positive NK cells exhibit a hierarchy of inhibition by HLA-Bw4 subtypes that limits their capacity for lysing leukemia. Retrospective analysis of patients with AML who underwent HLA-matched HCT revealed that strong inhibiting KIR3DL1/HLA-Bw4 combinations predict for higher relapse and mortality, whereas weak or /noninhibiting combinations are protective. The ideal NK effector against AML is educated for reactivity but is insensitive to inhibition by the patient’s HLA, a combination found with KIR3DL1-N+HLA-Bw4.
The observation that Bw4-80I allotypes are stronger inhibitors of KIR3DL1-h–positive NK cells is consistent with their known binding preference13,28; however, for KIR3DL1-l subtypes, specificity between the two HLA-Bw4 subtypes is less dichotomous. By comparing inhibition of primary KIR3DL1-l–positive NK cells by different allotypes, we found that KIR3DL1-l–positive NK cells are more inhibited by Bw4-80T–positive targets than Bw4-80I–positive targets, in contrast to a recent report that found no discernible difference.16,28 The discrepant results likely underscore the importance of considering the educating HLA of the NK cell in inhibition assays; to approximate an autologous or HLA-matched setting, both the NK and target cell should express the same KIR ligand. The mechanisms that underlie the association between receptor expression and differences in inhibitory sensitivity to HLA-Bw4 subtypes are not fully known, and amino acid residues that are important for receptor-ligand affinity do not dictate expression.17 Assignment of strong versus weak inhibition on the basis of receptor expression phenotype and HLA-Bw4 dimorphism may be an oversimplification of a more nuanced system. Nevertheless, the associations of subtype combinations with inhibitory dichotomy, AML cytotoxicity, and relapse protection are consistent.
Proinflammatory cytokines that are present during immune reconstitution lower the threshold for NK activation but upregulate HLA on leukemia cells.6,7,41,42 The protection from relapse that is conveyed by weak or noninhibiting KIR3DL1/HLA-B subtypes reflects a relative insensitivity to HLA-Bw4 that favors cytotoxicity over inhibition. In direct contrast to a suggestion that KIR3DL1-n donors should be avoided,47 our results demonstrate a distinct advantage of KIR3DL1-N+HLA-Bw4: Intracellular receptor sequestration separates NK education from inhibitory susceptibility.
The benefit of KIR3DL1/HLA-B subtype combinations on HCT outcomes persists even after correcting for KIR2DS1 with HLA-C1.31 Combined, KIR3DL1 and KIR2DS1 exhibit the added benefit of minimizing inhibition while maximizing activation. Together, these studies indicate that consideration of HLA and KIR allele typing in donor selection to enable the antileukemic benefits of NK alloreactivity in HCT is warranted. In the frequent case in which a patient has more than one HLA-equivalent donor available, priority should be given to minimize KIR3DL1 inhibition over KIR2DS1 benefit.
In a pilot study, we performed KIR3DL1 subtyping for 941 HLA-matched donors who were screened for 252 patients, 115 of whom completed HCT.48 Weak or noninhibiting KIR3DL1 subtype donors were identified for 93% of 211 patients who had more than one donor available. Of importance, the random 27% risk of a donor exhibiting high inhibition was reduced to 4% upon evaluation of up to three additional donors. Patients with weak or noninhibiting KIR3DL1/HLA-B partnerships experienced higher 2-year disease-free survival after HCT compared with those with strong inhibiting donors (64% v 39%; P = .05). Whether screening for KIR3DL1 subtypes among HLA-matched HCT donors will effect superior outcomes, especially in the high-risk HCT patients who encode all three KIR ligands, forms the basis of a larger, prospective clinical trial (NCT02450708).
The hallmark complications of HCT are GVHD, infection, and relapse. Whereas advances in allograft manipulation, HLA genetics, and antimicrobial therapies have improved the prevention of GVHD and the treatment of infection, disease relapse remains high. KIR and HLA titrate NK inhibition in a predictable, subtype-specific manner, which translates to hierarchical leukemia control. Therefore, refining donor selection algorithms to include KIR3DL1/HLA-B subtype analysis to avoid strong inhibition donors may reduce relapse and improve survival.
ACKNOWLEDGMENT
We thank James Young, MD, and the Memorial Sloan Kettering Cancer Center (MSKCC) Hematologic Oncology Tissue Bank for providing primary leukemia blasts and peripheral blood mononuclear cells. We thank Richard O’Reilly, MD (MSKCC), for providing the AML cell line SET-2, Peter Parham, MD (Stanford University), for providing the 721.221 BLCL transfectants, and Steven Nimer, MD (University of Miami), for providing all other AML cell lines.
Appendix
Fig A1.
HLA class I is expressed on acute myelogenous leukemia (AML) blasts and cell lines and upregulated in response to interferon-gamma (IFN-γ). (A and B) Peripheral blood mononuclear cells (PBMCs) from healthy donors or primary AML blasts from 12 patients were stained for CD33 and assessed for (A) total HLA class I expression or (B) for Bw4 expression at rest and after stimulation with IFN-γ. Three patients with AML and three HLA-B–matched healthy donors matched for HLA-B subtypes are shown and are representative of three to four samples per HLA-B subtype analyzed. Values indicate the mean fluorescence intensities of HLA-ABC or Bw4 staining among CD33+ and CD33− populations. (C) Nine cell lines of differing HLA-B epitope backgrounds are stained for total HLA class I expression and Bw4 at rest (yellow histograms) and after stimulation with IFN-γ (blue dashed histograms). Control (unstained) cells are shown as filled light gray histograms. The B lymphoid cell line, 721.221, which does not express HLA, is shown for comparison. The cell lines ML-2, OC-1, MO-91, and Kasumi-1 exhibit HLA-A epitopes that contain Bw4 motifs. Data represent two independent trials and numbers indicate mean fluorescent intensities.
Fig A2.
Optimization of staining for intracellular KIR3DL1-n. To enable assessment of intracellular KIR3DL1-n, cells were permeabilized and stained with anti-KIR3DL1 clone 177407. (A–C) Staining was optimized on natural killer (NK) cells from (A) KIR3DL1-n homozygous donors and verified on NK cells from donors exhibiting (B) KIR3DL1-n + KIR3DL1-h or (C) homozygous for KIR3DL1-h. Blue histograms indicate FMO control staining and yellow histograms indicate NK cells stained with anti-KIR3DL1 clone 177407.
Fig A3.
Donors who were heterozygous for KIR3DL1-l+h exhibit cytotoxicity most similar to KIR3DL1-L. (A) Cytotoxicity of the Bw4-80T+ acute myelogenous leukemia (AML) cell line SET-2 by peripheral blood mononuclear cells (PBMCs) from Bw4-80T+ healthy donors coexpressing KIR3DL1-l and KIR3DL1-h (l+h) or exhibiting only one of either KIR3DL1-h or KIR3DL1-l. (B) Cytotoxicity of the Bw4-80I+ AML cell line KG-1 by PBMCs from healthy Bw4-80I+ donors coexpressing KIR3DL1-l and KIR3DL1-h (l+h) or exhibiting only one of either KIR3DL1-h (H) or KIR3DL1-l (L). Bars represent means ± standard error of the mean and a minimum of seven independent donors and three independent trials. Means are compared by one-way ANOVA using Tukey’s post hoc test.
Table A1.
Donor, Recipient, and Transplant Characteristics According to Disease and KIR3DL1/HLA-B Subtypes
Table A2.
Donor KIR3DL1 and HLA-B Subtype Distributions
Table A3.
Alleles Comprising KIR3DL1 Subtype Groups and Their Differential Primer Binding Sites
Table A4.
Antibody Clones and Sources Used for Flow Cytometry
Table A5.
Combined Benefits Mediated by KIR3DL1 and KIR2DS1
Footnotes
Supported by National Institutes of Health (NIH) Grants No. R01-HL088134, U01-AI069197, and P01-CA23766 (to K.C.H.) and NIH Cancer Center Core Grant No. P30-CA008748 (to Memorial Sloan Kettering Cancer Center).
AUTHOR CONTRIBUTIONS
Conception and design: Jeanette E. Boudreau, Fabio Giglio, Katharine C. Hsu
Provision of study materials or patients: Brian C. Shaffer
Collection and assembly of data: Jeanette E. Boudreau, Fabio Giglio, Jean-Benoît Le Luduec, Brian C. Shaffer, Raja Rajalingam, Lihua Hou, Carolyn Katovich Hurley, Harriet Noreen, Elaine F. Reed, Neng Yu, Cynthia Vierra-Green, Michael Haagenson, Mari Malkki, Effie W. Petersdorf, Stephen Spellman, Katharine C. Hsu
Data analysis and interpretation: Jeanette E. Boudreau, Fabio Giglio, Ted A. Gooley, Philip A. Stevenson, Katharine C. Hsu
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
KIR3DL1/HLA-B Subtypes Govern Acute Myelogenous Leukemia Relapse After Hematopoietic Cell Transplantation
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Jeanette E. Boudreau
Patents, Royalties, Other Intellectual Property: A patent application has been submitted for the method used to detect KIR3DL1 subtypes (CA2907068A1; Inst)
Fabio Giglio
Travel, Accommodations, Expenses: Amgen, Alexion Pharmaceuticals
Ted A. Gooley
Stock or Other Ownership: Johnson & Johnson
Consulting or Advisory Role: Nohla Therapeutics
Philip A. Stevenson
No relationship to disclose
Jean-Benoît Le Luduec
No relationship to disclose
Brian C. Shaffer
No relationship to disclose
Raja Rajalingam
No relationship to disclose
Lihua Hou
No relationship to disclose
Carolyn Katovich Hurley
Patents, Royalties, Other Intellectual Property: Intellectual property related to an HLA typing workflow which will be marketed through One Lambda, Thermo Fisher Scientific Life Sciences.
Travel, Accommodations, Expenses: Thermo Fisher Scientific Life Sciences
Harriet Noreen
Travel, Accommodations, Expenses: One Lambda
Elaine F. Reed
Honoraria: Genentech, One Lambda
Consulting or Advisory Role: Genentech
Research Funding: Immuncor (Inst), CSL Behring (Inst)
Travel, Accommodations, Expenses: One Lambda, Genentech
Neng Yu
No relationship to disclose
Cynthia Vierra-Green
No relationship to disclose
Michael Haagenson
No relationship to disclose
Mari Malkki
No relationship to disclose
Effie W. Petersdorf
No relationship to disclose
Stephen Spellman
Travel, Accommodations, Expenses: Astellas Pharma
Katharine C. Hsu
Patents, Royalties, Other Intellectual Property: A patent application on the KIR3DL1 multiplex PCR assay that is used for subtyping in this manuscript (Inst), patent applications pertaining to the selection of hematopoietic cell donors on the basis of KIR and HLA for hematopoietic cell transplantation (Inst)
REFERENCES
- 1.Duval M, Klein JP, He W, et al. : Hematopoietic stem-cell transplantation for acute leukemia in relapse or primary induction failure. J Clin Oncol 28:3730-3738, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Souza A, Zhu X: Current uses and outcomes of hematopoietic cell transplantation (HCT): CIBMTR summary slides, 2016. http://www.cibmtr.org
- 3.Parham P: MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol 5:201-214, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Kim S, Poursine-Laurent J, Truscott SM, et al. : Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436:709-713, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Anfossi N, André P, Guia S, et al. : Human NK cell education by inhibitory receptors for MHC class I. Immunity 25:331-342, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Niederwieser D, Herold M, Woloszczuk W, et al. : Endogenous IFN-gamma during human bone marrow transplantation. Analysis of serum levels of interferon and interferon-dependent secondary messages. Transplantation 50:620-625, 1990 [DOI] [PubMed] [Google Scholar]
- 7.Brouwer RE, van der Heiden P, Schreuder GMT, et al. : Loss or downregulation of HLA class I expression at the allelic level in acute leukemia is infrequent but functionally relevant, and can be restored by interferon. Hum Immunol 63:200-210, 2002 [DOI] [PubMed] [Google Scholar]
- 8.Ruggeri L, Capanni M, Urbani E, et al. : Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 295:2097-2100, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Cooley S, Weisdorf DJ, Guethlein LA, et al. : Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 116:2411-2419, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu KC, Keever-Taylor CA, Wilton A, et al. : Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood 105:4878-4884, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu KC, Gooley T, Malkki M, et al. : KIR ligands and prediction of relapse after unrelated donor hematopoietic cell transplantation for hematologic malignancy. Biol Blood Marrow Transplant 12:828-836, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Yawata M, Yawata N, Draghi M, et al. : MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood 112:2369-2380, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boudreau JE, Mulrooney TJ, Le Luduec J-B, et al. : KIR3DL1 and HLA-B density and binding calibrate NK education and response to HIV. J Immunol 196:3398-3410, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr WH, Pando MJ, Parham P: KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol 175:5222-5229, 2005 [DOI] [PubMed] [Google Scholar]
- 15.O’Connor GM, Guinan KJ, Cunningham RT, et al. : Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol 178:235-241, 2007 [DOI] [PubMed] [Google Scholar]
- 16. doi: 10.1084/jem.20051884. Yawata M, Yawata N, Draghi M, et al: Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med 203:633-645, 2006 [Erratum: J Exp Med 203:1131, 2006] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saunders PM, Pymm P, Pietra G, et al. : Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med 213:791-807, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu KC, Pinto-Agnello C, Gooley T, et al. : Hematopoietic stem cell transplantation: Killer immunoglobulin-like receptor component. Tissue Antigens 69:42-45, 2007. (suppl 1) [DOI] [PubMed] [Google Scholar]
- 19.Forlenza CJ, Boudreau JE, Zheng J, et al. : KIR3DL1 allelic polymorphism and HLA-B epitopes modulate response to anti-GD2 monoclonal antibody in patients with neuroblastoma. J Clin Oncol 34:2443-2451, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen M, Linn YC, Ren EC: KIR-HLA profiling shows presence of higher frequencies of strong inhibitory KIR-ligands among prognostically poor risk AML patients. Immunogenetics 68:133-144, 2016 [DOI] [PubMed] [Google Scholar]
- 21.Tarek N, Le Luduec J-B, Gallagher MM, et al. : Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest 122:3260-3270, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parham P, Norman PJ, Abi-Rached L, et al. : Variable NK cell receptors exemplified by human KIR3DL1/S1. J Immunol 187:11-19, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivian JP, Duncan RC, Berry R, et al. : Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature 479:401-405, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halfpenny IA, Middleton D, Barnett YA, et al. : Investigation of killer cell immunoglobulin-like receptor gene diversity: IV. KIR3DL1/S1. Hum Immunol 65:602-612, 2004 [DOI] [PubMed] [Google Scholar]
- 25.Gardiner CM, Guethlein LA, Shilling HG, et al. : Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol 166:2992-3001, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Gumperz JE, Valiante NM, Parham P, et al. : Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J Exp Med 183:1817-1827, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillespie GMA, Bashirova A, Dong T, et al. : Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res Hum Retroviruses 23:451-455, 2007 [DOI] [PubMed] [Google Scholar]
- 28.O’Connor GM, Vivian JP, Widjaja JM, et al. : Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide sensitivity. J Immunol 192:2875-2884, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin MP, Qi Y, Gao X, et al. : Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39:733-740, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooley S, Weisdorf DJ, Guethlein LA, et al. : Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol 192:4592-4600, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venstrom JM, Pittari G, Gooley TA, et al. : HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med 367:805-816, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu KC, Liu X-R, Selvakumar A, et al. : Killer Ig-like receptor haplotype analysis by gene content: Evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol 169:5118-5129, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Vilches C, Castaño J, Gómez-Lozano N, et al. : Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens 70:415-422, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Belle I, Hou L, Chen M, et al. : Investigation of killer cell immunoglobulin-like receptor gene diversity in KIR3DL1 and KIR3DS1 in a transplant population. Tissue Antigens 71:434-439, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Lebedeva TV, Ohashi M, Zannelli G, et al. : Comprehensive approach to high-resolution KIR typing. Hum Immunol 68:789-796, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Levinson RD, Du Z, Luo L, et al. : Combination of KIR and HLA gene variants augments the risk of developing birdshot chorioretinopathy in HLA-A*29-positive individuals. Genes Immun 9:249-258, 2008 [DOI] [PubMed] [Google Scholar]
- 37.Boudreau JE, Le Luduec J-B, Hsu KC: Development of a novel multiplex PCR assay to detect functional subtypes of KIR3DL1 alleles. PLoS One 9:e99543, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vierra-Green C, Roe D, Hou L, et al. : Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS One 7:e47491, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.EMBL-EBI : Immuno polymorphism database. https://www.ebi.ac.uk/ipd/
- 40.Miller JS, Cooley S, Parham P, et al. : Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood 109:5058-5061, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spranger S, Spaapen RM, Zha Y, et al. : Up-regulation of PD-L1, IDO, and Tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci Transl Med 5:200ra116, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gajewski TF, Schreiber H, Fu Y-X: Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 14:1014-1022, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pando MJ, Gardiner CM, Gleimer M, et al. : The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol 171:6640-6649, 2003 [DOI] [PubMed] [Google Scholar]
- 44.Taner SB, Pando MJ, Roberts A, et al. : Interactions of NK cell receptor KIR3DL1*004 with chaperones and conformation-specific antibody reveal a functional folded state as well as predominant intracellular retention. J Immunol 186:62-72, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boudreau JE, Liu X-R, Zhao Z, et al. : Cell-extrinsic MHC class I molecule engagement augments human NK cell education programmed by cell-intrinsic MHC class I. Immunity 45:280-291, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wayne AS, Giralt S, Kröger N, et al. : Proceedings from the National Cancer Institute’s Second International Workshop on the Biology, Prevention, and Treatment of Relapse after Hematopoietic Stem Cell Transplantation: Introduction. Biol Blood Marrow Transplant 19:1534-1536, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alicata C, Pende D, Meazza R, et al. : Hematopoietic stem cell transplantation: Improving alloreactive Bw4 donor selection by genotyping codon 86 of KIR3DL1/S1. Eur J Immunol 46:1511-1517, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaffer BC, Heller G, Le Luduec J-B, et al. : Selection of unrelated allogeneic hematopoietic cell donor based on KIR3DL1 allotypes is feasible and results in improved disease-free survival in transplant recipients with MDS and AML. Blood 128:990, 2016 [Google Scholar]