Abstract
Objectives
Molecular medicine raised expectations for strategically targeted biologics in systemic lupus erythematosus (SLE), but clinical trials have been disappointing and difficult to interpret. Most studies add investigational agents to various, often effective, standard of care (SoC) immunosuppressants used at baseline, with unknown treatment interactions. Eliminating polypharmacy in trials of active lupus patients remains controversial. The BOLD study tested immunosuppressant withdrawal as a novel approach to interpretable SLE trials.
Methods
In 41 patients with active, non-organ threatening SLE flare (Group A), temporary steroids were given while background immunosuppressants were withdrawn. Time to loss of disease suppression (“flare”) and safety were evaluated; SoC was immediately resumed when symptoms recurred. Immunologic impacts of SoC treatments were studied at baseline by multiplex assay, ELISA, and mRNA array in Group A plus 62 additional patients donating a single sample (Group B).
Results
Patients with lower or higher baseline disease had median times-to-flare of 71 or 45 days, respectively; 98% (40/41) flared by six months. All flares were treated and resolved within six weeks. No serious adverse events occurred from flare or infection. Type I interferon, TH17, and BLyS pathways tracked together. Baseline immunosuppressants had distinct impacts on TH17 and BLyS, depending on interferon signature.
Conclusion
Trials in active, non-organ-threatening SLE can safely withdraw background treatments if patients who flare are designated non-responders and returned to SoC. Immunologic effects of SoC vary between interferon-defined subsets. These findings provide a strategy for minimizing or optimizing treatment combinations in lupus trials and clinical care.
Keywords: systemic lupus erythematosus, chemokines, DMARDS (synthetic)
Systemic Lupus Erythematosus (SLE) is a complex autoimmune disorder characterized by unpredictable flares of organ-threatening inflammation (1). Outlines of a common pathology have emerged (2, 3), involving innate and adaptive immunity, defective immune clearance (4–9), and various combinations of multifactorial genetic risk variants (10, 11), However, since patients may develop similar features via different routes in a circuitous and redundant immune system, it seems unlikely that any one treatment will work for all patients, and certainly not at a single dose.
With only one approved treatment in 60 years and 25 years of disappointing clinical trial programs (12–19), the standard of care for SLE remains, predominantly, empiric use of unapproved immunosuppressive combinations which often fail to adequately control disease for long periods of time. Attempts to restore immunologic homeostasis with targeted biologics might be served by considering the complexity and heterogeneity of the patients. Current treatment development programs universally ignore this issue. With few exceptions (18), most trials have failed to focus on patient subsets with immunopathology even relevant to the mechanism being addressed. Still, exploratory analyses of ambiguous study results have repeatedly uncovered, after the fact, subgroups of patients for whom each treatment might have succeeded better (14–19).
Standard of care (SoC) treatment varies greatly between patients. Most clinical trials in SLE are add-on studies, continuing whatever variegated background immunosuppressants and steroids are being taken at entry. This background polypharmacy obscures the interpretation of pharmacodynamic data and likely contribute to apparently high “placebo” response rates, which actually reflect SoC temporarily optimized by rescue regimens. Some recent trials have been marginally more successful by limiting the aggressiveness of rescue interventions while allowing patients to continue a variety of immunosuppressants (14–16). However, suggestions to completely eliminate confounding background medications in SLE trials have been extremely controversial, even though patients have appreciable rates of serious infection when entering add-on trials with aggressive background medications.
In addition to obscuring pharmacodynamics and true response rates, background polypharmacy inevitably superimposes unknown immunomodulatory variables (14–19) that could have synergistic, additive or inhibitory impact on a treatment under study. However, the impact of SoC on the mechanistic pathways of targeted, investigational biologics has never been studied in SLE. It follows that the true degree of heterogeneity innate to SLE cannot be known until the impact of therapeutic cross-talk on immunologic variables is better defined.
To provide an avenue for addressing these knowledge gaps, the Biomarkers of Lupus Disease (BOLD) study was designed to test several hypotheses: First, if patients with active but not organ-threatening SLE are given temporary relief by steroid injections, withdrawal of background immunosuppressants can be accomplished safely. Second, improvement from intramuscular steroids will gradually wane, ensuring low response rates after a few months in the absence of additional effective treatment. Third, upon flare, patients can be designated non-responders and immediately treated with reasonable safety. The BOLD study also sought to generate hypotheses based on gene expression and protein data that background treatments may have disparate impacts on different patient immunophenotypes. If the above hypotheses are correct, then withdrawing SoC immunosuppressants in trials could also help eliminate potential interactions between investigational and background treatments that may confound pathologic characterization of patients and impact the interpretability of trial results.
PATIENTS AND METHODS
Patient enrollment
This study was performed in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the Oklahoma Medical Research Foundation. Patients provided written informed consent prior to beginning study-specific procedures.
Patients in Group A (n=41) completed both cross-sectional and prospective substudies. Group B (n=62) participated in the cross-sectional study only. Inclusion criteria for patient Groups A and B included diagnosis and classification of SLE (20, 21); treatment with ≤20 mg prednisone (or equivalent) daily; active, symptomatic disease despite SoC, defined as at least two British Isles Lupus Assessment Group (BILAG) B (moderate activity) scores, or at least one BILAG A (severe activity) or Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) of at least six; and clinical state warranting intervention equivalent to the steroids offered to Group A. Additional inclusion criteria for Group A were ability and willingness to stop any immunosuppressants (e.g., azathioprine, methotrexate, or mycophenolate mofetil). Exclusion criteria included active infection at screening; known previous HIV, hepatitis B or C; pregnancy or inadequate birth control in women of childbearing potential; cancer (except basal cell or cervical carcinoma) within five years; and any medical condition that would interfere with the protocol or compromise patient safety, including but not limited to the investigator’s opinion of risk for organ-threatening disease.
Cross-sectional substudy
One hundred three patients with active SLE (Groups A and B) underwent clinical assessments and provided blood specimens (Baseline). To improve interpretability of gene expression studies (see below), fifty-five healthy control subjects (Group C) were matched to participants in either Group A or Group B by race, sex and age (within five years; Table 1). Group C was used as a representative control population for scaling the principal component values and establishing normal gene expression ranges. Control subjects provided two samples at two different visits. High quality samples were available for gene expression studies from 99 of those visits (see “Gene Expression Analysis”, below).
Table 1.
Participant demographics.
| Group A1 | Group B2 | Groups A + B | Group C (Controls) | |
|---|---|---|---|---|
| Total number | 41 | 62 | 103 | 55 |
| Females, n (%) | 39 (95.1%) | 56 (88.7%) | 94 (91.3%) | 53 (96.4%) |
| Age in years, mean (SD) | 42.3 (11.9) | 40.87 (11.3) | 41.3 (11.5) | 40.8 (12.1) |
| Race, n (%) | ||||
| Caucasian | 25 (61.0%) | 37 (59.7%) | 62 (60.2%) | 43 (78.2%) |
| African | 11 (26.8%) | 12 (19.4%) | 23 (22.3%) | 10 (18.2%) |
| Native American | 4 (9.8%) | 10 (16.1%) | 14 (13.6%) | 1 (1.8%) |
| Asian | 1 (2.4%) | 3 (4.8%) | 4 (3.9%) | 1 (1.8%) |
SLE patients participating in cross-sectional and prospective substudy.
SLE patients participating in cross-sectional substudy only.
Prospective substudy
After the Baseline blood draw, Group A patients withdrew immunosuppressants and received steroid injections (up to 640 mg depomedrol within two weeks; maximum of four injections allowed). To continue in the study, patients had to demonstrate clinical improvement. Six patients required 640 mg, 19 required 480 mg, and 16 required ≤320 mg. Withdrawal of hydroxychloroquine at baseline was optional and could be over-ridden by patient or clinician. Patients who entered on low dose oral steroids continued at the same dose. Scheduled monthly disease activity assessments and adverse event collection were performed, and blood samples were drawn at each visit for safety monitoring, biomarker assessments, and coded storage in a repository. Upon clinical improvement (Improving Visit), patients were followed monthly until disease activity increased (Flare Visit). As a safety mitigation strategy that is necessary for a trial of this design, patients were instructed to call or return to clinic (without requiring an appointment) at any time they developed worsening symptoms. At the Flare Visit, SoC treatments were re-initiated. Six patients with no improvement within two weeks of Baseline were dropped from Group A and immediately treated with SoC. Their baseline samples, with their consent, were used in Group B (Final Group A=41 patients, Group B= 62).
Clinical assessments
The following were evaluated at every visit: hybrid SLEDAI, which is identical to the Safety of Estrogens in Lupus National Assessment (SELENA) SLEDAI (22, 23) except for the proteinuria definition from SLEDAI 2K (24, 25); the SELENA SLEDAI Flare Index (SFI) with Physicians Global Assessment (PGA) (22, 23); the BILAG 2004 index (26); the Cutaneous Lupus Activity Score Index (CLASI) (27); and tender and swollen joint counts. The SLEDAI and BILAG are commonly evaluated over the previous month (12–16), but when patients improved within two weeks of the entry visit, a two-week evaluation using SLEDAI and BILAG clinical templates was performed. In these cases, the BILAG definition requiring two weeks of improvement was modified to one week.
Clinical outcomes
The primary endpoint was time to flare from Baseline in Group A patients (n=41), comparing patients with moderate disease at Baseline (BILAG B in three or fewer organs, no BILAG A, and SLEDAI score ≤10) to those with significant disease (more than three BILAG B scores, at least one BILAG A, SLEDAI >10, or severe flare by clinical SFI descriptors). Kaplan-Meier analysis and the log-rank test were used to compare time to flare in the primary endpoint in subpopulations based on disease severity, steroid dose, and race.
Clinical improvement (Improving Visit) required clinician’s opinion of significant improvement with no intent to increase treatment, along with at least one grade of improvement by BILAG in at least one organ or SLEDAI decrease of at least four points. The Flare Visit was defined as clinician opinion of significant worsening and intention to treat, with at least one grade worsening by BILAG or four point increase in SLEDAI. The key secondary endpoint was a descriptive evaluation of adverse events.
Cytokine, chemokine and soluble receptor measurement
Blood samples collected at baseline and selected subsequent visits were assayed for cytokine, chemokine, and soluble receptor levels. The Serum Analyte and Biomarker Core at the Oklahoma Medical Research Foundation uses a standardized xMAP 50-plex assay (Affymetrix/eBioscience, Santa Clara, CA) on the BioPlex200® platform (Bio-Rad Technologies, Hercules, CA). This two-laser immunobead multiplex technology quantifies 50 cytokines in 250 μL of plasma (28). Serum BLyS (R&D Systems, Minneapolis, MN USA) and APRIL (eBioscience, San Diego, CA USA) could not be multiplexed and were analyzed by ELISA.
Gene expression analysis
mRNA expression levels in whole blood samples were measured using TaqMan® Low Density Arrays (Applied Biosystems, Grand Island, NY USA) that included probe sets for 347 transcripts, and normalized to the median of endogenous controls. Log2-scale gene expression (dCt) values for eleven IFN-related genes (GBP5, HERC5, IFI27, IRF7, ISG15, LY6E, MX1, OAS2, OAS3, RSAD2, USP18) were mean-centered, and then subjected to principal component analysis using R version 2.15.2 (www.r-project.org) (29). The first principal component (PC1) for each patient visit captured the majority of variance, providing a summarized expression measure for the eleven IFN-related genes. To make the arbitrary-scale PC1 values more interpretable, the dCt values from 99 healthy volunteer (HV) visits from which high-quality samples were available were projected onto PC1, and PC1 values for both patients and HV were scaled linearly, where HV visits had mean zero and unit variance, determining an “IFN index”. The Mclust R package was used to fit a 2-Gaussian equal-variance mixture model to the IFN index to define a single dividing value to separate "IFN Low" from "IFN High". Samples from each patient visit were classified as “IFN High” or “IFN Low”, and patients were classified as “IFN High” or “IFN Low” based on the predominant visit-level assignment. Gene expression data will be available at the NCBI GEO database (accession number GSE92776) as of December 22, 2017.
Statistical analyses
Relationships between gene expression, IFN group (high versus low expression of interferon-inducible genes), and baseline immunosuppressants were examined using ANCOVA with covariates of RIN, assay batch, and percent neutrophils, based on our analysis of variable impacts. Relationships between protein concentrations (pg/mL), IFN group, and baseline immunosuppressants were evaluated using ANOVA. For comparisons between subpopulations, gene expression and disease activity scores were analyzed by t-test, autoantibody-positive frequencies by Fishers exact test, and protein concentrations by ANOVA. Exploratory biomarker assessments were hypothesis-driven based on known pathology of SLE, but were not adjusted for multiple comparisons.
RESULTS
Population
Participants were 91.3% female with a mean age of 41.3 years, of Caucasian, African American, Native American, and Asian race (Table 1). Demographics were similar between groups, but Group A had fewer Native Americans (Table 1). The mean (SD) cumulative BILAG score was 15.2 (5.73) and SLEDAI 8.8 (3.73). Baseline lupus treatments included steroids, hydroxychloroquine, and immunosuppressants (32.1% azathioprine, 17.5% mycophenolate, 25.2% methotrexate) (Table 2). Overall, medications were comparable in Groups A and B, with methotrexate slightly more common in Group A and mycophenolate in Group B (Table 2). More Group B patients used steroids and had low complement (Table 2), suggesting Group B patients were sicker. However, disease activity scores did not differ between groups.
Table 2.
Baseline characteristics of the population.
| Group A1 | Group B2 | Groups A + B | |
|---|---|---|---|
| Baseline disease activity | |||
| BILAG 2004, mean (SD) | 15.0 (4.32) | 15.3 (6.52) | 15.2 (5.73) |
| SLEDAI, mean (SD) | 8.1 (2.73) | 9.3 (4.21) | 8.8 (3.73) |
| PGA, mean (SD) | 1.9 (0.33) | 1.9 (0.33) | 1.89 (0.33) |
| CLASI activity, mean (SD) | 5.6 (7.50) | 4.6 (5.18) | 5.0 (6.20) |
| Tender joints, mean (SD) | 14.3 (8.06) | 11.7 (8.25) | 12.7 (8.22) |
| Swollen joints, mean (SD) | 10.7 (7.05) | 9.9 (7.28) | 10.2 (7.15) |
| Low C3, n (%) | 3 (7.3%) | 14 (22.6%) | 17 (16.5%) |
| Low C4, n (%) | 8 (19.5%) | 21 (33.9%) | 29 (28.2%) |
| Baseline Rx, n (%) | |||
| Azathioprine | 12 (29.3%) | 21 (33.9%) | 33 (32.1%) |
| Mycophenolate mofetil | 6 (14.6%) | 12 (19.3%) | 18 (17.5%) |
| Methotrexate | 12 (29.3%) | 14 (22.6%) | 26 (25.2%) |
| Any immunosuppressant3 | 30 (73.2%) | 47 (75.8%) | 77 (74.8%) |
| Antimalarial | 30 (73.2%) | 45 (72.6%) | 75 (72.8%) |
| Prednisone (or equivalent) | 8 (20.0%) | 22 (35.5%) | 30 (29.1%) |
| Steroid dose, mg/day4 mean(SD) | 8.6 (5.46) | 13.9 (10.03) | 12.5 (9.26) |
SLE patients participating in cross-sectional and prospective substudy.
SLE patients participating in cross-sectional substudy only.
Azathioprine, mycophenolate mofetil, or methotrexate.
In prednisone equivalents, one patient entered on 20 mg/day; all others received 10 mg or less/day.
Changes in Disease Activity (Group A)
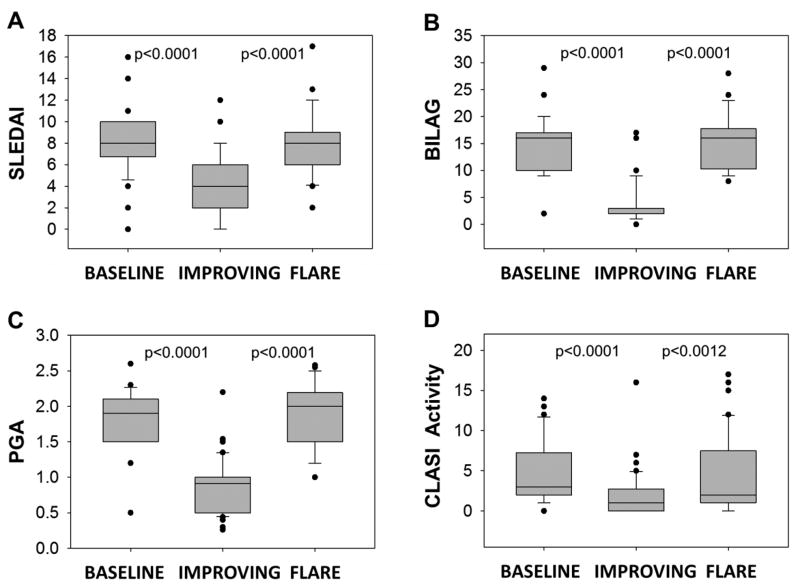
Supporting the clinician-weighted definitions of improvement and flare, scores of BILAG 2004, SLEDAI, CLASI, PGA, and joint counts decreased significantly at the Improving Visit and increased significantly at the Flare Visit for Group A (p<0.0001 in all cases except p=0.0012 for CLASI Improving vs. Flare) (Figure 1; Supplementary Figure 1).
Figure 1. Standardized disease activity measures at Baseline, Improving, and Flare Visits.
Improving and Flare Visits in Group A were designated based on clinician’s opinion with minimal input from the (A) SLEDAI or (B) BILAG tool, and no input from the (C) PGA, (D) CLASI, or Joint Counts (see Supplementary Figure 1). All disease activity measures significantly decreased from Baseline to Improving Visit and significantly increased from Improving Visit to Flare Visit. Disease activity measures are defined in detail in the Methods section. Changes from Baseline to Improving Visit and from Improving to Flare Visit were evaluated using the nonparametric Wilcoxon’s rank sum test.
The pre-specified primary endpoint was time to flare from Baseline. All steroid injections were administered within the first two weeks of Baseline. Within six months of Baseline, 40 of the 41 Group A patients exhibited flare, suggesting that analogous trials would have an extremely low placebo response rate at a six month milestone.
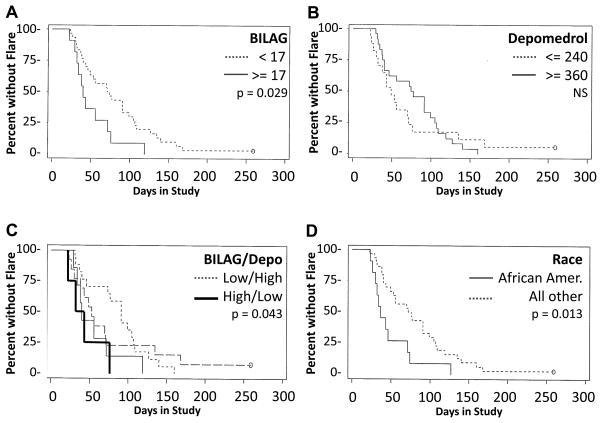
Time to flare did not differ between patients grouped by a priori definitions of moderate (n=25) vs high (n=16) disease activity (p=0.44). However, in exploratory evaluations seeking prognostic indicators, time to flare was significantly shorter in patients with Baseline cumulative BILAG scores ≥17 compared to patients with scores <17 (p=0.029). African Americans exhibited flare sooner than others (p=0.013) (Figure 2). Further, flare occurred sooner in patients with BILAG scores >17 who received <240 mg total depomedrol compared to those with BILAG scores <17 who received >320 mg total depomedrol (p=0.043). Thus, sicker patients requiring less depomedrol for an initial satisfactory response are likely to exhibit flares earlier than other patients, providing potentially useful parameters for clinical trial design (Figure 2). Low dose steroid or antimalarial use during the trial, or withdrawal of different baseline immunosuppressants had no discernable impact on time to flare; however this study was not powered to draw firm conclusions about these variables.
Figure 2. Time from Baseline to Flare Visit.
Flare occurred in 40 of 41 Group A patients (97.6%) within six months of Baseline. (A) Time to flare was reduced in patients with high BILAG scores (≥17; n=16) compared to those with low BILAG scores (<17; n=25). (B) Time to flare was not distinctly different in patients receiving high (≥360 mg) total depomedrol vs those receiving low (≤240 mg) depomedrol. (C) Flare was significantly delayed in patients with low BILAG scores who received high depomedrol compared to patients with high BILAG scores who were treated with low depomedrol. (D) Time to flare was reduced in African-American patients compared to all others. Comparisons were made by Kaplan-Meier analysis and log-rank test.
Safety
Adverse events were a pre-specified, descriptive secondary endpoint. There were no adverse events in Group B (single blood donation). Thirty-one adverse events were reported in Group A; all resolved after evaluation and/or treatment. Nineteen adverse events were grade one or two infections. The two grade three adverse events (Supplementary Table 1) included one serious (bleeding ulcer) and one non-serious event (anal abscess that responded to oral antibiotics). Both patients recovered and remained in the study. There were no serious adverse events from lupus flare or infection.
As another safety measure, disease severity was compared at Baseline versus Flare Visits (Supplementary Table 1). The percentage of patients with BILAG A (severe activity) did not differ between Baseline (29%) and Flare Visit (29%). Five patients (12.2%) who entered with BILAG B (moderate activity) exited with BILAG A, and five (12.2%) who entered with BILAG A ended with BILAG B. No severe flares were organ-threatening; no flares involved nephritis, CNS, serious hematologic features, or solid organs. One patient was followed for a year and had no flare. All end of study flares were treated with SoC and resolved within six weeks.
SLE immunopathology
A large percentage of patients with SLE are characterized by an elevated type I interferon-inducible gene signature,(8) but whether interferon signals reliably discriminate optimal treatment groups remains unclear. At Baseline, patients (Groups A and B) with high type I interferon signals (IFN High) were compared to those without the signature (IFN Low) (Table 3). IFN High patients displayed markedly higher gene expression of BLyS (TNFSF13B, p=0.005) and interleukin 17 receptor A (IL17RA, p=0.009), confirming previous observations that activated BLyS and IL17 pathways are associated with type I IFN activity in SLE (7–9). IL23A gene expression was not increased in IFN High patients. Protein levels of BLyS (p=0.0001) and IL23 (IL23p19) (p=0.01) were also higher in IFN High patients. More IFN High patients had antibodies to dsDNA (p=0.0001), SSA/Ro (p=0.0007), RNP (p=0.0001), and Sm (p=0.01) (Table 3).
Table 3.
Gene and protein expression relevant to IL17 and BLyS pathways and other clinical variables in SLE patient subsets with or without a type I interferon signature.
| IFN High n=49 |
IFN Low n=46 |
p-value1 | |
|---|---|---|---|
| Gene expression (mean ΔCt vs. healthy controls)2 | |||
| TNFSF13b (BLyS) | 1.41 | 2.55 | 0.003 |
| IL17RA | 1.37 | 1.59 | 0.01 |
| IL23A | 2.23 | 2.27 | 0.77 |
| Inflammatory mediators (mean pg/mL) | |||
| BLyS | 1539.4 | 908.2 | 0.0001 |
| IL23p19 | 0.229 | 0.019 | 0.01 |
| Autoantibodies (% positive) | |||
| anti-dsDNA | 30 | 2 | 0.0001 |
| anti-Ro | 30 | 4 | 0.0007 |
| anti-Sm | 13 | 0 | 0.01 |
| anti-RNP | 37 | 0 | 0.0001 |
| Disease Activity (mean) | |||
| SLEDAI | 9.73 | 7.70 | 0.009 |
| BILAG | 16.5 | 14.0 | 0.04 |
Gene expression was compared in 95 patients with evaluable RNA samples by ANCOVA with batch, RIN, and % neutrophils as covariates, with t-test to compare IFN high to IFN low. Other values were compared by ANOVA (protein concentration), Fishers Exact Test (autoantibodies), or t-test (disease activity scores).
Lower number indicates higher gene expression.
Impact of SoC on TH17 and BLyS pathways in IFN High and IFN Low patients
Whether categorization of patients by interferon signatures could clarify understanding of SoC effects has not been previously studied. Baseline IL17RA gene expression was modestly decreased in all patients on immunosuppressants compared to those who were not (20% lower on methotrexate, p=0.059; 41% lower on mycophenolate, p=0.0039; 9% lower on azathioprine, p=0.33) (Table 4, Supplementary Figure 2); however, the impact of these medications was more evident when IFN High and IFN Low patients were examined separately. Mycophenolate and methotrexate reduced IL17RA expression in the IFN Low group only (Table 4). Hydroxychloroquine treatment was associated with substantially lower TNFSF13B (BLyS) gene expression in IFN High patients, but not in IFN Low patients (Table 4).
Table 4.
Impact of standard SLE medications on gene expression in patient subsets with or without a type I interferon signature.
| Gene (RNA) | Rx | All Patients1 (n=95) | IFN High2 (n=49) | IFN Low (n=46) | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| % different from No Rx | p-value | % different from No Rx | p-value | % different from No Rx | p-value | ||
| IL17RA | MTX | 19.6 | 0.059 | 9.3 | 0.35 | 30.8 | 0.069 |
| MMF | 40.9 | 0.0039 | 1.2 | 0.15 | 96.2 | 0.0015 | |
| AZA | 8.8 | 0.33 | 14.7 | 0.15 | 3.2 | 0.93 | |
| TNFSF13B (BLyS) | HCQ | 48.9 | 0.22 | 204.1 | 0.0068 | −27.1 | 0.52 |
Gene expression was analyzed in 95 patients with evaluable RNA samples by ANCOVA with batch, RIN, and % neutrophils as covariates, with t-test to compare % difference between those taking or not taking a designated treatment. The overall population, IFN High subset, and IFN Low subset were analyzed separately. Note that despite lower numbers of patients in each defined subset, differences, where apparent, have greater statistical significance, emphasizing the increased clarity that might be derived from analysis of meaningful patient types.
The type I IFN signature was defined by GBP5, HERC5, IFI27, IRF7, ISG15, LY6E, MX1, OAS2, OAS3, RSAD2, and USP18 (see Methods).
MTX=methotrexate, MMF= mycophenolate mofetil, AZA= azathioprine, HCQ=hydroxychloroquine
Individual immunosuppressants had no consistent effects on protein or gene expression when comparing Baseline (active disease on treatment) and Flare Visits (off treatment) in Group A patients, but the numbers were small in each treatment group (data not shown). However, BlyS RNA (but not protein) expression increased 2.4-fold from Baseline to Flare Visit in all IFN Low patients, regardless of Baseline treatments (p=0.0003), reaching levels equivalent to those seen in IFN High patients (data not shown).
DISCUSSION
The BOLD data suggest that future SLE clinical trials could achieve very low placebo response rates without unacceptably compromising patient safety by enrolling SLE patients with active, non-organ threatening disease, providing temporary steroids, and withdrawing SoC immunosuppressants. Risk of serious infections might even be reduced by eliminating excess immunosuppressants at baseline, but this study was not designed to test that hypothesis.
Most SLE clinical trials continue various, often effective SoC background medications being taken by patients at entry (12–19), based on assumptions that this minimizes risk of serious flares and that immunologic interference is minimal. However, these two assumptions are not evidence-based. Moreover, they are inconsistent with each other, since flares are only prevented by modulating immunologic pathways. In fact, data from trials indicate that patients on SoC alone have almost as many infections as those on SoC plus biologics (12–16); it therefore seems likely that SoC is as risky or more so than some biologics. Other analyses suggest that in trials with a polypharmacy design, efficacy can only be distinguished in subsets of patients with higher grade disease activity (15, 16). Since these patients are at highest risk for serious flares, it may be fortunate that interpretable studies can be performed for them without requiring treatment withdrawal.
Does this mean that clinical trials in SLE (and subsequent regulatory approvals) should be restricted to polypharmacy protocols in patients with the most severe disease? A recent shift in Phase III plans for at least one agent (15) suggests this notion is gaining traction, risking disenfranchisement of patients with chronic, moderate SLE from access to targeted biologics. There is a significant unmet need in this population, as even with good disease control, chronic, smoldering SLE leads to progressive organ damage, long-term morbidity, and mortality (30, 31). Our data suggest that immunosuppressants can be safely withdrawn in these patients to support a safe and interpretable trial design. This approach might provide early proof of efficacy in less vulnerable patients while helping define pharmacodynamic variables without confounding immunomodulatory signals. In turn, later trials of more vulnerable patients would be armed with better information to minimize risk of choosing untoward treatment combinations.
The need to account for confounding medications is highlighted by our exploratory observation that certain SoC treatments might have unique effects on IFN High vs IFN Low patients. Although they are used interchangeably in clinic and in clinical trials, immunosuppressants may have disparate and even opposing effects in SLE subgroups; our preliminary observations support this hypothesis. If not addressed, this could confound clinical trials and selection of optimal treatment combinations in the clinic. Therefore, larger, prospective studies with prespecified biomarker endpoints and adjustments for multiple comparisons are needed to rigorously test the immunologic effects of antimalarials and immunosuppressants in different patient subsets. Our incidental finding that IFN Low patients exhibit uncharacteristically high BLyS RNA expression at the time of acute flare also warrants further exploration. We previously described BLyS as a robust marker for SLE disease activation (7), and various alternative immunologic routes can induce BLyS (32). Thus, BLyS could represent a common pathway for discrete SLE subpopulations at the time of flare.
Given the heterogeneity of lupus it is likely that exceptions to the broad themes inspected in this small study will arise, drawing attention to antagonistic or synergistic pathways in smaller patient subsets. However, better knowledge of immunologic effects of SoC lupus medications on major identifiable subgroups of lupus will aid more rational testing and more successful use of targeted biologics.
Supplementary Material
Acknowledgments
In addition to the authors, many collaborators at Pfizer contributed their research time, reagents and their laboratory infrastructure. We also thank Dr. Jill P. Buyon (New York University, New York, USA) for serving as the safety monitor and contributing helpful discussions to the design of the study, Dr. Rebecka Bourn (Oklahoma Medical Research Foundation, Oklahoma City, OK) for editorial assistance, and Dr. Angela Andersen (Life Science Editors) for comments on the manuscript.
Funding: This work was funded as a grant to the Oklahoma Medical Research Foundation as an investigator-initiated study by Pfizer. FI, TZ, MW, AH, M O’T, PR, MH, and SS were employees of Pfizer at the time this study was conducted. The sponsor had no role in the conduct of the clinical study, but many Pfizer colleagues participated in biomarker data collection, sample assays, data analysis, and the writing of the paper. These contributions are detailed above. This project did not include the study of any Pfizer products. Biomarker evaluation was also supported by grants from the NIH, including U19AI082714, U01AI101934, P30AR053483, and Institutional Development Awards (IDeA) from NIGMS (U54GM104938, P30GM103510). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial Support: This work was funded as an investigator-initiated study by a grant to the Oklahoma Medical Research Foundation from Pfizer. This project did not include the study of any Pfizer products. The sponsor had no role in the study design, but Pfizer colleagues participated in data collection, data analysis, and writing the paper (see Acknowledgements). Fred Immermann, Maryann Whitley, Tianhui Zhou, Andrew Hill, Margot O’Toole, Padmalatha Reddy, Marek Honczarenko, and Sudhakar Sridharan were employees of Pfizer in the course of the study. Fred Immermann, Maryann Whitley, Andrew Hill, and Padmalatha Reddy report Pfizer stock ownership and/or options. Joan T Merrill, Aikaterini Thanou, Joe Rawdon, Joel M Guthridge, and Judith A James report no conflicts of interest. Biomarker evaluation was also supported by grants from the NIH, including U19AI082714, U01AI101934, P30AR053483, and Institutional Development Awards (IDeA) from NIGMS (U54GM104938, P30GM103510).
Footnotes
ClinicalTrials.gov Identifier: NCT00987831 (See full protocol at https://omrf.org/wp-content/uploads/2015/09/Protocol-for-BOLD-Study-Version-5-26Jan2012-CLEAN-1-26-12.pdf)
Author Contributions: JTM, JAJ and SS developed the study concept and JTM designed the study and led the clinical trial. The other authors assisted with data acquisition, either in the clinic (JR and KT), by providing interim analyses and final statistical support (FI, TZ, AH, and MW), or by participating in biomarker measurements (all others). The authors had access to all the study data, take responsibility for the accuracy of the analysis, and had authority over manuscript preparation and the decision to submit the manuscript for publication.
References
- 1.Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358(9):929–39. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 2.Cervera R, Khamashta MA, Font J, Sebastiani GD, Gil A, Lavilla P, et al. Systemic lupus erythematosus: clinical and immunologic patterns of disease expression in a cohort of 1,000 patients. The European Working Party on Systemic Lupus Erythematosus. Medicine (Baltimore) 1993;72(2):113–24. [PubMed] [Google Scholar]
- 3.Font J, Cervera R, Ramos-Casals M, Garcia-Carrasco M, Sents J, Herrero C, et al. Clusters of clinical and immunologic features in systemic lupus erythematosus: analysis of 600 patients from a single center. Semin Arthritis Rheum. 2004;33(4):217–30. doi: 10.1053/s0049-0172(03)00133-1. [DOI] [PubMed] [Google Scholar]
- 4.Yao Y, Richman L, Higgs BW, Morehouse CA, de los Reyes M, Brohawn P, et al. Neutralization of interferon-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009;60(6):1785–96. doi: 10.1002/art.24557. [DOI] [PubMed] [Google Scholar]
- 5.Mohan C, Datta SK. Lupus: key pathogenic mechanisms and contributing factors. Clin Immunol Immunopathol. 1995;77(3):209–20. doi: 10.1006/clin.1995.1146. [DOI] [PubMed] [Google Scholar]
- 6.Kotzin BL. Systemic lupus erythematosus. Cell. 1996;85(3):303–6. doi: 10.1016/s0092-8674(00)81108-3. [DOI] [PubMed] [Google Scholar]
- 7.Ritterhouse LL, Crowe SR, Niewold TB, Merrill JT, Roberts VC, Dedeke AB, et al. B lymphocyte stimulator levels in systemic lupus erythematosus: higher circulating levels in African American patients and increased production after influenza vaccination in patients with low baseline levels. Arthritis Rheum. 2011;63(12):3931–41. doi: 10.1002/art.30598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brkic Z, Corneth OB, van Helden-Meeuwsen CG, Dolhain RJ, Maria NI, Paulissen SM, et al. T-helper 17 cell cytokines and interferon type I: partners in crime in systemic lupus erythematosus? Arthritis Res Ther. 2014;16(2):R62. doi: 10.1186/ar4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolff S, Quandt D, Wilde B, Feldkamp T, Hua F, Cai X, et al. Increased expression of costimulatory markers CD134 and CD80 on interleukin-17 producing T cells in patients with systemic lupus erythematosus. Arthritis Res Ther. 2010;12(4):R150. doi: 10.1186/ar3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sestak AL, Furnrohr BG, Harley JB, Merrill JT, Namjou B. The genetics of systemic lupus erythematosus and implications for targeted therapy. Ann Rheum Dis. 2011;70(Suppl 1):i37–43. doi: 10.1136/ard.2010.138057. [DOI] [PubMed] [Google Scholar]
- 11.Bentham J, Morris DL, Cunninghame Graham DS, Pinder CL, Tombleson P, Behrens TW, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47(12):1457–64. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navarra SV, Guzman RM, Gallacher AE, Hall S, Levy RA, Jimenez RE, et al. Efficacy and safety of belimumab in patients with active systemic lupus erythematosus: a randomised, placebo-controlled, phase 3 trial. Lancet. 2011;377(9767):721–31. doi: 10.1016/S0140-6736(10)61354-2. [DOI] [PubMed] [Google Scholar]
- 13.Furie R, Petri M, Zamani O, Cervera R, Wallace DJ, Tegzova D, et al. A phase III, randomized, placebo-controlled study of belimumab, a monoclonal antibody that inhibits B lymphocyte stimulator, in patients with systemic lupus erythematosus. Arthritis Rheum. 2011;63(12):3918–30. doi: 10.1002/art.30613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace DJ, Kalunian K, Petri MA, Strand V, Houssiau FA, Pike M, et al. Efficacy and safety of epratuzumab in patients with moderate/severe active systemic lupus erythematosus: results from EMBLEM, a phase IIb, randomised, double-blind, placebo-controlled, multicentre study. Ann Rheum Dis. 2014;73(1):183–90. doi: 10.1136/annrheumdis-2012-202760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furie RA, Leon G, Thomas M, Petri MA, Chu AD, Hislop C, et al. A phase 2, randomised, placebo-controlled clinical trial of blisibimod, an inhibitor of B cell activating factor, in patients with moderate-to-severe systemic lupus erythematosus, the PEARL-SC study. Ann Rheum Dis. 2015;74(9):1667–75. doi: 10.1136/annrheumdis-2013-205144. [DOI] [PubMed] [Google Scholar]
- 16.van Vollenhoven RF, Petri MA, Cervera R, Roth DA, Ji BN, Kleoudis CS, et al. Belimumab in the treatment of systemic lupus erythematosus: high disease activity predictors of response. Ann Rheum Dis. 2012;71(8):1343–9. doi: 10.1136/annrheumdis-2011-200937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrill JT, Erkan D, Buyon JP. Challenges in bringing the bench to bedside in drug development for SLE. Nat Rev Drug Discov. 2004;3(12):1036–46. doi: 10.1038/nrd1577. [DOI] [PubMed] [Google Scholar]
- 18.Merrill JT, Buyon JP. Connective tissue diseases: What does the death of Riquent hold for the future of SLE? Nat Rev Rheumatol. 2009;5(6):306–7. doi: 10.1038/nrrheum.2009.99. [DOI] [PubMed] [Google Scholar]
- 19.Merrill JT. Emergence of targeted immune therapies for systemic lupus. Expert Opin Emerg Drugs. 2005;10(1):53–65. doi: 10.1517/14728214.10.1.53. [DOI] [PubMed] [Google Scholar]
- 20.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 21.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 22.Buyon JP, Petri MA, Kim MY, Kalunian KC, Grossman J, Hahn BH, et al. The effect of combined estrogen and progesterone hormone replacement therapy on disease activity in systemic lupus erythematosus: a randomized trial. Ann Intern Med. 2005;142(12 Pt 1):953–62. doi: 10.7326/0003-4819-142-12_part_1-200506210-00004. [DOI] [PubMed] [Google Scholar]
- 23.Petri M, Kim MY, Kalunian KC, Grossman J, Hahn BH, Sammaritano LR, et al. Combined oral contraceptives in women with systemic lupus erythematosus. N Engl J Med. 2005;353(24):2550–8. doi: 10.1056/NEJMoa051135. [DOI] [PubMed] [Google Scholar]
- 24.Gladman DD, Ibanez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol. 2002;29(2):288–91. [PubMed] [Google Scholar]
- 25.Touma Z, Urowitz MB, Gladman DD. SLEDAI-2K for a 30-day window. LUPUS. 2010;19(1):49–51. doi: 10.1177/0961203309346505. [DOI] [PubMed] [Google Scholar]
- 26.Yee CS, Farewell V, Isenberg DA, Griffiths B, Teh LS, Bruce IN, et al. The BILAG-2004 index is sensitive to change for assessment of SLE disease activity. Rheumatology (Oxford) 2009;48(6):691–5. doi: 10.1093/rheumatology/kep064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein R, Moghadam-Kia S, LoMonico J, Okawa J, Coley C, Taylor L, et al. Development of the CLASI as a tool to measure disease severity and responsiveness to therapy in cutaneous lupus erythematosus. Arch Dermatol. 2011;147(2):203–8. doi: 10.1001/archdermatol.2010.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munroe ME, Vista ES, Guthridge JM, Thompson LF, Merrill JT, James JA. Proinflammatory adaptive cytokine and shed tumor necrosis factor receptor levels are elevated preceding systemic lupus erythematosus disease flare. Arthritis Rheumatol. 2014;66(7):1888–99. doi: 10.1002/art.38573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill AA, Immermann FW, Zhang Y, Reddy PS, Zhou T, O’Toole M, et al. FRI0003 Determination of interferon (IFN) signatures for sle patients may be critical for optimal treatment selection but depends on the genes chosen: report from the bold (biomarkers of lupus disease) study. Ann Rheum Dis. 2013;72(Suppl 3):A369–A70. [Google Scholar]
- 30.Urowitz MB, Gladman DD, Tom BD, Ibanez D, Farewell VT. Changing patterns in mortality and disease outcomes for patients with systemic lupus erythematosus. J Rheumatol. 2008;35(11):2152–8. doi: 10.3899/jrheum.080214. [DOI] [PubMed] [Google Scholar]
- 31.Urowitz MB, Gladman DD, Ibanez D, Fortin PR, Bae SC, Gordon C, et al. Evolution of disease burden over five years in a multicenter inception systemic lupus erythematosus cohort. Arthritis Care Res (Hoboken) 2012;64(1):132–7. doi: 10.1002/acr.20648. [DOI] [PubMed] [Google Scholar]
- 32.Schneider P. The role of APRIL and BAFF in lymphocyte activation. Curr Opin Immunol. 2005;17(3):282–9. doi: 10.1016/j.coi.2005.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.