Abstract
Background
Advancing structural and functional maturation of stem cell-derived cardiomyocytes remains a key challenge for applications in disease modelling, drug screening, and heart repair. Here, we sought to advance cardiomyocyte maturation in engineered human myocardium (EHM) towards an adult phenotype under defined conditions.
Methods
We systematically investigated cell composition, matrix and media conditions to generate EHM from embryonic and induced pluripotent stem cell-derived cardiomyocytes and fibroblasts with organotypic functionality under serum-free conditions. We employed morphological, functional, and transcriptome analyses to benchmark maturation of EHM.
Results
EHM demonstrated important structural and functional properties of postnatal myocardium, including: (1) rod-shaped cardiomyocytes with M-bands assembled as a functional syncytium; (2) systolic twitch forces at a similar level as observed in bona fide postnatal myocardium; (3) a positive force-frequency-response; (4) inotropic responses to β-adrenergic stimulation mediated via canonical β1- and β2-adrenoceptor signaling pathways; and (5) evidence for advanced molecular maturation by transcriptome profiling. EHM responded to chronic catecholamine toxicity with contractile dysfunction, cardiomyocyte hypertrophy, cardiomyocyte death, and NT-proBNP release; all are classical hallmarks of heart failure. Additionally, we demonstrate scalability of EHM according to anticipated clinical demands for cardiac repair.
Conclusions
We provide proof-of-concept for a universally applicable technology for the engineering of macro-scale human myocardium for disease modelling and heart repair from embryonic and induced pluripotent stem cell-derived cardiomyocytes under defined, serum-free conditions.
Keywords: Tissue Engineering, Stem Cells, Disease Modelling, Heart Failure, Regeneration
Journal Subject Terms: Translational Studies, Stem Cells, Cell Therapy, Biomarkers, Contractile Function, Myocardial Regeneration
Introduction
The availability of human embryonic stem cells (ESCs)1 as well as human induced pluripotent stem cells (iPSCs)2 and the scalability of their directed differentiation into bona fide cardiomyocytes3–7 have facilitated the rapid evolution of myocardial tissue engineering. Early tissue engineering studies in chick embryo and rodent models have established electromechanical stimulation as an important engineering paradigm8–10, which has now been translated to human models11–16. The accumulating evidence for advanced maturation in three-dimensional versus monolayer cultures provides a solid rationale for applications in phenotypic screens11 and heart repair17, 18. As the use of myocardial tissue engineering increases in academia and industry it is essential to establish conditions readily adaptable to current good manufacturing practice (cGMP). To achieve this goal, it is imperative to define the essential elements required for the structural and functional maturation of tissue engineered myocardium under defined, serum-free conditions. Finally, robust and reproducible utility in ESC- and iPSC-based models is of pivotal importance.
In this study, we report a systematic approach for the design of engineered human myocardium (EHM) with structural and functional properties observed in the postnatal heart. Unbiased transcriptome profiling provided evidence for advanced maturation in EHM as compared to parallel monolayer cultures. In order to demonstrate the applicability of EHM for the modelling of “human heart failure in the dish” we introduce a catecholamine overstimulation protocol with outcomes similar to what is typically observed in clinical heart failure. Finally, we provide proof-of-concept for the scalability and in vivo applicability of defined EHM as an important step towards clinical translation of tissue engineered heart repair.
Methods
Human pluripotent stem cell (PSC) lines
We utilized: H9.219; HES3 (Embryonic Stem Cell International, Singapore) including the transgenic derivative HES3-ENVY20; HES2 (Embryonic Stem Cell International, Singapore) including the transgenic derivative HES2-RFP21; H71 (WiCell, Madison, WI); hiPS-G1 (generated in-house using Sendai Virus reprogramming, Cytotune Kit, Thermo Fisher); hiPS-BJ (Dr. Toshiyuki Araki, New York) - approval according to the German Stem Cell Act by the Robert-Koch-Institute to W.-H.Z.: permit #12; reference number: 1710-79-1-4-16.
Cardiomyocyte differentiation and purification
Differentiated embryoid bodies (H9.2, HES3, HES3-ENVY, HES2, hiPS-BJ) were shipped to Göttingen at room temperature and arrived within 72–96 hrs. Cardiomyocytes from H7 (L. A. Couture, City of Hope) were shipped at −80°C. Frozen human cardiomyocytes were stored at −152°C. Most experiments were performed with HES2-RFP and hiPS-G1 lines differentiated in monolayers according to Hudson et al.22 with modifications. Briefly, PSCs were plated at 5×104–1×105 cells/cm2 on 1:30 Matrigel in PBS coated plates and cultured in Knock-out DMEM, 20% Knock-out Serum Replacement, 2 mmol/L glutamine, 1% non-essential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin (all Life Technologies) mixed 1:1 with irradiated human foreskin fibroblast (HFF)-conditioned medium with 10 ng/ml FGF2 or TeSR-E8 (STEMCELL Technologies). After 1 day the cells were rinsed with RPMI medium and then cultured in RPMI, 2% B27, 200 μmol/L L-ascorbic acid-2-phosphate sesquimagnesium salt hydrate (Asc., Sigma-Aldrich), 9 ng/ml Activin A (R&D Systems), 5 ng/ml BMP4 (R&D Systems), 1 μmol/L CHIR99021 (Stemgent), and 5 ng/ml FGF-2 (Miltenyi Biotec) for three days. Following another wash with RPMI medium cells were cultured from day 4–13 in RPMI, 2% B27, and 200 μmol/L Asc with 5 μmol/L IWP4 (Stemgent). Where indicated cardiomyocytes were metabolically purified by glucose deprivation23 from day 13–17 in RPMI without glucose and glutamine (Biological Industries), 2.2 mmol/L sodium lactate (Sigma-Aldrich), 100 μmol/l β-mercaptoethanol (Sigma-Aldrich), 100 U/ml penicillin, and 100 μg/ml streptomycin. Please refer to Supplementary Table 1 for an overview of the different cardiac differentiation protocols3, 17, 19, 22, 24 used in this study.
Engineered Heart Muscle (EHM) generation
An overview of the protocols to generate human EHM is displayed in Table 1. Details can be found in the Supplemental Material.
Table 1.
Overview of EHM protocols.
| Component | Starting Protocol | Matrix Protocol | Serum-free Protocol | |
|---|---|---|---|---|
| EHM reconstitution medium | Collagen rat (research grade) | 0.4 mg/EHM | ||
| Collagen bovine (medical grade) | 0.4 mg/EHM | 0.4 mg/EHM | ||
| Matrigel® | 10% v/v | |||
| Reconstitution Medium | DMEM | DMEM | RPMI | |
| Horse Serum | 10% | |||
| Chick Embryo Extract | 2% | |||
| Fetal Bovine Serum | 40% | |||
| B27 (without insulin) | 4% | |||
| EHM culture medium | Base Medium | Iscove | Iscove | Iscove* |
| Fetal Bovine Serum | 20% | 20% | ||
| B27 (without insulin) | 4% | |||
| IGF-1 | 100 ng/mL | |||
| FGF-2 | 10 ng/mL | |||
| VEGF165 | 5 ng/mL | |||
| TGF-β1 | 5 ng/mL | |||
| Non-Essential Amino Acids | 1% | 1% | 1% | |
| Glutamine | 2 mmol/L | 2 mmol/L | 2 mmol/L | |
| Penicillin | 100 U/mL | 100 U/mL | 100 U/mL | |
| Streptomycin | 100 μg/mL | 100 μg/mL | 100 μg/mL | |
| β-Mercaptoethanol | 100 μmol/L | 100 μmol/L |
Alternatively other basal medium with ≥1.2 mmol/L calcium
Analyses of contractile function
Contraction experiments were performed under isometric conditions in organ baths at 37°C in gassed (5% CO2/95% O2) Tyrode’s solution (containing: 120 NaCl, 1 MgCl2, 0.2 CaCl2, 5.4 KCl, 22.6 NaHCO3, 4.2 NaH2PO4, 5.6 glucose, and 0.56 ascorbate; all in mmol/L). Spontaneous beating frequency was determined at 2 mmol/L calcium after 10 min equilibration of EHMs. EHM were electrically stimulated at 1.5–2 Hz with 5 ms square pulses of 200 mA. EHMs were mechanically stretched at intervals of 125 μm until the maximum systolic force amplitude (force of contraction; FOC) was observed according to the Frank-Starling law. Responses to increasing extracellular calcium (0.2–4 mmol/L), increasing stimulation frequencies (1, 2, 3 Hz), and adrenergic stimulation with isoprenaline (1 μmol/L) followed by functional antagonism by the muscarinergic agonist carbachol (10 μmol/L) at ~EC50 calcium of individual EHM were investigated. Where indicated an isoprenaline concentration response curve was performed in the presence or absence of specific β-1 adrenoceptor antagonist CGP-20712A (300 nmol/L, Sigma-Aldrich) or specific β-2 adrenoceptor antagonist ICI-118551 (50 nmol/L, Sigma-Aldrich). Post-rest potentiation was assessed after 2 min stimulation at 1.5–2 Hz and pauses of 10 sec. The last stimulated beat amplitude was compared to the first stimulated beat amplitude after the pause. Only EHM without spontaneous contractions during the stimulation pause were included in the analysis.
EHM “heart failure” model
L-Norepinephrine hydrochloride (NE) and endothelin-1 (ET-1) were prepared in distilled water containing 200 μmol/L Asc (all from Sigma-Aldrich). EHM were treated with indicated concentrations for 7 days. NT-proBNP was measured using the Elecsys® kit (Roche Diagnostics).
EHM dissociation
To isolate single cells, EHMs were incubated in collagenase 1 solution (2 mg/ml in calcium-containing PBS in the presence of 20% fetal bovine serum) at 37°C for 60–90 mins. EHM were washed with PBS (without calcium) and further incubated in Accutase (Millipore), 0.0125% Trypsin (Life Technologies), 20 μg/ml DNAse (Calbiochem) for 30 mins at room temperature. Cells were then mechanically separated and transferred into PBS with 5% fetal bovine serum for live cell flow cytometry. To preserve rod-shaped morphology of EHM-derived cardiomyocytes, 30 mmol/L 2,3-butanedione monoxime (BDM) was added to the collagenase solution and the final cell suspension was quickly transferred to 4% formaldehyde (Histofix, Roth). EHM-derived cells were spread out on glass slides (Superfrost plus, Menzel-Gläser) in distilled water and air-dried.
Human samples
Human fetal heart tissue (3 biopsies from a single donation) was obtained after elective abortion (vacuum aspiration) without medical indication following informed consent. The collection of fetal material was approved by the Ethical Committee of the Leiden University Medical Center (MEC-P08.087). Human heart samples were collected from the left ventricles of non-failing donor hearts (n=4 donor hearts) not suitable for transplantation as approved by the Ethical Committee of the University Medical Center Göttingen (31/9/00). Gingiva samples were obtained from otherwise healthy donors during elective periodontal surgical treatment as approved by the Ethical Committee of the University Medical Center Göttingen (16/6/09). Cardiac fibroblasts were purchased from Lonza. The study was conducted in accordance with the Declaration of Helsinki by the World Medical Association.
RNA sequencing
RNA was prepared using Trizol (Life Technologies) following the manufacturer’s instruction. RNA integrity was assessed with the Agilent Bioanalyzer 2100. Total RNA was subjected to library preparation (TruSeq Stranded Total RNA Sample Prep Kit from Illumina) and RNA-sequencing on an Illumina HighSeq-2000 platform (SR 50 bp; >25 Mio reads /sample). Sequence images were transformed with the Illumina software BaseCaller to bcl files, which were demultiplexed to fastq files with CASAVA (v1.8.2). Fastq files were mapped to GRCh38/hg38 using STAR 2.4 or TopHat225 and Reads Per Kilobase of transcript per Million (RPKM) were calculated based on the Ensembl transcript length as extracted by biomaRt (v2.24). We only considered “protein_coding” transcripts for further analysis. Gene ontology (GO) analysis was performed through DAVID26. To determine cardiomyocyte and fibroblast transcriptomes the following algorithm was applied: (1) counts (>10) of purified PSC-derived cardiomyocytes (HES2, iCELL, hiPS-G1; n=3 from each line) and fibroblasts from three different sources (heart, skin, gingiva; n=3 from each source) were pooled and the differentially expressed genes (DEG, p<0.05, corrected by Benjamini-Hochberg method for multiple testing27) between cardiomyocyte and fibroblast pools determined using edgeR28; (2) log2 changes of DEG were calculated and genes omitted with a log2 difference lower than mean log2 of all cardiomyocyte genes; (3) resulting cardiomyocyte- and fibroblast-enriched genes were screened for RPKM values in adult healthy heart and all genes with RPKM <1 in adult heart were omitted.
3D printing of flexible holders for EHM patches
Flexible holders for the EHM patch construction were printed on a Connex350 (Stratasys) 3D printer using the biocompatible MED610 polymer as stiff base and TangoBlack polymer for the flexible poles. Support material was sprayed off using a Balco Powerblast waterjet. Holders were incubated for 15 min in isopropanol to dissolve traces of support material, sprayed again, rinsed, and soaked in water for at least 5 days to bleed-out left-overs from the polymerization process. Holders were then sterilized by plasma cleaning for 30 sec (Harrick Plasma).
Imaging of EHM patch function
For each measurement plates were recorded inside a 37°C climate chamber for at least two minutes at 50 fps resolution using a Basler acA2000 8 bit monochrome camera with a Kowa 35 mm lens from ~45cm distance. Back light was set to bleach background pixels and facilitate video analysis by a custom made Matlab code. The region of interest was manually adjusted to patch size, non-background pixel were selected by an intensity cutoff, and the Matlab imdilate and imfill commands were used to close gaps and fill holes in the tissue leaving pixels belonging to patch and poles white (1) and everything else black (0). The number of white pixel represents the surface area sa at time t and was converted to fractional area change (FAC) by division through the maximum sa of a contraction cycle. Contraction peaks, i.e., the time-points of maximal FAC, were identified automatically; FAC and beating frequency, determined from peak-to-peak intervals, were averaged over 2 minutes.
Implantation of human patches
EHM patches were epicardially implanted into immunosuppressed athymic (nude) rats (Charles River) as described previously17.
Flow cytometry and immunofluorescence staining
Quantitative and qualitative analyses were performed after antibody labelling (Supplementary Table 2) as described previously29.
Statistical analyses
Data are presented as mean ± standard error of the mean. Statistical differences between two groups were tested by two-sided unpaired or paired Student’s t-tests. In case of 3 and more groups 1-way or 2-way unrepeated or repeated-measures ANOVA with appropriate post-hoc testing was performed. The performed tests are specified in the respective figure legends. Statistical testing was performed with GraphPad Prism 6.
Additional methods are described in the Supplemental Material.
Results
Definition of Cell Composition in EHM
EHM formation comprises two macroscopically distinguishable phases (Figure 1a): (i) EHM consolidation in casting molds with an onset of spontaneous beating in variably sized areas within 24 h, and (ii) EHM maturation with coordinated and rhythmic contractions of the whole tissue after 3 days and onwards (Supplementary Video 1). In pilot experiments we defined 1.5x106 ESC (H9.2 and HES3)-derived cardiomyocytes suspended in 500 μl collagen type I/Matrigel hydrogels as the optimal condition for the construction of force-generating EHM (Supplementary Table 1).
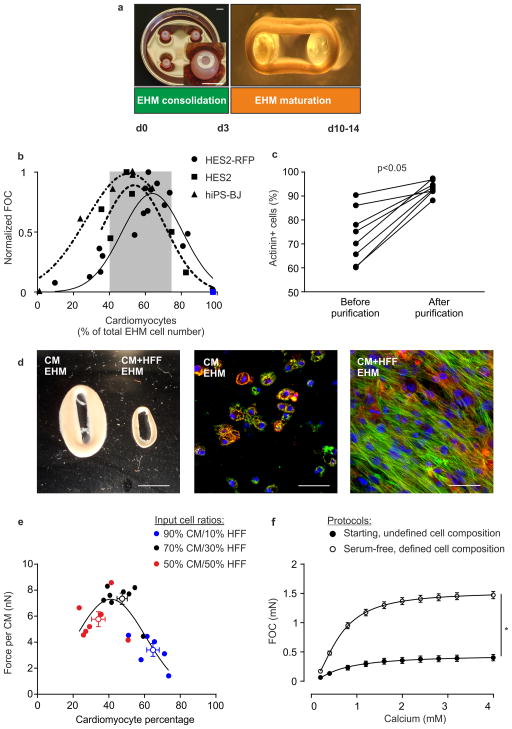
Figure 1. Defining human EHM.
(a) EHM generation is characterized by two phases: EHM consolidation for 3 days (left panel: casting mold with 4 EHMs; inset: magnification of EHM in mold) and EHM maturation for at least 7 days under mechanical load (right panel: EHM on flexible PDMS holders). Bars: 5 mm (left panel), 1 mm (right panel). (b) Force of contraction (FOC; normalized to maximal FOC) in relation to output cardiomyocyte percentage (actinin+ cells) of EHM made from HES2-RFP, HES2, and hiPS-BJ lines. Blue square indicates an EHM sample constructed from SIRPA2A-selected30 cardiomyocytes. Grey area indicates optimal cardiomyocyte percentage across indicated lines (mean±SD). (c) Purification of cardiomyocytes for defined EHM generation. Quantification of cardiomyocyte purity (actinin+ cells) before and after enrichment by metabolic selection; n=8, p<0.05 by two-tailed, paired Students t-test. (d) Macroscopic appearance of EHM with >92% CM (CM EHM) and EHM with >92% CM supplemented with HFF (70:30% CM+HFF EHM). Immunostaining for actinin (green), f-actin (red), and nuclei (blue) in CM EHM (middle panel) and CM+HFF EHM (right panel). Bars: 5 mm (left panel), 50 μm (middle and right panels). (e) Titration of the optimal CM:HFF-ratio. Output CM percentage and force per CM in 2 week old EHM made with indicated input cell ratios of purified CMs and HFFs. Colors indicate the input CM:HFF ratio of respective EHMs (each circle represents one individual EHM with an additional empty circle indicating the mean±SEM of the respective groups). (f) Force of contraction (FOC) recorded under increasing calcium concentrations and electrical stimulation at 1.5 Hz in 4 week EHM constructed according to the undefined Starting Protocol (n=19; Table 1) and defined, Serum-free Protocol (n=59; Table 1); pooled data from EHM generated from different ESC and iPSC lines (please refer also to Supplementary Figure 4 for detailed information); *p<0.05 by 2-way ANOVA with Tukey’s multiple comparisons post hoc test.
Using HES2 and hiPSC-BJ cardiac differentiation cultures with different cardiomyocyte content, we found that EHM containing a cardiomyocyte:non-myocyte composition of 1:1 at the time of force assessment developed maximal contractile forces (Figure 1b). This was in agreement with recent reports15 and our own experience in rodent models31 on the critical role of non-myocytes for the engineering of force-generating myocardium. We next formally tested the effect of the cardiomyocyte:non-myocyte ratio using metabolic selection23 for cardiomyocyte purification (Figure 1c) and human foreskin fibroblasts (HFF); EHM constructed directly from enriched cardiomyocyte populations did not condense and contained mostly rounded cardiomyocytes (Figure 1d; Supplementary Video 2). The addition of HFFs at a 70%/30% cardiomyocyte/fibroblast input ratio was optimal for the construction of force-generating EHM loops with a cardiomyocyte:fibroblast output ratio of ~1:1 (Figure 1e), confirming our initial findings (Figure 1b).
By defining the non-myocyte input, we observed advanced cardiomyocyte maturation with reduced variability in the functional maturation of EHM (Supplementary Fig. 1a) and a higher mean actinin fluorescence intensity per cell, indicating higher sarcomeric protein content per individual cardiomyocyte (Supplementary Fig. 1b). Further, the classical inotropic and lusitropic (relaxation) responses to isoprenaline were enhanced in defined EHM (Supplementary Fig. 1c). The number of immature ventricular cardiomyocytes (defined by simultaneous expression of MLC2A and MLC2V) was greatly reduced by EHM culture with more pronounced ventricular maturation in defined EHM (Supplementary Fig. 1d,e). Defining the non-myocyte population in EHM not only reduced intraline (Supplementary Fig. 1a), but also interline variability (Supplementary Fig. 1f). Moreover, expression of pluripotency associated genes and cell cycle activity in cardiomyocytes and non-myocytes were markedly reduced in defined EHM (Supplementary Fig. 2). Taken together, we conclude that defining the non-myocyte cell fraction increases robustness of the EHM protocol also with respect to its organotypic contractile function and ventricular fate.
Development of a defined, serum-free EHM construction protocol towards cGMP
The EHM Starting Protocol, which was devised from our original rodent tissue engineering protocol29, included a variety of undefined matrix (Matrigel) and serum (horse serum, fetal calf serum, chick embryo extract) components (Table 1). We first defined the matrix components and observed that EHM could be constructed from medical grade bovine collagen without Matrigel (Matrix Protocol), without any reduction in functionality Supplementary Fig. 3a–b). The addition of laminin (5 μg/EHM) or fibronectin (5 μg/EHM) to the Matrix Protocol did not further improve EHM function (Supplementary Fig. 3c). Factorial screens, including the assessment of the B27 supplement, were performed next with the aim to replace all animal culture medium components. To expedite the initial screens, we used simple HES2-cardiomyocyte aggregate cultures (Supplementary Fig. 3d) and subsequently tested putative cardio-instructive factors in EHM. We first selected a particular B27 medium supplementation (4% with insulin) based on cell viability. Subsequently, we selected growth factors (FGF-2, IGF-1, TGF-β1, VEGF165) for EHM testing according to the following criteria: [1] neutral or enhanced cell viability, and [2] enhanced cardiomyocyte actinin content or cardiomyocytes size. Finally, we confirmed that the combination of FGF-2, IGF-1, TGF-β1, and VEGF165 was maximally effective in supporting the formation of force-generating EHM (Supplementary Fig. 3e). In agreement with the important role of extracellular matrix (ECM) remodeling in early EHM cultures29, we found that TGF-β1 treatment in the consolidation phase (day 0–3) was necessary for enhanced EHM function (Supplementary Fig. 3e). Interestingly, we observed that antioxidants were not critical for EHM function and that omitting insulin (B27 minus insulin) enhanced EHM function compared to insulin-containing B27 (Supplementary Fig. 3f). This led to the definition of a minimal Serum-free Protocol containing 4% B27 without insulin plus TGF-β1, IGF-1, FGF-2, VEGF165 (Table 1). Finally, testing of the basal media identified calcium supplementation to physiological concentrations (1.2 mmol/L) as a critical parameter for optimal outcome (Supplementary Fig. 3g). Collectively, these experiments established a defined, Serum-free Protocol with markedly enhanced contractile performance compared to the undefined Starting Protocol (Figure 1f; Supplementary Video 3) and applicability to various ESC- and iPSC-EHM models (Supplementary Figure 4).
Evidence for Structural and Functional Maturation of EHM
We next investigated whether the defined, Serum-free Protocol supports EHM maturation. Enzymatic dispersion of EHM revealed cardiomyocytes with an elongated phenotype with sarcomeres in registry (Figure 2a, Supplementary Figure 5a). Compared to serum-containing EHM cultures and in line with the functional outcome (Figure 1f), intact rod-shaped cardiomyocytes from EHM constructed according to the Serum-free Protocol presented with a larger volume (12,101±1,240 vs. 5,649±1,410 μm3), but similar aspect ratio (7.6±0.4 vs. 6.7±0.9; n=28/10). Compared to 2D monolayer cardiomyocyte cultures and EHM constructed according to the Starting Protocol sarcomere size was larger in EHM constructed according to the defined, Serum-free Protocol (1.93±0.01 vs. 1.81±0.01 vs. 1.84±0.01 μm; n>120 sarcomeres from 12/8/10 cardiomyocytes). The low cardiomyocyte volume (20,000–35,000 μm3 reported in adult human cardiomyocytes32) was mainly due to a smaller cell width in EHM (width: 13±0.5 vs. 20–35 μm; length: 92±4 vs. 60–150 μm in adult human cardiomyocytes32, 33). Note that cardiomyocytes in EHM exhibited a similar width as observed in 6 week old infant heart (4–12 μm33). Ultrastructural analyses revealed that cardiomyocytes in EHM displayed a remarkable degree of sarcomere organization with clearly distinguishable Z-, I-, A-, H-, and M-bands (Figure 2b, Supplementary Figure 4b). Consistent with earlier reports on largely absent M-bands even in extended stem cell-derived cardiomyocyte monolayer cultures34 and tissue engineered models11–13, 15 we found little organization of M-bands in monolayer cardiomyocytes, but a high degree of organization in EHM-derived cardiomyocytes (Supplementary Figure 4c). Also, enhanced MLC2V organization and presence of n-cadherin+ intercalated disk-like structures were observed in EHM cardiomyocytes (Supplementary Figure 4b,c).
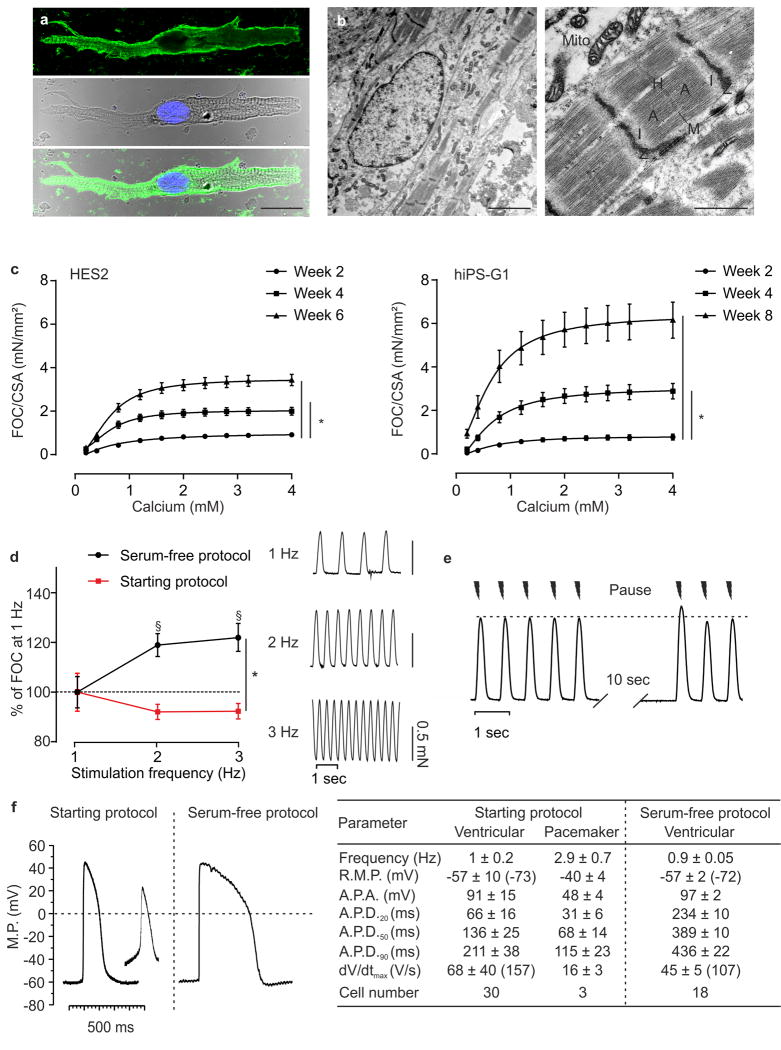
Figure 2. Morphological and functional maturation of EHM.
(a) Immunostaining of isolated cardiomyocyte from 4 week EHM (hiPS-G1); top panel: myosin heavy chain (green); middle panel: brightfield image with nucleus labelled with Hoechst (blue; bottom panel: overlay; bar: 20 μm. (b) Electron micrographs of 4 week EHM (hiPS-G1), low power (left panel, bar: 2.5 μm) and high power magnification (right panel: characteristic sarcomere structures are labelled; Mito: mitchondria; bar: 1 μm). (c) FOC per cross sectional area (CSA) of serum-free EHM from HES2 and hiPS-G1 at the indicated time points in culture; n=12/14/8 for weeks 2/4/6 in HES2 EHM and n=7/10/8 for weeks 2/4/8 in hiPS-G1 EHM *p<0.05 by 2-way ANOVA with Tukey’s multiple comparison post hoc test. (d) Force-frequency-response of hiPS-G1-EHM (at 4 weeks in culture) generated according to the Starting Protocol (red; n=8) and defined, Serum-free Protocol (black; n=21). §p<0.05 vs 1 Hz of the respective group by 2-way ANOVA with Tukey’s multiple comparison post hoc test; *p<0.05 by 2-way repeated measures ANOVA followed by Sidak’s multiple comparison test. (e) Representative force traces recorded from hiPS-G1-EHM (at 4 weeks in culture) at 1.5 Hz stimulation with an intermittent stimulation pause (10 sec); enhanced FOC at the reintroduction of electrical stimulation, i.e., post-rest potentiation, is characteristic for cardiomyocytes with mature intracellular calcium storage and release (the dotted line marks pre-pause baseline maximal FOC. (f) Representative action potentials recorded by impaling electrode measurements in EHM developed under the Starting Protocol (HES3) and the Serum-free Protocol (HES2); the table summarizes data recorded from together 51 independent action potential recordings; values in parentheses indicate maximally negative RMP and fastest dV/dtmax recorded in the respective groups.
Functional maturation was a continuous process with enhanced inotropic responses to calcium in “older” EHM (Figure 2c); this was despite similar cardiomyocyte content (Supplementary Fig. 6a). Because EHM cross sectional area (CSA) decreased over time in culture (Supplementary Fig. 6b), we opted to correct FOC by CSA to allow a direct comparison of the different models (HES and iPSC) and their developmental stages (Figure 2c); uncorrected FOC is displayed in Supplementary Fig. 6c). The average maximal FOC developed by EHM after 8 weeks in culture (6.2±0.8 mN/mm2 at 1.5 Hz; n=8) exceeded the reported FOC (~1 mN/mm2 at 1 Hz) in papillary muscle from human infants (3–14 months post birth)35 markedly, but remained lower than the FOC recorded in adult non-failing myocardium (~25 mN/mm2 at ~1.5 Hz)36. Interestingly, a positive force-frequency behavior (Bowditch phenomenon), which is absent in newborns and present in infants35 was clearly developed in defined, serum-free EHM (+19±5% at 2 Hz, +22±6% at 3 Hz versus 1 Hz, studied at 4 weeks) in contrast to EHM constructed according to the undefined Starting Protocol (Figure 2d). In agreement with this finding, post-rest potentiation (enhanced FOC by +9±1% [n=7] in the first electrically stimulated beat after a stimulation pause) was observed, providing evidence for intracellular calcium storage and release by the sarcoplasmic reticulum (Figure 2e).
Electrophysiological studies revealed that EHM were comprised mainly of working myocardium-like cells without pronounced spontaneous phase 4 depolarization (Figure 2f). This suggests that the spontaneous contractions of EHM are under the control of a small portion of pacemaker cells in EHM (<10% of all cells analyzed).
Molecular maturation of EHM
We next sought to use an unbiased approach to determine whether signs of molecular maturation could be identified in line with the observed structural and functional maturation of EHM. Hence, we first determined the differential transcriptome in high purity (94±2% ACTN2+ by flow cytometry, n=9) pluripotent stem cell-derived cardiomyocytes (n=777 transcripts; HES2, hiPS-G1, hiPS-CDI; each n=3) and fibroblasts (n=200 transcripts; skin-, gingiva-, heart-derived fibroblasts; each n=3) by RNA-sequencing (Figure 3a). As anticipated, the cardiomyocyte (CM) transcriptome was highly enriched for sarcomeric transcripts, and the fibroblast transcriptome was enriched for transcripts encoding for ECM-associated proteins and proteins mediating cell-cell or cell-matrix interactions (Figure 3b, refer to Supplementary Table 3 and 4 for a full list of the identified cardiomyocyte and fibroblast transcriptomes including a comparison to fetal and adult heart expression levels).
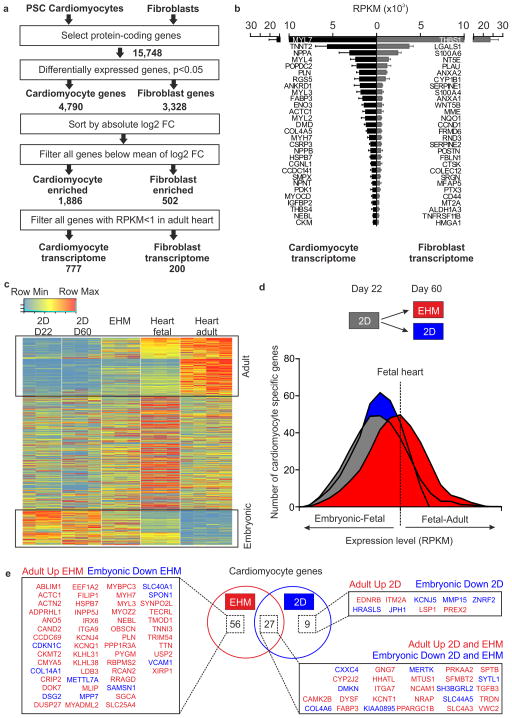
Figure 3. Molecular maturation of serum-free EHM.
(a) Strategy to determine cardiomyocyte and fibroblast transcriptomes from RNAseq data obtained from purified pluripotent stem cell-derived (PSC) cardiomyocytes (n=3 hES2 RFP, n=3 iCell CM, n=3 hiPS-G1) and primary fibroblasts (n=3 HFF, n=3 human cardiac fibroblasts, n=3 human gingiva fibroblasts). (b) RPKM values of the 29 most abundantly expressed transcripts in PSC-derived cardiomyocyte and primary fibroblasts. (c) Heatmap of cardiomyocyte transcripts in 22 day old cardiomyocyte monolayer cultures (2D D22), 60 day old cardiomyocyte monolayer cultures (2D D60), 6 week old EHM (note that cardiomyocyte “age” in these EHM was similar to 2D D60 cultures), fetal heart, and adult heart. Boxed areas indicate cardiomyocyte maturation genes; “adult”: increasing expression with development (upper box), and “embryonic”: decreasing expression with development (lower box). (d) Histogram of cardiomyocyte gene expression level (RPKM) compared to fetal heart as reference. Comparison of 22 day old cardiomyocyte monolayer cultures (2D, grey box) as starting point, 60 day old cardiomyocyte monolayer cultures (2D, blue box), and 6 week EHM cultures (red box). (e) Venn diagram and corresponding list of differentially expressed cardiomyocyte maturation genes with specific regulation in EHM, 60 day old cardiomyocyte monolayer cultures (2D), or both (overlap in Venn diagram; p<0.05 corrected for multiple testing by Benjamini-Hochberg method).
We next used the “cardiomyocyte transcriptome” to establish a temporal gene expression profile from embryonic to adult heart, taking embryonic cardiomyocytes (22 day old monolayer HES2 CM, n=3), fetal heart (n=3), and adult heart (n=4) as reference time points. We classified 3 gene clusters: [1] genes with continuous increase (“adult CM genes”; n=218; Figure 3c, upper box), [2] genes with continuous decrease (“embryonic CM genes”; n=128; Figure 3c, lower box), and [3)] genes without clear trajectory (n=431) in expression (refer to Supplementary Table 5 for a full list of genes in each of the 3 clusters). Transcriptional profiling revealed that transcription of 174 and 110 of the “adult CM genes” was enhanced and transcription of 94 and 72 of the “embryonic CM genes” was reduced in parallel EHM (6 weeks) and 2D (60 days) cultures, respectively. Direct comparison of the cardiomyocyte maturation gene transcripts (“embryonic plus adult CM genes”; n=346) showed higher frequencies of fetal-adult gene expression levels in EHM, indicating enhanced molecular maturation in EHM vs. parallel 2D cultures (Figure 3d). Compared to 2D cultures, but also to undefined EHM cultures, a significant upregulation of pivotal cardiomyocyte genes involved in “ventricular cardiac muscle tissue morphogenesis” (GO:0055010) further confirmed the cardiotypic development and a high degree of maturation in EHM constructed according to the defined, Serum-free Protocol (Figure 3e, Supplementary Figure 7).
Simulation of heart failure in EHM by neurohumoral overstimulation
The sympathetic nerve system controls heart function via the release of catecholamines and subsequent adrenoceptor activation. Experimentally, acute addition of isoprenaline (ISO; β1- and β2-adrenoceptor agonist) is used to simulate organotypic responses to catecholamine stimulation, including enhanced force development (inotropy), beating frequency (chronotropy), and relaxation (lusitropy); chronic application of norephinephrin (NE; α1-, α2-, β1- > β2-adrenoceptor agonist) is classically used to induce pathological cardiomyocyte hypertrophy.
While effects on chronotropy have been well established in human pluripotent stem cell-derived cardiomyocytes, there is so far little evidence for regular inotropic responses11, 13, 15, suggesting functional immaturity of the β-adrenergic signaling cascade. Transcriptome analyses revealed lower transcript abundance for most adrenergic receptors, including in particular the β1(ADRB1)- and β2(ADRB2)-adrenoceptors, in EHM versus adult myocardium (Supplementary Fig. 8a). Irrespective of the transcript levels, we observed a robust inotropic response of EHM to ISO, which was significantly enhanced in serum-free versus serum-containing cultures (Supplementary Fig. 8b,c). Interestingly, EHM displayed a similar sensitivity (EC50: 10±1 nmol/L; Supplementary Fig. 8d) to ISO as that reported for non-failing myocardium37. Classical pharmacological β1- and β2-adrenoceptor blocking experiments with CGP-20712A and ICI-118551, respectively, revealed that 32±6% of the acute inotropic effect in EHM were mediated via ADRB1 (Supplementary Fig. 8e).
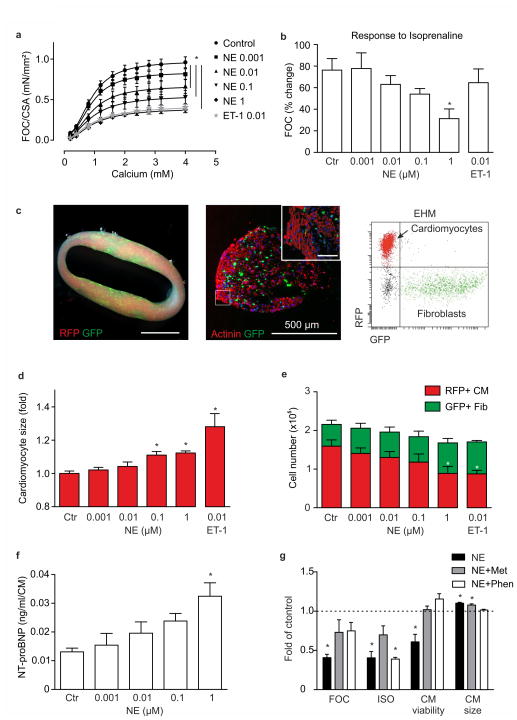
Chronic catecholamine overstimulation (serum levels in patients with heart failure: 1–10 nmol/L NE) contributes to heart failure development and progression38. In iPSC models results have been variable with recent reports demonstrating the need for defined media to elicit cardiomyocyte hypertrophy39. We asked whether EHM would exhibit the clinically observed heart failure phenotype, including β-adrenergic desensitization, cardiomyocyte hypertrophy, and the release of biomarkers (such as brain natriuretic peptide40). To recapitulate sympathetic overstimulation, we exposed EHM to NE at clinically relevant concentrations (0.001–1 μmol/L) for 7 days. We also included a group of EHM exposed to endothelin-1 (0.01 μmol/l), a well established inducer of cardiomyocyte hypertrophy via the alternative Gq-protein transduction pathway41. Similarly as observed in patients, chronic NE stimulation induced contractile dysfunction in a concentration dependent manner (Figure 4a) with desensitization to acute β-adrenergic stimulation (Figure 4b), which according to its underlying mechanism only occurred under NE and not ET-1. To enable a cell type specific analysis of cell size and cell composition, we developed a color-coded EHM model comprising RFP+-cardiomyocytes and GFP+-fibroblasts amenable to flow cytometry analyses (Figure 4c, Supplementary Video 4). This allowed us to confirm enhanced cardiomyocyte hypertrophy (Figure 4d, Supplementary Figure 9) and death (Figure 4e) in response to increasing NE concentrations. We also found the clinically relevant biomarker NT-proBNP released in a concentration dependent manner (Figure 4f) and a blunted force-frequency-response in serum-free, but not serum-containing EHM (Supplementary Figure 10a). A consistent observation was that the pathological phenotype was in general more pronounced in serum-free EHM (summarized in Supplementary Figure 10b) with a significantly reduced hypertrophic response in serum-containing EHM. This finding is consistent with earlier data on the hypertrophy-masking effects of serum in human PSC-derived cardiomyocytes39. Notably, the pathological phenotype could be partially or fully prevented by β1-adrenoreceptor and α1-adrenoreceptor blockade with metoprolol and phenoxybenzamine, respectively, demonstrating the applicability of EHM in the in vitro simulation of heart failure and its prevention by pharmacological means (Figure 4g).
Figure 4. Modeling heart failure in color-coded EHM.
(a) Effect of 7 day treatment with indicated concentrations (in μmol/L) of norepinephrine (NE) or endothelin-1 (ET-1) on FOC of EHM; *p<0.05 vs. Control by 2-way ANOVA with Tukey’s multiple comparison post hoc test, n=8–10 per Control and NE groups, n=4 for ET-1 group. (b) Inotropic response to acute isoprenaline (ISO) stimulation in EHM previously exposed to 7 day NE or ET-1 at the indicated concentrations (same EHM as in a); *p<0.05 vs. Ctr by 1-way ANOVA with Tukey’s multiple comparison post hoc test. (c) Left panel: macroscopic view of color-coded EHM (RFP+-CM: red, GFP+-Fib: green); scale bar: 1 cm; middle panel: cross section of color-coded EHM (red: actinin+-CM, green: GFP+-Fib); scale bar: 500 μm, inset: magnification - scale bar: 50 μm; right panel: flow cytometry of RFP+-CM and GFP+-Fib after enzymatic dispersion of color-coded EHM. (d) CM size measured by determination of RFP median fluorescence intensity (MFI, please refer to Supplementary Figure 9 for experimental details); *p<0.05 vs. Ctr by 1-way ANOVA with Tukey’s multiple comparison post hoc test. (e) Cell type distribution in color-coded EHM assessed by total cell quantification after enzymatic dispersion and subsequent flow cytometry for the separation of RFP+-CM and GFP+-Fib (from same EHM as in a); *p<0.05 for cardiomyocyte number vs. Ctr by 1-way ANOVA with Tukey’s multiple comparison post hoc test. (f) NT-proBNP secretion per CM into the culture medium (n=3/group). (g) Maximal FOC, response to ISO, CM viability, and CM size in comparison to control (dashed line) in EHM treated with 1 μmol/L NE with and without preincubation with 5 μmol/L metoprolol (Met) or 5 μmol/L phenoxybenzamine (Phen); *p<0.05 vs. Ctr by 1-way ANOVA with Tukey’s multiple comparison post hoc test (n=4–10/group).
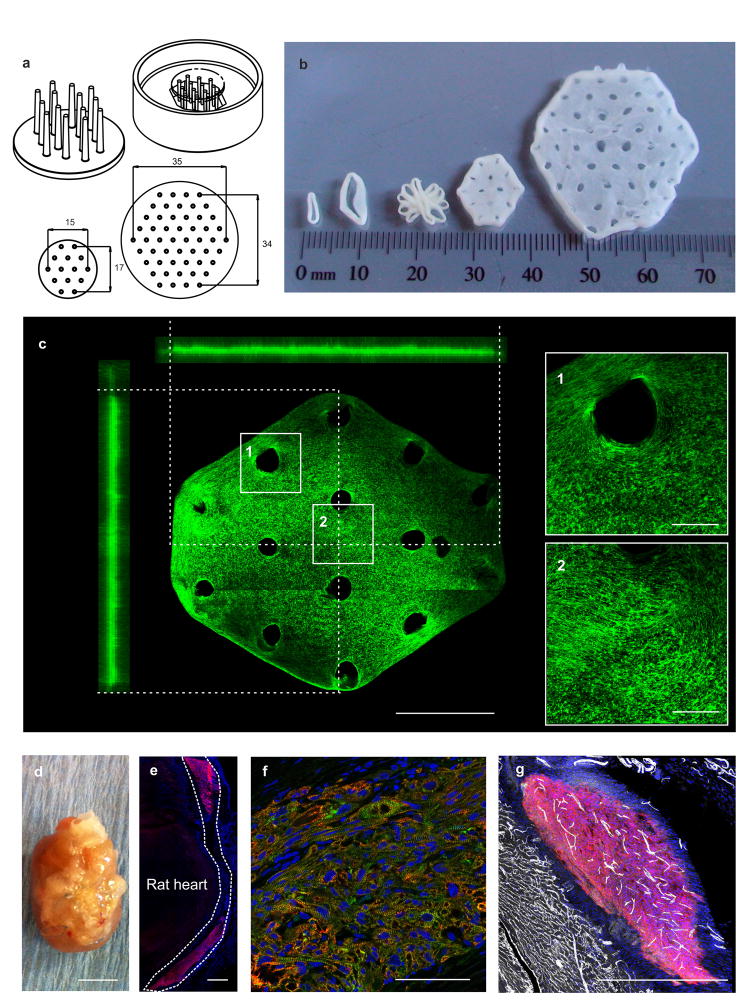
Scaling of EHM for heart repair
Remuscularization of myocardial scar tissue in the failing heart will require sizable muscle surrogates. Accordingly, we tested whether large EHM can be engineered under the defined, Serum-free EHM Protocol. We also reasoned that casting patches rather than loops would facilitate scaling towards clinical needs. Accordingly, we developed stamps with flexible tips by 3D-printing for the “penetration” of EHM mixtures cast into a size-adapted mold (Figure 5a). This allowed us to scale EHM patches variably, reaching sizes for clinical translation (15×17 mm and 35×34 mm containing 10×106 and 40×106 cardiomyocytes respectively; thickness: 0.5±0.1 mm, n=5; Figure 5b,c). Cells in EHM patches were homogenously distributed and structurally organized along traction force lines (Figure 5c). Importantly, EHM patches and loops contracted similarly (Supplementary Video 5). Because non-disruptive measurements will finally be essential to document EHM patch quality, we developed an optical force assessment strategy by correlating FOC recorded in individual EHM loops with fractional area change (FAC) in EHM patches from the same production runs. This analysis revealed a correlation of FOC and FAC recorded in EHM loops and patches, respectively (Supplementary Figure 11); further refinement of this measure will be required to account for homogeneity, shape, and force distribution of the different culture formats.
Figure 5. Scaling of EHM for heart repair.
(a) Technical drawings of the EHM patch manufacturing devices: (top left) 3D-printed patch holder with flexible poles; (top right) inverted patch holder positioned in hexagonal casting mold; (bottom) top view on patch holder for small and large EHM patch with dimensions in mm. (b) Display of different EHM designs (from left to right): small (1.5×106 cells/500 μl) and big (2.5×106 cells/900 μl) loops, fusion of five big loops according to technology reported earlier for rat42, small (10×106 cells/2 ml) and clinical-sized large (40×106 cells/8 ml) patch. (c) Overview and 90° projections of an immunostained (f-actin in green) small EHM patch (image stitched together from 24× 850×850 μm tiles); boxed areas magnified on right for a demonstration of cell orientation. Bars: 5 mm (overview) and 1 mm (magnifications). (d) Explanted rat heart 4 weeks after epicardial implantation of an EHM patch in a RNU rat; bar: 1 cm. (e) Overview of human EHM on rat heart, immunostaining of human MYH7 (red), dashed line outlines the human EHM; bar: 500 μm. (f) Immunostaining of human EHM 107 days after implantation, cardiac troponin T (red), sarcomeric actinin (green), nuclei (blue); bar: 100 μm. (g) Immunostaining of CD31 (white) and human specific β1-integrin (red); bar: 500 μm.
In continuation of a recently completed experimental series for the assessment of feasibility and safety of EHM grafting17, we now tested whether EHM patches would be retained after engraftment. In line with our recent study with EHM loops, we could demonstrate that EHM patches formed sizable and structurally highly developed grafts in RNU-rats (Figure 5d–f), which were progressively vascularized (Figure 5g).
Discussion
Our study demonstrates that differentiated, force-generating human heart muscle can be generated in vitro under defined, serum-free conditions for applications in heart failure modelling and tissue engineered heart repair. While the definition of cell composition and culture conditions reduced variability and procedural complexity, it also supported cardiomyocyte structural and functional maturation beyond the current state-of-the-art. The reported protocol is adaptable to cGMP and thus serves as the basis for highly standardized in vitro assay development and clinical translation of tissue engineered heart repair.
A number of factors have been previously identified to support maturation of human cardiomyocytes in tissue engineered heart muscle, such as mechanical stimulation13 and electrical stimulation12 as well as the co-culture of cardiomyocytes and fibroblasts15. In this study we systematically screened culture conditions and identified the minimal requirements for EHM formations under highly defined conditions (Table 1 – Serum-free Protocol). So far unrecognized were the need for an adaptation of extracellular calcium to “physiological levels” (1.2 mmol/L) and supplementation of TGFβ-1 during EHM consolidation. The requirement for calcium adaptations was identified serendipitously while testing different basal media with “normal” and reduced (RPMI 0.42 mmol/L) calcium. This observation is in agreement with the previously reported essential role of calcium for myofibrillogenesis in the mouse43. The mode of action of TGFβ-1 during EHM consolidation appears to be enhanced fibroblast mediated ECM remodeling, which was found earlier to be crucial in rodent EHM models29. Finally, addition of IGF-1, FGF-2, VEGF165 and B27 without insulin were sufficient to replace all serum supplements. The use of clinical grade bovine collagen instead of the widely used Matrigel supplemented hydrogels44 further assisted in defining culture conditions.
Using our highly defined EHM protocol we observed advanced structural, functional, and molecular maturation of cardiomyocytes. In fact, to our knowledge the following maturation characteristics have not been reported so far: [1] structural maturation with a rod-shaped cardiomyocyte morphology and sarcomers with distinguishable M-bands; both parameters are rarely observed even in extended (1 year) monolayer cultures45; [2] dominant ventricular structural and functional maturation evidenced by abundant Myl2 (MLC2V) positivity and characteristic action potential kinetics; [3] functional maturation with contractile forces and physiological responses such as a positive force-frequency behavior observed only in postnatal myocardium35, 46. While functional β1-adrenergic signalling is minute in immature pluripotent stem cell derived cardiomyocytes47 defined EHM displayed a robust β1-mediated inotropic response. The cardiotoxic effect of elevated norepinephrine levels further argues for relevant adrenergic signaling to model disease mechanisms of heart failure. Consistent with recent work the biomechanical stimulation of EHM may accelerate β-adrenergic maturation compared to monolayer cardiomyocytes47. Spontaneous contractions of EHM require specialized pacemaker cells. Random impalements with sharp electrodes for AP recordings did not identify bona fide pacemaker cells in defined, serum-free EHM. Optical imaging after loading with voltage sensitive dyes or the use of genetically encoded voltage sensors48 may help to better localize regions with pacemaker activity and guide detailed electrophysiological studies to define the underlying mechanisms of EHM automaticity.
Transcriptional profiling in 6 week EHM was in agreement with the structural and functional data, confirming an advanced degree of maturation compared to parallel monolayer cultures. However, reaching a fully adult phenotype remains a challenging task. In fact, unbiased global transcriptome profiling suggested that EHM are at large similar to fetal human heart at 13 weeks of gestation, despite some morphological (M-bands) and functional (Bowditch phenomenon) properties which develop postnatally. This suggests on the one hand that our defined, serum-free EHM protocol supports bona fide heart developement in the dish to a notable extent, and on the other hand introduces an unbiased approach for the benchmarking of tissue engineered myocardium.
Taken together, we conclude that the serum-free EHM protocol can serve as the foundation for the definition of specific biological, pharmacological or biophysical interventions controlling heart development. Whether in vitro interventions will finally enable the “speeding up” of heart development in a dish beyond the pace of natural cardiomyogenesis remains to be elucidated. The principle propensity for advanced maturation was further supported by long-term in vitro culture and in vivo implantation studies. We consider this an important prerequisite for applications of EHM in disease modelling, drug screens, and tissue engineered heart repair.
Supplementary Material
Clinical Perspective.
What is new?
Proof-of-concept for the engineering of scalable force-generating human myocardium from a variety of human pluripotent stem cells and biopsy-derived fibroblasts under defined, serum-free conditions.
Evidence for morphological, molecular, and functional maturation beyond the present state-of-the-art is demonstrated (e.g., positive force-frequency-response, sarcomere assembly with robust M-band formation).
Simulation of a human heart failure phenotype in the dish with (1) contractile dysfunction, (2) loss of a positive force-frequency-response, (3) adrenergic signal desensitization, (4) cardiomyocyte hypertrophy, and (5) biomarker release (NT-proBNP) by chronic catecholamine stimulation.
Implantability of scalable engineered human myocardium patches is demonstrated.
What are the clinical implications?
Robustness and readiness of defined, serum-free engineered human myocardium for applications in translational studies is demonstrated.
Advanced morphological, molecular, and functional maturation as well as organotypic responses to physiological (positive force-frequency response) and pathological (norepinephrine-induced heart failure) stimuli are key for the utility of engineered human myocardium in heart failure modelling.
Simulated heart failure in engineered human myocardium may be exploited for the development of novel heart failure therapeutics.
The reported defined, serum-free protocol will facilitate the engineering of human myocardium according to current good manufacturing practice for applications in tissue engineered heart repair.
Acknowledgments
The authors thank M. Hoch, I. Quentin, D. Reher, A. Schraut, and K. Sharkova for excellent technical assistance. The authors thank D. Ziebolz for providing gingiva samples, C. Rogge for preparing EHM during early phases of this study, and S. Lutz for sharing cardiac fibroblast cultures and antibodies. The authors also acknowledge B. Downie, T. Lingner, and G. Salinas from the Transcriptome and Genome Analysis Laboratory, University Medical Center Göttingen, for their support.
Funding Sources
This study was supported by the DZHK (German Center for Cardiovascular Research), the German Federal Ministry for Science and Education (BMBF FKZ 13GW0007A [CIRM-ET3]), the German Research Foundation (DFG ZI 708/7-1, 8-1, 10-1; SFB 937 TP18, SFB 1002 TPs B03, B04, C04, S1; IRTG 1618), a Lower Saxony–Israel grant (11-76251-99-30/09), the European Union FP7 CARE-MI, the Foundation Leducq, and the NIH (U01 HL099997). This study was also supported by the California Institute of Regenerative Medicine (CIRM DR2A-05394, CIRM TR3-05556, and CIRM RT3-07798). The collection and studies of fetal material is partly funded by the NIRM, the Netherlands Institute for Regenerative Medicine.
Footnotes
Conflict of Interest Disclosures
A patent concerning serum-free EHM generation for applications in drug screens and heart repair has been filed by the University of Goettingen with M.T., J.H., and W.H.Z. listed as inventors. W.H.Z. is founder and scientific advisor of myriamed GmbH and Repairon GmbH.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 4.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lian X, Bao X, Zilberter M, Westman M, Fisahn A, Hsiao C, Hazeltine LB, Dunn KK, Kamp TJ, Palecek SP. Chemically defined, albumin-free human cardiomyocyte generation. Nat Methods. 2015;12:595–596. doi: 10.1038/nmeth.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 7.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmermann WH, Fink C, Kralisch D, Remmers U, Weil J, Eschenhagen T. Three-dimensional engineered heart tissue from neonatal rat cardiac myocytes. Biotechnol Bioeng. 2000;68:106–114. [PubMed] [Google Scholar]
- 9.Radisic M, Park H, Shing H, Consi T, Schoen FJ, Langer R, Freed LE, Vunjak-Novakovic G. Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. Proc Natl Acad Sci U S A. 2004;101:18129–18134. doi: 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fink C, Ergun S, Kralisch D, Remmers U, Weil J, Eschenhagen T. Chronic stretch of engineered heart tissue induces hypertrophy and functional improvement. FASEB J. 2000;14:669–679. doi: 10.1096/fasebj.14.5.669. [DOI] [PubMed] [Google Scholar]
- 11.Schaaf S, Shibamiya A, Mewe M, Eder A, Stohr A, Hirt MN, Rau T, Zimmermann WH, Conradi L, Eschenhagen T, Hansen A. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS One. 2011;6:e26397. doi: 10.1371/journal.pone.0026397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Masse S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kensah G, Roa Lara A, Dahlmann J, Zweigerdt R, Schwanke K, Hegermann J, Skvorc D, Gawol A, Azizian A, Wagner S, Maier LS, Krause A, Drager G, Ochs M, Haverich A, Gruh I, Martin U. Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro. Eur Heart J. 2013;34:1134–1146. doi: 10.1093/eurheartj/ehs349. [DOI] [PubMed] [Google Scholar]
- 15.Zhang D, Shadrin IY, Lam J, Xian HQ, Snodgrass HR, Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials. 2013;34:5813–5820. doi: 10.1016/j.biomaterials.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soong PL, Tiburcy M, Zimmermann WH. Cardiac differentiation of human embryonic stem cells and their assembly into engineered heart muscle. Curr Protoc Cell Biol. 2012;55:23.8.1–23.8.21. doi: 10.1002/0471143030.cb2308s55. [DOI] [PubMed] [Google Scholar]
- 17.Riegler J, Tiburcy M, Ebert A, Tzatzalos E, Raaz U, Abilez OJ, Shen Q, Kooreman NG, Neofytou E, Chen VC, Wang M, Meyer T, Tsao PS, Connolly AJ, Couture LA, Gold JD, Zimmermann WH, Wu JC. Human Engineered Heart Muscles Engraft and Survive Long Term in a Rodent Myocardial Infarction Model. Circ Res. 2015;117:720–730. doi: 10.1161/CIRCRESAHA.115.306985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinberger F, Breckwoldt K, Pecha S, Kelly A, Geertz B, Starbatty J, Yorgan T, Cheng KH, Lessmann K, Stolen T, Scherrer-Crosbie M, Smith G, Reichenspurner H, Hansen A, Eschenhagen T. Cardiac repair in guinea pigs with human engineered heart tissue from induced pluripotent stem cells. Sci Transl Med. 2016;8:363ra148. doi: 10.1126/scitranslmed.aaf8781. [DOI] [PubMed] [Google Scholar]
- 19.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa M, Dottori M, Ng E, Hawes SM, Sourris K, Jamshidi P, Pera MF, Elefanty AG, Stanley EG. The hESC line Envy expresses high levels of GFP in all differentiated progeny. Nat Methods. 2005;2:259–260. doi: 10.1038/nmeth748. [DOI] [PubMed] [Google Scholar]
- 21.Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 22.Hudson J, Titmarsh D, Hidalgo A, Wolvetang E, Cooper-White J. Primitive cardiac cells from human embryonic stem cells. Stem Cells Dev. 2012;21:1513–1523. doi: 10.1089/scd.2011.0254. [DOI] [PubMed] [Google Scholar]
- 23.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Egashira T, Seki T, Muraoka N, Yamakawa H, Ohgino Y, Tanaka T, Yoichi M, Yuasa S, Murata M, Suematsu M, Fukuda K. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Graichen R, Xu X, Braam SR, Balakrishnan T, Norfiza S, Sieh S, Soo SY, Tham SC, Mummery C, Colman A, Zweigerdt R, Davidson BP. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76:357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiburcy M, Didie M, Boy O, Christalla P, Doker S, Naito H, Karikkineth BC, El-Armouche A, Grimm M, Nose M, Eschenhagen T, Zieseniss A, Katschinski DM, Hamdani N, Linke WA, Yin X, Mayr M, Zimmermann WH. Terminal differentiation, advanced organotypic maturation, and modeling of hypertrophic growth in engineered heart tissue. Circ Res. 2011;109:1105–1114. doi: 10.1161/CIRCRESAHA.111.251843. [DOI] [PubMed] [Google Scholar]
- 30.Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naito H, Melnychenko I, Didie M, Schneiderbanger K, Schubert P, Rosenkranz S, Eschenhagen T, Zimmermann WH. Optimizing engineered heart tissue for therapeutic applications as surrogate heart muscle. Circulation. 2006;114:I72–78. doi: 10.1161/CIRCULATIONAHA.105.001560. [DOI] [PubMed] [Google Scholar]
- 32.Severs NJ. The cardiac muscle cell. Bioessays. 2000;22:188–199. doi: 10.1002/(SICI)1521-1878(200002)22:2<188::AID-BIES10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Tracy RE, Sander GE. Histologically measured cardiomyocyte hypertrophy correlates with body height as strongly as with body mass index. Cardiol Res Pract. 2011;2011:658958. doi: 10.4061/2011/658958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiegerinck RF, Cojoc A, Zeidenweber CM, Ding G, Shen M, Joyner RW, Fernandez JD, Kanter KR, Kirshbom PM, Kogon BE, Wagner MB. Force frequency relationship of the human ventricle increases during early postnatal development. Pediatr Res. 2009;65:414–419. doi: 10.1203/PDR.0b013e318199093c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulieri LA, Hasenfuss G, Leavitt B, Allen PD, Alpert NR. Altered myocardial force-frequency relation in human heart failure. Circulation. 1992;85:1743–1750. doi: 10.1161/01.cir.85.5.1743. [DOI] [PubMed] [Google Scholar]
- 37.Flesch M, Kilter H, Cremers B, Lenz O, Sudkamp M, Kuhn-Regnier F, Bohm M. Acute effects of nitric oxide and cyclic GMP on human myocardial contractility. J Pharmacol Exp Ther. 1997;281:1340–1349. [PubMed] [Google Scholar]
- 38.Cohn JN, Levine TB, Olivari MT, Garberg V, Lura D, Francis GS, Simon AB, Rector T. Plasma norepinephrine as a guide to prognosis in patients with chronic congestive heart failure. N Engl J Med. 1984;311:819–823. doi: 10.1056/NEJM198409273111303. [DOI] [PubMed] [Google Scholar]
- 39.Dambrot C, Braam SR, Tertoolen LG, Birket M, Atsma DE, Mummery CL. Serum supplemented culture medium masks hypertrophic phenotypes in human pluripotent stem cell derived cardiomyocytes. J Cell Mol Med. 2014;18:1509–1518. doi: 10.1111/jcmm.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masson S, Latini R, Anand IS, Vago T, Angelici L, Barlera S, Missov ED, Clerico A, Tognoni G, Cohn JN, Val-He FTI. Direct comparison of B-type natriuretic peptide (BNP) and amino-terminal proBNP in a large population of patients with chronic and symptomatic heart failure: the Valsartan Heart Failure (Val-HeFT) data. Clin Chem. 2006;52:1528–1538. doi: 10.1373/clinchem.2006.069575. [DOI] [PubMed] [Google Scholar]
- 41.Carlson C, Koonce C, Aoyama N, Einhorn S, Fiene S, Thompson A, Swanson B, Anson B, Kattman S. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J Biomol Screen. 2013;18:1203–1211. doi: 10.1177/1087057113500812. [DOI] [PubMed] [Google Scholar]
- 42.Zimmermann WH, Melnychenko I, Wasmeier G, Didie M, Naito H, Nixdorff U, Hess A, Budinsky L, Brune K, Michaelis B, Dhein S, Schwoerer A, Ehmke H, Eschenhagen T. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med. 2006;12:452–458. doi: 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]
- 43.Li J, Puceat M, Perez-Terzic C, Mery A, Nakamura K, Michalak M, Krause KH, Jaconi ME. Calreticulin reveals a critical Ca(2+) checkpoint in cardiac myofibrillogenesis. J Cell Biol. 2002;158:103–113. doi: 10.1083/jcb.200204092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye L, Zimmermann WH, Garry DJ, Zhang J. Patching the heart: cardiac repair from within and outside. Circ Res. 2013;113:922–932. doi: 10.1161/CIRCRESAHA.113.300216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamakura T, Makiyama T, Sasaki K, Yoshida Y, Wuriyanghai Y, Chen J, Hattori T, Ohno S, Kita T, Horie M, Yamanaka S, Kimura T. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circ J. 2013;77:1307–1314. doi: 10.1253/circj.cj-12-0987. [DOI] [PubMed] [Google Scholar]
- 46.Molenaar P, Bartel S, Cochrane A, Vetter D, Jalali H, Pohlner P, Burrell K, Karczewski P, Krause EG, Kaumann A. Both beta(2)- and beta(1)-adrenergic receptors mediate hastened relaxation and phosphorylation of phospholamban and troponin I in ventricular myocardium of Fallot infants, consistent with selective coupling of beta(2)-adrenergic receptors to G(s)-protein. Circulation. 2000;102:1814–1821. doi: 10.1161/01.cir.102.15.1814. [DOI] [PubMed] [Google Scholar]
- 47.Jung G, Fajardo G, Ribeiro AJ, Kooiker KB, Coronado M, Zhao M, Hu DQ, Reddy S, Kodo K, Sriram K, Insel PA, Wu JC, Pruitt BL, Bernstein D. Time-dependent evolution of functional vs. remodeling signaling in induced pluripotent stem cell-derived cardiomyocytes and induced maturation with biomechanical stimulation. FASEB J. 2016;30:1464–1479. doi: 10.1096/fj.15-280982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang Liao ML, de Boer TP, Mutoh H, Raad N, Richter C, Wagner E, Downie BR, Unsold B, Arooj I, Streckfuss-Bomeke K, Doker S, Luther S, Guan K, Wagner S, Lehnart SE, Maier LS, Stuhmer W, Wettwer E, van Veen T, Morlock MM, Knopfel T, Zimmermann WH. Sensing Cardiac Electrical Activity With a Cardiac Myocyte--Targeted Optogenetic Voltage Indicator. Circ Res. 2015;117:401–412. doi: 10.1161/CIRCRESAHA.117.306143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.