Abstract
Cell packs a lot of genetic and regulatory information through a structure known as chromatin, i.e. DNA is wrapped around histone proteins and is tightly packed in a remarkable way. To express a gene in a specific coding region, the chromatin would open up and DNA loop may be formed by interacting enhancers and promoters. Furthermore, the mediator and cohesion complexes, sequence-specific transcription factors, and RNA polymerase II are recruited and work together to elaborately regulate the expression level. It is in pressing need to understand how the information, about when, where, and to what degree genes should be expressed, is embedded into chromatin structure and gene regulatory elements. Thanks to large consortia such as Encyclopedia of DNA Elements (ENCODE) and Roadmap Epigenomic projects, extensive data on chromatin accessibility and transcript abundance are available across many tissues and cell types. This rich data offer an exciting opportunity to model the causal regulatory relationship. Here, we will review the current experimental approaches, foundational data, computational problems, interpretive frameworks, and integrative models that will enable the accurate interpretation of regulatory landscape. Particularly, we will discuss the efforts to organize, analyze, model, and integrate the DNA accessibility data, transcriptional data, and functional genomic regions together. We believe that these efforts will eventually help us understand the information flow within the cell and will influence research directions across many fields.
Keywords: gene regulatory network, open chromatin, DNA accessibility, transcription factor colocalization, statistical model, data integration
INTRODUCTION
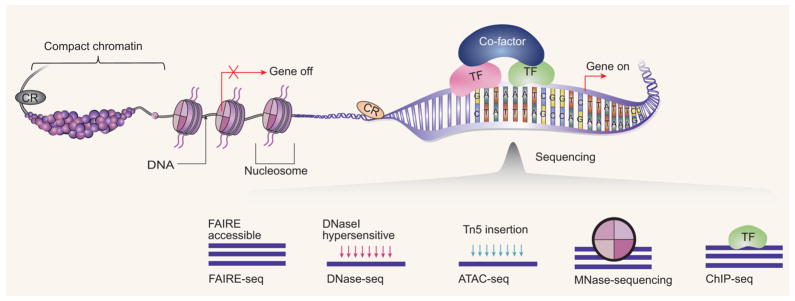
Information always needs to be coded and compressed to achieve efficient storage. In every human cell, the 2 meters long DNA molecule is packed so tightly that it actually fits into a nucleus just a few micrometers in diameter. This remarkable process involves wrapping DNA around special histone proteins and forming the nucleosome structure. The loosely linked nucleosomes are coiled into a more compact structure known as chromatin fiber. Finally, in closed chromatin regions or in metaphase chromosome the chromatin fiber is further compacted through the formation of higher order structures [1]. It is known that chromatin structure will affect the gene expression, protein expression, biological pathway, and eventually the complex phenotype. Specifically, in a specific cell type only some regions of the genome are accessible to transcription factors (TFs), RNA polymerases (RNAPs), and other cellular machines involved in gene expression, while other regions are compactly wrapped, sequestered, and unavailable to most cellular machinery. We named these two types of regions as open or closed regions. The basic concept is illustrated in Fig. 1. The open and close states are believed to be highly dynamic, i.e. the regions can change their states during important biological processes, such as during differentiation of progenitor cells to specific cell types [2,3].
Figure 1.
The basic concepts for open chromatin. Chromosomal DNA is compacted inside a nucleus by hierarchically folding DNA into certain chromatin structures. Most of the DNA fragments are compact chromatin and are tightly wrapped around histones (the cylinders are the core histones and around which the DNA is wrapped). Open chromatin are the DNA fractions which are accessible to the transcriptional machinery (including the bound TF and cofactors to regulate genes and the chromatin regulator (CR) modulating the chromatin state.) and further influences gene expression (turn the genes on or off). The rapidly developing sequencing methods, such as FAIRE-seq, DNase-seq, ATAC-seq, MNase-seq, and ChIP-seq, together provide the necessary information to decode the regulatory landscape inside cell. Those techniques utilize different mechanisms and provide complementary information. The FAIRE assay enriches for such open chromatin regions by differential solubility in phenol. The DNase I assay utilizes the fact that regions of the open chromatin are much more susceptible to DNase I digestion. ATAC assay integrates sequencing adaptors into regions of accessible chromatin by Tn5 transposase. MNase assay uses micrococcal nuclease to digest chromatin to study nucleosomes. The ChIP assay uses a specific antibody to enrich for DNA regions binding to a specific TF or a modified histone. Modeling the massive data generated by those technologies allows us to reveal the interplay among TF binding, active TSS, nucleosomes and nucleosome modifications, enhancers, and insulators in a wide variety of cell lines and tissue samples. Particularly causal regulatory network inference is promising by integrating the information from chromatin level with the gene expression data.
Chromatin offers the platform to store, read, and deliver the genomic information and is essential to understand intracellular regulatory landscape. The epigenome consists of signals from chemical modifications of histones, DNA methylation, noncoding RNA (ncRNA) expression, and TF that work in concert to determine the accessibility of the regulatory regions, so-called open regulatory region. In a simplified and global picture, chromatin regulates the accessibility of the DNA sequence information and carries the chemical modifications that may be established due to external signals. Together those information is organized in many functional regions in the chromatin [4]. Chromatin is the mediator for the epigenetics and genetics factors. It lies right in the middle of genome and epigenome and serves as the bridge. Therefore, the chromatin state is key to the understanding of the information flow in the cell and the regulatory mechanism among the functional molecules. The chromatin state can be quantified as open or close. The open regulatory regions serve as the site of action for TF, RNAPs, and other cellular regulatory machines to produce the final gene expression pattern.
Thanks to recent advances in sequencing technologies, the location and state of the functional regions now can be measured by sequencing following various assays. Each essay was designed to probe a specific aspect of the chromatin state. For example, chromatin immunoprecipitation (ChIP)-seq assays are used to profile whole genome TF–DNA binding and epigenomic states involving histone modifications. Chromatin capture methods such as Hi-C identify physical interactions between different parts of the genome. DNase I digestion combined with high-throughput DNA sequencing (DNase-seq), formaldehyde-assisted isolation of regulatory elements with sequencing (FAIRE-seq), and an assay for transposase-accessible chromatin using sequencing (ATAC-seq) are used to characterize DNA accessibility. Combinations of these emerging technologies have allowed groups to study gene regulation more comprehensively than previously possible. We will review the recent progresses in this direction.
Particularly, we will review progress on three areas in the study of gene regulatory network, namely the identification of (a) functional gene regulatory elements (b) and cognate regulators that act on these elements, and (c) the target genes whose expression is regulated by these elements. Furthermore, we will discuss the integration of data from chromatin level and transcriptome level for the modeling of causal regulatory network.
CHROMATIN BIOLOGY AND ASSAYS ON OPEN CHROMATIN
As mentioned above, chromatin ‘openness’ measures the accessibility of DNA to TF, RNAPs, and other cellular machines involved in gene expression. The recent revolution in high-throughput, genome-wide methods invented several biological assays for extracting open chromatin. We will briefly review the biological assays in current use for extracting open chromatin information: DNase Digestion, FAIRE-seq, and ATAC-seq (Fig. 1).
DNase I is a DNA-digestion enzyme. Treatment by DNase I causes degradation of accessible chromatin while leaving the closed regions largely intact. In a DNase I digestion assay, nuclei of samples are isolated and digested with DNase I for a short period of time. This generates many released DNA fragments and they are isolated and sequenced to identify the DNase I-hypersensitive regions. This assay allows the systematic identification of hundreds of thousands of DNase I-hypersensitive sites (DHS) per cell type, and this in turn has helped to delineate genomic regulatory compartments [5, 6].
FAIRE-Seq is a successor of DNase-seq for the genome-wide identification of accessible DNA regions in the genome. It takes advantage of the fact that formaldehyde cross-linking is more efficient in nucleosome-bound DNA than the nucleosome-depleted regions of the genome. There are three steps in this assay. In the first step, the cells are treated with formaldehyde and the cross-linked chromatin is extracted. In the second step, the covalently linked protein–DNA complexes between histones and DNA are sequestered. In the last step, the protein-free DNA fragments are extracted and then sequenced [7].
The third assay to be reviewed is the recently proposed ATAC-seq method [8]. Instead of relying on DNase I cleavage, it uses an engineered Tn5 transposase to integrate primer DNA sequences into the cleaved DNA fragments, which are largely generated from accessible regions. The sample preparation can be carried out in hours with little reagent consumption without the time-consuming steps of genomic fragmentation and ligation. In addition to the simplified protocol, the main advantage for ATAC-seq is resource efficiency. ATAC requires only about 10 000 cells, while the required numbers for DNase-seq and Faire-seq are 100 times higher [8]. Thus, ATAC-seq is particularly attractive for real clinical applications, where the materials are very limited. It is believed that the time and resource efficiency of ATAC-seq will make it a very useful tool in personalized medicine [9].
Even more excitingly, ATAC-seq has recently been shown to work at the single cell level [9,10]. They generate chromatin accessibility maps in several types of mammalian cells that allow to assess variation in accessibility across sets of genomic features and find particular TF associated with increased accessibility variation. This is a very timely technological advance after the recent establishment of single cell measurement of gene expression [11].
These three assays for chromatin accessibility generally produce consistent results, as shown in Ref. [8]. A study of seven diverse human cell types also shows that DNase-seq and FAIRE-seq produced consistent measurement with each cell type having 1%–2% of the human genome as open chromatin [7]. In addition to the three assays, Fig. 1 also illustrates two related technologies. ChIP-seq uses specific antibodies to extract DNA fragments that are bound to the TF or a complex that contains the target factor. Micrococcal nuclease digestion followed by sequencing (MNase-seq) identifies the nucleosome-binding region digested by an endo–exonuclease. Together, those technologies provide a genome-wide picture for the chromatin state.
Although promising, all the protocols for identifying open chromatin regions have biases depending on underlying sequence context. Such biases have been shown to confound the detection of subtle features such as the ‘footprint’ that signifies the binding of a particular TF. Removing those biases and enhancing the signal provide challenges to statistical modeling [6,12,13].
ACCESSIBILITY AND EXPRESSION DATA ARE AVAILABLE FOR DIVERSE CELL TYPES
These open chromatin assays with high-throughput sequencing have been applied to obtain massive data sets of whole genome open chromatin measurement in a wide variety of cell lines and tissue samples. Coupled with suitable computational analysis, this data provide invaluable information on areas of TF binding, active transcription start sites (TSS), enhancers, and insulators in diverse cellular context.
For example, the Encyclopedia of DNA Elements (ENCODE) project included as one of its aims the mapping all of the DHSs in the human genome with the intention of cataloging human regulatory DNA. The first extensive map of human DHSs in 125 diverse cell and tissue types is derived in Ref. [14]. Integration of this information with other datasets generated by ENCODE identified new relationships between chromatin accessibility, transcription, DNA methylation, and regulatory factor occupancy patterns [14]. In parallel, the NIH Roadmap Epigenomics Consortium published the largest collection of epigenomes characterized to date: 111 primary human tissues and cells profiled for his-tone modification patterns, DNA accessibility, DNA methylation, and gene expression. The findings provide remarkable insights into the complexity of the human epigenome [15]. In a similar effort, the Mouse ENCODE Consortium has mapped transcription, DNase I hypersensitivity, TF binding, chromatin modifications, and replication domains throughout the mouse genome in diverse cell and tissue types [16]. Open chromatin data is also available in other species. The high-resolution mapping of DHSs in the model plant Arabidopsis thaliana reported 38 290 and 41 193 DHSs in leaf and flower tissues [17]. As the newest technology, ATAC-seq is only beginning to generate data but the rate of increase is very rapid. GEO database already host many small-scale ATAC-seq datasets. There are 122 results for searching ‘ATAC-seq’ in the GEO DataSets. The samples are collected from Mus musculus, Homo sapiens, and Saccharomyces cerevisiae.
It is challenging to organize the huge data sources for integrative analysis across projects, and only limited progress has been made so far. UCSC genome browser coordinated data for the ENCODE Consortium. The ENCODE Project Portal also hosts ENCODE data from the first production phase, additional ENCODE access tools, and ENCODE project pages. ENCODE data and tracks can be viewed, searched, and visualized in the UCSC Genome Browser. There are also some databases based on the ENCODE data. RegulomeDB is a database that annotates single-nucleotide polymorphisms (SNPs) with known and predicted regulatory DNA elements that include regions of DNase hypersensitivity, binding sites of TF, and promoter regions that have been biochemically characterized to regulate transcription [18].
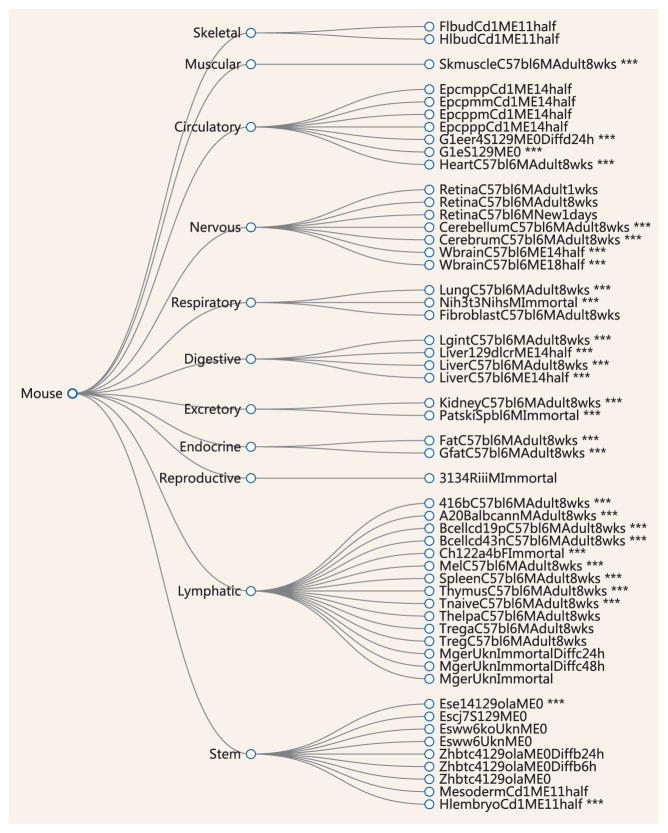
In addition to chromatin accessibility, large consortia such as ENCODE and Roadmap also provided important knowledge of the transcriptome data across many cell types. For a subset of the samples, matched transcriptome data and chromatin accessibility data are both available. Table 1 lists the current available matched samples in the major datasets. As an example, Fig. 2 illustrates the hierarchical tree for the ENCODE mouse experiments with DNase-seq data. A total of 28 out of 59 samples have matched DNase-seq and RNA-seq data (labeled with ***). It is our expectation that in the near future such matched data will be available for a diverse set of samples covering many cell and tissue types, and from many developmental, physiological, and disease contexts. This rich data resource will offer exciting opportunities for discoveries of gene regulatory mechanisms through computational analysis under novel interpretative frameworks and integrative models. In the following, we will outline the opportunity for modeling regulatory network.
Table 1.
Overview of the major datasets providing matched open chromatin and gene expression data. The Amit2014 dataset is obtained from Ref. [32]. The Pritchard2012 dataset is from Ref. [50]. The experiments are matched in biosample level means the two libraries used for sequencing are from the same sample. The experiments are matched in cell type level means the two libraries have the same cell type.
| Dataset | Mouse
|
Human
|
|||
|---|---|---|---|---|---|
| Encode | Amit2014 | Encode | Roadmap | Pritchard2012 | |
| No of DNase-seq experiments | 59 | 213 | 356 | 70 | |
| No of ATAC-seq experiments | 10 | ||||
| No of RNA-seq experiment | 147 | 16 | 425 | 257 | 70 |
| No of matched experiments in biosample level | 25 | 9 | 19 | 147 | 70 |
| No of matched experiments in cell type level | 56 | 9 | 550 | 189 | 70 |
| No of biosamples | 254 | 16 | 1772 | 889 | 70 |
| No of cell types | 54 | 16 | 356 | 188 | 70 |
Figure 2.
The mouse ENCODE experiments with DNase-seq data are organized in a hierarchical tree. The experiments labeled by ‘***’ have matched DNase-seq and RNA-seq data.
FOOTPRINTING MAY REVEAL TF OCCUPANCY REGULATORY REGIONS
Chromatin accessibility data is useful for the identification of functional regions by various peak calling algorithms. For example, ATAC-seq currently allows inference of accessible chromatin, TF occupancy, and nucleosome positions in regulatory regions in a single experiment. Those regions mark transcriptionally active regions and are tissue specific. About 5% are in TSS regions and 95% are in intronic and intergenic regions. The regions highlight the great complexity regulating the genetic expression in the human genome and the quantity of elements that control this regulation [14].
Those openness signals, as read out with base-specific sensitivity, can be used to footprint-binding sites of some TF within open chromatin regions. The rationale is that the enzyme (for example DNase I) is known to cleave DNA preferentially in accessible regions and is sterically hindered by DNA-bound proteins such as TFs. This leads to the creation of ‘footprints’ in which the presence of a bound TF diminishes cleavage at its binding site. This can be formulated as a computational problem to detect the shape from the read count data. CENTIPEDE is a pioneering work and presents a computational framework for predicting protein-binding sites based on a multinomial model for the distribution of reads to model for signal patterns [19]. PIQ is a recent method that uses Gaussian process for read count modeling, and that integrates time-series experiments to learn the cross-experiment structure as a Gaussian graphical model using L1 regularization [20]. Further by the footprint patterns, TFs are grouped as pioneer TFs, settler TFs, and migrant TFs. Pioneers TFs are capable of opening closed chromatin and is quantified by a pioneer index to measure the motif-specific local increase in chromatin opening activity (DNase I accessibility) from one time point to the next in developmental time course [21]. Settlers TFs require open chromatin to bind and the genomic binding is principally governed by proximity to open chromatin. Migrants TFs require both open chromatin to bind and the presence of coregulators. Together, these TFs support a hierarchical TF-binding model, which illustrates when and how the TFs interact with chromatin state [20].
In addition to single TF study, a comprehensive analysis on the footprint of DNase shows that footprint is both common and informative [22]. Transcriptional regulation is a pivotal process that confers cellular identity and modulates the biological activities within a cell. Footprint data derived in vivo can define TF regulatory networks by observing TFs that bind near the promoters of other regulatory genes. DNase I footprints have been utilized to assemble an extensive core human regulatory network comprising connections among 475 sequence-specific TFs across 41 diverse cell and tissue types [23]. The dynamics of these connections indicates that human TF networks are highly cell selective. All cell-type regulatory networks independently converge on a common architecture that closely resembles the topology of living neuronal networks [23].
Though conceptually appealing, at present DNase hypersensitivity analysis is still plagued by many difficulties such as against enzyme cut bias, non-homogeneous read count distribution, factor dynamics, GC content, and sequencing artifact [12]. Recent studies discussed the limitations of the DNase-based genomic footprinting approach and indicated a confounding factor as the scope of detectable protein occupancy, especially for TFs with short-lived chromatin binding. A distinct correlation was observed between footprint depth and the reported residence time of DNA binding for a compilation of TFs with in vivo DNA-binding dynamics data available in the literature [24]. Key practical experimental considerations crucial to the success of a genomic footprinting experiment, including library quality, complexity and sequencing depth, and single-versus paired-end sequencing, are discussed in Ref. [25]. Taken together, deep sequencing of a high-quality DNase I library to obtain a high proportion of all mapped reads and analytical strategies and considerations should be combined for a successful genomic footprinting experiment. These must be addressed by careful and through statistical research before footprinting information can be extracted in a robust manner. More high-quality data and novel analysis statistical methods are in pressing need.
TF COLOCALIZATION AND DYNAMICS BY CHROMATIN ACCESSIBILITY
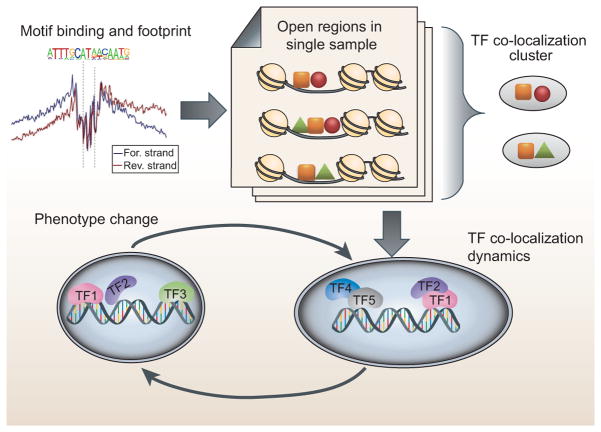
TFs collaborate and act in concert at distinct loci to perform accurate regulation of their target genes. Chromatin accessibility can be used to reveal the TF colocalization pattern. It has been demonstrated that DHSs can identify 95% of TF binding when pooled across a large number of cell types by the ENCODE data. This fact indicates open chromatin and motif binding can mimic ChIP-seq. This is true over thousand TFs and multiple cell types [26]. A large amount of ChIP-seq data from ENCODE has been used to study TF colocalization. The results include studying 76 TFs in K562 cells [27] and 128 TFs in three human cell types [28]. Given the fact that one open chromatin data can identify binding sites for many TFs, the large scale of open chromatin data will greatly broaden the scope of this type of study. Figure 3 illustrates the ideas and procedure to study TF colocalization by DNA sequence motif and open chromatin data. As the first step, the open regions can be identified from DNA accessibility data as the active regulatory regions in chromatin. Then those regions are scanned for the motif occurrence. Finally, the co-occurrence pattern of motifs in open region can be modeled to recover the TF colocalization cluster. The feasibility of this strategy has been demonstrated as the downstream analysis of footprint prediction and the colocalization was detected by a two-sample Poisson test [19]. There is still plenty of room to better model the co-occurrence pattern. Furthermore, TF dynamics can be examined and correlated with phenotype changes since open chromatin data can easily capture the condition-specific information.
Figure 3.
Illustration of the principle to use open chromatin data to study the transregulatory elements. Integrating open chromatin data with the available DNA sequence motif data can infer the TF colocalization patterns. As the first step, the open regions are derived from DNA accessibility data as the active regulatory regions in chromatin. Then those regions are scanned for the motif occurrence and quantify the binding strength of TF motifs. Finally, motif’s co-occurrence pattern in open region can be modeled to recover the TF colocalization cluster. Furthermore, the dynamics of TF colocalization clusters can be revealed by comparing open chromatin data in different conditions and even time series.
CORRELATION OF OPENNESS TO GENE EXPRESSION MAY REVEAL TARGET GENES OF REGULATORY ELEMENTS
In addition to finding the upstream TF regulator in an open region, we can also correlate the chromatin state with expression and find the downstream target genes. Specifically when looking at tissues or samples that constitute sequential temporal or developmental snapshots of a biological process, we can correlate changes in accessible chromatin, TF binding, and network topology with changes in gene expression. This provides powerful insight into relationships between epigenetic regulatory landscape and phenotype as reported by gene expression profiling.
In a previous work, we have found that in mESC a remarkably high proportion of variation in gene expression (65%) can be explained by the binding signals of 12 TFs [29]. Since open chromatin data can be used to determine chromatin accessibility and state, nucleosome positioning as well as TF-binding sites, we believe that ATAC-seq data can be used to predict gene expression. There are a number of works studying the correlation between open chromatin regions and gene expression. A colocalization between DNase-hypersensitive exons with promoters and distal regulatory elements leads to a new thinking about gene expression at DHSs [30]. A pipeline is presented for predicting cell-type-specific gene expression in Ref. [31]. First, the observation that cell-type-specific genes have different DNase sensitivity profiles is shown. Then the information is employed to determine whether a gene is cell-type specific or not [31]. We next present a general framework for this type of analysis.
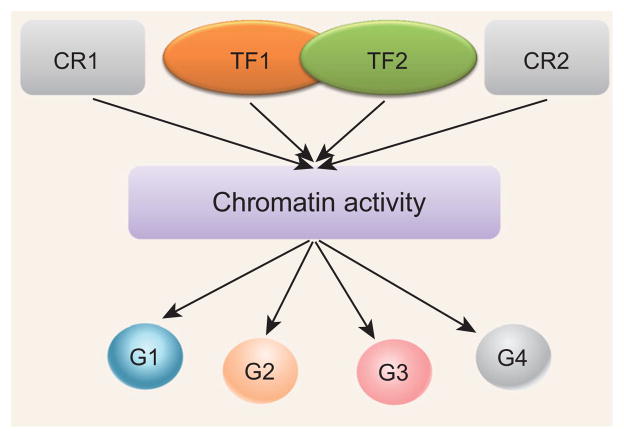
Figure 4 illustrates the basic idea to link the upstream regulatory factors and downstream target genes to interpret the chromatin activity. Annotated promoters and enhances will be included in the analysis automatically. The result is a large set of numerical or categorical features that we can associate to each gene across cell types, which we can use to learn a model for predicting gene expression. More formally, for each gene and in each cell type, we have a multidimensional data point (y, z) where y is the expression level of the gene measured by RNA-seq, and z is a vector of features computed as above from open chromatin data in the surrounding genomic region. We will have plenty of data points to learn the model. The chromatin activity can be a hidden variable. There are many statistical learning methodologies that can be developed to address the challenge.
Figure 4.
Illustration of the idea to use open chromatin data to annotate the cis-regulatory elements. An integrative model can put open chromatin data and RNA-seq data together to identify the upstream TFs and downstream genes for a given chromatin region. This local regulatory network will help us to better understand the regulatory role of active regulatory region, i.e. the mechanism that TFs bind to the genome, displace nucleosomes, and thereby expose the DNA and making it more sensitive to cleavage by enzymes. Since the cis-regulatory element is widely defined and it spans from the single base variant to Kb level promoter region and Mb wide topological domain. Particularly, it is useful to interpret the genomic variants and mutations given the fact the large amount of data will be available in near future.
Figure 4 is not limited to a static picture. When we have multiple samples across conditions or time points, we have more chance to reveal causal regulations. For example, in Ref. [32] the authors generate the open chromatin and expression data from cells undergoing hematopoietic differentiation. Correlating significant changes in chromatin accessibility, nucleosome positioning in regulatory regions, TF occupancy, chromatin state change, and gene expression using can identify causal enhancer.
In addition to gene expression, the study of open chromatin data is ready to be combined with other information, which allows analysis of regulatory network. This information includes but not limited to TF, chromatin regulator, DNA methylation patterns, promoter chromatin signature, and promoter/enhancer connections.
MODELS AND ALGORITHMS FOR MATCHED SAMPLE
Cells have evolved multilayer gene regulatory networks that allow them to maintain a stable cellular state and response to external stimuli or signals. To study these networks and their implication on the systems-level properties of the cell, it is necessary to go beyond individual regulator, target and cis-element, to study the cross-regulatory relationships among the regulators. Matched open chromatin and gene expression data allow us to go beyond single layer to study the interplay of two regulatory levels.
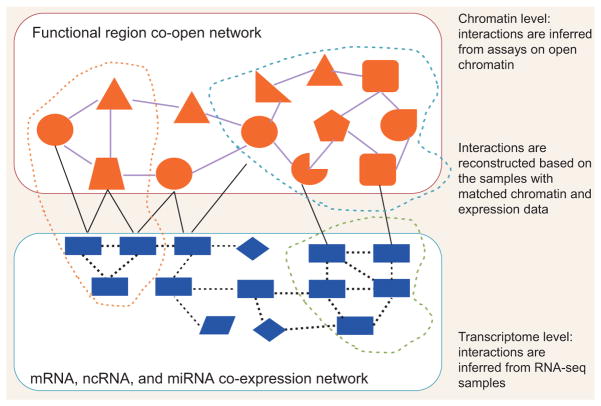
Figure 5 illustrates a network perspective to integrate the DNA accessibility and transcriptome data. We show that joint modeling of open chromatin and gene expression data can be represented by a two-layer network. First, the large amount of RNA-seq samples can be utilized to reconstruct the regulatory network among mRNAs, microRNA (miRNA), and long non-coding RNA (lncRNA). There are many existing models and algorithms that can be used to quantify the coexpression relationships and to reveal the coexpression modules [33]. Furthermore, assays on open chromatin provide the coopening relationships among functional regions. Here the term ‘functional regions’ is used to denote any genomic location or region with potential regulatory relevance, such as SNP loci, somatic mutation locis, TF-binding sites, promoter, enhancer, histone modification sites, conserved region, and topological domains. By measuring the covariation of two functional regions’ degrees of openness, we may identify regions that interact with each other to affect gene expression. For example, distal cis-regulatory elements, such as enhancers, can modulate the activity of the promoters. Thus, the distal cis-regulatory elements tend to synchronize its cooperating with their promoter in the cellular context in which the element of is activated. Correlations between openness can identify promoter/enhancer connections. In this way, a map of candidate enhancers controlling specific genes may be created.
Figure 5.
A network perspective to integrate the DNA accessibility and transcriptome data. We show that joint modeling open chromatin and gene expression data can be achieved by a two-layer network. Cell is organized into two layers as chromatin level and transcriptome level. First, the large amount of RNA-seq samples can be utilized to reconstruct the regulatory network among mRNAs, ncRNAs, and miRNAs (denoted by rectangle, diamond, and parallelogram). There are many existing models and algorithms that can be borrowed to quantify the coexpression relationships and reveal the coexpression modules (green circle). Furthermore, assays on open chromatin provide the coopening relationships among functional regions. Here functional regions are used to denote any genomic location or region with potential regulatory relevance, such as SNP loci, somatic mutation loci, TF binding sites, promoter, enhancer, histone modification sites, conserved region, and topological domains (denoted by different shapes). Then sophisticated modeling is in pressing need to quantify the openness of those regions and their correlations. The coopening module in this network can be identified and studied (the blue circle). Most interestingly, the connections between the chromatin level and gene expression level can be reconstructed by crossing those samples with matched gene expression and chromatin level data. With those connections as bridges, many causal relationships can be modeled by borrow information across different layers. In fact, the cross-network module represents exactly the general biological machinery performing specific function (the orange circle). This module can provide annotation for a specific region by finding downstream genes.
Most interestingly, as shown in Fig. 5, the connections between the chromatin and gene expression level can be reconstructed by utilizing those samples with matched gene expression and open chromatin data. With those connections as bridges, many causal relationships can be modeled by borrowing information across different layers. In fact, the cross-network module represents exactly is the general biological machinery performing specific function.
One advantage of the two-layer network representation is that it can be easily integrated with genotype and ontology information. Generally speaking, a group of functional regions may share the same annotations. For example, genome-wide association study (GWAS) usually identify a group of SNP variants connecting a disease type. Similarly, the gene set may be annotated by enriched GO function, pathway, and other annotations. Bridging by the cross-network module, those annotations can be cross-compared, and consistent ones can be used to annotate the module itself.
In general, we assume that we have two matrices X (n × p) and Y (n × m) for DNA accessibility data with p functional regions and transcriptome data with m genes, measured for the same n samples. We further assume that both X and Y have been columnwise standardized (zero mean, unit variance). Two layer data integration involves solving the problem to maximize certain combination of ΣXY, ΣXX, and ΣYY by selecting a single pair of variables for functional regions and genes. Here ΣXX and ΣYY are the covariance matrices of X and Y, respectively. ΣXY is the covariance matrix of X and Y. To study this two layer dataset in this framework, many existing methods can be utilized. Those methods are developed for many applications to identify disease-drug-gene module, integrate miRNA-gene-methylation, combine CNV and gene expression, and prioritize disease genes. The authors in Ref. [34] noticed that high-throughput technologies can be used to generate more than one type of data from the same biological samples. To properly integrate such data, they propose the drug-gene comodules, which describe coherent patterns across paired data sets, and conceive the ping-pong algorithm for their identification. Multivariate methods based on canonical correlation analysis (CCA), called sparse CCA, have been proposed for integrating paired genetic datasets [35,36]. Matrix decomposition framework is also useful in this task. The miRNA and gene expression profiles are jointly analyzed in a multiple non-negative matrix factorization framework, and additional network data are simultaneously integrated in a regularized manner [37]. This optimization framework is also used to study the drug-gene-disease comodule by treating the genes as the matched dimension [38]. Zhao et al. proposed a Bayesian partition method to identify drug-gene-disease comodules underlying the gene closeness data [39]. Linear regression and random walk also provide elegant solution for this task utilizing some reliable information as gold standard positives [40–42].
Recently, we casted the matched data integration into a multivariate regression framework and proposed a new method, T-SVD [43]. The application example is to analyze miRNA and lncRNA data from The Cancer Genome Atlas (TCGA) consortium. We formulated a statistical model for the regulation of global gene expression by multiple regulatory programs and propose a thresholding singular value decomposition regression method for learning such a model from data. Extensive simulations demonstrate that this method offers improved computational speed and higher sensitivity and specificity over competing approaches. The analysis on TCGA yields previously unidentified insights into the combinatorial regulation of gene expression by ncRNAs, as well as findings that are supported by evidence from the literature. The advantage of T-SVD is that the sparsity-inducing modeling and inference approach is effective in extracting the regulatory relations among very high-dimensional responses and predictors, even when the sample size is much lower. This is exactly the case in Fig. 5, in which functional regions in chromatin are in really high dimension.
Another recent effort to relate DHS to gene expression levels across multiple cell types is presented in Ref. [44]. A new statistics, called ARS, was proposed to characterize the relationship between chromatin accessibility and gene expression in a cell-type-specific manner and applied in a dataset on genome-wide, high-resolution chromatin accessibility measurements for 20 distinct human primary and culture cell lines. To deal with the challenge from variation across cell types and the non-linear chromatin and gene expression relationship, they placed the measurement units on a common scale and then ARS was computed as the product of the normalized distances and the angular penalties.
INCORPORATING CHROMATIN ACCESSIBILITY INTO THERMODYNAMICS-BASED MODELS
Since ATAC-seq is resource efficient, it is particularly attractive for dynamic biological processes, where the materials are very limited in time course. It is expected that the time and resource efficiency of ATAC-seq will make it a very useful tool in modeling causal gene regulation. It helps to quantitatively understand the precise relationship between gene expression and regulatory sequences, especially enhancers. We know enhancers as the cis-regulatory modules in some contexts are sequences ~1 kbp long that harbor DNA-binding sites or one or more TFs that act together to regulate a gene’s expression pattern. DNA accessibility data provide the measurement of open/close state for those enhancers and open a new door for detailed modeling.
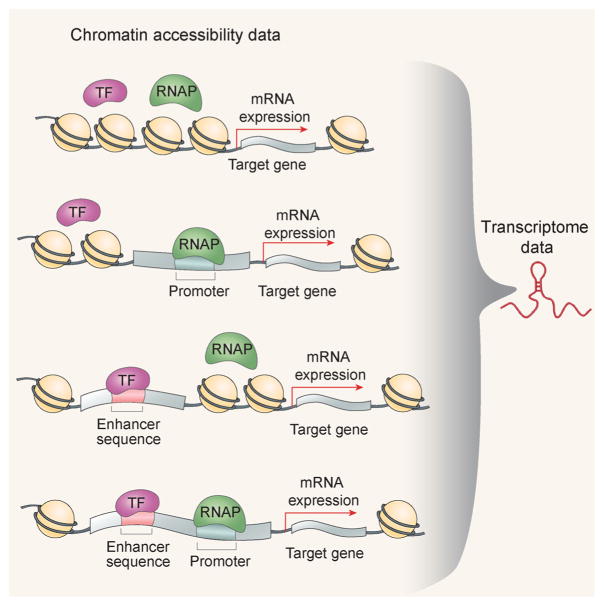
As shown in Fig. 6, thermodynamics-based sequence-to-expression models have proven capable of producing highly accurate fits to complex gene expression patterns. The models are built around molecular interactions involving TF proteins, DNA, and the basal transcriptional machinery, RNAP [45,46]. It uses the language of statistical thermodynamics to map combinations of interactions, both strong and weak, to gene expression levels. Fits of these models to sequence and expression data capture underlying mechanistic details of gene regulation.
Figure 6.
Illustration of incorporating DNA accessibility in thermodynamics model. It shows the major components of transcriptional regulation and their interactions in thermodynamic equilibrium. A simplified transcriptional system where the enhancer contains a single binding site for a TF, with the TF bound or not bound at its site and the RNAP bound or not bound at the promoter. Arrows indicate TF-DNA, RNAP-DNA, and TF-RNAP interactions. Thermodynamics model allows us to predict the level of gene expression in the dynamical system by modeling the energies associated with these interactions. Considering the DNA accessibility of those regulatory elements, one can quantitatively describe the TF-DNA binding energy at functional regions changes according to the open chromatin state.
Based on the thermodynamics scheme, we can model gene expression regulation as a dynamical system. Let x ∈ Rm represent RNA concentrations and y ∈ Rm represent protein concentrations corresponding to a set of n genes. The production rate of the RNA transcript xi of gene i is assumed to be proportional to the probability f(y) that RNAP is bound to the promoter and enhancer. By assuming that RNA transcription occurs at a rate whenever RNAP is bound to the promoter, the probability that RNAP is bound to the promoter as a non-linear function f of y, since RNAP binding is regulated by a set of TFs. The non-linear form of f(y) can be deduced from the thermodynamics of RNAP and TF binding, as shown in Bintu’s equation [45].
The above representation is useful to propose an experimental design and associated statistical method for inferring an unknown gene network by fitting the ODE-based Bintu gene regulation model. Our recent study shows how to design a sequence of experiments to collect the data and how to use it to fit the parameters of the Bintu model, leading to a set of ODEs that quantitatively characterize the regulatory network [47]. The required data are gene expression measurements at a set of perturbed steady states induced by gene knockdown and overexpression.
Now it is right time to incorporate DNA accessibility data in thermodynamics model. We expect the time course measurement of the chromatin state, especially general accessibility patterns, of cis-regulatory regions correlates with expression and with regulatory events leading to expression. One pilot study built and evaluated a quantitative model that maps regulatory DNA sequence to the expression of the regulated gene while integrating DNA accessibility data [48]. Figure 6 illustrates the major components of transcriptional regulation and their interactions in thermodynamic equilibrium. Considering the DNA accessibility of those regulatory elements, one can quantitatively describe the TF-DNA binding energy at functional regions changes according to the open chromatin state. The improved thermodynamics model allows us to predict the level of gene expression in the dynamical system by modeling the energies associated with these interactions. It provides an elegant and detailed way to integrate chromatin accessibility and transcriptome data and to model the causal regulatory network.
BIOLOGICAL APPLICATIONS OF OPEN CHROMATIN DATA
Open chromatin data identifies a large number of functional regions and can be used to interpret genetic variants. The rationale is based on the fact that variants of biological significance are expected to be enriched in open regions. Maurano et al. found that 57.1% of the non-coding GWAS-identified SNPs associated with 207 diseases and 447 quantitative traits were found within DHS regions and a further 19.5% in complete linkage disequilibrium [49]. This information can also be used to interpret expression quantitative trait loci (eQTLs). eQTLs are stretches of DNA that regulate gene transcription and expression and contribute to a particular phenotypic trait. eQTL mapping is an important tool for linking genetic variation to changes in gene regulation, but identifying the causal variants underlying eQTLs and the regulatory mechanisms involved remains a challenge. Degner et al. used DNaseI sequencing to measure genome-wide chromatin accessibility in 70 Yoruba lymphoblastoid cell lines to produce genome-wide maps of chromatin accessibility for each individual. They intersect these regions with eQTL’s derived from these samples, to identify variants, called DNaseI sensitivity quantitative trait loci. The implication is that changes in chromatin accessibility or transcription factor binding occur at many gene loci and are likely to be important contributors to phenotypic variation [50].
In addition to the genetic variants, open chromatin data may enable better interpretation of the somatic mutations identified in cancer samples. A recent effort discovered a remarkable change in the accessible regulatory landscape between these two tissues, with several thousand regions becoming more accessible in the cancer tissue [51]. Two TFs are identified to be involved in cancer (AP-1 and Stat92E), which regulate these newly accessible regulatory regions.
Finally, chromatin structure dynamics plays a fundamental role during development and cell differentiation. Open chromatin data provides valuable insight into this fundamentally process. Recently, Lara-Astiaso et al. developed a high-sensitivity indexing first ChIP approach to identify 48 415 enhancer regions and characterize their dynamics [32]. Combining their enhancer catalog with genome-wide open chromatin sites from ATAC-seq data and gene expression profiles, they elucidated the TF network controlling chromatin dynamics and lineage specification in hematopoiesis. Integrating chromatin and expression levels provides a comprehensive model of chromatin dynamics during development.
FUTURE GOALS AND CHALLENGES
Open chromatin data has the potential to generate many types of useful information. This approach has the important advantages of being cost-effective and time-efficient, as many types of information is generated from a single, relatively simple assay. We already discussed some existing problems and challenges in analyzing the data and integrating the data from gene regulatory network’s perspective. Here we will end the perspective with some general discussions.
Organizing the massive data will require new information technologies. Single open chromatin data will identify over 300 000 hotspots. The number of potential open regions will increase very rapidly. This massive data give rise to challenges in storage, analysis, and modeling, which must be met with the development of new computational and statistical approaches. It is expected that these methods will be built on top of modern distributed computation platforms such as Hadoop distributed file system, map-reduce computation model, and ‘big table’ style data management. The distributed file system, map reduce, big table ideas from Google should be borrowed. This combination of method and architecture will address the distinct and complementary need to effectively utilize the massive data.
Both RNA-seq and ATAC-seq can be pushed to single cell level [11,52]. The intrinsically noisy single cell data will pose new difficulties in modeling. An additional challenge relates to the need to combine and integrate different sets of ‘omics’ data from the same cell. For example, a method has been developed for simultaneous sequencing of genomic DNA and mRNA in a single cell [53]. Such methodology will help elucidate the correlation between molecular variability and phenotypic diversity—a fundamental question in biology. We will expect the measurement of chromatin state and mRNA expression for the same cell in near future. Then this integrative study will bring more insights and will impact many fields.
Because of its simplicity, ATAC-seq technology is a promising assay to measure the epigenomic state of individual patients and monitor the therapeutic intervention changes in real time. In this scenario, the challenge is to develop methods to integrate the diverse epigenomic measurements to infer causal regulators and specific regulatory elements affecting gene expression, reveal key dynamic and disease-specific features based on temporal data of the same individuals. To achieve precision, these regulatory insights should be iteratively refined to ensure comprehensiveness and accuracy.
Acknowledgments
We apologize to the authors of original work not cited. The authors thank the Center of Excellence in Genomic Science (CEGS) for Personal Dynamic Regulome at Stanford University with Grant P50HG007735.
FUNDING
This work was partially supported by NIH grants P50HG007735 and R01HG007834. This research was conducted when YW and RJ were visiting WW at Stanford. Their visits were partially supported by funds from the Chinese Academy of Sciences and Tsinghua University respectively. The authors thank the Center of Excellence in Genomic Science (CEGS) for Personal Dynamic Regulome at Stanford University. YW is also supported by NSFC under grant number 11422108. RJ is also supported by NSFC under grant number 61573207.
References
- 1.Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding–wrapping up transcription. Science. 2002;297:1824–7. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 2.Niwa H. Open conformation chromatin and pluripotency. Genes Dev. 2007;21:2671–6. doi: 10.1101/gad.1615707. [DOI] [PubMed] [Google Scholar]
- 3.Gaspar-Maia A, Alajem A, Meshorer E, et al. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellis M, Wold B, Snyder MP, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA. 2014;111:6131–8. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford GE, Holt IE, Whittle J, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–31. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vierstra J, Wang H, John S, et al. Coupling transcription factor occupancy to nucleosome architecture with DNase-FLASH. Nat Methods. 2014;11:66–72. doi: 10.1038/nmeth.2713. [DOI] [PubMed] [Google Scholar]
- 7.Giresi PG, Kim J, McDaniell RM, et al. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–85. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buenrostro JD, Giresi PG, Zaba LC, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenleaf WJ. Assaying the epigenome in limited numbers of cells. Methods. 2015;72:51–6. doi: 10.1016/j.ymeth.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cusanovich DA, Daza R, Adey A, et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–4. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang F, Barbacioru C, Bao S, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell stem cell. 2010;6:468–78. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer CA, Liu XS. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet. 2014;15:709–21. doi: 10.1038/nrg3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung MH, Guertin MJ, Baek S, et al. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell. 2014;56:275–85. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roadmap Epigenomics C. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue F, Cheng Y, Breschi A, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–64. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Zhang T, Wu Y, et al. Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell. 2012;24:2719–31. doi: 10.1105/tpc.112.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pique-Regi R, Degner JF, Pai AA, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–55. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherwood RI, Hashimoto T, O’Donnell CW, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–8. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–41. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neph S, Vierstra J, Stergachis AB, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neph S, Stergachis AB, Reynolds A, et al. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–86. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung MH, Baek S, Hager GL. Genome-wide footprinting: ready for prime time? Nat Methods. 2016;13:222–8. doi: 10.1038/nmeth.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vierstra J, Stamatoyannopoulos JA. Genomic footprinting. Nat Methods. 2016;13:213–21. doi: 10.1038/nmeth.3768. [DOI] [PubMed] [Google Scholar]
- 26.Blatti C, Kazemian M, Wolfe S, et al. Integrating motif, DNA accessibility and gene expression data to build regulatory maps in an organism. Nucleic Acids Res. 2015;43:3998–4012. doi: 10.1093/nar/gkv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerstein MB, Kundaje A, Hariharan M, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie D, Boyle AP, Wu L, et al. Dynamic trans-acting factor colocalization in human cells. Cell. 2013;155:713–24. doi: 10.1016/j.cell.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci USA. 2009;106:21521–6. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mercer TR, Edwards SL, Clark MB, et al. DNase I-hypersensitive exons colocalize with promoters and distal regulatory elements. Nat Genet. 2013;45:852–9. doi: 10.1038/ng.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natarajan A, Yardımcı GG, Sheffield NC, et al. Predicting cell-type specific gene expression from regions of open chromatin. Genome Res. 2012;22:1711–22. doi: 10.1101/gr.135129.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, et al. Chromatin state dynamics during blood formation. Science. 2014;345:943–9. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter SL, Brechbühler CM, Griffin M, et al. Gene co-expression network topology provides a framework for molecular characterization of cellular state. Bioinformatics. 2004;20:2242–50. doi: 10.1093/bioinformatics/bth234. [DOI] [PubMed] [Google Scholar]
- 34.Kutalik Z, Beckmann JS, Bergmann S. A modular approach for integrative analysis of large-scale gene-expression and drug-response data. Nature Biotechnol. 2008;26:531–9. doi: 10.1038/nbt1397. [DOI] [PubMed] [Google Scholar]
- 35.Parkhomenko E, Tritchler D, Beyene J. Sparse canonical correlation analysis with application to genomic data integration. Stat Appl Genet Mol Biol. 2009;8:1–34. doi: 10.2202/1544-6115.1406. [DOI] [PubMed] [Google Scholar]
- 36.Witten DM, Tibshirani RJ. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat Appl Genet Mol Biol. 2009;8:1–27. doi: 10.2202/1544-6115.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S, Liu C-C, Li W, et al. Discovery of multi-dimensional modules by integrative analysis of cancer genomic data. Nucleic Acids Res. 2012;40:9379–91. doi: 10.1093/nar/gks725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Wang Y, Hu Q, et al. Systematic analysis of new drug indications by drug-gene-disease coherent subnetworks. CPT Pharmacometrics Syst Pharmacol. 2014;3:e146. doi: 10.1038/psp.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao S, Li S. A co-module approach for elucidating drug-disease associations and revealing their molecular basis. Bioinformatics. 2012;28:955–61. doi: 10.1093/bioinformatics/bts057. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Wu X, Jiang R. Integrating human omics data to prioritize candidate genes. BMC Med Genomics. 2013;6:57. doi: 10.1186/1755-8794-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Jiang R, Zhang MQ, et al. Network-based global inference of human disease genes. Mol Syst Biol. 2008;4:189. doi: 10.1038/msb.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang R. Walking on multiple disease-gene networks to prioritize candidate genes. J Mol Cell Biol. 2015;7:214–30. doi: 10.1093/jmcb/mjv008. [DOI] [PubMed] [Google Scholar]
- 43.Ma X, Xiao L, Wong WH. Learning regulatory programs by threshold SVD regression. Proc Natl Acad Sci USA. 2014;111:15675–80. doi: 10.1073/pnas.1417808111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marstrand TT, Storey JD. Identifying and mapping cell-type-specific chromatin programming of gene expression. Proc Natl Acad Sci USA. 2014;111:E645–54. doi: 10.1073/pnas.1312523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bintu L, Buchler NE, Garcia HG, et al. Transcriptional regulation by the numbers: models. Curr Opin Genet Dev. 2005;15:116–24. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia HG, Kondev J, Orme N, et al. Thermodynamics of biological processes. Methods Enzymol. 2011;492:27–59. doi: 10.1016/B978-0-12-381268-1.00014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arwen Meister YHL, Choi Bokyung, Wong Wing Hung. Learning a nonlinear dynamical system model of gene regulation: A perturbed steady-state approach. Ann Appl Stat. 2013;7:1311–33. [Google Scholar]
- 48.Peng PC, Hassan Samee MA, Sinha S. Incorporating chromatin accessibility data into sequence-to-expression modeling. Biophys J. 2015;108:1257–67. doi: 10.1016/j.bpj.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Degner JF, Pai AA, Pique-Regi R, et al. DNase sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davie K, Jacobs J, Atkins M, et al. Discovery of Transcription Factors and Regulatory Regions Driving In Vivo Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling. PLoS Genet. 2015:11. doi: 10.1371/journal.pgen.1004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buenrostro JD, Wu B, Litzenburger UM, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–90. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dey SS, Kester L, Spanjaard B, et al. Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol. 2015;33:285–9. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]