Abstract
Introduction
Genetic constitution and inflammation both contribute to development of coronary artery disease (CAD). Several CAD-associated single-nucleotide polymorphisms (SNPs) have recently been identified, but their functions are largely unknown. We investigated the associations between CAD-associated SNPs and five CAD-related inflammatory biomarkers.
Methods
We genotyped 45 CAD-associated SNPs in 701 stable CAD patients in whom levels of high-sensitivity C-reactive protein (hsRCP), interleukin-6, calprotectin, fibrinogen and complement component 3 levels had previously been measured. A genetic risk score was calculated to assess the combined risk associated with all the genetic variants. A multiple linear regression model was used to assess associations between the genetic risk score, single SNPs, and the five inflammatory biomarkers.
Results
The minor allele (G) (CAD risk allele) of rs2075650 (TOMM40/APOE) was associated with lower levels of high-sensitivity C-reactive protein (effect per risk allele: -0.37 mg/l [95%CI -0.56 to -0.18 mg/l]). The inflammatory markers tested showed no association with the remaining 44 SNPs or with the genetic risk score.
Conclusions
In stable CAD patients, the risk allele of a common CAD-associated marker at the TOMM40/APOE locus was associated with lower hsCRP levels. No other genetic variants or the combined effect of all variants were associated with the five inflammatory biomarkers.
Introduction
Inflammation is of major importance for the development of coronary artery disease (CAD) [1]. Inflammatory cells and signaling molecules contribute to the disease process by modulating the arterial wall, promoting lipoprotein retention, plaque formation and possibly destabilization [1]. Accordingly, several inflammatory biomarkers have been shown to predict cardiovascular outcome [2–5].
Over the past 10 years, large-scale genome-wide association studies (GWAS) have identified a large number of single-nucleotide polymorphisms (SNPs) associated with CAD [6]. Combined into genetic risk scores (GRS), these SNPs predict adverse cardiovascular events in various populations with and without prior cardiovascular disease [7–10]. Although the majority of loci identified seem to act through induction of atherosclerosis, little is known about the underlying mechanisms.
The majority of CAD-associated SNPs are located in non-coding regions of the genome. Expression quantitative trait loci (eQTL) data indicate that the loci primarily exert their effect through regulation of nearby gene expression, but a large proportion of these genes have not previously been linked to CAD or risk factors for CAD [11,12]. A functional network analysis performed by the CARDIoGRAMplusC4D Consortium (as part of the largest GWAS meta-analysis available at the time of initiation of our study) suggested that SNPs related to the APOA1, IL6R, MRAS, and PLG genes may act on CAD development by affecting pathways of acute phase response signaling [11]. However, it remains unknown whether these SNPs are associated with commonly used outcome-associated biomarkers of inflammation such as high-sensitivity C-reactive protein (hs-CRP), interleukin 6 (IL-6), calprotectin, fibrinogen, and complement component 3 (C3) [2–5].
Therefore, our primary aim was to investigate the association between APOA1-, IL6R-, MRAS-, and PLG-linked SNPs, and hs-CRP, IL-6, calprotectin, fibrinogen, and C3. Secondarily, we explored the individual associations of other CAD-associated risk SNPs and the effect of all SNPs combined using a genetic risk score (GRS).
Methods
Design and study population
This was a cross-sectional study including patients with stable CAD. The entire cohort has previously been described in detail [13]. Briefly, 900 patients were recruited from the Western Denmark Heart Registry between November 2007 and January 2011, and all patients had CAD as verified by coronary angiography. At the time of enrollment, where blood samples were obtained, patients were considered stable (i.e. no cardiovascular events or revascularization procedures within the last 12 months).
From the entire cohort, substudies on inflammatory biomarkers were performed comprising hs-CRP [14], IL-6 [14], calprotectin [14], fibrinogen [15,16], and C3 [15,16]. Patients included in these substudies were younger, more often had diabetes, prior MI and prior coronary revascularization, whereas renal failure and antihypertensive medication was less common, compared with patients not included. In total, one or more inflammatory markers were measured in 713 patients, and DNA was available in 704 patients. All patients provided informed written consent. The project was approved by The Central Denmark Region Committees on Health Research Ethics (record number: 1-10-72-210-15) and by the Danish Data Protection Agency (record number: 1-16-02-400-15).
Inflammatory marker measurements
Standardized blood sampling was performed in the outpatient clinic between 8 a.m. and 3 p.m. Blood was sampled from the antecubital vein with patients in supine position after 30 minutes of rest using vacuum tubes, a large bore needle (19 G), and a minimum of stasis [13]. Blood for hs-CRP analysis was analysed using the KoneLab 30i (ILS Laboratories Scandinavia, Allerød, Denmark). The measurement range for hs-CRP was 0.2–10.0 mg/l. In 27 patients, hs-CRP values were outside this range (maximum CRP-value of 35.8 mg/l). These patients were excluded in order to avoid bias from patients with subclinical infections which could potentially affect the levels of the inflammatory markers measured. IL-6 analyses were performed using the Cobas® 6000 analyser, E module (Roche, Mannheim, Germany). Serum calprotectin was measured using enzyme-linked immunosorbent assay (ELISA) (MRP 8/14 Calprotectin, Bühlmann, Schönenbuch, Switzerland). Fibrinogen was measured by the clotting method of Clauss using a KC 10TM coagulometer (Henrich Amelung GmbH, Lemgo, Germany). Complement C3 was determined by ELISA according to the manufacturer’s instructions (GenWay Biotech, Inc., San Diego, CA, USA). The coefficient of variance was <5% for both calprotectin and C3 ELISA assays.
SNP selection and genotyping
A thorough literature review of CAD risk loci was used to select the lead SNPs or relevant proxies of 46 loci genome-wide significantly associated with CAD and/or myocardial infarction (MI) in populations of European ancestry [6]. This included CAD-associated lead SNPs previously linked to the APOA1, IL6R, MRAS, and PLG genes by either eQTL data or physical proximity [11].
DNA was obtained from whole blood and direct genotyping was performed on a Fluidigm Biomark HD as previously described [17]. One SNP (rs17114036) failed on all chips and three samples with less than 50% of SNPs successfully genotyped were excluded. Therefore, the final dataset consisted of 45 SNPs in 701 patients. Overall call rate was excellent (31376/31545 = 99.5%) and consistent for all SNPs, except for rs964184 (call rate: 570/701 = 81.3%). All genotypes were successfully called in 559/701 = 79.7% of samples, whereas ≥43 SNPs where successfully called in 697/701 = 99.4% of samples.
Statistical analysis
Patient characteristics are reported as mean ± standard deviation (SD), median (interquartile range [IQR]) or numbers (percentage). Each SNP was coded as 0, 1, or 2 depending on the number of CAD risk alleles in the patient. Under the assumption of additive genetic effects, a multivariable linear regression model was used to test the association between the individual SNPs and hs-CRP, IL-6, calprotectin, fibrinogen, and C3, respectively. Predefined covariates (age, sex, diabetes, prior MI, current smoking, body mass index [BMI], and renal failure defined as estimated glomerular filtration rate ≤60 ml/min) were simultaneously added to the model. Therefore, the beta coefficient of a SNP corresponds to the adjusted average effect per risk allele on the inflammatory biomarker.
To test the combined effect of all CAD-associated SNPs a weighted GRS was calculated as previously reported [17]. The GRS was calculated as the sum of the number of risk alleles in each individual, weighted by the log of the odds ratio for CAD obtained from the original discovery GWAS papers. In the rare case of a missing genotype, the average of the cohort (a number of 0–2) for that SNP was used to calculate the GRS (in order to avoid a value of zero). For statistical analyses, GRS was standardized meaning that the beta coefficient of the GRS corresponds to the adjusted effect on the inflammatory marker per SD increase in GRS.
In the primary analyses, we considered a conservative Bonferroni-corrected p-value <0.0025 as statistically significant (threshold: p = 0.05 / [4 SNPs × 5 inflammatory biomarkers]). When evaluating the remaining CAD-associated SNPs and the GRS, the level of significance was adjusted accordingly (threshold: p = 0.05 / [46 × 5] = 2.2×10−4). All analyses were performed using STATA version 13.1 (StataCorp, 4905 Lakeway Dr, College Station, TX, USA).
Results
Patient characteristics
A total of 701 patients were included in data analyses. Patient characteristics and numbers included in each analysis are displayed in Table 1. Mean age was 64 ± 9 years (range: 32–85 years) and 558 (80%) were males. Prior MI, diabetes and renal failure were present in 627 (89%), 218 (31%), and 102 (15%) of the patients, respectively.
Table 1. Patient characteristics.
| hs-CRPa (n = 484) |
IL-6b (n = 563) |
Calprotectinb (n = 543) |
Fibrinogenc (n = 700) |
C3c (n = 698) |
|
|---|---|---|---|---|---|
| Age | 64 ± 9 | 64 ± 9 | 64 ± 9 | 65 ± 9 | 65 ± 9 |
| Male sex | 386 (80) | 446 (79) | 430 (79) | 558 (80) | 557 (80) |
| Prior MI | 442 (91) | 518 (92) | 499 (92) | 626 (89) | 624 (89) |
| Prior PCI/CABG | 471 (97) | 544 (97) | 524 (97) | 672 (96) | 670 (96) |
| Prior Stroke | 25 (5) | 25 (4) | 25 (5) | 37 (5) | 37 (5) |
| Diabetes | 107 (22) | 140 (25) | 135 (25) | 218 (31) | 218 (31) |
| Renal failured | 66 (14) | 77 (14) | 76 (14) | 102 (15) | 102 (15) |
| Antihypertensive treatment | 437 (90) | 509 (91) | 493 (91) | 636 (91) | 635 (91) |
| Statin treatment | 448 (93) | 519 (92) | 501 (92) | 635 (91) | 634 (91) |
| Current smoking | 98 (20) | 117 (21) | 113 (21) | 150 (21) | 150 (22) |
| Systolic BP (mmHg) | 142 ± 20 | 142 ± 21 | 142 ± 20 | 142 ± 21 | 142 ± 21 |
| Diastolic BP (mmHg) | 83 ± 11 | 83 ± 11 | 83 ± 11 | 83 ± 11 | 83 ± 11 |
| Body mass index (kg/m2) | 27.6 ± 4.3 | 27.7 ± 4.4 | 27.8 ± 4.4 | 27.9 ± 4.3 | 27.9 ± 4.4 |
| Creatinine (mM) | 81 (71–93.5) | 81 (71–93) | 81 (71–93) | 81 (71–93.5) | 81 (71–94) |
Data are presented as mean ± SD, median (IQR), or n (%).Abbreviations: BMI, body mass index; BP, blood pressure; CABG, coronary artery bypass graft surgery; MI, myocardial infarction; PCI, percutaneous coronary intervention.
a Data on prior stroke, antihypertensive treatment, statin treatment, BP, and BMI were missing in 3, 1, 1, 25, and 2 individuals, respectively.
b Data on prior stroke, antihypertensive treatment, statin treatment, current smoking, BP, and BMI were missing in 4, 1, 1, 1, 28, and 2 individuals, respectively.
c Data on prior stroke, antihypertensive treatment, statin treatment, current smoking, BP, and BMI were missing in 6, 2, 3, 1, 30, and 2 individuals, respectively.
d Estimated glomerular filtration rate ≤60 ml/min.
Presumed inflammation-related SNPs and inflammatory proteins
The association between presumed inflammation-related SNPs and hs-CRP, IL-6, calprotectin, fibrinogen, and C3 is presented in Table 2. A weak association was observed between rs4845625 (IL6R) and C3 (mean adjusted effect per risk allele: 0.03 (95% CI 0.00–0.06) mg/ml, p = 0.04), but it did not meet the Bonferroni-corrected threshold of significance. Neither rs4252120 (PLG) rs964184 (APOA1), nor rs9818870 (MRAS) significantly affected the inflammatory markers measured.
Table 2. Associations between presumed inflammation-related SNPs and inflammatory proteins.
| Locus | SNP | Nearby genes | Call rate (%) |
RAF | hs-CRP Beta (95% CI) |
p | Interleukin-6 Beta (95% CI) |
p | Calprotectin Beta (95% CI) |
p | Fibrinogen Beta (95% CI) |
p | Complement C3 Beta (95% CI) |
p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6q26 | rs4252120 | PLG | 100 | 0.70 | -0.01 (-0.16–0.15) | 0.92 | -0.56 (-1.13–0.02) | 0.061 | 0.06 (-0.01–0.14) | 0.10 | 0.02 (-0.09–0.14) | 0.69 | -0.01 (-0.04–0.03) | 0.72 |
| 1q21.3 | rs4845625 | IL6R | 99.6 | 0.44 | 0.10 (-0.04–0.25) | 0.17 | 0.14 (-0.40–0.68) | 0.61 | 0.00 (-0.07–0.07) | 0.98 | 0.09 (-0.02–0.19) | 0.12 | 0.03 (0.00–0.06) | 0.042 |
| 11q23.3 | rs964184 | APOA1 | 81.3 | 0.15 | 0.15 (-0.05–0.36) | 0.14 | 0.05 (-0.71–0.82) | 0.89 | 0.04 (-0.06–0.14) | 0.45 | -0.04 (-0.15–0.07) | 0.49 | 0.02 (-0.03–0.06) | 0.52 |
| 3q22.3 | rs9818870 | MRAS | 99.9 | 0.18 | -0.02 (-0.20–0.15) | 0.79 | -0.24 (-0.91–0.44) | 0.49 | -0.02 (-0.11–0.06) | 0.59 | -0.04 (-0.17–0.10) | 0.59 | -0.01 (-0.05–0.03) | 0.63 |
Beta is the adjusted mean difference measured per risk allele. Bold indicate that the SNP meets a nominal threshold of significance of p<0.05. Abbreviations: hs-CRP, high-sensitivity C-reactive protein; RAF, risk allele frequency; SNP, single nucleotide polymorphism.
Remaining CAD-related SNPs and inflammatory proteins
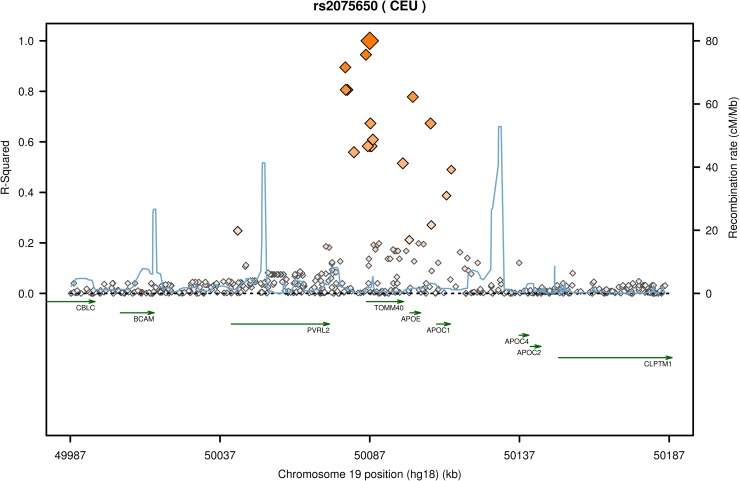
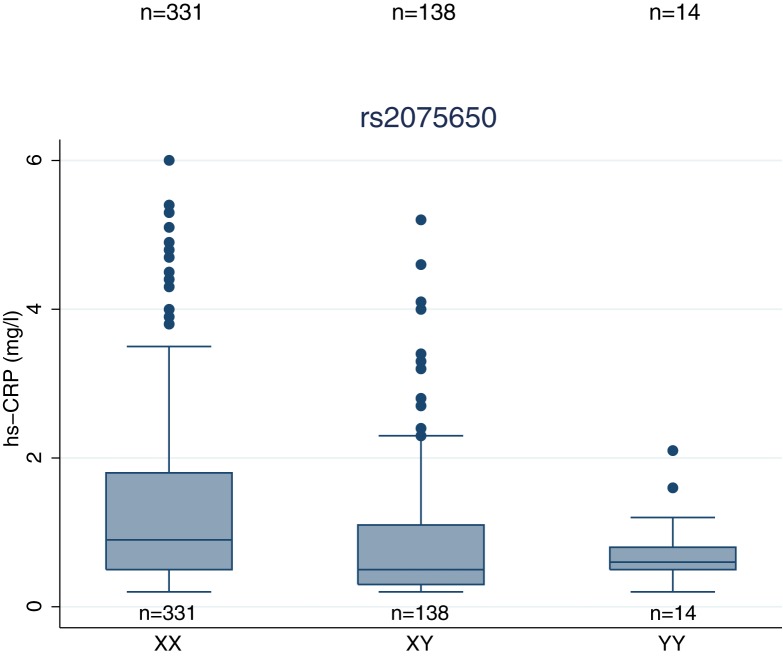
The evaluation of the remaining CAD-related SNPs is presented in Table 3. Of these, a nominally significant association was observed for rs1561198 (VAMP5/VAMP8) with IL-6 and calprotectin; rs17609940 (ANKS1A) with C3; rs2075650 (TOMM40, APOE) with hs-CRP and IL-6; rs264 (LPL) with C3; and finally rs599839 (SORT1) with fibrinogen. Only the association between rs2075650 and hs-CRP met the Bonferroni-corrected threshold of significance. The rs2075650 locus is displayed in Fig 1. Further analysis showed that mean level of hs-CRP in wildtype homozygous (A/A), heterozygous (A/G), and risk allele homozygous (G/G) patients were 1.38 mg/l (95% CI 1.25–1.52 mg/l), 0.96 mg/l (95% CI 0.79–1.13 mg/l), and 0.81 mg/l (95% CI 0.52–1.10 mg/l), respectively (Fig 2), demonstrating a gene-related dose-response effect.
Table 3. Associations between remaining CAD-related SNPs and inflammatory proteins.
| Locus | SNP | Nearby genes | Call rate (%) |
RAF | hs-CRP Beta (95% CI) |
p | IL-6 Beta (95% CI) |
p | Calprotectin Beta (95% CI) |
p | Fibrinogen Beta (95% CI) |
p | C3 Beta (95% CI) |
p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6p21.2 | rs10947789 | KCNK5 | 100 | 0.80 | 0.10 (-0.09–0.29) | 0.29 | -0.23 (-0.92–0.46) | 0.51 | 0.00 (-0.09–0.09) | 0.95 | 0.01 (-0.12–0.15) | 0.85 | 0.01 (-0.03–0.04) | 0.68 |
| 7q22 | rs10953541 | BCAP29 | 100 | 0.76 | -0.06 (-0.22–0.11) | 0.50 | 0.04 (-0.58–0.66) | 0.90 | -0.02 (-0.10–0.06) | 0.64 | -0.03 (-0.15–0.09) | 0.62 | 0.01 (-0.02–0.04) | 0.61 |
| 1p32.3 | rs11206510 | PCSK9 | 100 | 0.81 | -0.06 (-0.24–0.12) | 0.50 | 0.25 (-0.40–0.91) | 0.45 | 0.05 (-0.04–0.13) | 0.27 | 0.01 (-0.12–0.14) | 0.85 | 0.01 (-0.02–0.05) | 0.53 |
| 19p13.2 | rs1122608 | LDLR | 100 | 0.78 | -0.05 (-0.22–0.12) | 0.56 | 0.14 (-0.50–0.79) | 0.66 | 0.03 (-0.05–0.11) | 0.49 | 0.06 (-0.07–0.19) | 0.37 | 0.02 (-0.02–0.06) | 0.27 |
| 7q32.2 | rs11556924 | ZC3HC1 | 100 | 0.63 | -0.04 (-0.19–0.12) | 0.65 | 0.23 (-0.35–0.80) | 0.44 | 0.00 (-0.08–0.07) | 0.99 | -0.03 (-0.14–0.08) | 0.62 | 0.00 (-0.03–0.03) | 0.99 |
| 6q23.2 | rs12190287 | TCF21 | 100 | 0.65 | -0.13 (-0.28–0.03) | 0.10 | 0.00 (-0.58–0.57) | 0.99 | 0.02 (-0.05–0.10) | 0.52 | -0.02 (-0.14–0.09) | 0.70 | 0.01 (-0.02–0.04) | 0.59 |
| 10q24.3 | rs12413409 | CYP17A1 | 100 | 0.90 | -0.09 (-0.33–0.14) | 0.44 | 0.35 (-0.54–1.23) | 0.44 | -0.04 (-0.16–0.07) | 0.46 | -0.08 (-0.26–0.10) | 0.38 | 0.01 (-0.03–0.06) | 0.55 |
| 6p24.1 | rs12526453 | PHACTR1 | 99.9 | 0.69 | -0.05 (-0.20–0.10) | 0.53 | -0.12 (-0.69–0.45) | 0.67 | 0.04 (-0.04–0.11) | 0.33 | -0.01 (-0.12–0.10) | 0.87 | 0.02 (-0.01–0.05) | 0.29 |
| 17p11.2 | rs12936587 | RASD1. SMCR3. PEMT | 100 | 0.54 | 0.01 (-0.14–0.17) | 0.85 | 0.10 (-0.46–0.66) | 0.73 | -0.02 (-0.09–0.05) | 0.65 | -0.04 (-0.15–0.07) | 0.49 | 0.01 (-0.02–0.04) | 0.62 |
| 9p21.3 | rs1333049 | CDKN2BAS (ANRIL) | 99.9 | 0.51 | -0.06 (-0.21–0.08) | 0.40 | 0.15 (-0.38–0.69) | 0.58 | 0.01 (-0.06–0.08) | 0.83 | 0.07 (-0.04–0.17) | 0.21 | 0.00 (-0.03–0.03) | 0.92 |
| 10q23 | rs1412444 | LIPA | 99.9 | 0.34 | 0.00 (-0.15–0.14) | 0.98 | -0.25 (-0.79–0.30) | 0.38 | 0.00 (-0.07–0.07) | 0.92 | 0.01 (-0.10–0.12) | 0.89 | 0.02 (-0.01–0.05) | 0.22 |
| 2p11.2 | rs1561198 | VAMP5. VAMP8 | 100 | 0.48 | -0.05 (-0.19–0.09) | 0.47 | -0.58 (-1.10–-0.05) | 0.033 | -0.09 (-0.15–-0.02) | 0.014 | 0.00 (-0.11–0.10) | 1.00 | -0.02 (-0.05–0.01) | 0.12 |
| 1q41 | rs17465637 | MIA3 | 99.9 | 0.77 | -0.04 (-0.21–0.13) | 0.63 | -0.18 (-0.81–0.44) | 0.56 | 0.01 (-0.07–0.09) | 0.82 | -0.05 (-0.18–0.07) | 0.40 | 0.00 (-0.04–0.03) | 0.89 |
| 6p21.31 | rs17609940 | ANKS1A | 99.7 | 0.78 | 0.06 (-0.12–0.23) | 0.53 | 0.34 (-0.30–0.97) | 0.30 | -0.05 (-0.13–0.03) | 0.21 | 0.02 (-0.11–0.15) | 0.77 | -0.05 (-0.08–-0.02) | 0.0044 |
| 4q31.22 | rs1878406* | EDNRA | 100 | 0.15 | 0.20 (0.00–0.40) | 0.047 | -0.11 (-0.84–0.63) | 0.77 | 0.03 (-0.06–0.13) | 0.48 | 0.01 (-0.14–0.15) | 0.94 | -0.02 (-0.06–0.02) | 0.29 |
| 7p21.1 | rs2023938 | HDAC9 | 100 | 0.10 | 0.22 (-0.02–0.47) | 0.072 | 0.56 (-0.36–1.49) | 0.23 | -0.06 (-0.18–0.06) | 0.31 | 0.00 (-0.17–0.18) | 0.96 | 0.00 (-0.05–0.05) | 0.92 |
| 19q13 | rs2075650 | TOMM40, APOE | 99.9 | 0.17 | -0.37 (-0.56–-0.18) | 1.4×10−4 | -0.73 (-1.45–-0.02) | 0.046 | -0.06 (-0.15–0.03) | 0.19 | 0.05 (-0.10–0.19) | 0.54 | 0.00 (-0.04–0.04) | 0.94 |
| 17p13.3 | rs216172 | SMG6 | 99.7 | 0.38 | 0.00 (-0.15–0.15) | 0.99 | -0.02 (-0.44–0.40) | 0.93 | -0.04 (-0.11–0.03) | 0.26 | 0.06 (-0.05–0.17) | 0.31 | -0.03 (-0.06–0.00) | 0.061 |
| 2q22.3 | rs2252641 | ZEB2 | 100 | 0.48 | -0.11 (-0.26–0.04) | 0.14 | 0.30 (-0.26–0.85) | 0.30 | 0.01 (-0.06–0.08) | 0.75 | 0.05 (-0.06–0.16) | 0.38 | -0.02 (-0.05–0.01) | 0.20 |
| 10p11.23 | rs2505083 | KIAA1462 | 100 | 0.43 | 0.12 (-0.03–0.27) | 0.13 | 0.11 (-0.44–0.67) | 0.69 | 0.00 (-0.08–0.07) | 0.89 | 0.10 (-0.01–0.21) | 0.072 | -0.02 (-0.05–0.01) | 0.27 |
| 8p21.3 | rs264 | LPL | 99.6 | 0.86 | 0.16 (-0.05–0.38) | 0.13 | 0.40 (-0.20–1.00) | 0.19 | -0.03 (-0.13–0.07) | 0.54 | -0.05 (-0.20–0.10) | 0.53 | 0.04 (0.00–0.08) | 0.049 |
| 5q31.1 | rs273909 | SLC22A4 | 100 | 0.12 | -0.14 (-0.37–0.09) | 0.24 | -0.26 (-1.13–0.61) | 0.56 | -0.01 (-0.12–0.10) | 0.86 | -0.16 (-0.33–0.01) | 0.063 | 0.00 (-0.05–0.04) | 0.93 |
| 14q32.2 | rs2895811 | HHIPL1 | 99.9 | 0.47 | -0.02 (-0.17–0.12) | 0.76 | -0.32 (-0.73–0.10) | 0.13 | -0.01 (-0.08–0.06) | 0.79 | 0.03 (-0.08–0.14) | 0.55 | 0.01 (-0.02–0.04) | 0.69 |
| 8q24.13 | rs2954029 | TRIB1 | 100 | 0.52 | -0.14 (-0.29–0.01) | 0.061 | -0.40 (-0.95–0.14) | 0.15 | -0.05 (-0.12–0.02) | 0.18 | -0.04 (-0.15–0.07) | 0.50 | -0.01 (-0.04–0.02) | 0.49 |
| 12q24.12 | rs3184504 | SH2B3 | 99.0 | 0.56 | 0.01 (-0.13–0.16) | 0.89 | -0.25 (-0.78–0.28) | 0.35 | -0.05 (-0.12–0.02) | 0.16 | 0.00 (-0.10–0.11) | 0.93 | 0.00 (-0.03–0.03) | 0.96 |
| 6q25.3 | rs3798220 | LPA | 100 | 0.02 | 0.02 (-0.48–0.52) | 0.94 | -0.81 (-2.81–1.20) | 0.43 | -0.14 (-0.39–0.11) | 0.27 | -0.02 (-0.41–0.37) | 0.92 | -0.08 (-0.18–0.03) | 0.16 |
| 15q25.1 | rs3825807 | ADAMTS7 | 99.7 | 0.60 | 0.03 (-0.11–0.18) | 0.67 | 0.24 (-0.31–0.78) | 0.39 | -0.01 (-0.08–0.06) | 0.79 | 0.05 (-0.06–0.16) | 0.35 | 0.01 (-0.02–0.04) | 0.52 |
| 6p21.33 | rs3869109 | HLA-C. HLA-B | 99.7 | 0.59 | -0.09 (-0.23–0.05) | 0.22 | 0.18 (-0.35–0.71) | 0.50 | 0.06 (-0.01–0.12) | 0.10 | -0.10 (-0.21–0.00) | 0.062 | 0.02 (-0.01–0.04) | 0.29 |
| 17q21.32 | rs46522 | UBE2Z | 100 | 0.58 | -0.10 (-0.25–0.05) | 0.19 | -0.04 (-0.59–0.51) | 0.89 | 0.06 (-0.01–0.13) | 0.12 | 0.07 (-0.04–0.18) | 0.23 | 0.00 (-0.03–0.02) | 0.75 |
| 13q34 | rs4773144 | COL4A1 | 99.7 | 0.42 | 0.01 (-0.14–0.16) | 0.89 | -0.25 (-0.82–0.32) | 0.38 | -0.01 (-0.08–0.07) | 0.89 | 0.01 (-0.10–0.12) | 0.90 | -0.03 (-0.06–0.00) | 0.083 |
| 9q34 | rs495828 | AB0 | 99.7 | 0.23 | 0.09 (-0.08–0.26) | 0.29 | 0.40 (-0.23–1.03) | 0.21 | 0.01 (-0.07–0.09) | 0.87 | 0.00 (-0.13–0.12) | 0.98 | 0.01 (-0.02–0.05) | 0.46 |
| 10q11.1 | rs501120 | CXCL12 | 100 | 0.88 | -0.08 (-0.30–0.14) | 0.48 | -0.25 (-1.07–0.57) | 0.56 | 0.10 (-0.01–0.20) | 0.076 | -0.04 (-0.19–0.12) | 0.66 | 0.02 (-0.02–0.07) | 0.28 |
| 2p24.1 | rs515135 | APOB | 99.9 | 0.83 | 0.10 (-0.09–0.28) | 0.31 | 0.69 (-0.01–1.38) | 0.053 | 0.05 (-0.04–0.14) | 0.26 | -0.01 (-0.15–0.13) | 0.88 | -0.01 (-0.05–0.03) | 0.54 |
| 1p13 | rs599839 | SORT1 | 100 | 0.78 | 0.15 (-0.03–0.33) | 0.10 | 0.21 (-0.45–0.88) | 0.53 | 0.06 (-0.02–0.15) | 0.16 | 0.14 (0.01–0.28) | 0.034 | 0.01 (-0.02–0.05) | 0.54 |
| 2p21 | rs6544713 | ABCG8 | 100 | 0.30 | -0.04 (-0.20–0.12) | 0.62 | 0.06 (-0.53–0.66) | 0.83 | -0.05 (-0.12–0.03) | 0.22 | -0.02 (-0.13–0.10) | 0.80 | -0.01 (-0.04–0.02) | 0.54 |
| 2q33.1 | rs6725887 | WDR12 | 100 | 0.17 | -0.14 (-0.33–0.05) | 0.15 | 0.01 (-0.71–0.74) | 0.97 | 0.02 (-0.07–0.11) | 0.68 | -0.08 (-0.22–0.06) | 0.29 | 0.01 (-0.03–0.05) | 0.54 |
| 4q32.1 | rs7692387 | GUCY1A3 | 100 | 0.83 | -0.13 (-0.33–0.07) | 0.22 | 0.09 (-0.66–0.85) | 0.80 | 0.00 (-0.10–0.10) | 0.98 | -0.05 (-0.19–0.10) | 0.51 | -0.02 (-0.06–0.02) | 0.39 |
| 15q26.1 | rs8039305 | FURIN | 99.7 | 0.51 | -0.01 (0.16–0.13) | 0.85 | 0.27 (-0.28–0.82) | 0.33 | -0.01 (-0.08–0.06) | 0.87 | -0.03 (-0.14–0.08) | 0.56 | 0.03 (0.00–0.06) | 0.068 |
| 13q12.3 | rs9319428 | FLT1 | 99.7 | 0.33 | 0.00 (-0.15–0.16) | 0.96 | 0.24 (-0.33–0.82) | 0.41 | -0.03 (-0.11–0.04) | 0.37 | 0.00 (-0.11–0.12) | 0.95 | -0.01 (-0.04–0.02) | 0.66 |
| 11q22.3 | rs974819 | PDGFD | 100 | 0.27 | -0.04 (-0.20–0.12) | 0.62 | 0.18 (-0.42–0.79) | 0.55 | 0.03 (-0.05–0.11) | 0.45 | 0.02 (-0.10–0.14) | 0.75 | -0.02 (-0.05–0.01) | 0.24 |
| 21q22.1 | rs9982601 | MRPS6 | 99.9 | 0.14 | 0.14 (-0.07–0.35) | 0.18 | 0.22 (-0.57–1.01) | 0.59 | 0.06 (-0.04–0.16) | 0.24 | -0.02 (-0.17–0.14) | 0.84 | 0.03 (-0.01–0.07) | 0.16 |
Bold values indicate that the SNP meets a nominal threshold of significance of p<0.05.
*rs1878406 was genotyped as a C/T SNP.
Abbreviations: hs-CRP, high-sensitivity C-reactive protein; RAF, risk allele frequency; SNP, single nucleotide polymorphism.
Fig 1. Recombination rate and linkage disequilibrium at the rs2075650 locus in the CEU population.
Generated using SNAP (http://archive.broadinstitute.org/mpg/snap/ldplot.php) [18].
Fig 2. Distribution of hs-CRP levels stratified by the genotypes of rs2075650.
Boxes and whiskers indicate quartiles and adjacent values. Values outside the range of adjacent values are plotted as outliers. Abbreviations: hs-CRP, high sensitivity C-reactive protein.
GRS and inflammatory proteins
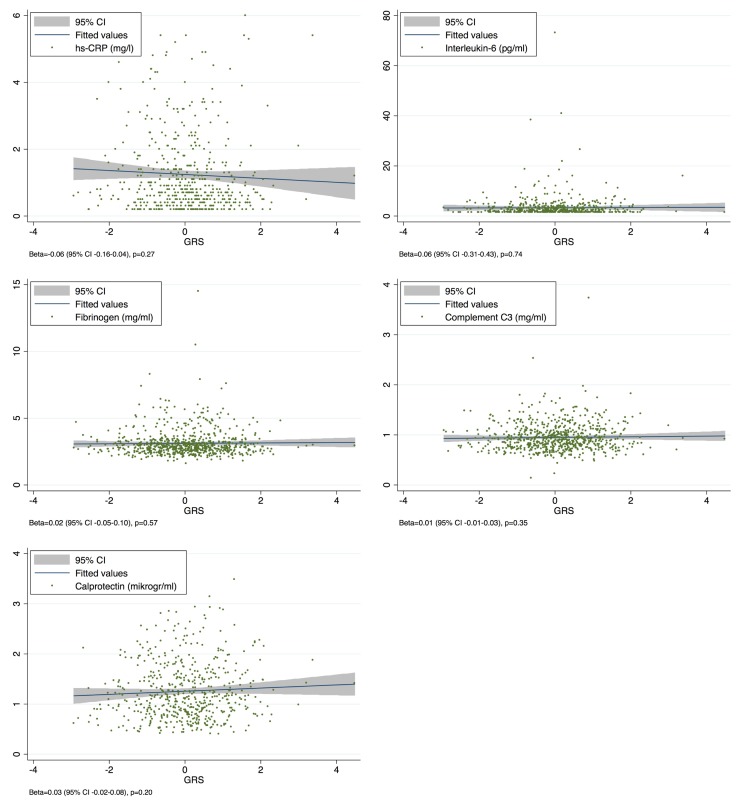
We found no associations between the GRS and hs-CRP, IL-6, calprotectin, fibrinogen, or C3 with results detailed in Fig 3.
Fig 3. Scatterplots of the association between inflammatory markers and the standardized GRS.
Abbreviations: GRS, genetic risk score; hs-CRP, high-sensitivity C-reactive protein.
Discussion
In the present study of patients with established CAD, we investigated the association between 45 lead SNPs from loci associated with CAD and five common biochemical markers of inflammation. The main findings were; 1) for SNP rs2075650 in the TOMM40, APOE locus, the established CAD-risk allele was significantly associated with lower hs-CRP levels, 2) No other CAD-related SNPs were associated with the inflammatory marker levels, either measured as individual SNPs or when combined into a GRS.
Previous GWASs have demonstrated a robust association between the rs2075650 G-allele and an increased risk of CAD [11]. In a recent study based on the present cohort, subanalysis also confirmed an association between rs2075650 and recurrent CAD events showing that an increase in the number of CAD risk alleles was associated with a hazard ratio of 1.40 (95% CI 1.00–1.97) of the primary endpoint composed of cardiovascular death, myocardial infarction and stable coronary revascularization [19]. Considering the well-established relationship between increasing levels of hs-CRP and adverse cardiovascular outcome [20], it may be surprising that the CAD risk allele of rs2075650 was associated with lower levels of hs-CRP in our sample. However, our findings are consistent with results from several previous large population-based cohort studies. In these studies, the same inverse relationship between the CAD risk allele and lower levels of hs-CRP has also been observed in European Americans [21], Australian twin families [22], Asians [23], and Hispanics [21] but not Afro-Americans [21]. Our work extends these findings by demonstrating an association in patients with established CAD. The mechanistic explanation for this inverse association is currently unknown. The marker rs2075650 is located in the TOMM40 gene, just upstream of APOE, and APOC1. The CAD risk allele (G) has been associated with a range of other phenotypes including reduced longevity [24], reduced BMI [25], increased low-density lipoprotein cholesterol (LDL-C) [22,26], and an increased risk of Alzheimer’s disease [27]. Because of the relatively strong linkage disequilibrium in the TOMM40/APOE locus, it has been suggested that the G-allele at rs2075650 is in fact tagging causal variation in the APOE gene. The APOE encodes the apolipoprotein E with three different isoforms (ε2, ε3, and ε4 defined by the combination of rs7412 and rs429358. Northwestern European ancestry (CEU): r2 = 0.02 and r2 = 0.20 with rs2075650, respectively), of which the ε4 isoform has long been known to associate with LDL-C, Alzheimer’s disease, and hs-CRP [28,29]. However, recent data suggest that the TOMM40/APOE locus is genetically complex [30], and therefore it is plausible that the G-allele is tagging different underlying causal variants with different effects on CAD risk and hs-CRP, a concept supported by Middleberg et al [22]. This would also be in line with the current understanding that hs-CRP is not causally related to cardiovascular risk [31].
Some previous GWASs have explored the association between CAD-associated risk variants and common inflammatory markers, of which the IL6R locus has been associated with several. In studies of hs-CRP, the IL6R locus (rs4129267) was consistently, though moderately, associated with hs-CRP levels (CEU: r2 = 0.54 with rs4845625) [32–34]. Furthermore, IL6R (rs4129267) has been associated with plasma levels of fibrinogen and IL-6 [35–37]. Although we observed a nominal association between the IL6R locus and C3, our study does not support a significant effect of IL6R on the inflammatory response. Other CAD-associated loci have also emerged in GWASs of inflammatory markers. A large study from the CHARGE (Cohorts for Heart and Aging Research in Genetic Epidemiology) consortium demonstrated a significant association between fibrinogen and variants near LIPA (rs2250644) and SH2B3 (rs7310615) [36]. Although these variants are in perfect linkage disequilibrium with the SNPs genotyped in our study (CEU: r2 = 1.00 for both), we did not find evidence of such association. Other GWASs have also demonstrated weak associations between variants at the AB0 locus (rs657152 and rs8176704; CEU: r2 = 0.46 and r2 = 0.02 with rs495828) and IL-6 [34,37], and a Chinese GWAS of C3 found an association with rs11575839 close to HLA-C (CEU: r2 = 0.02 with rs3869109) [38]. We were not able to confirm any of these associations. Importantly, our study was not powered to detect very small effect sizes. However, it is striking that none of our estimates indicated even a trend towards such relationships. Several explanations for these inconsistencies may exist. Some of the SNPs tagged in prior GWASs display different allele frequencies compared with the CAD-associated lead SNPs genotyped in our study and slightly different ancestral origins may possibly play a role as well. However, another important explanation may relate to the fact that we included patients with established CAD in contrast to prior studies performed in population-based cohorts without known cardiovascular disease. Patients with CAD have an increased inflammatory response compared with healthy subjects [2], either as the cause or as a consequence of CAD. Therefore, causal genetic variants might not associate with the levels of inflammatory biomarkers in cohorts where all patients are affected by CAD, although such an association may be evident in community-based populations, where some patients likely have subclinical CAD. In this context, it is important to note that we included stable CAD patients in our study. Ninety percent had previous MI occurring at least 12 months prior to inclusion, thus making it less likely that prior MIs influenced the levels of inflammatory biomarkers.
Calprotectin is suggested a new biomarker of CAD [39–41]. The expression of calprotectin has been found at the site of plaque rupture and in macrophages of atherosclerotic plaques and is considered an inflammatory marker of plaque instability [40,42]. To our knowledge, the present study is the first to explore the association between calprotectin and CAD-associated risk variants. Although none of the CAD-associated variants significantly affected calprotectin levels, a trend was observed for rs1561198. This SNP is located between the VAMP5 and VAMP8 genes, whose products are involved in different aspects of vesicle trafficking including cytokine release and phagocytosis [43]. Hence, a link between this locus and calprotectin levels may plausibly exist. However, further studies with larger number of individuals are needed to confirm this hypothesis.
Our study has limitations. Because of the number of statistical tests performed, we applied a conservative Bonferroni-corrected threshold of significance to reduce the risk of type I errors. This, together with the moderate sample size in the context of common complex diseases, reduces the power to detect small effect sizes, in particular for SNPs with low minor allele frequencies and for hs-CRP, IL-6, and calprotectin, which were not assessed in all patients. Therefore, our study should be considered as exploratory. We did not assess the presence of other inflammatory conditions, which may also affect the levels of inflammatory biomarkers. In case of bias, this would likely lead the associations towards the null, since no strong association between the genotyped SNPs and any such conditions has been reported. Moreover, we performed the statistical analyses assuming additive genetic effects of the risk alleles. Although this assumption may be reasonable for most of the genetic loci investigated, some might better fit a recessive model [44], which would affect the power of our analyses.
Conclusion
In the present study, a common CAD-associated variant at the TOMM40/APOE locus (rs2075650) was significantly associated with lower levels of hs-CRP in patients with stable CAD. Future studies using deep sequencing of the TOMM40/APOE locus in large clinical samples are warranted to determine if rs2075650 is truly causing opposite allelic effects on CAD and hs-CRP, or if the opposite association is explained by underlying linkage disequilibrium with several hidden functional variants of which some affect the development of CAD independent of hs-CRP. None of the remaining variants, both assessed independently or combined as a GRS, were associated with hs-CRP, IL-6, calprotectin, fibrinogen, or C3. Our findings may suggest that the effect of these CAD-loci on CAD development does not act through pathways significantly affecting these commonly used inflammatory biomarkers.
Supporting information
(PDF)
(DTA)
Acknowledgments
The authors are grateful to Lise Nielsen Wulff for assisting with patient inclusion. We also appreciate the help from Jakob Helin and Vivi Bo Mogensen for laboratory support and from Peter Nissen for administrating the biobank. Funding was received from the Novo Nordic Foundation (grant no. NNF14OC0008817 to SDK) and Pfizer (unrestricted research grant no. WS2632086 to HKJ).
Data Availability
All relevant data are within the paper.
Funding Statement
Funding was received from the Novo Nordic Foundation (grant no. NNF14OC0008817 to SDK) and Pfizer (unrestricted research grant no. WS2632086 to HKJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473: 317–325. doi: 10.1038/nature10146 [DOI] [PubMed] [Google Scholar]
- 2.Emerging Risk Factors Collaboration, Kaptoge S, Di Angelantonio E, Pennells L, Wood AM, White IR, et al. C-reactive protein, fibrinogen, and cardiovascular disease prediction. N Engl J Med. 2012;367: 1310–1320. doi: 10.1056/NEJMoa1107477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaptoge S, Seshasai SRK, Gao P, Freitag DF, Butterworth AS, Borglykke A, et al. Inflammatory cytokines and risk of coronary heart disease: new prospective study and updated meta-analysis. European Heart Journal. 2014;35: 578–589. doi: 10.1093/eurheartj/eht367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.JENSEN LJN, PEDERSEN S, BJERRE M, MOGELVANG R, JENSEN JS, FLYVBJERG A. Plasma calprotectin predicts mortality in patients with ST segment elevation myocardial infarction treated with primary percutaneous coronary intervention. J Interv Cardiol. 2010;23: 123–129. doi: 10.1111/j.1540-8183.2010.00532.x [DOI] [PubMed] [Google Scholar]
- 5.Muscari A, Bozzoli C, Puddu GM, Sangiorgi Z, Dormi A, Rovinetti C, et al. Association of serum C3 levels with the risk of myocardial infarction. Am J Med. 1995;98: 357–364. doi: 10.1016/S0002-9343(99)80314-3 [DOI] [PubMed] [Google Scholar]
- 6.McPherson R. Genome-Wide Association Studies of Cardiovascular Disease in European and Non-European Populations. Curr Genet Med Rep. 2014;2: 1–12. doi: 10.1007/s40142-014-0033-y [Google Scholar]
- 7.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376: 1393–1400. doi: 10.1016/S0140-6736(10)61267-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganna A, Magnusson PKE, Pedersen NL, de Faire U, Reilly M, Arnlöv J, et al. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler Thromb Vasc Biol. Lippincott Williams & Wilkins; 2013;33: 2267–2272. doi: 10.1161/ATVBAHA.113.301218 [DOI] [PubMed] [Google Scholar]
- 9.Abraham G, Havulinna AS, Bhalala OG, Byars SG, De Livera AM, Yetukuri L, et al. Genomic prediction of coronary heart disease. European Heart Journal. 2016;37: 3267–3278. doi: 10.1093/eurheartj/ehw450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaara S, Tikkanen E, Parkkonen O, Lokki M-L, Ripatti S, Perola M, et al. Genetic Risk Scores Predict Recurrence of Acute Coronary Syndrome. Circulation: Cardiovascular Genetics. Lippincott Williams & Wilkins; 2016;9: 172–178. doi: 10.1161/CIRCGENETICS.115.001271 [DOI] [PubMed] [Google Scholar]
- 11.CARDIoGRAMplusC4D Consortium, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45: 25–33. doi: 10.1038/ng.2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466: 714–719. doi: 10.1038/nature09266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen SB, Grove EL, Neergaard-Petersen S, Würtz M, Hvas A-M, Kristensen SD. Determinants of reduced antiplatelet effect of aspirin in patients with stable coronary artery disease. PLoS ONE. 2015;10: e0126767 doi: 10.1371/journal.pone.0126767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen SB, Grove EL, Pareek M, Kristensen SD, Hvas A-M. Calprotectin and platelet aggregation in patients with stable coronary artery disease. PLoS ONE. 2015;10: e0125992 doi: 10.1371/journal.pone.0125992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neergaard-Petersen S, Ajjan R, Hvas A-M, Hess K, Larsen SB, Kristensen SD, et al. Fibrin Clot Structure and Platelet Aggregation in Patients with Aspirin Treatment Failure. Eckle T, editor. PLoS ONE. 2013;8: e71150–8. doi: 10.1371/journal.pone.0071150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neergaard-Petersen S, Hvas AM, Kristensen SD, Grove EL, Larsen SB, Phoenix F, et al. The influence of type 2 diabetes on fibrin clot properties in patients with coronary artery disease. Thromb Haemost. 2014;112: 1142–1150. doi: 10.1160/TH14-05-0468 [DOI] [PubMed] [Google Scholar]
- 17.Christiansen MK, Nyegaard M, Pedersen LN, Larsen SB, Würtz M, Hjort J, et al. A 45-SNP genetic risk score is increased in early-onset coronary artery disease but independent of familial disease clustering. Atherosclerosis. 2017;257: 172–178. doi: 10.1016/j.atherosclerosis.2017.01.010 [DOI] [PubMed] [Google Scholar]
- 18.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24: 2938–2939. doi: 10.1093/bioinformatics/btn564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christiansen MK, Nyegaard M, Larsen SB, Grove EL, Würtz M, Neergaard-Petersen S, et al. A genetic risk score predicts cardiovascular events in patients with stable coronary artery disease. International Journal of Cardiology. Elsevier Ireland Ltd; 2017;: 1–6. doi: 10.1016/j.ijcard.2017.04.045 [DOI] [PubMed] [Google Scholar]
- 20.Emerging Risk Factors Collaboration, Kaptoge S, Di Angelantonio E, Lowe G, Pepys MB, Thompson SG, et al. C-reactive protein concentration and risk of coronary heart disease, stroke, and mortality: an individual participant meta-analysis. Lancet. 2010;375: 132–140. doi: 10.1016/S0140-6736(09)61717-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocarnik JM, Pendergrass SA, Carty CL, Pankow JS, Schumacher FR, Cheng I, et al. Multiancestral analysis of inflammation-related genetic variants and C-reactive protein in the population architecture using genomics and epidemiology study. Circulation: Cardiovascular Genetics. American Heart Association, Inc; 2014;7: 178–188. doi: 10.1161/CIRCGENETICS.113.000173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Middelberg RPS, Ferreira MAR, Henders AK, Heath AC, Madden PAF, Montgomery GW, et al. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med Genet. BioMed Central; 2011;12: 123 doi: 10.1186/1471-2350-12-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorajoo R, Li R, Ikram MK, Liu J, Froguel P, Lee J, et al. Are C-reactive protein associated genetic variants associated with serum levels and retinal markers of microvascular pathology in Asian populations from Singapore? PLoS ONE. 2013;8: e67650 doi: 10.1371/journal.pone.0067650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deelen J, Beekman M, Uh H-W, Helmer Q, Kuningas M, Christiansen L, et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10: 686–698. doi: 10.1111/j.1474-9726.2011.00705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y, Lanktree MB, Taylor KC, Hakonarson H, Lange LA, Keating BJ, et al. Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Human Molecular Genetics. 2013;22: 184–201. doi: 10.1093/hmg/dds396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371: 483–491. doi: 10.1016/S0140-6736(08)60208-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31: 1102–1110. doi: 10.1038/nbt.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261: 921–923. doi: 10.1126/science.8346443 [DOI] [PubMed] [Google Scholar]
- 29.Chasman DI, Kozlowski P, Zee RY, Kwiatkowski DJ, Ridker PM. Qualitative and quantitative effects of APOE genetic variation on plasma C-reactive protein, LDL-cholesterol, and apoE protein. Genes Immun. 2006;7: 211–219. doi: 10.1038/sj.gene.6364289 [DOI] [PubMed] [Google Scholar]
- 30.Bekris LM, Lutz F, Yu C-E. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J Hum Genet. 2012;57: 18–25. doi: 10.1038/jhg.2011.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C Reactive Protein Coronary Heart Disease Genetics Collaboration (CCGC), Wensley F, Gao P, Burgess S, Kaptoge S, Di Angelantonio E, et al. Association between C reactive protein and coronary heart disease: mendelian randomisation analysis based on individual participant data. BMJ. 2011;342: d548 doi: 10.1136/bmj.d548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridker PM, Paré G, Parker A, Zee RYL, Danik JS, Buring JE, et al. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am J Hum Genet. 2008;82: 1185–1192. doi: 10.1016/j.ajhg.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123: 731–738. doi: 10.1161/CIRCULATIONAHA.110.948570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah T, Zabaneh D, Gaunt T, Swerdlow DI, Shah S, Talmud PJ, et al. Gene-centric analysis identifies variants associated with interleukin-6 levels and shared pathways with other inflammation markers. Circulation: Cardiovascular Genetics. 2013;6: 163–170. doi: 10.1161/CIRCGENETICS.112.964254 [DOI] [PubMed] [Google Scholar]
- 35.Sabater-Lleal M, Huang J, Chasman D, Naitza S, Dehghan A, Johnson AD, et al. Multiethnic meta-analysis of genome-wide association studies in >100 000 subjects identifies 23 fibrinogen-associated Loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation. 2013;128: 1310–1324. doi: 10.1161/CIRCULATIONAHA.113.002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Vries PS, Chasman DI, Sabater-Lleal M, Chen M-H, Huffman JE, Steri M, et al. A meta-analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration. Human Molecular Genetics. 2016;25: 358–370. doi: 10.1093/hmg/ddv454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naitza S, Porcu E, Steri M, Taub DD, Mulas A, Xiao X, et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012;8: e1002480 doi: 10.1371/journal.pgen.1002480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X, Sun J, Gao Y, Tan A, Zhang H, Hu Y, et al. Genome-wide association study for serum complement C3 and C4 levels in healthy Chinese subjects. PLoS Genet. 2012;8: e1002916 doi: 10.1371/journal.pgen.1002916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Healy AM, Pickard MD, Pradhan AD, Wang Y, Chen Z, Croce K, et al. Platelet expression profiling and clinical validation of myeloid-related protein-14 as a novel determinant of cardiovascular events. Circulation. 2006;113: 2278–2284. doi: 10.1161/CIRCULATIONAHA.105.607333 [DOI] [PubMed] [Google Scholar]
- 40.Altwegg LA, Neidhart M, Hersberger M, Müller S, Eberli FR, Corti R, et al. Myeloid-related protein 8/14 complex is released by monocytes and granulocytes at the site of coronary occlusion: a novel, early, and sensitive marker of acute coronary syndromes. European Heart Journal. 2007;28: 941–948. doi: 10.1093/eurheartj/ehm078 [DOI] [PubMed] [Google Scholar]
- 41.Morrow DA, Wang Y, Croce K, Sakuma M, Sabatine MS, Gao H, et al. Myeloid-related protein 8/14 and the risk of cardiovascular death or myocardial infarction after an acute coronary syndrome in the Pravastatin or Atorvastatin Evaluation and Infection Therapy: Thrombolysis in Myocardial Infarction (PROVE IT-TIMI 22) trial. Am Heart J. 2008;155: 49–55. doi: 10.1016/j.ahj.2007.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ionita MG, Vink A, Dijke IE, Laman JD, Peeters W, van der Kraak PH, et al. High levels of myeloid-related protein 14 in human atherosclerotic plaques correlate with the characteristics of rupture-prone lesions. Arterioscler Thromb Vasc Biol. 2009;29: 1220–1227. doi: 10.1161/ATVBAHA.109.190314 [DOI] [PubMed] [Google Scholar]
- 43.Chaineau M, Danglot L, Galli T. Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking. FEBS Lett. 2009;583: 3817–3826. doi: 10.1016/j.febslet.2009.10.026 [DOI] [PubMed] [Google Scholar]
- 44.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Hólm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43: 333–338. doi: 10.1038/ng.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(DTA)
Data Availability Statement
All relevant data are within the paper.