Abstract
Background
The pathology of osteoarthritis (OA) is partly attributed to genetic factors; however, the role of ADAM12 polymorphism is still controversial. It is necessary to perform a meta-analysis to investigate this possible correlation.
Methods
Case–control studies on the association between OA susceptibility and ADAM12 polymorphism were comprehensively collected by searching PubMed, Embase, and Web of Science. Odds ratios (ORs) and 95% confidence intervals (CIs) were pooled to evaluate OA risk that was possibly conferred by ADAM12 variant. The analyses were performed not only among general population but also in male and female groups.
Results
A total of 8 studies with 10 populations were finally included in this meta-analysis. In the general population, 4 comparisons were carried out (C allele vs G allele, CC vs GG, GC + CC vs GG, and CC vs GC + GG) and found that ADAM12 rs3740199 polymorphism was not associated with increased OA vulnerability. On the other hand, the analyses stratified by gender made 5 comparisons (C allele vs G allele, CC vs GG, GC vs GG, GC + CC vs GG, and CC vs GC + GG). It was shown that rs3740199 polymorphism (GC + CC vs GG) was a risk factor for OA among male patients (OR =1.45, 95% CI =1.04–2.01). Sensitivity analysis indicated that it was an unstable outcome. No correlation was identified in women. Neither heterogeneity nor publication bias was detected in the analyses mentioned above.
Conclusion
ADAM12 rs3740199 polymorphism is likely to be associated with OA susceptibility among male patients, other than the general population. More studies are needed to confirm this observation. The mechanism by which ADAM12 variant plays a role in OA pathogenesis is also warranted and important for interpreting this possible correlation.
Keywords: osteoarthritis, meta-analysis, single nucleotide polymorphism, ADAM
Introduction
Osteoarthritis (OA) is a degenerative joint disease that results from breakdown of joint cartilages and bones. This disease is a major cause of disability worldwide, with poor functional outcomes and limited lifespan even after effective treatment. As a result, the focus is shifting to risk factor identification and disease prevention.1 A number of risk factors for OA exist, including physical activity, obesity, gender, aging, and genetic predisposition.2 In recent years, genetic risk factors associated with OA susceptibility have been extensively studied. At present, 21 independent OA susceptibility loci have been established from candidate gene studies, linkage studies, and genome-wide association studies (GWASs).3 Apart from that, more specific genes which conferred vulnerability of OA are still being identified. Evidence has been published for the association of OA predisposition with single-nucleotide polymorphisms (SNPs) of a member from disintegrin and metalloprotease (ADAM) family – ADAM12.4–6
The ADAM family comprises >30 zinc-dependent proteases that contribute to proteolysis, adhesion, fusion, and cell signaling.7 ADAM12 is one of the splice forms of ADAMs and the gene for human ADAM12 is located on chromosome 10q26.3.8 Two SNPs of the ADAM12 gene, rs3740199 and rs1871054, have been studied most frequently with OA susceptibility. It has been demonstrated that rs3740199 is associated with OA susceptibility among the general population,9 or certain groups of the studied participants.6,10 Apart from disease vulnerability, ADAM12 mutation has also been linked to OA severity.5 However, studies that did not observe any positive association of ADAM12 SNP with OA also exist among various races.11,12 As a result, until now, no consensus has been reached regarding the proposal that ADAM12 polymorphisms are related to disease predisposition, severity, or phenotype. In addition, the power of genetic association study is of importance. Individual study, especially with limited sample size, provides insufficient power. Therefore, it is necessary to collect the up-to-date evidence, perform a meta-analysis, and present with the latest and robust overview of this subject. To better understand the association of OA susceptibility with ADAM12 SNPs, a meta-analysis was conducted by a comprehensive literature search and synthesis of data from eligible case–control studies.
Materials and methods
Identification of eligible studies and data collection
The latest literature search was conducted in January 2017 on PubMed, Web of Science, and Embase. The following keywords were used: “osteoarthritis,” “OA,” “ADAM12,” and “polymorphism.” Case–control studies about the association of ADAM12 polymorphisms with OA susceptibility were collected. Relative references were further reviewed to identify potential extra studies. No restrictions of race and publication year were imposed. Published articles with full text, other than reviews or records from conference proceedings, were included. To exclude overlapping data observed from the same cohort, only the newest study or the study with the largest sample size was selected. Records were dropped to remove flawed studies when the genotypic distribution deviated from Hardy–Weinberg equilibrium (HWE).
Two reviewers extracted data independently, and disagreements were solved by discussion. The following data were collected: first author, publication year, race, characteristics of cases and controls, diagnosis criteria, SNP position, methods of genotyping, genotypic distributions, and odds ratios (ORs). Additional information regarding the characteristics of the participants was required if stratification analyses were needed.
Statistical analysis
Chi-square test was applied to assess whether the genotypic distribution was in concordance with HWE and whether the original article did not mention the outcome of the HWE test. Q-statistic was performed to evaluate the heterogeneity across studies.13 The magnitude of heterogeneity was measured by I2-value, indicating the proportion of the total variance across studies due to heterogeneity, other than chance. I2≥50%, 50%>I2≥25%, and 25%>I2 were defined as high, medium, and low heterogeneity, respectively.14 If no inconsistency was detected, ORs were aggregated by Mantel–Haenszel’s method in fixed-effect model. Otherwise, Dersimonian–Laird method in random-effect model was applied.15 Publication bias was detected by Egger’s linear regression test.13 Sensi-tivity analysis was performed by omitting one study in each turn and then checking statistical significance. All statistical analyses were conducted by Stata 9.0 (Stata Crop LP, College Station, TX, USA). All P-values were two-sided, with <0.05 being statistically significant.
Results
Characteristics of the eligible studies
The process of study inclusion is illustrated in Figure 1. Overall, 8 studies with 10 populations, including 8,553 cases and 6,349 controls, were eligible for this meta-analysis.4–6,9–12,16 The majority of the cases were individuals who were diagnosed with knee, hip, and hand OA from various races, according to the criteria of American College of Rheumatology (ACR).4–6,9,11,12 Four candidate gene positions (rs3740199, rs1871054, rs1278279, and rs1044122) were investigated; among which rs3740199 was the most frequently studied locus; therefore, the original data regarding the association of rs3740199 polymorphism with OA susceptibility were primarily extracted. The genotypic distributions from all the included studies did not deviate from HWE according to the descriptions from the corresponding articles (Table 1).
Figure 1.
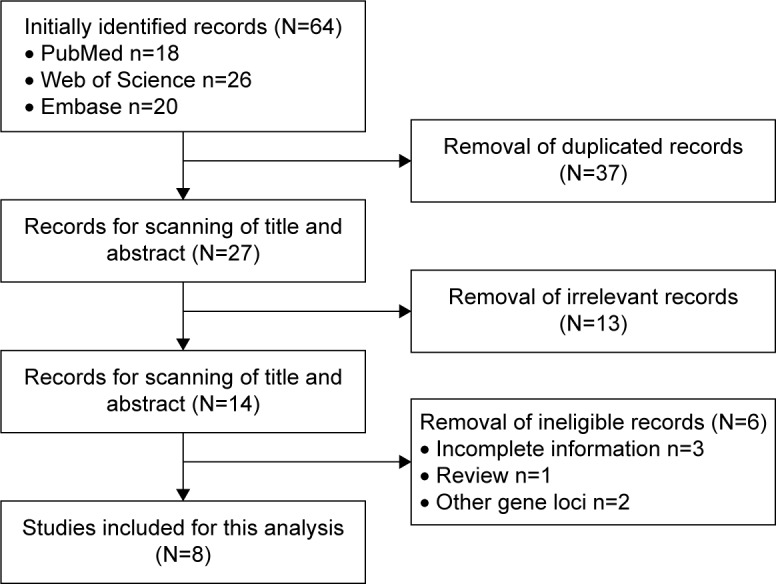
Flow diagram of the inclusion of the eligible studies.
Table 1.
Main characteristics of the included studies
| Study | Case population | Control population | Diagnosis criteria | SNPs | Genotyping method | Accordance with HWE |
|---|---|---|---|---|---|---|
| Poonpet et al6 | Thai patients aged from 51 to 91 years with primary knee OA (N=200) | Comparable individuals with no symptoms or signs of OA, arthritis, or joint disease (N=200) | Criteria of the ACR | rs3740199 | HRM analysis | Yes |
| Shin et al12 | Individuals with K/L score ≥2 from Korean cohort aged 50 years or older (N=725) | Individuals without knee disease from Korean cohort aged 50 years or older (N=1,737) | Criteria of the ACR | rs3740199 | TaqMan assay | Yes |
| Kerna et al10 | Individuals with knee OA from South-Estonian town of Elva (N=118) | Individuals without radiological features of knee OA from South- Estonian town of Elva (N=71) | Grading system of Nagaosa et al27 | rs3740199; rs1871054 | PCR-RFLP | Yes |
| Rodriguez- Lopez et al11 | Individuals with hand, knee, or hip OA from three recruitment sites of European Caucasians (N=4,870) | Individuals without any clinical manifestation of OA from three recruitment sites of European Caucasians (N=2,370) | Criteria of the ACR | rs3740199 | PCR | Yes |
| Valdes et al9 | Individuals from Chingford Study cohort28 (N=280) | Individuals without knee disease from Chingford Study cohort (N=469) | Criteria of the ACR | rs3740199 | PCR | Yes |
| Wang et al5 | Chinese Han individuals with knee OA (N=164) | Age-matched unrelated healthy Chinese Han individuals (N=200) | Criteria of the ACR | rs3740199; rs1871054; rs1278279; rs1044122 | iMLDR | Yes |
| Limer et al16 | Knee and hip OA patients from GOAL study (N=2,044) | Asymptomatic individuals from GOAL study (N=1,123) | Criteria of Zhang et al29 | rs3740199 | TaqMan assay | Yes |
| Lou et al4 | Chinese Han individuals with primary knee OA (N=152) | Age-matched healthy Chinese Han individuals with no signs or symptoms of OA (N=179) | Criteria of the ACR | rs3740199; rs1871054; rs1278279; rs1044122 | TaqMan assay | Yes |
Abbreviations: SNP, single-nucleotide polymorphism; ACR, American College of Rheumatology; HRM, high-resolution melting; HWE, Hardy–Weinberg equilibrium; K/L score, Kellgren/Lawrence score; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; iMLDR, improved multiplex ligase detection reaction; GOAL, genetics of OA and lifestyle; OA, osteoarthritis.
The information obtained from the studies revealed that the most common comparison was between carriers of rs3740199 C allele and carriers of rs3740199 G allele.4–6,10–12,16 Besides, the available genetic models that were introduced in the trials were CC versus GG, GC + CC versus GG, and CC versus GC + GG.4–6,9–12,16 Thus, the ORs with 95% confidence intervals (95% CIs) regarding these comparisons were extracted and pooled into the meta-analysis. Stratification analyses dependent on gender were performed among 6 populations6,10–12; thus, these data were also obtained (Table 2).
Table 2.
Main results of the studies about the association of OA susceptibility with rs3740199
| Study | Allelic frequencies*
|
Genotypic distribution*
|
Odds ratios with 95% confidence intervals
|
Stratification analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C allele | G allele | CC | GC | GG | C allele versus G allele | CC versus GG | GC + CC versus GG | CC versus GG + GC | ||
| Poonpet et al6 | 214/192 | 186/208 | 56/46 | 102/100 | 42/52 | 1.25 (0.94–1.66) | 1.57 (0.86–2.85) | 1.34 (0.84–2.13) | 1.29 (0.82–2.02) | By gender |
| Shin et al12 | 658/1,563 | 792/1,911 | 147/350 | 364/863 | 214/524 | 0.99 (0.87–1.14) | 1.01 (0.76–1.33) | 1.03 (0.85–1.25) | 1.01 (0.81–1.25) | By gender |
| Kerna et al10 (TFOA part) | 44/88 | 22/35 | 28/65 | 32/46 | 6/12 | 1.28 (0.52–4.35) | 0.85 (0.29–2.49) | 0.54 (0.17–1.75) | 0.66 (0.36–1.20) | By gender |
| Kerna et al10 (PFOA part) | 70/62 | 27/30 | 53/41 | 34/43 | 10/8 | 1.16 (0.75–1.82) | 1.01 (0.37–2.80) | 0.83 (0.31–2.20) | 1.50 (0.84–2.66) | By gender |
| Rodriguez-Lopez et al11 (TKR part) | NG | NG | NG | NG | NG | 0.92 (0.81–1.05) | NG | NG | NG | By gender |
| Rodriguez-Lopez et al11 (THR part) | NG | NG | NG | NG | NG | 1.08 (0.96–1.21) | NG | NG | NG | By gender |
| Valdes et al9 | NG | NG | NG | NG | NG | NG | NG | 1.84 (1.22–2.79) | NG | None |
| Wang et al5 | 47.6%/49.0% | 52.4%/51.0% | 36/47 | 84/102 | 44/51 | 0.93 (0.76–1.14) | 0.87 (0.48–1.61) | 0.92 (0.58–1.49) | 0.90 (0.54–1.52) | None |
| Limer et al16 | NG | NG | NG | NG | NG | 1.01 (0.90–1.13) | NG | NG | NG | By hip and knee OA |
| Lou et al4 | 46.7%/49.4% | 53.3%/50.6% | 32/42 | 78/93 | 42/44 | 0.89 (0.65–1.22) | 0.79 (0.41–1.50) | 0.88 (0.53–1.46) | 0.87 (0.52–1.47) | None |
Note:
Data are presented as case number/control number or case percentage/control percentage.
Abbreviations: TKR, total knee replacement; THR, total hip replacement; TFOA, tibiofemoral knee OA; PFOA, patellofemoral knee OA; NG, not given; OA, osteoarthritis.
ADAM12 rs3740199 polymorphism was not associated with OA susceptibility in the general population
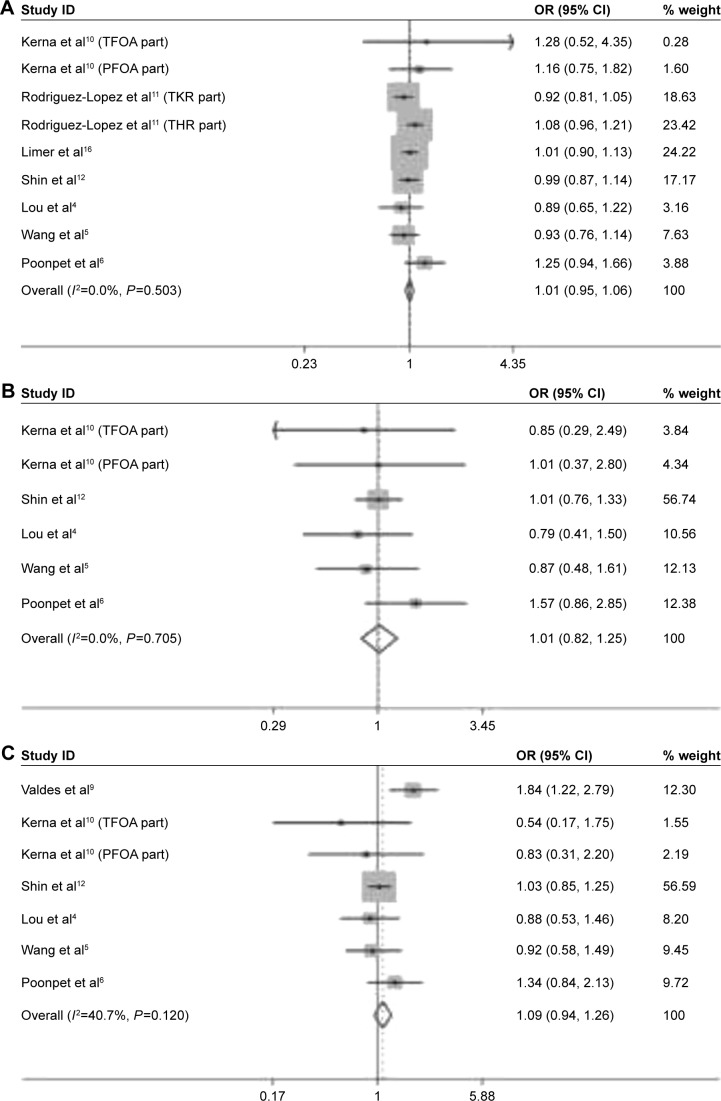
The comparison of risk for OA susceptibility between rs3740199 C allele carriers and G allele carriers was performed among 9 of 10 populations. As shown in Figure 2, no significant association was observed. Sensitivity analyses showed that all these insignificant results would not be influenced when any population was omitted. Neither heterogeneity across studies nor publication bias was detected. Similarly, the investigations on CC versus GG, GC + CC versus GG, and CC versus GG + GC genetic models did not yield any remarkable relationship, either. These results were also proven stable by sensitivity analyses. Heterogeneity and publication bias were not found, either (Table 3).
Figure 2.
Forest plots of the analyses on the association of OA susceptibility with rs3740199 polymorphism. (A) C allele versus G allele; (B) CC versus GG; (C) CC + GC versus GG; (D) CC versus GC + GG.
Abbreviations: OA, osteoarthritis; OR, odds ratio; CI, confidence interval; TFOA, tibiofemoral knee OA; PFOA, patellofemoral knee OA.
Table 3.
Main results of the meta-analysis
| Comparison | Number of studies | Estimated effects
|
Heterogeneity
|
Publication bias
|
Sensitivity analysis | ||||
|---|---|---|---|---|---|---|---|---|---|
| ORs (95% CI) | Z-value | P-value | I2 (%) | P-value | t-value | P-value | |||
| C allele versus G allele | 9 | 1.01 (0.95–1.06) | 0.20 | 0.843 | 0.0 | 0.503 | 0.55 | 0.602 | Stable |
| CC versus GG | 6 | 1.01 (0.82–1.25) | 0.13 | 0.897 | 0.0 | 0.705 | −0.19 | 0.862 | Stable |
| GC + CC versus GG | 7 | 1.09 (0.95–1.26) | 1.19 | 0.233 | 40.7 | 0.120 | −0.29 | 0.785 | Stable |
| CC versus GG + GC | 6 | 1.02 (0.87–1.19) | 0.20 | 0.844 | 6.8 | 0.373 | −0.13 | 0.906 | Stable |
Abbreviations: OR, odds ratio; CI, confidence interval.
ADAM12 rs3740199 polymorphism was likely to be associated with OA susceptibility among males
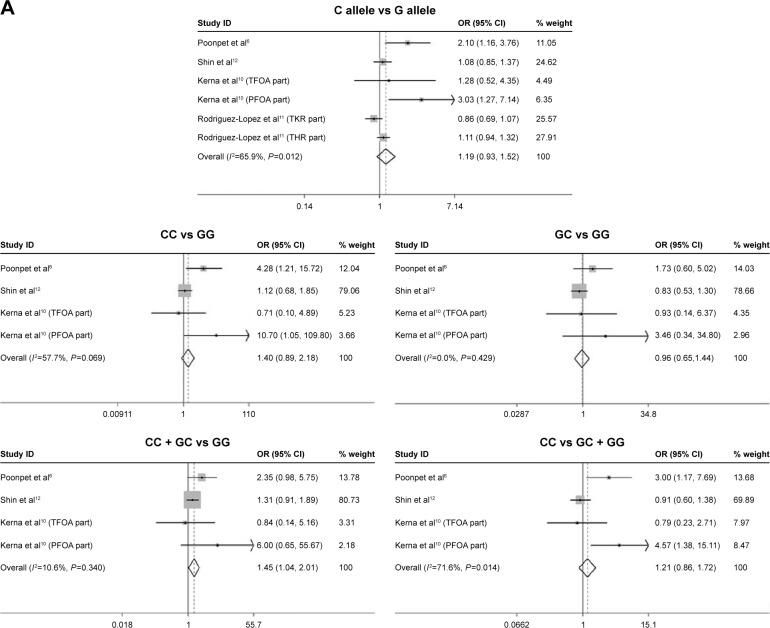
In some studies, the additional risk of OA conferred by rs3740199 polymorphism was exclusively observed in a specific group of people10,11,17; thus, further investigation into this hypothesis was done by analyzing data that were stratified by gender. As illustrated in Figure 3, the only significant association was identified among males, indicating that the risk of OA was 45% higher (95% CI: 1.04–2.01, P=0.028) in carriers of CG and CC, compared with carriers of GG. In this comparison, neither inconsistency nor publication bias was detected. However, sensitivity analysis demonstrated that the exclusion of study from Poonpet et al6 would reverse this relationship into insignificant.
Figure 3.
Forest plots of the analyses on the gender-dependent associations of OA susceptibility with rs3740199 polymorphism. (A) Different comparisons among males; (B) different comparisons among females.
Note: Weights are from random-effects analysis.
Abbreviations: TFOA, tibiofemoral knee OA; PFOA, patellofemoral knee OA; OA, osteoarthritis; CI, confidence interval.
No remarkable and stable correlation was found for the remaining comparisons among men. Specifically, in the study of C allele versus G allele, the exclusion of one population11 would result in a statistically significant association. The same scenario also happened when CC versus GG and CC versus GG + GC were studied, obtaining positive correlations between OA risk and rs3740199 polymorphism.
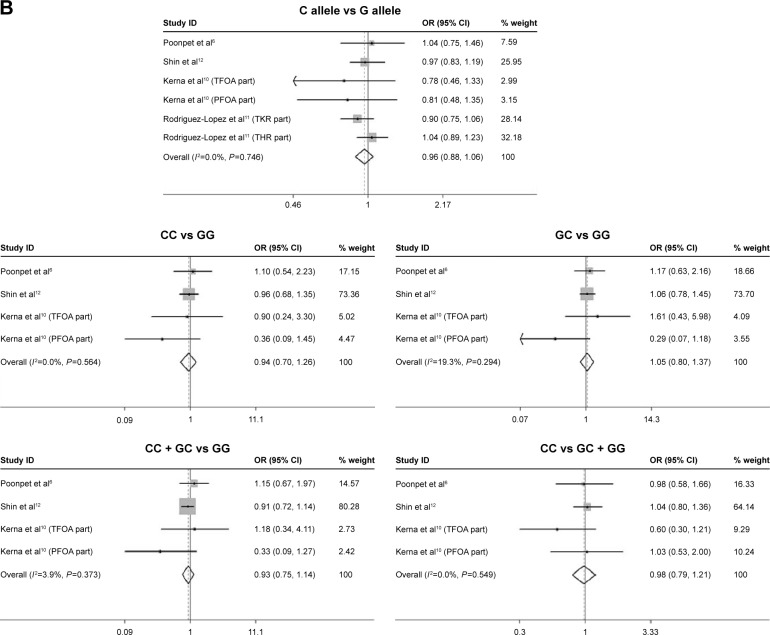
In women patients, this meta-analysis demonstrated that rs3740199 polymorphism was unlikely to confer extra risk for OA to this group of people, regardless of evaluating various genetic models. Sensitivity analyses confirm the stability of these outcomes. Neither heterogeneity nor publication bias was found (Table 4).
Table 4.
Results of the meta-analysis dependent on gender
| Comparison | Number of studies | Estimated effects
|
Heterogeneity
|
Publication bias
|
Sensitivity analysis | ||||
|---|---|---|---|---|---|---|---|---|---|
| ORs (95% CI) | Z-value | P-value | I2 (%) | P-value | t-value | P-value | |||
| Male | |||||||||
| C allele versus G allele | 6 | 1.19 (0.93–1.52) | 1.40 | 0.162 | 65.9 | 0.012 | 1.81 | 0.145 | Unstable |
| CC versus GG | 4 | 1.40 (0.90–2.18) | 1.47 | 0.142 | 57.7 | 0.069 | 1.15 | 0.368 | Unstable |
| GC versus GG | 4 | 0.97 (0.65–1.44) | 0.18 | 0.859 | 0.0 | 0.429 | 1.72 | 0.228 | Stable |
| GC + CC versus GG | 4 | 1.45 (1.04–2.01) | 2.20 | 0.028 | 10.6 | 0.340 | 0.93 | 0.451 | Unstable |
| CC versus GG + GC | 4 | 1.21 (0.86–1.72) | 1.09 | 0.274 | 71.6 | 0.014 | 1.30 | 0.322 | Unstable |
| Female | |||||||||
| C allele versus G allele | 6 | 0.96 (0.88–1.06) | 0.77 | 0.442 | 0.0 | 0.746 | −1.14 | 0.319 | Stable |
| CC versus GG | 4 | 0.94 (0.70–1.29) | 0.43 | 0.667 | 0.0 | 0.564 | −0.95 | 0.441 | Stable |
| GC versus GG | 4 | 1.05 (0.80–1.37) | 0.35 | 0.725 | 19.3 | 0.294 | −0.53 | 0.649 | Stable |
| GC + CC versus GG | 4 | 0.93 (0.75–1.14) | 0.74 | 0.460 | 3.9 | 0.373 | −0.34 | 0.766 | Stable |
| CC versus GG + GC | 4 | 0.98 (0.79–1.21) | 0.21 | 0.835 | 0.0 | 0.549 | −1.18 | 0.358 | Stable |
Notes: Bold values indicate the results were statistically significant.
Abbreviations: OR, odds ratio; CI, confidence interval.
Discussion
OA is a complex disease, induced by a combination of three main risk factors: genetics, environmental factors, and aging.17 Numerous candidate genes have been screened to identify susceptible loci of OA, and ADAM12 is one of them. This is, to the authors’ knowledge, the first meta-analysis to investigate the association between ADAM12 (rs3740199) polymorphism and OA predisposition. In this study, case–control studies on ADAM12 polymorphism and OA susceptibility were comprehensively collected and 8 trials with 10 populations were obtained. The most frequently researched gene locus on ADAM12 (rs3740199) was selected and analyzed. It was noteworthy that the unfavorable effect of rs3740199 polymorphism (GC + CC vs GG) was exclusively observed among male. Unfortunately, this result was unstable; therefore, the evidence was not strong. On the other hand, although comparisons concerning other genetic models did not yield significant correlations, these results were also unstable, heavily affected by the data from a single population. Therefore, until now, the available data indicated that ADAM12 rs3740199 polymorphism was likely to be associated with OA vulnerability among male patients, other than the general population, but more studies are needed to draw a stable and safe conclusion.
In addition to the SNPs of ADAM12, its haplotypes have been related to OA predisposition, with much more remarkable effect. In the genetic study conducted among Chinese Han individuals, haplotype analysis revealed that 5 haplotypes (TATC, TATG, TACG, CATG, and CGTC) were associated with the increased risk of OA.4 In line with that, haplotype CAAT has been found to elevate the risks for OA by as high as 6.10-fold among Caucasian women and 1.54-fold among Caucasian men.18 The sexual difference is also found in the haplotype study. It has been reported that CCAA haplotype increased the risk for tibiofemoral joint OA among males, and GCGG haplotype increased the risk for joint space narrowing among females.19 In contrast, some haplotypes may be protective against OA. rs3740199/rs1871054 GC decreased the risk for patellofemoral joint OA among male patients (OR =0.10) and the risk for osteophytes among overall people (OR =0.24).10 Besides, haplotype CGAT and CGGT were also related with smaller risk for knee OA predisposition.18
Apart from the genetic association studies about ADAM12 SNPs and OA, some clinical trials investigating the role of ADAM12 protein and mRNA also provide valuable supporting evidence. In overall, it has been observed that overexpression of ADAM12 is related with OA. The level of ADAM12-L mRNA expression was remarkably higher in OA cartilage than in normal samples. In situ hybridization demonstrated that clustered chondrocytes in OA cartilage were the main source of ADAM12-L mRNA. The expression of this protein was visualized on clustered chondrocyte membranes by immunohistochemistry.20 The expressions of ADAM12 mRNA and protein in synovium were significantly linked to the severity of histological synovitis.21 ADAM12-S protein in serum could be increased in some OA patients, correlating with the grade of this disease and osteophyte occurrence.22 However, the amount of ADAM12 protein may not be linked with gene polymorphism, and since it has been reported that the functional protein level is not determined by genetic variants (rs3740199, rs1871054, rs1278279, and rs1044122),23 the involvement of ADAM12 gene and protein in OA pathogenesis needs further research.
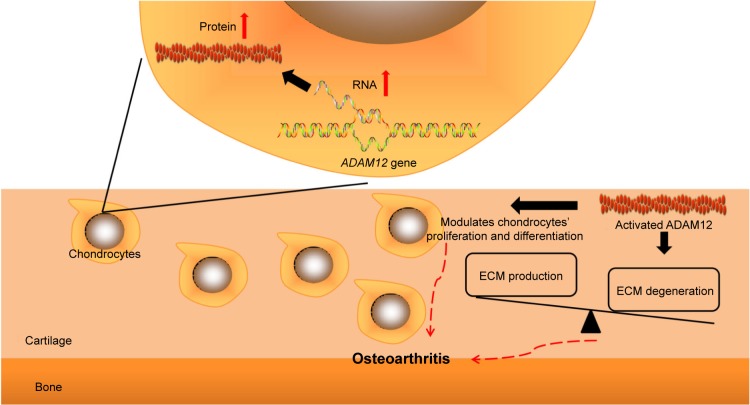
Despite the fact that so many observational studies have been conducted, little is known about the molecular biological role of ADAM12 on OA pathogenesis. The possible mechanism by which ADAM12 participates in OA is illustrated in Figure 4. Human ADAM12 consists of two forms produced by alternative splicing gene, which are secreted, short form (ADAM12-S) and larger, membrane-bounding form (ADAM12-L).8 Similar to the archetypical structure of ADAM proteins, the amino acid sequence of ADAM12 includes proprotease, metalloprotease, disintegrin, cysteine-rich domains, and in the case of L form, a transmembrane and cytoplasmic domains.8 The metalloprotease domain converts from latent state into functional state after cleavage of the prodomain, resulting in an ADAM12 protein with catalytic activity.24 Activated ADAM12 has the capacity to degrade extracellular matrix (ECM) components, including gelatin, fibronectin, and type IV collagen.25 The molecular interaction between ADAM12 and β1 integrins may change cell–matrix contacts in the growth plate and modulate proliferation and differentiation of chondrocyte.26 As it is known, articular cartilage is a narrow layer of specialized ECM that is elaborated and maintained by articular chondrocytes. In normal cartilage, chondrocytes maintain a balance between the production and the degeneration of ECM, resulting in stability of the tissue. This equilibrium is disturbed in degenerative joint diseases such as OA.20 Therefore, it is plausible that ADAM12 may contribute to the pathogenesis of OA by the influence on ECM and chondrocytes. Existing evidence has demonstrated that OA chondrocytes produce more ADAM12-L mRNA and protein after administration with TGF-β in a dose-dependent fashion.20 The overexpression of ADAM12-L could result in chondrocyte proliferation via insulin-like growth factor signaling.20
Figure 4.
Possible mechanism by which ADAM12 participates in OA pathogenesis.
Abbreviations: ADAM, disintegrin and metalloprotease; OA, osteoarthritis; ECM, extracellular matrix.
There are some limitations in this study, so the results should be treated with caution. The adverse impact of rs3740199 polymorphism was demonstrated by insufficient studies. Only 3 trials with 4 groups of individuals, involving 1,043 cases and 2,008 controls, were pooled together to address this issue. Furthermore, this positive correlation was proven to be unstable since it was remarkably influenced by the exclusion of study from Poonpet et al.6 In Poonpet’s study,6 limited male cases and controls were recruited. It was reported that CC male carriers had 328% higher risk than GG male carriers. Male patients with C allele had 110% higher risk than male patients with G allele. The extremely high OR values among males might be partly attributed to the insufficient sample size or selection bias. In addition, some of the extracted ORs with their 95% CIs originated from multivariate analyses and were adjusted for potential confounders.4,5,12,16 On the other hand, some ORs were given from the univariate model.6,10,18 Therefore, the aggregation of these heterogeneous ORs might distort the truth. Finally, the controversial association of OA with ADAM12 haplotype frequency was not investigated. It was mainly attributed to lack of evidence, different haplotypes, and different outcomes of interest.
In conclusion, ADAM12 rs3740199 polymorphism was likely to be associated with OA vulnerability among male patients, other than the general population, based on limited evidence. More studies are needed to further investigate this proposal. In addition, the biological mechanism by which ADAM12 gene and protein link with OA occurrence and progression needs to be elucidated.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Glyn-Jones S, Palmer AJ, Agricola R, et al. Osteoarthritis. Lancet. 2015;386(9991):376–387. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 2.Loeser RF, Collins JA, Diekman BO. Ageing and the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2016;12(7):412–420. doi: 10.1038/nrrheum.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramos YF, Meulenbelt I. Implementation of functional genomics for bench-to-bedside transition in osteoarthritis. Curr Rheumatolo Rep. 2015;17(8):53. doi: 10.1007/s11926-015-0528-x. [DOI] [PubMed] [Google Scholar]
- 4.Lou S, Zhao Z, Qian J, Zhao K, Wang R. Association of single nucleotide polymorphisms in ADAM12 gene with susceptibility to knee osteoarthritis: a case–control study in a Chinese Han population. Int J Clin Exp Pathol. 2014;7(8):5154–5159. [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Guo L, Tian F, Hao R, Yang T. Analysis of single nucleotide polymorphisms within ADAM12 and risk of knee osteoarthritis in a Chinese Han population. Biomed Res Int. 2015;2015:518643. doi: 10.1155/2015/518643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poonpet T, Tammachote R, Tammachote N, Kanitnate S, Honsawek S. Association between ADAM12 polymorphism and knee osteoarthritis in Thai population. Knee. 2016;23(3):357–361. doi: 10.1016/j.knee.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Giebeler N, Zigrino P. A Disintegrin and metalloprotease (ADAM): historical overview of their functions. Toxins (Basel) 2016;8(4):122. doi: 10.3390/toxins8040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilpin BJ, Loechel F, Mattei MG, Engvall E, Albrechtsen R, Wewer UM. A novel, secreted form of human ADAM 12 (meltrin alpha) provokes myogenesis in vivo. J Biol Chem. 1998;273(1):157–166. doi: 10.1074/jbc.273.1.157. [DOI] [PubMed] [Google Scholar]
- 9.Valdes AM, Hart DJ, Jones KA, et al. Association study of candidate genes for the prevalence and progression of knee osteoarthritis. Arthritis Rheum. 2004;50(8):2497–2507. doi: 10.1002/art.20443. [DOI] [PubMed] [Google Scholar]
- 10.Kerna I, Kisand K, Tamm AE, Lintrop M, Veske K, Tamm AO. Missense single nucleotide polymorphism of the ADAM12 gene is associated with radiographic knee osteoarthritis in middle-aged Estonian cohort. Osteoarthritis Cartilage. 2009;17(8):1093–1098. doi: 10.1016/j.joca.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Lopez J, Pombo-Suarez M, Loughlin J, et al. Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthritis Cartilage. 2009;17(3):321–327. doi: 10.1016/j.joca.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Shin MH, Lee SJ, Kee SJ, et al. Genetic association analysis of GDF5 and ADAM12 for knee osteoarthritis. Joint Bone Spine. 2012;79(5):488–491. doi: 10.1016/j.jbspin.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 15.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 16.Limer KL, Tosh K, Bujac SR, et al. Attempt to replicate published genetic associations in a large, well-defined osteoarthritis case–control population (the GOAL study) Osteoarthritis Cartilage. 2009;17(6):782–789. doi: 10.1016/j.joca.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 17.Sandell LJ. Etiology of osteoarthritis: genetics and synovial joint development. Nat Rev Rheumatol. 2012;8(2):77–89. doi: 10.1038/nrrheum.2011.199. [DOI] [PubMed] [Google Scholar]
- 18.Valdes AM, Van Oene M, Hart DJ, et al. Reproducible genetic associations between candidate genes and clinical knee osteoarthritis in men and women. Arthritis Rheum. 2006;54(2):533–539. doi: 10.1002/art.21621. [DOI] [PubMed] [Google Scholar]
- 19.Kerna I, Kisand K, Tamm AE, Kumm J, Tamm AO. Two single-nucleotide polymorphisms in ADAM12 gene are associated with early and late radiographic knee osteoarthritis in Estonian population. Arthritis. 2013;2013:878126. doi: 10.1155/2013/878126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada A, Mochizuki S, Yatabe T, et al. ADAM-12 (meltrin alpha) is involved in chondrocyte proliferation via cleavage of insulin-like growth factor binding protein 5 in osteoarthritic cartilage. Arthritis Rheum. 2008;58(3):778–789. doi: 10.1002/art.23262. [DOI] [PubMed] [Google Scholar]
- 21.Kerna I, Kisand K, Suutre S, et al. The ADAM12 is upregulated in synovitis and postinflammatory fibrosis of the synovial membrane in patients with early radiographic osteoarthritis. Joint Bone Spine. 2014;81(1):51–56. doi: 10.1016/j.jbspin.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Kerna I, Kisand K, Laitinen P, et al. Association of ADAM12-S protein with radiographic features of knee osteoarthritis and bone and cartilage markers. Rheumatol Int. 2012;32(2):519–523. doi: 10.1007/s00296-010-1717-6. [DOI] [PubMed] [Google Scholar]
- 23.Kerna I, Kisand K, Laitinen P, Tamm A, Tamm A. Association of metallopeptidase Domain 12 (Adam12) gene polymorphisms and Adam12 protein with the development of knee osteoarthritis. Osteoarthritis Cartilage. 2010;18:S170. [Google Scholar]
- 24.Loechel F, Gilpin BJ, Engvall E, Albrechtsen R, Wewer UM. Human ADAM 12 (meltrin alpha) is an active metalloprotease. J Biol Chem. 1998;273(27):16993–16997. doi: 10.1074/jbc.273.27.16993. [DOI] [PubMed] [Google Scholar]
- 25.Roy R, Wewer UM, Zurakowski D, Pories SE, Moses MA. ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J Biol Chem. 2004;279(49):51323–51330. doi: 10.1074/jbc.M409565200. [DOI] [PubMed] [Google Scholar]
- 26.Kveiborg M, Albrechtsen R, Rudkjaer L, Wen G, Damgaard-Pedersen K, Wewer UM. ADAM12-S stimulates bone growth in transgenic mice by modulating chondrocyte proliferation and maturation. J Bone Miner Res. 2006;21(8):1288–1296. doi: 10.1359/jbmr.060502. [DOI] [PubMed] [Google Scholar]
- 27.Nagaosa Y, Mateus M, Hassan B, et al. Development of logically devised line drawing atlas for grading of knee osteoarthritis. Ann Rheum Dis. 2000;59:587–595. doi: 10.1136/ard.59.8.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hart DJ, Spector TD. The relationship of obesity, fat distribution and osteoarthritis in the general population: the Chingford Study. J Rheumatol. 1993;20:331–335. [PubMed] [Google Scholar]
- 29.Zhang W, Robertson J, Doherty S, Liu JJ, Maciewicz RA, Muir KR, et al. Index to ring finger length ratio and the risk of osteoarthritis. Arthritis Rheum. 2008;58(1):137e44. doi: 10.1002/art.23237. [DOI] [PubMed] [Google Scholar]