Abstract
Purpose
Protease nexin-1 (PN-1), a serpin encoded by the SERPINE2 gene, has serine protease inhibitory activity and neurotrophic properties in the brain. PN-1 inhibits retinal angiogenesis; however, PN-1’s neurotrophic capacities in the retina have not yet been evaluated. Pigment epithelium-derived factor (PEDF) is a serpin that exhibits neurotrophic and antiangiogenic activities but lacks protease inhibitory properties. The aim of this study is to compare PN-1 and PEDF.
Methods
Sequence comparisons were performed using computer bioinformatics programs. Mouse and bovine eyes, human retina tissue, and ARPE-19 cells were used to prepare RNA and protein samples. Interphotoreceptor matrix lavage was obtained from bovine eyes. Gene expression and protein levels were evaluated with reverse-transcription PCR (RT–PCR) and western blotting, respectively. Recombinant human PN-1, a version of PN-1 referred to as PN-1[R346A] lacking serine protease inhibitory activity, and PEDF proteins were used, as well as synthetic peptides designed from PEDF and PN-1 sequences. Survival activity in serum-starved, rat-derived retinal precursor (R28) cells was assessed with terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling (TUNEL) cell death assays. Bcl2 levels were measured with RT–PCR.
Results
PN-1 is analogous in primary and tertiary structure to PEDF. A region in PN-1 shares homology with the neurotrophic active region of PEDF, a 17-residue region within alpha helix C. The native human retina, ARPE-19 cells, and murine RPE and retina expressed the gene for PN-1 (SERPINE2 and Serpine2 mRNA). The retina, ARPE-19 cell lysates, and bovine interphotoreceptor matrix contained PN-1 protein. The addition of PN-1, PN-1[R346A], or the 17mer peptide of PN-1 to serum-starved retina cells decreased the number of TUNEL-positive nuclei relative to the untreated cells, such as PEDF. PN-1, PN-1[R346A], and PN-1-17mer treatments increased the Bcl2 transcript levels in serum-starved cells, as seen with PEDF.
Conclusions
PN-1 and PEDF share structural and functional features, and expression patterns in the retina. These serpins’ mechanisms of action as cell survival factors are independent of serine protease inhibition. We have identified PN-1 as a novel factor for the retina that may play a neuroprotective role in vivo, and small peptides as relevant candidates for preventing retinal degeneration.
Introduction
Serpins (SERine Protease INhibitorS) comprise a superfamily of more than 3,000 proteins classified by a distinctive three-dimensional (3D) structure and mechanism of protease inhibition. Though primarily associated with the mediation of hemostasis and thrombolysis, serpins meet a wide range of additional cellular needs. The folded protein structures of serpins are composed of three beta sheets, eight to nine alpha helices, and a reactive center loop (RCL) that contains the P1 site, which confers protease specificity [1]. Upon binding to the protease target, the P1-P1’ bond of serpins is cleaved, after which the proteins undergo a characteristic stressed-to-relaxed (S to R) conformational change in a suicidal manner rendering the serpin and its targeted protease inactive and forming a serpin:protease complex [2]. Serpins with homologous protein conformation but with no demonstrable protease inhibitory activity are classified as non-inhibitory serpins.
Protease nexin-1 (PN-1), also known as glia-derived nexin, is a 43–50 kDa member of the serpin superfamily encoded by the human SERPINE2 gene (Gene ID: 5270 and OMIM 177010) [3,4]. PN-1 is highly expressed in the adult brain, bone, seminal vesicle, cauda epididymis, and ovary [5,6]. This secreted, pericellular serpin inhibits thrombin, plasmin, and plasminogen activators by forming complexes with these target proteases. Furthermore, the colocalization of PN-1 with fibronectin on the surfaces of fibroblasts prompted the observation that a minimum of 60–80% of cellular PN-1 resides in the extracellular matrix (ECM; [7]). PN-1 interacts with polysaccharides that accelerate thrombin inhibition by more than 100-fold [4]. These reactions are so prevalent that PN-1 has only been crystallized in complex with heparin and thrombin [2,8]. Aside from a primary role of thrombin inhibition in the blood, PN-1 serves as a neurotrophic factor in the brain, central nervous system (CNS), and peripheral nervous system (PNS) and prevents cell death in cases of brain injury, disturbances of the blood–brain barrier, and ischemia, all of which induce the upregulation of PN-1 [9]. Selbonne et al. were the first to report antiangiogenic properties associated with PN-1 after finding amplified angiogenic responses in vascular endothelial growth factor (VEGF)-mediated capillary formation in Serpine2-deficient mice [10]. They demonstrated that PN-1 is present in the mouse retina. However, the activity of PN-1 in the neural retina is unknown.
Pigment epithelium-derived factor (PEDF) is another serpin with neurotrophic and antiangiogenic activities present in ECMs. PEDF is a secreted 50 kDa glycoprotein encoded by the human SERPINF1 gene (Gene ID: 5176 and OMIM: 172860). PEDF is mainly secreted by the RPE into the interphotoreceptor matrix (IPM) toward the neural retina and is also found in the vitreous and aqueous humor [11,12]. Similar to PN-1, PEDF is found in cerebrospinal fluid and blood [13,14], and interacts with ECM components, such as glycosaminoglycans (heparin and heparan sulfate) and collagens [15-17]. In contrast to PN-1, PEDF does not have demonstrable inhibitory activity against serine proteases and belongs to a subgroup of serpins that are non-inhibitory to proteases [18]. The region that confers PEDF’s neurotrophic activity is located away from the homologous serpin active site [19,20], which interacts with the cell surface receptor PEDF-R shown to mediate the cytoprotective properties of PEDF in photoreceptors [21,22]. PEDF’s well-established neurotrophic, neuroprotective, gliastatic, antitumorigenic, antioxidant, and antiangiogenic effects in the retina have made PEDF a prime candidate for ocular therapeutic applications [12].
Given the emerging parallels between PEDF and PN-1, we performed structural, biochemical, and biologic comparisons of the two serpins. Here, we propose a retinoprotective role for PN-1 similar to that of PEDF and identify a small PN-1 fragment with retina cell survival activity.
Methods
Amino acid sequence alignments
Amino acid sequence alignments between human PEDF (human PEDF: pigment epithelium-derived factor precursor [Homo sapiens] NCBI Reference Sequence: NP_002606.3), human PN-1 (glia-derived nexin isoform a precursor [Homo sapiens] NCBI Reference Sequence: NP_006207.1), mouse PEDF (pigment epithelium-derived factor precursor [Mus musculus] NCBI Reference Sequence: NP_035470.3), and mouse PN-1 (glia-derived nexin precursor [Mus musculus] NCBI Reference Sequence: NP_033281.1) were performed using the ClustalW2 Multiple Sequence Alignment EMBL-EBI program.
Tertiary structure comparison
Tertiary structures of human PEDF (PDB: 1IMV_A) and human PN-1 (PDB: 4DY7_C) were evaluated by comparing their crystal structures and corresponding sequence alignments on the Cn3D macromolecular structure viewer (Version 4.3).
cDNA synthesis
RNA was obtained from RPE and retinas dissected from eyes of C57BL/6N [Crb1rd8] mice and human ARPE-19 cells and retinas (Clontech, Mountain View, CA). An oligo(dT) probe was used to reverse-transcribe mRNA from cell samples in a final volume of 20 µl using SuperScript first-strand synthesis system (Life Technologies, Frederick, MD) following the manufacturer’s instructions.
RT–PCR
Specific primers for screening the expression of human SERPINE2 were huPN-1-forward 5′-CCG CTG AAA GTT CTT GGC A-3′ and huPN-1-reverse 5′-CAG CAC CTG TAG GAT TAT GTC G-3′. Specific mouse Serpine2 primers were mPN-1-forward 5′-CAG TGT GAA GTG CAG AAT GTG A-3′ and mPN-1-reverse 5′-TTG GGG AAA GCA GAT TAT CAA-3′. Templates were cDNAs prepared from the cells and tissues in HotStarTaq DNA Polymerase reactions following the manufacturer’s instructions (Qiagen, Valencia, CA). Reverse-transcription PCR was performed in a final volume of 50 µl containing 2 µl of cDNA and 10 µM primers as listed above using a thermocycler (MJ Research PTC-200 DNA Engine; Bio-Rad, Hercules, CA) according to the following program: initial denaturation for 15 m at 95 °C, 60 cycles of denaturation for 1 m at 95 °C, annealing for 30 s at 58 °C, extension for 30 s at 72 °C, and a 5 m hold at 72 °C. PCR products were resolved by electrophoresis in E-Gel® Agarose Gels, 2%, containing ethidium bromide for visualization (Life Technologies). For quantitative RT–PCR, total RNA was purified using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Total RNA (0.1–1 μg) was used for reverse transcription using the SuperScript III First-Strand Synthesis System (Invitrogen). Bcl2 mRNA levels were normalized to 18S levels with quantitative RT–PCR using SYBR Green Mix (Applied Biosystems, Foster City, CA) in the Bio-Rad Chromo4 Real-Time PCR System. The primers used were as follows: rat Bcl2, 5′-TGG ACA ACA TCG CTC TGT GGA TGA-3′ (forward) and 5′-GGG CCA TAT AGT TCC ACA AAG GCA-3′ (reverse); and 18S, 5′-GGT TGA TCC TGC CAG TAG-3′ (forward) and 5′-GCG ACC AAA GGA ACC ATA AC-3′ (reverse).
Proteins and peptides
Human recombinant wild-type (WT) PN-1 and human recombinant PN-1[R346A] rendered inactive by a mutation of the reactive site were prepared from Escherichia coli containing expression vectors as previously described [10,23]. Briefly, the cDNA coding sequence for human PN-1 was inserted into pGEX-6P-1 vector resulting in a fusion gene construct that encoded glutathione S-transferase (GST)-PN-1. PN-1 mutagenesis was performed to change the arginine (R346) residue of the reactive site to alanine. The constructs were expressed in BL21(DE3) E. coli strain. After induction of expression of the recombinant gene, GST-PN-1 was purified from cell lysate by glutathione-Sepharose affinity chromatography. On-column cleavage of the protein from GST was performed to yield PN-1 and PN-1[R346A] proteins. Human recombinant PEDF was prepared from stably transfected baby hamster kidney cells containing a PEDF expression vector as previously described [24]. The recombinant protein was purified from culturing media of the cells by ammonium sulfate precipitation, cation exchange chromatography followed by anion exchange chromatography. Peptides 44mer and 17mer derived from PEDF were synthesized as previously described [19,20]. These peptides were chemically synthesized by a commercial source (Biosynthesis, Lewisville, TX). Peptide PN-1-17mer, a peptide designed from the homologous neurotrophic region of PEDF, corresponds to the human PN-1 polypeptide sequence Gly71-Gly87 (GRTKKQLAMVMRYGVNG) and was chemically synthesized (Biosynthesis, Lewisville, TX).
Cell culture
ARPE-19 cells, a spontaneously arising human RPE cell line (American Type Culture Collection (ATCC), Manassas, VA), were cultured in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12, HEPES, catalog # 11330057, ThermoFisher, Waltman, MA) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Cell line STR Validation analysis was performed by BioSYNTHESIS (catalog # CL1003, Lewisville, TX; Appendix 1). ARPE-19 cells in passage numbers 30 to 35 were used. R28 cells, an immortalized retinal progenitor cell line derived from the neonatal rat retina [25] (Kerafast, Boston, MA), were validated by IDEXX BioResearch (Columbia, MO; Appendix 2). Cells were cultured in DMEM with 10% FBS, 3% sodium bicarbonate, 1% MEM nonessential amino acids (Catalog # 11140050, ThermoFisher Scientific), 1% minimum essential medium (MEM) vitamins (Catalog # 11120029, ThermoFisher Scientific), 1% L-glutamine (Catalog # 25030-024, ThermoFisher Scientific), and 0.125% gentamicin (Sigma-Aldrich, Catalogue # G1397) at 37 °C with 5% CO2 and 95% humidity. Cells were revalidated at passage number 42, and all experiments were performed using cells in passage numbers 37 to 45. The cells were maintained in a T75 flask at 70–75% confluency.
Processing of ARPE-19 conditioned media and lysates
ARPE-19 cells were cultured in a six-well plate until confluent and serum-starved for 5 days. The media were collected and concentrated tenfold (from 1 ml to 100 µl) in Amicon Ultra Centrifugal Filters Ultracel-30K Membrane (Millipore, Temecula, CA) by centrifugation on the Beckman Allegra 6KR Kneewell Refrigerated Centrifuge (Analytical Instruments LLC, Minneapolis, MN) for 10 m at 4 °C. Wells containing the ARPE-19 cells were washed twice with cold PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4). Then 300 µl cold RIPA Lysis and Extraction buffer (Thermo Fisher Scientific, Catalogue # 89,900, Waltham, MA) was added to each well, and the plate was incubated on ice for 5 m. Cell lysates were collected in microcentrifuge tubes and sonicated for 20 s with a 50% pulse. Sonicated samples were centrifuged at maximum speed at 4 °C for 15 m to pellet any cell debris. Supernatants were concentrated as described above. All media and lysate samples were stored at −20 °C. Proteins were analyzed with western blotting against human serpin E2/PN1 antibody goat Ab-PN-1 (polyclonal goat IgG by R&D Systems, Catalogue # AF2980, Minneapolis, MN)) at 50 ng/ml (diluted in 1% bovine serum albumin/Tris-buffered saline/Tween 20 [BSA/TBST]) and anti-goat immunoglobulin (IgG) (H+L) antibody, human serum absorbed and peroxidase labeled secondary (KPL, Gaithersburg, MD, 14–13–06) diluted 1:200,000. Western blotting against anti-PEDF were incubated in rabbit AB-PEDF (BioProducts MD, Middletown, MD) at 1:100,000 in 1% BSA/TBST, followed by secondary antibodies peroxidase anti-rabbit IgG (KPL) diluted 1:200,000. Signaling was detected using West Dura Super Signal (ThermoFisher) and exposed to X-ray films.
Preparation of bovine IPM wash
All preparation procedures were performed at 4 °C. Adult bovine eyes were obtained from J. W. Trueth & Sons (Baltimore, MD). The soluble components of the IPM were obtained with the “no-cut” method described by Adler that ensures that the extracellular fluid is free of significant cellular contamination [26]. Each eye was dissected as follows: The anterior segment and the vitreous were removed from each eye, and a solution of PBS was gently introduced between the neural retina and the RPE with a needle at 0.5 ml per eye. The IPM wash was extracted and centrifuged at 1,000 xg for 10 min to remove cellular debris, and the supernatant was passed through a 0.45 µm syringe filter and stored at −80 °C. This filtrate constituted the IPM wash.
Bovine retinal extracts
All preparation procedures were performed at 4 °C as described by Aymerich et al. [27]. Fresh adult bovine eyes (J. W. Trueth & Sons) were dissected below the iris, the vitreous was removed from the inner retinal surface, and the neural retinas were gently separated from the pigment epithelium with forceps. Retinas were homogenized in a solution of cold 0.32 M sucrose in Tris-buffered saline (TBS; 20 mM Tris/HCL [pH 7.5] and 150 mM NaCl) containing protease inhibitors (1 mM aminoethyl-benzenesulfonyl fluoride hydrochloride [AEBSF], 5 µg/ml aprotinin, 1 µ/ml pepstatin, and 0.5 µg/ml leupeptin) at 7.5 ml per retina with a homogenizer (Polytron model 3000; Brinkman Instruments, Westbury, NY) set at 10,000 rpm for 20 s. The homogenized material was separated from tissue and cellular debris by centrifugation at 1,000 ×g for 10 m.
TUNEL assay
Rat retina R28 cells (1.5 × 104/well) were cultured in an eight-well Nunc chamber slide for 16 h in media containing 5% FBS. The media were then replaced with serum-free media containing effectors and incubated at 37 °C for 48 h. Cell death was evaluated at the end point by performing the terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling (TUNEL) assay using the ApopTag® Fluorescein in situ Apoptosis Detection Kit (Millipore, Billerica, MA) following the manufacturer’s protocol. The nuclei were counterstained with Hoechst, and images were taken using a Zeiss (Peabody, MA) Imager Z1 fluorescence microscope. Five different fields per treatment were imaged from two independent wells in each experiment. Cells were counted with ImageJ version 1.42 using fixed threshold values within each experiment to quantify based on intensity.
Statistical analyses
Data are shown as the mean ± standard deviation (SD). The data were analyzed with the two-tailed unpaired Student t test. A p value of less than 0.05 was considered statistically significant.
Results
Primary and tertiary structural comparison between PEDF and PN-1
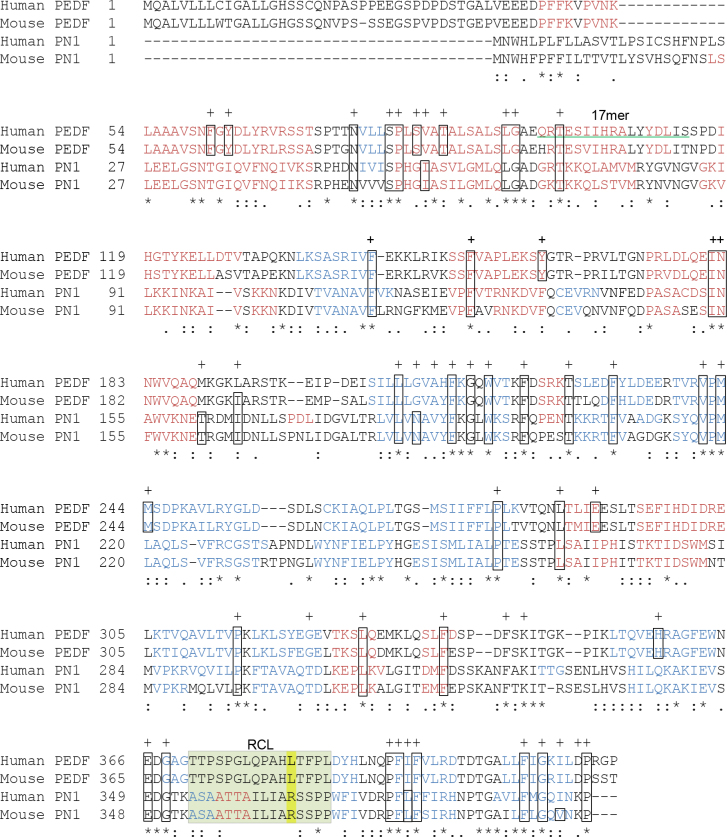
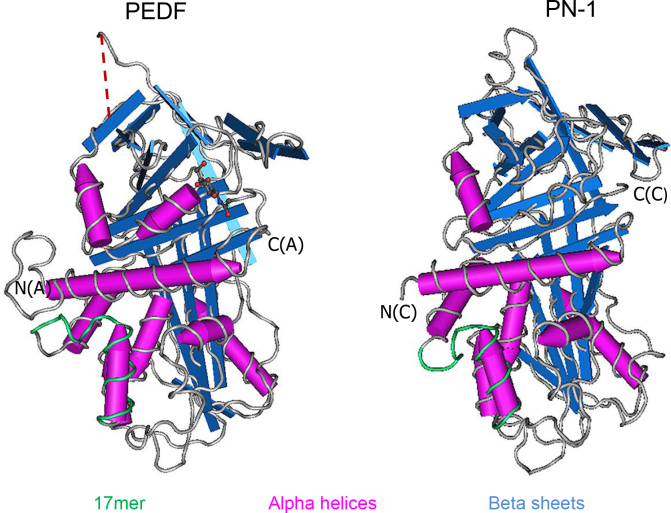
We used the ClustalW2 Multiple Sequence Alignment EMBL-EBI program to compare the amino acid sequences of human and mouse PEDF and PN-1 (Figure 1). With 25% identity at the primary level between PN-1 and PEDF, the sequences feature a conserved set of residues that predominantly governs the integrity of the spatial structure of serpins and biologic activity [28]. The presence of this common set, comprised of approximately 51 invariable residues, ensures proper folding of the native, metastable state protein [29]. Human and mouse PN-1 sequences contain 36 and 37 residues identical to those in this set, respectively, of which six residues have strongly similar properties to those in the set (Figure 1). Both proteins were crystallized, and the 3D structures have extensive overlap between the alpha helical and beta sheet regions (Figure 2). They contain exposed RCLs of 15 residues, in which P1, the protease specificity-determining site, is an arginine in PN-1 and a leucine in PEDF [30,31]. There is a degree of sequence similarity in the 17mer ligand-binding region of PEDF to its receptor PEDF-R [22]. Recent alanine scanning studies revealed that R99 of human PEDF within the 17mer region is crucial for receptor binding and survival activity, and an alteration of H105A increases the affinity for PEDF-R tenfold [20]. Alignment and comparison show that both requirements are present in the corresponding positions of human and mouse PN-1 (R/K at 72 and A at 78). Other residues crucial for receptor binding in PEDF (S102, I104, and Y110; [22]) are also present in human PN-1 as their semiconservative and conserved residues K, L, and Y, respectively. Overall, the distributions of alpha helices and beta sheets are consistent between the structures and are typical of the serpin protein superfamily. The PEDF 17mer and the corresponding region in PN-1 are visible in alpha helix C of the crystal structures presented in Figure 2 [32,33].
Figure 1.
Primary sequence comparison of PN-1 and PEDF. The alignment performed using the ClustalW2 Multiple Sequence Alignment EMBL-EBI program illustrates the degree to which the amino acid sequences of human and mouse PEDF (NP_002606.3 and NP_035470.3, respectively) and PN-1 (NP_006207.1 and NP_033281, respectively) are conserved. The secondary structures predicted based on the amino acid sequences are depicted in red (alpha helices) and blue (beta strands). The sequence of the 17mer fragment of human PEDF that binds to PEDF-R and displays neurotrophic activity in the retina is underlined with a green double line. Within the reactive center loop (RCL, highlighted in pale green), P1, the residue that confers protease specificity, is highlighted in yellow. Residue identity (*), residues with strongly similar properties scoring ˃0.5 in the Gonnet PAM 250 matrix (:) and residues with weakly similar properties scoring ˂0.5 on the Gonnet PAM 250 matrix (.) are indicated. The 51 conserved residues among serpins defined by Huber and Carrell [28] as key to maintaining the integrity of the serpin spatial structure are indicated with a “+,” and the boxes correspond to the amino acid found in the archetype serpin anti-trypsin.
Figure 2.
Tertiary structure comparison. Tertiary structures of PN-1 (PDB: 4DY7_C) and PEDF (PDB: 1IMV_A) were modeled on the Cn3D macromolecular structure viewer (Version 4.3). Alpha helices (pink) and beta sheets (blue) are highlighted. The 17mer region of PEDF (green) within alpha helix C exhibits neurotrophic activity. The corresponding peptide region in PN-1 is also shown in green. The dotted red line corresponds to the homologous serpin reactive center loop (RCL).
PN-1 in ocular tissue
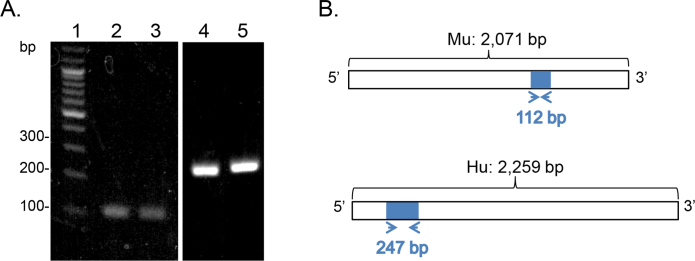
We assessed the presence of SERPINE2 transcripts in ocular tissues via RT–PCR. We used RNA from the mouse RPE and retina, human retina, and the spontaneously arising RPE cell line ARPE-19. Using specific primers for mouse Serpine2, the expected amplified fragment of 112 base pairs was detected with cDNA derived from murine RPE and retina in agarose gels (Figure 3). Similarly, the expected DNA fragments of 247 base pairs were amplified and detected from cDNAs of human ARPE-19 and from cDNAs of native human retina cDNA using specific primers for human SERPINE2 (Figure 3).
Figure 3.
SERPINE2 transcript in ocular tissue. A: Reverse-transcription PCR (RT–PCR) of SERPINE2 mRNA levels detected in the murine RPE, the murine retina, human ARPE-19 cells, and the human retina. Products were resolved in a 2% agarose gel and stained with ethidium bromide. A photo of the gel exposed to ultraviolet (UV) light is shown with DNA size markers (lane 1), and the cDNA templates for the reactions in the murine RPE (lane 2), the murine retina (lane 3), human ARPE-19 cells (lane 4), and the human retina (lane 5). The migration positions for some markers are indicated to the left. B: Schematics of the primer constructs used for PCR. The expected number of nucleotides for the product from the transcript NM_009255.4 encoding Mus musculus serine (or cysteine) peptidase inhibitor, clade E, member 2 (Serpine2), mRNA using the murine PN-1 primers is indicated. The expected number of nucleotides for the product from the transcript NM_006216.3 encoding Homo sapiens serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (SERPINE2), transcript variant 1, mRNA using the human PN-1 primers is indicated. Mu, murine; Hu, human; bp; base pairs.
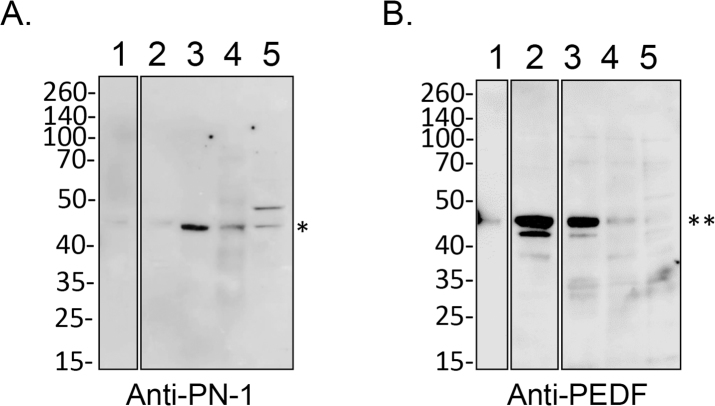
Selbonne et al. reported the presence of PN-1 in the mouse retina [34]. Western blots of protein extracts from the mouse retina with a specific antibody to PN-1 showed a main immunoreactive band for PN-1 protein (Figure 4) confirming what was previously reported [34]. An ARPE-19 cell culture was prepared with serum-free media conditioned for 5 days. After the conditioned media were removed, the cells were washed with 2 M NaCl in PBS to isolate proteins attached by ionic interactions to the cells and the ECM. The cells were also collected, and protein extracts were prepared. Western blots of conditioned media, the salt wash, and cell lysates showed PN-1-immunoreactive bands (50 kDa) only in the ARPE-19 cell lysates (Figure 4). The purified, bacterially derived, recombinant human PN-1 migrated slightly faster (40-kDa), as it corresponds to an unglycosylated form of the PN-1 polypeptide. The slower migration of ARPE-19 PN-1 is likely due to post-translational modification. PEDF was detected only in the conditioned media.
Figure 4.
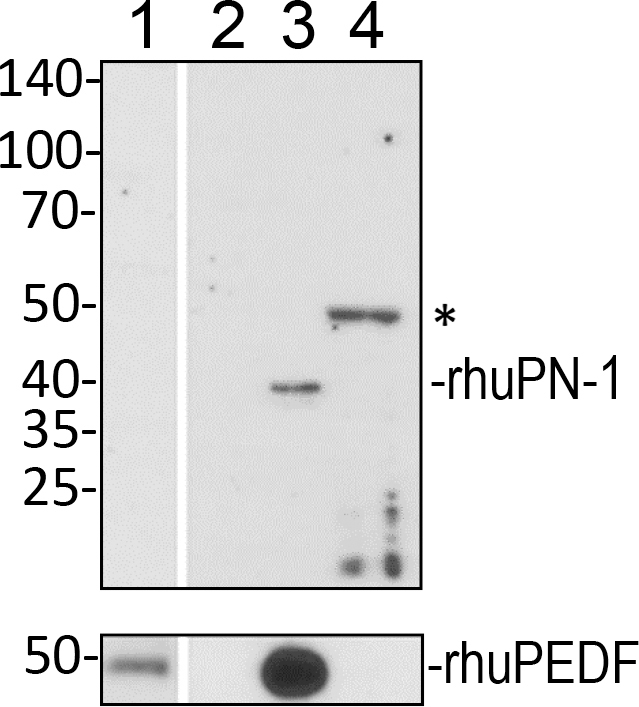
PN-1 protein in human ARPE-19 cell cultures. Protein samples were resolved in a polyacrylamide gel, transferred to nitrocellulose membranes, and immunostained with antibodies. ARPE-19 cells were cultured in serum-free media for 5 days. Conditioned media were removed, and a wash with PBS containing 2 M NaCl was performed. Cell lysates were prepared from the cells. Photos of blots with samples loaded in two gels in duplicate as follows: ARPE-19 conditioned media from serum-free media (lane 1), 2 M NaCl wash (lane 2), bacterially derived recombinant human PN-1 (rhuPN-1, lane 3 top), mammalian-derived, recombinant human PEDF (rhuPEDF, lane 3 bottom), and ARPE-19 cell lysate (lane 4). The top blot was immunostained with Ab-PN-1 and the bottom with Ab-PEDF. For both panels, the migration positions of the MW markers are shown to the left. The migration positions for rhuPN-1 and rhuPEDF are shown to the right. The asterisk points to the migration position of mature glycosylated PN-1.
As murine eye tissue yielded insufficient sample material to further evaluate physiologic PN-1, we implemented a previously established protocol describing the purification of PEDF from the bovine IPM in an effort to investigate the presence of the native ocular PN-1 protein [26]. Western blots of native bovine RPE and retina extracts revealed the presence of PN-1 bands with the expected migration patterns (Figure 5A). An additional slower migrating PN-1-immunoreactive band (approximately 50 kDa) was detected in the bovine retina, which was present also in the mouse retina but at a lower level. Given that PN-1 associates with ECM components via ionic interactions [4], we used 1 M NaCl to remove proteins bound to the IPM. We found PN-1 in IPM lavage with PBS, which increased in lavages with 1 M NaCl in PBS. The bacterially derived recombinant human PN-1 served as a positive control. To compare the distribution of PN-1 in the bovine ocular samples with those of PEDF, western blots of the same samples were performed with an antibody to PEDF (Figure 5B). PEDF was similarly detected in the bovine IPM lavage extracted with 1 M NaCl in PBS but was detected less intensely in the RPE and retina tissue extracts.
Figure 5.
PN-1 protein in the bovine retina. Bovine interphotoreceptor matrix (IPM) lavage and RPE and retina lysates were prepared. Protein samples were resolved in a polyacrylamide gel, transferred to nitrocellulose membranes, and immunostained with Ab-PN-1 (A) or Ab-PEDF (B). Photos of the blots with immunostaining are shown as indicated at the bottom. A: The samples were loaded as follows: fetal bovine serum (FBS; 10 μl of 10% FBS, lane 1), lavage of bovine IPM with PBS (15 μg, lane 2), lavage of bovine IPM with 1 M NaCl in PBS (20 μg, lane 3), bovine RPE lysates (10 μg, lane 4), and bovine retina lysate (20 μg, lane 5). B: The samples were loaded as follows: FBS (10 μl of 10% FBS, lane 1), lavage of bovine IPM with 1 M NaCl in PBS (20 μg, lane 2), lavage of bovine IPM with PBS (15 μg, lane 3), bovine RPE lysates (10 μg, lane 4), and bovine retina lysates (20 μg, lane 5). For each panel, the migration positions of the MW markers are shown to the left. The migration positions of PN-1 and PEDF are shown by * and **, respectively, to the right of each blot.
Cytoprotective activity of PN-1 on retina cells
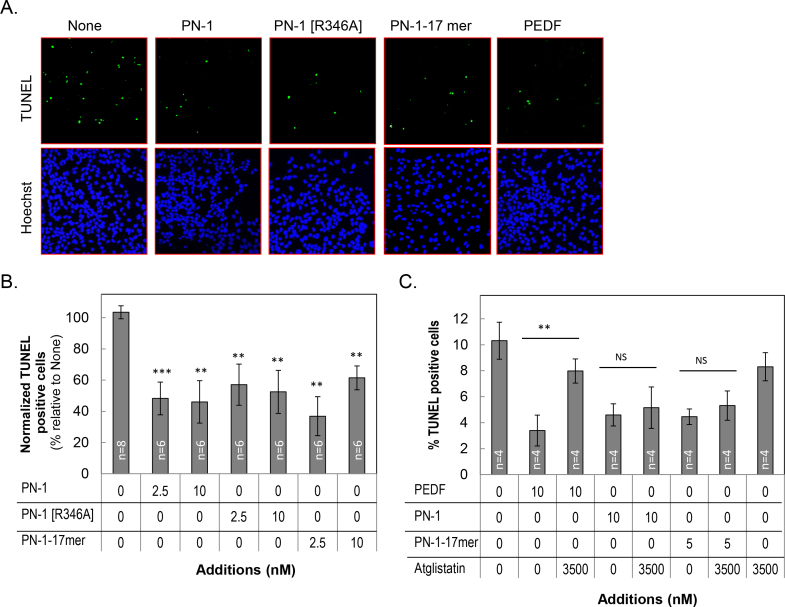
We have demonstrated that PEDF protects retina precursor R28 cells against serum deprivation-related injury [22]. To evaluate the retinoprotective activity of PN-1 on R28 cells, we set to determine the extent to which PN-1 can prevent cell death in serum-deprived R28 cells. Cells were treated with serpins upon serum removal, and death was followed by TUNEL labeling of the nuclei in the dying R28 cells, as previously described [24]. The treatment conditions employed were 2.5 nM and 10 nM of PN-1, as well as PEDF at concentrations known to exert a cytoprotective effect in the cells. Given that the cytoprotective action of PEDF is independent of its inhibitory potential against proteases, we tested PN-1 versions without inhibitory activity against serine proteases, such as PN-1[R346A] and PN-1-17mer. PN-1[R346A] is a recombinant PN-1 with a single residue alteration that abolishes the serine protease inhibitory capacity [10]. PN-1-17mer is a peptide designed from the homologous neurotrophic region of PEDF and distant from the RCL [20]. Figure 6A shows representative images of TUNEL-positive nuclei of R28 cells treated with PN-1 versions and PEDF at 2.5 nM and the cultures that remained untreated without serpins (None), as well as 4',6-diamidino-2-phenylindole (DAPI)-stained nuclei for the total number of cells. A lower number of TUNEL-positive nuclei was observed when the cells were treated with the PN-1 versions relative to the untreated cells, mirroring the protective effects of PEDF. Quantitation of the fluorescent images revealed a significant decrease in TUNEL-positive nuclei/total number of cells in cultures treated with PN-1 when compared to the cultures that were without serpins (None). Given that death by serum deprivation-mediated injury ranged between 4% and 10% from cell batch to cell batch, the number of TUNEL-positive nuclei/total cells was normalized to the cell death of those in untreated cultures (None) as 100% within each of three experiments, and the averages are plotted in Figure 6B. Wild-type PN-1 and PN-1[R346A], each at 2.5 nM or 10 nM, decreased the percentage of TUNEL-positive cells to 46–57% from those without effectors (100%). The cells treated with the 2.5 nM PN-1-17mer peptide exhibited an even further decrease in the percentage of dead cells (37%) and then less effectively with 10 nM peptide (62%). Cell death was also diminished to 32% in the presence of 10 nM PEDF (Figure 6C), a bit more effective than those with PN-1. We conclude that PN-1 with and without protease inhibitory capacity and even the 17mer of PN-1 protected serum-deprived R28 cells.
Figure 6.
Protective effects of PN-1 and PEDF proteins and peptides in cultured cells. R28 cells in serum-free media were treated with serpin proteins and peptides, as indicated, for 48 h. Cytoprotection was assayed by counting the terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling (TUNEL)-positive nuclei (green). Cells were fixed and processed for TUNEL staining and counterstained with Hoechst dye for the nucleus. A: Representative images from each condition show the TUNEL-positive nuclei (green) and the Hoechst-stained nuclei (blue, total cells). Concentrations of proteins and peptides were 2.5 nM. Bar = 20 µm. B: Quantification of the protective effects by PN-1 proteins treated as in A. Each bar corresponds to the average of the percentage of TUNEL-positive nuclei per the total number of cells normalized to the cells treated with serum-free medium (None) as 100%. Shown are data from three independent experiments each performed with duplicate wells ± standard error of the mean (SEM; error bars). C: Cells were pretreated with atglistatin (3.5 µM) for 1 h before a 48-h treatment with or without indicated concentrations of PEDF, PN-1, and PN-1-17mer. Quantification of TUNEL-positive nuclei is shown. Each bar corresponds to the average of the percentage of the TUNEL-positive nuclei per the total number of cells from two independent batches of cells and each performed with duplicate wells per assay ± standard deviation (SD) **, p<0.005; ***, p<0.0005; NS, not significant.
We have shown previously that atglistatin, a selective and competitive inhibitor of the lipase enzymatic activity of PEDF-R, attenuates the PEDF-mediated cytoprotective activity in R28 cells [20]. We used atglistatin to investigate its effects on PN-1-mediated cytoprotection. R28 cells were incubated in media with atglistatin before the effectors were added. Cell death among cell batches in three experiments ranged between 9% and 12%. As shown in Figure 6C, atglistatin completely abolished the survival activity of PEDF, but no effect on PN-1 and PN-1-17mer was observed. Atglistatin treatment alone did not affect the number of TUNEL-positive cells. Altogether, although PN-1 was similar to PEDF in that the protease inhibitory activity was dispensable for cytoprotection, PN-1 differed from PEDF as PN-1 promoted cell survival by a mechanism of action likely independent of the enzymatic activity of PEDF-R.
Antiapoptotic Bcl2 expression in retinal R28 cells
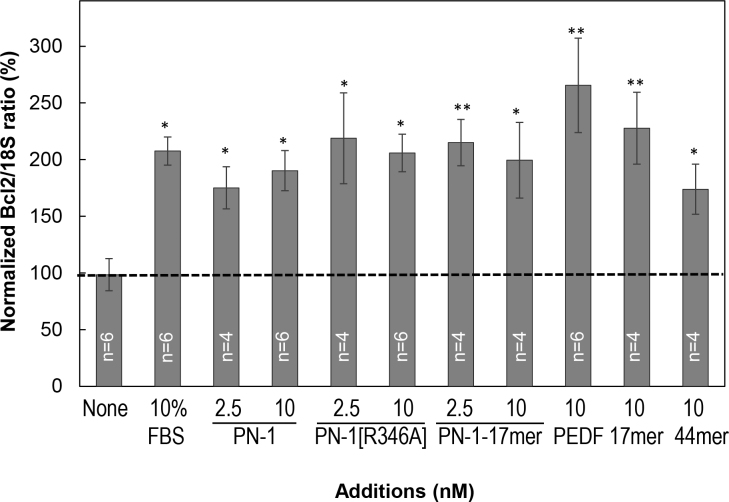
We and others have shown that PEDF protects retina R28 cells from serum-starvation injury by induction of the antiapoptotic Bcl2 gene [22,35]. We examined the Bcl2 expression of serum-starved R28 cells treated with PN-1. As shown in Figure 7, antiapoptotic Bcl2 expression increased upon treatment with PN-1, PN-1[R346A], and PN-1-17mer relative to untreated cells (None). FBS, PEDF, and the neurotrophic PEDF-derived peptides 44mer (a fragment spanning positions 78 and 121 of the human PEDF polypeptide) and 17mer were positive controls that induced Bcl2 by more than twofold. We conclude that similar to PEDF, PEDF-44mer, and PEDF-17mer, protection from serum-starvation injury was accompanied by induction of the antiapoptotic Bcl2 gene expression in R28 cells treated with PN-1 and PN-1-17mer.
Figure 7.
Antiapoptotic Bcl2 expression. Reverse-transcription PCR (RT–PCR) for Bcl2 in R28 cells treated for 6 h with PN-1, PN-1[R346A], PN-1-17mer, PEDF-44mer, PEDF-17mer, and PEDF at the indicated concentrations, as well as with 10% fetal bovine serum (FBS; positive control) and in serum-free medium (None, untreated control). Bar graphs depict the average Bcl2 transcript values obtained relative to their respective 18S RNA values. Shown is the average from three independent experiments (n = 2 for each experiment) each performed with a different batch of cells. The dotted line corresponds to the value of None (untreated control), as reference, ± standard error of the mean (SEM; error bars). *, p<0.05; **, p<0.005.
Discussion
We have identified PN-1 as a novel serpin expressed in ocular tissue. This protein is remarkably similar in tertiary structure to PEDF, a serpin referred to as an ocular guardian. Although some homology exists at the level of amino acid sequences, the crystal structures demonstrate nearly identical folded conformations with overlapping alpha helices, beta sheets, and RCLs. As structure directly governs function, we investigated the extent to which PN-1 and PEDF share functional activities. The recent detection of PN-1 in the retina by Selbonne et al. [34] provided the impetus to study the potential effects of PN-1, traditionally known for its role in the clotting process, as the most potent thrombin inhibitor, in the eye.
We report that the retinas and RPE from mice and humans express SERPINE2 transcripts and that the PN-1 protein product is produced in these tissues. The PN-1 protein is also found in bovine eyes and is secreted into the IPM, similar to PEDF. Naturally occurring PN-1 has an apparent molecular weight greater than that of the bacterially derived, recombinant human PN-1 protein, which does not contain post-translational modifications as in the mammalian-derived PN-1 protein. The observed discrepancy in molecular weight is likely due to post-translational modifications, such as glycosylation of the protein in the native samples. Although the attempts to deglycosylate these samples using peptide-N-glycosidase F (PNGase F) were unsuccessful, we reasoned that the PN-1 detected in western blots may be O-linked glycosylated and comigrate with the one found in serum (FBS), which serves as a positive control for the post-translationally modified form [36]. The PN-1 immunoreactive protein from bovine IPM, RPE, and retina aligned with the band from FBS samples, all of which were slightly greater in molecular weight than recombinant PN-1. Additionally, we noted that the migration patterns of bovine and mouse PN-1 were identical. The migration patterns of PN-1 proteins in FBS display additional bands of higher apparent molecular weight than that of monomeric PN-1 by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). They remind us of the close association of PN-1 with ECM components known to facilitate tissue remodeling [10]. It is known that PN-1 can form complexes with other proteins in the serum, such as thrombin, plasmin, plasminogen activator, or itself [37].
Considering the tendency of PN-1 to remain associated with the cell surface upon secretion [32], it is not surprising that the most intense bands are found in the bovine RPE and IPM components extracted with solutions of greater ionic strength (1 M NaCl). The increase in ionic strength serves to remove any PN-1 that adhered to the retina and/or RPE. The relative levels of PN-1 in the IPM, RPE, and retina suggest the protein is associated with cell surfaces and matrices, while PEDF is known to be secreted as a soluble, diffusible protein into the IPM (Figure 5 [26]). Previously characterized as a highly regulated protein critical to embryonic development and early postnatal nervous system development, PN-1 has not been typically associated with ocular tissue. Upon the initial discovery of PN-1, PN-1 expression was detected in the brain, large and small intestines, heart, kidney, liver, lung, skeletal muscle, olfactory bulb, skin, spleen, stomach, seminal vesicle, testis, thymus, and tongue [38]. Our findings confirm the novel identification of PN-1 in the eye and indicate the neuroprotective capacity of PN-1 in the ocular context, mirroring that of PEDF.
The neuroprotective abilities of PN-1 were originally considered byproducts of its protease inhibition; however, accumulating recent evidence of similarities to PEDF challenges this claim [39]. More recently, the reported ability of PN-1 to inhibit cancer cell proliferation [40] and PN-1’s antiangiogenic properties [10,34] similar to those of PEDF further defy this assumption. Cell survival assays suggest that PN-1 and PEDF are similarly potent in preventing cell death during serum starvation. Despite the structural similarity between PN-1 and PEDF, one clear biochemical difference is that PN-1 inhibits thrombin and other serine proteases, while PEDF is classified as a non-inhibitory serpin. A variant of PN-1 altered at a single amino acid position that yields a non-inhibitory PN-1 serpin, the PN-1[R346A] protein, retains its cytoprotective activity. These findings imply that the mechanism of action of PN-1 as a survival factor is independent of serine protease inhibition, as is the case for PEDF. Furthermore, even upon removal of most residues leaving a short peptide of 17 amino acids from helix C, the survival activity of PN-1 is retained. We observed that the PN-1-17mer peptide decreased its effectiveness from 37% to 62% cell death with a peptide concentration increase from 2.5 nM to 10 nM (Figure 6B). As previously reported by Smith-Swintosky et al. [41], PN-1 promoted the survival of rat hippocampal neurons in culture deprived of glucose in a concentration-dependent manner. The peak of protection occurred at 25 nM, and increasing concentrations were exceedingly less protective. It has been reported that high concentrations of PN-1, such as 2 µM, repressed prostate tumor PC3 cell numbers, perhaps showing opposed prodeath effects at high concentrations mediated by a different receptor than that for cell survival [40]. However, we have not tested higher concentrations of PN-1 versions on R28 cells to confirm such a reverse effect. Regardless, the finding of the efficacy of the PN-1 and PEDF fragments holds promising implications for retinal protection in vivo, which deserves further investigation. This small 17mer region toward the N-terminus confers the survival activity to the PN-1 polypeptide and is not expected to inhibit serine proteases, in agreement with a mechanism of action that is independent of protease inhibition.
Given these observations, we investigated the possible interaction of PN-1, PN-1[R346A], and its PN-1-17mer with the PEDF-R by performing numerous binding assays, such as pulldown of recombinant His6-tagged PEDF-R produced by in vitro protein synthesis and in E. coli, and binding to peptide-affinity resins containing the PEDF-R ligand-binding domain in various binding buffer conditions (data not shown). However, the biochemical experiments yielded inconclusive data regarding the interaction of PN-1 with PEDF-R. A more conclusive outcome was observed using an inhibitor of PEDF-R, showing that PN-1 does not require enzymatic activity of PEDF-R for preventing R28 cell death. Further exploration of the molecular mechanism of PN-1 as a survival factor for retina cells is warranted by these observations. As PEDF is known to exert its protective effects on injured retina R28 cells and cortical neurons by inducing Bcl2 [22,35,42], it is of interest to note the similarities observed with PN-1, its inactive version PN-1[R346A], and its 17mer peptide. Comparable to PEDF, these PN-1 versions protect R28 cells from serum-starvation injury and induce the antiapoptotic Bcl2 gene by a mechanism that is independent of serine protease inhibition. Thus, the observed effects with the 17mer region of these two serpins imply a consensus neurotrophic area in serpins. In conclusion, we have identified PN-1 as a novel factor for the retina that may play a neuroprotective role in vivo and small peptides as relevant candidates for cytoprotection with application in the retina.
Acknowledgments
This work was supported by the Intramural Research Program of the National Eye Institute, NIH. We thank Dr. Federica Polato for providing murine retinal and RPE protein lysates. Parts of data from this manuscript were presented at the 2015 and 2016 meetings of The Association for Research in Vision and Ophthalmology (ARVO) held in Denver, Colorado and Seattle, Washington, respectively. The authors have no disclosures and declare no conflicts of interest.
Appendix 1. Biosynthesis
To access the data, click or select the words “Appendix 1.”
Appendix 2. IDEXX
To access the data, click or select the words “Appendix 2.”
References
- 1.Silverman GA, Whisstock JC, Bottomley SP, Huntington JA, Kaiserman D, Luke CJ, Pak SC, Reichhart JM, Bird PI. Serpins flex their muscle: I. Putting the clamps on proteolysis in diverse biological systems. J Biol Chem. 2010;285:24299–305. doi: 10.1074/jbc.R110.112771. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20498369&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–6. doi: 10.1038/35038119. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11057674&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 3.Carter RE, Cerosaletti KM, Burkin DJ, Fournier RE, Jones C, Greenberg BD, Citron BA, Festoff BW. The gene for the serpin thrombin inhibitor (PI7), protease nexin I, is located on human chromosome 2q33-q35 and on syntenic regions in the mouse and sheep genomes. Genomics. 1995;27:196–9. doi: 10.1006/geno.1995.1025. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=7665170&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 4.Richard B, Bouton MC, Loyau S, Lavigne D, Letourneur D, Jandrot-Perrus M, Arocas V. Modulation of protease nexin-1 activity by polysaccharides. Thromb Haemost. 2006;95:229–35. doi: 10.1160/TH05-08-0546. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16493483&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 5.Djie MZ, Stone SR, Le Bonniec BF. Intrinsic specificity of the reactive site loop of alpha1-antitrypsin, alpha1-antichymotrypsin, antithrombin III, and protease nexin I. J Biol Chem. 1997;272:16268–73. doi: 10.1074/jbc.272.26.16268. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=9195929&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 6.Simpson CS, Johnston HM, Morris BJ. Neuronal expression of protease-nexin 1 mRNA in rat brain. Neurosci Lett. 1994;170:286–90. doi: 10.1016/0304-3940(94)90339-5. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8058202&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 7.Farrell DH, Wagner SL, Yuan RH, Cunningham DD. Localization of protease nexin-1 on the fibroblast extracellular matrix. J Cell Physiol. 1988;134:179–88. doi: 10.1002/jcp.1041340203. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=3279057&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 8.Li W, Huntington JA. Crystal structures of protease nexin-1 in complex with heparin and thrombin suggest a 2-step recognition mechanism. Blood. 2012;120:459–67. doi: 10.1182/blood-2012-03-415869. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22618708&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 9.Nitsch C, Scotti AL, Monard D, Heim C, Sontag KH. The glia-derived protease nexin 1 persists for over 1 year in rat brain areas selectively lesioned by transient global ischaemia. Eur J Neurosci. 1993;5:292–7. doi: 10.1111/j.1460-9568.1993.tb00496.x. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8261109&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 10.Selbonne S, Azibani F, Iatmanen S, Boulaftali Y, Richard B, Jandrot-Perrus M, Bouton MC, Arocas V. In vitro and in vivo antiangiogenic properties of the serpin protease nexin-1. Mol Cell Biol. 2012;32:1496–505. doi: 10.1128/MCB.06554-11. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22331468&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becerra SP, Fariss RN, Wu YQ, Montuenga LM, Wong P, Pfeffer BA. Pigment epithelium-derived factor in the monkey retinal pigment epithelium and interphotoreceptor matrix: apical secretion and distribution. Exp Eye Res. 2004;78:223–34. doi: 10.1016/j.exer.2003.10.013. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=14729355&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 12.Becerra SP, Notario V. The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential. Nat Rev Cancer. 2013;13:258–71. doi: 10.1038/nrc3484. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23486238&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuncl RW, Bilak MM, Bilak SR, Corse AM, Royal W, Becerra SP. Pigment epithelium-derived factor is elevated in CSF of patients with amyotrophic lateral sclerosis. J Neurochem. 2002;81:178–84. doi: 10.1046/j.1471-4159.2002.00813.x. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12067231&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 14.Petersen SV, Valnickova Z, Enghild JJ. Pigment-epithelium-derived factor (PEDF) occurs at a physiologically relevant concentration in human blood: purification and characterization. Biochem J. 2003;374:199–206. doi: 10.1042/BJ20030313. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12737624&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alberdi E, Hyde CC, Becerra SP. Pigment epithelium-derived factor (PEDF) binds to glycosaminoglycans: analysis of the binding site. Biochemistry. 1998;37:10643–52. doi: 10.1021/bi9802317. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=9692954&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 16.Alberdi EM, Weldon JE, Becerra SP. Glycosaminoglycans in human retinoblastoma cells: heparan sulfate, a modulator of the pigment epithelium-derived factor-receptor interactions. BMC Biochem. 2003;4:1. doi: 10.1186/1471-2091-4-1. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12625842&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becerra SP, Perez-Mediavilla LA, Weldon JE, Locatelli-Hoops S, Senanayake P, Notari L, Notario V, Hollyfield JG. Pigment epithelium-derived factor binds to hyaluronan. Mapping of a hyaluronan binding site. J Biol Chem. 2008;283:33310–20. doi: 10.1074/jbc.M801287200. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18805795&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becerra SP, Sagasti A, Spinella P, Notario V. Pigment epithelium-derived factor behaves like a noninhibitory serpin. Neurotrophic activity does not require the serpin reactive loop. J Biol Chem. 1995;270:25992–9. doi: 10.1074/jbc.270.43.25992. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=7592790&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 19.Alberdi E, Aymerich MS, Becerra SP. Binding of pigment epithelium-derived factor (PEDF) to retinoblastoma cells and cerebellar granule neurons. Evidence for a PEDF receptor. J Biol Chem. 1999;274:31605–12. doi: 10.1074/jbc.274.44.31605. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10531367&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 20.Kenealey J, Subramanian P, Comitato A, Bullock J, Keehan L, Polato F, Hoover D, Marigo V, Becerra SP. Small Retinoprotective Peptides Reveal a Receptor-binding Region on Pigment Epithelium-derived Factor. J Biol Chem. 2015;290:25241–53. doi: 10.1074/jbc.M115.645846. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26304116&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notari L, Baladron V, Aroca-Aguilar JD, Balko N, Heredia R, Meyer C, Notario PM, Saravanamuthu S, Nueda ML, Sanchez-Sanchez F, Escribano J, Laborda J, Becerra SP. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J Biol Chem. 2006;281:38022–37. doi: 10.1074/jbc.M600353200. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17032652&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 22.Subramanian P, Locatelli-Hoops S, Kenealey J, DesJardin J, Notari L, Becerra SP. Pigment epithelium-derived factor (PEDF) prevents retinal cell death via PEDF Receptor (PEDF-R): identification of a functional ligand binding site. J Biol Chem. 2013;288:23928–42. doi: 10.1074/jbc.M113.487884. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23818523&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francois D, Venisse L, Marchal-Somme J, Jandrot-Perrus M, Crestani B, Arocas V, Boulton MS. Increased expression of protease nexin-1 in fibroblasts during idiopathic pulmonary fibrosis regulates thrombin activity and fibronectin expression. Lab Invest. 2014;94:1237–46. doi: 10.1038/labinvest.2014.111. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25199049&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 24.Subramanian P, Deshpande M, Locatelli-Hoops S, Moghaddam-Taaheri S, Gutierrez D, Fitzgerald DP, Guerrier S, Rapp M, Notario V, Becerra SP. Identification of pigment epithelium-derived factor protein forms with distinct activities on tumor cell lines. J Biomed Biotechnol. 2012;xxx:425907. doi: 10.1155/2012/425907. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22701303&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seigel GM, Salvi RJ. A Microarray Dataset of Genes Expressed by the R28 Retinal Precursor Cell Line. Dataset Papers in Neuroscience. 2013;xxx:3. [Google Scholar]
- 26.Wu YQ, Notario V, Chader GJ, Becerra SP. Identification of pigment epithelium-derived factor in the interphotoreceptor matrix of bovine eyes. Protein Expr Purif. 1995;6:447–56. doi: 10.1006/prep.1995.1060. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8527930&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 27.Aymerich MS, Alberdi EM, Martinez A, Becerra SP. Evidence for pigment epithelium-derived factor receptors in the neural retina. Invest Ophthalmol Vis Sci. 2001;42:3287–93. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11726635&dopt=Abstract [PubMed] [Google Scholar]
- 28.Huber R, Carrell RW. Implications of the three-dimensional structure of alpha 1-antitrypsin for structure and function of serpins. Biochemistry. 1989;28:8951–66. doi: 10.1021/bi00449a001. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=2690952&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 29.Whisstock JC, Bottomley SP. Molecular gymnastics: serpin structure, folding and misfolding. Curr Opin Struct Biol. 2006;16:761–8. doi: 10.1016/j.sbi.2006.10.005. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17079131&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 30.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–804. doi: 10.1021/cr010170+. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12475206&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 31.Rashid Q, Kapil C, Singh P, Kumari V, Jairajpuri MA. Understanding the specificity of serpin-protease complexes through interface analysis. J Biomol Struct Dyn. 2015;33:1352–62. doi: 10.1080/07391102.2014.947525. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25052369&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 32.Huntington JA. Thrombin inhibition by the serpins. Journal of thrombosis and haemostasis. JTH. 2013;11(Suppl 1):254–64. doi: 10.1111/jth.12252. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23809129&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 33.Simonovic M, Gettins PG, Volz K. Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor. Proc Natl Acad Sci USA. 2001;98:11131–5. doi: 10.1073/pnas.211268598. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11562499&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selbonne S, Francois D, Raoul W, Boulaftali Y, Sennlaub F, Jandrot-Perrus M, Bouton MC, Arocas V. Protease nexin-1 regulates retinal vascular development. Cellular and molecular life sciences. Cell Mol Life Sci. 2015;72:3999–4011. doi: 10.1007/s00018-015-1972-5. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26109427&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami Y, Ikeda Y, Yonemitsu Y, Onimaru M, Nakagawa K, Kohno R, Miyazaki M, Hisatomi T, Nakamura M, Yabe T, Hasegawa M, Ishibashi T, Sueishi K. Inhibition of nuclear translocation of apoptosis-inducing factor is an essential mechanism of the neuroprotective activity of pigment epithelium-derived factor in a rat model of retinal degeneration. Am J Pathol. 2008;173:1326–38. doi: 10.2353/ajpath.2008.080466. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18845835&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boulaftali Y, Adam F, Venisse L, Ollivier V, Richard B, Taieb S, Monard D, Favier R, Alessi MC, Bryckaert M, Arocas V, Jandrot-Perrus M, Bouton MC. Anticoagulant and antithrombotic properties of platelet protease nexin-1. Blood. 2010;115:97–106. doi: 10.1182/blood-2009-04-217240. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19855083&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 37.Bouton MC, Boulaftali Y, Richard B, Arocas V, Michel JB, Jandrot-Perrus M. Emerging role of serpinE2/protease nexin-1 in hemostasis and vascular biology. Blood. 2012;119:2452–7. doi: 10.1182/blood-2011-10-387464. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22234688&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 38.Mansuy IM, van der Putten H, Schmid P, Meins M, Botteri FM, Monard D. Variable and multiple expression of Protease Nexin-1 during mouse organogenesis and nervous system development. Development. 1993;119:1119–34. doi: 10.1242/dev.119.4.1119. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8306878&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 39.Onuma Y, Asashima M, Whitman M. A Serpin family gene, protease nexin-1 has an activity distinct from protease inhibition in early Xenopus embryos. Mech Dev. 2006;123:463–71. doi: 10.1016/j.mod.2006.04.005. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16797167&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 40.McKee CM, Ding Y, Zhou J, Li C, Huang L, Xin X, He J, Allen JE, El-Deiry WS, Cao Y, Muschel RJ, Xu D. Protease nexin 1 induces apoptosis of prostate tumor cells through inhibition of X-chromosome-linked inhibitor of apoptosis protein. Oncotarget. 2015;6:3784–96. doi: 10.18632/oncotarget.2921. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25686839&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith-Swintosky VL, Zimmer S, Fenton JW, 2nd, Mattson MP. Protease nexin-1 and thrombin modulate neuronal Ca2+ homeostasis and sensitivity to glucose deprivation-induced injury. J Neurosci. 1995;15:5840–50. doi: 10.1523/JNEUROSCI.15-08-05840.1995. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=7643224&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez A, Tripathy D, Yin X, Luo J, Martinez J, Grammas P. Pigment epithelium-derived factor (PEDF) protects cortical neurons in vitro from oxidant injury by activation of extracellular signal-regulated kinase (ERK) 1/2 and induction of Bcl-2. Neurosci Res. 2012;72:1–8. doi: 10.1016/j.neures.2011.09.003. https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21946416&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]