Abstract
We are performing whole genome sequencing (WGS) of families with Autism Spectrum Disorder (ASD) to build a resource, named MSSNG, to enable the sub-categorization of phenotypes and underlying genetic factors involved. Here, we report WGS of 5,205 samples from families with ASD, accompanied by clinical information, creating a database accessible in a cloud platform, and through an internet portal with controlled access. We found an average of 73.8 de novo single nucleotide variants and 12.6 de novo insertion/deletions (indels) or copy number variations (CNVs) per ASD subject. We identified 18 new candidate ASD-risk genes such as MED13 and PHF3, and found that participants bearing mutations in susceptibility genes had significantly lower adaptive ability (p=6×10−4). In 294/2,620 (11.2%) of ASD cases, a molecular basis could be determined and 7.2% of these carried CNV/chromosomal abnormalities, emphasizing the importance of detecting all forms of genetic variation as diagnostic and therapeutic targets in ASD.
Introduction
Autism is a term coined about a century ago, derived from the Greek root referring to “self”, and describes a wide range of human interpersonal behaviors1. Autistic tendencies may be recognized in many individuals as part of human variation2, but these features can be severe and, therefore, disabling3–5. The most recent Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-5) uses this single omnibus classification “autism spectrum disorder” (ASD) to encompass what once were considered several distinct diagnostic entities (such as autistic disorder, Asperger disorder, and pervasive developmental disorder, not otherwise specified). The spectrum concept reflects both the diversity among individuals in severity of symptoms, from mild to severe, and the recognition of overlap among a collection of clinically-described disorders6–8. Features of this neurodevelopmental disorder, which has a worldwide population prevalence of ~1%, typically include impaired communication and social interaction, repetitive behaviors, and restricted interests; these may also be associated with psychiatric, neurological or physical co-morbidities, and/or intellectual disability.
Despite the unitary diagnostic classification, ASD is a heterogeneous spectrum, both in clinical presentation and in terms of the underlying etiology. Individuals with ASD are increasingly seen in clinical genetics services, and ~10% have an identifiable genetic condition4, 9. In fact, there are over 100 genetic disorders that can exhibit features of ASD (e.g. Rett and Fragile X syndromes)10. Clearly, ASD is strongly associated with genetic factors. Dozens of susceptibility genes (e.g. SHANK and NRXN family genes)11, 12 and copy number variation (CNV) loci (e.g. 16p11.2 deletion and 15q11-q13 duplication) facilitate a molecular diagnosis in ~5–40% of ASD cases. The variation largely depends on the cohort examined (e.g. syndromic or idiopathic), and the technology used (i.e. karyotyping, microarray, whole-exome sequencing)5, 9, 13–17.
In fact, the genetic diathesis towards ASD may be different for almost every individual18, making this a prime candidate for the coming age of precision medicine6, 7, 19. The first beneficiaries of a genetic diagnosis are young children, in whom formal diagnosis based on early developmental signs can be challenging, but who benefit most from earlier behavioral intervention3, 8. Understanding the genetic subtypes of ASD can also potentially inform prognosis, medical management, and assessment of familial recurrence risk, and in the future, it may facilitate pharmacologic-intervention trials through stratification based on pathway profiles14. The vast heterogeneity also means that meticulous approaches are needed to catalogue all the genetic factors that contribute to the phenotype, and to consider how these interact with one another and with non-genomic elements.
To move forward towards the goal of understanding all of the genetic factors involved in ASD, we recognize the need to scan the genome in its entirety using whole genome sequencing (WGS)14, 18, 20, 21 on thousands, if not tens of thousands (or more), of samples13, 22–24. Risk variants that remain undiscovered to this point are expected to be individually rare9, 18, 22, possibly involved in complex combinations18, 25, and to include single nucleotide variants (SNVs), small insertions/deletions (indels), more complex CNVs14, 18, 20, and other structural alterations15, 26. Some will reside in the ~98% non-coding genome largely unexplored by other microarray and exome sequencing studies21, 27, 28. Abundant genome sequences may help to resolve the role of common variants in ASD2, 29, and integrating these data with those on rare variants will aid understanding of the issues of penetrance, variable expressivity, and pleiotropic effects4, 6.
Such research brings us to the realm of “big data”: massive sequence data sets from multitudes of individuals, requiring fast and intensive searches for meaningful patterns30. This is where cloud-based computing excels. Its capacity for bulk data storage, with efficient processing and built-in security, is ushering in a new paradigm for data sharing, enabling access and collaboration across continents31, 32.
In our MSSNG initiative (where omission of letters from the name, represents the information about autism that is missing and yet to be uncovered), we are collecting whole genome sequences and detailed phenotypic information from individuals with ASD and their families, and making these data widely available to the research community (Figure 1). Here, we describe the MSSNG infrastructure, new analyses of the first 5,205 genomes, and examples of how to use the resource.
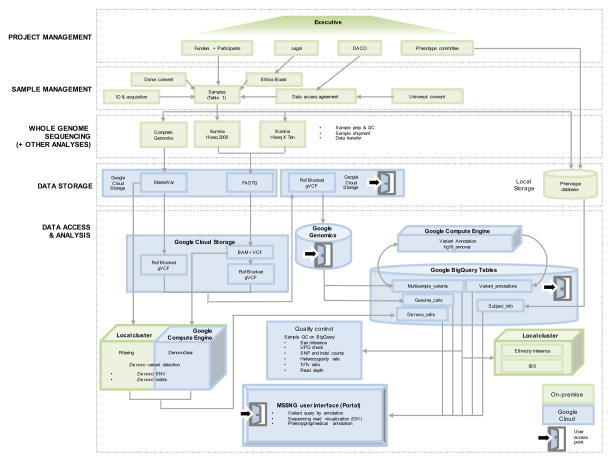
Figure 1. Schematic of sample and data processing in MSSNG.
An executive committee oversees the project. The parameters for DNA sample selection and (genetic and phenotypic) data are managed by the committee, including consenting and ethics protocols. Coded identifiers’ for samples selected for WGS are posted as they are identified at MSSNG portal (http://mss.ng/research), so the ASD research community can monitor progress. Phenome data include subject information (identity number, year of birth, sex), family code (proband, parent, sibling), results of diagnostic tests (e.g. ADOS, ADI-R, age at diagnosis, functional assessments, intelligence tests, body measurements and dysmorphic features). The database accommodates as much of this information as is available for each sample, but that varies widely. Future plans include incorporation of fields for co-morbidities, related conditions, exposures, extended family history, interventions, and other parameters that become apparent. WGS technologies were Complete Genomics and Illumina HiSeq (2000 and X). WGS data are transferred to Google Genomics for data processing through the Google Cloud. Ref-blocked gVCFs were generated and stored in Google Cloud Storage, which were also processed for de novo mutation detection in the local cluster (for Complete Genomics data using filtering method) and Google Compute Engine (for Illumina data using DenovoGear). The Ref-blocked gVCFs and the de novo mutations were annotated through the Google Compute Engine (using Annovar), which can be accessed through the BigQuery tables. Quality controls of the genomic data were performed in the local cluster and the Google Cloud. The processed genetic data and the phenotypic data are accessible through the MSSNG Portal interface. The MSSNG database is designed to support incremental addition of data without changes in architecture, scaling to at least tens of thousands. New WGS and phenotype data are continually added to MSSNG as new batches of 1,000 samples are processed. DACO: Data access committee; UPD: Uniparental disomy; Ti/Tv: Transition to transversion ratio; IBS: Identity by state.
Results
Samples and phenotypes
Our pilot work14, 18, 21 established four principles to guide the prioritization of the samples selected for WGS (Table 1), (1) DNA from whole blood is preferred for detecting de novo mutations (especially for the proband) rather than DNA from cell lines, which may acquire variants in vitro. (2) For a comprehensive ASD resource, it is important to sample families with different genetic characteristics in order to delineate the full spectrum of relevant variation (for example, heritable variants may differ from those arising de novo, and ascertainment biases can influence the frequency of genetic variants identified). (3) Families with extensive phenotype data who are accessible to participate in further study are most informative for ASD. (4) To participate in ASD genomic research on this scale, consent must be in place, or obtainable, for WGS and for the data to be stored in a cloud-based platform.
Table 1.
ASD studies contributing samples for WGS.
| Cohort | SPX | MPX | Illumina HiSeq20008 | Complete Genomics9 | Illumina HiSeqX10 | Total Genomes | Total ASD Genomes |
|---|---|---|---|---|---|---|---|
| AGRE1 | 452 | 952 | 0 | 0 | 1,404 | 1,404 | 730 |
| AGRE1; Autism Treatment Network2 | 321 | 20 | 0 | 0 | 341 | 341 | 192 |
| ASD: Genomes to Outcomes Study3 | 1,763 | 961 | 261 | 931 | 1,532 | 2,724 | 1,421 |
| Baby Siblings Research Consortium4 | 59 | 32 | 9 | 55 | 27 | 91 | 43 |
| Baby Siblings Research Consortium4; The Autism Simplex Collection5 | 15 | 3 | 12 | 6 | 0 | 18 | 6 |
| Infant Sibling Study6 | 52 | 75 | 9 | 81 | 37 | 127 | 62 |
| Infant Sibling Study6; The Autism Simplex Collection5 | 6 | 4 | 3 | 7 | 0 | 10 | 4 |
| Pathways in ASD7 | 35 | 25 | 0 | 58 | 2 | 60 | 24 |
| The Autism Simplex Collection5 | 397 | 33 | 267 | 95 | 68 | 430 | 144 |
|
| |||||||
| Total | 3,100 | 2,105 | 561 | 1,233 | 3,411 | 5,20511 | 2,626 |
AGRE, PMID: 11452364, www.agre.org;
Autism Treatment Network, http://www.asatn.org/;
ASD: Genomes to Outcomes Study, PMID: 23275889;
Baby Siblings Research Consortium, PMID: 21844053, https://www.autismspeaks.org/science/research-initiatives/high-risk-baby-sibs;
The Autism Simplex Collection, PMID: 25392729;
Infant Sibling Study, PMID: 15749241;
Pathways in ASD, PMID: 20405194, http://www.asdpathways.ca/;
Average coverage: 34X, Average read length: 100bp;
Average coverage: 54.5X, average read length: 35bp;
Average coverage: 36.4X, Average read length: 150bp
Of these 5,205 genomes, 1,745 of them are ASD probands, 879 are ASD affected siblings 1 is an affected father and 1 an affected grandfather. There are 1,282 fathers, 1,290 mothers, 1 grandmother, and 6 unaffected siblings involved reported in this study. Samples from some families are still being sequenced and are continually released in the MSSNG resource.
Here, we report on the WGS and analysis of 5,205 samples (5,193 unique individuals; 12 individuals were sequenced on two different platforms for technical replication, or were from different DNA sources). From nine collections, these samples included 2,626 from 2,620 individuals (2,066 unique families) diagnosed with ASD (Table 1). Of the total, 3,100 samples (3,090 individuals) are from simplex (one child with ASD) and 2,105 samples (2,103 individuals) from multiplex families (two or more affected siblings); 1,745 samples (1,740 individuals) are from probands and 879 samples (878 individuals) are from affected siblings (with the exception of two affected individuals within this cohort who are the father and paternal grandfather of a proband). The samples from individuals with ASD include 2,067 from males (2,062 individuals) and 559 from female (558 individuals) (3.7:1 male-to-female ratio). For 339 samples (46 ASD and 293 parents) only cell-line DNA was available. Based on self-reports and confirmed with genotypes from WGS or microarrays, the majority (72.6%) of participants are of European ancestry (Supplementary Table 1, Supplementary Figure 1). For all individuals, we obtained informed consent, as approved by the respective Research Ethics Boards (REBs). We have also developed a prospective consent form for WGS in ASD (Supplementary note).
An ASD diagnosis was of research quality when it met criteria on one (n=437) or both (n=1,361) of the diagnostic measures (Table 2): Autism Diagnostic Interview-Revised (ADI-R) and Autism Observation Schedule (ADOS) or considered a clinical diagnosis (n=819) when given by an expert clinician according to the DSM (IV or 5 editions). Additionally, many participants were assessed with standardized measures of intelligence (IQ), language and general adaptive function. Out of the 1,102 individuals with IQ data available, 216 (19.6%) had scores within the range for intellectual disability (Full Scale IQ<70). Physical measurements are also available for some individuals (n=1,022). Most samples of affected individuals (n=1,658) were genotyped on high-resolution microarrays (see below) and some by karyotyping or gene-specific assays.
Table 2.
Sample Phenotype Summary.
|
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall Sample (n=2,196) | Males (n=1,722) | Females (n=474) | ||||||||||
| n | Mean | SD | Range | n | Mean | SD | Range | n | Mean | SD | Range | |
| Age of Diagnosis (years) | 2,091 | 8.7 | 4.6 | 1.5 – 39.3 | 1,642 | 8.6 | 4.6 | 1.5 – 39.3 | 449 | 8.8 | 4.8 | 1.7 – 34.8 |
|
| ||||||||||||
| ADI1 Diagnosis | ||||||||||||
|
| ||||||||||||
| Autism | 1,477 | - | - | - | 1,186 | - | - | - | 291 | - | - | - |
| Not Autism2 | 248 | - | - | - | 185 | - | - | - | 63 | - | - | - |
|
| ||||||||||||
| ADOS3 Diagnosis | ||||||||||||
|
| ||||||||||||
| Autism | 909 | - | - | - | 737 | - | - | - | 172 | - | - | - |
| ASD | 346 | - | - | - | 270 | - | - | - | 76 | - | - | - |
| Not Autism2 | 179 | - | - | - | 137 | - | - | - | 42 | - | - | - |
|
| ||||||||||||
| Vineland Adaptive Behaviour Scale | ||||||||||||
|
| ||||||||||||
| Adaptive Behaviour Standard Score | 1,501 | 67.5 | 17.8 | 19 – 119 | 1,190 | 67.4 | 18 | 19 – 119 | 311 | 68 | 17.1 | 19 – 114 |
|
| ||||||||||||
| Repetitive Behaviour Scale – Revised | ||||||||||||
|
| ||||||||||||
| Overall Score | 1,090 | 30.2 | 19.9 | 0 – 115 | 851 | 30.2 | 19.9 | 0 – 115 | 239 | 30.1 | 20 | 0 – 96 |
|
| ||||||||||||
| Repetitive Behaviour Scale | ||||||||||||
|
| ||||||||||||
| Overall Score | 262 | 29.3 | 19 | 1 – 97 | 215 | 30.1 | 19 | 1 – 97 | 47 | 25.6 | 18.9 | 2 – 88 |
|
| ||||||||||||
| Social Responsiveness Scale | ||||||||||||
|
| ||||||||||||
| Total T-Score | 764 | 83 | 16.3 | 40 – 123 | 616 | 81.1 | 15.3 | 40 – 119 | 148 | 90.5 | 18.1 | 44 – 123 |
|
| ||||||||||||
| Social Communication Questionnaire | ||||||||||||
|
| ||||||||||||
| Total Score | 572 | 18.9 | 7.7 | 0 – 37 | 437 | 19 | 7.6 | 0 – 37 | 135 | 18.7 | 8 | 1 – 37 |
|
| ||||||||||||
| Language: | ||||||||||||
| OWLS4: Total Standard Score | 622 | 79.5 | 23.5 | 40 – 142 | 487 | 79.7 | 22.8 | 40 – 138 | 135 | 79.1 | 25.9 | 40 – 142 |
| PLS-3/PLS-45: Total Standard Score | 162 | 63.5 | 20.2 | 50 – 135 | 136 | 64.5 | 21.1 | 50 – 135 | 26 | 58.6 | 14.2 | 50 – 96 |
|
| ||||||||||||
| IQ | ||||||||||||
|
| ||||||||||||
| Full Scale Standard Score | 1,062 | 88.9 | 24.3 | 36 – 161 | 854 | 89.4 | 24.2 | 36 – 161 | 208 | 86.9 | 24.4 | 36 – 155 |
| Non-Verbal Standard Score | 76 | 74 | 27.9 | 42 – 136 | 65 | 74.2 | 28.5 | 42 – 136 | 11 | 72.7 | 24.5 | 42 – 117 |
|
| ||||||||||||
| Child Behavior Checklist (T-Scores) | ||||||||||||
|
| ||||||||||||
| Internalize Problems | 613 | 61.4 | 9.8 | 33 – 86 | 474 | 61.3 | 9.7 | 33 – 86 | 139 | 61.5 | 10 | 39 – 85 |
| Externalize Problems | 613 | 51.2 | 10.9 | 28 – 89 | 474 | 57 | 11.1 | 32 – 87 | 139 | 57.7 | 10.3 | 28 – 89 |
| Total Problems | 613 | 62.8 | 9.7 | 31 – 90 | 474 | 62.5 | 9.9 | 31 – 90 | 139 | 63.6 | 9.3 | 34 – 90 |
|
| ||||||||||||
| Aberrant Behavior Checklist (Sum Scores) | ||||||||||||
|
| ||||||||||||
| Irritability | 370 | 12.3 | 9.2 | 0 – 40 | 291 | 11.7 | 8.7 | 0 – 39 | 79 | 14.3 | 10.5 | 0 – 40 |
| Lethargy | 370 | 10.1 | 7.9 | 0 – 37 | 291 | 9.3 | 7.3 | 0 – 37 | 79 | 13.3 | 9 | 0 – 34 |
| Stereotype | 370 | 5.1 | 4.7 | 0 – 21 | 291 | 5.1 | 4.7 | 0 – 21 | 79 | 5.2 | 4.6 | 0 – 20 |
| Hyperactivity | 370 | 16.4 | 10.8 | 0 – 45 | 291 | 16.1 | 10.9 | 0 – 45 | 79 | 17.4 | 10.6 | 0 – 41 |
| Inappropriate Speech | 370 | 3.6 | 3.1 | 0 – 12 | 291 | 3.6 | 3.1 | 0 – 12 | 79 | 3.8 | 3 | 0 – 11 |
A summary of the scores from a subset of the available measures for affected individuals.
Autism Diagnostic Interview;
For this particular assessment the individual did not meet criteria for ASD but has a diagnosis of ASD based on the other assessments and/or DSM criteria;
Autism Diagnostic Observation Schedule;
Oral and Written Language Scales;
Preschool Language Scale (third and fourth editions).
WGS
We used different WGS platforms as they became available and were tested to assess data quality characteristics. We present data generated using Complete Genomics (n=1,233), Illumina HiSeq 2000 (n=561) and HiSeq X (n=3,411). The different WGS approaches and tools used for mapping/variant calling yield data with different characteristics (Figure 2), but, all platforms reliably called SNV and smaller indels (up to 100bp; larger CNVs are described below). Relative to the human reference sequence (hg19), the average coverage across all samples on three platforms was 93%, with an average of 40.4X sequence depth (Table 1; Supplementary Table 1). On average, we detected 3,654,992 SNVs and 722,816 indels per sample (Supplementary Table 1).
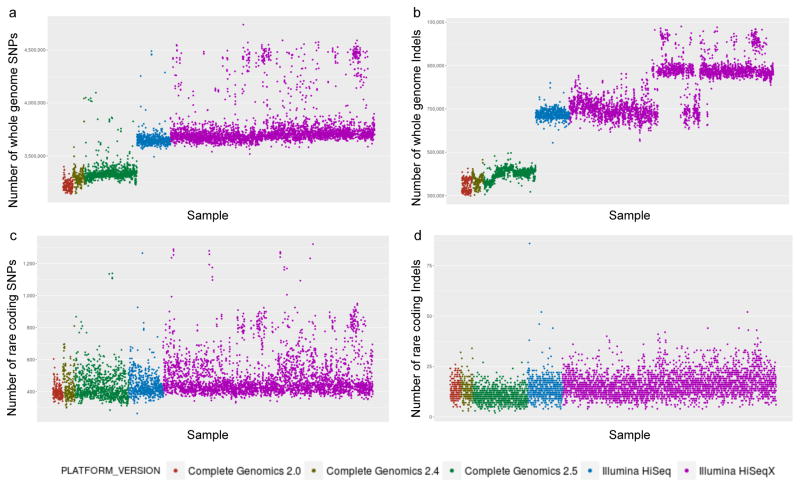
Figure 2. Characteristics and quality of WGS from different sequencing platforms.
(a) Number of SNVs detected per genome. (b) Number of indels detected per genome. (c) Number of rare coding SNVs detected per genome after quality filtering. (d) Number of rare coding indels detected per genome after quality filtering. Genomes sequenced by Complete Genomics with 2.0 pipeline version are colored in orange, by Complete Genomics with 2.4 pipeline version are colored in brown, by Complete Genomics with 2.5 pipeline version are colored in green, by Illumina HiSeq 2000 are colored in blue, and by Illumina HiSeq X are colored in purple. Details of quality for individual samples can be found in Supplementary Table 1.
Systematic detection of sequence-level de novo mutations and candidate ASD-risk genes
Identification of multiple de novo mutations occurring in the same gene from unrelated individuals highlighted candidate ASD-risk genes13, 22. Modifying our previous approaches14, 18, 21 (Methods), we studied those 1,239 families (1,627 parents-child trios) for which child and parental WGS data were available (excluding children whose DNA was derived from cell lines). We identified 86.4 spontaneous events per genome (73.8 SNVs and 12.6 indels) (Supplementary Table 2 and 3), including 1.3 de novo exonic variants per genome14, 18, 21. Experimental validation rates for selected de novo SNVs and indels were 88.2% (494/560) and 85.1% (103/121), respectively. Most (58.3%) of the non-validated calls were caused by false negative detection in the parents. In total, we detected 230 experimentally validated de novo LOF mutations (Supplementary Table 4 and 5).
To increase the power for ASD-risk gene identification, we combined our data with the de novo mutations detected from other large-scale systematic whole-exome or WGS studies, which included 2,864 de novo missense mutations and 599 de novo LOF mutations in 4,087 trios13, 23, 33–35. To identify candidate ASD-risk genes, we initially considered genes likely to be mutation-intolerant based on the ExAC database36 (with pLI> 0.9 for LOF mutations; with z-score of >0.2 for missense mutations), and higher than expected mutation rate (FDR<15%). This approach yielded 54 putative ASD-risk genes (Figure 3a).
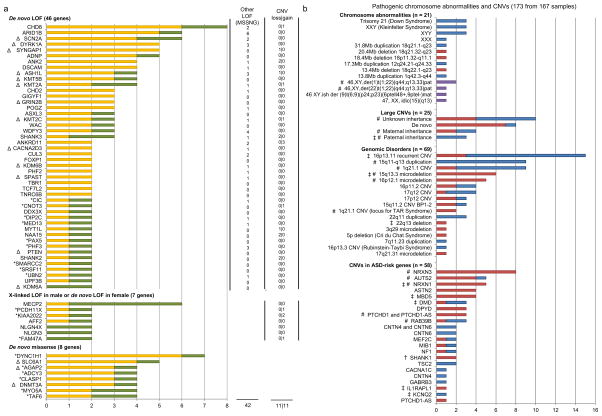
Figure 3. ASD-susceptibility genes/loci.
(a) ASD-risk genes with higher than expected mutation rate from MSSNG integrated with other large-scale high-throughput sequencing projects. ASD-risk genes are ranked in descending order of the number of mutations found for each gene. Other LOF mutations, including inherited LOF mutations and LOF mutations with unknown inheritance (where parents are unavailable for testing), and CNVs found in the MSSNG cohort are indicated (except for genes found by higher than expected de novo missense mutation rate). MSSNG data are in green and published data are in yellow. Novel putative ASD-risk genes identified in this study carry an asterisk. Δ indicate genes with druggable protein domains identified (Supplementary Table 6). (b) Pathogenic chromosomal abnormalities and CNVs identified falling into one of four categories: Chromosomal abnormalities; DECIPHER loci and other genomic disorders associated with ASD; large rare CNVs between 3–10Mb and CNVs disrupting ASD candidate genes not described above in Figure 3a. Deletions are in red, duplications are in blue and complex variants are in purple. # indicate CNVs shared between affected siblings; ‡ indicates a CNV carried by an individual with a second pathogenic CNV; † indicates a CNV shared between individuals within an extended pedigree. Details can be found in Supplementary Table 8. Examples of CNVs affecting the NRXN1 and CHD8 genes, and the PTCHD1-AS non-coding gene identified from the WGS are shown in Figure 4.
In addition to the de novo LOF mutations, we have also combined the de novo or maternally-inherited LOF mutations on the X chromosome in the affected males. We identified 7 genes (MECP2, AFF2, FAM47A, KIAA2022, NLGN3, NLGN4X and PCDH11X) with multiple LOF mutations and with pLI>0.65 (Figure 3a). Taken together, 112 of the 2,620 subjects (4.3%) bear de novo LOF or missense mutations or inherited LOF mutations in the 61 ASD-risk genes identified (Figure 3a; Supplementary Table 5). Among these, 43 were found as ASD-risk genes in a previous meta-analysis of exome sequencing24 or other CNV studies10, 15, 17.
Detection of CNVs and chromosome abnormalities
We examined CNVs detected from WGS using two calling algorithms for samples sequenced in Illumina platforms or provided by Complete Genomics, and for a subset of samples using additional microarray data (Methods). From the WGS derived CNVs, we detected 401.4 CNVs (>2kb) per genome. We validated these using laboratory-based methods and/or WGS read-depth comparisons (Methods, Figure 3b and Figure 4). We found that 189/2,620 (7.2%) of the subjects to carry one or more pathogenic chromosomal abnormalities (n=21), megabase CNVs (n=25), CNVs involving genomic disorder loci (n=69), or CNVs affecting previously reported ASD-risk genes (n=58), all determined by standard diagnostic reporting criteria16, 17, 37, many associated with known syndromes of which ASD can be a component feature5, 9, 10. There were also 22 CNVs that overlapped with the ASD-risk genes found in this study (Figure 3a). Three of the CNVs were around or less than 10kb, which were only detectable using WGS, and five were non-coding (Figure 4c).
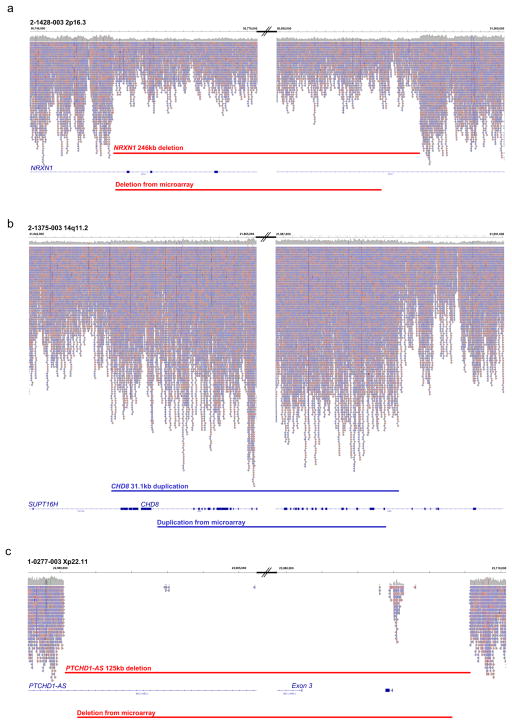
Figure 4. CNV characterization via WGS reads in the MSSNG Portal.
(a–c) Visualization of CNVs in WGS data. (a) A heterozygous 246kb deletion of three exons of NRXN1 at chromosome 2p16.3 in subject 2-1428-003 (average 50% decrease in sequence read-depth); (b) a 31.1kb duplication within CHD8 at chromosome 14q11.2 in subject 2-1375-003 (average of 50% increase in sequence read-depth) and (c) 125kb deletion of exon 3 of the non-coding gene PTCHD1-AS at Xp22.11 in male subject 1-0277-003 (no reads apparent, other than a small stretch of likely mis-aligned repetitive sequences). Left and right panels show the proximal and distal breakpoints of the CNVs respectively. Aligned reads viewed from the BAM files in the MSSNG browser are shown indicating the read depth. Genome co-ordinates are shown above and impacted genes below. The predicted CNVs visible from the WGS data and high-resolution microarray are shown by the red (deletion) and blue (duplication) bars. For 32 CNVs described in Figure 3b plus 17 additional CNVs, we derived a more accurate estimate of the breakpoints by visual inspection of read depth from the BAM file in the MSSNG browser. On average, the size difference between the CNV predicted by microarray data and the estimated size from WGS data was 6.9kb and for 31/49 (63%) CNVs, the size of the CNV was smaller in the microarray data than WGS. For four CNVs, the WGS-resolved breakpoints altered the exons of genes being annotated as deleted or duplicated. In another case, this resulted in a CNV from microarray no longer being classified as pathogenic as the revised breakpoints no longer included coding sequence.
Medical genetics and functional properties
Among these 61 ASD-risk genes with sequence-level mutations, 18 had not previously been reported in the literature (CIC, CNOT3, DIP2C, MED13, PAX5, PHF3, SMARCC2, SRSF11, UBN2, DYNC1H1, AGAP2, ADCY3, CLASP1, MYO5A, TAF6, PCDH11X, KIAA2022, and FAM47A). For two of these putative novel ASD-risk genes, mutations were found in at least three families from our data (Supplementary Figure 2); MED13, which is related to the intellectual disability gene MED13L38, carried putative damaging mutations in three families. PHF3 mutations, with PHF2 involved in ASD24, were found in four families; this gene encodes a PHD finger protein that regulates transcription by influencing chromatin structure39, a mechanism increasingly being implicated in ASD17, 21, 40. Some other mutation-intolerant genes were implicated in three or more families included PER2 and HECTD4 (Supplementary Figure 2). While they did not meet the statistical significance for Figure 3a, they may still potentially represent interesting functional candidates for the ASD or associated complications in these individuals.
Interestingly, of these 61 ASD susceptibility genes, 36 (59%) are associated with known syndromes/phenotypes in OMIM (with CHD8, SHANK2 and NLGN3 associated only with ASD). Most (78%) of the known syndromes/phenotypes involved were intellectual disability or other related disorders, which may highlight the pleiotropic effects of these genes (Supplementary Table 6). Combining the list of 61 genes with the CNV data identified in the WGS analysis yielded a framework map of ~100 ASD-linked loci or chromosomal abnormalities (all listed in Figure 3) for molecular diagnostic comparisons, accounting for 11.2% (294/2,620) of the subjects included in this study. Consistent with our previous findings18, ASD-relevant mutations were often different in affected siblings (Supplementary Figure 2).
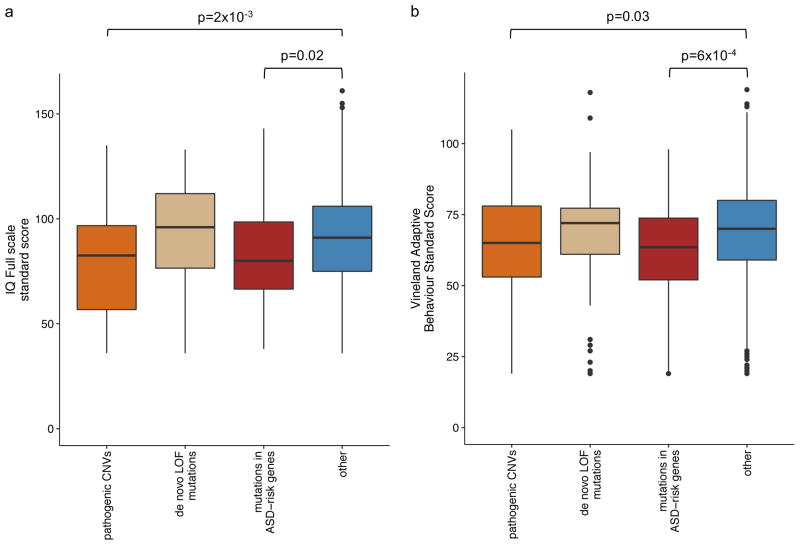
To assess the functional impact of genotypes, we compared the phenotypes (listed in Table 2) of participants with: de novo LOF mutations, mutations in the ASD-risk genes, pathogenic CNVs and no identified mutation in ASD-risk genes/CNV. Only the differences in Vineland Adaptive Behavior Score (FDR=0.04) and IQ Full Scale Standard Score (FDR=5×10−4) were significant after multiple testing corrections using Benjamini Hochberg approach. Consistent with the previous studies41, 42, we found that individuals with pathogenic CNVs have significantly lower IQ (p=2×10−3, −8.5, 95%CI: −16 to −3) (Figure 5a). Similarly, individuals with mutations in ASD-risk genes also have a trend of lower IQ (p=0.02, −11, 95%CI: −15 to −1.6×10−6). More strikingly, however, we found that those individuals carrying mutations in ASD-risk genes have significantly lower Vineland adaptive ability score (p=6×10−4, −6.5, 95%CI: −10 to −3) (Figure 5b). Given that Vineland adaptive score captures the adaptive functioning better than cognitive ability, it may suggest that the ASD-risk genes identified here are more specific to ASD behavioural traits than general cognitive deficits43.
Figure 5. Phenotype comparison for the samples with and without identified mutations.
Standard score of (a) IQ Full scale and (b) Vineland Adaptive Behavior were compared between samples with pathogenic CNVs, de novo LOF mutations, mutations in ASD-risk genes and other samples without any of these mutations.
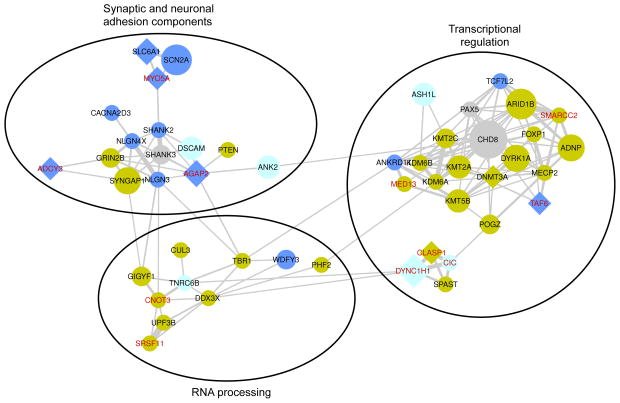
Many of the ASD-risk genes (80%; 49/61) identified connected into gene networks (Figure 6). These genes are enriched in synaptic transmission, transcriptional regulation and RNA processing functions, consistent with previous findings17, 21. We found genes associated with transcriptional regulation and RNA processing more often expressed in brain prenatally, while synaptic function related genes are expressed in brain throughout development44. Our extended gene network revealed more interactions of genes. The novel ASD-risk genes such as SRSF11 may closely interact with the known ASD-risk genes such as UPF3B (Figure 6).
Figure 6. Interaction similarity network of ASD-risk genes.
Connections represent gene similarity based on physical protein interactions and pathway interactions. Connection thickness is proportional to the fraction of interaction partners shared by the connected genes. The size of the node for each gene is proportional to the total mutation count (Figure 3). Genes associated with LOF mutations are in circle shape, while genes associated with missense mutations are in diamond shape. The node color corresponds to the BrainSpan brain expression Principal Component 1 (prenatal in yellow, postnatal in blue, balanced in light blue, undetermined in grey). The labels of novel ASD-risk genes are displayed in red. The network was visualized using Cytoscape.
Data access and processing
All data are available in the MSSNG Google Cloud or linked databases, with Autism Speaks as the MSSNG Data Custodian. A web-based portal was also developed (Figure 1 and 4, Supplementary Figure 3). Example queries include retrieving predicted damaging variants for one (or more) genes of interest, or all damaging de novo variants in a subject. In addition, variant annotations, sequence – read pile-ups (using the Integrative Genomics Viewer plugin) and psychometric measurements can be accessed. Researchers receive authorization from the MSSNG’s Data Access Committee via an online application (https://research.mss.ng/). Autism Speaks uses the Public Population Project in Genomics and Society (P3G) to independently recommend access according to guidelines established by Autism Speaks and P3G, based on consents provided by the data donors or on REB-approved waivers of consent for retrospective collections.
Discussion
Considering the breadth of data in our pilot WGS studies 14, 18, 21 and global impact of the ASDs, it became evident that an ‘Autism WGS Project’ encouraging use of data in a manner as unrestricted as possible, for wide-ranging research questions, would be beneficial. We could move quickly because investment to develop biosample repositories from individuals with ASD and their families, consented for genetic research, have already been made. The resources generated and managed through MSSNG support ASD research in three related areas, namely, (1) new gene discovery and diagnostics, (2) genetic disease pathways, mechanisms and pharmacologic development and trials, and (3) open-science queries of any type including exploring the significant heterogeneity underlying ASD, as well as the non-coding genome, most of which can only start to be conceptualized now that the resource exists.
First, using the statistical framework defining genes with higher than expected mutation rate, we have already identified 18 new candidate genes for ASD or associated complications (Figure 3 highlights ~100 diagnostic loci for ASD). Some of the newly detected mutations could reasonably be considered pathogenic and/or have possible implications for clinical management or genetic counselling for the subject or family members4, 8. Examples include screening for cardiac defects or maturity-onset diabetes of the young in cases with 1q21.1 or 17q12 deletions, respectively, secondary prevention to avoid the development of obesity in those with 16p11.2 microdeletion4, 45, and monitoring the use of growth factors (e.g. IGF-1) in PTEN mutation carriers who may react negatively8. In numerous other cases – including all instances of CNV and chromosomal abnormalities – detection of the mutation would lead to prioritization of these individuals for comprehensive clinical assessment and referral for earlier intervention3, 4, and end long-sought questions of causation8, 16. For other mutations, the role in ASD needs to be closely followed in the literature. Having the data accessible in the portal browser will continue to enable diagnosticians worldwide to remotely perform genotype-phenotype exploration of new testing results, against the latest WGS research data.
Second, 80% of the 61 ASD-risk genes on our refined list are connected in networks representing potential targets for pharmacologic intervention19. Sixteen genes contained subdomains that could be targeted by pharmaceutical intervention and 7 for which specific drugs-gene interactions are known (Supplementary Table 6). For example, individuals with mutations in SCN2A identify carriers as potential candidates for drug trials involving allosteric modulators of GABA receptors46. By extending the search to genes affected by CNV and/or to proteins that interact with or regulate these genes, the potential targets for modulating the pathways impacted in ASD expands. Additionally, the focus here was on gene products that could be pharmacologically modulated with small molecules, but the use of technologies such oligonucleotide-based therapeutics or gene therapy further increases the list of potential interventions that could be utilized in addressing the biological deficits created by the loss of function of these genes.
Third, solving the problem of the significant heterogeneity involved in ASDs will benefit by expanding this initiative, including partnering with other WGS projects and coordinating all information in a single open-science platform for which MSSNG provides a foundation. Regarding genotypic heterogeneity, using established criteria17, 47, in this study we were able to resolve a molecular basis in 11.2% of ASD cases and this tally should rise with more rare variant data to compare against22. An important message from our study was validation of the findings that CNV and chromosomal alterations contribute significantly in ASD (Figure 3b). These genetic alterations also often encompass multiple genes (Figure 3b), isoforms of single genes48, their regulatory elements27, 49, and can include non-coding genes such as the known ASD-risk gene PTCHD1-AS (Figure 4c), and combinations of mutations (7 cases have both ASD-relevant SNVs and CNVs), necessitating the use of a comprehensive technology like WGS.
Regarding phenotypic heterogeneity, our previous analysis of a subset of multiplex families in the MSSNG resource already showed that siblings with discordant mutations tended to demonstrate more within sib-ship variability than those who shared a risk variant18 Here, with the increased sample size and access to richer phenotypic measures, our data reveal that participants bearing mutations in ASD susceptibility genes had lower adaptive ability compared to participants without identified risk variants. Adaptive functioning as measured here using the Vineland Adaptive Behavior Score is, in fact, composed of estimates of socialization, communication, daily living and motor skills50. This finding needs to be further dissected to determine whether the association with risk variant is specific to one of these sub-domains or is more linked to the composite. The same is true for the association with IQ.
Large-scale computing, for this project, can be done from within the MSSNG cloud, and/or using the investigator’s local resources. Our intent, as we have already started, is that researchers will ‘move new code to the data’ (i.e. to access data for analysis with the cloud platform), in particular for massive WGS and phenotypic queries including performing meta-analyses incorporating their own data. Ultimately, the new information arising should then be broadly shared. MSSNG researchers, for example, can use open-standard tools supported by the Global Alliance for Genomics and Health Application Programming Interfaces (APIs)31, so that tools developed by individual groups can be applied to data published elsewhere. This kind of continued interactive participation in shared open-access research will continue to enable a better understanding of ASD, and set a course for other genomic initiatives in neuroscience.
Online Methods
Samples for whole-genome sequencing and data access policy
We collected 5,205 unique samples (5,193 individuals) from 2,066 unique families with children diagnosed with ASD. The cohort consists of 2,618 children with ASD (1,740 probands and 878 affected siblings). Details on the collections the samples were drawn from are described in Supplementary Table 7. Data collection and analysis were not performed blind to the conditions of the experiments”. We recruited other siblings and members of the family across generations whenever possible. We obtained informed consents, or waiver of consent, which were approved by the Western Institutional Review Board, Montreal Children’s Hospital – McGill University Health Centre Research Ethics Board, McMaster University – Hamilton Integrated Research Ethics Board, Eastern Health Research Ethics Board, Holland Bloorview Research Ethics Board and The Hospital for Sick Children Research Ethics Board. According to the consents or waiver of consent, participants agreed to make their coded genetic and phenotypic information available to researchers to help in the discovery of the DNA alterations underlying ASD, and ASD-related disorders. Their coded data were uploaded to the MSSNG Google Cloud database. Based on the current approved consent the genomic and phenotypic data can be submitted to this type of online database provided that all data is coded, that access to data is controlled and that specific data access policies are in place. The data access policy generated by the legal team at Autism Speaks was modelled on accepted practice in international research consortia such as the International Cancer Genome Consortium (ICGC). A researcher seeking to gain access to the data, and perform their analyses in the cloud-based environment or download the data to use their own analysis tools, will have to apply for access following the process outlined in the Data Access Policy (Supplementary note). Sequencing data is coded and access to the data is controlled and governed via an REB/IRB approved data access policy (Western Institution Review Board for use of AGRE data, and other Review Boards for specific sites contributing data). At the time or writing, 7,214 samples from individuals with ASD or their family members were analyzed by WGS and available. The goal of this project was to collect a large cohort of families to facilitate genetic analysis as previously described22. No statistical methods were used to pre-determine sample sizes.
Researchers can access data at multiple stages and levels of analysis: 1) through the MSSNG portal, which provides an interface for searching, filtering and browsing the final, curated variants, annotations and statistics via a web application; 2) using BigQuery tables (a petabyte-scale distributed data warehousing (storage) and analytics (query) service) under their own account to perform custom queries, which allows flexibility for development of new analyses and applications; 3) via user’s own Google Cloud Storage (GCS) bucket on request for raw sequencing data and results of primary mapping (BAM files) and variant calling (MasterVar, VCF, gVCF) processes. At the time of writing, 75 researchers from 17 institutions in four countries (Canada, South Korea, UK and USA) were approved access to MSSNG data.
WGS and data storage
We extracted DNA from whole-blood or lymphoblast-derived cell lines (LCLs). We assessed the DNA quality by PicoGreen and gel electrophoresis. We sequenced the 5,205 genomes using Complete Genomics (n=1,233), Illumina HiSeq 2000 (n=561) and HiSeq X (n=3,411) technology. WGS by Complete Genomics (Mountain View, CA) and Illumina HiSeq 2000 were performed as previously described14, 18, 21. For WGS by Illumina HiSeq X, we used between 100ng and 1ug of genomic DNA for genomic library preparation and whole genome sequencing. We quantified DNA samples using Qubit High Sensitivity Assay and checked sample purity using Nanodrop OD260/280 ratio. We used the Illumina TruSeq Nano DNA Library Prep Kit following the manufacturer’s recommended protocol. In brief, we fragmented the DNA into 350 bp average length using sonication on a Covaris LE220 instrument. The fragmented DNA was end-repaired, A-tailed and indexed TruSeq Illumina adapters with overhang-T added to the DNA. We validated the libraries on a Bioanalyzer DNA High Sensitivity chip to check for size and absence of primer dimers, and quantified it by qPCR using Kapa Library Quantification Illumina/ABI Prism Kit protocol (KAPA Biosystems). We pooled the validated libraries in equimolar quantities and sequenced the paired-end reads of 150-bases in length on an Illumina HiSeq X platform following Illumina’s recommended protocol.
For samples sequenced on Illumina platforms, raw reads are uploaded to GCS. For samples sequenced by Complete Genomics, only analyzed results from the Complete Genomics pipeline were uploaded to GCS. Results of variant calling and filtering pipelines were also uploaded to GCS for permanent archiving, sharing with MSSNG researchers, and processing into BigQuery tables for access via the portal.
Alignment and variant calling
Alignment and variant calling for genomes sequenced by Complete Genomics were performed as previously described18. For genomes sequenced by Illumina platforms, we processed them on Google Cloud using Google Genomics Application Programming Interfaces with a pipeline that follows the Best Practices recommended by the Broad Institute51. We inputted primarily the paired FASTQ files (with a few samples processed from Binary Alignment Maps/BAMs). We aligned the reads to the reference genome (build GRCh37) using the Burrows-Wheeler Aligner (BWA; version 0.7.10). We removed duplicated reads using Picard (version 1.117). We performed local realignment and quality recalibration with the Genome Analysis Toolkit (GATK; version 3.3) by each chromosome. We detected single SNVs and indels using GATK with HaplotypeCaller. We extracted non-variant segments (reference intervals) that were emitted by HaplotypeCaller using a custom Java program (NonVariantSiteFilter.jar). The output file was generated in the universal Variant Call Format (VCF). Both the VCF output by this process and the calls from Complete Genomics samples (MasterVar) were converted to separate variants and reference blocks in VCF and saved in GCS. The variants and reference blocks were imported into Google Genomics, then exported to a BigQuery table.
Sample quality controls
We performed quality control checks for samples utilizing codes from Google Genomics Codelab following the methodology developed previously52. We performed i) Duplicate Samples, ii) Samples per Platform, iii) Genome Call Rate, iv) Missingness Rate, v) Singleton Rate, vi) Heterozyosity Rate, vii) Homozygosity Rate, viii) Ti/Tv ratio, ix) Inbreeding Coefficient, and x) Sex Inference. To reduce the batch and cross-platform effects for analysis, we applied additional quality filters to remove variants caused by technical issues. We required variants to have genotype quality (GQ for Illumina; VAF for Complete Genomics) of at least 99. Since our analyses focused on rare variants, we required variants to be found in the population less than 1% of the time. We also required the variant to be called more than 95% of the time as a reference allele and less than 1% of time as a variant in the parents. Batch and cross-platform biases were substantially reduced after filtering (Figure 2). Detailed procedures and findings can be found in the Supplementary note.
Detection of de novo SNVs and indels
As described previously14, 18, 21, we considered a variant to be a potential de novo mutation when it is inconsistent with Mendelian inheritance (present in the offspring, but not in either parent). For a variant in the autosomal region, we considered it to be a potential de novo mutation when there was a heterozygous alternative genotype in the offspring, and homozygous reference genotypes in both parents. For a variant in the X chromosome, we considered male and female offspring with different criteria: in sex-specific regions of male offspring, we considered it to be a de novo variant when there was a hemizygous alternative genotype in the offspring and a homozygous reference genotype in the mother. We considered X-linked variants in female offspring and X-linked variants in pseudoautosomal regions in male offspring as for autosomal regions. We considered a variant in the Y chromosome to be de novo when a hemizygous alternative variant was present in the male offspring but absent at the same position in the father.
We performed de novo SNV and indel detection from Complete Genomics data as previously described18, except that here we considered both parents with each offspring in the same family as separate trios. We used DenovoGear (version 0.5.4) for de novo SNV and indel detection on genomes sequenced by Illumina platforms, running the program by each chromosome. We also extracted high quality variants (passed quality filter) that were inconsistent with Mendelian inheritance based on GATK with allelic frequency among parents less than 1%. We define a putative de novo SNV as if it has a pp_DNM from DenovoGear higher than 0.95 and overlap with GATK calls (GQ of at least 99). We define a putative de novo indel if it is found by both DenovoGear and GATK methods with the same start site. In addition, we have also performed a manual inspection on the quality of variants by inspecting reads from the BAM for the variants found to be de novo by DenovoGear or GATK for the ASD-risk genes.
We used Primer 3 to design primers to span at least 100 bp upstream and downstream of a putative variant. In designing primers, we avoided regions of repetitive elements, segmental duplication or known SNPs. We randomly selected putative de novo SNVs from the Illumina WGS data of two probands (2-1266-003 and 3-0141-000) in the trio families and from Complete Genomics WGS data of one proband (2-1292-003) in a quartet family for Sanger sequencing (Supplementary Table 3). In addition, by Sanger sequencing we validated all the de novo LOF SNVs and indels and reported pathogenic variants from all families, using DNA from whole blood. Candidate regions were amplified by PCR for all families and assayed by Sanger sequencing (Supplementary Table 3).
No substantial difference on de novo mutation detection rate (average number of de novo mutation for CG: 88.9; Illumina: 85.2) or distribution (Supplementary Figure 4) was found between platforms. There is a difference on the validation rate for de novo LOF mutations between two platforms (CG: 78%; Illumina: 92%), but samples from CG only constitute 23.7% of the total samples (Table 1). We found 27.9% of total exonic de novo mutations were contributed by CG, which is proportional to the given number of samples.
Identification ASD-risk genes
We performed a meta-analysis of de novo mutations for identification of ASD-risk genes. We concatenated the de novo mutations detected here with those detected from five other previous large-scale systematic whole-exome or WGS studies (from a total of 4,087 trios)13, 23, 33–35. To avoid sample duplication, we have checked through registry that none of the samples from MSSNG were studied in the previous large-scale exome or WGS studies. Since the raw data for the previous studies were not easily accessible, we could not identify duplicated sample based on the genotypes. However, we checked the possibility of duplicated samples based on the de novo mutation profiles given by each study. Focusing on exonic de novo mutations examined, there were only four pairs of samples sharing the same de novo variant out of the 4,087 trios examined. Two of these pairs were found within same studies. While these pairs could be derived from the same samples, they only constitute a small portion (<0.1%) of the cohort.
The variants were re-annotated using our custom annotation pipeline (see below). There were a total of 2,864 de novo missense mutations and 599 de novo LOF mutations reported. Combining with the de novo mutations detected in the present study (Supplementary Table 3), we performed a statistical analysis to identify genes with higher than expected mutation rate based on the model framework described previously47. Observed rate of de novo mutation for each gene was compared with its expected rate using binominal test. To address for the potential bias on mutation rate between observed data and expected simulation, we rescaled the statistics with a constant, k, which is derived from the ratio of overall de novo mutation rate observed to that expected. For LOF mutations, we required genes to have at least 2 de novo LOF mutations and a probability of loss-of-function intolerant rate (pLI) of >0.9. For missense mutations, we required genes to have at least 4 de novo missense mutations and a missense z-score of >0.2 (derived based on scores from known ASD-risk genes and comparable gene number distribution as pLI>0.9). We corrected p values with Benjamini-Hochberg procedure and defined candidate ASD-risk genes as having a false discovery rate (FDR) <0.15. We have also analyzed X-linked LOF mutations. We defined candidate ASD-risk genes as having at least two LOF mutations found in males or de novo LOF in females, and required genes to have pLI>0.65 (since X-linked genes and autosomal genes have different constraint, we derived the score for X-linked genes from the score of MECP2: pLI=0.69).
SNV and indel annotation
We annotated the variants on Google Cloud Engine using a custom pipeline based on Annovar as previously described14, 18, 21, 53. The annotation process infrastructure includes a separate internal portal, which automates distribution of annotation jobs in parallel over a dedicated Virtual Machine (VM)-based computing infrastructure cluster. The variant annotations were then exported to a BigQuery table. Variant information was downloaded from databases for the allele frequency (Exome aggregation Consortium36, 1000 Genomes54, NHLBI-ESP55 and internal Complete Genomics control databases), genomic conservation (UCSC PhyloP and phastCons for placental mammals and 100 vertebrates56), variant impact predictors (SIFT57, PolyPhen258, Mutation Assessor59 and CADD60), and implication in human genetic disorders (Human Phenotype Ontology61, Human Gene Mutation Database62 and Clinical Genomics Database63). Detailed descriptions of the annotation effort can be found in Supplementary note.
Genetic network construction
For each of the 61 ASD-risk genes, we retrieved top 200 closely interacting gene neighbors using GeneMANIA64. We generated an aggregate interaction network in GeneMANIA, based on physical protein interaction and pathways with the “Gene Ontology Biological Process” weighting option. We then computed a pairwise weighted jaccard index to model the similarity of the genes’ interacting neighborhoods, resulting in the final gene network (Figure 6). Finally, we performed hierarchical clustering, and manually optimized the weighted jaccard index cutoff for displaying the gene network in Cytoscape65, so that the gene clusters suggested by the network layout algorithm are similar to the clusters suggested by hierarchical clustering.
CNV analysis
For samples sequenced on Illumina platforms, we detected CNVs from WGS for each sample using two algorithms, CNVnator66 and ERDS67, as previously described14, 21. Algorithms were run using default parameters. We used 500bp as window size for CNVnator. For CNVnator, we removed calls with >50% of q0 (zero mapping quality) reads within the CNV regions (q0 filter), except for the homozygous autosomal deletions or hemizygous X-linked deletions in males (with normalized average read depth; NRD<0.03). We defined stringent calls as those that were called by both algorithms (with 50% overlap). For samples sequenced by Complete Genomics, CNV calls were taken as provided as described previously18. Sixty-five samples have a total number of CNVs >= 2 standard deviations of the average number, including 28 affected individuals. We retained CNVs with size >2kb. We defined a rare CNV as that found in less than 1% of the time in the parents, less than 0.1% in the population from microarray data and overlap with a region that is at least 75% copy number stable according to the copy number variation map of the human genome68. We have also performed a manual inspection on the quality of CNVs by inspecting reads from the BAM for confirmation.
We also analyzed CNV data for 1,658 affected individuals genotyped on one or more of the following microarrays: Illumina1M single or duo; Affymetrix CytoScan HD; Affymetrix single-nucleotide polymorphism 6.0; Illumina OMNI 2.5M; Agilent 1M CGH array; Affymetrix GeneChip Human Mapping 500K Array (Supplementary Table 8). We defined rare, stringent CNVs as previously described17, and also required them to overlap a region that is at least 75% copy number stable according to the copy number variation map of the human genome68.
We determined pathogenic CNVs as those resulting in chromosome abnormities; large rare CNVs between 3 and 10Mb in size; genomic disorders with recurrent breakpoints (including all DECIPHER loci and other loci known to be associated with ASD10, 17) and CNVs impacting coding exons of known ASD-risk genes or noncoding exons of PTCHD1-AS or MBD5. All pathogenic CNVs found by microarray were found by WGS, except CNVs that were filtered out based on the quality issues or size difference (Supplementary Table 8).
Statistical tests
We compared the scores for phenotype tests (Table 2) available for four groups of samples: (a) samples with pathogenic CNVs (n=177), (b) de novo LOF mutations (n=170), (c) mutations in ASD-risk genes (n=116) and (d) other samples without any of these mutations (2,153). Samples included in each category were mutually exclusive to each other and no replicates (randomization not applicable). Phenotype tests investigated included (i) Vineland Adaptive Behavior Standard Score, (ii) Repetitive Behaviour Scale Revised Overall Score, (iii) Repetitive Behaviour Scale Overall Score, (iv) Social Responsiveness Total T Score, (v) Social Communication Questionnaire Total Score, (vi) Language OWLS Total Standard Score, (vii) Language PLS Total Standard Score, (viii) IQ Full Scale Standard Score, and (ix) IQ Non-verbal Standard Score. Data distribution was assumed to be normal but this was not formally tested. We performed ANOVA for the mean difference of the 4 groups in each of these tests (degree of freedom (df)=3 in i, df=3 in ii, df=2 in iii, df=3 in iv, df=3 in v, df=3 in vi, df=3 in vii, df=3 in viii and df=1 in ix). The difference between samples with mutations and samples without mutations was further tested using Wilcoxon signed-rank Test (one-sided) since they were not normally distributed.
Gene-based drug targets
The 61 genes listed in Figure 3a were annotated utilizing D.A.V.I.D.69 for a number of gene ontology categories and structural elements including PFAM subdomains. The PFAM labels were compared to lists of protein families generally considered to be druggable70, 71. To identify previously validated gene-drug interactions, the 61 gene list was used to search the Drug Gene Interaction Database72 (http://dgidb.genome.wustl.edu/). Only those results with associated peer-reviewed publications were reported.
Acknowledgments
We thank the families for their participation in the study, The Centre for Applied Genomics and Google for their analytical and technical support, as well as staff at Autism Speaks for organizational and fundraising support. This work was funded by Autism Speaks, Autism Speaks Canada, the Canadian Institute for Advanced Research, the University of Toronto McLaughlin Centre, Genome Canada/Ontario Genomics Institute, the Government of Ontario, the Canadian Institutes of Health Research (CIHR), NeuroDevNet, Ontario Brain Institute, the Catherine and Maxwell Meighen Foundation, The Hospital for Sick Children Foundation. Special thanks to Bob and (the late) Suzanne Wright for their vision in helping to conceptualize and develop this project and to foundational philanthropic supporters Charles Dolan, Gordon Gund, Bernie Marcus, Vanessa and Jonathan Morgan and Steven Wise. R.K.C.Y. holds CIHR Postdoctoral Fellowship, NARSAD Young Investigator award and Thrasher Early Career Award. R.W. is funded by the Ontario Brain Institute and NeuroDevNet. M.U. holds the Banting Postdoctoral Fellowship. M.W. is supported by CIHR (Institute of Genetics) Clinical Investigatorship Award. L.Z. is supported by the Stollery Children’s Hospital Foundation Chair in Autism Research. P.S. holds the Patsy and Jamie Anderson Chair in Child and Youth Mental Health. B.M.K. holds the Canada Research Chair in Law and Medicine. S.W.S. holds the GlaxoSmithKline-CIHR Chair in Genome Sciences at the University of Toronto and The Hospital for Sick Children.
Footnotes
Accession Codes: Sequence data have been deposited at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/), which is hosted by the EBI, under accession number EGAS00001001023 and EGAS00001000850.
Data Availability: All sequence data can be accessed through the MSSNG database on Google Genomics (for access, see http://mss.ng/researchers).
Code availability: Codes used in MSSNG database can be found in the Supplementary note. Others are available upon request.
Author Contributions: R.K.C.Y. and S.W.S. conceived and designed the experiments. R.K.C.Y., D.M., M.B., B.T., R.V.P., J.Whitney, N.D., J.Bingham, Z.W., G.P. and S.W. processed and analyzed the whole genome sequencing data. S.W., L.D., A.J.S.C., S.K., T.P., E.J.H. and S.L. designed and performed experiments for variant characterization and validation. J.A.B., C.R.M., M.U., M.Z., E.D., S.L.P., W.E., K.H., W.L., J.R.M., T.N., W.W.L.S., F.J.T., J.Wei, L.X., W.V.E., S.T., B.J.F. J.T.R. and L.J.S helped perform different components of analysis and validation experiments. R.K.C.Y., M.B., J.L.H., R.H.R., D.G., M.T.P. and S.W.S. coordinated the whole genome sequencing experiments. R.K.C.Y., R.H.R., D.G., M.T.P. and S.W.S. conceived and coordinated the project. N.H., A-M.T., E.K., W.R., I.D., S.J., B.M.M., B.K., M.S., C.Cytrynbaum, R.W., L.Z., M.W-S., J.Brian, L.S, A.I., K.D-T., A.T., C.Chrysler, J.L., T.S-L., I.M.S., X.L., R.N., V.S., A.F., E.H.C., S.D., A.E., L.G., B.A.M., J.R.P., S.J.S., J.V., B.A.F., M.E., M.T.C., J.H., B.M.K., E.A. and P.S. managed, recruited, diagnosed and examined the recruited participants. R.K.C.Y. and S.W.S. wrote the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2014;383:896–910. doi: 10.1016/S0140-6736(13)61539-1. [DOI] [PubMed] [Google Scholar]
- 2.Robinson EB, et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet. 2016;48:552–555. doi: 10.1038/ng.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anagnostou E, et al. Autism spectrum disorder: advances in evidence-based practice. CMAJ. 2014;186:509–519. doi: 10.1503/cmaj.121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter MT, Scherer SW. Autism spectrum disorder in the genetics clinic: a review. Clin Genet. 2013;83:399–407. doi: 10.1111/cge.12101. [DOI] [PubMed] [Google Scholar]
- 5.Miles JH. Complex Autism Spectrum Disorders and Cutting-Edge Molecular Diagnostic Tests. JAMA. 2015;314:879–880. doi: 10.1001/jama.2015.9577. [DOI] [PubMed] [Google Scholar]
- 6.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16:551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 7.de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat Med. 2016;22:345–361. doi: 10.1038/nm.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scherer SW, Dawson G. Risk factors for autism: translating genomic discoveries into diagnostics. Hum Genet. 2011;130:123–148. doi: 10.1007/s00439-011-1037-2. [DOI] [PubMed] [Google Scholar]
- 9.Tammimies K, et al. Molecular Diagnostic Yield of Chromosomal Microarray Analysis and Whole-Exome Sequencing in Children With Autism Spectrum Disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 10.Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 11.Autism Genome Project, C., et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leblond CS, et al. Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments. PLoS Genet. 2014;10:e1004580. doi: 10.1371/journal.pgen.1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang YH, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller DT, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuen RK, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 19.Sahin M, Sur M. Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders. Science. 2015:350. doi: 10.1126/science.aab3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stavropoulos DJ, et al. Whole-genome sequencing expands diagnostic utility and improves clinical management in paediatric medicine. Npj Genomic Medicine. 2016;1:15012. doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuen RK, et al. Genome-wide characteristics of de novo mutations in autism. Npj Genom Medicine. 2016;1:16027. doi: 10.1038/npjgenmed.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buxbaum JD, et al. The autism sequencing consortium: large-scale, high-throughput sequencing in autism spectrum disorders. Neuron. 2012;76:1052–1056. doi: 10.1016/j.neuron.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders SJ, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leblond CS, et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noor A, et al. Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci Transl Med. 2010;2:49ra68. doi: 10.1126/scitranslmed.3001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong HY, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anney R, et al. Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Hum Mol Genet. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glazer D. Atoms, bits, and cells. Appl Transl Genom. 2015;6:11–14. doi: 10.1016/j.atg.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Global Alliance for, G. & Health. GENOMICS. A federated ecosystem for sharing genomic, clinical data. Science. 2016;352:1278–1280. doi: 10.1126/science.aaf6162. [DOI] [PubMed] [Google Scholar]
- 32.Stein LD. The case for cloud computing in genome informatics. Genome Biol. 2010;11:207. doi: 10.1186/gb-2010-11-5-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An JY, et al. Towards a molecular characterization of autism spectrum disorders: an exome sequencing and systems approach. Transl Psychiatry. 2014;4:e394. doi: 10.1038/tp.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michaelson JJ, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. 2016 doi: 10.1038/nature19057. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards CS, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 38.Utami KH, et al. Impaired development of neural-crest cell-derived organs and intellectual disability caused by MED13L haploinsufficiency. Hum Mutat. 2014;35:1311–1320. doi: 10.1002/humu.22636. [DOI] [PubMed] [Google Scholar]
- 39.Stender JD, et al. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciernia AV, LaSalle J. The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nat Rev Neurosci. 2016;17:411–423. doi: 10.1038/nrn.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mannik K, et al. Copy number variations and cognitive phenotypes in unselected populations. JAMA. 2015;313:2044–2054. doi: 10.1001/jama.2015.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ameis SH, et al. A Diffusion Tensor Imaging Study in Children With ADHD, Autism Spectrum Disorder, OCD, and Matched Controls: Distinct and Non-Distinct White Matter Disruption and Dimensional Brain-Behavior Relationships. Am J Psychiatry. 2016 doi: 10.1176/appi.ajp.2016.15111435. appiajp201615111435. [DOI] [PubMed] [Google Scholar]
- 44.Uddin M, et al. Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat Genet. 2014;46:742–747. doi: 10.1038/ng.2980. [DOI] [PubMed] [Google Scholar]
- 45.Jacquemont S, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hadley D, et al. The impact of the metabotropic glutamate receptor and other gene family interaction networks on autism. Nat Commun. 2014;5:4074. doi: 10.1038/ncomms5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corominas R, et al. Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat Commun. 2014;5:3650. doi: 10.1038/ncomms4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker S, Scherer SW. Identification of candidate intergenic risk loci in autism spectrum disorder. BMC Genomics. 2013;14:499. doi: 10.1186/1471-2164-14-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sparrow SS, Balla DA, Cicchetti DV, Harrison PL, Doll EA. Vineland adaptive behavior scales. 1984. [Google Scholar]
- 51.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 10 11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan C, et al. Interactive Analytics for Very Large Scale Genomic Data. 2015 bioRxiv. [Google Scholar]
- 53.Merico D, et al. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman Syndrome by disrupting minor intron splicing. Nat Commun. 2015;6:8718. doi: 10.1038/ncomms9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genomes Project, C., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 58.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohler S, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stenson PD, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 63.Solomon BD, Nguyen AD, Bear KA, Wolfsberg TG. Clinical genomic database. Proc Natl Acad Sci U S A. 2013;110:9851–9855. doi: 10.1073/pnas.1302575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu M, et al. Using ERDS to infer copy-number variants in high-coverage genomes. Am J Hum Genet. 2012;91:408–421. doi: 10.1016/j.ajhg.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 69.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 70.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 71.Liu K, Liu Y, Lau JL, Min J. Epigenetic targets and drug discovery Part 2: Histone demethylation and DNA methylation. Pharmacol Ther. 2015;151:121–140. doi: 10.1016/j.pharmthera.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 72.Wagner AH, et al. DGIdb 2.0: mining clinically relevant drug-gene interactions. Nucleic Acids Res. 2016;44:D1036–1044. doi: 10.1093/nar/gkv1165. [DOI] [PMC free article] [PubMed] [Google Scholar]