Summary
Candida albicans is well adapted to its host and is able to sense and respond to the nutrients available within. We have shown that C. albicans avidly utilizes amino acids as a carbon source, which allows this opportunistic pathogen to neutralize acidic environments, including the macrophage phagosome. The transcription factor Stp2 is a key regulator of this phenomenon, and we sought to understand the mechanism of activation of Stp2, focusing on the SPS sensor system previously characterized for its role in nitrogen acquisition. We generated deletion mutants of the three components, SSY1, PTR3 and SSY5 and demonstrated that these strains utilize amino acids poorly as carbon source, cannot neutralize the medium in response to these nutrients, and have reduced ammonia release. Exogenous amino acids rapidly induce proteolytic processing of Stp2 and nuclear translocation in an SPS-dependent manner. A truncated version of Stp2, lacking the amino terminal nuclear exclusion domain, could suppress the growth and pH neutralization defects of the SPS mutants. We showed that the SPS system is required for normal resistance of C. albicans to macrophages and that mutants defective in this system reside in more acidic phagosomes compared with wild type cells; however, a more equivocal contribution was observed in the murine model of disseminated candidiasis. Taken together, these results indicate that the SPS system is activated under carbon starvation conditions resembling host environments, regulating Stp2 functions necessary for amino acid catabolism and normal interactions with innate immune cells.
Introduction
The opportunistic fungal pathogen Candida albicans is a polymorphic yeast intimately associated with the human host. It is able to colonize diverse niches in the body including mucosal surfaces and the intestinal lumen, without causing any clinical symptoms (McManus and Coleman, 2014; Pérez and Johnson, 2013). However, several predisposing factors make the host susceptible to Candida infections, such as immunosuppression, disruption of anatomical barriers and disturbance of the normal microbiota (Brown et al., 2012; Fidel, 2006; Perlroth et al., 2007; Pfaller and Diekema, 2007).
The ability to utilize the nutrients available in the host to generate energy and biomass, so as to resist the host immune defenses, is an important criterion for any pathogen. C. albicans is metabolically flexible and capable of adapting to microenvironments in different anatomical sites (Wilson et al., 2009). In some of these environments, sugars are limiting, but organic acids such as lactate are available at relatively high concentrations, one example being the environment within the gut or the vaginal cavity (Brown et al., 2014). Other nutrient sources available in the host are proteins and peptides. To make use of these, C. albicans has an expanded family of secreted aspartic proteases and oligopeptide transporters that have a role in the acquisition of nutrients during colonization and infection (Hube et al., 1994; Naglik et al., 2003; Reuβ and Morschhäuser, 2006). In addition, several amino acid auxotrophs retain full virulence, indicating that a ready supply of these nutrients are found in the host (Kingsbury and McCusker, 2010; Noble and Johnson, 2005).
Professional phagocytes are one of the first lines of defense against C. albicans and employ diverse killing mechanisms to damage ingested fungal cells (Collette and Lorenz, 2011; Jiménez-López and Lorenz, 2013; Miramón et al., 2013), one of which is nutrient (specifically carbon) deprivation. Transcriptional and genetic data indicate that C. albicans adapts to this environment by upregulating metabolic pathways for alternative (non-sugar) carbon sources, such as the glyoxylate cycle, gluconeogenesis and fatty acid β-oxidation, as well as transport systems for oligopeptides, amino acids and carboxylic acids. Further, a number of these processes are required for virulence in cell culture and/or whole animal models (Barelle et al., 2006; Fradin et al., 2003; Fradin et al., 2005; Lorenz et al., 2004; Rubin-Bejerano et al., 2003).
C. albicans uses amino acids as a sole source of carbon with much greater avidity than other model fungi (Vylkova et al., 2011), and they appear to be a significant nutrient source for phagocytosed cells. In vitro, growth in glucose-poor, amino acid-rich conditions induces a dramatic increase in the media pH, which we have demonstrated is due to the excretion of amino acid-derived ammonia (Vylkova et al., 2011). Following phagocytosis, this adaptation allows C. albicans to interfere with the normal maturation and acidification of the phagosome, thereby promoting fungal survival (Danhof and Lorenz, 2015; Vylkova and Lorenz, 2014). Indeed, we have shown that cells lacking Stp2, a transcription factor (TF) known to regulate amino acid permease expression, fail to neutralize in vitro environments, occupy a more acidic phagolysosome and are impaired in multiple aspects of the Candida-macrophage interaction and in virulence in vivo (Vylkova and Lorenz, 2014; Vylkova et al., 2011).
Regulation of carbon utilization has been extensively studied in organisms such as Saccharomyces cerevisiae and Aspergillus nidulans (Turcotte et al., 2010). Glucose repression of genes required for utilization of non-fermentable carbon sources is mediated by Mig1 (CreA in filamentous fungi). When this repression is lifted, other TFs activate specific uptake and catabolic pathways to capitalize on the available nutrients. This structure is broadly conserved in C. albicans (Corvey et al., 2005; Zaragoza et al., 2000), although there has been some reassignment of TFs function (Ramírez and Lorenz, 2009) and intriguing adaptations in post-translational regulation (Sandai et al., 2012).
There is essentially no information on the regulatory networks that control utilization of amino acids as a carbon source, as model fungi have limited ability to do so. In contrast, more is known about their use as a source of nitrogen or for direct protein synthesis, which is regulated in part by Stp1 and Stp2, paralogous C2H2-type Zinc finger TFs. In S. cerevisiae, these TFs are activated by a proteolytic event regulated by the SPS system, comprised of Ssy1, Ptr3 and Ssy5 (Forsberg and Ljungdahl, 2001; Forsberg et al., 2001; Klasson et al., 1999). Ssy1 is an amino acid transporter homolog that senses amino acids in the extracellular environment, although it does not have transport activity. Ptr3 is a peripherally membrane-associated protein (Liu et al., 2008) that mediates the interaction of casein kinase I (CKI) with the last component of the system, Ssy5, a chymotrypsin-like serine endoprotease. This endoprotease becomes hyperphosphorylated in an inhibitory domain, leading to a proteolytic event that releases a catalytically active fragment (Abdel-Sater et al., 2004; Abdel-Sater et al., 2011; Omnus et al., 2011; Pfirrmann et al., 2010). In turn, Ssy5 processes Stp1 and Stp2 by removing a nuclear exclusion domain at the amino termini, thereby allowing translocation into the nucleus where they drive the expression of genes involved in amino acid utilization (Forsberg et al., 2001). Close homologs of these proteins exist in C. albicans where, in response to nitrogen starvation, they control the expression of two different set of genes: Stp1 induces the expression of genes involved in protein degradation and peptide transport, whereas Stp2 regulates the expression of genes involved in amino acid utilization (Martínez and Ljungdahl, 2005).
We have shown in C. albicans that Stp2 is required for the utilization and catabolism of amino acids as a carbon source and the resulting extrusion of ammonia (Vylkova et al., 2011), but the origin of these amino acids (from host or fungal pools) and the mechanism of activation of Stp2 under carbon starvation conditions has not been clear. To address these questions, we asked whether the SPS system also responds to carbon starvation and whether it, similar to Stp2, modulates the macrophage-fungal interaction. We demonstrate here that C. albicans requires a functional SPS system to grow on amino acids as a carbon source and to manipulate the extracellular pH. The SPS system regulates the proteolytic cleavage of a negative regulatory domain of Stp2 and expression of a truncated version of Stp2 lacking this domain suppresses the defects of mutants lacking a functional SPS system. The SPS system plays a major role in the interaction of C. albicans with macrophages, but it has a more modest role in the murine model of disseminated candidiasis. Our data suggest that the SPS system is a key player in the sensing of amino acids in the environment, regardless of eventual catabolic outcomes and that proper SPS function is important in facilitating fungal survival during phagocytosis.
Results
The SPS system mediates amino acid utilization and pH manipulation
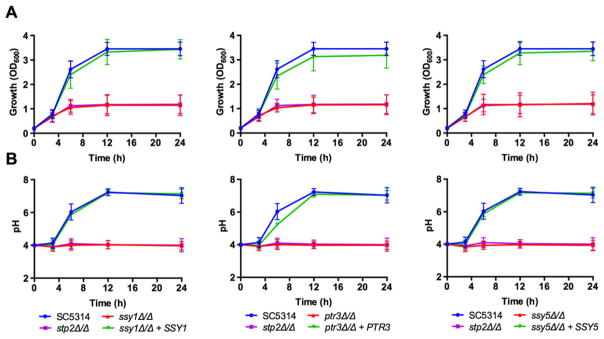
In order to investigate the role of the SPS system in amino acid utilization, we generated deletion strains lacking SSY1, PTR3 or SSY5, the three components of this system, in the SC5314 background as well as complemented strains. We first tested their ability to grow on amino acids as the sole carbon source in glucose-depleted conditions. As shown in Fig. 1A, the ssy1Δ/Δ, ptr3Δ/Δ and ssy5Δ/Δ strains grew poorly on YNB supplemented with 1% casamino acids; growth was similar to the stp2Δ/Δ strain. The ability to utilize this carbon source was restored to wild type levels upon reintegration of one allele of the respective gene. Similar to the stp2Δ/Δ strain, the SPS mutants were also unable to neutralize the initial acidic pH of the medium (Fig. 1B). The ssy1Δ/Δ, ptr3Δ/Δ and ssy5Δ/Δ strains behaved identically in growth and pH assays therefore we focused on ssy1Δ/Δ in further experiments, which lacks the transmembrane sensor protein.
Fig. 1.
The SPS system is required for amino acid utilization and environmental pH manipulation. Wild type SC5314 Candida albicans, ssy1Δ/Δ (CaPM07), ptr3Δ/Δ (CaPM15) and ssy5Δ/Δ (CaPM23), and complemented strains were grown at 37°C on yeast nitrogen base supplemented with 1% casamino acids, with an initial pH of 4.0. The stp2Δ/Δ (SVC17) strain was included for comparison purposes.
A. Growth was determined by measuring the OD600 at indicated time points.
B. Aliquots of the cultures were used to measure the pH of the medium at the same time points. Graphs show mean and standard deviation from three independent replicates.
To exclude the possibility that the inability to manipulate the pH of the medium is due to a general fitness defect, we supplemented the medium with glycerol, a carbon source that does not interfere with the ability to neutralize the pH in response to amino acids (Vylkova et al., 2011). As shown in Fig. S1A, glycerol greatly improves the growth of the three SPS mutant strains, reaching higher culture densities compared with medium without glycerol (Fig. S1A). Because the wild type and reconstituted strains are able to utilize both glycerol and amino acids, their culture densities remain higher than the SPS mutants. Despite the improved ability to grow in medium supplemented with glycerol, the SPS mutants were unable to neutralize the initial acidic pH (Fig. S1B), indicating that a functional SPS system is required to manipulate the external pH in response to amino acids.
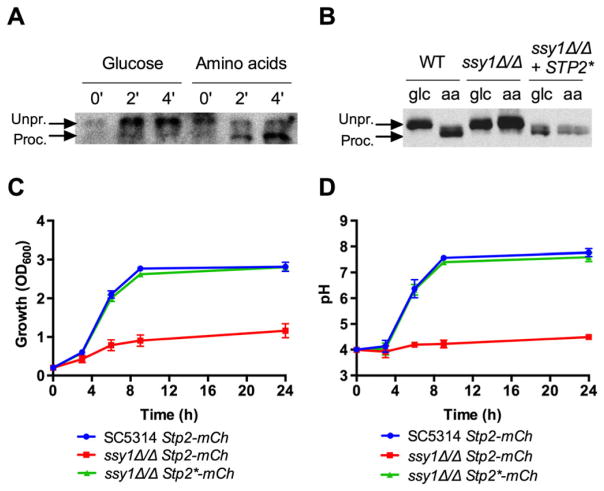
Cells growing in amino acid-rich media excrete ammonia derived from the amino and side chain amines and this drives the neutralization of the environment. Standard yeast media contains ammonium sulfate, and we have established that this can complicate assessment of ammonia generation (Danhof and Lorenz, 2015). Thus, we replaced the ammonium sulfate with allantoin. This nitrogen source supports optimal growth and pH changes, which are again impaired in the SPS mutants (Figs. 1A and B and data not shown). This confirms that the neutralization of the environmental pH is independent of the nitrogen source available in the medium. To demonstrate that ammonia is being excreted during growth on amino acids, we determined the release of ammonia by SC5314, ssy1Δ/Δ and reconstituted + SSY1 strains growing on solid YNBA medium (YNB-Allantoin) supplemented with casamino acids. As shown in Fig. 2C, SC5314 cells continuously released ammonia over the course of the experiment (72 h). In contrast, the ssy1Δ/Δ strain failed to release any detectable ammonia, which correlates with its inability to neutralize the environmental pH. As discussed later, a truncated allele of STP2 (STP2*) that mimics the proteolytically processed form suppresses all phenotypes of the ssy1Δ/Δ strain.
Fig. 2.
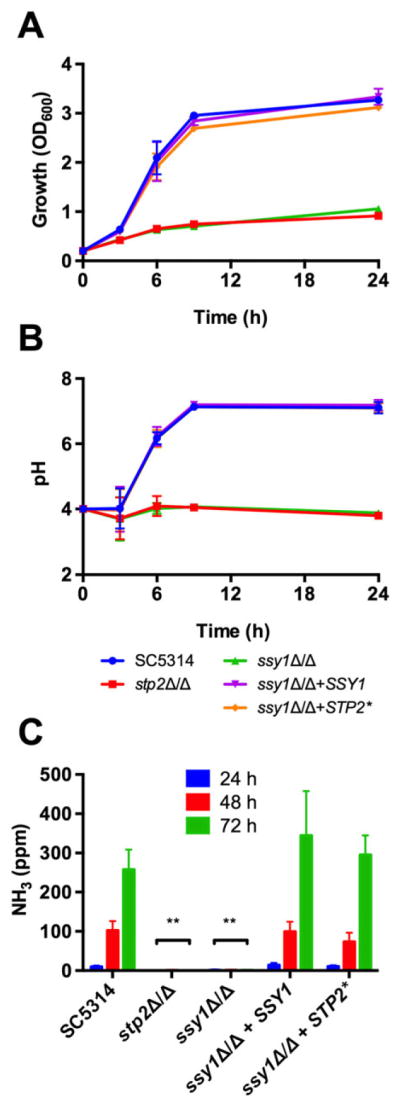
Growth and pH manipulation correlate with ammonia release. Wild type Candida albicans SC5314, ssy1Δ/Δ (CaPM07), ssy1Δ/Δ + SSY1 (CaPM25) and ssy1Δ/Δ + STP2* (CaPM36) strains were grown at 37°C on yeast nitrogen base allantoin (allantoin present as nitrogen source) with 1% casamino acids at an initial pH of 4.0. The stp2Δ/Δ (SVC17) strain was included for comparison purposes.
A. Growth was determined by measuring the OD600 at indicated time points.
B. Aliquots of the cultures were used to measure the pH of the medium at the same time points.
C. Ammonia excreted as byproduct of amino acid catabolism was collected in an acid trap and quantified with the Nessler’s reagent. Graphs show mean and standard deviation from three independent replicates.
The SPS system is responsible for Stp2 processing
The expression of genes necessary to utilize amino acids is controlled by the TF Stp2 (Martínez and Ljungdahl, 2005), which translocates to the nucleus because of a proteolytic cleavage that occurs in response to extracellular amino acids. We sought to determine the activation of Stp2 in wild type cells as well as in cells with a defective SPS system in the context of utilization of amino acids as carbon source. We generated a tagged version of Stp2 fused to mCherry at the carboxyl terminus to allow the identification of the protein by Western blotting and fluorescence microscopy.
To demonstrate activation of Stp2, cells were grown on glucose and amino acids. As shown previously in carbon replete conditions (Martínez and Ljungdahl, 2005), Stp2 is converted to a lower molecular weight form (from 94 kDa to ~82 kDa). This processing is extremely rapid when grown on amino acids, with a portion of Stp2 seen in the processed form within 2 min and the entire sample within 5 min, while Stp2 remained unprocessed even after 1 h incubation on glucose (Figs. 3A and S2A). Processing of Stp2 requires the SPS system, as ssy1Δ/Δ cells express only the full-length form, regardless of the carbon source (Fig. 3B). In agreement with Martínez and Ljungdahl (2005), Stp2 is processed only by a subset of amino acids (Fig. S2B), showing a certain degree of specificity.
Fig. 3.
Processing of Stp2 in response to amino acids depends on the SPS system. A. Western blot analysis to detect Stp2 processing in response to amino acids. Candida albicans cells expressing a single STP2 allele tagged with mCherry (CaPM41) were grown for 4 h in yeast nitrogen base supplemented with 2% glucose at 30°C, at an initial OD600 of 0.2. Cells were then exposed to yeast nitrogen base + 2% glucose or 1% casamino acids for the indicated time. Cell extracts were blotted and probed with an anti-mCherry antibody (see Materials and methods for details). The higher band corresponds to the unprocessed Stp2 (‘Unpr.’), whereas the lower one represents the processed version (‘Proc.’). B. Processing of Stp2 was not observed in ssy1Δ/Δ expressing STP2-mCherry (CaPM43) after exposure to glucose or amino acids for 1 h. As a control, an ssy1Δ/Δ strain expressing a truncated version STP2*-mCherry (CaPM45) lacking the N-terminal domain was included. Western blots were performed three independent times from biological replicates. Representative images are shown. (C) Growth and (D) pH was determined as stated in Fig. 1. Graphs show mean and standard deviation from three independent replicates.
Nuclear localization of Stp2 is SPS-dependent
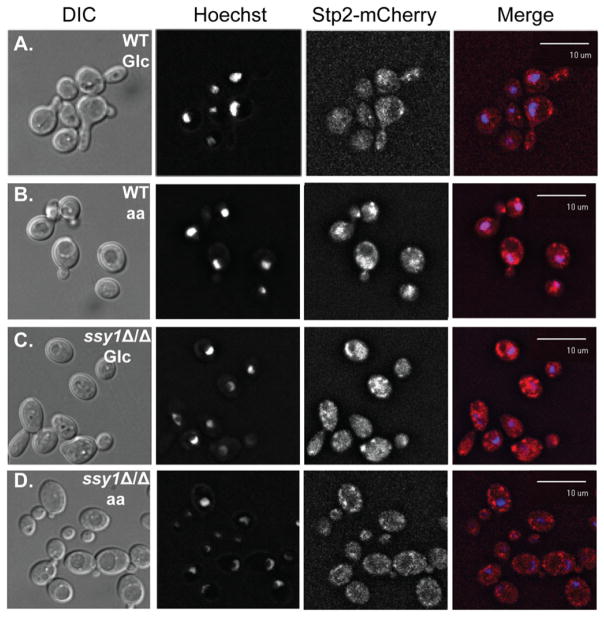
We next investigated the subcellular localization of Stp2 upon amino acid sensing. When wild type cells expressing Stp2-mCherry were grown on glucose, diffuse red fluorescence was detected in the cytoplasm with no apparent nuclear localization (Fig. 4A). In contrast, upon incubation with amino acids, red fluorescence co-localized with the nucleic acid-staining dye Hoechst 33342 (Fig. 4B), revealing translocation of the Stp2-mCherry into the nucleus. This nuclear localization was not observed in the ssy1Δ/Δ cells after incubation with glucose or amino acids (Fig. 4C and D), demonstrating that the SPS system is needed for processing and nuclear translocation of Stp2.
Fig. 4.
Nuclear translocation of Stp2 upon amino acid sensing relies on a functional SPS system. Wild type (CaPM41) and ssy1Δ/Δ cells expressing the STP2-mCherry fusion (CaPM43) were exposed to glucose or amino acids for 1 h at 37°C. Cells were fixed and stained with Hoechst 33342. Cells were visualized with fluorescence microscopy to detect DAPI and mCherry fluorescent signal in the corresponding channels. A) Wild type cells exposed to glucose. B) Wild type cells exposed to amino acids. C) ssy1Δ/Δ cells exposed to glucose or D) ssy1Δ/Δ cells exposed to amino acids. Experiments were performed three independent times. Representative images are shown.
Stp2 lacking the N-terminal domain is able to restore amino acid utilization
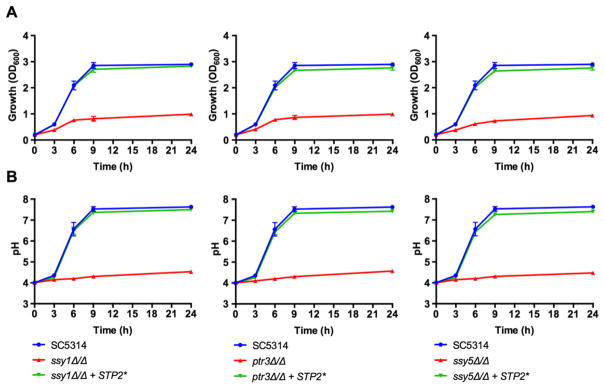
Once we demonstrated that the SPS system is required for activation of Stp2, and thus, for amino acid utilization and pH manipulation, we investigated whether the expression of a constitutively active form of Stp2 is able to suppress the defects observed in the SPS mutants. For this purpose, we created an STP2 allele lacking the first 100 residues containing the nuclear exclusion domain (Andreasson and Ljungdahl, 2004) and placed it under the control of the ACT1 promoter, similar to a strategy used previously (Martínez and Ljungdahl, 2005). The truncated STP2* allele induces the expression of genes involved in amino acid utilization in the absence of a functional SPS system (Martínez and Ljungdahl, 2005). As shown in Fig. 5A, expression of Stp2* is sufficient to suppress the growth defect observed in the three ssy1Δ/Δ, ptr3Δ/Δ and ssy5Δ/Δ mutant strains when growing on amino acids as carbon source. Moreover, the truncated STP2* allele also recovers the ability to neutralize the acidic pH of the medium (Fig. 5B). The truncated STP2* allele restores growth, the ability to neutralize the pH and ammonia production to the ssy1Δ/Δ mutant in YNBA (Fig. 2A–C).
Fig. 5.
Expression of a truncated STP2 allele lacking the N-terminal domain restores growth and neutralization ability in the SPS mutants. A truncated allele of STP2 lacking the first 100 residues (Stp2*), corresponding to the N-terminal nuclear exclusion domain, was expressed in the Candida albicans ssy1Δ/Δ (CaPM36), ptr3Δ/Δ (CaPM38) and ssy5Δ/Δ (CaPM40) strains. Cultures were grown at 37°C on yeast nitrogen base supplemented with 1% casamino acids, with an initial pH of 4.0.
A. Growth was determined by measuring the OD600 at indicated time points.
B. An aliquot of the cultures was used to measure the pH of the medium at the same time points. Graphs show mean and standard deviation from three independent replicates.
We also tagged the truncated STP2* allele expressed in ssy1Δ/Δ with mCherry. As shown in Fig. 3B, the truncated Stp2* expressed by glucose-grown or amino acids-grown ssy1Δ/Δ cells has a similar mobility pattern as the processed Stp2 in amino acids-grown wild type cells. The Stp2*-mCherry fusion is fully functional, because its expression suppresses the growth defect on amino acids and pH neutralization ability of ssy1Δ/Δ (Fig. 3C and D). In addition, the truncated Stp2* is constitutively localized to the nucleus under both glucose and amino acids when expressed in ssy1Δ/Δ (Fig. S3).
Candida albicans requires a functional SPS system for normal interaction with macrophages
Our group has shown that C. albicans lacking a functional Stp2 cannot efficiently escape from macrophage killing (Vylkova and Lorenz, 2014). We investigated whether the absence of a functional SPS system has an impact on the normal interaction of C. albicans with these phagocytes. First, we tested whether ssy1Δ/Δ cells are more susceptible to macrophage killing using an accepted end-point dilution assay (Miramón et al., 2014; Rocha et al., 2001). During coincubation with RAW264.7 murine macrophages, the ssy1Δ/Δ strain was significantly less robust than wild-type strains (Fig. 6A), suggesting increased susceptibility. The resistance to macrophage killing was restored to wild-type levels in the reconstituted strain, and partially restored in strains expressing the truncated STP2* allele.
Fig. 6.
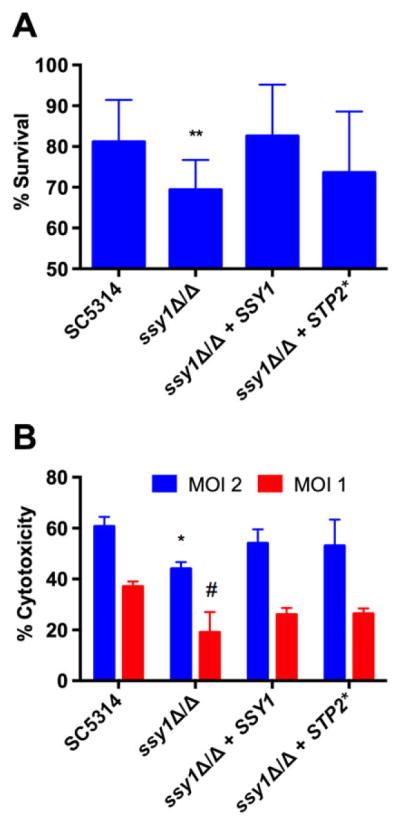
The SPS system is required for normal resistance towards macrophage attack and cytotoxicity. RAW264.7 murine macrophages were infected with wild type (SC5314), ssy1Δ/Δ (CaPM07), reconstituted (CaPM25) and STP2*-expressing (CaPM36) cells.
A. An end point dilution assay coupled with an XTT metabolic assay was performed to determine the sensitivity of these strains towards macrophage killing. Bars show mean and standard deviation of five independent experiments. **P < 0.01.
B. Macrophage cytotoxicity was determined by measuring the release of lactate dehydrogenase after coincubation of phagocytes with fungal cells for 15 h at two different multiplicities of infection (MOI). Bars show mean and standard deviation of three independent experiments. *P < 0.05 (compared with WT at MOI 2); #P < 0.05 (compared with WT at MOI 1).
We next determined the capacity of these strains to damage phagocytes by means of a cytotoxicity assay that detects release of the cytosolic enzyme lactate dehydrogenase (LDH) into the media as a proxy for macrophage membrane damage. As shown in Fig. 6B, ssy1Δ/Δ cells induced less cytotoxicity at two different multiplicities of infection (2:1 and 1:1), and the phenotype was at least partially restored in both the reconstituted strain as well as in the strain expressing the truncated STP2* allele. Thus, a functional SPS system is necessary for normal resistance towards macrophage attack as well as for normal killing of phagocytes.
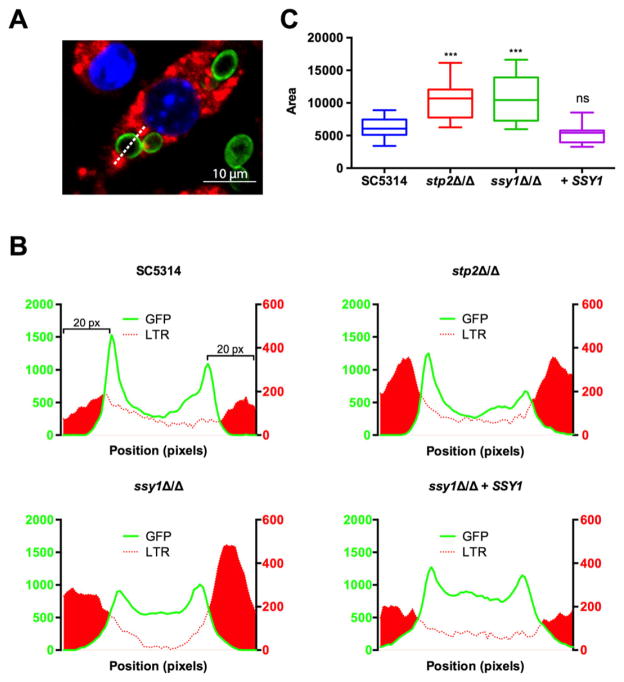
Because the SPS mutants cannot neutralize acidic environments, we tested whether the increased susceptibility of these mutants is due to their inability to block acidification of the phagosome. To this end, LysoTracker Red LTR, Invitrogen-loaded RAW264.7 macrophages were infected with SC5314, ssy1Δ/Δ and ssy1Δ/Δ + SSY1 cells expressing a Pma1-GFP fusion (Pma1 localizes to the plasma membrane). Using the microscope software Slidebook 6.0, we measured the fluorescence signal of GFP and LTR along a line drawn through the fungal cell (Fig. 7A). Plotting the GFP signal outlines the fungal cell shape, whereas the LTR signal delineates acidic sites. We quantified the LTR signal in the immediate vicinity of the ingested fungal cells (20 pixels, or 2 μm, from the maximum GFP peak, on both sides) and determined the total area under the curve as an estimation of the degree of acidification of the phagosomal compartment (Fig. 7B). Similar to the stp2Δ/Δ mutant, cells of the ssy1Δ/Δ mutant reside in a more acidic organelle than wild type or the reconstituted strain, as indicated by the intensity of LTR staining in the lumen of the vesicle. These data demonstrate that C. albicans requires the SPS system to avoid phagosomal acidification, which suggests that there is a source of amino acids in the lumen of this organelle.
Fig. 7.
The SPS system is required for neutralization of the phagosome. LysoTracker Red (LTR)-loaded RAW264.7 macrophages were infected with SC5314 (CaPM57), stp2Δ/Δ (CaPM61), ssy1Δ/Δ (CaPM62) and ssy1Δ/Δ + SSY1 (CaPM66) each expressing a C-terminal Pma1-GFP fusion.
A. Quantification of the LTR signal was performed using Slidebook 6.0. GFP and LTR signals were measured by drawing a line traversing the fungal cell (dashed line).
B. Representative profiles from each strain. GFP signal is plotted on the left axis, LTR signal plotted on the right axis. Area under the curve (filled in red) was calculated in Prism (GraphPad Software).
C. Graph shows box and whiskers (minimum to maximum values), representing the area under the curve from the LTR intensity. A minimum of 30 cells were analyzed per strain. The experiment was performed in triplicate.
The SPS system has limited effect on virulence in the murine model of disseminated candidiasis
Previous work revealed that a C. albicans mutant lacking Stp2 (stp2Δ/Δ) has a moderate but significant attenuation in virulence in the murine model of disseminated infection. We tested the three SPS mutant strains in this animal model to investigate the impact of amino acid sensing in virulence. The animals infected with the wild type strain (SC5314) succumbed within 6 days post infection whereas the survival curves for animals infected with the SPS mutants was slightly shifted toward longer survival, although it was not significantly different from the wild type strain (Fig. S4), suggesting that the SPS system has a limited role in the murine model of hematogenously disseminated candidiasis.
Discussion
The fungal regulatory networks that govern the utilization of disparate compounds including sugars, alcohols, carboxylic acids and fatty acids have been extensively studied in several model systems in which there is a conserved framework that involves glucose repression/derepression and subsequent activation of catabolic pathways for specific alternate nutrients. The same cannot be said for the utilization of amino acids as a source of carbon for the simple reason that this is disfavored in many fungi. C. albicans, in contrast, eagerly catabolizes amino acids, which appear to be an important and available nutrient in vivo. We have previously shown that amino acid metabolism has been adapted in this species to promote survival during contact with phagocytes and virulence and that Stp2, a TF with a known role in amino acid metabolism, compromises interactions with the host (Martínez and Ljungdahl, 2005; Vylkova and Lorenz, 2014; Vylkova et al., 2011). Thus, it is important to understand the regulatory mechanisms that underlie these adaptations.
In yeast, utilization of exogenous amino acids for synthesis of proteins or other nitrogenous compounds requires the SPS system, a three-protein complex (Ssy1, Ptr3 and Ssy5) that senses extracellular amino acids and results in the proteolytic processing and activation of Stp2 (Forsberg and Ljungdahl, 2001; Forsberg et al., 2001; Klasson et al., 1999). Available evidence suggests this works similarly in C. albicans (Martínez and Ljungdahl, 2004; Martínez and Ljungdahl, 2005), but whether this was important in the context of glucose repression/derepression was unknown. In this study, we have shown that the SPS system is also required for the ability to utilize amino acids as a carbon source with deletion of any single component (SSY1, PTR3 or SSY5) conferring identical phenotypes. Further, although wild type and complemented strains rapidly raise extracellular pH, the mutants cannot do so. This is independent of the growth defect, because addition of glycerol restores growth without ameliorating the inability to manipulate the pH, which results from a failure to excrete ammonia. The defects of SPS mutants in amino acid utilization, pH modulation and ammonia release occur regardless of the nitrogen source and in several different media (Figs 1 and 2, and data not shown).
We next explored the contribution of amino acid sensing during the interaction of C. albicans with macrophages. Our group has shown that stp2Δ/Δ strains are more susceptible to macrophage killing and upon phagocytosis, they reside in an acidified phagosome in contrast to wild-type cells (Vylkova and Lorenz, 2014). Consistent with this, the ssy1Δ/Δ cells exhibited increased susceptibility to RAW264.7 murine macrophages. In addition, this mutant germinates less readily in macrophages and is less cytotoxic to the phagocytes, indicating that these effects require a functional amino acid sensing mechanism. Indeed, ssy1Δ/Δ cells reside in a more acidic compartment inside the macrophage, demonstrating that the SPS system is required for normal inhibition of the phagosomal acidification.
The function of the SPS sensor is mediated through Stp2, which is also required for utilization of amino acids and the associated pH changes (Vylkova et al., 2011). We showed that Stp2 is cleaved in an SPS-dependent manner extremely rapidly, within minutes of a switch to media containing amino acids as the carbon source and that this promotes nuclear localization. Finally, expression of a constitutively nuclear isoform (Andreasson and Ljungdahl, 2004), created through an amino terminal truncation (Stp2*), suppresses all SPS phenotypes in vitro.
Overall, our data reveal that the SPS sensor in C. albicans has a similar architecture as in S. cerevisiae (Bernard and André, 2001; Didion et al., 1998; Forsberg and Ljungdahl, 2001; Forsberg et al., 2001; Iraqui et al., 1999; Klasson et al., 1999; Poulsen et al., 2005), which had been suggested by earlier characterization of Ssy1 (Csy1) in C. albicans by Ljungdahl and colleagues (Brega et al., 2004; Martínez and Ljungdahl, 2004; Martínez and Ljungdahl, 2005). Importantly, however, the SPS complex had not been investigated for a role in responses to carbon starvation, and our data indicate that this system is critical for amino acid sensing under these conditions as well. This has important implications, as increasing evidence has shown that peptides and protein may be important sources of carbon during interaction with the host (Barelle et al., 2006; Fernández-Arenas et al., 2007; Fradin et al., 2003; Fradin et al., 2005; Lorenz et al., 2004; Rubin-Bejerano et al., 2003). This adaptation has potentially selected for specialization of SPS targets: in C. albicans, Stp2 controls the expression of genes involved in amino acids utilization such as amino acid permeases, whereas its paralogue Stp1 controls the expression of genes involved in protein degradation (Martínez and Ljungdahl, 2005; Ramachandra et al., 2014). This is in contrast to S. cerevisiae, where these TFs have redundant roles (Eckert-Boulet et al., 2004; Forsberg and Ljungdahl, 2001; Forsberg et al., 2001).
Because the STP2* allele suppressed the SPS mutants, we took a step further and explored whether constitutive expression of three known or suspected Stp2 targets, GAP2 (general amino acid permease), CAN2 (basic amino acid permease) and DIP5 (dicarboxylic amino acid permease) (Martínez and Ljungdahl, 2005; Vylkova et al., 2011) could suppress the SPS growth phenotypes on amino acids and found that they could not (data not shown). This suggests that targets of Stp2 extend beyond the permeases, potentially to catabolic enzymes or other factors that are also necessary for growth under these conditions. Indeed, we have recently described a large family of putative ammonia/acetate transporters ATOs, some of which are Stp2-regulated, that mediate neutralization. Strikingly, over-expression of individual ATO genes in an stp2Δ/Δ strain confers significant growth defects thereby reinforcing the idea that efficient utilization of amino acids and active manipulation of pH requires coordinate regulation of multiple processes (Danhof and Lorenz, 2015).
Our previous work left unanswered questions of whether the source of the amino acids in the phagolysosome is from the host or the pathogen. Conceivably, protein recycling in the fungus may contribute to release peptides and amino acids to support growth. Indeed, as part of the response to phagocytes, C. albicans upregulates the expression of vacuolar proteases (Fradin et al., 2005; Lorenz et al., 2004). However, the survival of a mutant defective in autophagy, atg9Δ/Δ, is not affected following macrophage phagocytosis (Palmer et al., 2007), suggesting that protein recycling plays only a minor role in the nutrient acquisition during the interaction with phagocytes. In stark contrast, we have shown that the ability to properly sense extracellular amino acids is crucial for the normal resistance of C. albicans towards macrophages. This strongly suggests that the amino acids originate from the host. These may come from the hydrolysis of phagosomal proteins and peptides given the expression of multiple secreted proteases in phagocytosed cells (Fradin et al., 2005; Lorenz et al., 2004). Indeed, the only assay in which the constitutively active STP2* allele does not fully suppress ssy1Δ/Δ phenotypes is in the macrophage, which may indicate a role for Stp1 in proteolysis and import of luminal proteins. Alternatively, others have shown that C. albicans actively modifies the composition of the vesicles through its trafficking inside the cell (Bain et al., 2014; Fernández-Arenas et al., 2009; Okai et al., 2015), which may result in the fungal cell inhabiting a compartment distinct from a classical phagolysosome.
Inhibition of the phagosomal acidification is a strategy that fungal pathogens have exploited extensively. Histoplasma capsulatum has been appreciated to neutralize the phagolysosome for many years (Eissenberg et al., 1993), while we and others have shown that C. albicans also occupies a neutral phagolysosome (Bain et al., 2014; Fernández-Arenas et al., 2009; Vylkova and Lorenz, 2014) and have proposed a metabolism-driven mechanism. The distantly related pathogenic yeast Candida glabrata also inhibits phagosomal acidification in a process that depends on Golgi-localized α-mannosyl transferases (Kasper et al., 2014). Recently, Cryptococcus neoformans was shown to efficiently block phagosomal acidification when phagocytosed by macrophages (Smith et al., 2015). Further characterization of the phagosomal composition upon phagocytosis of strains defective in neutralization will shed light on the mechanisms by which C. albicans and other fungal pathogens are able to hijack and alter phagosomal maturation.
In contrast to findings with the stp2Δ/Δ mutant, in which virulence in a mouse model is attenuated (Vylkova and Lorenz, 2014), the ssy1Δ/Δ, ssy5Δ/Δ and ptr3Δ/Δ strains have only a small shift towards delayed mortality in the animals, a difference that was not statistically significant. There may be an SPS-independent pathway that could activate Stp2 in vivo or other SPS-independent functions of Stp2. Alternatively, SPS sensing may be less important in the intravenous injection model, which replicates only the late stages of the human disease, and may have a more striking role in other aspects of the host-pathogen interaction, such as during adhesion to epithelia and tissue invasion. In fact, genes important for amino acid transport and ammonia extrusion (ATO) are upregulated when C. albicans interacts with oral epithelial cells (Wächtler et al., 2011). Therefore, there may be a more prominent role for the SPS system in biofilm-related models, such as in the oral cavity (Dongari-Bagtzoglou et al., 2009; Solis and Filler, 2012), the urogenital tract (Harriott et al., 2010) or on abiotic surfaces (Andes et al., 2004; Nett et al., 2010; Ricicova et al., 2010; Wang and Fries, 2011). Amino acids (or proteins) are also likely a key nutrient in the gut and may be important for colonization in this native niche (Koh, 2013; Koh et al., 2008; Samonis et al., 1990; Wiesner et al., 2001). Thus, further study is needed to define the nutritional parameters of infection in diverse host niches.
Experimental procedures
Media, culture conditions and strains
Strains were routinely grown on YPD medium (1% Yeast extract, 2% Peptone and 2% Dextrose) at 30 °C in a roller incubator. Neutralization assays were performed in yeast nitrogen base (YNB) medium (0.17% YNB and 0.05% ammonium sulfate) or YNBA (0.17% yeast nitrogen base and 0.25% allantoin) supplemented with 1% casamino acids and 2% glycerol, when specified; pH was adjusted to 4.0 with HCl. When needed, agar was added at 2% final concentration.
Deletion of SSY1, PTR3 and SSY5 was performed using the SAT1-flipper method (Reuβ et al., 2004), in the wild type strain SC5314 (Gillum et al., 1984). In brief, fragments of ~350 bp upstream and downstream relative to the open reading frame of interest were polymerase chain reaction (PCR) amplified and cloned between the KpnI and XhoI (upstream fragment) or the SacII and SacI (downstream fragment) in pSFS2. The deletion cassette was PCR amplified using Phusion High-Fidelity DNA polymerase (NEB) and used for electroporation of the recipient C. albicans strain. Transformants were selected on YPD with nourseothricin (200 μg/mL; Werner Bioagents, Jena, Germany). Clones were screened by colony-PCR to confirm correct integration of the cassette. In order to excise the deletion cassette, positive clones were grown on YPM (1% yeast extract, 2% peptone and 2% maltose) overnight at 30°C in a roller incubator and spread on YPD. Correct excision of the SAT1-FLP1 cassette was verified by PCR. A second round of deletion was carried out to generate the homozygous strains described in Table S1. Reintegration of one allele of the respective gene was performed using a CIp10-based plasmid (Murad et al., 2000), where the URA3 marker has been substituted with the SAT1 gene (Jiménez-López et al., 2013). open reading frames including the upstream (promoter) and downstream (terminator) intergenic regions were cloned in the MCS of CIp10-SAT1. The resulting plasmids were linearized with StuI and used for transformation of the homozygous strains. Correct integration was verified by PCR.
Deletion of the first 100 residues of Stp2 was performed using a pSFS2-based plasmid (Reuβ et al., 2004), containing the ACT1 promoter (1012 bp) cloned in the NotI site. A 369 bp fragment upstream STP2 was PCR amplified and cloned between the KpnI and XhoI sites, and a 329 bp fragment (+321 to +629, as measured from the ATG codon) was amplified and cloned between the SacII and SacI of pSFS2-ACT1p; this fragment contains an initiation codon. The deletion cassette was PCR-amplified as described previously and used for electroporation of the SPS mutant strains. Correct integration and excision of the cassette was performed as described previously.
Tagging of one allele of STP2 was performed using a newly generated pFA-mCherry-SAT1 plasmid, using pFA-MYC-HIS1 (Lavoie et al., 2008) as backbone. First, the SAT1 gene was PCR-amplified from pSFS2 and cloned between the AscI and PmeI sites, replacing the HIS1 marker. Then, the mCherry gene (Keppler-Ross et al., 2008) was PCR amplified and cloned between the XmaI and AscI sites. A 447 bp STP2 fragment (+1306 to +1752, from ATG codon) was cloned in the XmaI site in frame with mCherry and 353 bp fragment downstream STP2 was cloned in the PmeI site of pFA-mCherry-SAT1. The tagging cassette was PCR-amplified and used to transform the recipient strains. Correct integration was verified by PCR.
Pma1-GFP strains were generated by cloning the GFP sequence (Barelle et al., 2004) in the ApaI site of pSFS2. A 366 bp fragment (+2320 to +2685) of PMA1 was cloned in the KpnI site, in frame with the GFP sequence. The PMA1 3′ UTR fragment (431 bp) was cloned between SacII and SacI. The tagging cassette was PCR amplified and used to transform the recipient strains. Correct integration was verified by PCR.
Ammonia release assay
We determined ammonia released by C. albicans growing on casamino acids by means of citric acid traps, as described previously (Danhof and Lorenz, 2015; Vylkova et al., 2011). Briefly, cells were grown overnight in YPD and diluted to an OD600 of 1. Cells (5 μL) were spotted onto solid YNBA with casamino acids, pH 4. Citric acid traps (120 μL of 10% citric acid) were placed in microtube caps attached to the lid of the dish, directly underneath the spotted inoculum. The plates were incubated at 37°C, and 20 μL samples were drawn every 24 h over a period of 3 days. Ammonia concentration was determined using the Nessler’s reagent. Experiments were performed in triplicate.
Western blot analysis
Strains were grown overnight on YPD at 30°C in a roller incubator. Cells were diluted and grown for a further 4 h at 30°C starting at an OD600 of 0.2 in YNB + 2% glucose. Cells were collected at 4°C and resuspended in prewarmed (37 °C) YNB + 1% casamino acids. To investigate Stp2 processing in response to specific amino acids, cells were incubated for 15 min in YNBA in presence of the respective amino acid (10 mM), pH 4.0, at 37°C. Cells were collected by centrifugation at 4 °C and washed with TEGN-TX100 (20 mM Tris-HCl pH 7.9, 0.5 mM EDTA, 10% glycerol, 50 mM NaCl, 0.1% Triton X-100) lysis buffer with protease inhibitors (Complete ULTRA, Roche, 1 tablet per 10 mL). Cell pellets were resuspended in TEGN-TX100 with protease inhibitors and mechanically disrupted using acid-washed glass beads. Samples were subjected to six rounds of 1.5 min of bead beating in a vortex mixer and 1.5 min on ice. Protein extracts were clarified by centrifugation and protein quantified by the Pierce BCA protein assay (ThermoFisher Scientific). Protein samples (50 μg) were separated by SDS-PAGE in a 6% acrylamide gel, and transferred to a 0.2 μm nitrocellulose membrane (BioRad). Membranes were blocked overnight at 4°C with 5% milk powder in 1× TBST (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20). Blots were probed with a primary rabbit anti-mCherry antibody (1:5000, Rockland) and a secondary goat anti-rabbit horseradish peroxi-dase (HRP)-conjugated antibody (1:3000, BioRad). Blots were developed using the Pierce ECL Western blotting substrate (ThermoFisher Scientific) and visualized in an ImageQuant LAS4000 mini apparatus (GE).
Visualization of Stp2 nuclear translocation
To visualize the cellular localization of Stp2-mCherry in response to amino acids, cells were exposed to glucose or amino acids as described previously. Cells were collected and fixed in 2.7% paraformaldehyde (pH 7.5) for 15 min at 37 °C, washed twice with 1× PBS and incubated in 500 μL 1× PBS with Hoechst 33342 (1 drop of NucBlue, Molecular Probes) overnight at room temperature. Cells were collected and resuspended in 20 μL 1× PBS and visualized by fluorescence microscopy. Images were captured and processed using Slidebook 6.0. Z-stacks were analyzed by deconvolution using the nearest-neighbor algorithm, and a projection was generated as the final image.
End point dilution assay
To determine the sensitivity of the fungal strains to macrophage killing, we performed the end point dilution assay (Rocha et al., 2001). In brief, RAW264.7 macrophages in RPMI 1640 with phenol red supplemented with 10% fetal bovine serum (RPMI-10) were seeded in wells in a 96-well plate at a concentration of 2.5 × 105 cells mL−1, 100 μL per well, incubated overnight at 37°C and 5% CO2. Fungal cells were grown overnight in YPD at 30°C, collected and washed twice with 1× PBS, and resuspended at a concentration of 5 × 105 cells mL−1. Prior to infection, the medium was replaced with 150 μL RPMI without phenol red. The first well in each column was inoculated with 50 μL of the fungal suspension, and then serially diluted 1:4 six times. Media controls were prepared in the same manner, and incubated 15 h at 37°C and 5% CO2. Quantification of fungal biomass (microcolonies developed at the bottom of the wells) was performed by means of the XTT assay as previously described (Miramón et al., 2014). In brief, media was carefully removed from each well and 150 μL of XTT/Coenzyme Q0 were added in each well and incubated at 37°C for 60 min. An aliquot of 100 μL was removed from each well and transferred to a new plate. Absorbance was read at 451 nm in a microplate reader. Experiments were performed five independent times.
Lactate dehydrogenase release assay
Cytotoxicity of the fungal strains on macrophages was determined by detecting the release of LDH after coincubation, using the CytoTox96 Non-Radioactive Cytotoxicity assay (Promega) according to manufacturer’s instructions (Vylkova and Lorenz, 2014). Briefly, RAW264.7 macrophages in RPMI-10 were seeded in a 96-well plate at a concentration of 2.5 × 105 cells mL−1, 100 μL per well, incubated overnight at 37°C and 5% CO2. Fungal suspensions were prepared as described previously, at 2 × 106 (for multiplicity of infection of 2) and 1 × 106 cells mL−1 (for MOI of 1). Prior to infection, the medium was replaced with 50 μL RPMI without phenol red. Macrophages were infected with 50 μL of the fungal suspensions and incubated for 15 h at 37°C and 5% CO2.
LDH release was normalized to chemically lysed macrophages. Experiments were performed in triplicate.
Phagosome acidification assay
Phagosomal acidification was investigated using the acidotropic fluorescent probe LysoTracker® Red DND-99, which accumulates in acidic organelles. Macrophages were seeded in glass-bottom μ-Slides (Ibidi) at a concentration of 1.5 × 105 cells mL−1 and loaded for 2 h with 50 nM LTR in RPMI without phenol red. Medium was replaced prior to infection. Overnight cultures of Pma1-GFP C. albicans strains were regrown in YPD for 5 h at 30°C, cells were collected and washed in 1× PBS, and diluted to a concentration of 1.5 × 107 cells mL−1. Macrophages were infected with 10 μL of the C. albicans suspensions and incubated for 1 h at 37°C, 5% CO2. Cells were fixed in paraformaldehyde, washed with 1× PBS, and nuclei were stained with NucBlue for visualization purposes. Images were captured and analyzed using Slidebook 6.0. To quantify the accumulation of LTR in the phagosome, we obtained the signal intensity profiles of GFP and LTR along a line extending beyond the edges of the fungal cell. We quantified the area under the curve of the LTR signal for a region of 20 pixels (2 μm) from each side of the C. albicans cell, outlined by the GFP signal. Experiments were performed in triplicate; a minimum of 30 fungal cells were analyzed per strain in each experiment.
Murine model of disseminated candidiasis
The murine model of hematogenously disseminated candidiasis was performed as previously described (Vylkova and Lorenz, 2014). Female ICR mice (10 individuals per strain, housed in groups of five) were inoculated via the tail vein with 100 μL of fungal suspensions in 1× PBS at a concentration of 5 × 106 cells mL−1. Animals were monitored twice daily and humanely terminated when moribund. Protocols were approved by the Animal Welfare Committee of the University of Texas Health Science Center at Houston.
Statistical analyses
Statistics were performed in GraphPad Prism 6. Tukey’s multiple comparisons test was used to analyze the ammonia release assay. Two-tailed paired Student’s T-test was used to analyze the fungal survival data (end point dilution assay). Macrophage cytotoxicity data (LDH assay) and phagosomal acidification (LTR assay) were analyzed using Dunn’s multiple comparisons test. Survival proportions were evaluated by Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests. P values≤0.05 were considered statistically significant.
Supplementary Material
Mutants lacking the SPS system fail to neutralize the environmental pH despite glycerol-supported growth. C. albicans ssy1Δ/Δ (CaPM07), ptr3Δ/Δ (CaPM15), ssy5Δ/Δ (CaPM23), and complemented strains + SSY1 (CaPM25), +PTR3 (CaPM27), and + SSY5 (CaPM29) were grown at 37 °C on YNB supplemented with 1% casamino acids and 2% glycerol, with an initial pH of 4.0. A) Growth and B) pH was determined as stated in Fig. 1. Graphs show mean and standard deviation from three independent replicates.
Stp2 is processed in response to amino acids but not glucose. Wild type cells expressing the Stp2-mCherry fusion (CaPM41) were exposed to glucose or amino acids. Cells were collected at the indicated time points and cell extracts were prepared. Samples were blotted and probed with an anti-mCherry antibody (see Materials and Methods for details). A) Glucose-exposed cells showed limited Stp2 processing, whereas cells exposed to amino acids rapidly processed Stp2 within the first 5 min. B) Stp2 processing is activated differentially in response to single amino acids. C. albicans CaPM41 cells were exposed for 15 min to the respective amino acid (10 mM) in YNBA at 37 °C. Basic amino acids (lysine, histidine and arginine) as well as aspartic acid and glutamine, readily induced Stp2 processing. Glutamic acid and serine induced partial activation. Asparagine and threonine exerted limited activation.
Stp2*-mCherry constitutively localizes in the nucleus in ssy1Δ/Δ cells. Cells expressing the STP2*-mCherry fusion in the ssy1Δ/Δ background (CaPM45) were exposed to A) glucose or B) amino acids for 1 h at 37 °C. Cells were washed and fixed with paraformaldehyde for 15 min at 37 °C and washed with 1× PBS. Nuclei were stained with Hoechst 33342 overnight at room temperature. Cells were visualized with fluorescence microscopy to detect DAPI and mCherry fluorescent signals in the corresponding channels. Experiments were performed three independent times. Representative images are shown.
Fig. S4. Virulence of the SPS mutants is only marginally affected in the murine model of systemic candidiasis. Female ICR mice (n = 10) were infected via the lateral tail vein with 5 × 105 fungal cells in 100 μl 1× PBS, with wild type (SC5314), ssy1Δ/Δ (CaPM07), ptr3Δ/Δ (CaPM15), and ssy5Δ/Δ (CaPM23) strains. Animals were monitored twice per day and humanely euthanized when moribund. The survival curves of the SPS mutants were not significantly different from that of the wild type under the tested conditions.
Table S1. Strains used in this study.
Acknowledgments
We thank Per Ljungdahl (Stockholm University) for kindly providing strains and reagents. We are grateful to Slavena Vylkova, Heather Danhof, Elisa Vesely and Carrie Graham for helpful discussions and advice. This work was funded by U.S. Public Health Service award R01AI075091 (to ML) and CONACYT Fellowship 232104 (to PM).
References
- Abdel-Sater F, Bakkoury ME, Urrestarazu A, Vissers S, André B. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol Cell Biol. 2004;24:9771–9785. doi: 10.1128/MCB.24.22.9771-9785.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Sater F, Jean C, Merhi A, Vissers S, André B. Amino acid signaling in yeast: activation of Ssy5 protease is associated with its phosphorylation-induced ubiquitylation. J Biol Chem. 2011;286:12006–12015. doi: 10.1074/jbc.M110.200592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson C, Ljungdahl PO. The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full Ssy1p-Ptr3p-Ssy5p sensor control. Mol Cell Biol. 2004;24:7503–7513. doi: 10.1128/MCB.24.17.7503-7513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain JM, Louw J, Lewis LE, Okai B, Walls CA, Ballou ER, et al. Candida albicans hypha formation and mannan masking of β-glucan inhibit macrophage phagosome maturation. mBio. 2014;5:1–17. doi: 10.1128/mBio.01874-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelle CJ, Manson CL, MacCallum DM, Odds FC, Gow NAR, Brown AJP. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast. 2004;21:333–340. doi: 10.1002/yea.1099. [DOI] [PubMed] [Google Scholar]
- Barelle CJ, Priest CL, MacCallum DM, Gow NAR, Odds FC, Brown AJP. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard F, André B. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol Microbiol. 2001;41:489–502. doi: 10.1046/j.1365-2958.2001.02538.x. [DOI] [PubMed] [Google Scholar]
- Brega E, Zufferey R, Mamoun CB. Candida albicans Csy1p is a nutrient sensor important for activation of amino acid uptake and hyphal morphogenesis. Eukaryot Cell. 2004;3:135–143. doi: 10.1128/EC.3.1.135-143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Brown AJP, Brown GD, Netea MG, Gow NAR. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 2014;22:614–622. doi: 10.1016/j.tim.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collette JR, Lorenz MC. Mechanisms of immune evasion in fungal pathogens. Curr Opin Microbiol. 2011;14:668–675. doi: 10.1016/j.mib.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Corvey C, Koetter P, Beckhaus T, Hack J, Hofmann S, Hampel M, et al. Carbon source-dependent assembly of the Snf1p kinase complex in Candida albicans. J Biol Chem. 2005;280:25323–25330. doi: 10.1074/jbc.M503719200. [DOI] [PubMed] [Google Scholar]
- Danhof HA, Lorenz MC. The Candida albicans ATO gene family promotes neutralization of the macrophage phagolysosome. Infect Immun. 2015;83:4416–4426. doi: 10.1128/IAI.00984-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion T, Regenberg B, Jorgensen MU, Kielland-Brandt MC, Andersen HA. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:643–650. doi: 10.1046/j.1365-2958.1998.00714.x. [DOI] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. Characterization of mucosal Candida albicans biofilms. PLoS One. 2009;4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert-Boulet N, Nielsen PS, Friis C, dos Santos MM, Nielsen J, Kielland-Brandt MC, Regenberg B. Transcriptional profiling of extracellular amino acid sensing in Saccharomyces cerevisiae and the role of Stp1p and Stp2p. Yeast. 2004;21:635–648. doi: 10.1002/yea.1120. [DOI] [PubMed] [Google Scholar]
- Eissenberg LG, Goldman WE, Schlesinger PH. Histoplasma capsulatum modulates the acidification of phagolysosomes. J Exp Med. 1993;177:1605–1611. doi: 10.1084/jem.177.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Arenas E, Cabezón V, Bermejo C, Arroyo J, Nombela C, Diez-Orejas R, Gil C. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics. 2007;6:460–478. doi: 10.1074/mcp.M600210-MCP200. [DOI] [PubMed] [Google Scholar]
- Fernández-Arenas E, Bleck CKE, Nombela C, Gil C, Griffiths G, Diez-Orejas R. Candida albicans actively modulates intracellular membrane trafficking in mouse macrophage phagosomes. Cell Microbiol. 2009;11:560–589. doi: 10.1111/j.1462-5822.2008.01274.x. [DOI] [PubMed] [Google Scholar]
- Fidel PLJ. Candida-host interactions in HIV disease: relationships in oropharyngeal candidiasis. Adv Dent Res. 2006;19:80–84. doi: 10.1177/154407370601900116. [DOI] [PubMed] [Google Scholar]
- Forsberg H, Ljungdahl PO. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol Cell Biol. 2001;21:814–826. doi: 10.1128/MCB.21.3.814-826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg H, Gilstring CF, Zargari A, Martínez P, Ljungdahl PO. The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol Microbiol. 2001;42:215–228. doi: 10.1046/j.1365-2958.2001.02627.x. [DOI] [PubMed] [Google Scholar]
- Fradin C, Kretschmar M, Nichterlein T, Gaillardin C, d’Enfert C, Hube B. Stage-specific gene expression of Candida albicans in human blood. Mol Microbiol. 2003;47:1523–1543. doi: 10.1046/j.1365-2958.2003.03396.x. [DOI] [PubMed] [Google Scholar]
- Fradin C, Groot PD, MacCallum D, Schaller M, Klis F, Odds FC, Hube B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol. 2005;56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Harriott MM, Lilly EA, Rodríguez TE, Fidel PLJ, Noverr MC. Candida albicans forms biofilms on the vaginal mucosa. Microbiology. 2010;156:3635–3644. doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube B, Monod M, Schofield DA, Brown AJ, Gow NA. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- Iraqui I, Vissers S, Bernard F, de Craene JO, Boles E, Urrestarazu A, André B. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-López C, Lorenz MC. Fungal immune evasion in a model host-pathogen interaction: Candida albicans versus macrophages. PLoS Pathog. 2013;9:e1003741. doi: 10.1371/journal.ppat.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-López C, Collette JR, Brothers KM, Shepardson KM, Cramer RA, Wheeler RT, Lorenz MC. Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryot Cell. 2013;12:91–100. doi: 10.1128/EC.00290-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper L, Seider K, Gerwien F, Allert S, Brunke S, Schwarzmüller T, et al. Identification of Candida glabrata genes involved in pH modulation and modification of the phagosomal environment in macrophages. PLoS One. 2014;9:e96015. doi: 10.1371/journal.pone.0096015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler-Ross S, Noffz C, Dean N. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics. 2008;179:705–710. doi: 10.1534/genetics.108.087080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, McCusker JH. Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase (ilv2Δ) mutants is influenced by the carbon source and rapamycin. Microbiology. 2010;156:929–939. doi: 10.1099/mic.0.034348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson H, Fink GR, Ljungdahl PO. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol Cell Biol. 1999;19:5405–5416. doi: 10.1128/mcb.19.8.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh AY. Murine models of Candida gastrointestinal colonization and dissemination. Eukaryot Cell. 2013;12:1416–1422. doi: 10.1128/EC.00196-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh AY, Kohler JR, Coggshall KT, Rooijen NV, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 2008;4:e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics. 2008;9:578. doi: 10.1186/1471-2164-9-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Thornton J, Spirek M, Butow RA. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol Cell Biol. 2008;28:551–563. doi: 10.1128/MCB.00929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Ljungdahl PO. An ER packaging chaperone determines the amino acid uptake capacity and virulence of Candida albicans. Mol Microbiol. 2004;51:371–384. doi: 10.1046/j.1365-2958.2003.03845.x. [DOI] [PubMed] [Google Scholar]
- Martínez P, Ljungdahl PO. Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol Cell Biol. 2005;25:9435–9446. doi: 10.1128/MCB.25.21.9435-9446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus BA, Coleman DC. Molecular epidemiology, phylogeny and evolution of Candida albicans. Infect Genet Evol. 2014;21:166–178. doi: 10.1016/j.meegid.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Miramón P, Kasper L, Hube B. Thriving within the host: Candida spp. interactions with phagocytic cells. Med Microbiol Immunol. 2013;202:183–195. doi: 10.1007/s00430-013-0288-z. [DOI] [PubMed] [Google Scholar]
- Miramón P, Dunker C, Kasper L, Jacobsen ID, Barz D, Kurzai O, Hube B. A family of glutathione peroxidases contributes to oxidative stress resistance in Candida albicans. Med Mycol. 2014;52:223–239. doi: 10.1093/mmy/myt021. [DOI] [PubMed] [Google Scholar]
- Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Marchillo K, Spiegel CA, Andes DR. Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun. 2010;78:3650–3659. doi: 10.1128/IAI.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okai B, Lyall N, Gow NAR, Bain JM, Erwig LP. Rab14 regulates maturation of macrophage phagosomes containing the fungal pathogen Candida albicans and outcome of the host-pathogen interaction. Infect Immun. 2015;83:1523–1535. doi: 10.1128/IAI.02917-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omnus DJ, Pfirrmann T, Andréasson C, Ljungdahl PO. A phosphodegron controls nutrient-induced proteasomal activation of the signaling protease Ssy5. Mol Biol Cell. 2011;22:2754–2765. doi: 10.1091/mbc.E11-04-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer GE, Kelly MN, Sturtevant JE. Autophagy in the pathogen Candida albicans. Microbiology. 2007;153:51–58. doi: 10.1099/mic.0.2006/001610-0. [DOI] [PubMed] [Google Scholar]
- Pérez JC, Johnson AD. Regulatory circuits that enable proliferation of the fungus Candida albicans in a mammalian host. PLoS Pathog. 2013;9:1–3. doi: 10.1371/journal.ppat.1003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–346. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfirrmann T, Heessen S, Omnus DJ, Andreasson C, Ljungdahl PO. The prodomain of Ssy5 protease controls receptor-activated proteolysis of transcription factor Stp1. Mol Cell Biol. 2010;30:3299–3309. doi: 10.1128/MCB.00323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P, Wu B, Gaber RF, Kielland-Brandt MC. Constitutive signal transduction by mutant Ssy5p and Ptr3p components of the SPS amino acid sensor system in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1116–1124. doi: 10.1128/EC.4.6.1116-1124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandra S, Linde J, Brock M, Guthke R, Hube B, Brunke S. Regulatory networks controlling nitrogen sensing and uptake in Candida albicans. PLoS One. 2014;9:e92734. doi: 10.1371/journal.pone.0092734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez MA, Lorenz MC. The transcription factor homolog CTF1 regulates β-oxidation in Candida albicans. Eukaryot Cell. 2009;8:1604–1614. doi: 10.1128/EC.00206-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuβ O, Morschhäuser J. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol Microbiol. 2006;60:795–812. doi: 10.1111/j.1365-2958.2006.05136.x. [DOI] [PubMed] [Google Scholar]
- Reuβ O, Vik A, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Ricicova M, Kucharikova S, Tournu H, Hendrix J, Bujdakova H, Eldere JV, et al. Candida albicans biofilm formation in a new in vivo rat model. Microbiology. 2010;156:909–919. doi: 10.1099/mic.0.033530-0. [DOI] [PubMed] [Google Scholar]
- Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, et al. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Fraser I, Grisafi P, Fink GR. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc Natl Acad Sci USA. 2003;100:11007–11012. doi: 10.1073/pnas.1834481100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonis G, Anaissie EJ, Rosenbaum B, Bodey GP. A model of sustained gastrointestinal colonization by Candida albicans in healthy adult mice. Infect Immun. 1990;58:1514–1517. doi: 10.1128/iai.58.6.1514-1517.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandai D, Yin Z, Selway L, Stead D, Walker J, Leach MD, et al. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. mBio. 2012;3:e00495–12. doi: 10.1128/mBio.00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Dixon EF, May RC. The fungal pathogen Cryptococcus neoformans manipulates macrophage phagosome maturation. Cell Microbiol. 2015;17:702–713. doi: 10.1111/cmi.12394. [DOI] [PubMed] [Google Scholar]
- Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nat Protoc. 2012;7:637–642. doi: 10.1038/nprot.2012.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B, Liang XB, Robert F, Soontorngun N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010;10:2–13. doi: 10.1111/j.1567-1364.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vylkova S, Lorenz MC. Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog. 2014;10:e1003995. doi: 10.1371/journal.ppat.1003995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio. 2011;2:e00055–11. doi: 10.1128/mBio.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtler B, Wilson D, Haedicke K, Dalle F, Hube B. From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS One. 2011;6:e17046. doi: 10.1371/journal.pone.0017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fries BC. A murine model for catheter-associated candiduria. J Med Microbiol. 2011;60:1523–1529. doi: 10.1099/jmm.0.026294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner SM, Jechorek RP, Garni RM, Bendel CM, Wells CL. Gastrointestinal colonization by Candida albicans mutant strains in antibiotic-treated mice. Clin Diagn Lab Immunol. 2001;8:192–195. doi: 10.1128/CDLI.8.1.192-195.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, Thewes S, Zakikhany K, Fradin C, Albrecht A, Almeida R, et al. Identifying infection-associated genes of Candida albicans in the postgenomic era. FEMS Yeast Res. 2009;9:688–700. doi: 10.1111/j.1567-1364.2009.00524.x. [DOI] [PubMed] [Google Scholar]
- Zaragoza O, Rodríguez C, Gancedo C. Isolation of the MIG1 gene from Candida albicans and effects of its disruption on catabolite repression. J Bacteriol. 2000;182:320–326. doi: 10.1128/jb.182.2.320-326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutants lacking the SPS system fail to neutralize the environmental pH despite glycerol-supported growth. C. albicans ssy1Δ/Δ (CaPM07), ptr3Δ/Δ (CaPM15), ssy5Δ/Δ (CaPM23), and complemented strains + SSY1 (CaPM25), +PTR3 (CaPM27), and + SSY5 (CaPM29) were grown at 37 °C on YNB supplemented with 1% casamino acids and 2% glycerol, with an initial pH of 4.0. A) Growth and B) pH was determined as stated in Fig. 1. Graphs show mean and standard deviation from three independent replicates.
Stp2 is processed in response to amino acids but not glucose. Wild type cells expressing the Stp2-mCherry fusion (CaPM41) were exposed to glucose or amino acids. Cells were collected at the indicated time points and cell extracts were prepared. Samples were blotted and probed with an anti-mCherry antibody (see Materials and Methods for details). A) Glucose-exposed cells showed limited Stp2 processing, whereas cells exposed to amino acids rapidly processed Stp2 within the first 5 min. B) Stp2 processing is activated differentially in response to single amino acids. C. albicans CaPM41 cells were exposed for 15 min to the respective amino acid (10 mM) in YNBA at 37 °C. Basic amino acids (lysine, histidine and arginine) as well as aspartic acid and glutamine, readily induced Stp2 processing. Glutamic acid and serine induced partial activation. Asparagine and threonine exerted limited activation.
Stp2*-mCherry constitutively localizes in the nucleus in ssy1Δ/Δ cells. Cells expressing the STP2*-mCherry fusion in the ssy1Δ/Δ background (CaPM45) were exposed to A) glucose or B) amino acids for 1 h at 37 °C. Cells were washed and fixed with paraformaldehyde for 15 min at 37 °C and washed with 1× PBS. Nuclei were stained with Hoechst 33342 overnight at room temperature. Cells were visualized with fluorescence microscopy to detect DAPI and mCherry fluorescent signals in the corresponding channels. Experiments were performed three independent times. Representative images are shown.
Fig. S4. Virulence of the SPS mutants is only marginally affected in the murine model of systemic candidiasis. Female ICR mice (n = 10) were infected via the lateral tail vein with 5 × 105 fungal cells in 100 μl 1× PBS, with wild type (SC5314), ssy1Δ/Δ (CaPM07), ptr3Δ/Δ (CaPM15), and ssy5Δ/Δ (CaPM23) strains. Animals were monitored twice per day and humanely euthanized when moribund. The survival curves of the SPS mutants were not significantly different from that of the wild type under the tested conditions.
Table S1. Strains used in this study.