Abstract
Increased glucose consumption is a hallmark of cancer cells. The increased consumption and subsequent metabolism of glucose during proliferation creates the need for a constant supply of NAD, a co-factor in glycolysis. Regeneration of the NAD required to support enhanced glycolysis has been attributed to the terminal glycolytic enzyme, lactate dehydrogenase (LDH). However, loss of glucose carbons to biosynthetic pathways early in glycolysis reduces the carbon supply to LDH. Thus, alternative routes for NAD regeneration must exist to support the increased glycolytic rate while allowing for the diversion of glucose to generate biomass and support proliferation. Here we demonstrate, using a variety of cancer cell lines as well as activated primary T cells, that cytosolic malate dehydrogenase 1 (MDH1) is an alternative to LDH as a supplier of NAD. Moreover, our results indicate that MDH1 generates malate with carbons derived from glutamine, thus enabling utilization of glucose carbons for glycolysis and for biomass. Amplification of MDH1 occurs at an impressive frequency in human tumors and correlates with poor prognosis. Together, our findings suggest proliferating cells rely on both MDH1 and LDH to replenish cytosolic NAD and therapies designed at targeting glycolysis must consider both dehydrogenases.
Keywords: Cancer, metabolism, glycolysis, lactate, malate
Introduction
Cancer cells rewire their metabolism in a variety of ways to support de novo synthesis of macromolecules needed for proliferation. They increase their consumption of glucose but uncouple glycolysis from the citric acid cycle (TCA), diverting glucose carbon into biosynthetic pathways that support growth and proliferation(1). A constant supply of cytosolic NAD, which serves as an electron acceptor in the reaction catalyzed by glyceraldehyde-3-phosphate dehydrogenase (GAPDH), is required to sustain the enhanced glycolysis associated with proliferation. The cytosolic pool of NAD/NADH is independent of the mitochondrial NAD/NADH pool involved in the electron transport chain. The regeneration of cytosolic NAD from NADH has been largely attributed to the production of lactate from pyruvate by the lactate dehydrogenase (LDH) enzyme(1, 2). However, given that diversion of glucose carbons for biomass reduces the flow of carbons to pyruvate, it is evident that LDH activity alone cannot satisfy the increased need for cytosolic NAD in these cells(3). Under these circumstances, how do cancer cells resupply GAPDH with its cofactor NAD at a rate conducive to maintaining the accelerated glycolysis required for proliferation?
In this study we set out to identify alternative reactions that could support the sustained glycolytic rate exhibited by proliferating cells. We report the generation of malate through malate dehydrogenase 1 (MDH1) supports lactate dehydrogenase to regenerate NAD during proliferation. MDH1 deletion in cancer cells slowed proliferation and glucose consumption. In human tumors, MDH1 amplification is a prominent genomic aberration and correlates with poor prognosis. Furthermore, we demonstrate that reductive metabolism of glutamine provides carbon for the MDH1 reaction. Overall, our results suggest MDH1 works with LDHA during Warburg metabolism in proliferating cells and that therapies targeting glycolysis in cancer cells must consider targeting MDH1.
Results
Malate dehydrogenase activity helps regenerate cytosolic NAD in proliferating cells
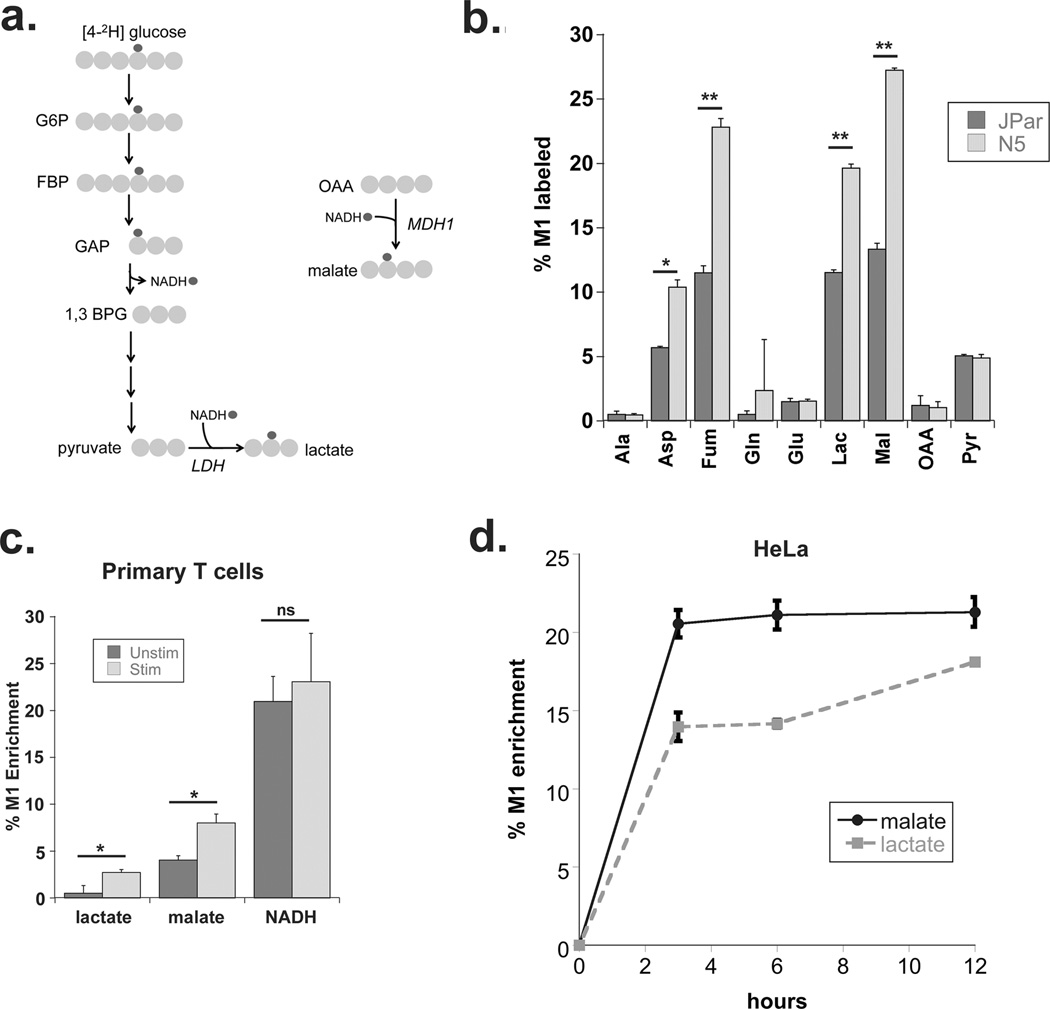
We previously demonstrated that stable over-expression of the Bcl-2 family member Noxa increased glucose consumption, extracellular acidification and promoted greater dependence on the pentose phosphate pathway (PPP) in Jurkat leukemia cells. At the same time, the Noxa over-expressing (N5) cells showed lower glycolysis completion rates suggesting reduced flux of glucose carbons to lactate(4). We used this isogenic model to trace the flow of deuterium from the glucose isotopomer, [4-2H]-glucose, to cytosolic NADH, and thence to metabolites generated from NADH-dependent dehydrogenase activity (Figure 1a). We assayed M1 enriched metabolites by gas chromatography-coupled mass spectrometry (GC-MS) following 24 hours of labeling with [4-2H] glucose. As expected, the highest concentration of M1 labeled metabolite was lactate (Supplementary Figure 1a). However, we detected increased M1 enrichment of additional metabolites in N5 cells, suggesting other dehydrogenase(s) in addition to lactate dehydrogenase were involved in regenerating cytosolic NAD during Warburg metabolism (Figure 1b, Supplementary Figure 1a). While lactate production and accumulation is well documented in cancer cells (reviewed in (5)), most other M1-labeled metabolites we detected are substrates for other reactions, which made direct comparison of the concentration (peak area) of M1 metabolites difficult. Instead, we focused on the M1 enrichment levels of the individual metabolites in N5 cells as a consequence of increased glycolysis (Figure 1b). The M1 malate pool showed the highest increase in N5 cells over parental cells, followed by aspartate and fumarate. Fumarate is likely to be an additional indicator of malate enrichment given that it is not directly associated with a dehydrogenase and can be generated from malate via cytosolic fumarase. M1 labeled aspartate is also likely to be derived from fumarate which, as a symmetrical molecule could retain the M1 hydrogen label as it returns to malate through fumarase and then OAA on its way to aspartate synthesis by aspartate transaminase. An alternative explanation for M1 labeled aspartate is aspartate dehydrogenase (ASPDH), which generates aspartate from OAA using NADH and free ammonia, has been reported in humans based on homology(6). However, NMR analysis indicated that the deuterium from NADH was associated with the β-carbon of aspartate (Supplementary Figure 1b) and not the α-carbon predicted from the ASPDH activity reported in other species(7). These findings suggest that the increased M1 labeled aspartate and fumarate we observe in N5 cells originates from malate. Thus, we focused on the cytosolic malate dehydrogenase 1 (MDH1) enzyme, a component of the malate aspartate shuttle, which moves electrons in the form of NADH from the cytosol into the mitochondria through malate (Reviewed by Menzies et al.(8)). While MDH1 is recognized as a source of NAD in differentiated cells, its role in proliferation and cancer has not been described.
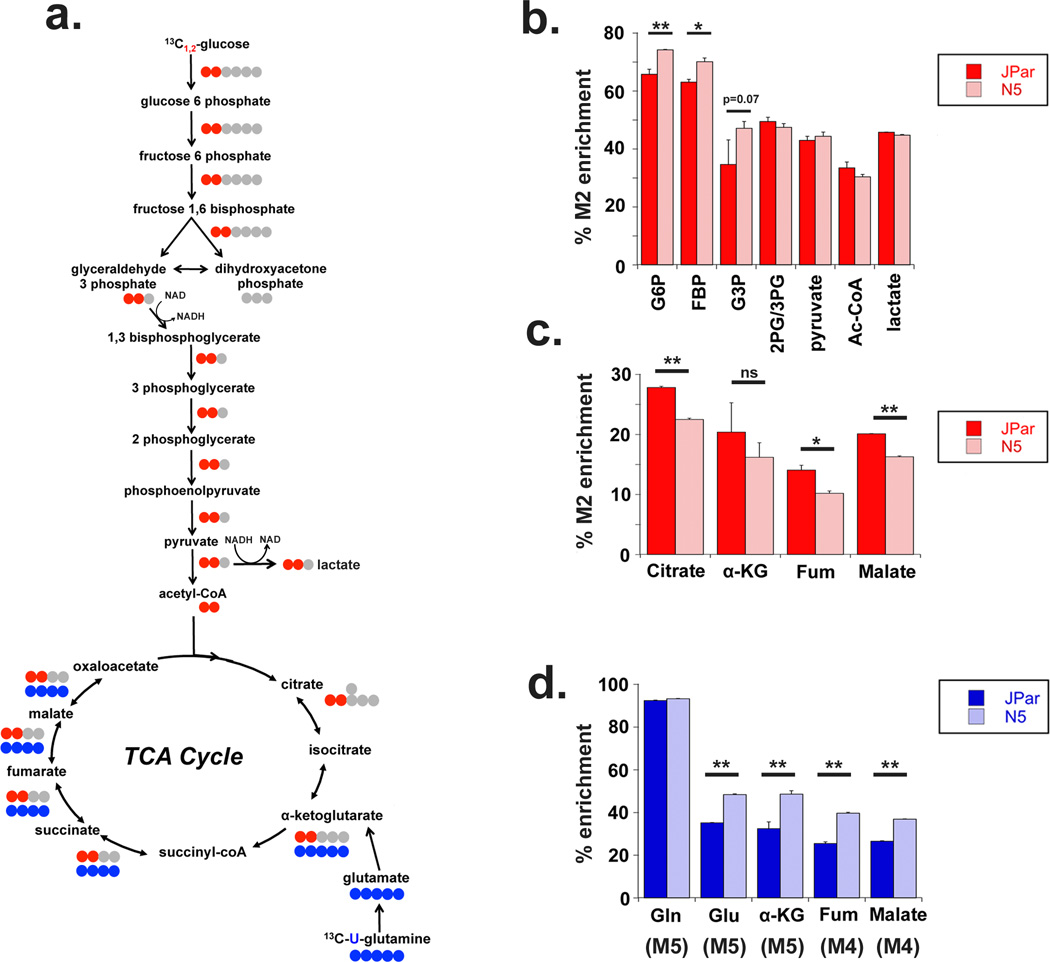
Figure 1. Malate dehydrogenase regenerates cytosolic NAD in cancer cells and activated primary T cells.
(a) Schematic of [2H] labeling used to determine cytosolic NAD regeneration. (b) Enrichment of M1 labeled metabolites from [4-2H] glucose 24 hours after labeling as measured by GC-MS in JPar (baseline glucose consumption) and N5 (high glucose consumption) cells. (c) Enrichment of M1 labeled lactate, malate and NADH from primary human T cells incubated with [4-2H] glucose in the presence or absence of co-stimulation for 24 hours with anti-CD3/anti-CD28 antibodies. (d) Enrichment of M1 malate and M1 lactate in HeLa cells incubated with [4-2H] glucose for 3, 6 and 12 hours (as determined by GC-MS) to assess transfer of [2H] into malate and lactate. Data shown are the average and standard deviation of triplicate samples from a representative experiment repeated at least twice. Significance was calculated using the student’s T test (* p<0.01, ** p<0.001).
To further investigate the involvement of malate in the regeneration of cytosolic NAD in proliferating cells, primary human T cells were stimulated to proliferate using anti-CD3/anti-CD28 based receptor co-stimulation. Upon activation, primary T cells switch to a glycolytic phenotype and increase their glucose consumption(9, 10). We assessed the transfer of deuterium from [4-2H] glucose to malate and lactate in primary CD3+ T cells purified from normal human donor PBMCs following 24 hours of stimulation. Consistent with published studies, levels of M1 labeled lactate increased following stimulation (Figure 1c). However, the enrichment of M1 malate also increased demonstrating that malate synthesis through MDH1 is a significant route for regenerating cytosolic NAD in activated T cells (Figure 1c). To better understand the contribution of malate synthesis to the regeneration of cytosolic NAD, we examined malate and lactate flux in HeLa cells, commonly used as models for Warburg metabolism for their highly glycolytic phenotype(11–13). Intracellular M1 enrichment of malate occurred rapidly, reaching steady state at 6 hours after labeling with [4-2H] glucose (Figure 1d). MDH1 activity kinetics mirror that of LDHA further demonstrating a relationship between MDH1 activity and glucose consumption and suggesting, moreover, that OAA, the substrate for the MDH1 reaction, is readily available in the cytosol. The results in Figure 1 demonstrate the contribution of malate to glycolysis in both non-adherent and adherent cancer cell lines. Interestingly, M1 malate also increased over time in the medium (Supplementary Figure 1c) suggesting malate that accumulates in the cytosol during proliferation is in excess of amounts the malate aspartate shuttle is able to process. Overall, these data point to a significant role for malate dehydrogenase in replenishing the cytosolic NAD required to support glycolysis in proliferating cells.
Malate dehydrogenase 1 supports proliferation and glucose consumption in cancer cells
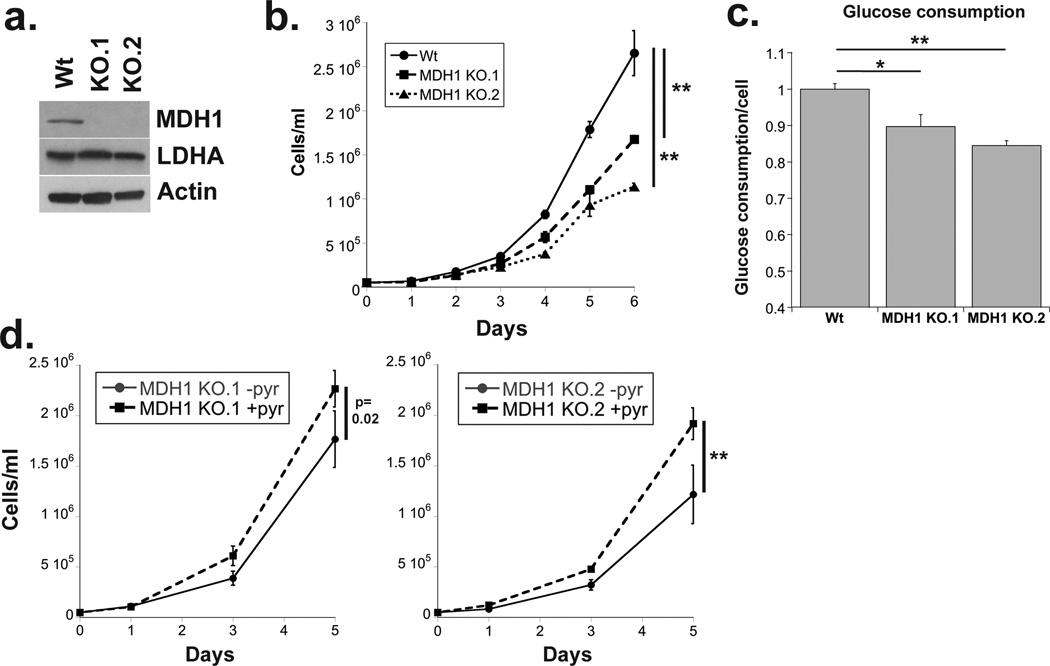
To evaluate the contribution of MDH1 to the proliferative state we utilized Jurkat cells knocked-out for MDH1(14) (Figure 2a). Figure 2b shows that MDH1 KO cells proliferate at a significantly lower rate. They also consume less glucose per cell than their wild-type counterparts (Figure 2c). MDH1 support of glycolysis would predict that less pyruvate is available for NAD regeneration via LDHA in the KO cells and that addition of exogenous pyruvate should allow MDH1 KO cells to increase their proliferation rate. Indeed, MDH1 KO cells were rescued by pyruvate (Figure 2d). However, glucose consumption by these cells was not affected by pyruvate addition (data not shown). These results are consistent with recent studies that suggest the cellular NAD/NADH ratio must be maintained to sustain proliferation and may serve a purpose in addition to that of supplying GAPDH and glycolysis(15, 16).
Figure 2. Loss of MDH1 slows proliferation and glucose consumption.
(a) Western blot showing protein expression of MDH1 and LDHA in control and MDH1 KO Jurkat cells (b) Graph shows cell number over time for indicated cell lines. Cells were plated at 50,000 cells per ml and counted every 24 hours. (c) Glucose levels in the spent medium were determined on day 3 and normalized to cell number. (d) MDH1 KO.1 and KO.2 cells were counted at 1, 3 and 5 days after addition of 1mM sodium pyruvate added to RPM1-1640, a pyruvate deficient medium. Data are the average and standard deviation of samples from experiments repeated at least three times. Significance was calculated using the student’s T test (* p<0.01, ** p<0.001).
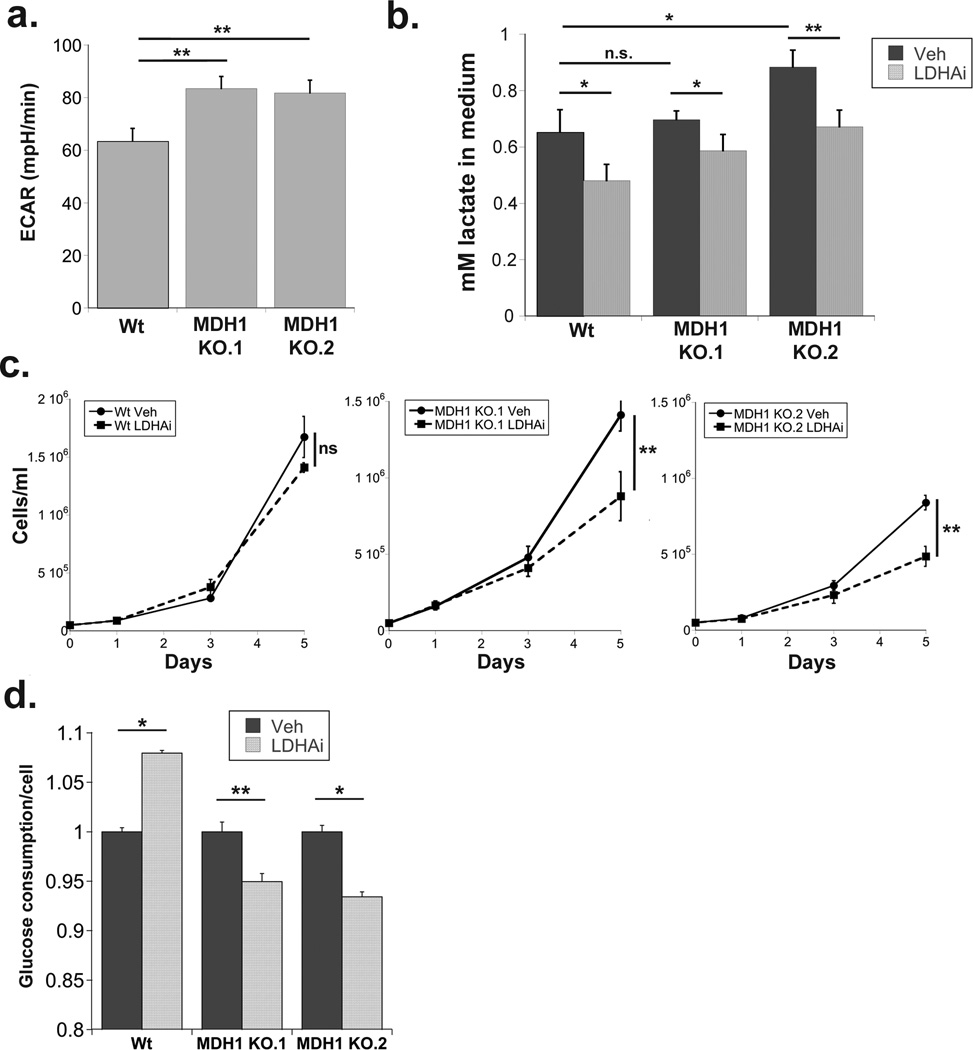
We also surmised that cells lacking MDH1 should exhibit greater dependence on LDHA to sustain glycolysis. Consistent with our hypothesis, the basal extracellular acidification rate (ECAR) of the MDH1 KO cells was significantly higher than that of wild-type (Figure 3a). LDHA inhibition has been shown to be effective at inhibiting aerobic glycolysis in several cancer cell types and is under consideration as a clinical therapeutic(17–20). We hypothesized that LDHA inhibitors would be less effective at suppressing proliferation in cells that utilize MDH1 for NAD regeneration than in cells that lacked the enzyme. We used the LDHA inhibitor GSK2837808A(17, 20) to inhibit lactate production in the MDH1 knock-out cell model (Figure 3b). LDHi had no effect on the growth of MDH1 wild-type cells, but significantly suppressed proliferation of MDH1 KO cells without inducing apoptosis (Figure 3c, Supplementary Figure 2). LDHA inhibition also caused a substantial decrease in glucose consumption in the MDH1 KO cells (Figure 3d). Interestingly, MDH1 wild-type cells treated with LDHAi increased glucose consumption (Figure 3d) suggesting that shifting the task of NAD regeneration to other dehydrogenases, such as MDH1, may enhance the efficiency of glycolysis.
Figure 3. Loss of MDH1 sensitizes cells to LDH inhibition.
(a) Extracellular acidification rate measured by Seahorse in Wt and MDH1 knock-out Jurkat cells. (b) Extracellular lactate levels in the medium of DMSO (Veh) or GSK2837808A (LDHAi) treated cells 48 hours after administration and normalized to cell number (c) Cell concentrations on the indicated days following treatment with either DMSO (Veh) or GSK2837808A (LDHAi). (d) Glucose levels in the spent medium from day 3, normalized to cell number and shown relative to levels detected in vehicle treated cell medium. Data are the average and standard deviation of samples from experiments repeated at least twice. Significance was calculated using the student’s T test (* p<0.01, ** p<0.001).
Malate dehydrogenase 1 is amplified in human tumors
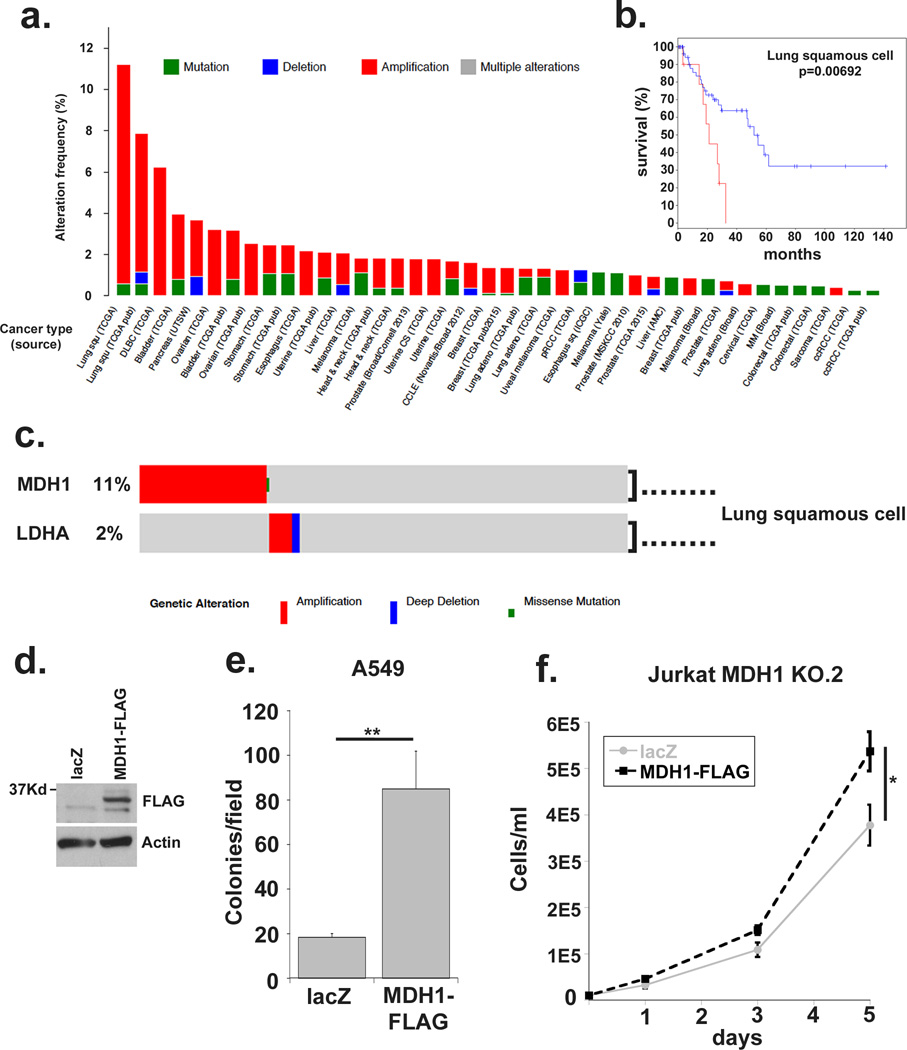
If MDH1 activity is critical for supporting proliferative metabolism, overexpression of the wild-type enzyme should impart a growth advantage and be selected for in cancers and specifically in tumors that arise in nutrient limited environments. The Cancer Genome Atlas (TCGA) database and cBioPortal(21, 22) showed MDH1 to be mutated in less than 1% of tumor samples queried (Supplementary Figure 4a). Moreover, amplification of this enzyme was preferred to deletion across tumor types (Figure 4a). MDH1 was amplified in almost 11% of one squamous cell lung carcinoma dataset, and a second MDH1 amplified squamous cell lung carcinoma subset showed approximately 50% reduction in disease free survival rate (Figure 4b). Notably MDH1 was more frequently amplified than LDHA in the TCGA database and the amplifications were almost always mutually exclusive (Figure 4c, Supplementary Figure 3b) which suggests a level of redundancy among these dehydrogenases and highlights the need to target both. Notably, the genomic locus of MDH1 (2p13) is not a commonly reported amplification event suggesting selection for MDH1 amplification is not a driver or passenger aberration, rather may be selected for its ability to impart a proliferative advantage.
Figure 4. MDH1 is amplified in human cancers and correlates with poor prognosis.
(a) The Cancer Genome Atlas online database was queried using cBioportal software to query MDH1 expression aberrations, including amplification, deletions and mutations in human tumors. (b) The lung squamous cell carcinoma subset was further interrogated to correlate MDH1 amplification with disease free survival. (c) MDH1 and LDHA queried together for genomic aberrations in the lung squamous cell carcinoma dataset. (d) Western blot demonstrating expression of MDH1-FLAG in stably transfected A549 cell lines. (e) Colony counts after 2-week soft agar colony formation assays. The average number of colonies from ten low-magnification microscopic fields from triplicate wells is shown. (f) MDH1 KO.2 Jurkat cells were stably transfected with MDH1-FLAG or lacZ and plated at 10,000 cells per ml. Cell concentration was plotted on days 1, 3 and 5. Data represented are the average and standard deviation of triplicate samples from a representative experiment repeated at least twice. Significance was calculated using the student’s T test (* p<0.01, ** p<0.001).
A hallmark of epithelial cancer cell behavior is the loss of contact inhibition, which can be measured by the ability to form non-adherent colonies in soft agar suspensions. We looked at soft agar colony formation following MDH1 over-expression in A549 lung cancer cells, which are highly glycolytic and particularly sensitive to lactate inhibition(20, 23). Over-expression of MDH1 in A549 lung carcinoma cells resulted in significantly higher soft agar colony formation compared to lacZ expressing controls (Figure 4d, Figure 4e and Supplementary Figure 3c). Furthermore, stable re-expression of MDH1-FLAG in Jurkat MDH1 knock-out cells increased their proliferation rate (Figure 4f). Together, these data point to a significant role for MDH1 in supporting proliferation in human tumor cells, and underscore the potential for MDH1 inhibition as a therapeutic strategy.
Reductive carboxylation of glutamine supports malate dehydrogenase 1 activity during proliferation
The existing scenario designating lactate dehydrogenase as the predominant source of cytosolic NAD regeneration presents two problems. First, the enhanced glycolysis associated with the proliferative state requires a one-to-one stoichiometry between glucose uptake and lactate production, not accounting for diversion of glucose carbons to biomass production. Second, the carbons that move through glycolysis to lactate are eventually secreted and lost, compounding the inefficiency. Evidence presented thus far points to MDH1 activity as a major alternative source of cytosolic NAD. Unlike lactate, which is largely disposed of as waste, malate can be further utilized for energy generation or for biomass. Additionally, the possibility that MDH1 substrate, OAA, derives from a carbon source other than glucose must also be considered. We returned to our Noxa over-expressing Jurkat cell model to test this possibility.
We used targeted metabolomics to trace the fate of carbons derived from [1,2-13C] D-glucose through glycolysis and into the TCA cycle (Figure 5a). N5 cells showed increased enrichment of glycolytic metabolites through glyceraldehyde 3-phosphate (G3P), but not beyond phosphoglycerate (Figure 5b). Glucose carbon enrichment in the pentose phosphate pathway intermediate S7P, indicated a fraction of glucose carbon was diverted to the PPP (Supplementary Figure 4a, b). Correspondingly, glucose contribution to TCA cycle intermediates was reduced in N5 cells (Figure 5c). Importantly, enrichment of pyruvate and lactate in N5 cells was similar to controls (Figure 5b), lending support to our previous observation that the increased glucose carbon consumed by N5 cells was diverted prior to enolase(4). These data also argue for the existence of an alternative source other than lactate that regenerates NAD to support the increased glucose consumption we observe in these cells(4).
Figure 5. Noxa over-expressing cells increase glycolysis but not lactate production from glucose.
(a) Schematic tracing the incorporation of [13C] label from [1,2-13C] glucose (red) through glycolysis and the TCA cycle, and [U-13C] glutamine (blue) into TCA cycle metabolites. (b) Glycolysis metabolites labeled from [1,2-13C] glucose following 24 hours of labeling, shown as a percentage of total metabolite measured using LC/MS. G6P: glucose 6 phosphate, FBP: fructose 1,6 bisphosphatase, G3P: glyceraldehyde 3 phosphate, 2PG/3PG: phosphoglycerate. (c) TCA metabolites labeled from glucose carbon measured via LC/MS following 24 hours of labeling. (d) Enrichment of glutaminolytic and TCA metabolites derived from [U-13C] glutamine following 24 hours of labeling, detected using LC/MS. Gln: glutamine, Glu: glutamate, aKG: alpha-ketoglutarate, Fum: fumarate. Number of labeled carbons is indicated in parentheses. Data represented are the average and standard deviation of triplicate samples from a representative experiment repeated at least twice. Significance was calculated using the student’s T test (* p<0.01, ** p<0.001).
Glutamine serves as an alternative source of carbon for anaplerosis of the oxidative TCA cycle during proliferation(24). Glutaminolysis produces alpha-ketoglutarate (αKG) which is metabolized to succinate following the release of CO2 as it enters the TCA cycle (Figure 5a). Targeted studies with uniformly labeled [U-13C] glutamine showed increased enrichment of glutamine carbons in the TCA cycle metabolites of N5 cells compared to controls (Figure 5d). Glutamine contribution to lactate was, however, insignificant (Supplementary Figure 4c).
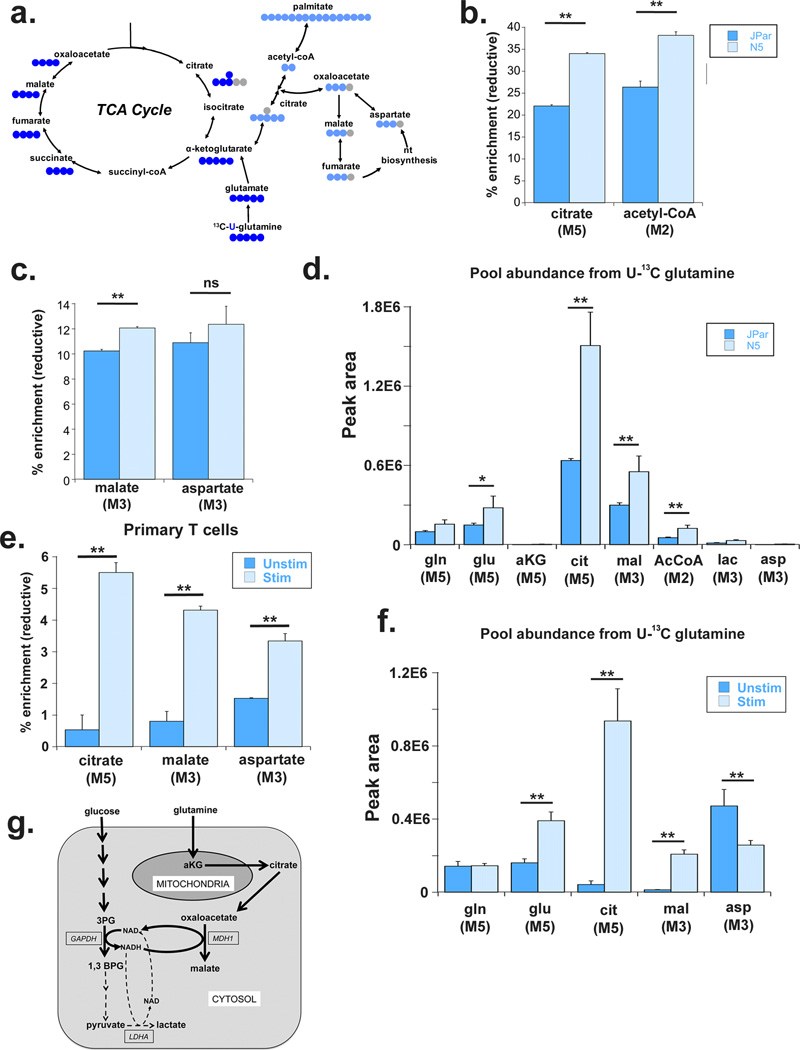
Glutamine is also metabolized through reductive carboxylation where αKG is directly converted to isocitrate and citrate, which is then split into acetyl-CoA and oxaloacetate (OAA) in the cytosol(25) (Figure 6a). While reductive carboxylation has been suggested as the preferred metabolic path for channeling glutamine carbons via acetyl-CoA into palmitate for de novo lipid synthesis(25, 26) (Figure 6a), the fate of the cytosolic OAA has been less clear. We observed a significant increase in the enrichment of reductively generated citrate and acetyl-CoA (Figure 6b), and MDH1 generated M3 malate, but not of M3 aspartate in the N5 cells (Figure 6c). Overall, the pool abundance of metabolites generated from reductive metabolism of glutamine, including MDH1 generated malate, significantly increased in N5 cells (Figure 6d). We had shown in Figure 1c that enrichment of M1 malate increased following co-stimulation of primary human T cells incubated with [4-2H] glucose. We also detected a significant increase in the reductive carboxylation of glutamine and production of M3 malate after stimulation both by enrichment percentage as well as pool abundance (Figure 6e and 6f). Interestingly, despite the increase in percent enrichment of reductively generated aspartate, the M3 aspartate pool decreased after stimulation suggesting the generation of aspartate through the reductive pathway was independent of the proliferative state. Taken together these data suggest that the increased MDH1 activity helps stem the flux of glucose carbons to LDHA by using glutamine as a carbon source to supply cytosolic NAD for glycolysis.
Figure 6. Noxa over-expressing cells increase reductive carboxylation of glutamine to support cytosolic malate production.
(a) Schematic tracing the incorporation of [13C] label from [U-13C] glutamine metabolized in oxidative (royal blue) and reductive (light blue) directions. (b) Percent enrichment of citrate and acetyl-CoA derived from reductive carboxylation of [U-13C] glutamine 24 hours after labeling, determined via LC/MS. The number of labeled carbons is indicated in parentheses below each metabolite. (c) Percent enrichment of malate and aspartate derived from reductive carboxylation of [U-13C] glutamine 24 hours after labeling. (d) Pool abundance of indicated metabolites in JPar and N5 cells quantified by peak area and normalized to sample protein concentration. Number of labeled carbons indicated in parenthesis below metabolites. (e) Percent enrichment of malate and aspartate derived from reductive carboxylation of [U-13C] glutamine in primary human T cells 24 hours after labeling and activation. (f) Pool abundance of indicated metabolites in unstimulated and stimulated primary human T cells quantified by peak area and normalized to sample protein concentration. Isotopomer label indicated in parenthesis below metabolites. Data presented are the average and standard deviation of triplicate samples from a representative experiment repeated at least twice. Significance was calculated using the student’s T test (* p<0.01, ** p<0.001). (g) Simplified model to show how malate dehydrogenase 1 and glutamine help support the increased glucose uptake and glycolysis by replenishing the NAD consumed by the GAPDH catalyzed step, a function primarily attributed to LDH activity in highly glycolytic cells. Our studies suggest that, in proliferating cells, MDH1 is a significant alternative source of cytosolic NAD, utilizing the carbons from the reductive carboxylation of glutamine for the NADH dependent synthesis of malate.
Discussion
This study focuses on pathways proliferating cells utilize to provide GAPDH with a constant supply of its co-factor NAD to sustain accelerated glycolysis. The regeneration of cytosolic NAD from NADH has been largely attributed to the production of lactate through LDH activity(1–3). However, a number of studies have reported that glucose carbons are lost to biosynthetic pathways highlighting the need for a supplementary reaction and carbon source to regenerate cytosolic NAD(1). This study shows MDH1 plays a critical role in replenishing cytosolic NAD to support increased glycolysis during proliferation (Figure 6g).
We initially identified MDH1 as a major source of cytosolic NAD using a strategy of labeling cells with [4-2H] glucose and tracing the deuterium from glucose into NADH, and then into [2H]-labeled metabolites (Figure 1). The use of [4-2H] glucose to trace the utilization of NADH is a relatively novel strategy that has previously been used to investigate specific pathways(27–29). Here we took an unbiased approach to identify novel sources of NAD regeneration during proliferation. This strategy revealed malate is a significant hydrogen acceptor during proliferation not only in our Jurkat model but also in activated primary human T cells and in a variety of cancer cell lines, underscoring the physiological relevance of the pathway for replenishing the NAD supply in the cytosol. Moreover, MDH1 KO cells proliferate more slowly and consume less glucose compared to wild-type cells while MDH1 over-expressing cells exhibit increased anchorage independence growth. This is the first study to reveal a strong association of MDH1 with proliferative metabolism and glycolysis.
These results do not contradict the numerous reports of increased lactate and LDH activity as a major supporter of the enhanced glycolysis in proliferating cells. Rather, the data suggest that malate synthesis likely serves as an alternative, and arguably more efficient, route for replenishing the NAD turning over rapidly in the cytosol. The efficiency arises from the fact that malate can be further metabolized for biomass, energy generation or ROS control, whereas lactate is primarily secreted into the extracellular environment as waste. A recent study has also demonstrated the importance of MDH1 activity in KRAS driven pancreatic ductal carcinomas to control ROS by providing malate as a substrate for malic enzyme and NADPH production(30). Moreover, LDHA inhibition did not significantly repress growth in Jurkat cells unless MDH1 was knocked out. Surprisingly, LDHA inhibition increased glucose consumption when MDH1 was present (Figure 2e). We suggest that blocking the flow of carbons to LDHA in the presence of an active MDH1, capable of replenishing cytosolic NAD using glutamine carbons, increases the availability of glucose carbons for biomass. Thus, inhibition of LDHA could have a beneficial effect on these cells. Similar effects of inhibiting the downstream flow of glucose carbons have been described previously with a variant of pyruvate kinase (PKM2) expressed in proliferating cells(31). We did not detect increased expression of MDH1 in N5 cells compared to JPar (Supplementary Figure 4D). Additional cell culture models also seemed to express relatively similar levels of MDH1 (Supplementary Figure 4E). However, TCGA database revealed MDH1 amplification in solid tumor models (Figure 4) suggesting that, unlike in cell culture models, the availability of substrate drives selection for MDH1 over-expression. A recent study also revealed MDH1 amplification and increased MDH1 activity in human PDAC samples(30). In demonstrating the importance of MDH1-mediated malate synthesis to cytosolic NAD regeneration in highly glycolytic cells, this study offers fresh insights into metabolic alterations aimed at meeting the demands of growth and division and highlights the need for alternative strategies to target the increased consumption and utilization of glucose in cancers.
Materials and Methods
Cell Culture
Jurkat, HL60, and K562 (ATCC) cells were maintained in RPMI-1640 medium supplemented with 10% FBS, 100 U/ml penicillin, 100ug/ml streptomycin, NEAA, and 4mM L-glutamine. HEK293, A549 and HeLa (ATCC) cells were maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100ug/ml streptomycin. JPar and N5 cells have been previously described(4). MDH1 KO cells were a generous gift from Kivanc Birsoy and David Sabatini(14). The LDHA inhibitor, GSK 2837808A (Tocris Bioscience, Bristol UK) was solubilized in DMSO, added to culture medium at the time of plating and maintained at 10uM throughout the experiment. Concentration of glucose in the medium was measured via the EnzyChrom Glucose Assay Kit (BioAssay Systems, Hayward, CA USA) according to the manufacturer’s protocol, and then normalized to cell counts. Concentration of lactate in the medium was measured via the Lactate Assay kit (Sigma-Aldrich, St. Louis, MO USA) following filtration through a 10,000Kd MWCO spin filter. Lactate measurements were then normalized to cell number.
Plasmid Constructs
The wild-type MDH1 vector was a kind gift from Hong-Duk Youn(32). The MDH1 insert was amplified from the source vector to carry a 5’ FLAG tag and cloned into pcDNA 3.1 and verified by sequencing. The pcDNA 3.1 lacZ Myc/His construct was used as a vector control. Transfections were carried out in 5E6 cells A549 cells using 1ug of plasmid and 5ul of FuGENE HD (Promega, Madison, WI USA) reagent in 200ul of antibiotic free medium. After a 15-minute incubation at ambient temperature, the DNA/FuGENE complex was added to a sub-confluent 10cm plate containing 10ml of antibiotic free medium. After 24 hours of transfection, medium was replaced and supplemented with 1mg/ml of G418 (Geneticin) for one week. For Jurkat transfections, 10ug of plasmid was electroporated using a Neon Transfection System (Invitrogen, Carlsbad, CA, USA) into 10E6 cells in a 100ul Neon tip using 1350V at a pulse width of 10ms for 3 pulses. Cells were returned to antibiotic free medium for 24 hours. After 24 hours, medium was supplemented with G418 for 10 days. Live cells were recovered using a Ficoll density gradient.
Western Blot
Cells were washed 1X with cold PBS, resuspended in Radio immunoprecipitation (RIPA) buffer supplemented with protease and phosphatase cocktails. Lysates were incubated on ice for 10 min and spun at 10,000 rpm at 4°C in a microcentrifuge. The supernatant was collected for further analysis. After BCA protein assay, equal amounts of protein (10–50µg) were resolved by SDS-PAGE, transferred to nitrocellulose and probed with antibodies specific for Noxa (Santa Cruz, Santa Cruz, CA USA; SC-56169), beta actin (Santa Cruz; SC-69879), MDH1 (Abcam, Cambridge, MA USA; Ab180152), LDHA (Cell Signaling Technology, Danvers, MA USA; #3582) or the FLAG epitope (Sigma-Aldrich, St. Louis, MO USA; F7425).
Seahorse Extracellular Flux Assay
Seahorse medium was supplemented with 10 mM glucose, 1mM sodium pyruvate and 4mM L-glutamine and adjusted to pH 7.4 with NaOH. Cells were plated at 250,000 cells per well and adhered to the Seahorse 96 well culture plate using CellTak (Corning, Oneonta, NY USA). Extracellular acidification rate was measured using the Seahorse XFe96 (Agilent, Santa Clara, CA USA).
Primary T cell experiments
Human peripheral blood mononuclear cells (PBMCs) were obtained from a healthy donor via Memorial Blood Center (St. Paul, MN USA). PBMCs were purified from red blood cells using Histopaque. CD3+ T cells were isolated from PBMCs using the negative selection MACS magnetic separation system (Miltenyi Biotec, San Diego, CA USA) and allowed to recover in RPMI-1640 supplemented with 10% FBS, 100 U/ml penicillin, 100µg/ml streptomycin, NEAA, and 4mM L-glutamine overnight. At 18 hours, live T cells were recovered by Ficoll density gradient centrifugation and allowed to rest for 4 hours before being washed in glucose-free medium and starved of glucose for 1 hour. For labeling, T cells were moved to culture dishes coated with 5µg/ml anti-CD3 antibody (Biolegend, San Diego, CA USA). 5ug/ml of anti-CD-28 antibody (Biolegend) was then added along with 10mM [4-2H] glucose. Cells were harvested 18 hours later, resuspended in 150µl of cold −20°C methanol, snap frozen and stored at −80°C until required for metabolomics analysis.
Metabolic labeling experiments in cell culture
Live, healthy cells were recovered by Ficoll density gradient centrifugation within 24 hours prior to experiment. At T0 10E6-20E6 cells per sample were pelleted and washed in glucose/glutamine free medium. For glucose labeling experiments, cells were resuspended at 1E6 cells/ml in complete glucose free medium supplemented with 10% dialyzed FBS, NEAA, and 4mM L-glutamine for 1 hour. After starvation, cells were supplemented with 10mM [13C] or [2H] labeled glucose for the time indicated in each experiment. For glutamine labeling experiments cells were washed and then resuspended in complete glutamine free medium supplemented with 10% dialyzed FBS and 10mM glucose for 3–4 hours. Following depravation, 4mM [U-13C] glutamine was added to the medium and cells were incubated for 24 hours. Cells were then pelleted and washed 1X in ice cold PBS. Pellets were then resuspended in 100–200µl −20°C methanol, snap frozen and stored at −80C. Liquid chromatography/mass spectrometry (LC/MS) and gas chromatography/mass spectrometry (GC/MS) were used for identification and quantification of labeled metabolites of all samples. [1,2-13C] glucose and [U-13C] glutamine were purchased from Cambridge Isotopes (Tewksbury, MA USA) and [4-2H] glucose was purchased from Omicron Biochemicals (South Bend, IN USA).
Central Carbon Metabolism sample preparation
Microtubes containing cell pellets were removed from −80°C storage and maintained on wet ice throughout the processing steps. To initiate protein precipitation, 0.3 mL of a chilled mixture of methanol, chloroform and water (8:1:1) (EMD) was added to each sample, the mixture vortexed briefly, and allowed to incubate on ice for 10 mins. Post-incubation, the vortex step was repeated, and samples centrifuged at 14,000 RPM, for 10 mins in 4°C. Post-centrifugation, 100 µL of supernatant was transferred to an autosampler vial for LC-MS analysis. From the remaining supernatant from each sample, a small aliquot was transferred to a new microtube to create a pooled sample for quality control purposes. The remaining supernatant from each sample was transferred to an autosampler vial, and brought to dryness. To each sample, 50 µL of 20 mg/mL of a solution of methoxyamine hydrochloride in pyridine was added. Samples were vortexed briefly and the samples incubated at 37°C for 90 mins. Samples were removed from heating and allowed to cool to room temperature. Addition of 50 µL of MTBSTFA with 1% tBDCMS (Regis Technologies, Morton Grove, IL USA) and samples were incubated at 70°C for one hour. Samples were allowed to cool and GC-MS was performed. A series of calibration standards were prepared along with samples for quality control of instrument performance and chromatography for both LC and GC analysis.
LC-MS analysis
LC-MS analysis was performed on an Agilent system consisting of a 1260 UPLC module coupled with a 6520 Quadrupole-Time-of-flight (QTOF) mass spectrometer (Agilent Technologies, Santa Clara, CA USA) Metabolites were separated on a 150×1mm Luna NH2 hydrophilic interaction chromatography column (Phenomenex, Torrance, CA USA) using 10 mM ammonium acetate in water, adjusted to pH 9.9 with ammonium hydroxide, as mobile phase A, and acetonitrile as mobile phase B. The flow rate was 0.075 mL/min and the gradient was linear 20% to 100% A over 15 mins, followed by isocratic elution at 100% A for 5 minutes. The system was returned to starting conditions (20% A) in 0.1 min and held there for 10 minutes to allow for column re-equilibration before injecting another sample. The mass spectrometer was operated in ESI- mode according to previously published conditions(33).
GC-MS analysis
GC-MS analysis was performed on an Agilent 69890N GC −5975 MS detector with the following parameters: a 1µL sample was injected splitlessly on an HP-5MS 15m column (Agilent Technologies) with a helium gas flow rate of 1.4 mL/min. The GC oven initial temperature was 60°C and was increased at 10°C per minute to 300°C, and held at 300°C for 5 mins. The inlet temperature was 250°C and the MS-source and quad temperatures were 230°C and 150°C respectively.
Metabolite Data Analysis
For LC-MS analysis, metabolite peaks (both molecular ion and stable isotopes) were identified by matching the retention time and mass (+/− 10 ppm) to authentic standards. For GC-MS, metabolite peaks (both molecular ion and stable isotopes) were identified by matching the retention time and mass (+/− 0.2 Da) to authentic standards. Metabolite masses (M-H for LC-MS, TBDMS derivative for GC-MS) are indicated in Supplementary Table 1, along with the qualifying ions used for the GC-MS identification. Isotope peaks (natural abundance and stable isotope labeling) were identified as increments of +1.0034 Da and +1 Da, respectively, for LC-and GC-MS, from the masses shown in the Supplementary Table 1. Presence of stable isotope labeling above the natural abundance was verified by absence of the same MS peak in the unlabeled preparation of the same condition. Peak areas were integrated using MassHunter Quantitative Analysis vB.07.00 (Agilent Technologies) Peak areas were corrected for natural isotope abundance [using an in-house written software package based on the method described previously28 and the residual isotope signal was reported. Data were normalized to cell protein content prior to analysis of metabolite fluxes for Central Carbon metabolites.
Soft Agar Colony Assays
Soft agar plates were prepared by mixing 3.2% low melt agarose with DMEM culture medium to yield 0.8% agarose at 38.5C. 1ml was added per well of a 6 well plate and allowed to solidify. 11,500 cells per ml were then mixed with 3.2% agarose to yield 0.48% agarose in DMEM culture medium and allowed to solidify. 1ml of DMEM supplemented with 1mg/ml G418 was added to the top of the agarose. Each condition was plated in triplicate. Plates were incubated for 15 days. Colonies were stained with 0.005% crystal violet in PBS and 4% formaldehyde. Images were captured on an inverted microscope using QCapture software. Ten microscopic fields were captured and counted per condition. Colonies were counted using ImageJ software.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. David Sabatini and Kivanc Birsoy for the MDH1 KO Jurkat cells, and Hong-Duk Youn for the MDH1 construct. We would also like to thank Todd Rappe and University of Minnesota Nuclear Magnetic Resonance Core for help with NMR analysis. We are grateful to Dr. David Bernlohr, Michael Downey, Yan Yan and Dr. Jenna Benson for stimulating discussions. We also thank Drs. Lucas Sullivan and Sandra Armstrong for valuable suggestions. This study was supported by a Seed Grant and an Infrastructure grant from the University of Minnesota Academic Health Center (to AK), National Institutes of Health (NIH) grant R01 CA157971, P/F grant, 3002751553, through the NIH-funded RCMC in Michigan (to AK) and by T32 training fellowship, CA009138, and F31 award, CA177119 (to EAH). The study utilized Metabolomics Core Services supported by grant U24 DK097153 of NIH Common Funds Project to the University of Michigan.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supplementary information is available at the Oncogene website.
References
- 1.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. Epub 2011/10/12. [DOI] [PubMed] [Google Scholar]
- 2.Goldman RD, Kaplan NO, Hall TC. Lactic Dehydrogenase in Human Neoplastic Tissues. Cancer Res. 1964;24:389–399. Epub 1964/04/01. [PubMed] [Google Scholar]
- 3.Liberti MV, Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci. 2016;41(3):211–218. doi: 10.1016/j.tibs.2015.12.001. Epub 2016/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowman XH, McDonnell MA, Kosloske A, Odumade OA, Jenness C, Karim CB, et al. The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Molecular cell. 2010;40(5):823–833. doi: 10.1016/j.molcel.2010.11.035. Epub 2010/12/15. [DOI] [PubMed] [Google Scholar]
- 5.Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer research. 2011;71(22):6921–6925. doi: 10.1158/0008-5472.CAN-11-1457. Epub 2011/11/16. [DOI] [PubMed] [Google Scholar]
- 6.Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89(3):307–315. doi: 10.1016/j.ygeno.2006.11.012. Epub 2007/01/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Savchenko A, Yakunin A, Zhang R, Edwards A, Arrowsmith C, et al. Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643. J Biol Chem. 2003;278(10):8804–8808. doi: 10.1074/jbc.M211892200. Epub 2002/12/24. [DOI] [PubMed] [Google Scholar]
- 8.Menzies KJ, Zhang H, Katsyuba E, Auwerx J. Protein acetylation in metabolism - metabolites and cofactors. Nature reviews Endocrinology. 2016;12(1):43–60. doi: 10.1038/nrendo.2015.181. Epub 2015/10/28. [DOI] [PubMed] [Google Scholar]
- 9.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185(2):1037–1044. doi: 10.4049/jimmunol.0903586. Epub 2010/06/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16(6):769–777. doi: 10.1016/s1074-7613(02)00323-0. Epub 2002/07/18. [DOI] [PubMed] [Google Scholar]
- 11.Pike Winer LS, Wu M. Rapid analysis of glycolytic and oxidative substrate flux of cancer cells in a microplate. PloS one. 2014;9(10):e109916. doi: 10.1371/journal.pone.0109916. Epub 2014/11/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg EB, Colowick SP. The Role of Glycolysis in the Growth of Tumor Cells. 3. Lactic Dehydrogenase as the Site of Action of Oxamate on the Growth of Cultured Cells. The Journal of biological chemistry. 1965;240:2786–2790. Epub 1965/07/01. [PubMed] [Google Scholar]
- 13.Calvo MN, Bartrons R, Castano E, Perales JC, Navarro-Sabate A, Manzano A. PFKFB3 gene silencing decreases glycolysis, induces cell-cycle delay and inhibits anchorage-independent growth in HeLa cells. FEBS letters. 2006;580(13):3308–3314. doi: 10.1016/j.febslet.2006.04.093. Epub 2006/05/16. [DOI] [PubMed] [Google Scholar]
- 14.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162(3):540–551. doi: 10.1016/j.cell.2015.07.016. Epub 2015/08/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gui DY, Sullivan LB, Luengo A, Hosios AM, Bush LN, Gitego N, et al. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell metabolism. 2016;24(5):716–727. doi: 10.1016/j.cmet.2016.09.006. Epub 2016/10/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162(3):552–563. doi: 10.1016/j.cell.2015.07.017. Epub 2015/08/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Billiard J, Dennison JB, Briand J, Annan RS, Chai D, Colon M, et al. Quinoline 3-sulfonamides inhibit lactate dehydrogenase A and reverse aerobic glycolysis in cancer cells. Cancer Metab. 2013;1(1):19. doi: 10.1186/2049-3002-1-19. Epub 2013/11/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Augoff K, Hryniewicz-Jankowska A, Tabola R. Lactate dehydrogenase 5: an old friend and a new hope in the war on cancer. Cancer Lett. 2015;358(1):1–7. doi: 10.1016/j.canlet.2014.12.035. Epub 2014/12/22. [DOI] [PubMed] [Google Scholar]
- 19.Rani R, Kumar V. Recent Update on Human Lactate Dehydrogenase Enzyme 5 (hLDH5) Inhibitors: A Promising Approach for Cancer Chemotherapy. J Med Chem. 2015 doi: 10.1021/acs.jmedchem.5b00168. Epub 2015/09/05. [DOI] [PubMed] [Google Scholar]
- 20.Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, et al. Targeting lactate dehydrogenase--a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014;19(5):795–809. doi: 10.1016/j.cmet.2014.03.003. Epub 2014/04/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. Epub 2013/04/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. Epub 2012/05/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu PP, Liao J, Tang ZJ, Wu WJ, Yang J, Zeng ZL, et al. Metabolic regulation of cancer cell side population by glucose through activation of the Akt pathway. Cell death and differentiation. 2014;21(1):124–135. doi: 10.1038/cdd.2013.131. Epub 2013/10/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21(9):1037–1049. doi: 10.1101/gad.1529107. Epub 2007/04/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. Epub 2011/11/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, et al. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat Commun. 2013;4:2236. doi: 10.1038/ncomms3236. Epub 2013/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510(7504):298–302. doi: 10.1038/nature13236. Epub 2014/05/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Molecular cell. 2014;55(2):253–263. doi: 10.1016/j.molcel.2014.05.008. Epub 2014/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Shah S, Fan J, Park JO, Wellen KE, Rabinowitz JD. Malic enzyme tracers reveal hypoxia-induced switch in adipocyte NADPH pathway usage. Nature chemical biology. 2016;12(5):345–352. doi: 10.1038/nchembio.2047. Epub 2016/03/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YP, Zhou W, Wang J, Huang X, Zuo Y, Wang TS, et al. Arginine Methylation of MDH1 by CARM1 Inhibits Glutamine Metabolism and Suppresses Pancreatic Cancer. Molecular cell. 2016;64(4):673–687. doi: 10.1016/j.molcel.2016.09.028. Epub 2016/11/15. [DOI] [PubMed] [Google Scholar]
- 31.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. Epub 2008/03/14. [DOI] [PubMed] [Google Scholar]
- 32.Lee SM, Kim JH, Cho EJ, Youn HD. A nucleocytoplasmic malate dehydrogenase regulates p53 transcriptional activity in response to metabolic stress. Cell death and differentiation. 2009;16(5):738–748. doi: 10.1038/cdd.2009.5. Epub 2009/02/21. [DOI] [PubMed] [Google Scholar]
- 33.Lorenz MA, Burant CF, Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Analytical chemistry. 2011;83(9):3406–3414. doi: 10.1021/ac103313x. Epub 2011/04/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.