Summary
To investigate the role of immune cells in lung regeneration, we used a unilateral pneumonectomy model that promotes the formation of new alveoli in the remaining lobes. Immunofluorescence and single cell RNA- sequencing found CD115+ and CCR2+ monocytes and M2-like macrophages accumulating in the lung during the peak of type 2 alveolar epithelial stem cell (AEC2) proliferation. Genetic loss of function in mice and adoptive transfer studies revealed that bone marrow-derived macrophages traffic to the lung through a CCL2-CCR2 chemokine axis and are required for optimal lung regeneration, along with Il4ra-expressing leukocytes. Our data suggest that these cells modulate AEC2 proliferation and differentiation. Finally, we provide evidence that group 2 innate lymphoid cells are a source of IL13 that promotes lung regeneration. Together, our data highlight the potential for immunomodulatory therapies to stimulate alveologenesis in adults.
Keywords: Lung regeneration, pneumonectomy, type 2 alveolar pneumocyte, macrophage, monocyte, ILC2
Introduction
The past few decades have brought many discoveries about the identities and behaviors of epithelial stem cells in the lung (Hogan et al., 2014; Kotton and Morrisey, 2014; Rock and Hogan, 2011). These data have intensified efforts toward stimulating the regeneration of the human lung from endogenous progenitors to ameliorate declining function associated with aging and progressive end stage disease. However, the realization of this goal requires a more complete knowledge of the molecular and cellular events mediating lung homeostasis, pathological remodeling and regeneration.
In some species, partial pneumonectomy (PNX), the surgical removal of one or more lung lobes, stimulates compensatory growth in the remaining lobes that restores alveolar surface area and diffusing capacity (Buhain and Brody, 1973; Dane et al., 2013; Fehrenbach et al., 2008; Ravikumar et al., 2013; Voswinckel et al., 2004). This compensatory growth requires the coordinated proliferation and rearrangement of numerous epithelial and stromal cell types, including type 2 alveolar epithelial cells (AEC2s), an epithelial stem cell population capable of self-renewal and differentiation into type 1 alveolar epithelial cells (AEC1s) (Barkauskas et al., 2013; Brody et al., 1978; Desai et al., 2014; Ding et al., 2011; Hoffman et al., 2010; Li et al., 2013; Nolen-Walston et al., 2008; Rock et al., 2011a). Recent efforts have elucidated some components of the regenerative AEC2 niche, but the roles of recruited and resident lung macrophages during adult alveologenesis post-PNX remain unclear (Chamoto et al., 2012 Chamoto et al., 2013; Chen et al., 2012; Ding et al., 2011; Rafii et al., 2015).
Tissue macrophages in different organs are heterogeneous with respect to origin, marker expression, and functions. Recent work has helped to delineate these parameters for macrophages in the lung (Tan and Krasnow, 2016). Three main subpopulations of lung macrophages have been characterized by their ontogeny, mode of maintenance, and location within the tissue. Two of these, “primitive” interstitial macrophages and alveolar macrophages, are derived from hematopoietic progenitors arising from the yolk sac and fetal liver, respectively. The third population of “definitive” interstitial macrophages is derived from circulating bone marrow-derived monocytes and replaces the “primitive” interstitial macrophages over time. Recent data suggest that lung macrophages are maintained independently by self-renewal under steady-state conditions (Guilliams et al., 2013; Hashimoto et al., 2013; Hoeffel and Ginhoux, 2015; Hoeffel et al., 2015; Jakubzick et al., 2013; Perdiguero et al., 2015; Schulz et al., 2012; Suzuki et al., 2014; Yona et al., 2013). Bone marrow-derived and circulating monocytes replenish and supplement resident macrophages only after tissue macrophage depletion and during some inflammatory/regenerative states (Hashimoto et al., 2013; Yona et al., 2013), often through a mechanism involving the chemokine receptor CCR2 (Willenborg et al., 2012; Lee et al., 2015; Nishiyama et al., 2015; Ramachandran et al., 2012). Both resident and recruited macrophages are capable of proliferation at their sites of action (Davies et al., 2013).
Mounting evidence supports a model in which macrophages play essential roles in the regeneration of organs including limbs, intestines, liver, kidney, and heart (Aurora and Olson, 2014; Aurora et al., 2014; Boulter et al., 2012; Dutta et al., 2015; Epelman et al., 2014; Gibbons et al., 2011; Godwin et al., 2013; Lin et al., 2010; Pull et al., 2005; Ramachandran et al., 2012). However, while their roles in lung host defense are relatively well established (Aggarwal et al., 2014; Herold et al., 2011), including how epithelial-macrophage crosstalk regulates lung immunity (Westphalen et al., 2014), little is known about how macrophages might modulate lung regeneration, including adult alveologenesis. The pathology of bleomycin-induced lung fibrosis in mice was worsened when inflammatory monocytes were adoptively transferred during the fibrotic stage, but also when macrophages were depleted during the recovery stage (Gibbons et al., 2011). These data illustrate a dynamic requirement for macrophages in lung maintenance and repair. Increased numbers of alveolar macrophages have been reported in the context of PNX-induced lung regeneration (Chamoto et al., 2012 Chamoto et al., 2013), and parabiosis experiments demonstrated that this results from local proliferation rather than recruitment from circulation (Chamoto et al., 2012). Whether these cells affect lung regeneration was not addressed. This same group showed that Cd11b is required for optimal regeneration (Chamoto et al., 2013). However, the nonspecific expression of Cd11b by a number of leukocyte subpopulations, both interstitial and in the airspace, leaves many unanswered questions regarding the mechanisms by which resident and recruited cells promote lung regeneration.
Once macrophages arrive at their site of action, they are polarized along a spectrum by microenvironmental cues (Martinez and Gordon, 2014; Murray et al., 2014). Generally speaking, classically activated macrophages (M1) are associated with inflammatory conditions, while “alternatively activated” macrophages (M2) have been implicated in wound repair and tissue regeneration (Aurora and Olson, 2014; Aurora et al., 2014; Gibbons et al., 2011; Gordon and Martinez, 2010). Cytokines associated with type 2 immunity, such as IL-4 and IL-13 signaling through IL4-Rα heterodimers, can induce M2 macrophage polarization and variably activate transcription of Arg1, Fizz1, Ym1 (Chil3), and Mrc1 (Stein et al., 1992). M2 macrophages are best defined functionally. For example, M2 macrophages may dampen inflammation through IL-10 secretion, secrete growth factors that directly affect stem cell behavior, or remodel matrix to reverse fibrosis (De’Broski et al., 2004; Ruffell et al., 2009; Shiraishi et al., 2016). M2-polarized macrophages may arise from either tissue resident or monocyte-derived macrophages (Arnold et al., 2007; Egawa et al., 2013; Shechter et al., 2013). The polarization state of macrophages in the regenerating lung following PNX has not been reported.
Here, we use flow cytometry, immunofluorescence, and both population level and single cell RNA sequencing to characterize the dynamics of macrophage subpopulations following PNX. Myeloid cells in the regenerating lung were most abundant at the peak of AEC2 stem cell proliferation. We use genetic loss of function and adoptive transfer experiments to demonstrate that recruited CCR2 monocytes and Th2 cytokine signaling are critical components of the regenerative niche required for lung regeneration post-PNX. Our data suggest that monocytes and macrophages stimulate AEC2 progenitor cell behaviors. Finally, we provide evidence that type 2 innate lymphoid cells are a source of IL13 in the regenerating lung. These data establish important roles myeloid cells in promoting regenerative alveologenesis.
Results
Increased numbers of macrophages in the regenerating lung post-PNX
Immunofluorescence on sections of lungs from Csf1r-GFP reporter mice, in which macrophages, monocytes and some dendritic cells express green fluorescent protein (GFP) (Rae et al., 2007), showed a peak in the number of GFP+ cells at 7d post-PNX and a return to steady state by 14d (Figure 1A). These cells were localized throughout the tissue, in close proximity to AEC2s and at the periphery of the lung where the majority of alveologenesis is thought to occur (Figure S1) (Konerding et al., 2012). Flow cytometry showed that CSF1R-GFP+, F4/80+ lung macrophages were significantly increased 7d post-PNX (18.6% +/− 2.8% of total live cells) compared to 7d post-sham littermate controls (14.4% +/− 2.1%) (Figure 2B). These data are consistent with a previous report demonstrating an increase in lung macrophages after PNX (Chamoto et al., 2012). Both interstitial macrophages/circulating monocytes (F4/80+, CD11b+) and alveolar macrophages (F4/80+, CD11c+) were significantly increased at 7d after PNX (29.1% +/− 3.2% of CD45+ and 15.4% +/− 2.1% of CD45+, respectively) compared to sham surgery (19.9% +/− 2.7% and 9.6% +/− 1.6%) (Figures 1C, 1D, 1E, and 1F). We also observed a transient increase in neutrophils (CD45+, CD11b+, Ly6G+) 4d after PNX, but this subsided by 7d after PNX when macrophage numbers and epithelial proliferation peaked (Figure S1).
Figure 1.

Increased numbers of myeloid cells post-PNX. (A) Immunofluorescent staining of lung sections after PNX shows increased number of CSF1R-GFP+ cells (green, myeloid lineages) compared to unoperated animals. Scale bar = 100 um. (B) Flow cytometry shows increased numbers of CSF1R-GFP+,F4/80+ macrophages 7d post-PNX compared to sham operated animals. n=3 animals per group. Data shown are mean +/− SD. (C) Flow cytometry shows increased numbers of CD45+,,F4/80+,CD11b interstitial macrophages and monocytes 7d post-PNX compared to sham operated animals. (D) Flow cytometry shows increased numbers of CD45+,F4/80+,CD11c alveolar macrophages and monocytes 7d post-PNX compared to sham operated animals. (E, F) Quantification of data in C and D. n≥3 animals per group. Data shown are mean +/− SD. (G) Hierarchical clustering of 68 CD45+,CSF1R-GFP+,F4/80+,Ly6G-macrophages isolated from mouse lungs 7d post-PNX. At least 6 cell groups (x-axis) were defined by expression of 4 gene groups (y-axis). (H) Subpopulations of lung macrophages post-PNX included those which expressed high levels of monocyte markers (orange bar) and those which expressed high levels of M2-like macrophage markers (red bar). See also Figure S1.
Figure 2.

CCR2+ monocytes are recruited to the lung post-PNX. (A) A cytokine protein array shows increased CCL2 7d post-PNX compared to sham operated animals. (B) Immunoflourescence shows increased numbers of Ccr2RFP+ monocytes 7d post-PNX compared to sham operated mice. (C) Flow cytometry shows increased numbers of CD45+,CD11b+,Ccr2FP+ cells 7d post-PNX compared to unoperated littermate controls. (D) Quantification of flow cytometry data shows increased numbers of CCR2+ monocytes after both PNX and sham operation compared to unoperated littermate controls. (E) Immunofluorescence on lungs from Ccr2RFP/RFP mice show decreased RFP+ monocytes 7d post-PNX compared to heterozygous mice (see B above). (F) Flow cytometry of dissociated lungs from Ccr2−/− mice shows decreased numbers of CD45+,F4/80+,CD11b+ interstitial monocytes and macrophages 7d post-PNX compared to wild type littermate controls. (G) Flow cytometry of dissociated lungs from Ccr2−/− mice shows that the number of CD45+,F4/80+,CD11c+ alveolar macrophages 7d post-PNX is not different from wild type littermate controls. For all experiments, n≥3 animals per group and data shown are mean +/− SD. Scale bars = 100 um. See also Figure S2.
Next, we isolated resident and recruited macrophages (CD45+, CSF1R-GFP+, F4/80+, Ly6G−) by FACS 7d post-PNX. Negative selection for Ly6G eliminated neutrophils that can express GFP in Csf1r-GFP reporter mice (Sasmono et al., 2007). We performed single cell RNA sequencing of 68 individual cells. Unsupervised hierarchical clustering revealed subpopulations of cells defined by distinct but partially overlapping transcriptional profiles (Figures 1G and 1H). These included CCR2+;Ly6c+ recruited monocytes (orange) and CD206+;Chil3+ “M2-like” macrophages (red). We performed additional single cell RNA sequencing on macrophages from sham operated mice to allow comparison of macrophage subpopulations in response to PNX. Markers of monocytes (CCR2) and M2-like macrophages (CD206 and Arg1) were enriched 7d post-PNX compared to sham (Figure S1).
CCR2-dependent recruitment of monocytes into the lung post-PNX
Because a subset of lung macrophages post-PNX was characterized by the expression of CCR2, we hypothesized that a CCL2-CCR2 recruitment axis drives the accumulation of macrophages during regeneration. Increased levels of CCL2 protein have been reported in lungs following PNX (Chamoto et al., 2012, 2013a; Chen et al., 2012; Ding et al., 2011; Rafii et al., 2015), with increased Ccl2 transcription in AEC2 7d post-PNX (Chamoto et al., 2013a) and PDGFRA+ lung fibroblasts 4d post-PNX (data not shown). We detected a 4.2-fold increase in CCL2 protein in whole lung lysate 7d post-PNX compared to sham operated control (Figures 2A and S2). To assess the recruitment of CCL2-responsive cells, we used Ccr2-RFPTm2.1Ifc mice in which red fluorescent protein (RFP) is knocked into the Ccr2 locus to mark inflammatory monocytes, further functioning as a null allele of CCR2. Immunofluorescence on lungs from Ccr2RFP/+ mice showed increased numbers of CCR2+ cells throughout the lung tissue, with concentrations around large vessels, a likely site of entry from circulation, and at the periphery of the regenerating lung lobes (Figures 2B and S2). We crossed the Ccr2-RFPtm2.1Ifc allele to mice carrying Sftpc-CreER; Rosa-fGFP alleles and administered tamoxifen to adult animals, resulting in the expression of GFP in AEC2 and RFP in CCR2+ monocytes. Analysis confirmed increased numbers of CCR2+ cells 7d post-PNX compared to sham operated animals and demonstrated that these cells were frequently in close proximity to lineage labeled AEC2s (Figure S2). Flow cytometry showed a significant increase in recruited monocytes (CCR2-RFP+, CD11b+) 7d post-PNX (18.5% +/− 5.6% of live, CD45+ cells) compared to unoperated littermate controls (10.9% +/− 2.7%) (Figures 2B, 2C, and 2D). This suggested that circulating monocytes are recruited into regenerating lung tissue by CCL2. However, we also noticed a significant increase in CCR2+ monocytes 7d post-sham (17.1% +/− 3.6%), suggesting that monocyte mobilization into lung tissue is a response to thoracic surgery. Of note, sham operations similar to those performed here stimulate proliferation in the lung pleura, possibly a conserved function of recruited monocytes (Brody et al., 1978). Another population of non-inflammatory monocytes expresses CX3CR1 (Geissmann et al., 2003) and has also been implicated in tissue regeneration (Arnold et al., 2007). However, using Cx3Cr1tm1Litt reporter mice, we did not detect an increase in Cx3cr1+ monocytes post-PNX (Figure S2). Thus, the major monocyte population post-PNX is defined by expression of CCR2.
To determine whether CCR2 is required for the increase in macrophages and monocytes post-PNX, we performed PNX in two CCR2-deficient mouse strains: Ccr2RFP/RFP mice (described above) and Ccr2tm1Mae null mice (hereafter Ccr2−/−). Lung tissue from Ccr2RFP/RFP mice contained fewer CCR2-RFP+, CD11b+ cells 7d post-PNX compared to Ccr2RFP/+ controls (Figure 2 B,E). Flow cytometry on lungs of Ccr2−/− mice showed a significant decrease in interstitial macrophages/recruited monocytes (F4/80+, CD11b+) 7d post-PNX (18.1% +/− 1.9% of CD45+) compared to wild type littermate controls (25.3% +/− 1.0%) (Figure 2G). In contrast, the number of alveolar macrophages (F4/80+, CD11c+) 7d post-PNX was unaffected by CCR2-deficiency (15.5% +/− 5.3% of CD45+) compared to wild type littermate controls (15.2% +/− 2.4%) (Figure 2H). This is consistent with data demonstrating that circulating monocytes rarely contribute to lung resident alveolar macrophages, as these cells are capable of self-renewal (Jakubzick et al., 2013; Suzuki et al., 2014).
Previous work has shown that local proliferation is an additional mechanism by which macrophage numbers increase post-PNX (Chamoto et al. 2012). We performed short-term EdU pulse labeling and confirmed an increase in the proliferation of CSF1R-GFP+ macrophages immediately after PNX that peaked at 4d post-PNX, and subsided by 7d post-PNX (Figure S2). Closer examination of these proliferating macrophages revealed localization in alveolar airspaces and a rounded morphology — characteristics of alveolar macrophages. Together, our data suggest that both local proliferation of lung macrophages and CCR2-dependent recruitment of monocytes contribute to the increased number CSF1R-GFP+ lung macrophages in the lung post-PNX. Because previous studies have focused on airspace macrophages (Chamoto et al., 2012, 2013a), but recruited CCR2+ monocytes have not been studied in the context of PNX-induced lung regeneration, we sought to determine their potential role in compensatory lung growth through disruption of the CCL2-CCR2 axis.
CCR2+ monocytes promote lung regeneration post-PNX
After PNX, all of the remaining right lobes grow, but the response is most profound in the accessory lobe (Konerding et al., 2012). Whole mount images show that the right accessory lobes of Ccr2−/− mice are smaller than littermate controls 21d after PNX, when lung regeneration is complete (Figure 3A). The generation of new alveoli post-PNX in wild type mice is associated with a 48% increase in the dry weight of the remaining right lobes compared to those of unoperated mice (Figure 3B). In contrast, PNX only induced a 23% increase in dry weight in Ccr2−/− mice, suggesting a significant impairment of lung regeneration (Figure 3B). To determine if this was permanent or reflected a delay in regeneration, we measured lung dry weights at 35d post-PNX. CCR2−/− lungs were significantly smaller than wild type littermate control lungs (Figure S3). Because AEC2s are the most proliferative epithelial progenitor cells post-PNX (Nolen-Walston et al., 2008), we hypothesized that reduced AEC2 proliferation contributes to the regenerative defect in CCR2−/− mice. Flow cytometry showed a significant reduction in EdU incorporation in AEC2 in CCR2−/− mice 7d post-PNX (5.4% +/− 1.3%) compared to wild type littermate controls (7.4% +/− 1.6%) (Figures 3C and 3D). As new alveoli are formed post-PNX, AEC2 are the major source of new AEC1 (Jain et al., 2015). To determine whether CCR2-deficiency affects the differentiation of AEC2 into AEC1, we generated Ccr2+l+ and Ccr2−/− mice that also carry Sftpc-CreER and Rosa-tomato alleles for lineage tracing experiments. We administered tamoxifen to adult animals, resulting in the heritable expression of RFP in AEC2 (Barkauskas et al., 2013; Rock et al., 2011a). Two weeks later we performed PNX and analyzed tissue sections for lineage traced AEC1s (RFP+, RAGE+) 21d post-PNX, looking specifically at the periphery of accessory lobes (Figure 3E). Ccr2−/− mice had significantly fewer lineage traced AEC1s (9.1% +/− 2.9%) compared to wild type littermate controls (43.7 % +/− 11.4%) (Figure 3F). Together, these data suggest that the CCR2-dependent recruitment of myeloid cells is critical for lung regeneration post-PNX, at least partly through the direct or indirect modulation of AEC2 behaviors.
Figure 3.
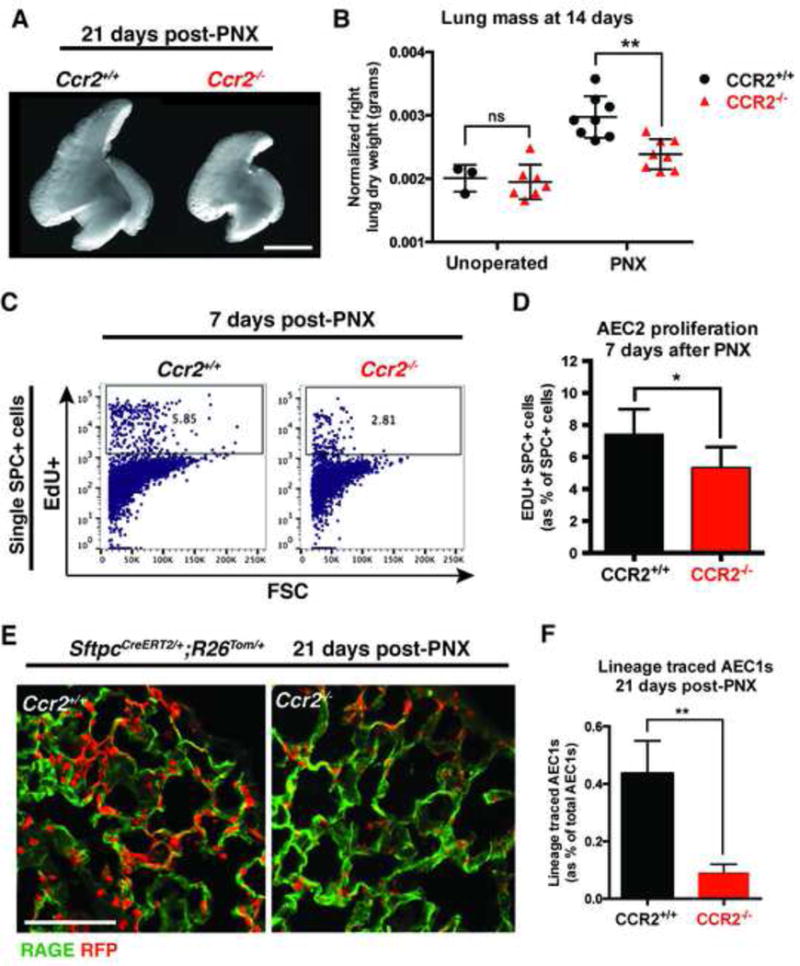
CCR2+ monocytes are required for lung regeneration post-PNX. (A) Whole mount images of right accessory lobes 21d post-PNX show impaired regeneration in Ccr2−/− mice compared to wild type littermate controls. Scale bar = 5 mm. (B) Ccr2−/− mice show impaired lung regeneration assessed by dry weight of remaining right lobes 14d post-PNX compared to wild type littermates. (C, D) Flow cytometry shows decreased incorporation of EdU (24 h pulse) in AEC2 7d post-PNX in Ccr2−/− mice compared to wild type littermate controls. (E,F) Immunofluorescence on lung sections Sftpc-CreER;Rosa-tomato mice 21d post-PNX shows impaired differentiation of lineage labeled AEC2 (red) into AEC1 (RAGE, green) in Ccr2−/− mice compared to littermate controls. Scale bar = 100 um. For all experiments, n≥3 animals per group and data shown are mean +/− SD. See also Figure S3.
Transfer of CCR2+ bone marrow promotes lung regeneration in CCR2-deficient mice
Bone marrow-derived monocytes express high levels of CCR2 protein, and when stimulated, this receptor promotes monocytes to enter the circulation and regenerating tissues (Gordon and Taylor, 2005). Flow cytometry revealed that approximately 10% of all bone marrow cells are CCR2+, CD11b+ monocytes (Figure S4). We isolated whole bone marrow from Csf1r-GFP mice and injected 1×107 cells via tail vein into Ccr2−/− mice that were either unoperated or 6d post-PNX. We timed this transfer to coincide with the increased CCL2 and peak influx of recruited monocytes we observed in wild type animals post-PNX (Figure 1). Flow cytometry 24 h later revealed CSF1R-GFP+ cells in the lungs of recipients, with more accumulating in PNX lungs (1.7% +/− 0.3%) compared to unoperated controls (0.5% +/− 0.1%) (Figure S4). To determine how CCR2 signaling affects monocyte accumulation in regenerating lungs, we injected 1×107 whole bone marrow cells from wild type Csf1r-GFP or Ccr2−/−; Csf1r-GFP mice into Ccr2−/− mice 6d post-PNX. The next day, we dosed mice with APC-CD45 by retro-orbital injection to specifically label cells in the pulmonary vasculature that had not extravasated into the interstitial tissue or airspace. Five minutes later, the mice were sacrificed and lungs were dissociated and incubated with FITC-CD45 to label all leukocytes, regardless of localization. Flow cytometry showed the proportion of CSF1R-GFP+, FITC-CD45+ cells that was APC-CD45 negative (interstitial and airspace cells) in recipients of Ccr2+/+ donor bone marrow was nearly 2-fold higher than this population in the lungs of recipients of Ccr2−/− donor bone marrow cells (Figure S4). This suggests that CCR2 plays an active role in the extravasation and retention of adoptively transferred bone marrow in regenerating lungs. Furthermore, analysis of Ccr2−/− recipients of Ccr2+/+; Csf1r-GFP+ donor bone marrow demonstrated that adoptively transferred cells persist until at least 14d post-PNX (Figure S4).
Next we sought to determine whether Ccr2+/+ bone marrow-derived cells were sufficient to rescue the defect in lung regeneration in Ccr2−/− mice. We delivered 1×107 whole bone marrow cells from Ccr2+/+ littermate controls to Ccr2−/− mice by tail vein injection 4d and 7d post-PNX (Figure 4A). Ccr2−/− recipients of wild type bone marrow had significantly increased lung dry weights (46% increase from unoperated) compared to Ccr2−/− mice that had not been injected (23% increased from unoperated) or recipients of Ccr2−/− bone marrow (28% increase from unoperated) (Figure 4B). Importantly, the regenerated lung mass of Ccr2−/− mice receiving Ccr2−/− bone marrow was not different from uninjected Ccr2−/− mice. Furthermore, adoptive transfer of Ccr2+/+ bone marrow cells into Ccr2−/− mice increased the number of lineage traced AEC1s at 21d post-PNX (27.2% +/− 0.08%) compared to uninjected Ccr2−/− mice (9.1% +/− 2.9%) (Figure S4). Together, these data suggest that CCR2 plays an active role in the recruitment of monocytes and macrophages into lungs post-PNX where they promote regeneration.
Figure 4.

Macrophages support AEC2. (A) Ccr2−/− mice were given whole bone marrow from Ccr2+/+ or Ccr2−/− mice 4 and 7 d post-PNX by tail vein injection. Recipient mice were analyzed 14d post-PNX by lung dry weights. (B) Transfer of wild type bone marrow, but not bone marrow from Ccr2−/− mice, rescues the lung regeneration defect in CCR2-deficient mice. n≥3 mice per group, data shown are mean +/− SD. * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001 (C) Heat map and selected genes from RNA sequencing of pooled CD45+,F4/80+ macrophages isolated by FACS from wild type mice 7d post-PNX or sham operation. (D) Lineage labeled AEC2 were isolated by FACS from heterozygous Sftpc-CreER; Rosa-tomato mice and cultured for 14 days in 3-dimensions in varying ratios with primary CD45+,CSF1R-GFP+,F4/80+ macrophages from wild type mice; all wells contained 10,000 AEC2s, and either no macrophages (A), 5,000 macrophages (B), 10,000 macrophages (C), or 20,000 macrophages (D). (E, F) Macrophages have a dose-dependent effect on the colony formation of lineage labeled AEC2 in co-culture. N≥5 co-cultures for each ratio. Data shown are mean +/− SD. (G) Representative image of immunofluorescence on a histological section of a co-culture from condition D shows an F4/80+ macrophage in close association with a pneumosphere. Scale bar = 100 um. See also Figure S4.
Macrophages support AEC2 in culture
To better understand how macrophages might regulate AEC2 behavior, we modified an organoid assay in which AEC2s are sorted and cultured in Matrigel and supplemented growth medium (Barkauskas et al., 2013). Cultured alone under these conditions, AEC2s rarely form colonies. However, the addition of PDGFRA+ fibroblasts, a putative component of the alveolar stem cell niche, greatly enhances the ability of AEC2 to form three-dimensional “alveolospheres” (Barkauskas et al., 2013). We isolated RFP+ lineage labeled AEC2s and CD45+;F4/80+ macrophages by FACS and co-cultured them in varying ratios without the addition of fibroblasts or endothelial cells (Figures 4D and S5). As expected, AEC2 alone rarely formed colonies (colony forming efficiency, CFE < 0.1%), but the addition of macrophages had a dose-dependent effect on the formation of organoids that we call pneumospheres (CFE = 1.61% +/− 0.51%) (Figures 4E and 4F). Pneumospheres had clear lumens and contained both lineage labeled epithelial cells and F4/80+ macrophages (Figure 4G). Macrophages persisted in co-cultures with pneumospheres at least 14d, but also survived when cultured alone (Figure S4). Partial overlap of the AEC2 marker SPC and AEC1 marker RAGE suggests that these cultured AEC2s may be reverted to a bipotential state in vitro (Treutlein et al., 2014)(Figure S4). AEC2 co-cultured in a 1:1 ratio with splenic macrophages formed far fewer spheres (CFE = 0.20% +/− 0.6%) compared to lung macrophages at the same ratio (CFE = 1.38% +/− 0.22%) (Figures 4F and S4). To determine whether bone marrow-derived macrophages (BMDMs) can also support AEC2-pneumospheres, we derived macrophages using an established protocol whereby whole bone marrow is cultured with MCSF-enriched media for at least 7d. BMDM cultured in a 1:1 ratio with AEC2s also promoted sphere formation (CFE = 1.01% +/− 0.10) (Figure S4). We conclude that lung macrophages can support formation of organoids from AEC2s and might directly promote AEC2 survival and proliferation.
Matrix-remodeling and M2-like macrophage gene signature in regenerating lungs
Our single cell RNA sequencing data suggested that a subset of the accumulating macrophages in post-PNX lungs express markers of M2-polarization (Figures 1G and 1H). To further understand how these cells might contribute to the regenerative process, we performed population level RNA sequencing on macrophages sorted from lungs 7d after PNX and sham operation (Figure 4C). These data confirmed robust expression of markers associated with M2-like macrophages, including Arginase1 (up 2.3 fold), Retnla/Fizz1 (up 2.9 fold), and Chil3/Ym1 (up 1.5 fold) in macrophages 7d post-PNX compared to sham controls. We also observed increased expression of genes associated with extracellular matrix and matrix remodeling including matrix metalloproteinases-2, 3, 12, 14, and 19 and collagens-1a1, 1a2, 3a1, 5a1, and 14a1. Macrophages from the lungs of PNX mice expressed lower levels of inflammatory cytokines TNFα, IL-1β, and IL-6, further suggesting an anti-inflammatory M2-like phenotype. Using qPCR, we confirmed that the expression of Arginase1 is increased in CD45+;F4/80+ macrophages isolated from the lungs of mice 7d after PNX compared to sham operation (Figure 5S).
Decreased M2-like macrophages and impaired lung regeneration in Il4ra-deficient mice
Because M2-like macrophages are present during alveolar formation during postnatal lung development (Jones and Williams, 2013; Saluzzo et al., 2017), modulate tissue inflammation (Gibbons et al., 2011, Herbert et al., 2004), and promote progenitor cell activity in muscle and heart (Ruffel et al., 2009, Shiraishi et al., 2016), we hypothesized that this subset of macrophages promotes alveologenesis post-PNX. We performed PNX in Arg1Tm1Lky/J mice (hereafter YARG), in which yellow fluorescent protein is knocked into the Arginase-1 locus (Van Dyken and Locksley, 2013; Reese et al., 2007). Flow cytometry and immunofluorescence showed few YARG+;F4/80+ M2-like macrophages in sham operated controls (0.4% +/− 0.1%). However, the number of these cells increases more than 2-fold post-PNX, with a peak at 4d post-PNX (1.4% +/− 0.4%) (Figures 5A, 5B, and 5C). Consistent with previous reports showing that M2-like macrophages proliferate in “Th2-type” microenvironments (Jenkins et al., 2011), EdU pulse experiments revealed that some YARG+ M2-like macrophages proliferate 4d and 7d post-PNX (Figure S5).
Figure 5.

IL4RA signaling is required for lung regeneration. (A) Immunofluorescent stains on sections from lungs of Arginase-YFP (YARG) mice show increased numbers of YARG+,F4/80+ M2-like macrophages (arrowheads) 4 and 7d post-PNX compared to sham. (B, C) Flow cytometry shows increased numbers of YARG+,F4/80+ M2-like macrophages 4 and 7d post-PNX compared to sham operated mice. (D–F) Immunofluorescence and flow cytometry on lungs from Il4ra−/− mice show rare YARG+; F4/80+ M2-like macrophages (arrowhead) 4d post-PNX compared to wild type YARG littermate controls (see A above). (G) Il4ra−/− mice show impaired lung regeneration assessed by dry weight of the right accessory lobe 14d post-PNX. ** = p ≤ 0.01. (H) Immunoflourescence on sections of lungs from Ccr2RFP/+;YARG mice show YARG+ M2-like macrophages that are both CCR2-RFP+ (arrows) and CCR2-RFP-(arrowheads). (I) Quantification of flow cytometry on dissociated lungs from Ccr2RFP/RFP; YARG mice show decreased numbers of YARG+, F4/80+ M2-like macrophages 4d post-PNX compared to wild type YARG controls. (J) CD45.1 wild type mice were lethally irradiated and given bone marrow from either wild type or Il4ra−/− (CD45.2) mice by tail vein injection. 10 weeks were allowed for hematopoietic reconstitution. PNX was performed on chimeras and they were analyzed by lung dry weight 14d later. (K) Il4ra−/− bone marrow recipients show impaired lung regeneration compared to recipients of wild type bone marrow. * = p ≤ 0.05. For all experiments, n≥3 animals per group and data shown are mean +/− SD. Scale bars = 100 um. See also Figure S5.
IL4RA is a shared receptor subunit for both IL4 and IL13, cytokines that promote the polarization of M2-like macrophages. Consistent with a role for M2-like macrophages in lung regeneration, both Il4ra and a co-receptor Il13ra1 were expressed at higher levels in macrophages 7d post-PNX compared to sham controls (Figure S1). To determine whether loss of IL4/13 signaling would negatively impact lung regeneration in vivo, we crossed the YARG reporter allele onto an Il4ra−/− background. Il4ra−/− mice had fewer M2-like macrophages 4d post-PNX (1.1% +/− 0.4%) compared to wild type littermate controls (2.2% +/− 0.4%) (Figures 5D–F). Moreover, Il4ra−/− mice showed impaired compensatory growth by lung mass 14d post-PNX compared to wild type littermate controls (Figure 5G). Finally, when AEC2 were lineage traced as described above in Il4ra−/− mice, there were fewer lineage traced AEC1 21d post-PNX (13.4% +/− 4.5%) compared to wild type controls (39.7% +/− 6.3%) (Figure 5S).
We performed bone marrow chimera experiments to confirm that the lung regeneration defect in Il4ra−/− mice is attributable to loss of IL4/13 signaling in leukocytes and not another cell type (Figure 5J). Wild type CD45.1 mice were lethally irradiated at 6 weeks of age and transplanted with bone marrow from CD45.2 wild type or Il4ra−/− mice. After 10 weeks to allow for hematopoietic reconstitution, we performed PNX. At the time of surgery, resected left lobes were dissociated for flow cytometric assessment of hematopoietic reconstitution from donor-derived cells. Lung monocytes and alveolar macrophages were ~99% and ~97% donor-derived, respectively (Figure S5). Mice reconstituted with Il4ra−/− bone marrow had impaired lung regeneration assessed by dry weight 14d post-PNX compared to littermate controls reconstituted with wild type bone marrow (Figure 5K). These data demonstrate that IL4RA signaling is required within the hematopoietic compartment for optimal lung regeneration.
Our single cell RNA sequencing revealed a subset of myeloid cells that expresses markers of both monocytes and M2-like macrophages, raising the possibility that CCR2+ monocytes could be a source of M2-like macrophages. We observed CCR2-RFP+ and CCR2-RFP-YARG+ M2-like macrophages in tissue sections of lungs from YARG;Ccr2RFP/+ mice 7d post-PNX (Figure 5H). However, these alleles are transcriptional reporters rather than heritable marks for lineage tracing, so we cannot draw conclusions about lineage relationships between monocytes and M2-like macrophages.
We sought to determine whether CCR2+ monocytes influence the generation of M2-like macrophages post-PNX, regardless of their origin. We detected some YARG+, F4/80+ M2-like macrophages by immunofluorescence in YARG;CCR2RFP/RFP mice 7d post-PNX (Figure S5), but flow cytometry showed that Ccr2RFP/RFP mice had significantly fewer YARG+, F4/80+ M2-like macrophages (91,000 +/− 22,000 cells, 1.1% +/− 0.2% of CD45+ cells) compared to wild type mice (158,000 +/− 26,000 cells, 1.9% +/− 0.5%) 4d post-PNX (Figures 5I and 5S). Based on these data and the fact that loss of CCR2 did not affect Arg1 expression in macrophages (Figure 5S), we conclude that CCR2+ recruited monocytes are not an essential source of M2-like macrophages post-PNX. However, we note that the absence of CCR2+ monocytes leads less efficient generation of M2-like macrophages.
Innate lymphoid cells are a source of IL13 in regenerating lungs
Macrophage polarization towards an M2-like phenotype can be induced by IL4 and IL13 in the microenvironment. Sources of these cytokines include CD4 helper T cells, basophils, and eosinophils (DeNardo et al., 2009; Egawa et al., 2013; Herbert et al., 2004; Loke et al., 2007; Wu et al., 2011). To determine whether helper T cells may play a role in PNX-induced lung regeneration, either directly or indirectly by regulating macrophage activity, we performed PNX in Rag1Tm1Mom null mice that lack all functional T and B cells. There was no difference in lung mass 14d post-PNX between Rag1−/− mice and wild type littermate controls (Figure S6). Furthermore, we did not detect an increase in IL4-expressing basophils, ILC2s, CD4+ T cells, nor NKT cells in the lungs of KN2 mice, a high-fidelity reporter for IL4 (Mohrs et al., 2005; Sullivan et al., 2011) (data not shown).
Type 2 innate lymphoid cells (ILC2s) are a major source of IL13 that polarizes lung macrophages toward an M2-like phenotype in certain inflammatory models (Van Dyken et al., 2014; Molofsky et al., 2013). Immunofluorescent stains of lungs from mice in which RFP is knocked into the Il5 locus as a highly specific marker for ILC2s (Nussbaum et al., 2013) demonstrated that ILC2s were present post-PNX near small conducting airways (central) and at the periphery of the lung (Figure 6A). We performed flow cytometry to quantify ILC2s based on cell surface markers (Lin−Thy1+ST2+) and by reporter activity (Lin−Red5+) and found that the proportion and absolute numbers of ILC2s was significantly increased 7d post-PNX (1.46% +/− 0.21%; 39,300 +/− 17,200) compared to sham-operated mice (0.64% +/− 0.33%; 23,700 +/− 9,200) (Figure 6B,C). Similar to steady state lungs, almost all Red5+ cells in the lung post-PNX were ILC2s (Figure S6C) (Nussbaum et al., 2013). Lastly, to determine if ILC2s are a source of IL13 post-PNX, we FACS purified ILC2s 4d post-PNX or sham for qRT-PCR analysis. ILC2s 4d post-PNX had a significant increase in IL5 and IL13 transcripts, but not in IL4 transcripts, compared to sham operated controls (Figure 6S). We analyzed lungs of Smart13 reporter mice, in which a modified human CD4 is knocked into the Il13 locus (Liang et al., 2012), to evaluate IL13 expression in vivo. There was a significant increase in the proportion of ILC2s (Lin−Thy1+ST2+) that were Smart13+ by flow cytometry 4d post-PNX when compared to sham-operated littermate controls (Figure 6D,E). Finally, using Ki67-based flow, we found a significant increase in proliferating ILC2s 7d post-PNX compared to sham-operated littermate controls (Figure 6S). Together, these data demonstrate an expansion of ILC2s post-PNX and implicate ILC2s as a source of IL13 during lung regeneration post-PNX.
Figure 6.

ILC2s accumulate and produce IL-13 post-PNX. (A) Immunofluorescent stains on sections from lungs of IL5RFP/+ (Red5) mice show ILC2s near small airways and in alveolar spaces post-PNX. Scale bar = 100 um. (B, C) Flow cytometry of dissociated lungs from Il5RFP/+ mice shows increased numbers of CD45+Lin−Thy1+IL5+ ILC2s 7d post-PNX compared to sham operated animals. (D, E) Flow cytometry of dissociated lungs from Smart13 mice shows increased numbers of IL-13 producing ILC2s 4d post-PNX compared to sham operated animals. n≥3 animals for each group. Data are represented as mean +/− SD. (F) Proposed model demonstrating the roles of CCR2+ monocytes, M2-like macrophages, and ILC2s in PNX-induced lung regeneration. See also Figure S6.
Discussion
Single cell RNA sequencing reveals macrophage heterogeneity in the lung post-PNX
Macrophages are usually categorized as classically or alternatively activated based on the expression of a panel of markers. Here, we took the unbiased approach of single cell sequencing to determine the extent of heterogeneity amongst myeloid cells in the regenerating lung. As expected, and consistent with data from other regeneration models, our single cell transcriptional profiling revealed a population of cells that expressed markers of M2-like macrophages post-PNX. In addition, this unbiased approach led us to the unexpected discovery that CCR2+ Ly6c+ inflammatory monocytes constitute a component of the myeloid compartment in the regenerating lung following PNX. These data do not exclude the possibility of additional myeloid subsets at other stages during regeneration or subsets that were below the detection threshold of our analysis.
CCR2+ monocytes and interstitial macrophages are required for regeneration post-PNX
Recent evidence suggests that bone marrow derived and circulating monocytes rarely contribute to resident macrophage populations during adult homeostasis, but replace macrophages following their depletion or during inflammation (Gordon and Taylor, 2005; Hashimoto et al., 2013; Yona et al., 2013). In many contexts, such as adult myocardial infarction, pulmonary fibrosis, and influenza infection, CCR2+ monocytes prolong and exacerbate injury (Gibbons et al., 2011; Herold et al., 2008; Nahrendorf et al., 2007). However, recruitment of CCR2+ monocytes is essential for tissue regeneration in other contexts, including the resolution liver fibrosis and regeneration of the heart and skin (Aurora et al., 2014; Gibbons et al., 2011; Ramachandran et al., 2012; Willenborg et al., 2012).
CD18, expressed on monocytes and several other leukocyte populations, is required for lung regeneration post-PNX (Chamoto et al., 2013a). We show that bone marrow-derived monocytes enter the lung through a CCL2-CCR2 axis. CCR2 deficiency impairs accumulation of interstitial monocytes and macrophages, but not alveolar macrophages, in the lung post-PNX. CCR2-deficient mice have impaired capacity for compensatory lung growth following PNX and adoptive transfer of wild type, but not CCR2-deficient, bone marrow was able to rescue the regeneration defect in CCR2-deficient mice. Together, these data unequivocally demonstrate for a requirement for recruited monocytes in lung regeneration.
Our bone marrow chimera experiments demonstrate that IL4RA is required specifically on leukocytes for the optimal regeneration. Although not conclusive, these data are consistent with a role for M2-like macrophages in lung regeneration post-PNX. Interestingly, human peripheral blood monocytes show M2-like polarization in response to CCL2 (Roca et al, 2009). Consistent with this, we observed fewer M2-like macrophages in the lungs of pneumonectomized CCR2-deficient mice compared to controls. However, M2-like macrophages were not completely absent in the lungs of CCR2-deficient mice. Together, our data suggest that recruited monocytes promote the generation of M2-like macrophages in the regenerating lung, either as a direct source or through modulation of the microenvironment. Future lineage tracing experiments are required to determine the relative contributions of recruited and resident cells to the M2-like macrophage population.
Macrophages are a component of the regenerative AEC2 niche
We show that the number of myeloid cells in the regenerating lung peaks at the time of maximal AEC2 proliferation. Consistent with these cells constituting a component of the regenerative niche, we frequently observed CSF1R-GFP+ and CCR2+ cells adjacent to AEC2 stem cells during regeneration. Moreover, resident lung macrophages and bone marrow-derived macrophages were able to support the growth of AEC2-derived pneumospheres in vitro, suggesting a direct interaction between these populations. These in vitro data and our analysis of AEC2 stem cell behaviors in vivo are consistent with a model in which macrophages and monocytes promote the proliferation and differentiation of AEC2 (Figure 6F). Our transcriptional profiling of macrophages also revealed a number of mechanisms by which myeloid cells might indirectly promote lung regeneration post-PNX. These include the secretion of pro-angiogenic factors and matrix remodeling enzymes (including MMP14), both of which have been shown to promote lung regeneration post-PNX (Chamoto et al., 2013a; Ding et al., 2011).
Our data implicate ILC2s as a source of IL-13 in a sterile model of lung regeneration. This cytokine could promote the polarization of M2-like macrophages and modulate their biochemical functions, including anti-inflammation, collagen synthesis and remodeling of the extracellular matrix (Chiaramonte et al., 1999; Knipper et al., 2015; Madala et al., 2010; Martinez et al., 2009 Pellicoro et al., 2012; Pouladi et al., 2004; Zheng et al., 2000). Alternatively or in addition, IL-13 could affect other cell types including lung epithelium (Kuperman et al., 2002). Further studies will characterize the ontogeny, activation and functions of M2-like macrophages and ILC2s in lung homeostasis and repair.
There is at least one report of adult alveologenesis in humans over 15 years following pneumonectomy (Butler et al., 2012). Our data further support the development of macrophage-based therapies to enhance the efficiency and kinetics of this regenerative capacity as a therapeutic approach for lung disease. This will require a deeper understanding of the manner in which monocytes, macrophages and other populations — including epithelial stem cells - interact in the context of the healthy, diseased and regenerating lung.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal (clone M1/70) anti-mouse CD11b (purified, used at 1:150) | Biolegend | Cat#101201; RRID: AB_312784 |
| Hamster monoclonal (clone N418) anti-mouse CD11c (purified, used at 1:150) | Biolegend | Cat#117301; RRID:AB_313770 |
| Rat monoclonal (clone C1:A3-1) anti-mouse F4/80 (purified, used at 1:150) | Biorad/AbD Serotec | Cat#MCA497G; RRID:AB_872005 |
| Hamster monoclonal (1A8) anti-mouse Ly6G (purified, used at 1:150) | Biolegend | Cat#127607; RRID:AB_1186104 |
| Hamster monoclonal (30-F11) anti-mouse CD45 (purified, used at 1:200) | BD Bioscience | Cat#550539; RRID:AB_2174426 |
| Rat monoclonal (clone MEC 13.3) anti-mouse PECAM (purified, used at 1:200) | BD Bioscience | Cat#550274; RRID:AB_393571 |
| Rat monoclonal (clone 175410) anti-mouse RAGE (purified, used at 1:200) | R&D Systems | Cat#MAB1179; RRID:AB_2289349 |
| Rabbit polyclonal anti-mouse PECAM (used at 1:250) | MIllipore | Cat#AB3786; RRID:AB_91588 |
| Hamster monocloncal (8.1.1) anti-mouse T1α (used at 1:900) | Developmental Studies Hybridoma Bank, Univ. of Iowa | RRID: AB_531893 |
| Chicken polyclonal anti-GFP (used at 1:500) | Aves | Cat#GFP-1020; RRID:AB_10000240 |
| Rabbit polyclonal anti-RFP (used at 1:250) | Rockland | Cat#600-401-379; RRID:AB_2209751 |
| Chicken polyclonal anti-RFP (used at 1:250) | Rockland | Cat#600-901-379; RRID:AB_10704808 |
| Rat monoclonal (clone 30-F11) anti-CD45-Apc-Cy7 (used at 1:200) | Biolegend | Cat#103116; RRID:AB_312981 |
| Rat monoclonal (M1/70) anti-CD11b-PerCP-Cy5 (used at 1:200) | Tonbo | Cat#65-0112 also OWL-A11257; RRID:AB_2621885 |
| Rat monoclonal (N418) anti-CD11c-APC (used at 1:200) | Biolegend | Cat#117309; RRID:AB_313778 |
| Rat monoclonal (BM8) anti-F4/80-PE (used at 1:200) | eBioscience | Cat#12-4801-82; RRID:AB_465923 |
| Rat monoclonal (1A8) anti-Ly6G-PE-Cy7 (used at 1:400) | Biolegend | Cat#127617; RRID:AB_1877262 |
| Goat anti-Chicken IgG (H+L) Secondary Antibody, Alexa Fluor 488 (used at 1:500) | Thermo Fisher Scientific | Cat#A-11039 also A11039; RRID:AB_142924 |
| Donkey anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 488 (used at 1:500) | Thermo Fisher Scientific | Cat#A-21206 also A21206; RRID:AB_2535792 |
| Donkey anti-Rat IgG (H+L) Secondary Antibody, Alexa Fluor 488 (used at 1:500) | Thermo Fisher Scientific | Cat#A-21208 also A21208; RRID:AB_141709 |
| Cy3-AffiniPure F(ab′)2 Donkey anti-Chicken IgG (H+L) Secondary Antibody (used at 1:500) | Jackson ImmunoResearch Labs | Cat#703-166-155; RRID:AB_2340364 |
| Donkey anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 555 (used at 1:500) | Thermo Fisher Scientific | Cat#A-31572 also A31572; RRID:AB_162543 |
| Goat anti-Rat IgG (H+L) Secondary Antibody, Alexa Fluor 555 (used at 1:500) | Thermo Fisher Scientific | Cat#A-21434 also A21434; RRID:AB_141733 |
| Goat anti-Hamster IgG (H+L) Secondary Antibody, Alexa Fluor 647 (used at 1:500) | Thermo Fisher Scientific | Cat#A-21451; RRID:AB_2535868 |
| Goat anti-Rat IgG (H+L) Secondary Antibody, Alexa Fluor 647 (used at 1:500) | Thermo Fisher Scientific | Cat#A-21247 also A21247; RRID:AB_141778 |
| Rat monoclonal (clone 30-F11) anti-CD45-BUV395 (used at 1:100) | BD Biosciences | Cat# 564279 |
| Rat monoclonal (clone 17A1) anti-CD3-PE-Cy7 (used at 1:100) | Biolegend | Cat# 100205; RRID:AB_312662 |
| Rat monoclonal (clone RM4-5) anti-CD4-BV711 (used at 1:100) | Biolegend | Cat# 317439; RRID:AB_11219404 |
| Rat monoclonal (clone 6D5) anti-CD19-BV650 (used at 1:100) | Biolegend | Cat#115541; RRID:AB_11204087 |
| Rat monoclonal (clone PK136) anti-NK-1.1-BV650 (used at 1:100) | Biolegend | Cat#108735; RRID:AB_11147949 |
| Rat monoclonal (clone RB6-8C5) anti-Ly6-G/Ly6-C-BV650 (used at 1:100) | Biolegend | Cat#108441; RRID:AB_2562401 |
| Rat monoclonal (clone M1/70) anti-CD11b-BV650 (used at 1:100) | Biolegend | Cat#101239; RRID:AB_11125575 |
| Rat monoclonal (clone N418) anti-CD11c-BV785 (used at 1:100) | Biolegend | Cat# 117335 RRID:AB_11219204 |
| Rat monoclonal (clone E50-2440) anti-SiglecF-APC (used at 1:100) | BD Biosciences | Cat#562680 |
| Rat monoclonal (PC61) anti-CD25-APC-Cy7 (used at 1:100) | Biolegend | Cat#102025; RRID:AB_830744 |
| Rat monoclonal (53-2.1) anti-CD90.2-BV605 (used at 1:100) | Biolegend | Cat#140317; RRID:AB_11203724 |
| Rat monoclonal (DJ8) anti-T1/ST2-PE (used at 1:100) | MD Biosciences | Cat#101001PE |
| Rat monoclonal (2F1) anti-KLRG1/MAFA-PerCP-eFluor710 (used at 1:100) | eBioscience | Cat#46-5893-80 also 46-5893; RRID:AB_10671072 |
| Rat monoclonal (MAR-1) anti-FcεRIα Pacific Blue (used at 1:100) | Biolegend | Cat#134313; RRID:AB_10612933 |
| Rat monoclonal (TAWJ) anti-Gata-3-eFluor660 (used at 1:20) | eBioscience | Cat#50-9966-41 also 50-9966; RRID:AB_10597909 |
| Rat monoclonal (SolA15) anti-Ki67-FITC (used at 1:100) | eBioscience | Cat#11-5698-80; RRID:AB_11151689 |
| Rat monoclonal (RPA-T4) anti-hCD4-APC (used at 1:100) | eBioscience | Cat#17-0049-41; RRID:AB_1272118 |
| Rat monoclonal (A20) anti-hCD45.1-PE (used at 1:200) | Biolegend | Cat#110707; RRID:AB_313496 |
| Rat monoclonal (104) anti-hCD45.2-APC (used at 1:200) | Biolegend | Cat#109813; RRID:AB_389210 |
| CD16/CD32 Fc Receptor Block | BD Biosciences | Cat# 553142 RRID:AB_394657 |
| Rat monoclonal (clone 30-F11) anti-CD45-Apc (used 2 ug/mouse for in vivo labeling) | Biolegend | Cat#103112 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Buprenex (0.3 mg/mL) | Reckitt Benckiser | Cat# NDC 12496-0757-5 |
| Lidocaine (5 mg/mL) | Phoenix | Cat# NDC 57319-533-05 |
| Bupivacaine (2.5 mg/mL) | Hospira | Cat# NDC 0409-1159-01 |
| Donkey Serum | Sigma-Aldrich | Cat#D9663-10ML |
| Goat Serum | Sigma-Aldrich | Cat#G9023-10ML |
| Bovine Serum Albumin | Fisher | Cat#BP1600-100 |
| Tamoxifen | Sigma-Aldrich | Cat#T5648 |
| EdU (5-ethynyl-2′-deoxyuridine) | Thermo Fisher Scientific | Cat#E10415 |
| Collagenase Type I (used at 450 U/mL) | Gibco Life Tech | Cat##17100–017 |
| Elastase (used at 4 U/mL) | Worthington Biochemical Corporation | Cat##LS002279 |
| Dispase (used at 5 U/mL) | BD Biosciences | Cat#354235 |
| DNaseI (used at 0.33 U/mL) | Roche | Cat#10104159001 |
| Red Blood Cell Lysis Buffer | Biolegend | Cat#420301-BL |
| Liberase TM | Roche | Cat#05401119001 |
| Sytox Blue | Life Tech | Cat#S34857 |
| AbC Anti-Mouse Bead kit | Molecular Probes | Cat#A10344 |
| Fix and Perm Cell Permeabilization Kit | Thermo Fisher Scientific | Cat#GAS003 |
| DMEM/F12 + GlutaMAX | Gibco Life Tech | Cat#10565-018 |
| Matrigel (Growth Factor Reduced) | BD Biosciences | Cat#356230 |
| Characterized Fetal Bovine Serum (used at 10%) | HyClone | Cat#SH30071 |
| Insulin-Transferrin-Selenium (used at 1x) | Gibco Life Tech | Cat#41400-045 |
| Epidermal Growth Factor (used at 20 ng/mL) | R&D Systems | Cat#2028-EG-200 |
| Fibroblast Growth Factor 2 or Basic (used at 25 ng/mL) | Gibco Life Tech | Cat#PMG0033 |
| Keratinocyte Growth Factor or Fibroblast Growth Factor 7 (used at 10 ng/mL) | Gibco Life Tech | Cat#PHG0094 |
| Hepatocyte Growth Factor (used at 10 ng/mL) | Gibco Life Tech | Cat#PHG0254 |
| RIPA Lysis and Extraction Buffer | Thermo Fisher Scientific | Cat#89900 |
| Complete, Mini Protease Inhibitor Cocktail | Roche | Cat#11-836-153-001 |
| Protein Assay Dye Reagent Concentrate | Bio-Rad | Cat#500-0006 |
| Protector RNase Inhibitor | Roche | Cat#03-335-402-001 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 647 Imaging Kit (for flow cytometry) | Invitrogen | Cat#C10340 |
| Click-iT Plus EdU Alexa Fluor 647 Imaging Kit (for tissue histology) | Invitrogen | Cat#C10640 |
| Proteome Profiler Mouse Cytokine Array Kit, Panel A | R&D Systems | Cat#ARYOO6 |
| RNAeasy Plus Micro Kit | Qiagen | Cat#74O34 |
| SuperScript IV VILO Master Mix | Invitrogen Life Tech | Cat#11756050 |
| SYBR GreenER qPCR Supermix Universal | Invitrogen Life Tech | Cat#1176202K |
| Agilent RNA 600 Pico Kit | Agilent Technologies | Cat#5067-1513 |
| C1 Single-Cell Reagent Kit for mRNA Seq | Fluidigm | Cat#100-6201 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131-1096 |
| Quant-iT™ PicoGreen® dsDNA Assay Kit | Thermo Fisher Scientific | Cat#P7589 |
| Deposited Data | ||
| Raw data files for RNA sequencing (PNX) | NCBI GEO | GEO: GSE96104 |
| Raw data files for RNA sequencing (sham) | NCBI GEO | GEO: GSE96105 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: Csf1r-GFP : B6N.Cg-Tg(Csf1r-EGFP)1Hume/J | Jackson Laboratories | Stock No. 018549 |
| Mouse: Ccr2−/− : cCR2tm1Mae | Kuziel et al., 1997 | |
| Mouse: Ccr2RFP/+ : B6.129(Cg)-Ccr2ím2.1,fc/J | Jackson Laboratories | Stock No. 017586 |
| Mouse: Cx3cr1GFP/+: B6.129P-C×3cr1tm1Ltt/J | Jackson Laboratories | Stock No. 005582 |
| Mouse: Sftpc-CreER: Sftpctm1(cre/ERT2)Blh | Jackson Laboratories | Stock No. 028054 |
| Mouse: Rosa-dtomato : Gt(ROSA)26Sortm2(CAG-tdTomato)Fawa | Rock et al., 2011 | |
| Mouse: Rosa-fGFP : Gt(ROSA)26Sortm1(CAG-EGFP)Blh | Rawlins et al., 2009 | |
| Mouse: YARG : B6.129S4-Arg1tm1Lky/J | Jackson Laboratories | Stock No. 015857 |
| Mouse: Il4ra−/− : BALB/c-Il4ratm1Sz/J | Jackson Laboratories | Stock No. 003514 |
| Mouse: ll5tm11(Cre)Lky | Nussbaum et al., 2013 | |
| Mouse: lL13tm2.1Lky: C.129S4(Cg)-Il13ím2.1Lky/J | Jackson Laboratories | Stock No. 018869 |
| Mouse: CD45.1 : B6-Ly5.1/Cr | Charles River | Stock No. 564 |
| Oligonucleotides | ||
| Genotyping primers-See Supplemental Table 1 | This paper | |
| qRT-PCR primers-See Supplemental Table 2 | This paper | |
| Recombinant DNA | ||
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| FlowJo (v10.1r5) | TreeStar Inc. | https://www.flowjo.com/ |
| GraphPad Prism 6.0 | GraphPad Software Inc. | http://www.graphpad.com/scientificsoftware/prism/ |
| Tophat (v2.1.0) | Kim et al., 2013; Trapnell et al., 2012 | |
| Bowtie2 (v2.2.8) | Kim et al., 2013; Trapnell et al., 2012 | |
| Cufflinks (v2.2.1) | Kim et al., 2013; Trapnell et al., 2012 | |
| Picard tools | http://broadinstitute.github.io/picard/ | |
| HTSeq-count | Anders et al., 2015 | http://www-huber.embl.de/users/anders/HTSeq/doc/count.html |
| Trimmatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| RSEM | Li and Dewey, 2011 | |
| Other | ||
CONTACT FOR REAGENTS
Further information and requests for resources and reagents should be directed to, and will be fulfilled by, the Lead Contact, Jason Rock (Jason.rock@edu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice were bred and maintained in a specific-pathogen-free barrier facility with free access to food and water. Both male and female mice were used between 8–12 weeks of age for all experiments. Tg(Csf1r-EGFP)1Hume/J (also referred to as Csf1r-GFP) mice, CCR2tm1Mae (also referred to as Ccr2−/−) mice, Ccr2tm2.1Ifc (also referred to as Ccr2RFP/+) mice, and Cx3Cr1tm1Litt (also referred to as Cx3cr1GFP/+) mice were used for macrophage and monocyte studies (Kuziel et al., 1997; Jung et al., 2000; Saederup et al., 2010; Sasmono et al., 2003) Ccr2−/−(pure-bred BALB/c strain) and Ccr2RFP/RFP (pure-bred C57Bl/6 strain) mice were used to assess the contribution of CCR2+ cells in PNX-induced lung regeneration. Mice containing Sftpc-CreER and Rosa-dtomato alleles for lineage tracing AEC2s were backcrossed to Ccr2tm1Mae BALB/c mice for at least 5 generations. Arg1tm1Lky/J (also referred to as YARG) mice were used to assess M2-like polarization of macrophages and Il4ratm1Sz mice were used to test a requirement for M2-like macrophages in PNX-induced lung regeneration (Noben-Trauth et al., 1997; Reese et al., 2007). Rag1tm1Mom were used to assess the requirement for lymphoid cells in PNX-induced lung regeneration (Mombaerts et al., 1992). B6-Ly5.1/Cr were obtained from Charles River Laboratories International, Inc. and used to distinguish host vs. donor-derived hematopoietic cells in bone marrow chimera experiments. Il5m1.1(Cre)Lky (also referred to as Red5) mice were used to assess the contribution of ILC2s in PNX-induced lung regeneration (Nussbaum et al., 2013). IL13tm2.1Lky (also referred to as Smart13) mice were used to measure IL-13 protein production from ILC2s (Liang et al., 2012). All studies were approved and performed according to the guidelines issued by the University of California, San Francisco (UCSF) Institutional Animal Care and Use Committees (IACUC).
METHOD DETAILS
Pneumonectomy procedure
Prior to surgery, adult mice (8–12 weeks old) were weighed, given Buprenex (0.3 mg/mL, Reckitt Benckiser, catalog #NDC 12496-0757-5) at 20g/kg, and shaved at the surgical site on the left lateral side. Mice were anesthetized with 2% isofluorane and intubated for ventilation using a Harvard mini-vent ventilator. Mice were anesthetized and ventilated throughout the pneumonectomy procedure with 200 μL of stroke volume at 200 strokes per minute. The surgical site was sterilized with ethanol and beta-iodine and then a 2 cm long incision was made on the left lateral side of the skin. Left-sided thoracotomy was performed with a 1 cm long incision at the 5th intercostal space to expose the left lung lobe underneath. The ribs were spread by retraction and the left pulmonary vasculature and mainstem bronchus were ligated with a titanium clip. The left lobe was resected, and the ribs and skin closed. An angiocath port was inserted to evacuate the void space and reestablish negative intrathoracic pressure. Sham control surgeries were performed as thoracotomy procedure without lung removal. After closure, topical analgesics were applied at the surgical sites (Lidocaine, 5 mg/mL, Phoenix, catalog #NDC 57319-533-05; Bupivacaine, 2.5 mg/mL, Hospira, catalog #NDC 0409-1159-01). Anesthesia was discontinued and mice remained intubated until autonomous breathing recovered. Mice were placed on a warming pad and monitored until awake. A second Buprenex dose was given 4–6 hours later and monitored to ensure that a full recovery was made.
Tissue harvest and fixation
Mice were euthanized by CO2 exposure until respiration ceased. Mice were dissected to expose the diaphragm, which was cut to cause pneumothorax. To perfuse the pulmonary vasculature, the descending aorta was cut and a 21 gauge needle was inserted into the right ventricle to flush at least 20 mL of cold PBS until lungs had blanched. The trachea was exposed and a small incision was made at the pharyngeal cartilage to allow inflation with cold 4% paraformaldehyde. Lungs were inflated to 20 cm H2O pressure and fixed in situ for 10 minutes. Lungs were then removed en bloc and then submerged 4% paraformaldehyde for 1 hour at 4°C. Individual lung lobes were separated and then washed three times for 15 minutes in cold PBS. In preparation for embedding, lungs were submerged in 30% sucrose for 24 hours, then moved to a 50:50 mixture of 30% sucrose/O.C.T medium (Optimal Cutting Temperature Medium, Tempura) for 24 hours, and then finally embedded in 100% O.C.T. medium and immediately frozen on dry ice and stored at −20°C.
Immunostaining for histology
Tissue blocks were sectioned at 12−13 μm and were left to dry on a slide warmer overnight. Wax slides were dewaxed and rehydrated. Tissue sections were permeabilized with 0.5% TritonX-100 in PBS for 5 minutes. Tissue sections were blocked in 5% donkey serum (Sigma, catalog #D9663-10ML), 5% goat serum (Sigma, catalog #G9023-10ML), 3% bovine serum albumin (Fisher, catalog #BP1600-100) and 0.1% TritonX-100 for 1 hour at room temperature. Primary antibodies were diluted in block at concentrations listed in the Key Resources Table and incubated overnight at 4°C. All fluorophore-conjugated secondary antibodies were diluted in block at 1:500. EdU staining was performed according to manufacturer recommendations (Invitrogen, see Proliferation Studies). DAPI nuclear staining was performed according to manufacturer recommendations. Tissue sections were washed 5× with PBS for 15 minutes after/between antibody incubations. Images of sections were captured on a Zeiss Imager M2 AxioCam MRm microscope. For pneumonectomy studies, imaging was focused exclusively on the accessory lobe for all mice. For quantification of cells, at least 10 randomly selected images were counted per animal. Images were processed with ImageJ/FIJI (version 2.0.0, NIH).
Lung dissociation for flow cytometry and FACS
Mice were sacrificed as described in the tissue harvest protocol. After perfusion, lungs were inflated intratracheally with 1 mL of protease solution containing Collagenase Type I (Gibco, catalog #17100–017, used at 450 U/mL), Elastase (Worthington Biochemical Corporation, catalog #LS002279, used at 4 U/mL;), Dispase (BD Biosciences, catalog #354235, used at 5 U/mL;) and DNaseI (Roche, catalog #10104159001, used at 0.33 U/mL) solubilized in DMEM/F12. The lungs were resected en bloc, separated into individual lobes and cut into small pieces (<2 mm2) by scalpel blade. Diced tissues were incubated in 4–5 mL protease solution for 30 minutes at 37 °C with frequent agitation until completely dissociated into single cells. Cells were washed with serum media made from 10% FBS in DMEM/F12. Cells were incubated with 2 mL of red blood cell lysis buffer (Biolegend, catalog #420301-BL) for 4 minutes at room temperature and the washed with serum media. Cells were filtered through a 40-μm strainer, centrifuged, and resuspended in flow cytometry/FACS buffer (5% FBS in HANKS buffer or HBSS, modified without Ca2+ or Mg2+). Flow cytometry was performed on a BD LSR II and sorting was performed on a BD FACSARIA II. Specifically for ILC2 flow cytometry, lungs were dissociated with GentleMACS protocol “m_lung_01_01” (Miltenyi Biotec), digested for 30 minutes with Liberase TM (Roche, catalog #05401119001) & DNase I (Sigma-Aldrich), further homogenized using gentleMACS protocol “m_lung_01_02,” and treated with Pharm Lyse (BD Biosciences). Data was analyzed with FACS Diva (BD Biosciences) and FlowJo (version 10.1r5, FLOWJO LLC).
Immunostaining for flow cytometry & FACS
Single isolated cells were kept on ice and stained in flow cytometry/FACS buffer (5% FBS in HANKS buffer or HBSS, modified without Ca2+ or Mg2+). Cells were blocked with CD16/CD32 Fc receptor block (BD Biosciences, catalog #553142) at 1:100 for 20 minutes and washed. Fluorophore-conjugated antibodies were used according to the Key Resources Table and cells were stained for 30 minutes - 1 hour. For live cell staining and sorting, a cell viability dye was added before sample acquisition (Sytox Blue at 1:1000, Life Tech, catalog #S34857 or DAPI at 1:2000 of 1 mg/mL). For intracellular flow cytometry (detection of SPC, Gata-3, or Ki67), cells were prepped using a cell fixation and permeabilization kit (Invitrogen, catalog #GAS003). EDU-based proliferation was assessed using the Click-iT® EdU Alexa Fluor® 647 Imaging Kit (Invitrogen, catalog #C10340) as per the manufacturer’s instructions. Flow cytometry was performed on a BD LSRII or BD LSRFortessa DUAL and cell sorting was performed on BD FACSARIA II or Sony SH800S Cell Sorter. Compensation was performed with single antibody/channel controls using a AbC Anti-Mouse Bead kit (Molecular Probes, catalog #A10344). Gating controls for each channel were established with samples containing all the staining antibodies except the antibody in that channel (fluorescence minus one). To determine gating controls for reporter gene expression, mice lacking the reporter or protein were used. Data were analyzed with FACS Diva (BD Biosciences) and FlowJo (v10.1r5, FLOWJO LLC).
Intravascular CD45 Labeling
To label immune cells in the pulmonary vasculature, we injected mice with CD45 bound to high molecular weight fluorophore by intravenous route shortly before sacrifice. 2 μg of CD45-APC (10 μL, Biolegend, cat# 103112) were diluted in 100 μL of sterile saline and loaded into a 28.5 gauge insulin syringe. Mice were put in a sealed container containing a cotton ball soaked in isofluorane and monitored closely. When breathing slowed, mice were removed from the container. Gentle pressure was applied around the peri-orbital area to induce eye protrusion. The syringe needle was inserted in the retro-orbital space and contents injected. Mice were allowed to regain consciousness and were sacrificed by CO2 asphyxiation 5 minutes later.
Dry weight lyophilization
Murine lungs were perfused as previously described, isolated into individual lobes, and flash frozen in liquid N2 immediately after harvest. Lobes were placed into microcentrifuge tubes and transferred to a freeze/dry flask attached to a Labconco Freeze Dry System. Lung lobes were lyophilized for twenty-four hours and immediately weighed. Lung lobe weights were normalized to mouse height (anus to snout).
Proliferation studies
For assessing cell proliferation in vivo, EdU (Thermo Fisher Scientific, Catolog #E10415) was resuspended at 5 mg/mL in sterile PBS and administered to mice by intrapertoneal injection at 50 mg of EdU per kg of mouse weight. For flow cytometry, EdU incorporation was detected using the Click-iT EdU Alexa Fluor 647 Imaging Kit (Invitrogen, catalog #C10340). For tissue histology, EdU incorporation was detected using the Click-iT Plus EdU Alexa Fluor 647 Imaging Kit (Invitrogen, catalog #C10640). To assess AEC2 proliferation by flow cytometry, EDU was given twenty-four hours prior to sacrifice. To assess local macrophage proliferation, EdU was given 3.5 hours prior to sacrifice.
Lineage labeling
Tamoxifen (Sigma-Aldrich, catalog #T5648) was dissolved in corn oil and administered to mice via intraperitoneal injection at 0.25 mg of Tamoxifen per kg of mouse weight. Three full doses of TMX were given to each animal at eight weeks every other day. To provide a tamoxifen “wash-out” period, we waited at least 14 days after the last tamoxifen dose before any study intervention.
Adoptive transfer of bone marrow cells
Mice were sacrificed by CO2 asphyxiation. Long bones (femur and humerus) were isolated and flushed with 10% FBS in DMEM to clear bone marrow cells. Bone marrow cells were filtered through a 40 μm strainer and resuspended in sterile saline for injection. Cells were manually counted using a Fisher Scientific Hemacytometer (catalog #0267110) and a Zeiss Primo Vert Inverted Microscope. 10 million bone marrow cells were resuspended in 200 μL of sterile saline for intravenous lateral tail vein injection. Prior to injection, mouse tails were sterilized and gently warmed.
Generation of bone-marrow derived macrophages
Bone marrow cells were isolated as described in adoptive transfer protocol, and plated on non-tissue treated petri dishes with macrophage-differentiation media. This media contained 10% F.B.S, 1× penicillin/streptomycin, and 10% MCSF-enriched media harvested from MCSF secreting-3T3 fibroblasts. Bone marrow-derived macrophages were cultured for at least 7 days, with media changes every 3–4 days, until use in co-culture 3D-organoid in vitro pneumosphere assay.
Pneumosphere in vitro assay
FACS sorted cells were plated in the upper chamber of 0.4 μm transwells nestled in 24-well plates and grown in a 5% CO2, 37 °C incubator. Cells were seeded in 75 μL of growth factor reduced Matrigel (BD Biosciences, catalog #356230) and 75 μL of pneumosphere media to form a cell-mixture plug. This plug was pipetted at the bottom of the transwell and 1000-900 μL of pneumosphere media was placed in the lower chamber. Pneumosphere media contained 10% F.B.S. (Hyclone, catalog #SH30071), 1x Insulin-Transferrin-Selenium (Gibco Life Tech, Catalog #41400-045), 1x penicillin/streptomycin, EGF 20 ng/mL (R&D Systems, catalog #2028-EG-200), FGF2 25 ng/mL (Gibco Life Tech, catalog #PMG0033), KGF 10 ng/mL (Gibco Life Tech, catalog #PHG0094), HGF 10 ng/mL (Gibco Life Tech, #PHG0254) in DMEM/F12 + GlutaMAX (Gibco Life Tech, catalog #10565-018). Pneumosphere media was changed every two-three days after plating. Pneumospheres were counted on day 14 by bright-field microscopy.
Bone marrow chimeras
At 6 weeks of age, co-housed littermate Ly5.1 (CD45.1) B6 mice were lethally irradiated with 2 does of 550 rads separated by 4 hours, followed by a whole bone marrow transplant several hours later. Bone marrow was harvested from wild type or Il4rα-deficient mice, counted, and divided amongst the recipient mice. Bone marrow was re-suspended in sterile saline and injected by intravenous tail vein route. Animals were carefully monitored over the next 10 weeks to allow turn-over and reconstitution of lung myeloid and bone marrow-derived monocytes.
Protein Cytokine Array
Tissues were harvested as above, except lungs were not fixed. Whole accessory lobes were isolated and weighed ex vivo. Accessory lobes were then flash frozen in liquid nitrogen and pulverized with a chilled glass dounce. RIPA lysis and extraction buffer (ThermoFischer Scientific, catalog #89900) containing Complete, Mini protease inhibitor cocktail (Roche, catalog #11-836-153-001) was added to each sample at 1 mL of solution per 60 mg of tissue. Tissue were further homogenized by dounce and then briefly sonicated. Samples were then centrifuged at 14,000g for 15 minutes and supernatants were isolated. A standardized Bradford assay was performed to measure lysate protein concentration (Protein Assay Dye Reagent Concentrate, Biorad, catalog #500-0006; Bio-Rad SmartSpec Plus Spectrophotometer, catalog #170-2525). Purified protein lysates (200 μg) were applied to the Proteome Profiler Mouse Cytokine Array Kit, Panel A (R&D Systems, catalog #ARY006). The chemiluminescence reaction was measured with a ChemiDoc XRS+ System (Biorad). Densitometry quantitation was performed in FIJI/ImageJ (NIH).
qRT-PCR
Single cells isolated by FACS were lysed using the QIAshredder (Qiagen, catalog #79654) or immediately sorted into RLT buffer. RNA was purified using the RNeasy Plus Micro Kit (Qiagen, catalogue #74034). RNA quantity and quality was measured using a Nanodrop (Thermo Scientific). cDNA was made using the Superscript IV VILO cDNA Mastermix Kit (Invitrogen, catalog #11756050). qRT-PCR was performed using the SYBR GreenER qPCR Supermix Universal Kit (Invitrogen, catalog #11762100). Reactions were run in triplicate on a ViiA7 Real-Time PCR System (Thermo Fischer). Expression levels were measured using the 2ˆ− (ΔΔCT) method in which n≥3 PNX mice were compared to n≥3 sham mice and normalized to house-keeping genes.
RNA sequencing
For bulk population level RNA sequencing, RNA was obtained as described in qRT-PCR methods. Quality control was performed with an Agilent RNA 600 Pico Kit (catalog #5067-1513) using an Agilent 2100 Bioanalyzer (Agilent Technologies). Purified RNA was submitted to the UCSF Genomics Core for high throughput RNA sequencing. The library was prepared with TruSeq Stranded mRNA and sequencing was done on an Illumina HiSeq 4000 with 50 bp single-end reads. Sequencing yielded ~230 million reads with an average read depth of 28.7 million reads/sample. Reads were then aligned to the mouse genome (Ensemble Mouse GRCm38.78) using STAR_2.3.2a. Those that mapped uniquely to known mRNAs were used to assess differential expression.
For single cell RNA sequencing, lung macrophages were isolated by FACS. Fluidigm C1 and C1 integrated fluidics circuits (IFCs) were used to capture live cells, lyse, convert polyA+RNA into full length cDNA, amplify cDNA and generate cDNA according to their detailed protocol (“Using C1 to Generate Single-Cell cDNA Libraries for mRNA Sequencing”, Fluidigm, PN 100–7168). 88 single CD45+, CSF1R-GFP+, F4/80+, Ly6G-cells were captured on a C1 Single-Cell DNA seq IFC, 5–10 μm (Fluidigm, catalog #100–5759) using the C1 Single-Cell Reagent Kit for mRNA Seq (Fluidigm, catalog #100–6201). Library preparation for sequencing was performed following the modified Illumina Nextera XT DNA library preparation protocol using the Nextera XT DNA Library Preparation Kit (Illumina, Catalog #FC-131-1096). The concentration of cDNA was determined using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies, catalog #P7589). Sequencing was performed by Elim BioPharm (Oakland, CA) on one lane of an Illumin HiSeq2500 Flow cell in rapid mode with 50bp paired end reads. De-multiplexed files were created using CASAVA 1.8 (Illumina).
Reads were aligned and mapped using Tophat (v2.1.0) and Bowtie2 (v2.2.6) and Cufflinks (v2.2.1) software (Kim et al., 2013; Trapnell et al., 2012) 141 million reads were aligned with an average of 1.66 million reads per cell per end. All of the accepted hits in bam files output from cufflinks were processed using Picard tools: FixMateInformation (http://broadinstitute.github.io/picard/). Counts were compiled using HTSeq-count (Anders et al., 2015; http://www-huber.embl.de/users/anders/HTSeq/doc/count.html) and aligned with UCSC mm10 mouse assembly. All of the gene counts for each cell were compiled into a single file. Filtering was then performed to remove any cell that did not have at least a 50% alignment rate. Genes that did not have a least one read aligned in at least 3 cells were removed. Fluidigm’s R package Singular was used to generate an unbiased hierarchical clustering heat map.
During revisions, we performed additional single cell RNA sequencing on macrophages harvested 7 days after sham surgery to compare with our initial dataset of PNX-macrophages. These cells and their RNA were isolated as before, but the downstream analysis differed as follows:
Sequencing was performed by UCSF Genomics Core on one lane of an Illumin HiSeq2500 Flow cell in rapid mode with 100bp paired end reads. De-multiplexed files were created using CASAVA 1.8 (Illumina). Each sample was filtered using Trimmatic v0.36 (Bolger et al., 2014; http://www.usadellab.org/cms/?page=trimmomatic) with the settings LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:70 and only paired matches were kept. Counts were compiled with RSEM aligner (v1.3.0, Li and Dewey, 2011) and aligned with genome Ensembl GRC m38 release 87. The gene count matrix was then created by running the rsem command “rsem-generate-data-matrix” on all of the genes.results files for each cell.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters, including the number of samples (n), descriptive statistics (mean and standard deviation), and significance are reported in the figures and figure legends. In general, at least n = 3 mice were used for each time point and for each intervention. For each metric analyzed, we performed an unpaired, two-tailed, Student’s t test between treated and control groups. When quantifying cell numbers in histological sections, counters were blinded, microscopy fields randomly selected, and at least 10 images were scored per animal. GraphPad Software version 6.0 was used for all mathematical statistical analysis.
DATA AND SOFTWARE AVAILABILITY
Data Resources
The accession number for the RNA-seq data reported in this paper is NCBI GEO: GSE96104 (PNX) and GSE96105 (sham).
Supplementary Material
Acknowledgments
We would like to thank Zena Werb, Anthony DeFranco, Michael Matthay, Eric Dang, Jen Bando, Jakob von Moltke, Steve Van Dyken, Zhi-En Wang, Christoph Schneider, Amy-Jo Casbon, Nancy McNamara, Ari Molofsky, Trevor Burt, Chris Allen, and Brigid Hogan for helpful discussions and reagents. We would like to thank Gorica Amidzic, Molly Kastner, Raymond Ho, and Tyler Kim for technical assistance. AJL was supported by UCSF MSTP Grant T32 GM007618 and the Ruth L. Kirshstein National Research Service Award 1F30HL131198-01. JRR is supported by R01HL127002.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information includes six figures and two tables and can be found with this article online at:
AUTHOR CONTRIBUTIONS
Conceptualization, AL, JR; Methodology and Investigation, AL, ID, JL, CC, and AN; Reagents, RL; Data Analysis and Interpretation, AL, ID, JL, RL, JR; Writing, Review and Editing, AL, JR; Supervision, Project Administration, and Funding Acquisition, RL, JR.
References
- Aggarwal NR, King LS, D’Alessio FR. Diverse macrophage populations mediate acute lung inflammation and resolution. Am J Physiol Lung Cell Mol Physiol. 2014;306:L709–25. doi: 10.1152/ajplung.00341.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora A, Olson E. Immune Modulation of Stem Cells and Regeneration. Cell Stem Cell. 2014;15:1425. doi: 10.1016/j.stem.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, Sadek HA, Olson EN. Macrophages are required for neonatal heart regeneration. J Clin Invest. 2014;124:1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BL. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barner M, Mohrs M, Brombacher F, Kopf M. Differences between IL-4Rα-deficient and IL-4-deficient mice reveal a role for IL-13 in the regulation of Th2 responses. Curr Biol. 1998;8:669–672. doi: 10.1016/s0960-9822(98)70256-8. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, Ridgway RA, Seo SS, Spee B, Van Rooijen N, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18:572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JP, Loring SH, Patz S, Tsuda A, Yablonskiy DA, Mentzer SJ. Evidence for adult lung growth in humans. N Engl J Med. 2012;367:244–247. doi: 10.1056/NEJMoa1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamoto K, Gibney BC, Ackermann M, Lee GS, Lin M, Konerding MA, Tsuda A, Mentzer SJ. Alveolar macrophage dynamics in murine lung regeneration. J Cell Physiol. 2012;227:3208–3215. doi: 10.1002/jcp.24009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamoto K, Gibney BC, Lee GS, Ackermann M, Konerding MA, Tsuda A, Mentzer SJ. Migration of CD11b+ accessory cells during murine lung regeneration. Stem Cell Res. 2013a;10:267–277. doi: 10.1016/j.scr.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamoto K, Gibney BC, Ackermann M, Lee GS, Konerding MA, Tsuda A, Mentzer SJ. Alveolar epithelial dynamics in postpneumonectomy lung growth. Anat Rec (Hoboken) 2013b;296:495–503. doi: 10.1002/ar.22659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Acciani T, Le Cras T, Lutzko C, Perl A-KTK. Dynamic regulation of platelet-derived growth factor receptor α expression in alveolar fibroblasts during realveolarization. Am J Respir Cell Mol Biol. 2012;47:517–527. doi: 10.1165/rcmb.2012-0030OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dane DM, Yilmaz C, Estrera AS, Hsia CC. Separating in vivo mechanical stimuli for postpneumonectomy compensation: physiological assessment. J Appl Physiol. 2013;114:99–106. doi: 10.1152/japplphysiol.01213.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies L, Rosas M, Jenkins S, Liao CT, Scurr M, Brombacher F, Fraser D, Allen J, Jones S, Taylor P, et al. Distinct bone marrow-derived and tissue-resident macrophage lineages proliferate at key stages during inflammation. Nature Communications. 2013 doi: 10.1038/ncomms2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De’Broski RH, Hölscher C, Mohrs M, Arendse B. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity. 2004 doi: 10.1016/s1074-7613(04)00107-4. [DOI] [PubMed] [Google Scholar]
- Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding BSS, Nolan DJ, Guo P, Babazadeh AO, Cao Z, Rosenwaks Z, Crystal RG, Simons M, Sato TN, Worgall S, et al. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell. 2011;147:539–553. doi: 10.1016/j.cell.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta P, Sager HB, Stengel KR, Naxerova K, Courties G, Saez B, Silberstein L, Heidt T, Sebas M, Sun Y, et al. Myocardial Infarction Activates CCR2(+) Hematopoietic Stem and Progenitor Cells. Cell Stem Cell. 2015;16:477–487. doi: 10.1016/j.stem.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Locksley RM. Interleukin-4-and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol. 2013;31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa M, Mukai K, Yoshikawa S, Iki M, Mukaida N, Kawano Y, Minegishi Y, Karasuyama H. Inflammatory monocytes recruited to allergic skin acquire an anti-inflammatory M2 phenotype via basophil-derived interleukin-4. Immunity. 2013;38:570–580. doi: 10.1016/j.immuni.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40:91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbach Voswinckel, Michl Mehling, Fehrenbach Seeger, Nyengaard Neoalveolarisation contributes to compensatory lung growth following pneumonectomy in mice. Eur Respir J. 2008;31:515–522. doi: 10.1183/09031936.00109407. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Jung S, Littman D. Blood Monocytes Consist of Two Principal Subsets with Distinct Migratory Properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- Gibbons MA, MacKinnon AC, Ramachandran P, Dhaliwal K, Duffin R, Phythian-Adams AT, van Rooijen N, Haslett C, Howie SE, Simpson AJ, et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am J Respir Crit Care Med. 2011;184:569–581. doi: 10.1164/rccm.201010-1719OC. [DOI] [PubMed] [Google Scholar]
- Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proc Natl Acad Sci USA. 2013;110:9415–9420. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Martinez F. Alternative Activation of Macrophages: Mechanism and Functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Gordon S, Taylor P. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, Mack M, Kuziel WA, Corazza N, Brunner T, et al. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med. 2008;205:3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Mayer K, Lohmeyer J. Acute lung injury: how macrophages orchestrate resolution of inflammation and tissue repair. Front Immunol. 2011;2:65. doi: 10.3389/fimmu.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Ginhoux F. Ontogeny of Tissue-Resident Macrophages. Front Immunol. 2015;6:486. doi: 10.3389/fimmu.2015.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AM, Shifren A, Mazan MR, Gruntman AM, Lascola KM, Nolen-Walston RD, Kim CF, Tsai L, Pierce RA, Mecham RP, et al. Matrix modulation of compensatory lung regrowth and progenitor cell proliferation in mice. Am J Physiol Lung Cell Mol Physiol. 2010;298:L158–68. doi: 10.1152/ajplung.90594.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Barkauskas CE, Takeda N, Bowie EJ, Aghajanian H, Wang Q, Padmanabhan A, Manderfield LJ, Gupta M, Li D, et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat Commun. 2015;6:6727. doi: 10.1038/ncomms7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, Ivanov S, Duan Q, Bala S, Condon T, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 2013;39:599–610. doi: 10.1016/j.immuni.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, Allen JE. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konerding MA, Gibney BC, Houdek JP, Chamoto K, Ackermann M, Lee GS, Lin M, Tsuda A, Mentzer SJ. Spatial dependence of alveolar angiogenesis in post-pneumonectomy lung growth. Angiogenesis. 2012;15:23–32. doi: 10.1007/s10456-011-9236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotton DN, Morrisey EE. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med. 2014;20:822–832. doi: 10.1038/nm.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuziel WA, Morgan SJ, Dawson TC, Griffin S, Smithies O, Ley K, Maeda N. Severe reduction in leukocyte adhesion and monocyte extravasation in mice deficient in CC chemokine receptor 2. Proc Natl Acad Sci USA. 1997;94:12053–12058. doi: 10.1073/pnas.94.22.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YG, Jeong JJ, Nyenhuis S, Berdyshev E, Chung S, Ranjan R, Karpurapu M, Deng J, Qian F, Kelly EA, et al. Recruited alveolar macrophages, in response to airway epithelial-derived monocyte chemoattractant protein 1/CCl2, regulate airway inflammation and remodeling in allergic asthma. Am J Respir Cell Mol Biol. 2015;52:772–784. doi: 10.1165/rcmb.2014-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. Available at: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rossen N, Sinn P, Hornick A, Steines B, Karp P, Ernst S, Adam R, Moninger T, Levasseur D, et al. Integrin α6β4 Identifies Human Distal Lung Epithelial Progenitor Cells with Potential as a Cell-Based Therapy for Cystic Fibrosis Lung Disease. PLoS ONE. 2013 doi: 10.1371/journal.pone.0083624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SLL, Li B, Rao S, Yeo EJJ, Hudson TE, Nowlin BT, Pei H, Chen L, Zheng JJ, Carroll TJ, et al. Macrophage Wnt7b is critical for kidney repair and regeneration. Proc Natl Acad Sci USA. 2010;107:4194–4199. doi: 10.1073/pnas.0912228107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014 doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JLL, Libby P, Weissleder R, Pittet MJ. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama K, Nakashima H, Ikarashi M, Kinoshita M, Nakashima M, Aosasa S, Seki S, Yamamoto J. Mouse CD11b+Kupffer Cells Recruited from Bone Marrow Accelerate Liver Regeneration after Partial Hepatectomy. PLoS ONE. 2015;10:e0136774. doi: 10.1371/journal.pone.0136774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth N, Shultz LD, Brombacher F, Urban JF, Gu H, Paul WE. An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc Natl Acad Sci USA. 1997;94:10838–10843. doi: 10.1073/pnas.94.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen-Walston RD, Kim CF, Mazan MR, Ingenito EP, Gruntman AM, Tsai L, Boston R, Woolfenden AE, Jacks T, Hoffman AM. Cellular kinetics and modeling of bronchioalveolar stem cell response during lung regeneration. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1158–65. doi: 10.1152/ajplung.00298.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn M, Geissmann F, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518 doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci USA. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae F, Woods K, Sasmono T, Campanale N, Taylor D, Ovchinnikov DA, Grimmond SM, Hume DA, Ricardo SD, Little MH. Characterisation and trophic functions of murine embryonic macrophages based upon the use of a Csf1r-EGFP transgene reporter. Dev Biol. 2007;308:232–246. doi: 10.1016/j.ydbio.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Rafii S, Cao Z, Lis R, Siempos II, Chavez D, Shido K, Rabbany SY, Ding B-SS. Platelet-derived SDF-1 primes the pulmonary capillary vascular niche to drive lung alveolar regeneration. Nat Cell Biol. 2015;17:123–136. doi: 10.1038/ncb3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Pellicoro A, Vernon MA, Boulter L, Aucott RL, Ali A, Hartland SN, Snowdon VK, Cappon A, Gordon-Walker TT, et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci USA. 2012;109:E3186–95. doi: 10.1073/pnas.1119964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar P, Yilmaz C, Bellotto DJ, Dane DM, Estrera AS, Hsia CC. Separating in vivo mechanical stimuli for postpneumonectomy compensation: imaging and ultrastructural assessment. J Appl Physiol. 2013;114:961–970. doi: 10.1152/japplphysiol.01394.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Liang HEE, Tager AM, Luster AD, Van Rooijen N, Voehringer D, Locksley RM. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature. 2007;447:92–96. doi: 10.1038/nature05746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Hogan BL. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol. 2011;27:493–512. doi: 10.1146/annurev-cellbio-100109-104040. [DOI] [PubMed] [Google Scholar]
- Rock JR, Barkauskas CE, Cronce MJ. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proceedings of the …. 2011a doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Barkauskas CE, Cronce MJ, Xue Y, Harris JR, Liang J, Noble PW, Hogan BL. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci USA. 2011b;108:E1475–83. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez R, Rosenthal N, Nerlov C. A CREB-C/EBPβ cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proceedings of the National Academy of Sciences. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup N, Cardona AE, Croft K, Mizutani M, Cotleur AC, Tsou CLL, Ransohoff RM, Charo IF. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS ONE. 2010;5:e13693. doi: 10.1371/journal.pone.0013693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano F, Ueda K, Murakami J, Hayashi M, Nishimoto A, Hamano K. Enhanced tumor growth in the remaining lung after major lung resection. Journal of Surgical Research. 2015 doi: 10.1016/j.jss.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wainwright BJ, Ostrowski MC, Himes SR, Hume DA. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101:1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- Sasmono RT, Ehrnsperger A, Cronau SL, Ravasi T, Kandane R, Hickey MJ, Cook AD, Himes SR, Hamilton JA, Hume DA. Mouse neutrophilic granulocytes express mRNA encoding the macrophage colony-stimulating factor receptor (CSF-1R) as well as many other macrophage-specific transcripts and can transdifferentiate into macrophages in vitro in response to CSF-1. J Leukoc Biol. 2007;82:111–123. doi: 10.1189/jlb.1206713. [DOI] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Shechter R, Miller O, Yovel G, Rosenzweig N, London A, Ruckh J, Kim KWW, Klein E, Kalchenko V, Bendel P, et al. Recruitment of beneficial M2 macrophages to injured spinal cord is orchestrated by remote brain choroid plexus. Immunity. 2013;38:555–569. doi: 10.1016/j.immuni.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi M, Shintani Y, Shintani Y, Ishida H, Saba R, Yamaguchi A, Adachi H, Yashiro K, Suzuki K. Alternatively activated macrophages determine repair of the infarcted adult murine heart. J Clin Invest. 2016 doi: 10.1172/JCI85782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Arumugam P, Sakagami T, Lachmann N, Chalk C, Sallese A, Abe S, Trapnell C, Carey B, Moritz T, et al. Pulmonary macrophage transplantation therapy. Nature. 2014;514:450–454. doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SY, Krasnow MA. Developmental origin of lung macrophage diversity. Development. 2016 doi: 10.1242/dev.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voswinckel Motejl, Fehrenbach Wegmann, Mehling Fehrenbach, Seeger Characterisation of post-pneumonectomy lung growth in adult mice. Eur Respir J. 2004;24:524–532. doi: 10.1183/09031936.04.10004904. [DOI] [PubMed] [Google Scholar]
- Westphalen K, Gusarova GA, Islam MN, Subramanian M, Cohen TS, Prince AS, Bhattacharya J. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature. 2014;506:503–506. doi: 10.1038/nature12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenborg S, Lucas T, van Loo G, Knipper JA, Krieg T, Haase I, Brachvogel B, Hammerschmidt M, Nagy A, Ferrara N, et al. CCR2 recruits an inflammatory macrophage subpopulation critical for angiogenesis in tissue repair. Blood. 2012;120:613–625. doi: 10.1182/blood-2012-01-403386. [DOI] [PubMed] [Google Scholar]
- Yona S, Kim KWW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


