Abstract
Megacystis microcolon intestinal hypoperistalsis syndrome (MMIHS) is a congenital disorder characterized by loss of smooth muscle contraction in the bladder and intestine. To date, three genes are known to be involved in MMIHS pathogenesis: ACTG2, MYH11, and LMOD1. However, for approximately 10% of affected individuals, the genetic cause of the disease is unknown, suggesting that other loci are most likely involved. Here, we report on three MMIHS-affected subjects from two consanguineous families with no variants in the known MMIHS-associated genes. By performing homozygosity mapping and whole-exome sequencing, we found homozygous variants in myosin light chain kinase (MYLK) in both families. We identified a 7 bp duplication (c.3838_3844dupGAAAGCG [p.Glu1282_Glyfs∗51]) in one family and a putative splice-site variant (c.3985+5C>A) in the other. Expression studies and splicing assays indicated that both variants affect normal MYLK expression. Because MYLK encodes an important kinase required for myosin activation and subsequent interaction with actin filaments, it is likely that in its absence, contraction of smooth muscle cells is impaired. The existence of a conditional-Mylk-knockout mouse model with severe gut dysmotility and abnormal function of the bladder supports the involvement of this gene in MMIHS pathogenesis. In aggregate, our findings implicate MYLK as a gene involved in the recessive form of MMIHS, confirming that this disease of the visceral organs is heterogeneous with a myopathic origin.
Keywords: MMIHS, MYLK, smooth muscle contractility, bladder and intestinal obstruction
Main Text
Contraction of smooth muscle cells (SMCs) results from an elaborate molecular process that involves the cross-bridge interaction between thin actin filaments and thick myosin filaments.1 Loss of any of the proteins involved in this process is likely to affect cell physiology, resulting in impaired contractility. Recently, loss of SMC contraction has been found to underlie the development of megacystis microcolon intestinal hypoperistalsis syndrome (MMIHS [OMIM: 155310]), a rare congenital disease of the visceral organs, mainly characterized by bladder distension and the presence of a microcolon.2, 3 This link was made from the identification of pathogenic variants in three muscle-related genes: actin, gamma 2, smooth muscle, enteric (ACTG2 [OMIM: 102545]),4, 5, 6, 7, 8 myosin heavy chain 11 (MYH11 [OMIM: 160745]),9 and leimodin 1 (LMOD1 [OMIM: 602715]).10 De novo variants in ACTG2 are implicated in the autosomal-dominant form of MMIHS,4, 5, 6, 7, 8 whereas homozygous variants in MYH11 and LMOD1 cause a recessive form of the disease.9, 10 Although the identification of pathogenic variants in these genes explains approximately 90% of all MMIHS cases characterized by bladder and intestinal problems and for which genetic information is available,4, 5, 6, 7, 8, 9, 10 for some affected individuals, the causative mutation and associated gene have yet to be identified.
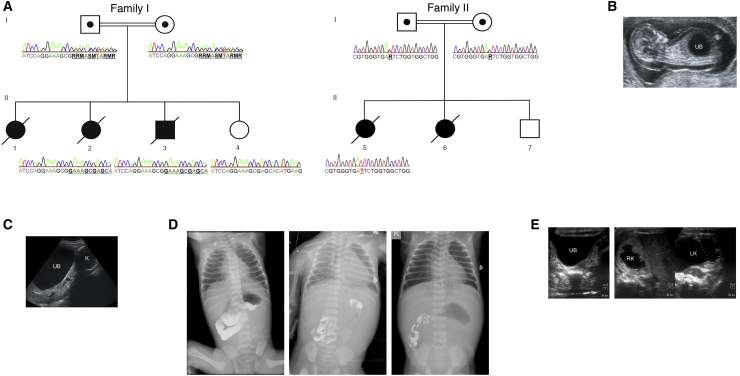
Here, we describe three MMIHS subjects from two independent families and for whom no variants in the known MMIHS-associated genes were found (Figure 1A). Subject 1 (II-2 in Figure 1A) was born to consanguineous parents of North African origin. Prenatal ultrasound performed at 13 weeks of gestation identified the presence of a distended bladder and a generalized subcutaneous edema (Figure 1B). Severe oligohydramnios was also reported. Autopsy of the fetus confirmed the diagnosis of MMIHS (the pregnancy was terminated at 15 weeks of gestation). Subject 2 (II-3 in Figure 1A) was the younger brother of subject 1. Distension of the bladder was observed on prenatal ultrasound (Figure 1C), and anhydramnios was detected. Labor occurred prematurely at 31 weeks of gestation. The neonate experienced respiratory distress and died. Further anamnesis revealed the presence of a distended bladder in the older sister (II-1 in Figure 1A) of subjects 1 and 2 before her intrauterine death at 30 weeks of gestation. Subject 3 (II-5 in Figure 1A) was the first child of a consanguineous couple of Indian origin. The antenatal period was complicated with polyhydramnios, but the baby was born at term by normal delivery. No neonatal complications were reported. At 2 days of age, she was admitted back to the hospital with bilious vomiting. Barium enema confirmed intestinal obstruction, which was most likely caused by malrotation of the intestine (Figure 1D). Surgery was performed 1 day later to place an ileostomy and correct the intestinal malrotation. During surgery, a distal microcolon was revealed, and the bladder was catheterized. Histopathological analysis of the mid-ileum identified the presence of ganglia. Abdominal ultrasound revealed distension of the bladder and bilateral hydroureteronephrosis (Figure 1E), allowing the diagnosis of MMIHS to be made. Further follow-up of the family revealed that a younger sister (II-6 in Figure 1A) of subject 3 had also been diagnosed with MMIHS.
Figure 1.
Genetic Analysis and Description of the Three MMIHS Subjects Included in This Study
(A) Pedigrees of the two consanguineous families analyzed by Sanger sequencing show the presence of homozygous variants in MYLK. Subject 1 (II-2) and subject 2 (II-3) carry a 7 bp duplication in exon 23 (c.3838_3844dupGAAAGCG [p.Glu1282_Glyfs∗51]), whereas their unaffected sister (II-4) shows no mutant allele. Subject 3 (II-5) has a putative splice-site variant affecting exon 23 (c.3985+5C>A). Both variants are present in a heterozygous state in the parents.
(B) Prenatal ultrasonography of subject 1 (II-2) at 13 weeks of gestation revealed a distended bladder and generalized subcutaneous edema.
(C) Prenatal ultrasonography of subject 2 (II-3) at 24 weeks revealed a distended bladder, hydronephrosis, and severe oligohydramnios.
(D) Barium enema performed in subject 2 (II-3) suggested intestinal obstruction and malrotation.
(E) Neonatal ultrasonography of subject 3 (II-5) revealed bladder distension and bilateral hydronephrosis.
Abbreviations are as follows: UB, urinary bladder; K, kidney; RK, right kidney; and LK, left kidney.
To identify the genetic cause of the disease in these subjects, we used a combined strategy of homozygosity mapping and whole-exome sequencing (WES). Written informed consent was obtained from the parents, and this study was approved by the Erasmus Medical Center ethics committee (Medisch Ethische Toetsings commissie 2011/148, Algemeen Beoordelings en Registratieformulier form NL35920.042.11) and by the institutional review board at Baylor College of Medicine (Baylor Hopkins Centers for Mendelian Genomics protocol H-29697). Absence of heterozygosity (AOH), copy-number variants (CNVs), and exonic variants with deleterious changes predicted in silico (single-nucleotide variants [SNVs] and insertions or deletions) were determined as previously described.10 Because consanguinity is known to increase the number of regions of shared haplotypes and is therefore associated with the occurrence of autosomal-recessive disorders,11, 12 we analyzed the filtered variants by identifying regions that are presumed to be shared identically by descent in extended genomic intervals of AOH. These identical-by-descent (IBD) regions are enriched with potential deleterious variants fulfilling Mendelian expectations13 and were the starting point of our genetic analysis. For subject 1 (II-2 in Figure 1A), 55 homozygous regions of at least 1 Mb in length and harboring at least 50 probes were identified by SNP arrays. Of these 55 regions, 38 were present in the parents in a heterozygous state (Figure S1 and Table S1) and were likely to be IBD regions. Analysis of the SNP arrays did not identify any deleterious large CNVs. WES was also performed for subject 1, and the filtering criteria followed are described in Figure S2. Out of 2,848 rare variants found (minor allele frequency below 1%), only 20 were proven to be recessively inherited and predicted to be deleterious (Table 1). From this list, one gene stood out because of its known biological function in the contraction of SMC: myosin light chain kinase (MYLK [OMIM: 600922]). In this gene, subject 1 carried an exon 23 duplication that led to a frameshift and to the appearance of an early stop codon at the beginning of exon 24 (c.3838_3844dupGAAAGCG [p.Glu1282Glyfs∗51] [GenBank: NM_053025.3]). It is unlikely that this duplication resulted from secondary structure mutagenesis given that the surrounding sequence is not palindromic. MYLK was also located in an AOH region of 3 Mb (Table S1). Previous studies have shown that mice lacking the smooth muscle Mylk isoform have severe gut dysmotility and abnormal function of the bladder,14 a phenotype reminiscent of that described for individuals affected by MMIHS. Moreover, according to data from the Human Integrated Protein Expression Database and the Genotype-Tissue Expression project15 in GeneCards,16 MYLK was the only gene in this list with expression in human fetal gut and bladder, the two major organs affected in MMIHS. On the basis of this evidence, we considered MYLK to be the best candidate gene for this family.
Table 1.
Prioritized Rare Recessive Variants Present in AOH Regions and Predicted to Be Deleterious in Subject 1
| Gene | Type | Exon | Effect | HGVS Genomic Change (GenBank ID) | cDNA | Protein | CADD Phred Score |
|---|---|---|---|---|---|---|---|
| DENND4B | Ins | 18 | F | g.153907306_ 153907307insGCTGCTGC (NC_000001.10) | c.2702_2703insGCAGCAGC | p.Gln904Hisfs∗48 | 32 |
| LENEP | SNV | 1 | MS | g.154966258C>G (NC_000001.10) | c.175C>G | p.Leu59Val | 23.6 |
| SPTA1 | SNV | 14 | SP | g.158639351C>G (NC_000001.10) | c.1680G>C | – | 23.1 |
| ITLN1 | SNV | 8 | MS | g.160846459G>A (NC_000001.10) | c.937C>T | p.Arg313Cys | 24 |
| MCM6 | SNV | 17 | MS | g.136598443A>G (NC_000002.11) | c.2428T>C | p.Tyr810His | 24.2 |
| MYLK | Ins | 23 | F | g.123383093_ 123383099dup (NC_000003.11) | c.3838_3844dupGAAAGCG | p.Glu1282Glyfs∗51 | 36 |
| AFF1 | SNV | 5 | SP | g.88011230C>T (NC_000004.11) | c.1170+8C>T | – | 4.535 |
| ADAMTS16 | SNV | 16 | MS | g.5239937C>T (NC_000005.9) | c.2422C>T | p.Arg808Trp | 25.7 |
| ID4 | SNV | 1 | MS | g.19838067C>G (NC_000006.11) | c.82C>G | p.Leu28Val | 25.7 |
| ZNF596 | Del | 4 | CSP | g.193799_ 193812delCTGCAAGGTGAGCT (NC_000008.10) | c.217_223+7delCTGCAAGGTGAGCT | – | 23.2 |
| C8orf34 | SNV | 13 | S, SP | g.69728122T>C (NC_000008.10) | c.1551T>C | – | 6.825 |
| PKHD1L1 | SNV | 71 | MS | g.110523018A>G (NC_000008.10) | c.11408A>G | p.His3803Arg | 24.2 |
| TRPM6 | SNV | 18 | MS | g.77411729C>G (NC_000009.11) | c.2319G>C | p.Gln773His | 26.1 |
| CCDC81 | SNV | 4 | MS | g.86103688C>T (NC_000011.9) | c.404C>T | p.Ser135Leu | 28.3 |
| METTL7B | SNV | 1 | MS | g.56075840C>T (NC_000012.11) | c.302C>T | p.Pro101Leu | 33 |
| EXD1 | SNV | 8 | MS | g.41488231G>C (NC_000015.9) | c.539C>G | p.Ala180Gly | 24.5 |
| DUOX2 | SNV | 30 | SP | g.45388019C>T (NC_000015.9) | c.4080+7G>A | – | 5.471 |
| AARS | SNV | 6 | MS | g.70304215G>A (NC_000016.9) | c.700C>T | p.Pro234Ser | 28.9 |
| NSF | SNV | 20 | MS | g.44832731G>A (NC_000017.10) | c.2209G>A | p.Gly737Arg | 33 |
| USP29 | SNV | 4 | MS | g.57642673G>A (NC_000019.9) | c.2630G>A | p.Gly877Glu | 24.4 |
Abbreviations are as follows: AOH, absence of heterozygozity; Ins, insertion; Del, deletion; SNV, single-nucleotide variant; MS, missense variant; F, frameshift variant; S, synonymous variant; SP, putative splice variant; ncRNA, non-coding RNA; and CSP, canonical splice site.
WES investigations of another independent cohort of individuals consisting of 42 probands with visceral myopathy, MMIHS, and prune-belly phenotypes5 led to the identification of one subject, subject 3 (II-5 in Figure 1A), in whom a homozygous putative splice variant affecting exon 23 of MYLK (c.3985+5C>A [GenBank: NM_053025.3]) had also been identified. Analysis of the AOH regions present in this subject showed that stretches of homozygosity composed 8% of her genome and that MYLK was located within a 9 Mb genomic region of AOH (Figure S3 and Table S2). It is also worth noting that combined analysis of the WES data generated for subject 1 (Table 1) and subject 3 (Table 2) showed that MYLK was the only shared gene in which recessive variants predicted to be deleterious had been identified (Figure S2). Sanger sequencing performed with specific primers (MYLKF and MYLKR; Table S3) confirmed the presence of both variants in a homozygous state in these subjects and showed that they were inherited from heterozygous parents (Figure 1A). Subject 2 (II-3 in Figure 1A) was also found to carry the same homozygous duplication detected in subject 1 (II-2 in Figure 1A), whereas their unaffected sister showed no mutant allele (Figure 1A). No genetic information was available for the older sister of subjects 1 and 2 or for the younger sister and brother of subject 3 as a result of DNA unavailability.
Table 2.
Prioritized Rare Recessive Variants Present in Subject 3
| Gene | Type | Exon | Effect | HGVS Genomic Change (GenBank ID) | cDNA | Protein | CADD Phred Score |
|---|---|---|---|---|---|---|---|
| THUMPD2 | SNV | 3 | MS | g.39996996C>A (NC_000002.11) | c.526G>T | p.Asp176Tyr | 25.4 |
| NBEAL2 | SNV | 32 | MS | g.47043984C>T (NC_000003.11) | c.5275C>T | p.Arg1759Cys | 26.8 |
| RNF123 | SNV | 24 | MS | g.49743006G>A (NC_000003.11) | c.2198G>A | p.Arg733Gln | 24.4 |
| IFRD2 | SNV | 2 | MS | g.50328075T>C (NC_000003.11) | c.286A>G | p.Ser96Gly | 25.4 |
| CACNA2D3 | SNV | 9 | MS | g.54615858C>G (NC_000003.11) | c.917C>G | p.Pro306Arg | 31 |
| IL17RD | SNV | 10 | MS | g.57136593A>G (NC_000003.11) | c.893T>C | p.Ile298Thr | 28.4 |
| MYLK | SNV | 23 | SP | g.123382947C>A (NC_000003.11) | c.3985+5G>T | – | 21.4 |
| SEC24D | SNV | 2 | SP | g.119754899C>T (NC_000004.11) | c. −41−7G>A | – | 9.701 |
| SLC26A5 | SNV | 16 | MS | g.103019758T>C (NC_000007.13) | c.1609A>G | p.Ile537Val | 23.6 |
| CNTLN | SNV | 22 | SP | g.17465974T>C (NC_000009.11) | c.3532−5T>C | – | 12.13 |
| EHMT1 | SNV | 12 | S | g.140671136C>A (NC_000009.11) | c.1858C>A | – | 20.8 |
| SIGLEC1 | Del | 21 | F | g.3669236delT (NC_000020.10) | c.5101delA | p.Thr1701Profs∗26 | 33 |
Abbreviations are as follows: SNV, single-nucleotide variant; MS, missense variant; F, frameshift variant; S, synonymous variant; and SP, putative splice variant.
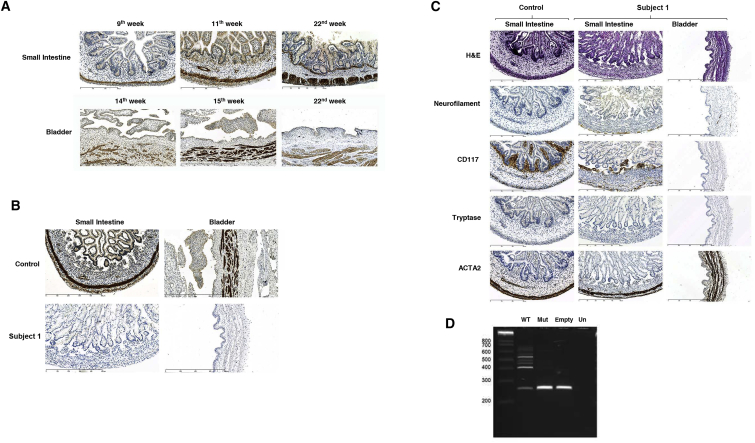
In humans, three major MYLK isoforms exist: a long isoform referred to as non-muscle MYLK, a short isoform known as smooth muscle MYLK, and a very small isoform called telokin.17 The variants identified in the families included in this study affect both the long and the short isoforms (Figure S4). To further evaluate the involvement of MYLK in MMIHS pathogenesis, we determined the presence of this protein at different stages of human embryonic development. Because the bladder and intestine are the two organs affected in MMIHS, we used a specific antibody to evaluate the presence of MYLK in these organs by immunohistochemistry. Formalin-fixed paraffin-embedded human small intestinal and bladder specimens obtained from control samples were retrieved from the Biobank of the Pathology Department of the Erasmus University Medical Center. We observed that, across all developmental stages included in this study, MYLK was localized in all SMCs that form all muscular layers of the intestine and bladder (Figure 2A). We also immunostained specimens collected from the small intestine and bladder of subject 1 (II-2 in Figure 1A) by the Department of Pathology of the Hôpital Universitaire Robert Debré and found no signal for MYLK. Because the recognition epitope of the antibody used is located before the homozygous duplication (amino acids 908–938), it is likely that MYLK expression is abolished in this subject, confirming loss of function and pathogenicity of the variant identified (Figure 2B). Surprisingly, no changes in the levels of neurofilament, c-Kit/CD117, tryptase, or ACTA2 were detected in this subject. Moreover, in comparison with age-matched control samples, the structure and cellular constituents of the bladder and intestine showed no apparent pathological abnormalities (Figure 2C). This suggests that although MYLK is instrumental for the proper function of SMCs, its presence is not required for maintaining the structural architecture of these organs.
Figure 2.
Evaluation of the Effect of the MYLK Variants Identified
(A) Immunohistochemistry performed in control specimens with a specific MYLK antibody (1:100; Thermo Fisher Scientific) showed that MYLK is present in SMCs that constitute muscular structures of the intestine and the bladder, including muscularis mucosa, blood vessels, and circular and longitudinal muscles of the muscularis propria. Similar patterns and expression levels were present throughout all developmental stages included in this study.
(B) MYLK is absent in the intestine and bladder of subject 1 (II-2 in Figure 1A).
(C) H&E staining and immunostainings performed in specimens derived from control samples and subject 1 with specific antibodies against neurofilament (1:600; Monosan), c-Kit/CD117 (1:200; Cell Marque), tryptase (1:1,600; Dako), and ACTA2 (ready to use; Dako) showed no significant differences.
(D) Splicing assays revealed impaired splicing of exon 23 of MYLK in subject 3 (II-5 in Figure 1A). SD6 and SA2 primers (Table S3) were used for evaluating transcripts produced by the wild-type (WT) vector, the mutant (Mut) vector containing the putative splice-site variant c.3985+5C>A, the empty vector (Empty), and untransfected cells (Un) according to a standard PCR protocol.
We also investigated the pathogenicity of the putative splice-site variant (c.3985+5C>A) found in subject 3 (II-5 in Figure 1A) by performing splicing assays as previously described.18 Wild-type (WT) and mutant constructs were generated with genomic DNA from control and affected subjects with a primer set designed to include the genomic sequence that stretches from the last part of intron 22 of MYLK to the beginning of intron 23 (MinigeneF and MinigeneR; Table S3). Whereas the WT construct generated a transcript with the expected size (414 bp), the mutant construct eliminated all transcription products seen in the WT situation and produced a band with the same size as the empty vector. This result indicates that the identified variant is indeed a splice variant likely leading to the skipping of exon 23 (Figure 2D). As a consequence, a frameshift will occur and lead to the appearance of an early stop codon at the beginning of exon 24, but in a different location than the one identified in subjects 1 and 2. Although tissue specimens were not available for subject 3 (II-5 in Figure 1A), we suspect that expression of MYLK is also impaired, given that the splice variant had an effect similar to that of the duplication described for subjects 1 and 2 and is therefore likely to result in loss of function. Interestingly, we noticed that several other transcripts were generated for exon 23 in the WT situation, suggesting that this exon might normally be targeted for alternative splicing.
MYLK encodes an important kinase required for phosphorylation of the regulatory light chain (RLC) of myosin, leading to its activation and subsequent interaction with actin filaments17 (Figure S5). This interaction is essential for SMC contraction, a mechanism that is already known to be defective in MMIHS.4, 7, 10 According to our results, MYLK expression is most likely impaired in the subjects included in this study, and we expect that myosin activation cannot occur because of a lack of phosphorylation of its RLC. As a consequence, myosin will be unable to interact with actin, resulting in impaired SMC contractility (Figure S5). Considering that MYLK is a ubiquitous protein, it is surprising that the phenotype seen in individuals affected with MMIHS, as well as in the conditional-knockout mouse,14 is restricted to the intestine and bladder. This might be related to the fact that the extra visceral phenotypes develop only later in life, and we were unable to detect it because none of our subjects survived longer than a few days after birth. This hypothesis is supported by previous reports linking heterozygous loss of MYLK to aortic vascular smooth muscle defects.19, 20 In these subjects, aneurysms often occur without preceding dilatation of the aorta, suggesting that it takes time for the reduction of MYLK to take its toll on major blood vessels. However, it could also be that other kinases are able to salvage SMC contraction of the extra-visceral organs. This hypothesis is supported by the fact that RLC phosphorylation could still be induced in aortic cells collected from mice where expression of all three Mylk isoforms is abolished.21 Considering that the heterozygous parents of the subjects included in this study do not show any cardiac problems as far as we know, the second hypothesis is perhaps more likely, but further studies need to be performed to bring new insight into this matter.
In conclusion, we provide evidence for MYLK involvement in MMIHS, confirming that this is a heterogeneous disease of the visceral organs where disruption of SMC contraction seems to be the major trigger for pathogenicity.
Conflicts of Interest
The Department of Molecular and Human Genetics at the Baylor College of Medicine derives revenue from molecular testing offered at Baylor Genetics Laboratories. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen Inc., is a member of the scientific advisory board of Baylor Genetics, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting.
Acknowledgments
The authors would like to thank the two families described in this manuscript for their cooperation and willingness to be part of this study. The authors would also like to thank Dr. Raj Kapur for his help with the analysis of the immunohistochemistry data and Tom de Vries Lentsch for making Figure S5. This work received funding from the National Human Genome Research Institute and the National Heart, Lung, and Blood Institute (UM1 HG006542) to the Baylor-Hopkins Center for Mendelian Genomics and from the Stitching Sophia Kinderziekenhuis Fonds (S15-30) to R.M.W.H. and M.M.A.
Published: June 8, 2017
Footnotes
Supplemental Data include five figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.05.011.
Contributor Information
Robert M.W. Hofstra, Email: r.hofstra@erasmusmc.nl.
Maria M. Alves, Email: m.alves@erasmusmc.nl.
Accession Numbers
The accession numbers for variants c.3838_3844dupGAAAGCG and c.3985+5C>A are ClinVar: SCV000299345 and SCV000574720, respectively.
Web Resources
ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/
OMIM, https://www.omim.org/
Supplemental Data
References
- 1.Kamm K.E., Stull J.T. Dedicated myosin light chain kinases with diverse cellular functions. J. Biol. Chem. 2001;276:4527–4530. doi: 10.1074/jbc.R000028200. [DOI] [PubMed] [Google Scholar]
- 2.Berdon W.E., Baker D.H., Blanc W.A., Gay B., Santulli T.V., Donovan C. Megacystis-microcolon-intestinal hypoperistalsis syndrome: a new cause of intestinal obstruction in the newborn. Report of radiologic findings in five newborn girls. AJR Am. J. Roentgenol. 1976;126:957–964. doi: 10.2214/ajr.126.5.957. [DOI] [PubMed] [Google Scholar]
- 3.Wymer K.M., Anderson B.B., Wilkens A.A., Gundeti M.S. Megacystis microcolon intestinal hypoperistalsis syndrome: Case series and updated review of the literature with an emphasis on urologic management. J. Pediatr. Surg. 2016;51:1565–1573. doi: 10.1016/j.jpedsurg.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Thorson W., Diaz-Horta O., Foster J., 2nd, Spiliopoulos M., Quintero R., Farooq A., Blanton S., Tekin M. De novo ACTG2 mutations cause congenital distended bladder, microcolon, and intestinal hypoperistalsis. Hum. Genet. 2014;133:737–742. doi: 10.1007/s00439-013-1406-0. [DOI] [PubMed] [Google Scholar]
- 5.Wangler M.F., Gonzaga-Jauregui C., Gambin T., Penney S., Moss T., Chopra A., Probst F.J., Xia F., Yang Y., Werlin S., Baylor-Hopkins Center for Mendelian Genomics Heterozygous de novo and inherited mutations in the smooth muscle actin (ACTG2) gene underlie megacystis-microcolon-intestinal hypoperistalsis syndrome. PLoS Genet. 2014;10:e1004258. doi: 10.1371/journal.pgen.1004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuzovic L., Tang S., Miller R.S., Rohena L., Shahmirzadi L., Gonzalez K., Li X., LeDuc C.A., Guo J., Wilson A. New Insights into the Genetics of Fetal Megacystis: ACTG2 Mutations, Encoding γ-2 Smooth Muscle Actin in Megacystis Microcolon Intestinal Hypoperistalsis Syndrome (Berdon Syndrome) Fetal Diagn. Ther. 2015;38:296–306. doi: 10.1159/000381638. [DOI] [PubMed] [Google Scholar]
- 7.Halim D., Hofstra R.M., Signorile L., Verdijk R.M., van der Werf C.S., Sribudiani Y., Brouwer R.W., van IJcken W.F., Dahl N., Verheij J.B. ACTG2 variants impair actin polymerization in sporadic Megacystis Microcolon Intestinal Hypoperistalsis Syndrome. Hum. Mol. Genet. 2016;25:571–583. doi: 10.1093/hmg/ddv497. [DOI] [PubMed] [Google Scholar]
- 8.Matera I., Rusmini M., Guo Y., Lerone M., Li J., Zhang J., Di Duca M., Nozza P., Mosconi M., Pini Prato A. Variants of the ACTG2 gene correlate with degree of severity and presence of megacystis in chronic intestinal pseudo-obstruction. Eur. J. Hum. Genet. 2016;24:1211–1215. doi: 10.1038/ejhg.2015.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauthier J., Ouled Amar Bencheikh B., Hamdan F.F., Harrison S.M., Baker L.A., Couture F., Thiffault I., Ouazzani R., Samuels M.E., Mitchell G.A. A homozygous loss-of-function variant in MYH11 in a case with megacystis-microcolon-intestinal hypoperistalsis syndrome. Eur. J. Hum. Genet. 2015;23:1266–1268. doi: 10.1038/ejhg.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halim D., Wilson M.P., Oliver D., Brosens E., Verheij J.B.G.M., Han Y., Nanda V., Lyu Q., Doukas M., Stoop H. Loss of LMOD1 impairs smooth muscle cytocontractility and causes megacystis microcolon intestinal hypoperistalsis syndrome in humans and mice. Proc. Natl. Acad. Sci. USA. 2017;114:E2739–E2747. doi: 10.1073/pnas.1620507114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamamy H., Antonarakis S.E., Cavalli-Sforza L.L., Temtamy S., Romeo G., Kate L.P., Bennett R.L., Shaw A., Megarbane A., van Duijn C. Consanguineous marriages, pearls and perils: Geneva International Consanguinity Workshop Report. Genet. Med. 2011;13:841–847. doi: 10.1097/GIM.0b013e318217477f. [DOI] [PubMed] [Google Scholar]
- 12.Hamamy H. Consanguineous marriages : Preconception consultation in primary health care settings. J. Community Genet. 2012;3:185–192. doi: 10.1007/s12687-011-0072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szpiech Z.A., Xu J., Pemberton T.J., Peng W., Zöllner S., Rosenberg N.A., Li J.Z. Long runs of homozygosity are enriched for deleterious variation. Am. J. Hum. Genet. 2013;93:90–102. doi: 10.1016/j.ajhg.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He W.Q., Peng Y.J., Zhang W.C., Lv N., Tang J., Chen C., Zhang C.H., Gao S., Chen H.Q., Zhi G. Myosin light chain kinase is central to smooth muscle contraction and required for gastrointestinal motility in mice. Gastroenterology. 2008;135:610–620. doi: 10.1053/j.gastro.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishilevich S., Zimmerman S., Kohn A., Iny Stein T., Olender T., Kolker E., Safran M., Lancet D. Genic insights from integrated human proteomics in GeneCards. Database (Oxford) 2016;2016:baw030. doi: 10.1093/database/baw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong F., Haldeman B.D., Jackson D., Carter M., Baker J.E., Cremo C.R. Biochemistry of smooth muscle myosin light chain kinase. Arch. Biochem. Biophys. 2011;510:135–146. doi: 10.1016/j.abb.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Der Werf C.S., Wabbersen T.D., Hsiao N.H., Paredes J., Etchevers H.C., Kroisel P.M., Tibboel D., Babarit C., Schreiber R.A., Hoffenberg E.J. CLMP is required for intestinal development, and loss-of-function mutations cause congenital short-bowel syndrome. Gastroenterology. 2012;142:453–462.e3. doi: 10.1053/j.gastro.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 19.Wang L., Guo D.C., Cao J., Gong L., Kamm K.E., Regalado E., Li L., Shete S., He W.Q., Zhu M.S. Mutations in myosin light chain kinase cause familial aortic dissections. Am. J. Hum. Genet. 2010;87:701–707. doi: 10.1016/j.ajhg.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannuksela M., Stattin E.L., Klar J., Ameur A., Johansson B., Sörensen K., Carlberg B. A novel variant in MYLK causes thoracic aortic dissections: genotypic and phenotypic description. BMC Med. Genet. 2016;17:61. doi: 10.1186/s12881-016-0326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somlyo A.V., Wang H., Choudhury N., Khromov A.S., Majesky M., Owens G.K., Somlyo A.P. Myosin light chain kinase knockout. J. Muscle Res. Cell Motil. 2004;25:241–242. doi: 10.1023/b:jure.0000038362.84697.c0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.