Abstract
A delayed fetal-to-adult hemoglobin (Hb) switch ameliorates the severity of β-thalassemia and sickle cell disease. The molecular mechanism underlying the epigenetic dysregulation of the switch is unclear. To explore the potential cis-variants responsible for the Hb switching, we systematically analyzed an 80-kb region spanning the β-globin cluster using capture-based next-generation sequencing of 1142 Chinese β-thalassemia persons and identified 31 fetal hemoglobin (HbF)-associated haplotypes of the selected 28 tag regulatory single-nucleotide polymorphisms (rSNPs) in seven linkage disequilibrium (LD) blocks. A Ly1 antibody reactive (LYAR)-binding motif disruptive rSNP rs368698783 (G/A) from LD block 5 in the proximal promoter of hemoglobin subunit gamma 1 (HBG1) was found to be a significant predictor for β-thalassemia clinical severity by epigenetic-mediated variant-dependent HbF elevation. We found this rSNP accounted for 41.6% of β-hemoglobinopathy individuals as an ameliorating factor in a total of 2,738 individuals from southern China and Thailand. We uncovered that the minor allele of the rSNP triggers the attenuation of LYAR and two repressive epigenetic regulators DNA methyltransferase 3 alpha (DNMT3A) and protein arginine methyltransferase 5 (PRMT5) from the HBG promoters, mediating allele-biased γ-globin elevation by facilitating demethylation of HBG core promoter CpG sites in erythroid progenitor cells from β-thalassemia persons. The present study demonstrates that this common rSNP in the proximal Aγ-promoter is a major genetic modifier capable of ameliorating the severity of thalassemia major through the epigenetic-mediated regulation of the delayed fetal-to-adult Hb switch and provides potential targets for the treatment of β-hemoglobinopathy.
Main Text
The human β-globin cluster, spanning a 70-kb region, is composed of five genes (5′-ε-γG-γA-δ-β-3′; HBE [MIM 142100]-HBG2 [MIM 142250]-HBG1 [MIM 142200]-HBD [MIM 142000]-HBB [MIM 141900]) and a distal regulatory element known as the locus control region (LCR).1 The clinical manifestations of β-thalassemia (MIM 613985) mainly depend on the mutation in the β-globin gene.2 However, individuals with identical β-thalassemia genotypes can exhibit variable clinical severities.3 Several genetic modulators4, 5, 6, 7, 8 and cis-regulatory elements8, 9, 10, 11, 12 involved in the regulation of human fetal hemoglobin (HbF), and concomitant α-thalassemia (MIM 604131) have been identified as ameliorators of β-thalassemia.13 These modifiers included HBG2: −158C>T (NC_000011.9: g.5276169G>A, rs7482144, or XmnI polymorphism) in the β-globin cluster identified to be linked to HBG1: +25G>A polymorphism (NC_000011.9:g.5271063C>T or rs368698783),12, 14 and the master genes involved in the regulation of fetal-to-adult hemoglobin (Hb) switching, including B cell CLL/lymphoma 11A (BCL11A [MIM 606557]),5, 8, 15 Kruppel-like factor 1 (KLF1 [MIM 600599]),6, 15 and MYB (MIM 189990).7, 15 Identifying regulators of Hb switching including these genetic variants could provide promising predictors of β-thalassemia severity and therapeutic targets for re-activating HbF production.15, 16 The proposed modes of Hb switching based on current discoveries rely on various epigenetic and transcriptional regulatory factors that could have interacting roles in this developmental event.17, 18, 19, 20 Recently, leukemia/lymphoma-related factor encoded by zinc finger and BTB domain containing 7A (ZBTB7A [MIM 605878]) was identified as a regulator that represents an alternative mechanism independent of the γ-globin repressor BCL11A, as it can occupy γ-globin genes directly.21 Ly1 antibody reactive (LYAR, HGNC:26021) is a zinc finger transcription factor (TF) that modulates Hb switching by binding the γ-globin gene and epigenetically silencing HbF.22 However, the underlying mechanisms driving epigenetic regulation of Hb switching involved in ameliorating β-thalassemia severity are largely unclear.
To explore the potential genetic cis-variants that ameliorate β-thalassemia severity, we performed capture-based next generation sequencing (NGS) analysis of an 80-kb region spanning the β-globin cluster (Table S1) from the genomic DNA of 1142 Chinese β-thalassemia individuals (The procedures followed were accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and proper informed consent was obtained), and discovered 271 common single-nucleotide polymorphisms (SNPs, Minor allele frequency [MAF] > 0.01) in the cluster. Taking the HbF Z score as dependent variable in single SNP association study by Plink, we identified 107 out of 271 SNPs associated with the HbF levels with Bonferroni correction (p < 0.0002; Table S2). Furthermore, seven linkage disequilibrium (LD) blocks containing 163 out of 271 common SNPs were identified in the 80-kb region (Figure S1) and 28 from 163 SNPs were selected as tag SNPs (r2 = 0.5 and p = 0.05) based on Haploview and Plink program. Haplotype-based association analysis showed that 31 haplotypes of the selected 28 tag SNPs in 7 LD blocks significantly associated with HbF levels (p < 0.05; Table S3). Among these tag SNPs, rs368698783 (p = 5.03E-18) from LD block 5 was found embedded within a highly conserved hexanucleotide LYAR-binding motif in the proximal promoter of HBG1.14, 22 To further validate the effect of this SNP on clinical severity of β-thalassemia persons, age at first blood transfusion was introduced as a dependent variable in Stepwise Cox proportional hazards analysis including SNP rs368698783, as well as β+-thalassemia mutations, hemoglobin subunit alpha (HBA [MIM 141800-141850]) defects and the known functional variants in KLF1, HBS1L-MYB (MIM 612450-189990) intergenic region and BCL11A, which had been verified previously in our cohort as ameliorating factors.6 The analysis showed that SNPs rs368698783 was identified as a critical variant of ameliorating factors (HR = 0.552, p = 3.029 × 10−14) after well-known mutations at KLF1 and HBB in a ranking of Hazard ratio (Table 1). These results together with rs368698783 embedded within a highly conserved hexanucleotide LYAR-binding motif required for methylation-related γ-globin gene silencing as previously described,22 suggested that rs368698783-mediated epigenetic regulation of HbF elevation might be involved in the amelioration of β-thalassemia severity.
Table 1.
Stepwise Cox Proportional Hazards Analysis for 1142 β-Thalassemia Individuals in Cohort C
| Ameliorating alleles | p | Hazard Ratio | 95% CI |
|---|---|---|---|
| KLF1 mutations | 2.294 × 10−7 | 0.219 | 0.123-0.389 |
| HBB mutations (β+) | 3.286 × 10−48 | 0.379 | 0.333-0.432 |
| HBG1: rs368698783 (A) | 3.029 × 10−14 | 0.552 | 0.473-0.643 |
| HBS1L-MYB: rs9399137 (C) | 1.175 × 10−10 | 0.710 | 0.640-0.788 |
| HBA mutations | 1.875 × 10−10 | 0.713 | 0.643-0.791 |
| BCL11A: rs4671393 (A) | 1.830 × 10−5 | 0.806 | 0.730-0.889 |
A backward stepwise Cox proportional hazards model in 1,142 β-thalassemia individuals in cohort C was conducted to evaluate the associations between putative ameliorating alleles and the age at first transfusion. The covariates were classified based on the number of copies of putative modifying allele. Motif-disrupting SNP rs368698783 alone with ten known modifiers (HBB mutations, HBA mutations, KLF1 mutations, HBS1L-MYB: rs9399137, rs4895441, rs9402686, rs1427407; BCL11A: rs4671393, rs11886868, and rs766432) were included in the analysis. The discriminative ability of the model was high (Harrell’s concordance index = 0.708, R2 = 0.274). The performance of the model was measured by Harrell’s concordance index by using the Hmisc and rms package in R version 3.3.1.
HBB genotype categories are defined as (β0): NM_000518(HBB_v001): c.126_129delCTTT, NM_000518(HBB_v001): c.52A>T, NM_000518(HBB_v001): c.316-197C>T, NM_000518(HBB_v001): c.216_217insA, NM_000518(HBB_v001): c.92+1G>T, NM_000518(HBB_v001): c.130G>T, NM_000518(HBB_v001):c.84_85insC, NM_000518(HBB_v001):c.91A>G, NM_000518(HBB_v001):c.45_46insC, NM_000518(HBB_v001):c.165_177delTATGGGCAACCCT, NM_000518(HBB_v001):c.315+1G>A, NM_000518(HBB_v001):c.287_288insA, NM_000518(HBB_v001):c.113G>A, NM_000518(HBB_v001):c.93-1G>C;
(β+): NM_000518(HBB_v001): c.-78A>G, NM_000518(HBB_v001): c.79G>A, NM_000518(HBB_v001): c.-79A>G, NM_000518(HBB_v001): c.315+5G>C, NM_000518(HBB_v001): c.-140C>T, NM_000518(HBB_v001): c.-81A>C, NM_000518(HBB_v001): c.92+5G>C.
HBA genotype categories are defined as (-α): NG_000006.1: g.34247_38050del, NC_000016.9: g.219817_(223755_224074)del; (–): NG_000006.1: g.26264_45564del19301, NG_000006.1: g.10664_44164del33501; (αTα): NM_000517.4(HBA2_v001): c.427T>C, NM_000517.4(HBA2_v001): c.369C>G, NM_000517.4(HBA2_v001):c.377T>C.
To evaluate the prevalence of rs368698783, the genotypes of this rSNP from NGS-analyzed 1,142 individuals, as well as another 1,596 consecutive individuals from two subpopulations determined by Sanger sequencing showed that the derived allele frequencies (DAFs) to be 0.089 and 0.789 for the Chinese and Thai participants, respectively (Figure S2), which are similar to those in previous reports (0.114-0.289) based on other ethnic subpopulations and confirmed the presence of a common allele.14, 23 In 2,170 individuals from southern China, HBG1-rs368698783 (G/A) was highly linked with HBG2-rs7482144 (C/T), except for 14 individuals with CT (HBG2)-GG (HBG1) combination genotypes of these two SNPs, which were almost completely linked in 568 homozygous hemoglobin E (HbEE) individuals from the Thai subpopulation with 31.7% for the heterozygous and 63.0% for the homozygous individuals (Figure S2). We also identified a total of 712 of 1,710 individuals with β-hemoglobinopathy who carried the A allele of rSNP rs368698783 and accounted for 41.6% of the individuals from the two subpopulations (Figure S2).
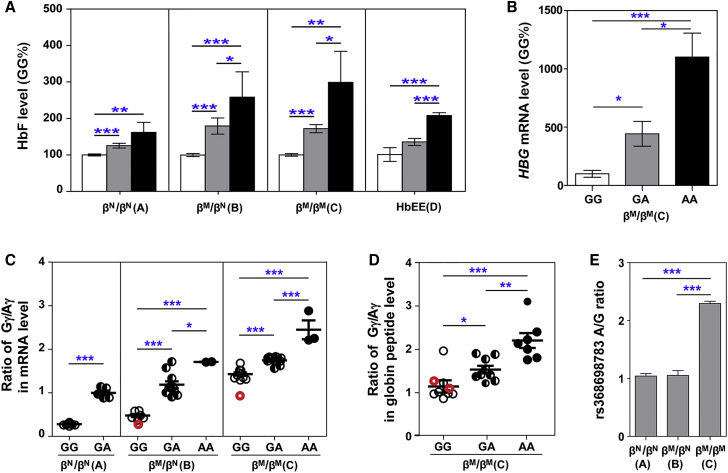
Effects of rs368698783 A allele on HbF levels examined from above two ethnic subpopulations showed that the genotypes GA and AA of HBG1-rs368698783 exhibited a significantly elevated level of HbF (p < 0.05) compared with the genotype GG in each of three cohorts from China (Figure 1A). The HBG1-rs368698783 AA genotype exhibited significantly elevated HbF levels compared with GA in thalassemia (cohorts B and C) and HbEE (cohort D) individuals, as well as in two unrelated Chinese families with β-thalassemia intermedia (Figure S3 and Table S4). Moreover, the genotypes AA and GA of HBG1-rs368698783 exhibited a significantly elevated HBG mRNA level (p < 0.05) compared with the genotype GG in bone marrow (BM)-derived CD235a+ erythroblasts from the individuals with β-thalassemia (Figure 1B). Univariate analysis after correction for HBB, HBA, KLF1, BCL11A, and HBS1L-MYB genotypes demonstrated that variant HBG1-rs368698783 could act as a modifier associated with elevated levels of HbF in β-thalassemia (p = 7.79 × 10−10) and HbEE (p = 1.34 × 10−7) and could ameliorate the severity of β-thalassemia in age at onset (p = 3.82 × 10−10) and transfusion requirements (p = 1.44 × 10−9) and reduce the risk of developing β-thalassemia major (p = 9.17 × 10−18; Table 2, S5–S8). Moreover, AA (p = 0.002) or GA (p < 0.001) genotypes had significantly increased age at first transfusion with β-thalassemia according to the Kaplan-Meier log-rank test (Figure S4). We also observed that the A allele significantly increased the Gγ/Aγ ratio through an increase in the production of γ-globin mRNA (Figure 1C) and peptide (Figure 1D), as well as allele-biased RNA expression in β-thalassemia individuals (βM/βM) with an A/G allelic ratio of 2.4 compared with non-thalassemic controls (βN/βN) or β-thalassemic traits (βM/βN) with an A/G allelic ratio of 1.1 (Figures 1E and S5). This evidence supports that the HbF level in β-thalassemia major is elevated by a variant-dependent activation of the γ-globin gene with an increasing Gγ/Aγ ratio expression.
Figure 1.
Ameliorating Effects of rs368698783 on β-Thalassemia Severity
(A) The effects of rs368698783 genotypes on the levels of HbF in peripheral red blood cells. Relative HbF level (GG%, g/L) determined as described6 from each of four cohorts (the numbers of samples for cohorts A = 513, B = 515, C = 1142 and D = 568, successively) are shown in columns (white, GG; gray, GA; black, AA) with standard errors indicated by bars.
(B) Relative HBG mRNA level in BM-derived CD235a+ erythroblasts from thalassemia individuals with the GG (n = 3), GA (n = 4), or AA (n = 2) genotypes at rs368698783 are quantified by qPCR ([2-ΔΔCt, SYBR Premix Ex Taq [Takara, China]) with β-actin as a reference gene and shown in columns with standard errors indicated by bars.
(C and D) The effects of rs368698783 on the ratio of Gγ/Aγ mRNA (C) as described in Figure S5 and globin protein (D) as described in Figure S3 expression in peripheral blood. Open circles, GG; half-filled circles, GA; full-filled circles, AA. The red circles indicate individuals with the HBG1-rs368698783 GG and HBG2-rs7482144 CT genotype. The numbers of samples for each genotype are as follows: in B: GG = 9 and GA = 5 in cohort A; GG = 7, GA = 12 and AA = 2 in cohort B; GG = 11, GA = 10 and AA = 3 in cohort C; in C: GG = 9, GA = 9 and AA = 7 in cohort C. Solid lines represent as the mean ± SEM.
(E) Measurements of allelic ratios in the peripheral blood of non-thalassemia controls (βN/βN = 5), thalassemia carriers (βM/βN = 12) and thalassemia persons (βM/βM = 10) as described in Figure S5. Data are represented as the mean ± SEM from three independent experiments with triplication. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 based on the Mann-Whitney U test (A) or the T-test (B–E). The genotype of βN/βN represents the normal HBB genotype for non-thalassemia controls, βN/βM (βM representing β0 or β+ genotype in HBB as illustrated in Table 1 legend) for β-thalassemia carriers, and βM/βM for β-thalassemia persons.
Table 2.
Univariate Analysis of HBG1-rs368698783 in 581 β-Thalassemia and 386 HbEE Individuals
| Characteristics | rs368698783 | p | ||
|---|---|---|---|---|
| βM/βMCohorta | GG (n = 500) | GA (n = 77) | AA (n = 4) | |
| Gender (Male: Female) | 326:174 | 48:29 | 2:2 | 0.733 |
| Age of onset (months), median (5th-95th percentile) | 6 (2.0-19.9) | 12 (2.0-36.0) | 30 (24.0-42.0) | 3.82 × 10−10 |
| Hb (g/L)c | 73.35 ± 22.82 | 73.74 ± 21.62 | 69.50 ± 21.42e | 0.891 |
| HbF (g/L)d | 9.14 ± 11.11 | 17.65 ± 16.57 | 37.52 ± 24.03 | 7.79 × 10−10 |
| Requirement for systematic transfusion (No.) | 479 (95.8%) | 62 (80.5%) | 2 (50.0%) | 1.44 × 10−9 |
| Category of anemia (No.) TI: TM | 60:440 | 38:39 | 3:1 | 9.17 × 10−18 |
| HbEE Cohortb | GG (n = 24) | GA (n = 128) | AA (n = 234) | |
| Gender (Male: Female) | 8:16 | 49:79 | 114:120 | 0.086 |
| Hb (g/L) | 118.83 ± 16.30 | 113.87 ± 13.80 | 115.82 ± 13.98 | 0.075 |
| HbE (g/L) | 107.35 ± 16.86 | 103.44 ± 16.24 | 104.01 ± 16.05 | 0.499 |
| HbF (g/L) | 4.12 ± 4.45 | 5.14 ± 4.99 | 8.41 ± 6.24 | 1.34 × 10−7 |
Univariate analysis was conducted according to our previous operation.6
The individuals with the similar genetic variants of β0/β0, αα/αα, KLF1 (WT), BCL11A-rs766432 (AA or AC), and HBS1L-MYB- rs9399137 (TT or CT) in βM/βM thalassemia cohort.
The individuals with the similar genetic variants of αα/αα, KLF1 (WT), BCL11A-rs4671393 (GG or GA), HBS1L-MYB-rs4895441 (AA or AG), and HBS1L-MYB-rs9399137 (TT or TC) in HbEE cohort.
Hemoglobin levels were untransfused or pre-transfusion data.
HbF (g/L) was calculated from total Hb level and HbF (%).
Lower hemoglobin levels in the individuals with the genotype of AA were most likely to be correlated with low frequency of transfusion. The odds ratio (95% CI) of requirement for systematic transfusion was 0.181 (0.089-0.370, p = 1.16 × 10−5) between the GA group and the GG group, 0.044 (0.006-0.327, p = 0.011) between the AA group and the GG group. The odds ratio (95% CI) of TI diagnosis was 0.140 (0.083-0.236, p = 4.47 × 10−16) between the GA group and the GG group, 0.045 (0.005-0.444, p = 0.007) between the AA and the GG group.
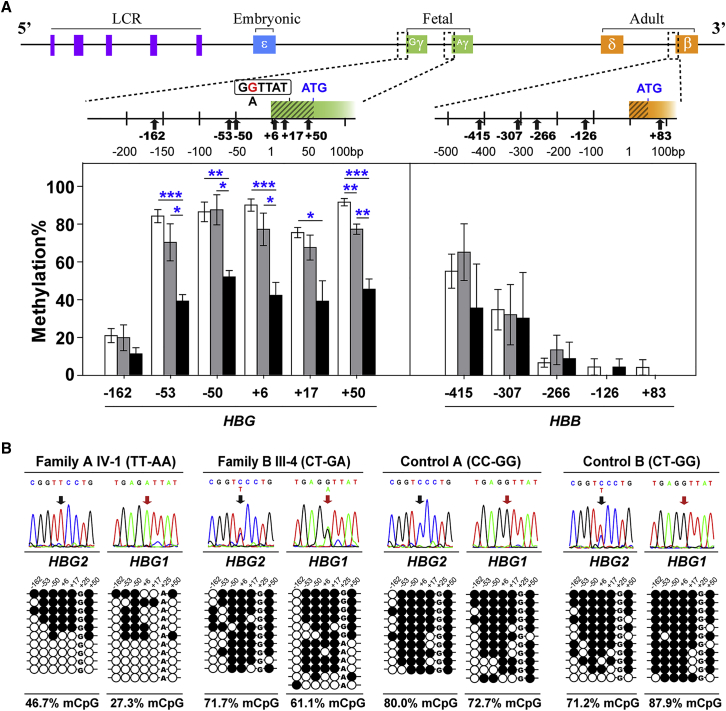
To explore whether the presence of rs368698783-A in β-thalassemia disrupts the LYAR-binding motif (GGTTAT) in the HBG1 promoter and can induce demethylation-mediated γ-globin elevation, we examined the relationship of the methylation levels of HBE1, HBG, HBD, and HBB loci in BM-derived CD235a+ erythroblasts from ten β0/β0-thalassemic subjects (GG = 4, GA = 3 and AA = 3) with respect to their genotypes (Figures 2A, S6, and S7). Participants with the AA genotype of HBG1-rs368698783 exhibited significant hypomethylation at CpG site positions −53, −50, +6, +17, and +50 in the HBG promoter regions compared with the GG genotypes (Figure 2A). Additionally, individuals with higher levels of HbF (> 90 g/L) exhibited significant hypomethylation at the HBG promoters than individuals with lower levels of HbF (< 5 g/L, Figure S7). This result demonstrated a motif-disrupting variant in HBG1 involved in the demethylation-mediated elevation of γ-globin expression.
Figure 2.
Genotype-Dependent Demethylation of HBG Promoters in β-Thalassemia
(A) The effects of the rs368698783 genotype on CpG methylation at HBG and HBB loci using CD235a+ BM-derived erythroblasts from ten β0/β0 thalassemia individuals (GG = 4, GA = 3, and AA = 3). The broken-line boxes represent the analyzed region compassing CpG methylation sites on the top diagram and their enlarged graphs depict the distribution of these sites, with indicated by black arrows. The boxed GGTTAT sequence indicates the LYAR-binding motif. The mean methylation frequencies are shown in column (white, GG; gray, GA; black, AA) with standard error indicated by bars. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 based on the T-test.
(B) The effects of two combined SNPs, HBG2-rs7482144 (C/T, black arrow), and HBG1-rs368698783 (G/A, red arrow), on HBG promoter methylation. Sequence variations of four individuals with the genotypes of TT-AA, CT-GA, CC-GG, or CT-GG at the two SNPs are shown in the middle of the sequencing chromatograph, and their relationships with HBG methylation levels obtained from BM-derived CD235a+ erythroblasts of individuals are shown at the bottom. Each row within a group represents a single bisulfite-treated clone with methylated CpGs (●) or unmethylated CpGs (○) from the G or A alleles of HBG1-rs368698783. See the Figures S6–S8 for the detailed procedures of methylation analysis.
To further examine the fine CpG methylation patterns in the proximal promoters of HBG2 and HBG1, we performed DNA methylation analysis of the two probands from two Chinese families (Figure S3 and Table S4) using CD235a+ BM-derived erythroblasts, and the AA genotype displayed different degrees of hypomethylation at the six CpG sites flanking the TSS in both HBG2 and HBG1 (−162 to +50) compared with the two distinct GG genotype controls (Figure 2B). This finding was further validated using the BS-seq method (Figure S8). These results suggested that the T allele of the HBG2-rs7482144 polymorphism alone could not reduce the levels of methylation in both HBG promoters and that the HBG1 promoter exhibited a higher level of methylation in this case. Thus, these results further indicated rs368698783-mediated hypomethylation responsible for efficient HbF elevation.
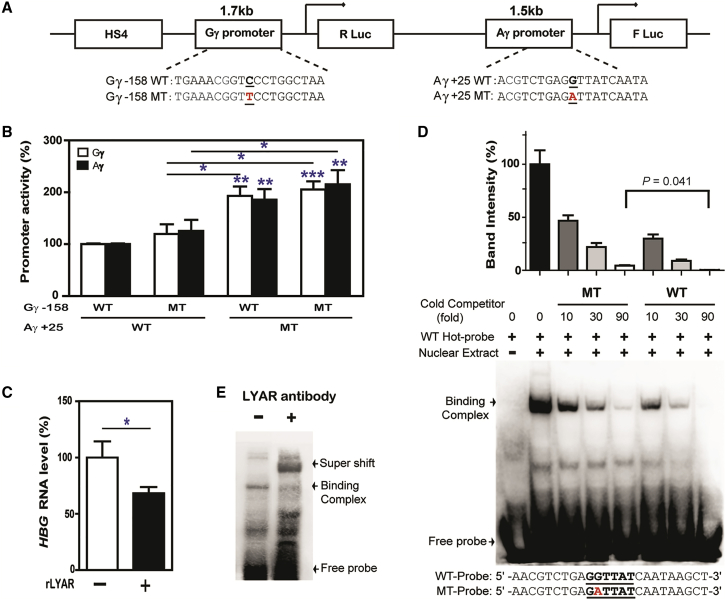
To test the de-repression of LYAR due to a motif-disrupting variant, we constructed a dual luciferase reporter gene containing DNase hypersensitive site 4 (HS4) and the full length of HBG1 and HBG2 promoters with wild-type or mutant allele at the two rSNP sites to evaluate the role of variant HBG1-rs368698783 alone and its combined effect with HBG2-rs7482144 in regulation of HBG transcriptional activity (Figure 3A). When transfected into K562 cells, the HBG1-rs368698783 mutant (Aγ+25MT) combined either with the HBG2-rs7482144 wild-type (Gγ-158WT) reporter or with the HBG2-rs7482144 mutant reporter had significantly increased promoter activity compared to the wild-type of both promoters or the HBG2-rs7482144 mutant-type alone (Figure 3B). When induced by transiently expressed LYAR, decreased endogenous HBG mRNA expression in K562 cells was also observed (Figure 3C), supporting the hypothesis that the motif-disrupting variant HBG1-rs368698783 led to the de-repression of γ-promoter activity via diminished binding activity of LYAR. Moreover, the results shown in Figure 3B indicated that both HBG promoters were obviously upregulated in the presence of this motif-disrupting variant, suggesting effect of HBG1-rs368698783A on the activation of γ-globin expression. This is consistent with the observation that the HBG2-rs7482144 polymorphism alone exhibited the lowest level of HBG expression in both RNA and protein production (Figures 1C and 1D), as well as hypomethylation can be dominated by rs368698783 rather than rs7482144 (Figure 2B).
Figure 3.
rs368698783-Mediated HBG1 Transcriptional Activation in K562 Cells
(A) Schematic representation of a Renilla luciferase (R Luc) reporter driven by the HBG2 promoter containing either the C allele (wild-type, Gγ-158WT) or the T allele (mutant, Gγ-158MT) of rs7482144 and a firefly luciferase (F Luc) reporter driven by the HBG1 promoter containing either the G allele (wild-type, Aγ+25WT) or the A allele (mutant, Aγ+25MT) of rs36869783. HS4 represents DNase hypersensitive site 4.
(B) Relative dual luciferase activities were measured using a Wallac Victor V 1420 Multilabel Counter (PerkinElmer, USA) 30 hr after K562 cells were transfected with four genotypes of the HBG2 and HBG1 promoters using the 4D-Nucleofector System (Lonza, Switzerland). The data from three independent experiments with triplications are shown in columns (white, Gγ; black, Aγ) and standard errors indicated by bars. The Aγ+25 MT promoter combined either with the Gγ-158 WT reporter or with the Gγ-158 MT reporter had significantly increased promoter activity compared to the WT of both promoters shown with two or three asterisks (∗). The promoter activity for the Aγ+25 WT promoter combined with the Gγ-158 MT reporter significant different from the Aγ+25 MT promoter was shown by asterisk above the indicated comparison. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 based on the T-test.
(C) Endogenous HBG mRNA expression in K562 cells co-transfected with (+) or without (−) the recombinant LYAR (rLYAR) cloned in the pcDNA 3.1 vector at the XhoI and BamHI sites were quantified by SYBR Premix Ex Taq (Takara) with β-actin as a reference gene. The data from three independent experiments with triplications are shown in columns and standard errors indicated by bars. ∗p < 0.05 based on the T-test.
(D) EMSA competition analysis with the indicated amounts (10-, 30-, or 90-fold molar excess) of cold wild-type or mutant competitors in K562 nuclear extract. Relative intensity of binding complex from three-independent experiments are shown in columns with standard errors indicated by bars and significant differences are marked by p value from the T-test above the indicated comparison (upper panel). DNA binding complex bands and a free-probe band are indicated by arrows and sequences of probes are shown at the bottom (lower panel).
(E) The gel super shift assay of K562 nuclear extracts with (+) or without (–) the anti-LYAR antibody. The super-shifted band is indicated.
We then used electrophoretic mobility shift assay (EMSA) with a 24-bp probe corresponding to the sequence flanking rSNP to test whether variant HBG1-rs368698783 could affect the binding activity of LYAR on the HBG promoters. We observed a significant LYAR-binding band, which could be easily competed with a cold wild-type probe but only weakly with a cold mutant probe using K562 cell nuclear extracts (Figure 3D) and LYAR expressed by TNT®Quik Coupled Transcription/Translation Systems (Figure S9), confirming that allele A in the mutant probe weakened the LYAR binding activity. Moreover, the indicated binding band was super-shifted by the addition of LYAR antibody in the gel-shift assay (Figure 3E). These results indicated that alterations in the DNA sequence containing the GGTTAT motif affect the binding of the LYAR-containing complex to the HBG promoters, which might directly influence the interactions between LYAR and epigenetic regulators that are required for γ-globin gene silencing.
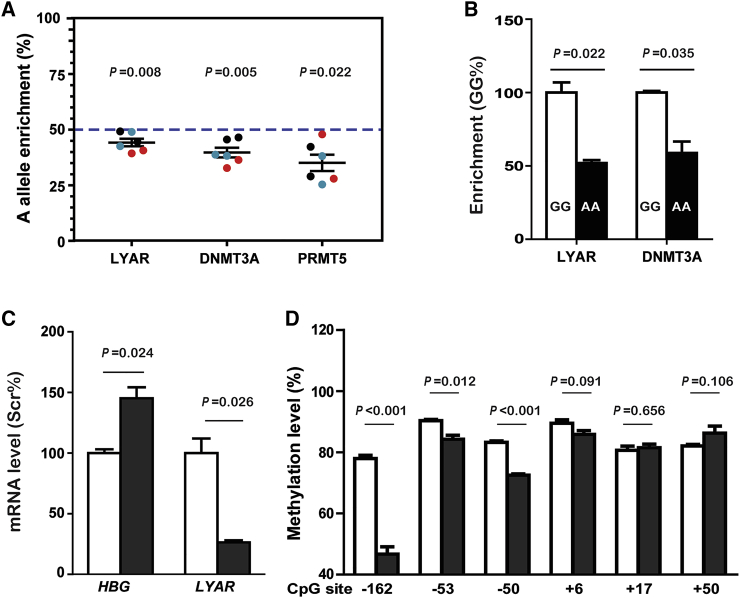
To ascertain whether epigenetic regulators displayed rs368698783 allelic-biased enrichment at the HBG promoters, we used chromatin immunoprecipitation (ChIP) assays to analyze the interactions between the reported epigenetic regulators24, 25 and specific HBG promoter regions containing rSNP using CD235a+ erythroblasts from β-thalassemia individuals. When the input DNA was normalized to contain equivalent amounts of both alleles for the individuals with the AA genotype (50% A-allele from HBG1 and 50% G-allele from HBG2, horizontal dashed line in Figure 4A), LYAR (44% A-allele, p = 0.008), DNA methyltransferases 3A (DNMT3A [MIM 602769]), 39%, p = 0.005), and protein arginine methyltransferase 5 (PRMT5 [MIM 604045]), 33%, p = 0.022) enrichments were less frequent, binding to the A- than G-alleles of rs368698783. Additionally, LYAR (p = 0.022) and DNMT3A (p = 0.035) displayed different recruitments in erythroblasts of the HBG1-rs368698783 AA individuals from the GG individuals (Figure 4B). Furthermore, we observed an elevated HBG mRNA level (Figure 4C) accompanied by the hypomethylation at CpG site positions −162, −53, and −50 in the HBG promoter regions (Figure 4D) in the LYAR knockdown erythroid progenitor cultured cells from BM of β-thalassemia individuals. These results demonstrate that the presence of the rs368698783 A-allele triggers the attenuation of LYAR and two repressive epigenetic regulators DNMT3A and PRMT5 from the HBG promoters, thereby resulting in demethylation-mediated elevation of γ-globin expression.
Figure 4.
Epigenetic Regulation of HBG Transcription in β-Thalassemia Primary Cells
(A) ChIP analysis of the CD235a+ erythroblasts of three βM/βM thalassemia individuals (red-, black-, and blue-filled circles) with the AA genotype for the A-allele in the HBG1 and the G-allele in the HBG2 at rs368698783 using EZ-ChIP kit (Upstate, USA). Three antibodies against LYAR (home-made),22 DNMT3A (Abcam, USA) or PRMT5 (Sigma-Aldrich, USA) used in this assay are indicated at the bottom. Because of highly similarity in promoter sequences between HBG1 and HBG2 in the PCR amplification region, qPCR products from ChIP-DNAs are the mixtures of the HBG1 and HBG2 promoter, and the A/G ratio of Input DNA is 1 for the AA HBG1-rs368698783 subject with 50% A-allele from HBG1 and 50% G-allele from HBG2. Abundance of the rs368698783 A-allele in the qPCR products containing a heterozygous A/G mixture from both HBG1 and HBG2 DNA was quantified by two TaqMan probes, one for G allele labeled with FAM-dye and another for A allele labeled with HEX-dye, in each reaction after the input DNA was normalized to contain equivalent amounts of both alleles (50% A-allele, horizontal dashed line). The data are shown as the mean ± SEM from two independent experiments (same color circles) with duplication. The p values compared to the input DNA were obtained from the T-test.
(B) ChIP analysis of the CD235a+ erythroblasts of two βM/βM thalassemia individuals with the GG genotype (white column) or the AA genotype (black column) of the HBG1-rs368698783. Enrichment of HBG promoter in the PCR products containing HBG1 and HBG2 from ChIP assay using anti-LYAR or anti-DNMT3A antibodies was quantified by qPCR with a TaqMan probe and a pair of primers targeting the common region of HBG1 and HBG2 promoter and GAPDH promoter as a reference gene. The data are shown as the mean ± SEM from two independent experiments with duplication. The p values between the indicated group were obtained from T-test.
(C) qPCR analysis of HBG and LYAR mRNA levels normalized to β-actin mRNA from LYAR-knockdown (LYAR-KD, black column) or negative scrambled control (Scr, white column) erythroid progenitor cells from β-thalassemia individuals. The data are shown as the mean ± SEM from two independent experiments with duplication. The p values between the indicated group were obtained from T-test.
(D) Methylation levels of HBG promoter in the LYAR-KD (black column) relative to the Scr (white column) erythroid progenitor cells from β-thalassemia individuals. The mean methylation levels of each of six CpG sites obtained by BS-seq method are shown in column with standard error indicated by bars. The p values between the indicated group were obtained from T-test. The procedure for LYAR-KD in the cultured erythroid progenitor cells was shown in Figure S8 legend.
In summary, we found a batch of HbF-associated genetic variants including haplotypes and SNPs from 1,142 Chinese β-thalassemia individuals by systematical analysis of an 80-kb region spanning the β-globin cluster based on NGS method. Then a common rSNP rs368698783, a LYAR-binding motif-disruptive SNP located in the HBG1 proximal promoter, was found to be a significant predictor of clinical severity by elevating HbF levels in β-thalassemia. Furthermore, univariate analysis using a matched case-controls and transfusion-free survival curve analysis, supported rs368698783 accounting for 41.6% of β-hemoglobinopathy individuals as an ameliorating factor for the clinical severity of β-thalassemia phenotype. Finally, we uncovered that the minor allele of the rSNP impairs the LYAR-binding activity and triggers the attenuation of repressive epigenetic regulators DNMT3A and PRMT5 from the HBG core promoter resulting in the demethylation of the promoter CpG sites and the elevation of γ-globin gene expression in erythroid progenitor cells from β-thalassemia individuals. In conclusion, this finding delineates the mechanism insights gained from the epigenetic regulation of the fetal-to-adult hemoglobin switch, which is driven by the rSNP rs368698783 in the HBG1 proximal promoter highly linked with another mechanism-unidentified well-known rSNP rs7482144 in the HBG2 proximal promoter, expands our knowledge of Hb switch regulation from theoretical and practical points of views and provides potential targets for the treatment of β-hemoglobinopathy.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (NSFC)-Guangdong Joint Fund (U1201222), the NSFC (31671314 and 31471291), the National Key Technology Research & Development Program of China (2012BAI09B01), the Doctoral Fund of Ministry of Education of China-Key Program of Priority Fields (20134433130001), and the Talents Program in Higher Education of Guangdong (2050205). We thank the individuals for their willingness to participate in this study; Drs. Feng Zhang, Hongyan Wang, Xin Xu, and Mingding Li for valuable advices and comments on this work; Yi Cheng, Qiang Zhang, Kui Hong, Qifa Liu, Juan Tang, Xiaowei Wu, Junneng Zhang, Yuan Yang, and others for assistance in collecting samples from β-thalassemia individuals and controls.
Published: June 29, 2017
Footnotes
Supplemental Data include nine figures and nine tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.05.012.
Contributor Information
Cunyou Zhao, Email: cyzhao@smu.edu.cn.
Xiangmin Xu, Email: gzxuxm@pub.guangzhou.gd.cn.
Web Resources
The URLs for data presented herein are as follows:
HUGO Gene Nomenclature Committee, http://www.genenames.org/
Mutalyzer, https://mutalyzer.nl/index
NCBI SNP, https://www.ncbi.nlm.nih.gov/snp/
OMIM, http://www.omim.org/
R statistical software, the Hmisc and rms package, http://www.r-project.org/
Sequence Variant Nomenclature, http://varnomen.hgvs.org/
Supplemental Data
References
- 1.Levings P.P., Bungert J. The human beta-globin locus control region. Eur. J. Biochem. 2002;269:1589–1599. doi: 10.1046/j.1432-1327.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 2.Higgs D.R., Engel J.D., Stamatoyannopoulos G. Thalassaemia. Lancet. 2012;379:373–383. doi: 10.1016/S0140-6736(11)60283-3. [DOI] [PubMed] [Google Scholar]
- 3.Thein S.L. Genetic association studies in β-hemoglobinopathies. Hematology (Am Soc Hematol Educ Program) 2013;2013:354–361. doi: 10.1182/asheducation-2013.1.354. [DOI] [PubMed] [Google Scholar]
- 4.Sankaran V.G., Menne T.F., Xu J., Akie T.E., Lettre G., Van Handel B., Mikkola H.K., Hirschhorn J.N., Cantor A.B., Orkin S.H. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 5.Zhou D., Liu K., Sun C.W., Pawlik K.M., Townes T.M. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat. Genet. 2010;42:742–744. doi: 10.1038/ng.637. [DOI] [PubMed] [Google Scholar]
- 6.Liu D., Zhang X., Yu L., Cai R., Ma X., Zheng C., Zhou Y., Liu Q., Wei X., Lin L. KLF1 mutations are relatively more common in a thalassemia endemic region and ameliorate the severity of β-thalassemia. Blood. 2014;124:803–811. doi: 10.1182/blood-2014-03-561779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadhouders R., Aktuna S., Thongjuea S., Aghajanirefah A., Pourfarzad F., van Ijcken W., Lenhard B., Rooks H., Best S., Menzel S. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Invest. 2014;124:1699–1710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer D.E., Kamran S.C., Lessard S., Xu J., Fujiwara Y., Lin C., Shao Z., Canver M.C., Smith E.C., Pinello L. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lettre G., Sankaran V.G., Bezerra M.A.C., Araújo A.S., Uda M., Sanna S., Cao A., Schlessinger D., Costa F.F., Hirschhorn J.N., Orkin S.H. DNA polymorphisms at the BCL11A, HBS1L-MYB, and β-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. USA. 2008;105:11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sankaran V.G., Xu J., Byron R., Greisman H.A., Fisher C., Weatherall D.J., Sabath D.E., Groudine M., Orkin S.H., Premawardhena A., Bender M.A. A functional element necessary for fetal hemoglobin silencing. N. Engl. J. Med. 2011;365:807–814. doi: 10.1056/NEJMoa1103070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell J.J., Sherva R.M., Chen Z.Y., Luo H.Y., Chu B.F., Ha S.Y., Li C.K., Lee A.C., Li R.C., Li C.K. A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood. 2011;117:4935–4945. doi: 10.1182/blood-2010-11-317081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilman J.G., Huisman T.H. DNA sequence variation associated with elevated fetal G gamma globin production. Blood. 1985;66:783–787. [PubMed] [Google Scholar]
- 13.Mettananda S., Gibbons R.J., Higgs D.R. α-Globin as a molecular target in the treatment of β-thalassemia. Blood. 2015;125:3694–3701. doi: 10.1182/blood-2015-03-633594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bianchi N., Cosenza L.C., Lampronti I., Finotti A., Breveglieri G., Zuccato C., Fabbri E., Marzaro G., Chilin A., De Angelis G. Structural and Functional Insights on an Uncharacterized Aγ-Globin-Gene Polymorphism Present in Four β0-Thalassemia Families with High Fetal Hemoglobin Levels. Mol. Diagn. Ther. 2016;20:161–173. doi: 10.1007/s40291-016-0187-2. [DOI] [PubMed] [Google Scholar]
- 15.Sankaran V.G., Weiss M.J. Anemia: progress in molecular mechanisms and therapies. Nat. Med. 2015;21:221–230. doi: 10.1038/nm.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulmovits B.M., Appiah-Kubi A.O., Papoin J., Hale J., He M., Al-Abed Y., Didier S., Gould M., Husain-Krautter S., Singh S.A. Pomalidomide reverses γ-globin silencing through the transcriptional reprogramming of adult hematopoietic progenitors. Blood. 2016;127:1481–1492. doi: 10.1182/blood-2015-09-667923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amaya M., Desai M., Gnanapragasam M.N., Wang S.Z., Zu Zhu S., Williams D.C., Jr., Ginder G.D. Mi2β-mediated silencing of the fetal γ-globin gene in adult erythroid cells. Blood. 2013;121:3493–3501. doi: 10.1182/blood-2012-11-466227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mabaera R., Richardson C.A., Johnson K., Hsu M., Fiering S., Lowrey C.H. Developmental- and differentiation-specific patterns of human γ- and β-globin promoter DNA methylation. Blood. 2007;110:1343–1352. doi: 10.1182/blood-2007-01-068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lessard S., Beaudoin M., Benkirane K., Lettre G. Comparison of DNA methylation profiles in human fetal and adult red blood cell progenitors. Genome Med. 2015;7:1. doi: 10.1186/s13073-014-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forster L., McCooke J., Bellgard M., Joske D., Finlayson J., Ghassemifar R. Differential gene expression analysis in early and late erythroid progenitor cells in β-thalassaemia. Br. J. Haematol. 2015;170:257–267. doi: 10.1111/bjh.13432. [DOI] [PubMed] [Google Scholar]
- 21.Masuda T., Wang X., Maeda M., Canver M.C., Sher F., Funnell A.P., Fisher C., Suciu M., Martyn G.E., Norton L.J. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016;351:285–289. doi: 10.1126/science.aad3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju J., Wang Y., Liu R., Zhang Y., Xu Z., Wang Y., Wu Y., Liu M., Cerruti L., Zou F. Human fetal globin gene expression is regulated by LYAR. Nucleic Acids Res. 2014;42:9740–9752. doi: 10.1093/nar/gku718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kersey P.J., Allen J.E., Christensen M., Davis P., Falin L.J., Grabmueller C., Hughes D.S., Humphrey J., Kerhornou A., Khobova J. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 2014;42:D546–D552. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Q., Rank G., Tan Y.T., Li H., Moritz R.L., Simpson R.J., Cerruti L., Curtis D.J., Patel D.J., Allis C.D. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rank G., Cerruti L., Simpson R.J., Moritz R.L., Jane S.M., Zhao Q. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood. 2010;116:1585–1592. doi: 10.1182/blood-2009-10-251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.