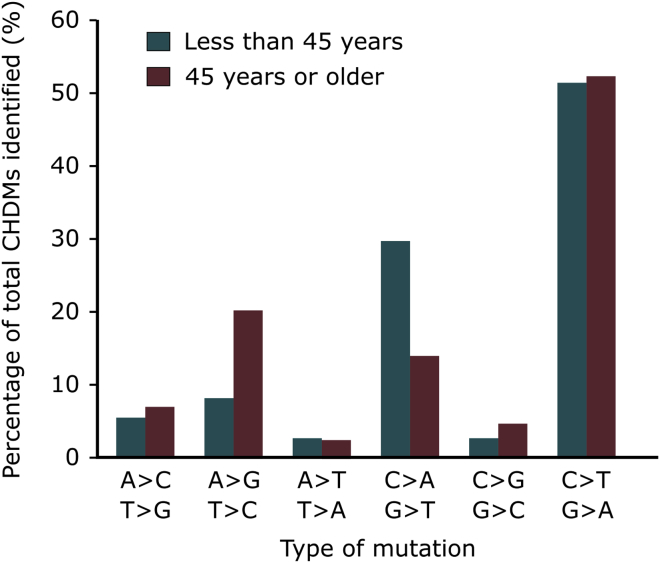Abstract
Clonal hematopoiesis results from somatic mutations in hematopoietic stem cells, which give an advantage to mutant cells, driving their clonal expansion and potentially leading to leukemia. The acquisition of clonal hematopoiesis-driver mutations (CHDMs) occurs with normal aging and these mutations have been detected in more than 10% of individuals ≥65 years. We aimed to examine the prevalence and characteristics of CHDMs throughout adult life. We developed a targeted re-sequencing assay combining high-throughput with ultra-high sensitivity based on single-molecule molecular inversion probes (smMIPs). Using smMIPs, we screened more than 100 loci for CHDMs in more than 2,000 blood DNA samples from population controls between 20 and 69 years of age. Loci screened included 40 regions known to drive clonal hematopoiesis when mutated and 64 novel candidate loci. We identified 224 somatic mutations throughout our cohort, of which 216 were coding mutations in known driver genes (DNMT3A, JAK2, GNAS, TET2, and ASXL1), including 196 point mutations and 20 indels. Our assay’s improved sensitivity allowed us to detect mutations with variant allele frequencies as low as 0.001. CHDMs were identified in more than 20% of individuals 60 to 69 years of age and in 3% of individuals 20 to 29 years of age, approximately double the previously reported prevalence despite screening a limited set of loci. Our findings support the occurrence of clonal hematopoiesis-associated mutations as a widespread mechanism linked with aging, suggesting that mosaicism as a result of clonal evolution of cells harboring somatic mutations is a universal mechanism occurring at all ages in healthy humans.
Keywords: age-related clonal hematopoiesis, somatic mutations, mosaicism, DNMT3A, ultra-sensitive sequencing, smMIPs
Introduction
Low-level mosaicism resulting from somatic mutations is frequent in healthy tissues,1 particularly in those with high turnover rates such as blood2, 3, 4, 5, 6 and skin.7, 8 Novel mutations may arise due to failure to repair DNA replication errors9 or secondary to DNA damage caused by exposure to endogenous and exogenous mutagens.10 While most somatic mutations are phenotypically silent in the cell in which they arise, some of them can lead to changes in cell behavior. For example, mutations abolishing the function of a gene can be detrimental or even lethal for the cell in which they arise. In contrast, a subset of mutations have the ability to promote cell proliferation and/or survival, granting mutant cells a growth advantage compared to wild-type ones.11, 12 This fitness advantage can allow a single mutant cell to grow into groups of identical daughter cells, which is known as “clonal expansion.”13, 14 Mutations driving clonal expansion can arise in all cell types including somatic stem cells, which are characterized by their longevity and continuous division. These two characteristics would allow somatic stem cells to undergo recurring cycles of acquisition of mutations and subsequent clonal expansion, leading to the accumulation and propagation of mutations over time.
A number of recurrent somatic mutations have been implicated in “clonal hematopoiesis,” a process in which a mutant hematopoietic stem cell (HSC) expands clonally and contributes to a significant and detectable fraction of circulating blood cells.2, 3, 4, 5 Somatic mutations involved in clonal hematopoiesis are often detected in blood-derived DNA at a variant allelic frequency (VAF) ranging from 0.008 to 0.1, suggesting that between 1.6% and 20% of nucleated cells circulating in blood are derived from mutant HSCs.2, 3, 4, 5 Clonal hematopoiesis driver mutations (CHDMs) most often disrupt genes such as DNMT3A (MIM: 602769), TET2 (MIM: 612839), and ASXL1 (MIM: 612990), which are associated with blood disorders like myelodysplasia and leukemia.2, 3, 4 Because of this link, the acquisition of CHDMs has been suggested to represent the earliest phase in the development of hematologic malignancies.15 Indeed, clonal hematopoiesis of indeterminate potential, defined by the presence of CHDMs with a VAF ≥ 0.02 in individuals without overt hematologic disease, is currently considered a pre-cancerous state carrying a risk of converting to leukemia of 0.5% to 1% per year.3, 16
Clonal hematopoiesis is thought to be rare in individuals younger than 50 years and increases in frequency with age, affecting at least 10% of individuals older than 65 years2, 4 and close to 20% of persons above 90 years.5 Some mutations involved in clonal hematopoiesis, such as JAK2 (MIM: 147796) or DNMT3A mutations, have been detected in blood of healthy adults of all ages and are thought to be able to cause clonal expansion of mutant HSCs throughout life. On the other hand, a subset of recurrent mutations has been observed only in individuals over the age of 70 years, such as mutations in SRSF2 (MIM: 600813) or SF3B1 (MIM: 605590), suggesting that clonal expansion of HSCs harboring these mutations is age dependent.5 This observation has led to the hypothesis that the aging cellular background may play a crucial role in the selection and expansion of mutant HSCs.5 Indeed, aging is accompanied by a decline in HSC function,17 a bias toward myeloid differentiation,18 and changes in the bone marrow niche.19 It is therefore possible that certain mutations provide a cellular advantage in the aging bone marrow environment, allowing for clonal expansion of mutant HSCs exclusively in this context.5, 20, 21 However, it is also possible that CHDMs associated with aging arise in young individuals but remain undetected due to technical limitations; because of the low VAF at which CHDMs are often present, the detection method used heavily influences the ability to identify these mutations.22 For instance, studies favoring a targeted approach to provide deep sequencing coverage in known hotspot regions have led to the identification of a number of CHDMs with low VAF which would have otherwise been missed by exome or genome sequencing.5, 6
In the present study, we aim to characterize the genetic profile and features of clonal hematopoiesis in individuals below the age of 70 years. To identify CHDMs with a VAF ≥ 0.002 in a cohort of more than 2,000 population control subjects between 20 and 69 years of age, we use single-molecule molecular inversion probes (smMIPs).23, 24, 25, 26 As a novel and highly flexible method for targeted enrichment of genomic regions of interest, we have made use of smMIPs to screen our cohort for somatic mutations in 40 established loci for CHDMs.
Furthermore, and unexpectedly, reference population databases for genetic variation have been found to contain pathogenic variants established to cause developmental disorders when present in the germline. A possible explanation for this surprising observation is that these mutations represent somatic mutations with elevated VAFs in blood due to their role as CHDMs.27 However, the extent of the genetic overlap between somatic mutations in clonal hematopoiesis and germline mutations in developmental disorders remains unclear. Therefore, we screened our cohort for somatic mutations in an additional set of 64 loci in which recurrent germline de novo mutations have been found to cause severe developmental disorders. Several of these loci have been previously implicated in paternal age effect disorders28 with the causative mutations shown to cause clonal expansion in spermatogonial stem cells.29, 30, 31, 32 We here aim to determine whether these loci may represent novel sites for CHDMs.
Material and Methods
Samples
This study was performed using data and biomaterial from the Nijmegen Biomedical Study (NBS). The NBS is a population-based study of 9,350 individuals, based on an age- and sex-stratified random sample from the register of the municipality of Nijmegen, a city in the eastern part of the Netherlands. Extensive questionnaire data on health and lifestyle were collected. Blood samples were collected in EDTA tubes and DNA was extracted by salt precipitation method.33 For this study, we obtained DNA samples and information on age and sex for 2,014 NBS participants via the Radboud Biobank.33 Approximately 400 samples equally distributed between men and women were obtained for each age group (400 for age group 20–29, 405 for age group 30–39, 404 for age group 40–49, 403 for age group 50–59, and 402 for age group 60–69 years of age; see Table S1). This study was approved by the Committee on Research involving Human Subjects (Commissie Mensgebonden Onderzoek) of the Radboudumc (CMO approval: 2015-2228) and informed consent was obtained from all participants. Due to restrictions in the research permit obtained to carry out this study, requiring preservation of anonymity within our cohort, only information concerning the sex and age of the individual at the moment when the blood sample was taken could be accessed. The quality of purified DNA was tested and each sample was normalized to 25 ng/μL by optical density measurement (Dropsense, Trinean).
Targeted Loci to Screen for Mutations
We performed a literature review to identify mutations observed recurrently in age-related clonal hematopoiesis. By combining the results from several published studies,2, 3, 4, 5 we collected 1,158 coding substitutions in 513 amino acid residues which were ranked by total number of substitutions identified per residue. We selected 35 loci with the largest number of coding mutations observed in clonal hematopoiesis, which corresponds to a total of 599 SNVs in 87 residues. Furthermore, five loci in which CHDMs have been previously identified and in which overlapping germline or postzygotic de novo mutations are known to cause developmental disorders were included.34, 35, 36, 37 In addition, we selected seven loci with nine residues in which substitutions are known to be causative for paternal age effect disorders and to lead to spermatogonial stem cell expansion.28 Finally, we included 57 additional loci in which recurrent identical de novo mutations have been found in developmental disorders.38, 39, 40 These loci are therefore candidates either for elevated mutation rates at that genomic site or for mutations leading to expansion of spermatogonial stem cells. This analysis resulted in 104 loci in total (Table S2).
smMIP Design
To screen for mutations in these 104 loci, MIPGEN software41 was used to design probes covering the regions of interest, followed by manual curation and selection. The smMIPs were 80 nucleotide-long DNA molecules consisting of an extension and ligation arm with a combined length of 40 nucleotides, separated by a linker sequence of 30 nucleotides (see Figure S1 for more details). All smMIPs contained a unique molecule identifier (UMI) or molecular tag consisting of 2 × 5 random nucleotides to identify each individual captured DNA molecule. These smMIPs were designed to target regions of 54 nucleotides. At least one smMIP on each of the sense and the antisense DNA strands were designed per locus. Probes targeting genomic regions containing a SNP with a population frequency above 1% were designed to have complementary arms targeting both alleles. In total, 231 smMIPs were designed to capture the 104 regions of interest. The smMIP oligonucleotides were produced by Integrated DNA Technologies (IDT) at 25 nmol scale and normalized to a concentration of 100 μM.
smMIPs Assay Setup
The smMIPs assay was set up with minor modifications to previously published protocols.23, 42 In brief, individual smMIPs were pooled equimolarly and phosphorylated using T4 polynucleotide kinase and 10× T4 DNA ligase buffer supplemented with 10 mM ATP (New England Biolabs). The smMIP capture was performed on 8 μL of input DNA (200 ng) supplied with 17 μL of capture mix containing 0.28 μL of a phosphorylated smMIP pool dilution at 3.12 nM, resulting in a ratio of 8,000 smMIP molecules per DNA molecule. The capture reaction was incubated for 18–22 hr at 60°C, after which the mix was cooled and treated with exonuclease. Each exo-treated sample was split in two technical replicates of 10 μL, which were then amplified and barcoded separately by PCR. The PCR products were run on gel, pooled, purified using AMPureXP Beads (Agencourt), and run on a Tapestation (Agilent) to verify the integrity of the sequencing library. Sequencing was performed on an Illumina NextSeq500 platform with 2 × 79-bp paired-end reads (i.e., each DNA insert being sequenced in both directions). A pilot smMIP experiment was performed on control DNA for the optimization of the smMIP library. The performance of each individual probe was evaluated by examining the sequencing coverage per probe, in order to identify under-performing and over-performing smMIPs. After excluding smMIPs with off-target capture, a new and rebalanced smMIP pool was prepared adjusting the volumes for each probe. The new pool was phosphorylated and an experiment was run on control DNA samples to verify pool rebalancing. The library was subsequently prepared and sequenced as described previously using blood DNA samples from the cohort. After sequencing, 18 smMIPs were shown to have an overall median coverage <20× and were excluded from further analysis. Seven samples for which both replicates had an average sequencing coverage below 100× were excluded due to poor quality or quantity of the input DNA. For additional information on sequencing coverage per smMIP and per age group, refer to Table S3.
Analysis
FASTQ files were obtained from the bcl files and demultiplexed using the sample barcode. Sequenced reads were mapped with BWA MEM, using a modified version of an in-house bioinformatics pipeline which allows trimming of the MIP extension and ligation arms (MIPVAR). To analyze unique DNA molecules, we detected and removed PCR duplicates of individual captured molecules per sample and per smMIP by identifying reads with the same UMI. From the regions selected for screening, 88 out of 104 loci had one or two mutated residues. For these loci, we analyzed the genomic region corresponding to the mutated residue(s) ±6 base pairs. Regions with more than two mutated residues (13 out of 104 loci) were analyzed so that we would examine the entire region encompassing all mutated residues within the locus ±6 bp. For TP53, we analyzed the entire region captured by the smMIPs (3 out of 104 loci). For a list of all 2,364 positions analyzed, refer to Table S4. Pileups were generated for all samples for these positions using SAMtools with the following filter settings: sequence quality ≥25 and a mapping quality ≥15.5 The coverage, each nucleotide change, and the presence of insertions and deletions were counted at each position for each sample replicate. Within each sample replicate, positions with a unique molecule sequencing coverage below 200× were excluded. The sequencing error was calculated for each position and nucleotide change (A, C, G, T, insertion, and deletion) based on all samples from the cohort. Additionally, the run-specific sequencing error for each position and nucleotide change was determined using only samples within the same sequencing run. Subsequently, we used a Poisson distribution to calculate a p value reflecting the probability to obtain a number equal or higher to the observed number of mutation reads per position for all sample replicates based on the sequencing error. This calculation was performed in parallel using each determined sequencing error in two independent analyses. For each sample, we extracted the p values for all nucleotide changes for all positions from only one of the replicates and performed Benjamini-Hochberg multiple test correction on these values. All nucleotide changes with an adjusted p value <0.05 and with ≥2 reads for SNVs and ≥5 reads for indels were included as “statistically significant nucleotide changes.” We then extracted from the second sample replicate the p values obtained for the statistically significant nucleotide changes in the first replicate and performed Benjamini-Hochberg multiple test correction. We used the same filtering criteria and obtained a list of potential mutations. Finally, the list of potential mutations obtained with the overall and the run-specific sequencing error were overlapped. We included only mutations in which both replicates have ≥2 reads for SNVs and ≥5 reads for indels, representing a statistically significantly higher number of mutations counts than expected based both on the overall and the run-specific sequencing error. For positions within a mutational hotspot in which more than five substitutions were identified (such as JAK2 p.Val617Phe and all DNMT3A hotspots), a separate analysis examining only nucleotide changes at those hotspots was performed. To exclude germline events, somatic mutations were defined as mutations with a VAF ≤ 0.35 and an allele frequency < 0.001 in ExAC.
Validation of DNMT3A p.Arg882Cys Mutations with Restriction Digestion
Recurrent mutations in DNMT3A leading to DNMT3A p.Arg882Cys and p.Arg882His were found to eliminate a recognition site for the restriction enzyme TauI.43 Substitutions in this residue were identified in 13 samples at different variant allele frequencies. These mutations were validated by PCR amplification (forward primer 5′-GAACTAAGCAGGCGTCAGAGGA-3′, reverse primer 5′-AAAAAGGGAAGGGGGAGGAAGG-3′) of a region of 577 bp surrounding the region of interest in DNMT3A, followed by restriction digestion. Amplicons of the wild-type sequence are digested by TauI in two fragments of 369 and 208 bp, while amplicons of either mutant allele fail to be recognized by the enzyme and remain undigested. Size analysis of the digested products was performed on a TapeStation. A subset of the digested products was selected for subsequent sequencing on an Ion Torrent platform.
Results
Sensitive and Specific Detection of Somatic Mutations in Blood by smMIPs
We sequenced 104 loci in 2,007 blood samples of population controls between 20 and 69 years of age with a median unique coverage of 845-fold per sample (see Figures S1 and S2). The median unique coverage corresponds to the number of unique DNA molecules sequenced per sample and per position after removal of PCR duplicates. Using an approach based on modeling sequencing error rates per targeted position, we identified 224 somatic SNVs and indels, of which 223 localize to the coding regions screened, with VAFs ranging between 0.0008 and 0.35 (average 0.015, median 0.0061). The median unique coverage of these mutation loci was 4103×.
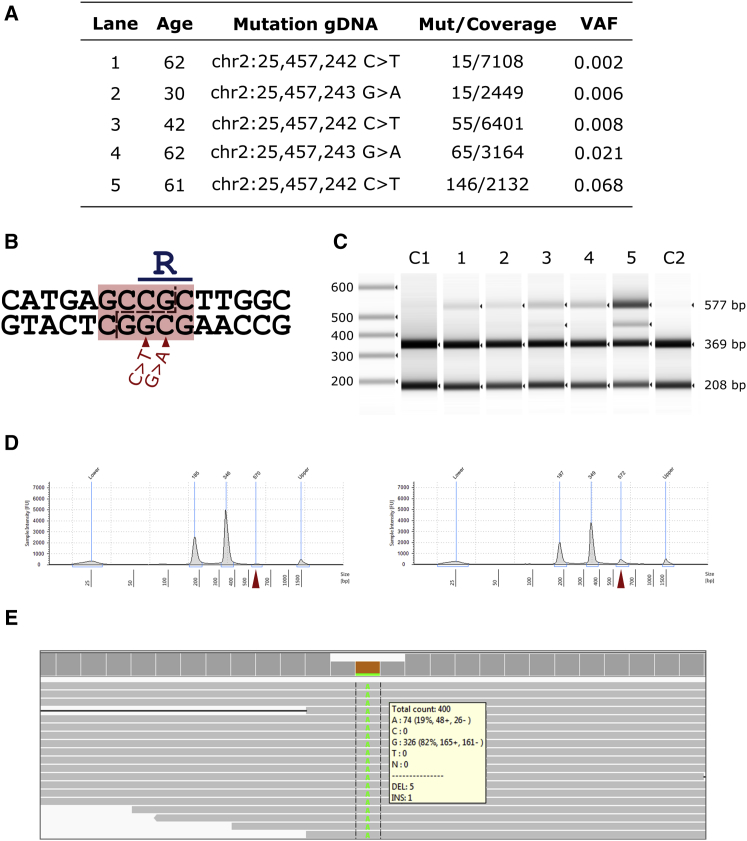
To validate the specificity of our method, we used restriction digestion to analyze five somatic mutations localizing to a hotspot of DNMT3A, in which mutations disrupt a restriction digestion site for TauI (see Figures 1A and 1B). For all five samples, a band of undigested DNA was observed at 577 bp proportional in size to the mutation VAF per sample (Figures 1C and 1D). We selected one sample and one independent control for sequencing after restriction digestion and identified an enrichment for reads corresponding to the DNMT3A mutation compared to the wild-type allele for the sample in which a mutation was detected (Figure 1E). These findings support that mutations identified with VAFs as low as 0.002 represent true mutations in the original DNA sample rather than false positives.
Figure 1.
Validation of Mutations by Restriction Digestion and Re-sequencing
(A) Mutations identified in DNMT3A Arg882 selected for additional validation by non-sequencing-based method.
(B) Scheme showing recognition site for restriction digestion enzyme TauI in the genomic sequence corresponding to DNMT3A Arg882. Mutations chr2:25457242C>T and chr2:25457243G>A (hg19) leading to p.Arg882His and p.Arg882Cys, respectively, are marked below with red arrows.
(C) Size analysis of restriction digestion of DNMT3A PCR products. Lanes 1 to 5 represent samples with mutations with different VAFs. C1 is a control with false positive signal for a G>A mutation at chr2:25457243, as determined statistically. C2 is a control with no DNMT3A mutation.
(D) Gel trace of size analysis of digestion, with sample 4 on the left and C2 on the right. The peak corresponding to the full-size product is marked with a red triangle for both samples. Note that C2 present a small peak at 577 bp, corresponding to undigested PCR products due to digestion enzyme saturation.
(E) Sequencing results of digested PCR product for sample 5. We obtain a higher ratio of mutation to wild-type reads than in the original sample due to digestion of the wild-type product.
Somatic Mutations in Known Clonal Hematopoiesis Driver Genes in Blood of Population Controls
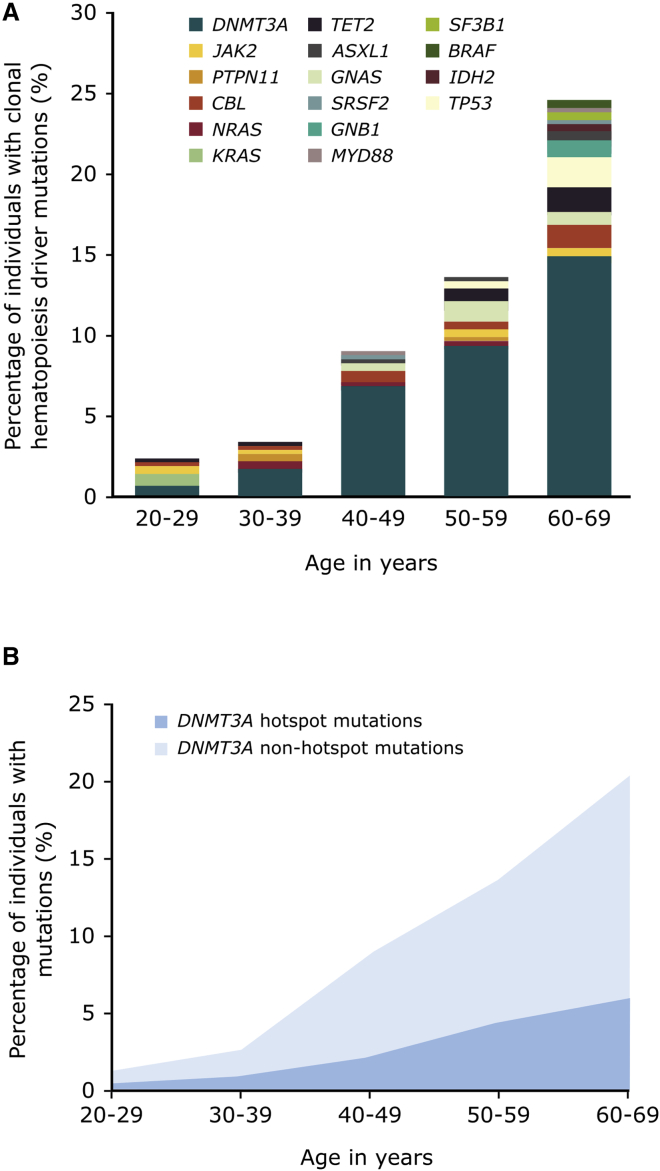
In total, 216 somatic mutations were identified in coding regions of known clonal hematopoiesis driver genes in our cohort (see Figure 2A). Among these, 170 were mutations previously identified in clonal hematopoiesis.2, 3, 4, 5 For instance, several known clonal hematopoiesis driver missense substitutions in genes such as DNMT3A, JAK2, GNAS (MIM: 139320), NRAS (MIM: 164790), SRSF2, and SF3B1 were detected in our cohort (see Table 1). Additionally, nonsense substitutions were identified in genes previously identified to harbor truncating mutations in clonal hematopoiesis such as ASXL1, DNMT3A, TET2, and TP53 (MIM: 191170) and a total of 20 indels involving DNMT3A and TET2 were detected (see Table 2). The most frequently mutated gene in our cohort is DNMT3A, for which a wide variety of mutations were observed including hotspot and non-hotspot missense substitutions, loss-of-function point mutations, and indels (see Tables 1, 2, and S5; Figure 2B). Hotspots in DNMT3A are here defined as residues in which five or more missense substitutions were detected in our cohort, including DNMT3A Arg326, Arg729, Tyr735, Arg736, Trp860, and Arg882.
Figure 2.
Prevalence of Clonal Hematopoiesis-Driver Mutations
(A) Prevalence and distribution of clonal hematopoiesis-driver mutations identified in healthy individuals aged between 20 and 69 years of age.
(B) Prevalence of mutations in DNMT3A per age group. Hotspot in DNTM3A are defined as residues in which five or more mutations were identified in our cohort and include Arg326, Arg729, Tyr735, Arg736, Trp860, and Arg882. All other missense, loss-of-function, and indels are included in the non-hotspot mutations.
Table 1.
Summary of Known Clonal Hematopoiesis Driver SNVs Identified
| Gene Name | Number of Individuals with Known SNVs | Age Range | Average Age | Number of Known SNVs Identified | Type of SNVs Identified |
|---|---|---|---|---|---|
| ASXL1 | 4 | 42–66 | 55.8 | 4 | stop |
| CBL | 5 | 24–63 | 47.4 | 5 | missense |
| DNMT3A | 93a | 25–69 | 54.9 | 97 | missense, stop, splice site |
| GNAS | 8 | 45–63 | 55.9 | 8 | missense |
| GNB1 | 4 | 60–69 | 64.3 | 4 | missense |
| IDH2 | 2 | 62 | 62 | 2 | missense |
| JAK2 | 7 | 22–65 | 45.1 | 7 | missense |
| MYD88 | 2 | 45–68 | 56.5 | 2 | missense |
| NRAS | 2 | 47–56 | 51.5 | 2 | missense |
| PTPN11 | 1 | 39 | 39 | 1 | missense |
| SF3B1 | 2 | 60–68 | 64 | 2 | missense |
| SRSF2 | 2 | 49–65 | 57 | 2 | missense |
| TET2 | 9 | 38–67 | 57.7 | 9 | stop |
| TP53 | 4 | 53–67 | 61.3 | 4 | missense, stop |
| U2AF1 | 1 | 60 | 60 | 1 | missense |
For a complete table containing mutations at genomic and cDNA level, GenBank accession numbers, and VAFs, please refer to Table S5.
93 individuals were found to carry 97 SNVs in DNMT3A, including 90 individuals with one SNV, two individuals with two SNVs, and one individual with three SNVs in DNMT3A.
Table 2.
Indels Identified in Known Clonal Hematopoiesis Driver Genes
| Age | Gene | VAF | Mutation (hg19 gDNA) | mRNA Changes | Protein Change |
|---|---|---|---|---|---|
| 33 | DNMT3A | 0.0167 | chr2:25458597del | c.2576del | p.Leu859Yfs |
| 42 | DNMT3A | 0.0405 | chr2:25463300dup | c.2193dup | p.Phe732fs |
| 48 | DNMT3A | 0.0476 | chr2:25458620_25458627dup | c.2546_2553dup | p.Met852fs |
| 49 | DNMT3A | 0.0256 | chr2:25463297dup | c.2196dup | p.Glu733∗ |
| 49 | DNMT3A | 0.0032 | chr2:25463196_25463197del | c.2297_2296del | p.Lys766fs |
| 57 | DNMT3A | 0.0045 | chr2:25458596dup | c.2577dup | p.Trp860fs |
| 57 | DNMT3A | 0.0070 | chr2:25458591delinsTT | c.2582delinsAA | p.Cys861∗ |
| 58 | DNMT3A | 0.1360 | chr2:25463196_25463197del | c.2297_2296del | p.Lys766fs |
| 60 | DNMT3A | 0.0586 | chr2:25463308del | c.2185del | p.Arg729fs |
| 61 | DNMT3A | 0.0717 | chr2:25463196_25463197del | c.2297_2296del | p.Lys766fs |
| 63 | DNMT3A | 0.0205 | chr2:25463312del | c.2181del | p.Gly728fs |
| 64 | DNMT3A | 0.0033 | chr2:25463196_25463197del | c.2297_2296del | p.Lys766fs |
| 65m | DNMT3A | 0.0063 | chr2:25463297dup | c.2196dup | p.Glu733∗ |
| 66q | DNMT3A | 0.0030 | chr2:25458605_25458606del | c.2568_2567del | p.Glu856fs |
| 66q | DNMT3A | 0.0090 | chr2:25463291del | c.2202del | p.Tyr735fs |
| 67 | DNMT3A | 0.0091 | chr2:25458608del | c.2565del | p.Glu856fs |
| 68r | DNMT3A | 0.0016 | chr2:25463190del | c.2303del | p.Asp768fs |
| 68 | DNMT3A | 0.0076 | chr2:25463196_25463197del | c.2297_2296del | p.Lys766fs |
| 68 | DNMT3A | 0.0012 | chr2:25458595del | c.2578del | p.Trp860fs |
| 64 | TET2 | 0.0044 | chr4:106157389dup | c.2290dup | p.Gln764fs |
Samples with more than one mutation are marked with a superscript letter for identification. For a complete table containing GenBank accession numbers, please refer to Table S5.
Furthermore, somatic mutations were identified in known clonal hematopoiesis driver genes which had not been previously reported as CHDMs (see Table 3). These consist of 46 SNVs in coding regions of BRAF (MIM: 164757), BRCC3 (MIM: 300617), CBL (MIM: 165360), DNMT3A, GNAS, KRAS (MIM: 190070), NRAS, PIK3CA (MIM: 171834), PTPN11 (MIM: 176876), TET2, and TP53. Several of these novel mutations were found adjacent to residues previously identified to harbor CHDMs. For instance, three individuals in our cohort were found to have KRAS p.Gly13Asp substitutions. These missense substitutions occur contiguous to Gly12, a recurrently mutated residue in which p.Gly12Cys, p.Gly12Arg, and p.Gly12Ser substitutions have been identified in clonal hematopoiesis.3, 5 Additionally, we detected mutations within specific genes with a different mechanism than those usually observed to drive clonal hematopoiesis. For instance, while loss-of-function somatic TET2 mutations are frequent in clonal hematopoiesis and leukemia, we detected one missense TET2 substitution in a 20-year-old. Comparing the VAF of the 170 known and the 46 novel mutations we identified in genes involved in clonal hematopoiesis reveals a statistically significant difference in the VAF of both groups of mutations (0.0069 versus 0.0038 for known versus novel, p = 0.0003, Wilcoxon rank sum test). This difference between both groups suggests that known mutations identified have a stronger clonal advantage than the novel mutations identified in our cohort, which may explain why they had not been detected previously.
Table 3.
Novel Somatic Mutations Identified in Coding Regions of Clonal Hematopoiesis Driver Genes
| Age | Gene Name | VAF | Predicted Protein Substitution | Mutation Type | PhyloP | CADD PHRED | Grantham Score |
|---|---|---|---|---|---|---|---|
| 67 | BRAF | 0.0009 | p.Phe595Leu | missense | 2.26 | 24.7 | 22 |
| 67 | BRAF | 0.0098 | p.Lys601Asn | missense | 1.12 | 21.3 | 94 |
| 54 | BRCC3 | 0.0294 | p.Asp88Gly | missense | 8.54 | 17.8 | 94 |
| 35 | CBL | 0.0017 | p.Pro395His | missense | 7.45 | 19.7 | 77 |
| 41 | CBL | 0.0158 | p.Cys384Tyr | missense | 9.42 | 18.4 | 194 |
| 52 | CBL | 0.0081 | p.His398Gln | missense | 0.95 | 14.8 | 24 |
| 58 | CBL | 0.0032 | p.Cys419Ser | missense | 9.48 | 21.4 | 112 |
| 65m | CBL | 0.0027 | p.His398Gln | missense | 0.95 | 14.8 | 24 |
| 65m | CBL | 0.0175 | p.Thr377Ile | missense | 7.45 | 18.1 | 89 |
| 68r | CBL | 0.0015 | p.Cys396Tyr | missense | 9.42 | 18.9 | 194 |
| 69 | CBL | 0.0097 | p.Ser376Pro | missense | 7.66 | 18.2 | 74 |
| 41b | DNMT3A | 0.0053 | p.Val328Phe | missense | 7.57 | 27.8 | 50 |
| 48 | DNMT3A | 0.0033 | p.Asp768Glu | missense | 5.68 | 24.7 | 45 |
| 49 | DNMT3A | 0.0021 | p.Met864Arg | missense | 9.23 | 24.9 | 91 |
| 49 | DNMT3A | 0.0036 | p.Thr862Ile | missense | 9.75 | 32 | 89 |
| 49 | DNMT3A | 0.0043 | p.Leu888Pro | missense | 9.34 | 18.7 | 98 |
| 51 | DNMT3A | 0.0011 | p.Val763Gly | missense | 7.33 | 25.5 | 109 |
| 51 | DNMT3A | 0.0090 | p.Asp765Gly | missense | 8.04 | 24.9 | 94 |
| 52 | DNMT3A | 0.0028 | p.Ala884Thr | missense | 6.09 | 28.7 | 58 |
| 54 | DNMT3A | 0.0028 | p.Arg885Ser | missense | 1.59 | 18.1 | 110 |
| 54 | DNMT3A | 0.0042 | p.Glu774Glu | synonymous | 7.78 | 15.6 | NA |
| 55 | DNMT3A | 0.0110 | p.Lys766Asn | missense | 2.56 | 19.6 | 94 |
| 57 | DNMT3A | 0.0127 | p.Ala884Val | missense | 9.86 | 28 | 64 |
| 59 | DNMT3A | 0.0016 | p.Arg866Met | missense | 7.73 | 35 | 91 |
| 61 | DNMT3A | 0.0052 | p.Thr862Ile | missense | 9.75 | 32 | 89 |
| 64 | DNMT3A | 0.0132 | p.Asp857Ala | missense | 7.95 | 28.4 | 126 |
| 64k | DNMT3A | 0.0026 | p.Glu863Gly | missense | 7.95 | 32 | 98 |
| 68v | DNMT3A | 0.0080 | p.Thr862Ile | missense | 9.75 | 32 | 89 |
| 68 | DNMT3A | 0.0035 | p.Leu889Val | missense | 7.99 | 19.8 | 32 |
| 68 | DNMT3A | 0.0038 | p.Asp768Glu | missense | 5.68 | 24.7 | 45 |
| 46 | GNAS | 0.0046 | p.Cys843Arg | missense | 7.55 | 21.2 | 180 |
| 62 | GNAS | 0.0042 | p.Leu846Pro | missense | 7.55 | 21.2 | 98 |
| 22 | KRAS | 0.0107 | p.Gly13Asp | missense | 7.74 | 27.8 | 94 |
| 25 | KRAS | 0.0037 | p.Gly13Asp | missense | 7.74 | 27.8 | 94 |
| 28 | KRAS | 0.0039 | p.Gly13Asp | missense | 7.74 | 27.8 | 94 |
| 35 | NRAS | 0.0008 | p.Glu62∗ | nonsense | 7.55 | 38 | NA |
| 37a | NRAS | 0.0012 | p.Glu62∗ | nonsense | 7.55 | 38 | NA |
| 23 | PIK3CA | 0.0008 | p.Glu39∗ | nonsense | 9.41 | 29 | NA |
| 37a | PTPN11 | 0.0016 | p.Leu65Leu | synonymous | 0.62 | 9.4 | NA |
| 56 | PTPN11 | 0.0011 | p.Thr73Ala | missense | 9.33 | 19.5 | 58 |
| 20 | TET2 | 0.0168 | p.Gln548Lys | missense | 0.57 | 4.4 | 53 |
| 60h | TP53 | 0.0008 | p.Phe113Val | missense | 7.39 | 24.6 | 50 |
| 63 | TP53 | 0.0052 | p.Met237Ile | missense | 7.56 | 22.8 | 10 |
| 69 | TP53 | 0.0039 | p.Val216Met | missense | 7.78 | 28.9 | 21 |
| 68s | TP53 | 0.0031 | p.Met237Lys | missense | 9.02 | 24 | 95 |
| 68t | TP53 | 0.0090 | p.Val216Met | missense | 7.78 | 28.9 | 21 |
Samples with more than one mutation are marked with a superscript letter for identification. For a complete table containing mutations at genomic and cDNA level and GenBank accession numbers, please refer to Table S5.
Clonal Hematopoiesis Can Arise throughout Adult Life
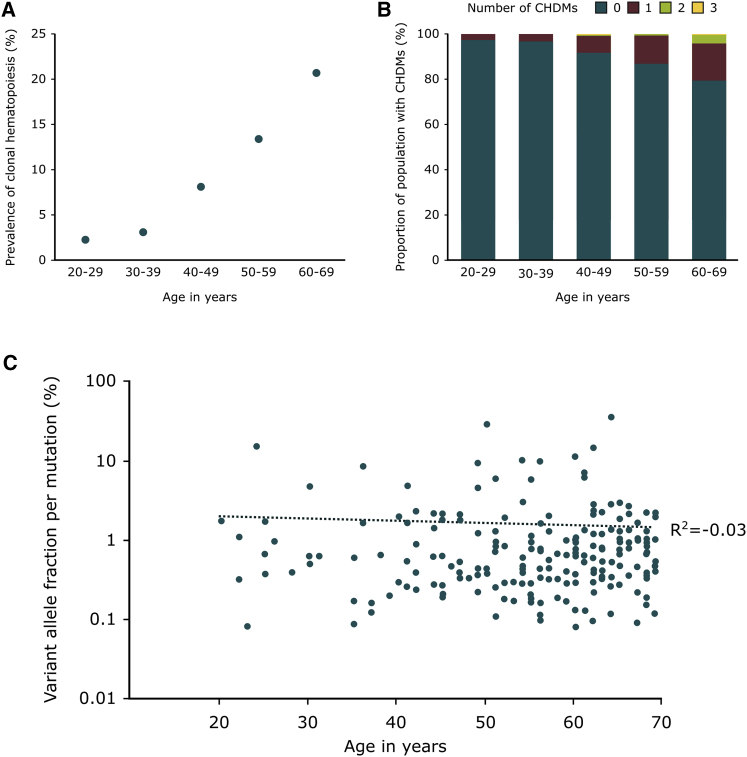
The abovementioned 216 somatic mutations in clonal hematopoiesis-driver genes were identified in 192 individuals between 20 and 69 years of age, representing an overall prevalence of 9.56% for CHDMs throughout our cohort. Individuals in our cohort with clonal hematopoiesis were significantly older than those without (median 57 versus 43 years of age, p < 2.2 × 10−16, Wilcoxon rank sum test). The prevalence of CHDMs increased in an exponential manner with age and at least one CHDM was observed in 2.5% of the 20- to 29-year-old group, in 3.2% of the 30- to 39-year-old group, in 8.2% of the 40- to 49-year-old group, in 13.2% of the 50- to 59-year-old group, and in 20.6% of the 60- to 69-year-old group (see Figure 3A). Two or more CHDMs were identified in 22 individuals in our cohort, who were significantly older than those in whom only one CHDM was identified (median of 65 versus 56 years of age, p < 0.01, Wilcoxon rank sum test). Sixteen of these individuals with more than one CHDM were 60 or older, representing close to 4% of the group of individuals between 60 and 69 years of age (see Figure 3B). No difference in the mutational status or number of mutations was observed between sexes.
Figure 3.
Clonal Hematopoiesis-Driver Mutations per Age Group
(A) Prevalence of clonal hematopoiesis per age group, defined as the frequency of individuals with one or more CHDMs per decade of age.
(B) Proportion of the population per age group with CHDMs.
(C) Mutation variant allele fraction per age of the individual in which the mutation was identified. The y axis is in logarithmic scale. No significant correlation is observed between the age of the individual and the VAF of the mutation identified.
Remarkably, while the prevalence and number of CHDMs in the population increased with age, we did not detect a correlation between the VAF of CHDMs and the age of the individual in which they were identified (R2 = −0.03, Pearson’s correlation; see Figure 3C). We examined a set of previously published CHDMs5 in which we also observed a lack of correlation between the VAF of mutations identified in individuals below the age of 70 and the age of the individual in which they were detected (R2 = 0.06, Pearson’s correlation). However, when analyzing the VAF of mutations in individuals 70 years of age or older, we observed a moderate correlation between these two parameters (R2 = 0.3, Pearson’s correlation). No statistically significant difference between the VAF of mutations identified in the abovementioned study in individuals below and above the age of 70 was observed (0.025 versus 0.027, p = 0.87, Wilcoxon rank sum test).
Age-Specific Patterns of Mutation in Clonal Hematopoiesis Driver Genes
More than 85% of all somatic SNVs in clonal hematopoiesis driver genes correspond to A>G/T>C, C>A/G>T, or C>T/G>A mutations (170 out of 196 mutations). The distribution of these types of mutations differs between the individuals below the age of 45 and those 45 or older in our cohort (p < 0.05, Pearson’s chi-square test, df = 2). While the most frequent CHMDs are C>T/G>A transitions, their frequency does not change with age, as they represent 51% and 52% of all SNVs in the younger and older age group. On the other hand, A>G/T>C mutations increase with age, corresponding to 20% of all SNVs in individuals 45 years or older in contrast with only 8% in individuals below this age (see Figure 4). This increase in frequency occurs at the expense of C>A/G>T mutations, which represent close to 30% of all SNVs in young individuals, as opposed to approximately 14% in individuals above the age of 45. This difference in patterns of mutations could not be attributed to mutations in any single gene.
Figure 4.
Type of Mutation Change per Age Group
Aging is associated with different patterns of clonal hematopoiesis and it has been proposed that DNMT3A and JAK2 mutations appear throughout life, while mutations in SRSF2 and SF3B1 arise only in individuals over the age of 70.5 Indeed, mutations in DNMT3A and JAK2 were observed throughout all age groups; the youngest individual in which an established CHDM was identified in our cohort was a 22-year-old woman with a JAK2 p.Val617Phe substitution at a VAF of 0.003. As with CHDMs in general, the frequency of DNMT3A mutations increased with age, reaching a prevalence of 13.7% in population controls between 60 and 69 years of age. The comparison of DNMT3A hotspot versus non-hotspot mutations reveals that the VAF of clones carrying DNMT3A hotspot mutations is significantly larger than that of clones with non-hotspot mutations (median 0.025 versus 0.009, Wilcoxon test, p < 0.01). For most genes in which five or more CHDMs were identified in our cohort, including CBL, DNMT3A, GNAS, JAK2, and TET2, no statistically significant difference was detected between the age of the carriers of mutations in any of these genes and individuals with one or more CHDMs in other genes. Although individuals with CHDMs in TP53 were older than carriers of CHDMs in other genes (63.2 versus 53.9 years of age, Wilcoxon test, p < 0.05), once we corrected for multiple testing, this observation lost statistical significance at p < 0.05. Some genes included in our screen were found to be mutated only in individuals over the age of 60, such as GNB1 (MIM: 139380), IDH2 (MIM: 147650), SF3B1, and U2AF1 (MIM: 191317). Although mutations in components of the spliceosome have been proposed to drive clonal hematopoiesis only in older individuals, SRSF2 mutations were identified in two individuals of 49 and 66 years of age. Remarkably, the mutation observed in the 49-year-old man was a p.Pro95Leu missense substitution in SRSF2 with a relatively high VAF of 0.09.
Somatic Mutations in Candidate Loci for Clonal Hematopoiesis Driver Mutations
Unexpectedly, mutations known to cause developmental disorders when arising in the germline can be found at low frequencies in population databases of genetic variation such as ExAC.27 It has been suggested that some of these findings may be explained by mutations which, in addition to causing developmental disorders when present in the germline, would also be involved in clonal hematopoiesis when arising in HSCs.27 A revision of published literature led to the identification of 1,158 somatic mutations in 90 genes involved in clonal hematopoiesis.2, 3, 4, 5 We compared this list of 90 genes with a subset of 464 genes involved in dominant and de novo developmental disorders from DDG2P44 to determine whether there was significant overlap between genes involved in clonal hematopoiesis when mutated somatically and genes involved in developmental disorders. In this way, we identified 29 genes in which germline mutations cause developmental disorders while somatic mutations are involved in clonal hematopoiesis. This represents a significant enrichment compared to the expectation (hypergeometric distribution with a universe of 22,285 protein-coding genes, p = 9.2 × 10−28).
In addition to the 40 known loci for CHDMs in our assay, we screened for somatic mutations in blood in 64 novel candidate loci for CHDMs. These included seven loci with nine residues in which recurrent missense substitutions are known to be causative for paternal age effect disorders, such as achondroplasia, when present in the germline.29, 30, 31, 45, 46 These mutations cause clonal expansion in spermatogonial stem cells when arising during spermatogenesis28 and we hypothesized that some may also undergo clonal expansion in other tissues. Therefore, we considered mutations in these loci as candidates for clonal expansion in HSCs during hematopoiesis. Furthermore, we selected 57 additional loci in which recurrent identical de novo mutations have been found in developmental disorders (see Table S2 for all candidate loci screened).38, 39, 40 Screening of these 64 candidate loci for CHDMs led to the identification of 7 somatic mutations, including 1 nonsense, 3 missense, and 3 synonymous substitutions with VAFs ranging from 0.0012 to 0.024 (average 0.0074, median 0.0025). Genes in which mutations were detected include ADNP (MIM: 611386), COL4A3BP (MIM: 604677), CUX2 (MIM: 610648), HECTD1, KCNQ3 (MIM: 602232), RHEB (MIM: 601293), and SMAD4 (MIM: 600993) (see Table 4). No mutations were identified that were identical to mutations involved in spermatogonial stem cell selection or found to be recurrently mutated in developmental disorders.
Table 4.
Somatic Mutations Identified in Candidate Clonal Hematopoiesis Driver Genes
| Age | Gene Name | VAF | Predicted Protein Substitution | Mutation Type | PhyloP | CADD PHRED | Grantham Score |
|---|---|---|---|---|---|---|---|
| 41 | ADNP | 0.0025 | p.Tyr719Tyr | synonymous | −1.96 | 0.02 | NA |
| 25 | COL4A3BP | 0.0012 | p.Ser260∗ | nonsense | 9.84 | 40 | NA |
| 58 | CUX2 | 0.0246 | p.Leu592Pro | missense | 8.04 | 21.2 | 98 |
| 59 | HECTD1 | 0.0012 | p.Ser2223Pro | missense | 8.96 | 12.6 | 74 |
| 43 | KCNQ3 | 0.0030 | p.Ser228Asn | missense | 5.91 | 19.7 | 46 |
| 48 | RHEB | 0.0173 | p.Tyr35Tyr | synonymous | 0.1 | 8.8 | NA |
| 60 | SMAD4 | 0.0017 | p.Cys499Cys | synonymous | 1.86 | 4.9 | NA |
For a complete table containing mutations at genomic and cDNA level and GenBank accession numbers, please refer to Table S5.
Discussion
Recurrent somatic mutations have been recently identified in blood-derived DNA of population controls.2, 3, 4 In this study, we established a smMIPs assay targeting 104 known or candidate loci for CHDMs and sequenced blood-derived DNA of 2,007 population controls between the ages of 20 and 69. With a median unique sequencing coverage of 845-fold, corresponding to the number of unique DNA molecules captured per position per sample, we identified 223 somatic SNVs and indels in the coding regions screened of which 216 affected known CH-driver genes. A high ratio for non-synonymous to synonymous somatic mutations was observed for loci screened within genes known to drive clonal hematopoiesis (214 non-synonymous versus 2 synonymous point substitutions). This observation suggests overall positive selection among a large proportion of the mutations identified in the screened loci.47
Both the prevalence of CH as well as the number of mutations increased with age, reaching a frequency over 20% for CHDMs in individuals between 60 and 69 years of age. This number is close to double that of previous reports mentioning a prevalence for CHDMs in 5% to 10% in healthy individuals older than 60.2, 3, 4 It is likely that the higher prevalence of CH detected in our study results from the increased sensitivity of smMIPs, compared to other sequencing methods used in previous studies.22 The use of a deep coverage smMIP approach has the advantage of allowing a large number of loci to be screened for mutations with high sensitivity for the identification of mutations with low VAFs. In contrast, exome sequencing has the advantage of allowing the identification of mutations throughout the coding region, but it may miss mutations present at low allelic frequencies when performed at current day standard coverage. Indeed, an average sequence of 84 reads sets the lower limit for detection of somatic mutations at an allele fraction of approximately 0.035.3 More than 90% of the CHDMs detected in our study (198 out of 216) are below this VAF and would most likely have been missed by average coverage exome sequencing. On the other hand, amplicon-based sequencing limits the detection of mutations to the targeted regions but provides deep coverage, which enables the identification of mutations with low VAF. For instance, one study detected mutations in 15 loci using a sequencing coverage above 1,000-fold, which lowered the limit for mutation detection to an allele fraction of 0.008.5 Due to the limitation in the number of regions that could be targeted by our assay while providing deep coverage, we screened only for mutations the genomic loci reported to be most frequently mutated in clonal hematopoiesis. This resulted in the exclusion of genes in which mutations have been identified throughout the sequence of the gene, such as loss-of-function mutations in PPM1D.2, 4 One of the main limitations for the detection of somatic mutations is the sequencing error rate of NGS platforms, ranging around 0.1%–1% for sequencing by synthesis approaches.48 Mutations introduced during the preparation of the sequencing library further limit the ability to distinguish true CHDMs present in blood from false positive signal.49 Some of these limitations have been bypassed in previous studies by using technologies based on the incorporation of random tags in an amplicon-based sequencing library.50, 51 Similarly, smMIPs include a random tag in each probe assigning a UMI to each individual DNA molecule captured,23 which allows multiple sequencing reads descending from the same DNA molecule to be traced in order to generate a true molecular count without PCR duplicates. Additionally, molecular tags can be used to create a consensus sequence of this DNA fragment, thus increasing the specificity for the detection of true somatic events.50, 51 To ensure specificity in our detection of CHDMs, we modeled the sequencing error to identify statistically significant mutation counts in two replicates from the same sample. Furthermore, at least two independent smMIPs were used to target each DNA strand within the screened loci. The mutations identified in our assay had a median VAF of 0.0061, ranging from 0.0008 to 0.35, and a subset of mutations were validated using restriction digestion, confirming that the deviations in number of mutation reads at specific positions detected through sequencing data reflected a true mutation present in the sample DNA.
Although the prevalence of CH increased with age, CHDMs were observed throughout all ages in our cohort. The youngest individual in which an established CHDM was identified in our cohort was a 22-year-old woman in whom we detected a JAK2 p.Val617Phe substitution with a VAF of 0.003. CHDMs were detected in 2.3% of population controls between 20 and 29 years of age, suggesting that CH is not a rare finding in early adult life. While age was a poor predictor for the presence and number of CHDMs at the individual level, when analyzing our findings by 10-year age groups, we observed that age explained 96% of the variation in the presence and number of CHDMs in the population. This suggests that while age is a major factor in the occurrence of CHDMs at the population level, stochasticity and inter-individual differences, such as genetic variation or environmental factors, may play a prominent role in the occurrence of CHDMs at the individual level. Notably, we observe that the increase in prevalence and number of CHDMs at the population level is not linear. This supports that factors associated with aging may accelerate the occurrence of CHDMs over time, due to either increased DNA damage or decreased DNA repair. An increase with age was observed in the proportion of A>G:T>C mutations detected in our cohort. This type of mutation has been linked to deamination of adenine and has been shown to be associated with transcription-coupled repair.52 The prevalence of CHDMs in population controls contrasts with the incidence of leukemia in the general population. Given this discrepancy, it is unclear whether all individuals with CHDMs have an increased risk for developing leukemia. The presence of CHDMs may place individuals at increased risk for development of hematologic malignancies but does not seem sufficient in itself to develop leukemia. It is therefore likely that the evolution from clonal hematopoiesis to leukemia is the result of many cycles of selection of mutant HSCs in which the cellular, tissue, and organism environment plays a role.
While some CHDMs, such as those in DNMT3A and JAK2, were found in individuals of all ages, some CHDMs were exclusively detected in individuals over the age of 60, including mutations in GNB1, IDH2, SF3B1, and U2AF1. Additionally, individuals with CHDMs in TP53 were older than those with CHDMs in other genes, although this result did not reach statistical significance. It has been suggested that some mutations may arise in HSCs in young individuals but can expand clonally only in the context of the aging bone marrow.5, 13, 20 Mutations in components of the spliceosome, such as SRSF2 and SF3B1, have been proposed to fall under this category.5 Interestingly, we identified a SRSF2 mutation with a VAF of 0.09 in a 49-year-old individual, suggesting that although rare, clonal expansion of spliceosome mutations can occur in younger individuals. Compared to HSCs in young individuals, aging HSCs show decline in replication capacity17, 22 and a bias toward myeloid differentiation.18 These changes may be specific to the aging cells or could result from alterations in the bone marrow niche.17, 19, 53 Cell-intrinsic alterations arising in aging HSCs include epigenetic alterations and upregulation of genes involved in myeloid differentiation, DNA repair, cell death, and genes linked to leukemia.14, 17, 53 However, computational modeling and studies in mouse models support that the aging microenvironment exerts strong selective pressure on HSCs.11, 22 Therefore, certain mutations involved in clonal hematopoiesis may confer aging HSCs a growth advantage over wild-type aging HSCs in which function is generally declining. In contrast, the same mutations in young HSCs may not result in increased cellular fitness compared to wild-type young HSCs and thus hamper expansion.54 Mutations in the genomic regions screened in our study have been implicated in various hematologic malignancies affecting different age groups, such as myelodysplasia, acute and chronic myeloid leukemia, and lymphocytic leukemia. The age of onset of these disorders may reflect the age-specific biological conditions which allow the clonal expansion of HSCs carrying mutations in certain genes implicated in these malignancies. As our cohort consists of population controls, we cannot exclude the possibility that some of the somatic mutations detected may originate from true hematologic malignancies developing in individuals in our cohort. For instance, this may be the case for some somatic mutations identified in genes such as CBL and JAK2 in relatively young individuals.55
The gene harboring most CHDMs in our study was DNMT3A, where we identified hotspot and non-hotspot missense substitutions, as well as truncating mutations throughout all five loci screened. Our assay covered approximately 8.5% of the coding sequence of DNMT3A and mutations in this gene have been identified throughout its coding sequence. Similarly, our assay only included close to 2.5% of the coding sequence of ASXL1 and TET2, which are disrupted by truncating mutations that may arise throughout the gene. Therefore, the detected prevalence of somatic mutations in DNMT3A, ASXL1, and TET2 in our study is likely an underestimation and more mutations may be found outside the regions screened. Nevertheless, our findings support DNMT3A as the most frequently mutated gene in CH.2, 3, 4, 5 Close to 60% of DNMT3A mutations identified in hematologic malignancies disrupt codon Arg882,56, 57 while substitutions involving codon Arg326 represent only 0.1% of DNMT3A mutations in hematologic cancer.58 In clonal hematopoiesis, we find that 9.6% of DNMT3A mutations identified in controls disrupt codon Arg882, similar to those disrupting Arg326 which represent 8.9% (13/135 and 12/135 substitutions for Arg882 and Arg326, respectively). We therefore estimate that the ratio of DNMT3A Arg882 to Arg326 mutations is approximately 600:1 in malignancy and observe that this ratio is 1.1:1 in clonal hematopoiesis. This supports the hypothesis that recurrent DNMT3A mutations grant an advantage to mutated cells, but the risk of progressing to malignancy is much higher in the presence of DNMT3A Arg882 mutations.56 This higher risk associated with these mutations may stem from the dominant negative effect of DNMT3A Arg882 mutants which severely affects DNMT3A function through homodimeric interactions with the wild-type protein.59 Mutations in DNMT3A affecting residues other than Arg882 are thought to have a smaller effect on the activity of the wild-type protein, which may not be sufficient to drive the development of cancer.59 Residues other than Arg882 can also be mutated in myeloid and lymphoid malignancies and the bi-allelic presence of this type of DNMT3A mutations has been identified in different forms of leukemia. In our cohort, we identified six individuals with more than one DNMT3A mutation but the VAF reflected the existence of clones of different sizes. It is unclear whether in these subjects the two mutations may be present in the same cell or whether they represent completely independent DNMT3A mutant clones.
No significant correlation was observed between the VAF of the CHDMs identified and the age of the individuals carrying the mutations (R2 = −0.03, Pearson’s correlation). This may result from an increase in the occurrence of mutations over time, leading to a higher number of mutant clones of small size in older individuals that would lower the average VAF in this group. However, the fact that examining only mutations with a VAF ≥ 0.02 also reveals a lack of correlation between age and VAF argues against this point. Another explanation lies in the possibility that the size of a mutant clone depends on the age of the clone rather than the age of the individual in which it is present. Our study consisted of a single measurement and as such, we are not able to follow the evolution of mutant clones over time. One paper analyzing multiple samples from the same individual followed the evolution of mutant clones over time and determined that all mutant clones were still present between 4 and 8 years after initial detection, with the vast majority of mutant clones increasing or remaining stable over time.3 These fluctuations may reflect actual changes in the size of mutant HSC clones, but could also be due to variation over time in the contribution of mutant clones of HSCs to blood. It is unclear at present whether the size of a mutant clone as reflected by the VAF of a mutation does in fact increase with the age of the clone. The presence of a CHDM in itself may not be sufficient to lead to clonal expansion over time, either because of a weak proliferative effect or because an additional factor may be required to allow for expansion. A higher correlation between VAF and age was observed for individuals over the age of 70 from a different study,5 which may suggest that aging HSCs and bone marrow environment may represent this additional factor that allows for clonal expansion of mutant HSCs.
A recent study highlighted the unexpected presence of mutations associated with developmental disorders at high frequency in 60,706 reference exomes in ExAC.27 For instance, 345 individuals were found to carry 56 different truncating ASXL1 mutations, an unexpected finding, considering that germline truncating ASXL1 mutations cause Bohring-Opitz syndrome (MIM: 605039), a severe developmental syndrome.27, 60 This study shows that these mutations had lower VAFs than expected for germline events and were present mainly in older individuals and therefore likely represented CHDMs. This suggests that the presence in ExAC of other mutations causative for developmental disorders may also reflect somatic events involved in clonal hematopoiesis rather than germline mutations.27 We therefore screened our cohort for somatic mutations in candidate genes for clonal hematopoiesis that cause developmental disorders when mutated in the germline. Similar to this previously published study, we identified somatic mutations in blood overlapping with germline mutations known to cause developmental disorders when present in the germline. In addition to four truncating mutations in ASXL1, we identified 13 missense substitutions in DNMT3A Arg882, which is known to be mutated in Tatton-Brown syndrome (MIM: 615879), a developmental disorder with overgrowth.61 Similarly, three somatic missense mutations in CBL overlapping with germline mutations leading to a Noonan-like phenotype (MIM: 613563) were identified in our cohort.62, 63 However, we failed to identify somatic mutations in blood in our candidate loci which overlapped with known developmental disease-causing mutations. This suggests that this genetic overlap between developmental disorders and clonal hematopoiesis may be restricted to specific mutations in known candidate genes for clonal hematopoiesis. Thus, the presence of other developmental disease-causing variants in reference databases remains unexplained and may also be due to other factors such as sequencing errors. We did, however, identify a somatic mutation in CBL leading to CBL p.Arg420Gln at a VAF of 0.15 in a 24-year-old man, with the result that 30% of circulating blood cells carry this mutation. The high VAF for this mutation is remarkable given the young age of the individual and the fact that this was the only somatic mutation identified in this individual. As such, it is unknown whether this mutation represents a somatic event arising in a HSC during postnatal life or a postzygotic de novo mutation arising in early embryogenesis. Interestingly, this mutation has been reported to cause Noonan-like syndrome when present in the germline62 and leukemia when present somatically.58 We could not access clinical information or additional samples from this individual to verify the presence of mosaicism in other tissues. It may be that pathogenic mutations such as the one identified in this individual or those present in these reference databases reflect genetic variation in resilient individuals.64
In summary, we have screened a cohort of population controls between 20 and 69 years of age to identify somatic mutations in blood implicated in clonal hematopoiesis. Our method provides high sensitivity which allowed for the identification of CHDMs at higher prevalence than previously reported despite studying a limited set of mutations. Somatic mutations were identified in individuals of all ages, with differences in the profile of genes mutated per age group and a strong increase in the number of mutations detected with age, with more than 20% of individuals between 60 and 69 years carrying at least one CHDM, while up to 3% of individuals younger than 30 years of age had at least one CHDM. Our findings support the occurrence of clonal hematopoiesis associated with somatic mutations as a widespread mechanism linked to aging, suggesting that clonal evolution of cells harboring somatic mutations is a universal mechanism occurring at all ages in humans.
Acknowledgments
The Nijmegen Biomedical Study is a population-based survey conducted at the Department for Health Evidence and the Department of Laboratory Medicine of the Radboud University Medical Center. Principal investigators of the Nijmegen Biomedical Study are L.A.L.M.K., A.L.M. Verbeek, D.W. Swinkels, and B. Franke. We thank the members of the Radboud Genomics Technology Center for their technical support in DNA normalization and sequencing. This work was in part financially supported by grants from the Netherlands Organisation for Scientific Research (918-15-667 to J.A.V.) and the European Research Council (ERC Starting grant DENOVO 281964 to J.A.V.). R.A.-H. was supported by a Radboudumc PhD grant.
Published: June 29, 2017
Footnotes
Supplemental Data include two figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.05.013.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
gene2phenotype, http://www.ebi.ac.uk/gene2phenotype/downloads
OMIM, http://www.omim.org/
Supplemental Data
Previous reports used include Xie et al.,4 Genovese et al.,2 Jaiswal et al.,3 and McKerrell et al.5
Samples with more than one mutation are marked with a superscript letter for identification.
References
- 1.Yadav V.K., DeGregori J., De S. The landscape of somatic mutations in protein coding genes in apparently benign human tissues carries signatures of relaxed purifying selection. Nucleic Acids Res. 2016;44:2075–2084. doi: 10.1093/nar/gkw086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genovese G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie M., Lu C., Wang J., McLellan M.D., Johnson K.J., Wendl M.C., McMichael J.F., Schmidt H.K., Yellapantula V., Miller C.A. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKerrell T., Park N., Moreno T., Grove C.S.S., Ponstingl H., Stephens J., Crawley C., Craig J., Scott M.A.A., Hodkinson C., Understanding Society Scientific Group Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–1245. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young A.L., Challen G.A., Birmann B.M., Druley T.E. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun. 2016;7:12484. doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martincorena I., Roshan A., Gerstung M., Ellis P., Van Loo P., McLaren S., Wedge D.C., Fullam A., Alexandrov L.B., Tubio J.M. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abyzov A., Mariani J., Palejev D., Zhang Y., Haney M.S., Tomasini L., Ferrandino A.F., Rosenberg Belmaker L.A., Szekely A., Wilson M. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Z., Wyman M.J., Sella G., Przeworski M. Interpreting the dependence of mutation rates on age and time. PLoS Biol. 2016;14:e1002355. doi: 10.1371/journal.pbio.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsey M.J., Moore D.H., 2nd, Briner J.F., Lee D.A., Olsen La., Senft J.R., Tucker J.D. The effects of age and lifestyle factors on the accumulation of cytogenetic damage as measured by chromosome painting. Mutat. Res. 1995;338:95–106. doi: 10.1016/0921-8734(95)00015-x. [DOI] [PubMed] [Google Scholar]
- 11.McKerrell T., Vassiliou G.S. Aging as a driver of leukemogenesis. Sci. Transl. Med. 2015;7:306fs38. doi: 10.1126/scitranslmed.aac4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams P.D., Jasper H., Rudolph K.L. Aging-induced stem cell mutations as drivers for disease and cancer. Cell Stem Cell. 2015;16:601–612. doi: 10.1016/j.stem.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhury S.S., Morison J.K., Gibson B.E.S., Keeshan K. Insights into cell ontogeny, age, and acute myeloid leukemia. Exp. Hematol. 2015;43:745–755. doi: 10.1016/j.exphem.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Corces-Zimmerman M.R., Majeti R. Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia. 2014;28:2276–2282. doi: 10.1038/leu.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steensma D.P., Bejar R., Jaiswal S., Lindsley R.C., Sekeres M.A., Hasserjian R.P., Ebert B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akunuru S., Geiger H. Aging, clonality, and rejuvenation of hematopoietic stem cells. Trends Mol. Med. 2016;22:701–712. doi: 10.1016/j.molmed.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beerman I., Bhattacharya D., Zandi S., Sigvardsson M., Weissman I.L., Bryder D., Rossi D.J. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. USA. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiger H., de Haan G., Florian M.C. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 2013;13:376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- 20.Rozhok A.I., DeGregori J. Toward an evolutionary model of cancer: Considering the mechanisms that govern the fate of somatic mutations. Proc. Natl. Acad. Sci. USA. 2015;112:8914–8921. doi: 10.1073/pnas.1501713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holstege H., Pfeiffer W., Sie D., Hulsman M., Nicholas T.J., Lee C.C., Ross T., Lin J., Miller M.A., Ylstra B. Somatic mutations found in the healthy blood compartment of a 115-yr-old woman demonstrate oligoclonal hematopoiesis. Genome Res. 2014;24:733–742. doi: 10.1101/gr.162131.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shlush L.I., Zandi S., Itzkovitz S., Schuh A.C. Aging, clonal hematopoiesis and preleukemia: not just bad luck? Int. J. Hematol. 2015;102:513–522. doi: 10.1007/s12185-015-1870-5. [DOI] [PubMed] [Google Scholar]
- 23.Hiatt J.B., Pritchard C.C., Salipante S.J., O’Roak B.J., Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neveling K., Mensenkamp A.R., Derks R., Kwint M., Ouchene H., Steehouwer M., van Lier B., Bosgoed E., Rikken A., Tychon M. BRCA testing by single-molecule molecular inversion probes. Clin. Chem. 2017;63:503–512. doi: 10.1373/clinchem.2016.263897. [DOI] [PubMed] [Google Scholar]
- 25.Eijkelenboom A., Kamping E.J., Kastner-van Raaij A.W., Hendriks-Cornelissen S.J., Neveling K., Kuiper R.P., Hoischen A., Nelen M.R., Ligtenberg M.J.L., Tops B.B.J. Reliable next-generation sequencing of formalin-fixed, paraffin-embedded tissue using single molecule tags. J. Mol. Diagn. 2016;18:851–863. doi: 10.1016/j.jmoldx.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Weren R.D.A., Mensenkamp A.R., Simons M., Eijkelenboom A., Sie A.S., Ouchene H., van Asseldonk M., Gomez-Garcia E.B., Blok M.J., de Hullu J.A. Novel BRCA1 and BRCA2 tumor test as basis for treatment decisions and referral for genetic counselling of patients with ovarian carcinomas. Hum. Mutat. 2017;38:226–235. doi: 10.1002/humu.23137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlston C.M., O’Donnell-Luria A.H., Underhill H.R., Cummings B.B., Weisburd B., Minikel E.V., Birnbaum D.P., Tvrdik T., MacArthur D.G., Mao R., Exome Aggregation Consortium Pathogenic ASXL1 somatic variants in reference databases complicate germline variant interpretation for Bohring-Opitz syndrome. Hum. Mutat. 2017;38:517–523. doi: 10.1002/humu.23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goriely A., Wilkie A.O.M. Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am. J. Hum. Genet. 2012;90:175–200. doi: 10.1016/j.ajhg.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maher G.J., McGowan S.J., Giannoulatou E., Verrill C., Goriely A., Wilkie A.O.M. Visualizing the origins of selfish de novo mutations in individual seminiferous tubules of human testes. Proc. Natl. Acad. Sci. USA. 2016;113:2454–2459. doi: 10.1073/pnas.1521325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon S.-R., Choi S.-K., Eboreime J., Gelb B.D., Calabrese P., Arnheim N. Age-dependent germline mosaicism of the most common noonan syndrome mutation shows the signature of germline selection. Am. J. Hum. Genet. 2013;92:917–926. doi: 10.1016/j.ajhg.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giannoulatou E., McVean G., Taylor I.B., McGowan S.J., Maher G.J., Iqbal Z., Pfeifer S.P., Turner I., Burkitt Wright E.M.M., Shorto J. Contributions of intrinsic mutation rate and selfish selection to levels of de novo HRAS mutations in the paternal germline. Proc. Natl. Acad. Sci. USA. 2013;110:20152–20157. doi: 10.1073/pnas.1311381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goriely A., Hansen R.M.S., Taylor I.B., Olesen I.A., Jacobsen G.K., McGowan S.J., Pfeifer S.P., McVean G.A.T., Rajpert-De Meyts E., Wilkie A.O.M. Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat. Genet. 2009;41:1247–1252. doi: 10.1038/ng.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galesloot T.E., Vermeulen S.H., Swinkels D.W., de Vegt F., Franke B., den Heijer M., de Graaf J., Verbeek A.L., Kiemeney L.A. Cohort Profile: The Nijmegen Biomedical Study (NBS) Int. J. Epidemiol. 2017:dyw268. doi: 10.1093/ije/dyw268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Champion K.J., Bunag C., Estep A.L., Jones J.R., Bolt C.H., Rogers R.C., Rauen K.A., Everman D.B. Germline mutation in BRAF codon 600 is compatible with human development: de novo p.V600G mutation identified in a patient with CFC syndrome. Clin. Genet. 2011;79:468–474. doi: 10.1111/j.1399-0004.2010.01495.x. [DOI] [PubMed] [Google Scholar]
- 35.Niihori T., Aoki Y., Narumi Y., Neri G., Cavé H., Verloes A., Okamoto N., Hennekam R.C.M., Gillessen-Kaesbach G., Wieczorek D. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 36.Tartaglia M., Martinelli S., Stella L., Bocchinfuso G., Flex E., Cordeddu V., Zampino G., Burgt Iv., Palleschi A., Petrucci T.C. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am. J. Hum. Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groesser L., Herschberger E., Ruetten A., Ruivenkamp C., Lopriore E., Zutt M., Langmann T., Singer S., Klingseisen L., Schneider-Brachert W. Postzygotic HRAS and KRAS mutations cause nevus sebaceous and Schimmelpenning syndrome. Nat. Genet. 2012;44:783–787. doi: 10.1038/ng.2316. [DOI] [PubMed] [Google Scholar]
- 38.Hoischen A., Krumm N., Eichler E.E. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat. Neurosci. 2014;17:764–772. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B.A., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 40.McRae J.F., Clayton S., Fitzgerald T.W., Kaplanis J., Prigmore E., Rajan D., Sifrim A., Aitken S., Akawi N., Alvi M. Prevalence, phenotype and architecture of developmental disorders caused by de novo mutation. bioRxiv. 2016 [Google Scholar]
- 41.Boyle E.A., O’Roak B.J., Martin B.K., Kumar A., Shendure J. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30:2670–2672. doi: 10.1093/bioinformatics/btu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mancini M., Hasan S.K., Ottone T., Lavorgna S., Ciardi C., Angelini D.F., Agostini F., Venditti A., Lo-Coco F. Two novel methods for rapid detection and quantification of DNMT3A R882 mutations in acute myeloid leukemia. J. Mol. Diagn. 2015;17:179–184. doi: 10.1016/j.jmoldx.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., DDD study Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goriely A., McVean G.A.T., Röjmyr M., Ingemarsson B., Wilkie A.O.M. Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science. 2003;301:643–646. doi: 10.1126/science.1085710. [DOI] [PubMed] [Google Scholar]
- 46.Choi S.-K., Yoon S.-R., Calabrese P., Arnheim N. Positive selection for new disease mutations in the human germline: evidence from the heritable cancer syndrome multiple endocrine neoplasia type 2B. PLoS Genet. 2012;8:e1002420. doi: 10.1371/journal.pgen.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L., Liu P., Evans T.C., Jr., Ettwiller L.M. DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science. 2017;355:752–756. doi: 10.1126/science.aai8690. [DOI] [PubMed] [Google Scholar]
- 50.Wong T.N., Ramsingh G., Young A.L., Miller C.A., Touma W., Welch J.S., Lamprecht T.L., Shen D., Hundal J., Fulton R.S. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518:552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi K., Wang F., Kantarjian H., Doss D., Khanna K., Thompson E., Zhao L., Patel K., Neelapu S., Gumbs C. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. Lancet Oncol. 2017;18:100–111. doi: 10.1016/S1470-2045(16)30626-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexandrov L.B., Jones P.H., Wedge D.C., Sale J.E., Campbell P.J., Nik-Zainal S., Stratton M.R. Clock-like mutational processes in human somatic cells. Nat. Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moehrle B.M., Geiger H. Aging of hematopoietic stem cells: DNA damage and mutations? Exp. Hematol. 2016;44:895–901. doi: 10.1016/j.exphem.2016.06.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mason C.C., Khorashad J.S., Tantravahi S.K., Kelley T.W., Zabriskie M.S., Yan D., Pomicter A.D., Reynolds K.R., Eiring A.M., Kronenberg Z. Age-related mutations and chronic myelomonocytic leukemia. Leukemia. 2016;30:906–913. doi: 10.1038/leu.2015.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24:1128–1138. doi: 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Link D.C., Walter M.J. 'CHIP’ping away at clonal hematopoiesis. Leukemia. 2016;30:1633–1635. doi: 10.1038/leu.2016.130. [DOI] [PubMed] [Google Scholar]
- 57.Grimwade D., Ivey A., Huntly B.J.P. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang L., Rau R., Goodell M.A. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoischen A., van Bon B.W.M., Rodríguez-Santiago B., Gilissen C., Vissers L.E.L.M., de Vries P., Janssen I., van Lier B., Hastings R., Smithson S.F. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat. Genet. 2011;43:729–731. doi: 10.1038/ng.868. [DOI] [PubMed] [Google Scholar]
- 61.Kosaki R., Terashima H., Kubota M., Kosaki K. Acute myeloid leukemia-associated DNMT3A p.Arg882His mutation in a patient with Tatton-Brown-Rahman overgrowth syndrome as a constitutional mutation. Am. J. Med. Genet. A. 2017;173:250–253. doi: 10.1002/ajmg.a.37995. [DOI] [PubMed] [Google Scholar]
- 62.Martinelli S., De Luca A., Stellacci E., Rossi C., Checquolo S., Lepri F., Caputo V., Silvano M., Buscherini F., Consoli F. Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am. J. Hum. Genet. 2010;87:250–257. doi: 10.1016/j.ajhg.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niemeyer C.M., Kang M.W., Shin D.H., Furlan I., Erlacher M., Bunin N.J., Bunda S., Finklestein J.Z., Sakamoto K.M., Gorr T.A. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat. Genet. 2010;42:794–800. doi: 10.1038/ng.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen R., Shi L., Hakenberg J., Naughton B., Sklar P., Zhang J., Zhou H., Tian L., Prakash O., Lemire M. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Previous reports used include Xie et al.,4 Genovese et al.,2 Jaiswal et al.,3 and McKerrell et al.5
Samples with more than one mutation are marked with a superscript letter for identification.