Abstract
We produced complete sequences and conducted comparative analysis of the maternally inherited chloroplast (cp) genomes and bi-parentally inherited 45S nuclear ribosomal RNA genes (nrDNA) from ten Araliaceae species to elucidate the genetic diversity and evolution in that family. The cp genomes ranged from 155,993 bp to 156,730 bp with 97.1–99.6% similarity. Complete 45S nrDNA units were about 11 kb including a 5.8-kb 45S cistron. Among 79 cp protein-coding genes, 74 showed nucleotide variations among ten species, of which infA, rpl22, rps19 and ndhE genes showed the highest Ks values and atpF, atpE, ycf2 and rps15 genes showed the highest Ka/Ks values. Four genes, petN, psaJ, psbF, and psbN, related to photosynthesis and one gene, rpl23, related to the ribosomal large subunit remain conserved in all 10 Araliaceae species. Phylogenetic analysis revealed that the ten species could be resolved into two monophyletic lineages, the Panax-Aralia and the Eleutherococcus-Dendropanax groups, which diverged approximately 8.81–10.59 million years ago (MYA). The Panax genus divided into two groups, with diploid species including P. notoginseng, P. vietnamensis, and P. japonicus surviving in Southern Asia and a tetraploid group including P. ginseng and P. quinquefolius Northern Asia and North America 2.89–3.20 MYA.
Introduction
The Araliaceae (also known as the ginseng family) and the Apiaceae are the major families in the order Apiales belonging to Asterid II1–3. The Araliaceae family comprises 55 genera and more than 1,500 plant species widely distributed in tropical, subtropical and temperate regions4, 5, many of which are used as oriental medicines, such as species in the genus Panax, Eleutherococcus and Aralia 6, 7. According to taxonomical studies, Araliaceae encompasses two large monophyletic groups: the Aralia-Panax group and the Asian Palmate group8. The Aralia-Panax group consists of the two closely-related genera, Aralia and Panax. Meanwhile, the Asian Palmate group is represented by the genera Eleutherococcus, Dendropanax, and Schefflera characterized as distinctive woody plants.
Although the conserved basic chromosome number was estimated to be x = 12 in Araliaceae family species based on diploid taxa (2n = 24), the chromosome numbers vary from 2n = 48 to 2n = 192 in polyploid species of the family9, 10. In the genus Panax, P. notoginseng, P. vietnamensis and P. japonicus are diploid with chromosome number of 2n = 24, while P. ginseng and P. quinquefolius are considered to be tetraploid with chromosome numbers of 2n = 48. The genus Aralia and Eleutherococcus are reported to have various chromosome numbers including 2n = 24, 36 or 48. (CCDB-chromosome count database; http://ccdb.tau.ac.il/).
Although many studies have reported taxonomical classification and divergence of Araliaceae species based on molecular data derived from a few chloroplast (cp) and nuclear sequences11–16, genetic diversity surveys and molecular phylogenetic classification of Panax and its relatives are still very limited. Cytoplasmic cp genomes and nuclear ribosomal DNA have widely been used to elucidate the evolution of plant species, owing to their characteristic highly conserved sequences, such that minor sequence divergences reflect evolutionary history17–20. Recently, we developed a de novo assembly method to obtain complete cp and nrDNA sequences using low-coverage whole-genome sequence (dnaLCW) and applied it to reveal the evolutionary history of various plant lineages such as Oryza AA genomes and Epimedium species21, 22, and also to identify intra-species diversity23.
In this study, we characterized cp genomes and 45S nrDNA sequences of ten Araliaceae species including Panax, Aralia, Eleutherococcus, and Dendropanax species and investigated genetic diversity among them to understand diversity and molecular evolution of the Araliaceae species.
Results
Complete chloroplast genomes and 45S nrDNA sequences
Novel complete cp genomes of five species, P. notoginseng, P. japonicus, P. vietnamensis, A. elata, and E. sessiliflorus, and 45S nrDNA sequences of seven species, P. quinquefolius, P. notoginseng, P. japonicus, P. vietnamensis, A. elata, and E. sesiliflorus, and D. morbifera, were characterized in this study (Table 1).
Table 1.
Cp genomes and 45S nrDNA sequences used for comparative analysis in this study.
| Genus | Species (Abbreviated name) | WGS reads used | Length (GenBank accession no.) | |||
|---|---|---|---|---|---|---|
| Amounts (Mb) | Cp Coverage (x) | 45S nrDNA Coverage (x) | Cp | 45S nrDNA (bp) | ||
| Panax | P. ginseng (PG) | 505 | 97 | 659 | 156,248 (KM088019) | 11,091 (KM036295)b |
| P. quinquefolius (PQ) | 1,010 | 127 | 533 | 156,088 (KM088018) | 11,169 (KM036297)c | |
| P. notoginseng (PN) | 2,811 | 246 | 2555 | 156,466 (KP036468) | 6,306 (KT380921)b | |
| P. japonicus (PJ) | 2,870 | 237 | 4295 | 156,188 (KP036469) | 6,275 (KT380920)b | |
| P. vietnamensis (PV) | 4,586 | 1,005 | 2267 | 155,993 (KP036470) | 7,280 (KT380922)b | |
| Aralia | A. elata (AE) | 505 | 90 | 267 | 156,220 (KT153023) | 6,073 (KT380919)b |
| A. undulata (AU) | NA | NA | NA | 156,333 (NC_022810)a | 610 (AF273540)d | |
| Eleutherococcus | E. sessiliflorus (ES) | 468 | 57 | 426 | 156,730 (KT153019) | 10,109 (KT380924)c |
| E. senticosus (ESen) | NA | NA | NA | 156,768 (NC_016430)a | 610 (AB570259.1)d | |
| Dendropanax | D. morbifera (DM) | 3,453 | 222 | 1124 | 156,366 (KR136270) | 9,332 (KT380923)b |
aCp sequences retrieved from GenBank. b45S nrDNA including full 45S transcription sequence (5.8 kb) and partial IGS sequence. c45S nrDNA including full 45S transcription sequence (5.8 kb) and full IGS sequence. dnrITS (ITS1-5.8S-ITS2) sequences retrieved from GenBank. NA: not available.
Complete lengths of the cp genomes ranged between 155,993 bp (P. vietnamensis) and 156,730 bp (E. sessiliflorus), with average read-mapping coverages of 57× to 1,005× (Table 1). The 45S nrDNA sequences were assembled into single contig for each of the seven species. The sequence lengths of the complete 45S nrDNA unit was around 11 kb including full intergenic spacer (IGS) sequences (Table 1). The 45S nrDNA sequences of P. quinquefolius and E. sessiliflorus were full-length sequences covering a 45S transcription unit (18S-ITS1–5.8S-ITS2-26S) and complete IGS sequences, whereas those of the other five species (P. notoginseng, P. japonicus, P. vietnamensis, A. elata and D. morbifera) consisted of 45S nrDNA transcription regions and partial IGS sequences (Table 1).
Diversity of cp genome sequences
In addition to the five cp genomes described above, we used cp genomes of three species, P. ginseng, P. quinquefolius, and D. morbifera that were obtained in our previous study23–25. Cp genome sequences for two Araliaceae species, A. undulata, E. senticosus, were also retrieved from GenBank for comparative analysis (Table 1). The gene order and gene content were highly conserved among the ten cp genomes (Fig. 1a), and the complete cp genome sequences showed 97.1~99.6% similarity among ten species (Supplementary Table S1). With regard to the quadripartite structure of the cp genome, nucleotide polymorphisms were lower in the inverted repeat regions (IRs) than in large single copy (LSC) and small single copy (SSC) regions (Fig. 1b). The cp genomes of five Panax species shared more than 98.9% similarity, among which P. ginseng and P. quinquefolius showed the highest similarity over 99.6% at both whole cp genome and cp coding sequence levels, respectively (Supplementary Table S1).
Figure 1.
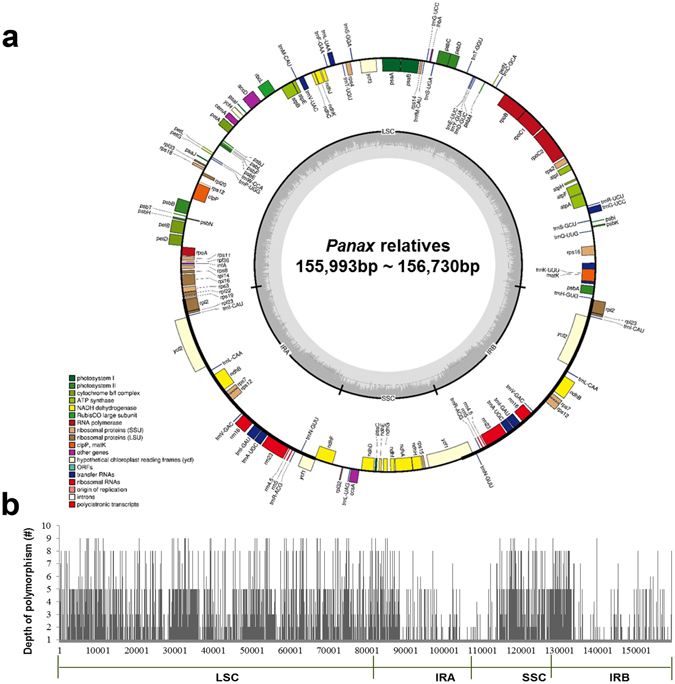
Gene map and nucleotide polymorphism of cp genomes in ten Araliaceae species. (a) Colored boxes are conserved chloroplast genes classified based on product function. The complete cp genome sequence was generated by the dnaLCW method and annotated using the DOGMA program (http://dogma.ccbb.utexas.edu/). The map was prepared using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/). Genes transcribed clockwise and counterclockwise are indicated on the outside and inside of the large circle, respectively. (b) The depth of polymorphisms found among cp genomes of ten Araliaceae species. Cp genome (KM088019) of P. ginseng was used as a reference for comparison.
The sequence polymorphism rates of conserved coding regions were from 0.4~1.8% among ten Araliaceae species, in which the rate between P. ginseng and P. quinquefolius was the lowest (0.4%). Five genes, namely petN, psaJ, psbF, psbN, and rpl23, showed no sequence polymorphism among the ten Araliaceae species (Fig. 2 and Supplementary Dataset S1, Supplementary Fig. S4). The ndhF genes in cp genomes of Aralia, Eleutherococcus and Dendropanax species were 97 bp, 36 bp and 18 bp farther from the border junction of IRa and SSC, compared to those in cp genomes of Panax species (Supplementary Fig. S1).
Figure 2.
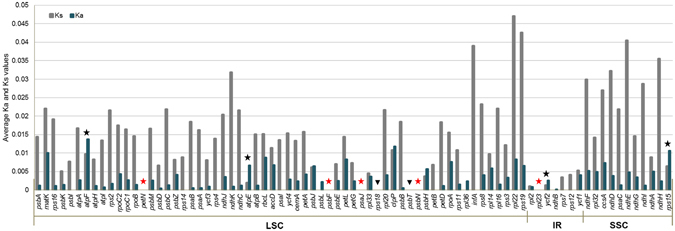
Summary of Ks and Ka values among cp genomes of ten species. The rates of non-synonymous substitution (Ka) and synonymous substitution (Ks) of 79 conserved protein-coding sequences were calculated and averaged using KaKs calculator. The average Ks and Ka values are indicated by grey and dark blue bars, respectively. Black stars indicate the genes evolved under positive selection pressure with over 1 for the value of Ka/Ks ratios. Red stars indicate five conserved genes without variations and black triangles indicate two conserved genes without SNP but with 21 and 6 bp InDel variations in Araliaceae family.
The average Ks values for all protein-coding genes except cp-rRNA genes were from 0.0014 to 0.0199 and 0.0014 to 0.0068 at the inter- and intra-genus levels, respectively. The lowest Ks value of 0.0014 was between the two closest Panax species, P. ginseng and P. quinquefolius. The average Ks value among five species within the Panax genus was lower (0.0056) than those between Aralia-Panax (0.0110) and Eleutherococcus-Dendropanax (0.0080) (Table 2). The highest Ks values were detected in infA, rpl22, rps19 and ndhE genes, while significantly high Ka/Ks ratios of more than 1 were detected in atpF, atpE, ycf2 and rps15 genes (P-value 8.3E-27–3.2E-2, Chi-square test) (Fig. 2).
Table 2.
Ks values and estimated divergence time of 10 Araliaceae species.
| Species | Divergence time (MYA)b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PG | PQ | PJ | PV | PN | AU | AE | Esen | ES | DM | ||
| Average Ks a | PG | 0.72 | 3.41 | 3.30 | 3.05 | 8.41 | 8.28 | 8.70 | 9.37 | 8.27 | |
| PQ | 0.0014 | 3.30 | 3.18 | 2.93 | 8.28 | 8.15 | 8.57 | 9.32 | 8.06 | ||
| PJ | 0.0068 | 0.0066 | 1.69 | 3.15 | 8.85 | 8.72 | 9.20 | 9.95 | 8.72 | ||
| PV | 0.0066 | 0.0064 | 0.0034 | 3.01 | 8.62 | 8.53 | 8.98 | 9.74 | 8.48 | ||
| PN | 0.0061 | 0.0059 | 0.0063 | 0.0060 | 8.46 | 8.26 | 8.82 | 9.58 | 8.38 | ||
| AU | 0.0168 | 0.0166 | 0.0177 | 0.0172 | 0.0169 | 3.20 | 8.33 | 9.13 | 8.11 | ||
| AE | 0.0166 | 0.0163 | 0.0174 | 0.0171 | 0.0165 | 0.0064 | 8.30 | 9.09 | 7.96 | ||
| ESen | 0.0174 | 0.0171 | 0.0184 | 0.0180 | 0.0176 | 0.0167 | 0.0166 | 2.98 | 4.10 | ||
| ES | 0.0187 | 0.0186 | 0.0199 | 0.0195 | 0.0192 | 0.0183 | 0.0182 | 0.0060 | 4.85 | ||
| DM | 0.0165 | 0.0161 | 0.0174 | 0.0170 | 0.0168 | 0.0162 | 0.0159 | 0.0082 | 0.0097 | ||
aAverage Ks values between common cp protein coding genes of each species calculated using KaKs calculator program. bDivergence time was estimated by Ks/2λ, where λ = 1.0 × 10−9. PG to DM indicate abbreviated species name (Table 1).
Although repeat motifs were diverse among cp genomes, conserved common repeat motifs were also present at both the intra-genus and inter-genus levels. Tandem repeat (TR) units with sizes of 6 to 12 bp were abundant among ten cp genomes (Supplementary Fig. S2). High copy number variation (CNV) for TRs was detected in the ycf1 protein-coding gene and in the intergenic regions between trnC-GCA and petN, trnS-GGA and rps4, trnT-UGU and trnL-UAA, and cemA and petA (Fig. 3 and Supplementary Table S2, Supplementary Fig. S3).
Figure 3.
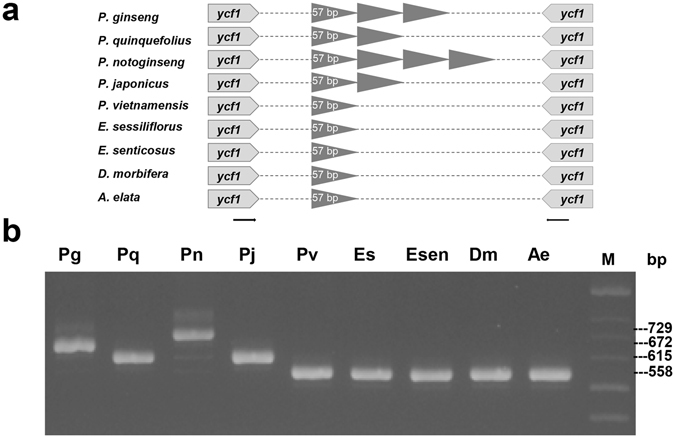
Validation of polymorphic sites with TR CNV in cp genomes of Araliaceae species. (a) Schematic diagram of CNV of TR units found in ycf1 genes among nine species, and triangles indicate TR unit of 57 bp. (b) Validation of CNV of 57-bp TR in nine species using genomic DNA PCR analysis with the pgycf01 primer set (Supplementary Table S2). Abbreviated species names (Table 1) are shown above the lanes. M indicates 100-bp DNA ladder.
Sequence variations of nrDNA sequences in Panax and relatives
In addition to seven 45S nrDNA sequences assembled in this study, 45S nrDNA sequence of P. ginseng and nrITS sequences of A. undulata and E. senticosus were retrieved from GenBank and used for comparative analysis (Table 1). When 45S nrDNA transcription sequences (18S-ITS1-5.8S-ITS2) were compared among eight species (excluding A. undulata and E. senticosus for which there was no available sequence), sequence polymorphisms were found in genic regions as well as in two ITS regions although the polymorphisms were more relatively frequent in two ITS regions (Fig. 4 and Supplementary Tables S3 and S4); 9, 4, 50, 42, and 44 SNPs in 18S (1,808 bp), 5.8S (160 bp), 26S (3,452 bp), ITS1 (223 bp) and ITS2 (233 bp) regions, respectively. The numbers of SNPs in the 45S nrDNA sequence were higher at the inter-genus level than at the intra-genus level (Supplementary Tables S3 and S4).
Figure 4.
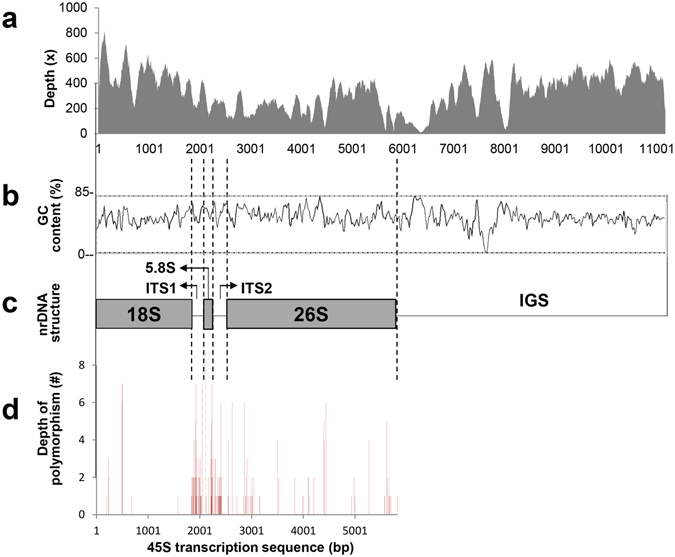
Assembly and comparison of 45S nrDNA sequences. (a–c) Full-length 45S nrDNA sequence of P. quinquefolius assembled in this study. Average read depth was 376× (a) and average GC content was 53.15% (b). (c) Schematic diagram of 45S nrDNA structure. (d) Comparison of 45S nrDNA transcription sequences of eight Araliaceae species. P. ginseng 45S nrDNA transcription sequence (KM036295) was used as reference for sequence comparison.
Phylogenetic analysis and divergence time estimation
A phylogenetic tree based on cp protein coding sequences showed two typical monophyletic lineages consisting of the Aralia-Panax group and the Eleutherococcus-Dendropanax group, which was also confirmed by a phylogenetic tree based on nrITS sequences (Fig. 5). Species of each genus, Panax, Aralia, Eleutherococcus, and Dendropanax, were grouped separately. The five Panax species were divided into two subgroups, in which one included P. ginseng and P. quinquefolius and the other included the remaining three Panax species (Fig. 5a). The topology of the tree based on nrITS sequences was almost identical to that in the cp sequence-derived tree, except for some differences among the Panax species (Fig. 5b).
Figure 5.
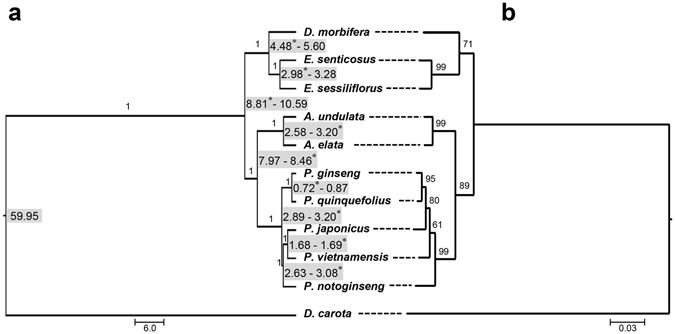
Phylogenetic tree and divergence time of 10 Araliaceae species. (a,b) Phylogenetic trees were generated based on cp protein-coding sequences (a) and nrDNA ITS sequences (ITS1-5.8S-ITS2) (b). Dashed lines connect the positions of each species in the two trees. Numbers next to nodes indicate average divergence time (*) based on Ks values (Table 2), and median divergence time using the BEAST program with 95% highest posterior density. The number above each branch refers to the Bayesian posterior probability (a) and ML bootstrap values (b).
Based on cp protein coding sequences, the divergence time between the Aralia-Panax group and the Eleutherococcus-Dendropanax group could be estimated at approximately 8.81–10.59 MYA (Fig. 5a). In the Eleutherococcus-Dendropanax clade, Dendropanax and Eleutherococcus were estimated to have diverged 4.48–5.60 MYA. In the Aralia-Panax clade, Aralia and Panax were estimated to have diverged 7.97–8.46 MYA, with subsequent speciation within these two genera likely to have occurred during a period approximately 2.89–3.20 and 2.58–3.20 MYA in Panax and Aralia, respectively. The two closest Panax species, P. ginseng and P. quinquefolius, were predicted to have diverged approximately 0.72–0.87 MYA.
Discussion
Genetic diversity in cp genomes of the Araliaceae family
Sequence variation was low throughout cp protein coding sequences and intergenic sequences among the ten Araliaceae species examined herein, although the cp protein coding sequences were more conserved (0.4~1.8% polymorphism) compared to intergenic sequences (0.4~2.9% polymorphism). Based on Ks values and divergence time inferred in this study, the substitution rate among cp genomes of Panax and relatives was approximately 1.0 × 10−9 per year, which seems to be consistent with a previous report26.
Based on analysis of non-synonymous and synonymous SNPs, average Ks and Ka values in LSC, SSC and IR regions among Panax relatives were low and ranged from 0.0022 to 0.0236, and from 0.0011 to 0.0046, respectively. Overall, genes in IR regions showed lower Ks values than did those in LSC and SSC regions. The ratio of the two parameters, Ka/Ks, is defined as the degree of evolutionary change in plants27. Among the protein-coding genes in the ten Araliaceae cp genomes, atpF, atpE, ycf2, and rps15 were identified as being under positive selection (Fig. 2). Some of these genes were also reported to be related to evolution under positive selection pressure in Orobanchaceae and Sesamun indicum 28, 29. Among these five genes, atpF is a group II intron-containing gene with a known mechanism of splicing and heterogeneity in intron sequence30, 31.
Five cp genes conserved in the Araliaceae family
We identified five identical genes which showed no sequence variation among all 10 Araliaceae species. Among the five conserved genes, four (petN, psaJ, psbF, and psbN) were related to photosynthesis and one (rpl23) was related to the ribosomal large subunit (Fig. 2 and Supplementary Dataset S1). We compared the five genes with those of Daucus carota belonging to the sister family Apiaceae in the Apiales order and to those of Arabidopsis thaliana in the Brassicales order (Supplementary Fig. S4). The psbF gene in the Araliaceae species was identical to that of D. carota but differed from that of A. thaliana, suggesting that the gene may be highly conserved in the Apiales order. Four of the five conserved genes showed sequence variation with orthologs of D. carota, indicating that these four genes are specifically conserved only in the Araliaceae species (Supplementary Fig. S4). It will be interesting to further elucidate the role of these five genes in the evolution of Araliaceae family.
Nucleotide variation in ribosomal genes
Comparative analysis of eight 45S nrDNA sequences revealed that nucleotide polymorphism was much higher in nrITS regions than in transcribed ribosomal genes, 18S, 5.8S and 26S, as observed in other species32, 33. Most genetic diversity studies in plants have focused on ITS1 and ITS2 regions34–38. However, our study demonstrates that there is sufficient genetic diversity in the ribosomal gene regions for evolutionary analysis (Fig. 4 and Supplementary Tables S3 and S4). Among ribosomal genes, 26S was more divergent than 18S and 5.8S among the 10 Araliaceae species. Consistent with this finding, the 26S rDNA sequences have been applied for phylogenetic and divergence studies in yeasts39. The inter-genus level polymorphism was approximately three times higher than the intra-genus polymorphism (Supplementary Tables S3 and S4). Overall, our analysis suggests that 26S ribosomal gene sequences, in addition to the widely used ITS regions, can also be good targets for genetic diversity analyses for broad range and over genus-level taxon identification in the plant kingdom.
Phylogeny based on cpDNA and nrDNA of Araliaceae
Two monophyletic groups, Aralia-Panax and Eleutherococcus-Dendropanax, were simultaneously confirmed with both cpDNA-based and nrITS-based trees. The finding that Aralia and Panax were the most closely related genus is in accordance with previous reports15. The Panax genus was clearly classified as two groups, with diploid species spread in Southern Asia including P. notoginseng, P. vietnamensis, and P. japonicas and a tetraploid group widely distributed in Northern Asia and North America including P. ginseng and P. quinquifolius (Fig. 5a).
The cpDNA-derived topology reflects the uniparental inheritance of cpDNA and agrees with the previously proposed models for evolution of these five Panax species. However, our nrITS-derived tree did not clearly distinguish between tetraploid and diploid groups (Fig. 5b). The different topology for the Panax genus between the cpDNA-based and nrITS-based trees may be caused by differences inherent to cpDNA and nrDNA in hybrid species including polyploids. According to previous reports, nrDNA (or nrITS)-based trees occasionally lack clear resolution in hybrid species (included polyploids) or closely related groups40 owing to low substitution rates in nrDNA. The nrDNA homogenized by inter-locus concerted evolution can give rise to multiple-heterogeneous types, especially in hybrid speciation. Under homogenization conditions, analysis based solely on nrDNA (or nrITS) sequence can produce misleading phylogenetic results in closely related species and polyploids (e.g. allopolyploids). Therefore, we recently suggested utilization of both cpDNA-based and nrDNA-based trees as a method conducive to determining well-resolved taxonomical positions of inter-subspecies hybrids among Oryza AA genomes22.
Molecular clocks for speciation of Panax relatives
Based on previous reports, Araliaceae and Apiaceae diverged from a common ancestor in the Apiales order approximately 60.2 MYA41–44. In this study, we demonstrated that the Araliaceae family diverged into two monophyletic lineages approximately 8.81–10.59 MYA, followed by divergence of genera. Aralia and Panax, the two closest-related genera, diverged around 7.97–8.46 MYA, and Dendropanax and Eleutherococcus diverged after that (4.48–5.60 MYA).
In the Panax genus, we inferred speciation to have occurred more recently, around 2.89–3.20 MYA. This speciation step seems, like other plant speciation events, to be related to an Asian temperate climate change at that time period corresponding to uplift of the Himalaya-Tibetan plateau (in the late Cenozoic era)45–47. The Himalayas and southwestern China were suggested to be the base of speciation of Panax 8, and also reported to have a high rate of polyploidy because of the appearance of diverse species in the widespread alpine environment in these regions48. Earlier studies showed that P. ginseng underwent a recent whole-genome duplication 2.3 MYA49, followed by divergence of P. ginseng and P. quinquefolius 50. Our molecular clock estimation using complete cp coding genes supports the previous estimate and provides stronger evidence for that evolution scenario. P. ginseng might have evolved from allotetraploidization of diploid ancestors triggered by a large Asian temperate climate change. The diploid ancestors would have been isolated to the Northern hemisphere by uplift of the Himalaya-Tibetan plateau 2.89–3.20 MYA. We estimated divergence time between P. ginseng and P. quinquefolius to be approximately 0.72–0.87 MYA, i.e., following the recent genome duplication event at 2.3 MYA49. P. quinquefolius was suggested to have settled in North America by migration of the P. ginseng seeds via glacial movement of the Bering land bridge from eastern Asia to northern America and disjunction by geographical isolation 0.9–2.3 MYA51.
Conclusion
The complete sequences of cp genomes and 45S nrDNA obtained in this study will contribute toward further understanding of evolution in the Apiales order. Phylogenetic and evolutionary analysis of cp genomes indicated that the evolutionary divergence of Araliaceae family produced two monophyletic lineages, followed by diversification of genera and speciation. Our findings demonstrated that simultaneous utilization of cp genomes and nrDNAs support fine-scale resolution of the evolutionary and taxonomical relationships of Panax and relatives in the Araliaceae.
Materials and Methods
Plant materials
Plant samples of five Panax species (P. ginseng, P. quinquefolius, P. notoginseng, P. vietnamensis and P. japonicus) and four relative species (Aralia elata, Eleutherococcus sessiliflorus, E. senticosus and Dendropanax morbifera) were used in this study (Table 1). Leaves were harvested from P. ginseng, P. quinquefolius, and D. morbifera plants grown in a ginseng research field (Seoul National University, Suwon, Korea), and from P. vietnamensis wild plants collected from Dak To district, Kon Tum province, Vietnam. Roots of P. notoginseng and P. japonicus were collected from Dafang County, Guizhou province and Enshi County, Hubei province, China, respectively. Leaves of A. elata, E. sessiliflorus and E. senticosus were from the farm of the Susinogapy Corporation (Cheonan, Korea, www.susinogapy.com).
Genomic DNA isolation and whole-genome shotgun sequencing
Total genomic DNAs were isolated from tissue samples of the nine species using a modified cetyltrimethylammonium bromide (CTAB) method52 and examined using a spectrometer and agarose-gel electrophoresis. Illumina paired-end (PE) libraries were constructed with 300-bp insert size for each of eight species and sequenced using MiSeq or NextSeq platform by LabGenomics (Seongnam, Korea, http://www.labgenomics.co.kr/). Sequences of five species, P. notoginseng, P. vietnamensis, P. japonicus, A. elata, and E. sessiliflorus, were newly obtained in this study.
De novo assembly and annotation of cp genomes and 45S nrDNA sequences
Complete cp genomes and 45S nrDNA sequences were assembled by de novo assembly with the low-coverage whole-genome sequence (dnaLCW) method22. Assembly errors and gaps were manually corrected by mapping of raw PE reads22
Structural features and genes in cp genomes were predicted using the DOGMA program (http://dogma.ccbb.utexas.edu/) and manual curation based on BLAST searches. Circular maps of cp genomes were made using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/). The structures of 45S nrDNA sequences were predicted by comparison with reported P. ginseng 45S nrDNA sequence (KM036295) and analyses using RNAmmer (http://www.cbs.dtu.dk/services/RNAmmer/), and BLAST searches.
Comparative analysis of cp genomes and 45S nrDNA sequences
Among the ten species, complete cp genomes of two species, A. undulata 12 (NC_022810) and E. senticosus 53 (NC_016430.1) were retrieved from GenBank. In addition, ITS1-5.8S-ITS2 (hereafter, nrITS) sequences of two species, A. undulata (AF273540) and E. senticosus (AB570259) were also retrieved from GenBank. To identify inter-species polymorphism, complete cp genome sequences of ten species were aligned and compared using MAFFT (http://mafft.cbrc.jp/alignment/server/) and mVISTA (http://genome.lbl.gov/vista/mvista/submit.shtml). For 45S nrDNA comparison, nrDNA sequences were extracted and compared among eight species, as for cp genomes. Tandem repeats (TRs) present on cp genomes of ten species were investigated using Tandem Repeat Finder (https://tandem.bu.edu/trf/trf.html).
Validation of polymorphic sites
Among polymorphic sites found in cp genomes, those with copy number variation (CNV) of TRs and InDels were selected for validation. PCR primer pairs were designed based on the flanking sequences of the selected polymorphic sites using Primer 3 program (http://bioinfo.ut.ee/primer3-0.4.0/) and used for genomic DNA PCR analyses. Amplified fragments were analyzed by separation in agarose gels and ethidium bromide staining.
Phylogenetic analysis and estimation of divergence time
Divergence time was first calculated based on Ks value. Ka and Ks values represent the number of non-synonymous and synonymous substitution per site, respectively. To estimate divergence time of these ten species in the Araliaceae family, 79 protein-coding sequences from each cp genome of the ten species were extracted and concatenated. Mean Ka and Ks values were calculated by pair-wise comparison of nucleotide substitution among the common cp protein-coding genes of the ten species, using the PAML program54. Divergence time (T) was given by T = Ks/2λ, where λ is approximately 1.0 × 10−9 substitutions per site per year26.
Second, divergence time was calculated using the Bayesian Inference (BI) method. A phylogenetic tree was generated based on BI analysis using BEAST version 1.8.155 with 95% highest posterior density. The analysis was conducted with the data of 79 chloroplast protein-coding gene sequences using a strict clock approach, with Yule prior on the tree, general time reversible (GTR + I + Γ) as a substitution model and the default priors for generating a random starting tree. The maximum probability clade trees were calculated using TreeAnnotator (version 1.8.1) and root age was constrained to be 60.2 million years ago (MYA) based on previously reported divergence time between Araliaceae and Apiaceae in the order Apiales41–44. The final tree was visualized in FigTree v1.3.1.
The nrITS-based tree was constructed using the ML method of MEGA6.0 with 1,000 bootstrap replicates. As outgroup sequences for phylogenetic analysis, cp genome56 (NC_008325.1) and nrITS sequence (AY552527.1) of Daucus carota (carrot) belonging to the Apiaceae family were used.
Electronic supplementary material
Acknowledgements
This work is supported by Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01100801), Rural Development Administration, the Bio & Medical Technology Development Program (NRF-2015M3A9A5030733) of the National Research Foundation (NRF) which was funded by the Ministry of Science, ICT, and Future Planning, and a grant (16172MFDS229) from Ministry of Food and Drug Safety in 2016, Republic of Korea.
Author Contributions
Conceived and designed the experiments: K.K., V.B.N., T.J.Y. Prepared the samples and performed the experiments: K.K., V.B.N., J.Z.D., Y.W. Analyzed the data: K.K., V.B.N., J.Y.P., T.J.Y. Wrote the paper: K.K., V.B.N., S.C.L., T.J.Y.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Kyunghee Kim and Van Binh Nguyen contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05218-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Group, T. A. P. An ordinal classification for the families of flowering plants. Annals of the Missouri Botanical Garden, 531–553 (1998).
- 2.Chase, M. W. et al. Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Annals of the Missouri Botanical Garden, 528–580 (1993).
- 3.Olmstead, R. G., Bremer, B., Scott, K. M. & Palmer, J. D. A parsimony analysis of the Asteridae sensu lato based on rbcL sequences. Annals of the Missouri Botanical Garden, 700–722 (1993).
- 4.Sanderson, M. J. & Donoghue, M. J. Shifts in diversification rate with the origin of angiosperms. Science-New York Then Washington, 1590–1590 (1994). [DOI] [PubMed]
- 5.Plunkett, G. M., Soltis, D. E. & Soltis, P. S. Higher level relationships of Apiales (Apiaceae and Araliaceae) based on phylogenetic analysis of rbcL sequences. American Journal of Botany, 499–515 (1996).
- 6.Tang, W. & Eisenbrand, G. Panax ginseng CA Mey, (Springer, 1992).
- 7.Davydov M, Krikorian A. Eleutherococcus senticosus (Rupr. & Maxim.) Maxim.(Araliaceae) as an adaptogen: a closer look. Journal of Ethnopharmacology. 2000;72:345–393. doi: 10.1016/S0378-8741(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 8.Wen J, Zimmer EA. Phylogeny and biogeography of Panax L. (the ginseng genus, Araliaceae): inferences from ITS sequences of nuclear ribosomal DNA. Molecular phylogenetics and evolution. 1996;6:167–177. doi: 10.1006/mpev.1996.0069. [DOI] [PubMed] [Google Scholar]
- 9.Yi T, Lowry PP, Plunkett GM. Chromosomal evolution in Araliaceae and close relatives. Taxon. 2004;53:987–1005. doi: 10.2307/4135565. [DOI] [Google Scholar]
- 10.Waminal, N. E. et al. A refined Panax ginseng karyotype based on an ultra-high copy 167-bp tandem repeat and ribosomal DNAs. Journal of Ginseng Research (2016). [DOI] [PMC free article] [PubMed]
- 11.Lee, Y. S. et al. Comparative analysis of the transcriptomes and primary metabolite profiles of adventitious roots of five Panax ginseng cultivars. Journal of Ginseng Research (2016). [DOI] [PMC free article] [PubMed]
- 12.Li R, Ma PF, Wen J, Yi TS. Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications. PloS one. 2013;8:e78568. doi: 10.1371/journal.pone.0078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C, Wen J. Phylogeny of Panax using chloroplast trnC–trnD intergenic region and the utility of trnC–trnD in interspecific studies of plants. Molecular phylogenetics and evolution. 2004;31:894–903. doi: 10.1016/j.ympev.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Wen J, Shi S, Jansen R, Zimmer E. Phylogeny and biogeography of Aralia sect. Aralia (Araliaceae) American Journal of Botany. 1998;85:866–866. doi: 10.2307/2446422. [DOI] [PubMed] [Google Scholar]
- 15.Wen J, Plunkett GM, Mitchell AD, Wagstaff SJ. The evolution of Araliaceae: a phylogenetic analysis based on ITS sequences of nuclear ribosomal DNA. Systematic Botany. 2001;26:144–167. [Google Scholar]
- 16.Plunkett GM, Wen J, Lowry IPP. Infrafamilial classifications and characters in Araliaceae: Insights from the phylogenetic analysis of nuclear (ITS) and plastid (trnL-trnF) sequence data. Plant Systematics and Evolution. 2004;245:1–39. doi: 10.1007/s00606-003-0101-3. [DOI] [Google Scholar]
- 17.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim NH, Choi HI, Kim KH, Jang W, Yang TJ. Evidence of genome duplication revealed by sequence analysis of multi-loci expressed sequence tag–simple sequence repeat bands in Panax ginseng Meyer. Journal of Ginseng Research. 2014;38:130–135. doi: 10.1016/j.jgr.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proceedings of the National Academy of Sciences. 1989;86:6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reboud X, Zeyl C. Organelle inheritance in plants. Heredity. 1994;72:132–140. doi: 10.1038/hdy.1994.19. [DOI] [Google Scholar]
- 21.Zhang, Y. et al. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Frontiers in plant science7 (2016). [DOI] [PMC free article] [PubMed]
- 22.Kim, K. et al. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Scientific reports5 (2015). [DOI] [PMC free article] [PubMed]
- 23.Kim, K. et al. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PloS one10, e0117159 (2015). [DOI] [PMC free article] [PubMed]
- 24.Kim K, et al. The complete chloroplast genome sequence of Panax quinquefolius (L.) Mitochondrial DNA A DNA MappSeq Anal. 2016;27:3033–4. doi: 10.3109/19401736.2015.1063121. [DOI] [PubMed] [Google Scholar]
- 25.Kim K, Lee SC, Yang TJ. The complete chloroplast genome sequence of Dendropanax morbifera (Léveillé) Mitochondrial DNA Part A. 2016;27:2923–2924. doi: 10.3109/19401736.2015.1060442. [DOI] [PubMed] [Google Scholar]
- 26.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, Liu F, Wang L, Huang S, Yu J. Nonsynonymous substitution rate (Ka) is a relatively consistent parameter for defining fast-evolving and slow-evolving protein-coding genes. Biol Direct. 2011;6:6150–6. doi: 10.1186/1745-6150-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samigullin TH, Logacheva MD, Penin AA, Vallejo-Roman CM. Complete Plastid Genome of the Recent Holoparasite Lathraea squamaria Reveals Earliest Stages of Plastome Reduction in Orobanchaceae. PloS one. 2016;11:e0150718. doi: 10.1371/journal.pone.0150718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Li C, Miao H, Xiong S. Insights from the complete chloroplast genome into the evolution of Sesamum indicum L. PloS one. 2013;8:e80508. doi: 10.1371/journal.pone.0080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel F, Kazuhiko U, Haruo O. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 31.Kelchner SA. Group II introns as phylogenetic tools: structure, function, and evolutionary constraints. American Journal of Botany. 2002;89:1651–1669. doi: 10.3732/ajb.89.10.1651. [DOI] [PubMed] [Google Scholar]
- 32.Iwen P, Hinrichs S, Rupp M. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Medical Mycology. 2002;40:87–109. doi: 10.1080/mmy.40.1.87.109. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto Y, Yanase T, Tsuda T, Noda H. Characterization of Internal Transcribed Spacer (ITS1)-ITS2 region of ribosomal RNA gene from 25 species of Culicoides biting midges (Diptera: Ceratopogonidae) in Japan. Journal of medical entomology. 2009;46:1099–1108. doi: 10.1603/033.046.0517. [DOI] [PubMed] [Google Scholar]
- 34.Rampersad SN. ITS1, 5.8 S and ITS2 secondary structure modelling for intra-specific differentiation among species of the Colletotrichum gloeosporioides sensu lato species complex. SpringerPlus. 2014;3:1. doi: 10.1186/2193-1801-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hausner G, Wang X. Unusual compact rDNA gene arrangements within some members of the Ascomycota: evidence for molecular co-evolution between ITS1 and ITS2. Genome. 2005;48:648–660. doi: 10.1139/g05-037. [DOI] [PubMed] [Google Scholar]
- 36.Gerbi S. Evolution of ribosomal DNA. Molecular evolutionary genetics. 1985;35:419–517. doi: 10.1007/978-1-4684-4988-4_7. [DOI] [Google Scholar]
- 37.GI-YOUNG K, HA MG, Lee TH, Lee JD. Chemosystematics and molecular phylogeny of a new bioflocculant-producing Aspergillus strain isolated from Korean soil. Journal of microbiology and biotechnology. 1999;9:870–872. [Google Scholar]
- 38.Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Molecular phylogenetics and evolution. 2003;29:417–434. doi: 10.1016/S1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 39.Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/A:1001761008817. [DOI] [PubMed] [Google Scholar]
- 40.Hamby, R. K. & Zimmer, E. A. Ribosomal RNA as a phylogenetic tool in plant systematics. in Molecular systematics of plants 50–91 (Springer, 1992).
- 41.Shi FX, et al. The impacts of polyploidy, geographic and ecological isolations on the diversification of Panax (Araliaceae) BMC plant biology. 2015;15:1. doi: 10.1186/s12870-014-0410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magallón S, Gómez‐Acevedo S, Sánchez‐Reyes LL, Hernández‐Hernández T. A metacalibrated time‐tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist. 2015;207:437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- 43.Court, W. E. The Genus Panax. in Ginseng, the Genus Panax 13–21 (CRC Press, 2000).
- 44.Tank DC, et al. Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytologist. 2015;207:454–467. doi: 10.1111/nph.13491. [DOI] [PubMed] [Google Scholar]
- 45.Thompson JD, Lumaret R. The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends in ecology & evolution. 1992;7:302–307. doi: 10.1016/0169-5347(92)90228-4. [DOI] [PubMed] [Google Scholar]
- 46.Yamane K, Yasui Y, Ohnishi O. Intraspecific cpDNA variations of diploid and tetraploid perennial buckwheat, Fagopyrum cymosum (Polygonaceae) American Journal of Botany. 2003;90:339–346. doi: 10.3732/ajb.90.3.339. [DOI] [PubMed] [Google Scholar]
- 47.Chen G, Sun WB, Sun H. Ploidy variation in Buddleja L.(Buddlejaceae) in the Sino‐Himalayan region and its biogeographical implications. Botanical Journal of the Linnean Society. 2007;154:305–312. doi: 10.1111/j.1095-8339.2007.00650.x. [DOI] [Google Scholar]
- 48.Nie, Z. L., Wen, J., Gu, Z. J., Boufford, D. E. & Sun, H. Polyploidy in the flora of the Hengduan Mountains hotspot, southwestern China. Annals of the Missouri Botanical Garden, 275–306 (2005).
- 49.Choi HI, et al. Major repeat components covering one‐third of the ginseng (Panax ginseng CA Meyer) genome and evidence for allotetraploidy. The Plant Journal. 2014;77:906–916. doi: 10.1111/tpj.12441. [DOI] [PubMed] [Google Scholar]
- 50.Kim JH, et al. Diversity and evolution of major Panax species revealed by scanning the entire chloroplast intergenic spacer sequences. Genetic resources and crop evolution. 2013;60:413–425. doi: 10.1007/s10722-012-9844-4. [DOI] [Google Scholar]
- 51.Choi HI, et al. Evolutionary relationship of Panax ginseng and P. quinquefolius inferred from sequencing and comparative analysis of expressed sequence tags. Genetic resources and crop evolution. 2013;60:1377–1387. doi: 10.1007/s10722-012-9926-3. [DOI] [Google Scholar]
- 52.Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nature protocols. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 53.Yi DK, Lee HL, Sun BY, Chung MY, Kim KJ. The complete chloroplast DNA sequence of Eleutherococcus senticosus (Araliaceae); comparative evolutionary analyses with other three asterids. Molecules and cells. 2012;33:497–508. doi: 10.1007/s10059-012-2281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z. PAML: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 55.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruhlman T, et al. Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms. BMC genomics. 2006;7:1. doi: 10.1186/1471-2164-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


